Abstract
The human pharyngeal microbiome, which resides at the juncture of digestive and respiratory tracts, may have an active role in the prevention of respiratory tract infections, similar to the actions of the intestinal microbiome against enteric infections. Recent studies have demonstrated that the pharyngeal microbiome comprises an abundance of bacterial species that interacts with the local epithelial and immune cells, and together, they form a unique micro-ecological system. Most of the microbial species in microbiomes are obligate symbionts constantly adapting to their unique surroundings. Indigenous commensal species are capable of both maintaining dominance and evoking host immune responses to eliminate invading species. Temporary damage to the pharyngeal microbiome due to the impaired local epithelia is also considered an important predisposing risk factor for infections. Therefore, reinforcement of microbiome homeostasis to prevent invasion of infection-prone species would provide a novel treatment strategy in addition to antibiotic treatment and vaccination. Hence continued research efforts on evaluating probiotic treatment and developing appropriate procedures are necessary to both prevent and treat respiratory infections.
Keywords: Respiratory tract infections, Pharyngeal microbiome, Homeostasis, Probiotics, Restoration balance of microbiome
Introduction
Respiratory tract infections (RTIs) continue to be a leading cause of morbidity and mortality worldwide, despite the emergence of antibiotics. According to a recent report by the World Health Organization (WHO), RTI-related mortality remains high, second only to that of cancers and cardio-cerebrovascular diseases (World Health Statistics 2013, www.who.int). RTIs often result from new invasion and abnormal propagation of specific pathogens into airways. Apart from the number and virulence of such invasive pathogens, host defenses also govern the occurrence and severity of infection. Over the past several decades, advancements in the understanding of adaptive immunity — a major protective mechanism against pathogenic infection — have greatly influenced medical practice and are invaluable to the development of effective vaccines against infections by many lethal pathogens. In accordance, further elucidation of local mucosal and non-specific immunity is also important for the development of therapies to prevent pathogenic invasion [1]. Recently, increasing lines of evidence have indicated that the human microbiome (complicated bacterial communities residing in specific anatomic sites of the human body, see Box 1) is extensively involved in infections and other pathogen-related diseases, and has an important role in the maintenance of overall health [2]. Additionally, probiotic supplementation to restore microbiome balance may be useful as an adjuvant treatment against infections and has therefore gained much attention in current clinical research and practice especially in intestinal infections, because the overall effectiveness of antibiotics continues to decrease due to the emergence of drug-resistant pathogens [3]. We hypothesize that the pharyngeal microbiome, which resides at the juncture of the digestive and respiratory tracts, may share common features with intestinal microbiomes and could serve as a key player in the development of respiratory diseases. Here, we provide our perspective on potentially similar protective roles of the pharyngeal microbiome and discuss the strategy of using probiotics as an adjuvant therapy for treatment of RTIs based on an in-depth review of the current literature.
Box 1 Glossary.
Metagenome: A composite of genomes or genes of heterogeneous taxa from a defined environment. Metagenomic analysis is directly performed on samples based on high-throughput sequencing which makes it possible to characterize all microbial compositions, including uncultivable organisms, and to analyze the collective properties of the community as a whole and its microbial interactions in situ.
Pangenome: A superset of genes or genomic information of all strains of a species (mainly applicable to bacteria or archaea, which have significantly variable gene contents among strains). Pangenomic analysis delivers a whole picture of species genomic properties in the context of global distribution or evolution.
Microbiome: Species surveyed at different coverages for a microbial community in a defined environment. In recent years, microbial communities within human body have been studied by using metagenomic tools and methodology.
Microecosystem: A system includes the microbial community and its defined environmental components within limited spacing and is organized in general through a network of nutrient exchanges and energy flows. In the human body, the microecosystem includes microbiome, immunity, epithelial lining and other local physiochemical factors (such as pH, temperature and nutrients).
