Abstract
Acinetobacter baumannii is an important opportunistic pathogen in hospital, and the multidrug-resistant isolates of A. baumannii have been increasingly reported in recent years. A number of different mechanisms of resistance have been reported, some of which are associated with plasmid-mediated acquisition of genes. Therefore, studies on plasmids in A. baumannii have been a hot issue lately. We have performed complete genome sequencing of A. baumannii MDR-TJ, which is a multidrug-resistant isolate. Finalizing the remaining large scaffold of the previous assembly, we found a new plasmid pABTJ2, which carries many phage-like elements. The plasmid pABTJ2 is a circular double-stranded DNA molecule, which is 110,967 bp in length. We annotated 125 CDSs from pABTJ2 using IMG ER and ZCURVE_V, accounting for 88.28% of the whole plasmid sequence. Many phage-like elements and a tRNA-coding gene were detected in pABTJ2, which is rarely reported among A. baumannii. The tRNA gene is specific for asparagine codon GTT, which may be a small chromosomal sequence picked up through incorrect excision during plasmid formation. The phage-like elements may have been acquired during the integration process, as the GC content of the region carrying phage-like elements was higher than that of the adjacent regions. The finding of phage-like elements and tRNA-coding gene in pABTJ2 may provide a novel insight into the study of A. baumannii pan-plasmidome.
Keywords: A. baumannii, Plasmid, tRNA gene, Phage-like elements
Introduction
Acinetobacter baumannii is an important opportunistic pathogen, which causes serious nosocomial infections in hospital, especially in the intensive care unit (ICU) [1]. Unfortunately, the multidrug-resistant isolates of A. baumannii have been increasingly reported in recent years, which largely limited the performance of antibiotics against its infection [2]. Studies on plasmids in A. baumannii have been a hot issue lately, as the presence of some plasmids was closely related with drug resistance [3]. Most clinically significant resistance to carbapenem in this species has been associated with plasmid-mediated acquisition of genes encoding either class B metallo-β-lactamases or carbapenem-hydrolyzing class D OXA-type β-lactamases (CHDLs) [4]. Plasmids are extrachromosomal DNA molecules that are capable of autonomous replication, and they can be inherited both vertically and horizontally in a prokaryotic community [5]. As a mobile element, plasmid may accelerate the dissemination of genetic determinants, such as the spread of resistance genes [6].
We previously reported the complete genome sequence of the multidrug-resistant A. baumannii strain MDR-TJ, which was collected in Tianjin, China in 2012 [7]. The genome of A. baumannii MDR-TJ consists of a circular chromosome and a circular plasmid pABTJ1. Using bioinformatics tools, we further finalized the remaining large scaffold of the previous assembly [8]. Interestingly, we found that this newly-assembled circular DNA carried many phage-like elements, which was suspected to be a prophage. This circular DNA molecule was not described when we submitted the previous draft genome. Further scrutinization of the experimental procedures showed that no cellular lysis was performed following mitomycin C or UV radiation treatment [9], suggesting that this DNA was not likely to be an active prophage. Moreover, elements related to plasmid replication and stabilization were identified in this new circular DNA. Therefore, we consider this DNA as a new circular plasmid and name it as pABTJ2. Here we report the complete nucleotide sequence of pABTJ2 and describe its structure briefly.
Results and discussions
The plasmid pABTJ2 was determined to be a 110,967 bp circular double-stranded plasmid with an average GC content of 41.61%. We annotated 125 coding sequences (CDSs) in pABTJ2, representing 88.28% of the complete sequence. Notably, a tRNAAsnGTT gene was also found in this plasmid. Among the protein-coding genes, only 44 CDSs were assigned to a known function. The map of pABTJ2 is shown in Figure 1.
Figure 1.

Graphical circular map of plasmid pABTJ2 DNA replication-related or phage-like CDSs (in blue) and the tRNA gene (in pink) were indicated in the forward (Circle 1) and reverse (Circle 4) strands, respectively; whereas the open reading frames (ORFs) were indicated in the forward (Circle 2) and reverse (Circle 3) strands, respectively. Circles are numbered 1 to 4 from the outside inward. Note that only ORFs containing more than 100 codons are shown. The locations of seven nucleotides that are different between pABTJ2 and pOIFC189-111 are put in brown and indicated by arrows, and the corresponding nucleotides on the pOIFC189-111 are marked in parentheses. Three out of these seven nucleotides are also different between pABTJ2 and pABUH4-111, which are marked with triangles. The origin of replication (oriV) is marked in green.