The pharyngeal microbiome and microecosystem
Complexity of the human pharyngeal microbiome
Hundreds of microbial species inhabit the human nasal, oral and pharyngeal cavities, including 25–40 families of bacteria, archaea, ameba and fungi, according to ample evidence from laboratory cultures [4]. The number of newly discovered species has considerably increased owing to the recent advances in metagenomic (Box 1) research techniques, especially high-throughput sequencing technology, that enable the discovery of non-culturable species [5–7]. In the pharynx, five major bacterial phyla have been identified thus far according to data released by the Human Microbiome Project (HMP): Firmicutes, Bacteroidetes, Proteobacteria, Actinobacteria and Fusobacteria (Figure 1A). The pharyngeal microbiome (referred as throat in HMP) is distinguished by having more Bacteroidetes than those found in other body sites, including the skin, intestines and vaginal cavity. Meanwhile, the pharyngeal microbiome comprises 27% Bacteroidetes and 10% Proteobacteria, whereas the nearby saliva microbiome contains 9% Bacteroidetes and 51% Proteobacteria (Figure 1B). These two phyla are important to human infections because the major causative pathogens of periodontitis are Bacteroidetes and the most common Gram-negative pathogens (e.g., Acinetobacter, Moraxella, Pseudomonas, Haemophilus, Klebsiella and Legionella spp.) in chronic respiratory diseases, such as cystic fibrosis (CF) and chronic obstructive pulmonary disease (COPD), are Proteobacteria (Figure 1C). Located at the lowest part of the respiratory track, the lung microbiome does not contain any particular set of species distinguishable from that of the pharynx, although there are variations in the proportions of the known species [10]. However, a healthy lung microbiome is characterized by a relatively reduced biomass, which is often 2–4 fold less than that of the pharynx [11].
Figure 1.
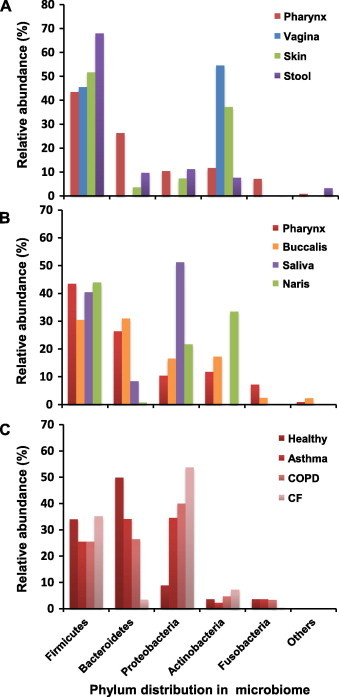
Relative abundance of major phyla in the microbiomes of different sites or conditions in the human body
A. Microbiomes in stool, skin, vagina and pharynx. B. Microbiomes in nares, saliva, buccal mucosa and pharynx. C. Microbiomes in the lung of healthy individuals and patients with asthma, COPD and CF. Microbial abundances (%) in panels A and B are calculated based on raw data from the Human Microbiome Project (http://hmpdacc.org/HMBSA), whereas microbial abundances (%) in panel C are calculated based on data described previously [8,9]. COPD, chronic obstructive pulmonary disease; CF, cystic fibrosis.
The pharyngeal microecosystem
In the human body, a microecosystem includes an anatomically defined microbiome together with local immune system and the epithelial lining of the host (Box 1). Studies of the intestinal and other microbiomes have shown that co-habitating microbes within a microbiome are not random assortments, but rather often exhibit various interactions, such as competing for or mutually utilizing metabolic resources in coordinated ways [12]. As such, microbiomes are always organized to maintain a homeostatic status, thereby increasing the demand for greater metabolic efficiency [12]. Consequently, all components of the microbiome are constrained in optimized proportions [12]. Meanwhile, the microbiome provides critical signals to promote maturation of immune cells and differentiation of the tissue lining to promote a protective entity to prevent dominance of aggressive members and infections from non-residential pathogens [13].
In the human pharynx, the most common genera in descending order are Prevotella, Capnocytophaga, Campylobacter, Veillonella, Streptococcus, Neisseria and Haemophilus, which account for 9.72%–1.26% of the members of the indigenous microbiome, according to the HMP data. Each of these genera includes species that are well-known fastidious bacteria hosted by the human body. Although it may not be easy to demonstrate a subset of bacterial species unique to the pharyngeal microbiome, it is possible to differentiate it from the corresponding gut microbiomes in terms of the proportions of constituents. Because of the limited number of studies on the pharyngeal microbiome, many interactions among microecosystem components remain unclear. However, homeostasis and interactions of microbes with local environmental factors are universal features of microbiome. Therefore, it is rather reasonable to infer that the pharyngeal microbiome also share such common features of all human microbiomes, although the understanding of which mainly comes from studies on intestinal microbiome.
Habitat-specific variability
It is necessary to clearly differentiate the inner and outer microbiomes of the human body, i.e., the human-associated microbiomes and the habitat-specific microbiomes of the host (Figure 2). Habitat-specific microbiomes can be characterized as either restricted or open. As an example of a restricted microbiome, consider the hospital environment where continuous antibiotic stress drives both species variability and content limitation. Moreover, species in hospital environments are often multi-drug resistant and may be opportunistic pathogens that cause severe infections especially in patients with chronic illness, often leading to hospital-acquired pneumonia (HAP). On the contrary, microbiomes within an ordinary human habitat are perceived as relatively open, where it is assumed not to be directed by any one specific stressor other than seasonal, geographic and biological variables [14]. Pathogens from such a microbiome may lead to infection even in healthy persons when there is sufficient competitiveness among species and toxicity to host, rendering a condition widely termed as community-acquired pneumonia (CAP).
Figure 2.
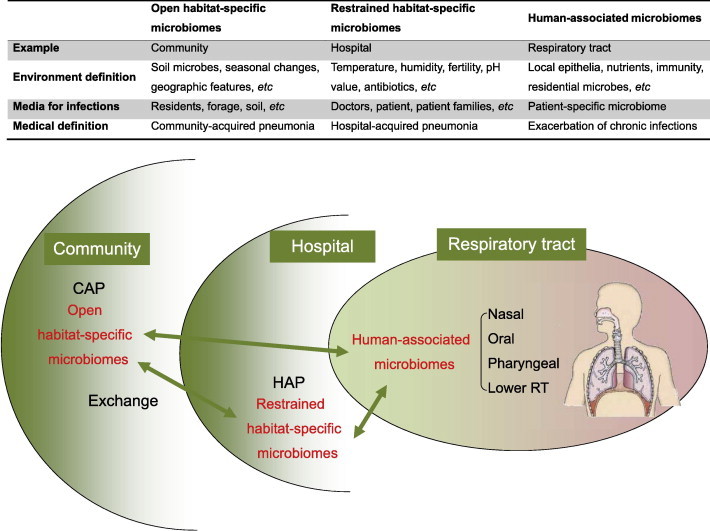
Open or restrained habitat-specific microbiomes and RTI
The human pharyngeal microbiome is unavoidably influenced by environmental microbiomes in the habitat where the host lives. Some of the habitat-specific microbiomes are open and stable, such as those of residential communities, and others are restrained, such as those in hospitals or even ICU rooms. Examples of infections related to such microbiomes are CAP and HAP. ICU, intensive care unit; CAP, community-acquired pneumonia; HAP, hospital-acquired pneumonia; RT, respiratory tract.
Potential roles of the pharyngeal microbiome in preventing RTIs
Living in a microbe-dominant world, all animals, including humans, must develop strategies to avoid unpredictable invasion of microbial pathogens. Hence, introduction of a handful of harmless microbes, as co-habitant defenders, presents an effective means to accomplish this requirement. Through the development of an efficient metabolic network, symbiotic commensal bacteria become established internally and externally and cooperate with the immune response of their hosts, thus excluding other foreign microbes (“strangers”) from establishing habitats. In the meantime, they release toxins or boost host immunity to kill other invasive species (“aggressors”) [15]. It has been well demonstrated that the intestinal microbiome plays an active role in the prevention of local infections [16]. Whether or not a pathogen can cause an infection depends on the balance between pathogenic expansion and microbiome homeostasis, which has been extensively studied in murine models [17]. Although the role of the pharyngeal microbiome in RTIs has not been thoroughly elucidated, there is some evidence suggesting similar protective effects to those of the gut microbiome as discussed below.
The pharyngeal microbiome has an essential role in the airway linings to protect against infection of air-transmitted pathogens, in addition to the host immunity, especially against the newly emerging infectious agents. The importance of the microbiome integrity is supported by the so-called herd effect in vaccination. That is, vaccination to a limited number of people offers protection from infection to others without vaccination. One well-known example is the epidemic of Haemophilus influenza type b in Finland, where a low vaccination rate of 50% resulted in a 95% reduction in the number of the invasive infections [18].
To prevent infection by indigenous species, it is necessary to maintain the microbiome homeostasis with beneficial abundance of each species. In the pharynx, many pathogenic species, including Streptococcus pneumonia, Staphylococcus aureus, H. influenza and Mycoplasma pneumonia, are well-adapted to the pharyngeal environment and become established in the resident microbiome, rendering the host asymptomatic [19–21]. Epidemiological studies suggested that the proportion of aforementioned resident pathogens in human hosts varies by season, as does the incidence of RTIs attributed to them [22]. Our recent study revealed that for each species in a set of common bacterial respiratory pathogens, there exists an abundance threshold, above which the risk of infection increases. Therefore, this threshold indicates the stringently-regulated homeostasis of a microbiome [23].