Replication modules
The putative origin of replication (oriV) region contained four imperfectly-conserved 22 bp direct repeats, which may be termed as “iterons” [10]. The rep gene lies next to the oriV region, encoding a 373-amino acid replicase (also known as the replication initiation protein). This replicase belongs to the Rep-3 superfamily (pfam0151), similar as the replicase of most A. baumannii plasmids [3]. A. baumannii PCR-based replicon typing (AB-PBRT) [3] is one of the most common methods used for plasmid typing in A. baumannii. The rep gene in pABTJ2 was compared with representatives of each group (GR) categorized by AB-PBRT using BLASTN to identify its closest matches. However, the rep gene in pABTJ2 showed low similarities with each of these groups. Therefore, we failed to categorize pABTJ2 into any identified GRs using this method. These data indicate that pABTJ2 would possess a novel replicon type.
The pABTJ2 also contained many genes that are potentially related to DNA metabolism and replication, such as dnaN, polA, nrdA and nrdB. These genes are normally located on the chromosome of bacteria or bacteriophages but rarely found in the plasmid. In bacteriophage T4, NrdA and NrdB, which are part of the primase replication complex, contribute to rapid phage DNA biosynthesis [11]. We speculate that the genes in pABTJ2 may also help improve the replication functions under stressful conditions, such as the high antibiotic environment or in the host [11].
tRNA gene
A gene encoding tRNA was identified in pABTJ2, which is uncommon. This tRNA gene is specific for the second most frequently used asparagine codon, GTT. tRNA genes are often observed in large DNA viruses or in bacterial genomes [12]. For viruses, tRNA may serve as a recruitment element to neutralize the compositional differences between the phage and the host genome, so as to adjust the translation capacity during infection [13]. However, this may not be the case for pABTJ2, since there are four copies of the tRNAAsnGTT present in the A. baumannii chromosome, suggesting that this tRNA may be dispensable for inheritance of the plasmid. tRNA loci commonly serve as insertion sites for mobile elements in the prokaryotic chromosomes, as they are highly conserved between bacteria and thus allow for a promiscuous process [13]. Therefore, the asn tRNA gene carried by pABTJ2 could also be utilized as the recognition site for recombination. As genetic elements could be probably exchanged between plasmids and the host chromosome, we did a BLASTN analysis between the tRNAAsnGTT gene in pABTJ2 and the ones on the chromosome to uncover the potential origin of the tRNAAsnGTT gene. However, the nucleotide sequence of the tRNAAsnGTT gene in pABTJ2 shares no significant similarity with the ones on the chromosome, suggesting that this gene may not originate from the host chromosome of A. baumannii MDR-TJ. The mechanism underlying the presence of tRNA gene in the plasmid remains obscure. One possibility is that plasmids may be able to integrate into the bacterial chromosome and pick up the chromosomal sequences, such as the tRNA gene, in the course of incorrect excision [14]. For instance, the asn tRNA gene in the plasmid pHCM2 was hypothesized to be acquired through this mechanism [14].
Phage-like elements
A cluster of genes encoding phage proteins were detected, which were scattered in a 30 kb region at the end of pABTJ2. Compared to larger cryptic prophages, these genes are isolated, which were thus proposed to be phage remnants. Nine phage-like genes were annotated (Table 1), which encode proteins related to packing/morphogenesis and host lysis (muraminidase). Among them, 7 genes share similarities with those of the Salmonella phage SSU5. However, compared to Salmonella phage SSU5, pABTJ2 lacks the genes required for the phage life cycle (i.e., receptor-recognizing phage tail fiber adhesion, superinfection exclusion protein and phage repressor) [15]. Instead, pABTJ2 mainly harbors some phage structural genes, which corresponds to the non-inducibility of pABTJ2 as mentioned above. Since many ORFs included in this region are annotated as hypothetical proteins, it is currently difficult to predict whether these phage-like elements would confer any fitness advantage to the strain.
Table 1.
Characteristics of phage-like elements in pABTJ2
| ORF | Putative function | Length | Amino acid similarity (%) |
|---|---|---|---|
| ABTJ_00102 | Phage terminase large subunit | 414 | 59.97 |
| ABTJ_00105 | Phage capsid family | 300 | 58.61 |
| ABTJ_00115 | Phage tail tape measure protein | 1862 | 10.86 |
| ABTJ_00116 | Phage minor tail | 108 | 31.25 |
| ABTJ_00117 | Phage minor tail | 231 | 46.81 |
| ABTJ_00118 | Cell wall-associated hydrolases | 249 | 36.80 |
| ABTJ_00119 | Phage tail component | 192 | 34.83 |
| ABTJ_00120 | Phage tail component | 3727 | 18.27 |
| ABTJ_00126 | Phage-related lysozyme | 212 | 42.40 |
Note: Length of protein is indicated by number of amino acid residues comprised; amino acid similarity (%) of individual ORFs is calculated against those in Salmonella phage SSU5, except for ABTJ_00115 (against Enterobacterial phage mEp390) and ABTJ_00126 (against Acinetobacter phage ZZ1).