Damages of the pharyngeal microbiome prior to RTIs
Although the microbiome often acts as a line of defense against infection, the strength of such defense is largely dependent on the state of the host, especially that of the local epithelia [24]. To maintain a highly efficient and mutual metabolic support network, a spectrum of microbial species in appropriate proportions becomes prerequisite. However, the epithelia of the respiratory tract, including the pharynx, can become damaged by many factors, such as air pollutants, smoking, immune injury and infection. In hosts with genetic defects or epithelial dysfunction, such as in CF or COPD, the defensive abilities of the microbiome may decrease and render the host susceptible to pathogenic invasion. Such conditions often lead to repeated and chronic infection, which, in turn, further damages the epithelia and deteriorates the protective characteristics of the microbiome over time, similar to what occurs in dental and intestinal infections [25,26].
Recent studies have shown that an unhealthy status of the respiratory microbiome persists in high-risk populations, such as infants, the elderly and patients with COPD, CF or bronchiectasis, in whom the microbial communities are much less diversified than those found in healthy individuals [27–32]. Diversity is a major factor that promotes system stability [33], loss of diversity thus lessens the protective effects of the microbiome and renders patients vulnerable to infection. In patients with chronic RTIs, the defending frontier retreats from the throat to the trachea or bronchia as indicated by increased number of microbial sequences found in bronchoalveolar lavage fluid (BALF) and sputum samples [27,29]. These ectopic microbiomes are abnormal and usually fragile, resulting in repeated infections. In addition, susceptible populations may share a reduced abundance threshold, thereby allowing specific pathogens to easily establish infection [23].
In addition to the loss of diversity, the loss of protective species is also detrimental to homeostasis. Generally, among patients with chronic RTIs, those with CF are most vulnerable to infection, followed by those with COPD and then asthma. Metagenomic studies indicate that the proportion of Bacteroidetes spp. in sputa gradually decreases in the sequence of asthma, COPD and CF, whereas Proteobacteria, which include most of the common respiratory pathogens and are abundant in saliva, gain their proportions (Figure 1C) [8,9]. Therefore, it seems that Proteobacteria gradually expand when Bacteroidetes retreat. Although the protective effect of Bacteroidetes spp. remains to be validated, it is still reasonable to believe that not all species bear equal weight in maintaining pharyngeal microbiome homeostasis.
Impaired pharyngeal microbiome homeostasis may also be involved in the onset of acute infection. Predisposed infection with influenza or other viruses is commonly observed in patients with bacterial pneumonia, often leading to intensive aggravation of disease [34]. The predisposed viral infection is reported to be able to alter the local microbial ecology, epithelial function and immunity [35]. In such cases, some dormant pathogens may disengage from homeostatic control, propagate abnormally, and invade downward into the lung, eventually causing secondary bacterial infections [36–39].
Restoration of the pharyngeal microbiome
In addition to antibiotics and vaccines, restoration of a stable and healthy microbiome is another strategy that should be considered in infection control and anti-infective therapy, since the microbiome plays a direct role in fighting pathogenic infection. This strategy capitalizes on the protective roles of intact microbiomes, as described in both large-scale pangenomic [40] and metagenomic [41] studies. To reinforce host defense during infection, methods for evaluating the stability and imbalances of the microbiomes are needed to identify the infection-prone status and the species that should be replenished as recently reported [42].
Currently, the stability of microbiomes can only be evaluated based on resident biodiversity. Although many diversity indices have been developed [43–45], most consider only species richness and equilibrium in a given community, but disregard different beneficial effects specific to each species. Therefore, next-generation evaluation methods are in demand to specify appropriate proportions of each species in specific microbiomes, especially those with protective roles, in order to define deficiency and to further develop appropriate treatment regimens.
Large-scale clinical trials have already demonstrated several protective effects of probiotics or microbiome transplantation in combating intestinal infections, especially in managing secondary pathogens, such as Clostridium difficile [46–50]. These treatments have been shown to modulate host immune status and improve host resistance to pathogens [51,52], and have also been applied to the treatment of tooth [53], urinary tract [54] and vaginal infections [55].
Regarding the respiratory tract, recent studies have suggested potential protective effects of probiotic supplementation against infections with influenza virus and Corynebacterium tuberculostearicum [56,57], as well as pneumococcal [58] and ventilator-associated pneumonia [59], although other studies disputed the efficacy of probiotics for treatment of RTIs [60]. It is of note that main management in all these studies was orally-administrated Lactobacillus which may not be a good choice because of the low proportion of Lactobacillus in pharyngeal or respiratory tract microbiomes [61]. In addition, effective administration methods that can detain probiotics in the pharynx and multiple probiotic regimes may further improve the overall treatment outcome [62]. Therefore, there are no straightforward conclusions regarding the efficacy of probiotics for treatment of RTIs, based on the findings of current reports.
Conclusions
The constituents of human microbiome, which form an impressive biomass, are regarded as “permanent guests” of the body [63]. Increasing evidence suggests that resident microbiomes act more as guards than guests, in consideration of the benefits to the host. In the scope of the respiratory tract, there have been clues to suggest a potential protective role of the pharyngeal microbiome against pathogens invading the lungs. However, our current knowledge on this friendly microbiome is rather limited. Metagenomic and pangenomic studies based on high-throughput sequencing technologies are thus warranted to obtain additional molecular details of the airway infections. We therefore are able to ultimately develop new treatments based on the possibility of restoring the pharyngeal microbiome.
Acknowledgements
This work was supported by the National High Technology Research and Development Program of China (863 Program, Grant No. 2006AA02Z4A9) and the National Basic Research Program of China (973 Program, Grant No. 2011CB944100).
Footnotes
Peer review under responsibility of Beijing Institute of Genomics, Chinese Academy of Sciences and Genetics Society of China.
References
- 1.Sato S., Kiyono H. The mucosal immune system of the respiratory tract. Curr Opin Virol. 2012;2:225–232. doi: 10.1016/j.coviro.2012.03.009. [DOI] [PubMed] [Google Scholar]
- 2.Cenit MC, Matzaraki V, Tigchelaar EF, Zhernakova A. Rapidly expanding knowledge on the role of the gut microbiome in health and disease. Biochim Biophys Acta 2014. http://dx.doi.org/10.1016/j.bbadis.2014.05.023. [DOI] [PubMed]
- 3.Morrissey I, Hackel M, Badal R, Bouchillon S, Hawser S, Biedenbach D. A review of ten years of the Study for Monitoring Antimicrobial Resistance Trends (SMART) from 2002 to 2011. Pharmaceuticals (Basel, Switzerland) 2013;6:1335–1346. [DOI] [PMC free article] [PubMed]
- 4.Pennisi E. A mouthful of microbes. Science. 2005;307:1899–1901. doi: 10.1126/science.307.5717.1899. [DOI] [PubMed] [Google Scholar]
- 5.Ahn J., Yang L., Paster B.J., Ganly I., Morris L., Pei Z. Oral microbiome profiles: 16S rRNA pyrosequencing and microarray assay comparison. PLoS One. 2011;6:e22788. doi: 10.1371/journal.pone.0022788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Belda-Ferre P., Alcaraz L.D., Cabrera-Rubio R., Romero H., Simon-Soro A., Pignatelli M. The oral metagenome in health and disease. ISME J. 2012;6:46–56. doi: 10.1038/ismej.2011.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Griffen A.L., Beall C.J., Firestone N.D., Gross E.L., Difranco J.M., Hardman J.H. CORE: a phylogenetically-curated 16S rDNA database of the core oral microbiome. PLoS One. 2011;6:e19051. doi: 10.1371/journal.pone.0019051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guss A.M., Roeselers G., Newton I.L., Young C.R., Klepac-Ceraj V., Lory S. Phylogenetic and metabolic diversity of bacteria associated with cystic fibrosis. ISME J. 2011;5:20–29. doi: 10.1038/ismej.2010.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hilty M., Burke C., Pedro H., Cardenas P., Bush A., Bossley C. Disordered microbial communities in asthmatic airways. PLoS One. 2010;5:e8578. doi: 10.1371/journal.pone.0008578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Morris A., Beck J.M., Schloss P.D., Campbell T.B., Crothers K., Curtis J.L. Comparison of the respiratory microbiome in healthy nonsmokers and smokers. Am J Respir Crit Care Med. 2013;187:1067–1075. doi: 10.1164/rccm.201210-1913OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Charlson E.S., Bittinger K., Haas A.R., Fitzgerald A.S., Frank I., Yadav A. Topographical continuity of bacterial populations in the healthy human respiratory tract. Am J Respir Crit Care Med. 2011;184:957–963. doi: 10.1164/rccm.201104-0655OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wintermute E.H., Silver P.A. Dynamics in the mixed microbial concourse. Genes Dev. 2010;24:2603–2614. doi: 10.1101/gad.1985210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chow J., Lee S.M., Shen Y., Khosravi A., Mazmanian S.K. Host-bacterial symbiosis in health and disease. Adv Immunol. 2010;107:243–274. doi: 10.1016/B978-0-12-381300-8.00008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Song SJ, Lauber C, Costello EK, Lozupone CA, Humphrey G, Berg-Lyons D, et al. Cohabiting family members share microbiota with one another and with their dogs. eLife 2013;2:e00458. [DOI] [PMC free article] [PubMed]