The mechanism underlying the presence of phage-like elements in the plasmid is not clear. According to the GC-Profile [16] (http://tubic.tju.edu.cn/GC-Profile/), the GC content of this region (70,050 nt −106,632 nt) (44.90%) is higher than that of the adjacent regions (38.44% and 39.06%), indicating that the phage-like elements may be acquired through horizontal transfer. Genetic exchanges between plasmids and their host chromosome are common [5]. Considering that chromosome usually harbors prophages, the phage-like elements in pABTJ2 may be acquired from the chromosome via homologous recombination [5]. Other site-specific integration processes may have been functional in the acquisition of mobile genetic elements as well [17]. The predicted asn tRNA gene in pABTJ2 may be used as an insertion site for integration of these phage-like elements. Generally, the site-specific integration of genetic elements through tRNA could generate two direct repeat sequences on both ends, namely attL and attR [18]. However, no direct repeat portions of the tRNA gene are found elsewhere to suggest that this region functions as an excisable island.
Through an NCBI BLASTN analysis, we found that two plasmids, pOIFC189-111 and pABUH4-111, shared 99% nucleotide sequence identity with pABTJ2. pOIFC189-111 was extracted from A. baumannii strain OIFC189 that was isolated from combat casualties in Iraq. It is 110,967 bp long with only seven nucleotides different from pABTJ2. On the other hand, 111,007 bp pABUH4-111 was extracted from A. baumannii strain UH2107, which was isolated from sputum sample of an inpatient in a US hospital [19]. Compared to pABTJ2, there exist 40-bp stretches of nucleotides in pABUH4-111, which turned out to be duplication located at both the head and the tail of the sequence. Given the distinct geographical origins, the presence of highly-identical plasmids is intriguing. It should be noted that, pABTJ2 is neither a conjugative plasmid, as it lacks the required transfer machinery, nor it is a mobilizable plasmid, as it lacks the mobilization regions either and it is much larger than the co-resident conjugative plasmid pABTJ1 [7,20]. Therefore, pABTJ2 should not be able to transfer independently. Natural transformation or phage transduction may be the alternative mechanism through which transmission is achieved [21]. However, whether these A. baumannii strains are able to engage in natural transformation remains unknown, and the presence of endogenous transducing phages in A. baumannii has not been demonstrated yet. Besides, due to the high similarity in the plasmid sequences, the genetic relationships of their hosts capture our interest. We thus constructed a single nucleotide polymorphism (SNP)-based phylogenetic tree using the genomes of 16 completely-sequenced A. baumannii strains and the draft of the A. baumannii OIFC189 and UH2107 published in the NCBI. It is shown that the A. baumannii MDR-TJ, A. baumannii UH2107 and A. baumannii OIFC189 are very closely related (Figure 2). Therefore, the close relationship among the bacterial strains may explain in part the high level of similarity observed among the plasmids they host. The presence of nearly-identical plasmids and closely-related bacterial strains isolated from geographically-widespread areas indicates the global dissemination of these organisms and elements.
Figure 2.
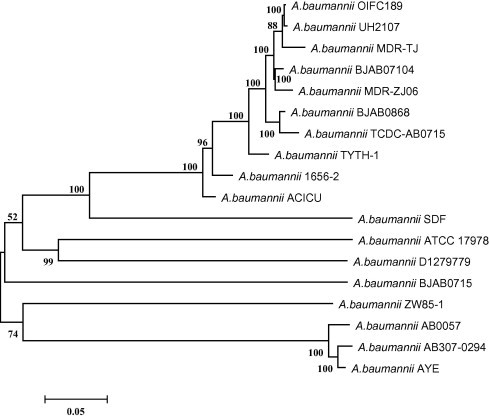
SNP-based phylogenetic tree of A. baumannii genomes 83,560 SNPs were identified on the basis of whole-genome alignment of 18 sequenced A. baumannii strains, which were used to construct a maximum-likelihood tree with 100 bootstrap replicates. The bar indicates 0.05 substitutions per nucleotide position. Bootstrap values >50 are shown on the branches.