- 15.Schmitz J.M., Durham C.G., Schoeb T.R., Soltau T.D., Wolf K.J., Tanner S.M. Helicobacter felis-associated gastric disease in microbiota-restricted mice. J Histochem Cytochem. 2011;59:826–841. doi: 10.1369/0022155411416242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Britton R.A., Young V.B. Role of the intestinal microbiota in resistance to colonization by Clostridium difficile. Gastroenterology. 2014;146:1547–1553. doi: 10.1053/j.gastro.2014.01.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Reeves A.E., Theriot C.M., Bergin I.L., Huffnagle G.B., Schloss P.D., Young V.B. The interplay between microbiome dynamics and pathogen dynamics in a murine model of Clostridium difficile infection. Gut Microbes. 2011;2:145–158. doi: 10.4161/gmic.2.3.16333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim T.H., Johnstone J., Loeb M. Vaccine herd effect. Scand J Infect Dis. 2011;43:683–689. doi: 10.3109/00365548.2011.582247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Watson K., Carville K., Bowman J., Jacoby P., Riley T.V., Leach A.J. Upper respiratory tract bacterial carriage in aboriginal and non-aboriginal children in a semi-arid area of Western Australia. Pediatr Infect Dis J. 2006;25:782–790. doi: 10.1097/01.inf.0000232705.49634.68. [DOI] [PubMed] [Google Scholar]
- 20.Quintero B, Araque M, van der Gaast-de Jongh C, Escalona F, Correa M, Morillo-Puente S, et al. Epidemiology of Streptococcus pneumoniae and Staphylococcus aureus colonization in healthy Venezuelan children. Eur J Clin Microbiol Infect Dis 2011;30:7–19. [DOI] [PMC free article] [PubMed]
- 21.Gnarpe J., Lundback A., Sundelof B., Gnarpe H. Prevalence of Mycoplasma pneumoniae in subjectively healthy individuals. Scand J Infect Dis. 1992;24:161–164. doi: 10.3109/00365549209052607. [DOI] [PubMed] [Google Scholar]
- 22.Bogaert D., Keijser B., Huse S., Rossen J., Veenhoven R., van Gils E. Variability and diversity of nasopharyngeal microbiota in children: a metagenomic analysis. PLoS One. 2011;6:e17035. doi: 10.1371/journal.pone.0017035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kang Y., Deng R., Wang C., Deng T., Peng P., Cheng X. Etiologic diagnosis of lower respiratory tract bacterial infections using sputum samples and quantitative loop-mediated isothermal amplification. PLoS One. 2012;7:e38743. doi: 10.1371/journal.pone.0038743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Larsson E., Tremaroli V., Lee Y.S., Koren O., Nookaew I., Fricker A. Analysis of gut microbial regulation of host gene expression along the length of the gut and regulation of gut microbial ecology through MyD88. Gut. 2012;61:1124–1131. doi: 10.1136/gutjnl-2011-301104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Santos A.L., Siqueira J.F., Jr., Rocas I.N., Jesus E.C., Rosado A.S., Tiedje J.M. Comparing the bacterial diversity of acute and chronic dental root canal infections. PLoS One. 2011;6:e28088. doi: 10.1371/journal.pone.0028088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.DuPont A.W., DuPont H.L. The intestinal microbiota and chronic disorders of the gut. Nat Rev Gastroenterol Hepatol. 2011;8:523–531. doi: 10.1038/nrgastro.2011.133. [DOI] [PubMed] [Google Scholar]
- 27.Erb-Downward J.R., Thompson D.L., Han M.K., Freeman C.M., McCloskey L., Schmidt L.A. Analysis of the lung microbiome in the “healthy” smoker and in COPD. PLoS One. 2011;6:e16384. doi: 10.1371/journal.pone.0016384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sibley C.D., Grinwis M.E., Field T.R., Eshaghurshan C.S., Faria M.M., Dowd S.E. Culture enriched molecular profiling of the cystic fibrosis airway microbiome. PLoS One. 2011;6:e22702. doi: 10.1371/journal.pone.0022702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Blainey P.C., Milla C.E., Cornfield D.N., Quake S.R. Quantitative analysis of the human airway microbial ecology reveals a pervasive signature for cystic fibrosis. Sci Transl Med. 2012;4:153ra30. doi: 10.1126/scitranslmed.3004458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zemanick E.T., Sagel S.D., Harris J.K. The airway microbiome in cystic fibrosis and implications for treatment. Curr Opin Pediatr. 2011;23:319–324. doi: 10.1097/MOP.0b013e32834604f2. [DOI] [PubMed] [Google Scholar]
- 31.Huang Y.J., Lynch S.V. The emerging relationship between the airway microbiota and chronic respiratory disease: clinical implications. Expert Rev Respir Med. 2011;5:809–821. doi: 10.1586/ers.11.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Whelan F.J., Verschoor C.P., Stearns J.C., Rossi L., Luinstra K., Loeb M. The loss of topography in the microbial communities of the upper respiratory tract in the elderly. Ann Am Thorac Soc. 2014;11:513–521. doi: 10.1513/AnnalsATS.201310-351OC. [DOI] [PubMed] [Google Scholar]
- 33.Chang J.Y., Antonopoulos D.A., Kalra A., Tonelli A., Khalife W.T., Schmidt T.M. Decreased diversity of the fecal microbiome in recurrent Clostridium difficile-associated diarrhea. J Infect Dis. 2008;197:435–438. doi: 10.1086/525047. [DOI] [PubMed] [Google Scholar]
- 34.Goffard A., Lambert V., Salleron J., Herwegh S., Engelmann I., Pinel C. Virus and cystic fibrosis: rhinoviruses are associated with exacerbations in adult patients. J Clin Virol. 2014;60:147–153. doi: 10.1016/j.jcv.2014.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Oshansky C.M., Gartland A.J., Wong S.S., Jeevan T., Wang D., Roddam P.L. Mucosal immune responses predict clinical outcomes during influenza infection independently of age and viral load. Am J Respir Crit Care Med. 2014;189:449–462. doi: 10.1164/rccm.201309-1616OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jacoby P., Watson K., Bowman J., Taylor A., Riley T.V., Smith D.W. Modelling the co-occurrence of Streptococcus pneumoniae with other bacterial and viral pathogens in the upper respiratory tract. Vaccine. 2007;25:2458–2464. doi: 10.1016/j.vaccine.2006.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.DeLeo F.R., Musser J.M. Axis of coinfection evil. J Infect Dis. 2010;201:488–490. doi: 10.1086/650304. [DOI] [PubMed] [Google Scholar]
- 38.Rizzo C., Caporali M.G., Rota M.C. Pandemic influenza and pneumonia due to Legionella pneumophila: a frequently underestimated coinfection. Clin Infect Dis. 2010;51:115. doi: 10.1086/653444. [DOI] [PubMed] [Google Scholar]
- 39.Moore H.C., Jacoby P., Taylor A., Harnett G., Bowman J., Riley T.V. The interaction between respiratory viruses and pathogenic bacteria in the upper respiratory tract of asymptomatic aboriginal and non-aboriginal children. Pediatr Infect Dis J. 2010;29:540–545. doi: 10.1097/INF.0b013e3181d067cb. [DOI] [PubMed] [Google Scholar]
- 40.Markowitz V.M., Chen I.M., Chu K., Szeto E., Palaniappan K., Grechkin Y. IMG/M: the integrated metagenome data management and comparative analysis system. Nucleic Acids Res. 2012;40:D123–D129. doi: 10.1093/nar/gkr975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tosh P.K., McDonald L.C. Infection control in the multidrug-resistant era: tending the human microbiome. Clin Infect Dis. 2012;54:707–713. doi: 10.1093/cid/cir899. [DOI] [PubMed] [Google Scholar]
- 42.Rogers G.B., Zain N.M., Bruce K.D., Burr L.D., Chen A.C., Rivett D.W. A novel microbiota stratification system predicts future exacerbations in bronchiectasis. Ann Am Thorac Soc. 2014;11:496–503. doi: 10.1513/AnnalsATS.201310-335OC. [DOI] [PubMed] [Google Scholar]
- 43.Shannon C.E. The mathematical theory of communication. 1963. MD Comput. 1997;14:306–317. [PubMed] [Google Scholar]
- 44.Tuomisto H. A consistent terminology for quantifying species diversity? Yes, it does exist. Oecologia. 2010;164:853–860. doi: 10.1007/s00442-010-1812-0. [DOI] [PubMed] [Google Scholar]
- 45.Tuomisto H., Ruokolainen K. Analyzing or explaining beta diversity? Understanding the targets of different methods of analysis. Ecology. 2006;87:2697–2708. doi: 10.1890/0012-9658(2006)87[2697:aoebdu]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 46.Cimperman L., Bayless G., Best K., Diligente A., Mordarski B., Oster M. A randomized, double-blind, placebo-controlled pilot study of Lactobacillus reuteri ATCC 55730 for the prevention of antibiotic-associated diarrhea in hospitalized adults. J Clin Gastroenterol. 2011;45:785–789. doi: 10.1097/MCG.0b013e3182166a42. [DOI] [PubMed] [Google Scholar]
- 47.Borody T.J., Khoruts A. Fecal microbiota transplantation and emerging applications. Nat Rev Gastroenterol Hepatol. 2012;9:88–96. doi: 10.1038/nrgastro.2011.244. [DOI] [PubMed] [Google Scholar]
- 48.Zihler A., Gagnon M., Chassard C., Lacroix C. Protective effect of probiotics on Salmonella infectivity assessed with combined in vitro gut fermentation-cellular models. BMC Microbiol. 2011;11:264. doi: 10.1186/1471-2180-11-264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lundell L. Use of probiotics in abdominal surgery. Dig Dis. 2011;29:570–573. doi: 10.1159/000332984. [DOI] [PubMed] [Google Scholar]
- 50.Gough E., Shaikh H., Manges A.R. Systematic review of intestinal microbiota transplantation (fecal bacteriotherapy) for recurrent Clostridium difficile infection. Clin Infect Dis. 2011;53:994–1002. doi: 10.1093/cid/cir632. [DOI] [PubMed] [Google Scholar]
- 51.Willing B.P., Vacharaksa A., Croxen M., Thanachayanont T., Finlay B.B. Altering host resistance to infections through microbial transplantation. PLoS One. 2011;6:e26988. doi: 10.1371/journal.pone.0026988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wells J.M. Immunomodulatory mechanisms of lactobacilli. Microb Cell Fact. 2011;10(Suppl 1):S17. doi: 10.1186/1475-2859-10-S1-S17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Culp D.J., Robinson B., Parkkila S., Pan P.W., Cash M.N., Truong H.N. Oral colonization by Streptococcus mutans and caries development is reduced upon deletion of carbonic anhydrase VI expression in saliva. Biochim Biophys Acta. 2011;1812:1567–1576. doi: 10.1016/j.bbadis.2011.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Amdekar S., Singh V., Singh D.D. Probiotic therapy: immunomodulating approach toward urinary tract infection. Curr Microbiol. 2011;63:484–490. doi: 10.1007/s00284-011-0006-2. [DOI] [PubMed] [Google Scholar]
- 55.Martin D.H. The microbiota of the vagina and its influence on women’s health and disease. Am J Med Sci. 2012;343:2–9. doi: 10.1097/MAJ.0b013e31823ea228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Youn H.N., Lee D.H., Lee Y.N., Park J.K., Yuk S.S., Yang S.Y. Intranasal administration of live Lactobacillus species facilitates protection against influenza virus infection in mice. Antiviral Res. 2012;93:138–143. doi: 10.1016/j.antiviral.2011.11.004. [DOI] [PubMed] [Google Scholar]
- 57.Abreu N.A., Nagalingam N.A., Song Y., Roediger F.C., Pletcher S.D., Goldberg A.N. Sinus microbiome diversity depletion and Corynebacterium tuberculostearicum enrichment mediates rhinosinusitis. Sci Transl Med. 2012;4:151ra24. doi: 10.1126/scitranslmed.3003783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tanaka A., Seki M., Yamahira S., Noguchi H., Kosai K., Toba M. Lactobacillus pentosus strain b240 suppresses pneumonia induced by Streptococcus pneumoniae in mice. Lett Appl Microbiol. 2011;53:35–43. doi: 10.1111/j.1472-765X.2011.03079.x. [DOI] [PubMed] [Google Scholar]
- 59.Bailey J.L., Yeung S.Y. Probiotics for disease prevention: a focus on ventilator-associated pneumonia. Ann Pharmacother. 2011;45:1425–1432. doi: 10.1345/aph.1Q241. [DOI] [PubMed] [Google Scholar]
- 60.Barraud D., Bollaert P.E., Gibot S. Impact of the administration of probiotics on mortality in critically ill adult patients: a meta-analysis of randomized controlled trials. Chest. 2013;143:646–655. doi: 10.1378/chest.12-1745. [DOI] [PubMed] [Google Scholar]
- 61.Forsythe P. Probiotics and lung diseases. Chest. 2011;139:901–908. doi: 10.1378/chest.10-1861. [DOI] [PubMed] [Google Scholar]
- 62.Osipov G.A., Verkhovtseva N.V. Study of human microecology by mass spectrometry of microbial markers. Benef Microbes. 2011;2:63–78. doi: 10.3920/BM2010.0017. [DOI] [PubMed] [Google Scholar]
- 63.Avila M., Ojcius D.M., Yilmaz O. The oral microbiota: living with a permanent guest. DNA Cell Biol. 2009;28:405–411. doi: 10.1089/dna.2009.0874. [DOI] [PMC free article] [PubMed] [Google Scholar]