Conclusion
In this article, we reported the complete sequence of pABTJ2, a newly-identified plasmid, in A. baumannii strain MDR-TJ. To the best of our knowledge, the existence of the tRNA gene and phage-like elements has rarely been reported in other A. baumannii plasmids. The tRNA gene is specific for asparagine codon, GTT, and this tRNA gene may be a small chromosomal sequence picked up through incorrect excision during plasmid formation. The phage-like elements are mainly phage structural genes, which may have been acquired during the integration process. The exact mechanism of the presence of tRNA gene and phage-like elements has yet to be clarified. pABTJ2 also encodes many genes that are potentially related to DNA metabolism and replication, which are speculated to improve replication function under stressful conditions. The presence of nearly-identical plasmids may indicate the global dissemination of plasmids similar to pABTJ2. Although underlying mechanisms need to be further explored, the finding of pABTJ2 may provide a novel insight into the research of A. baumannii pan-plasmidome.
Materials and methods
Genome annotation and bioinformatics analysis
The nucleotide sequence of pABTJ2 was obtained from the whole-genome shotgun sequencing of A. baumannii MDR-TJ [7]. Genome annotation was performed using IMG ER [22] and ZCURVE_V [23]. Phage-like elements were predicted according to the annotation results. tRNA genes were predicted using tRNAscan-SE [24]. The origin of replication (oriV) was identified using Ori-Finder [25]. A circular map of plasmid pABTJ2 was created using the CGView Server [26]. Protein sequences of phage-like elements of pABTJ2 were aligned using DNAMAN v6 software (Lynnon Biosoft, Quebec, Canada). Nucleotide differences between pABTJ2, pOIFC189-111 and pABUH4-111 were determined using BLASTN. The GenBank accession numbers for these three plasmid sequences were listed in Table S1.
Phylogenetic analysis
Whole genome sequences of 18 A. baumannii strains were obtained from NCBI. The whole-genome alignment was performed by using MUMmer and 83,560 SNPs were identified, which were used to construct a maximum-likelihood tree with 100 bootstrap replicates [27]. The SNP-based phylogenetic tree was constructed using snpTree [27]. The names of the 18 A. baumannii strains and GenBank accession numbers of their genome sequences were listed in Table S1.
Authors’ contributions
HH and FG conceived the idea and planned the project. DY and ZLY performed experiments and analyzed the sequence of pABTJ2. HL and XZ participated in bioinformatics analysis. DY drafted the manuscript. All authors edited the manuscript and approved the final manuscript.
Competing interests
The authors have declared that no competing interests exist.
Acknowledgements
We would like to thank Professor Chun-Ting Zhang for invaluable assistance and inspiring discussion. The present work was partially supported by the National Natural Science Foundation of China (Grant Nos. 90408028, 31171238, 30800642 and 10747150), the Program for New Century Excellent Talents in University of China (Grant No. NCET-12-0396) and Tianjin Municipal Natural Science Foundation of China (Grant No. 09JCZDJC17100).
Footnotes
Peer review under responsibility of Beijing Institute of Genomics, Chinese Academy of Sciences and Genetics Society of China.
Supplementary material
GenBank accession number of genome sequences analyzed in this study
References
- 1.Peleg A.Y., Seifert H., Paterson D.L. Acinetobacter baumannii: emergence of a successful pathogen. Clin Microbiol Rev. 2008;21:538–582. doi: 10.1128/CMR.00058-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.García-Quintanilla M., Pulido M.R., López-Rojas R., Pachón J., McConnell M.J. Emerging therapies for multidrug resistant Acinetobacter baumannii. Trends Microbiol. 2013;21:157–163. doi: 10.1016/j.tim.2012.12.002. [DOI] [PubMed] [Google Scholar]
- 3.Bertini A., Poirel L., Mugnier P.D., Villa L., Nordmann P., Carattoli A. Characterization and PCR-based replicon typing of resistance plasmids in Acinetobacter baumannii. Antimicrob Agents Chemother. 2010;54:4168–4177. doi: 10.1128/AAC.00542-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Poirel L., Nordmann P. Carbapenem resistance in Acinetobacter baumannii: mechanisms and epidemiology. Clin Microbiol Infect. 2006;12:826–836. doi: 10.1111/j.1469-0691.2006.01456.x. [DOI] [PubMed] [Google Scholar]
- 5.Fondi M., Bacci G., Brilli M., Papaleo M.C., Mengoni A., Vaneechoutte M. Exploring the evolutionary dynamics of plasmids: the Acinetobacter pan-plasmidome. BMC Evol Biol. 2010;10:59. doi: 10.1186/1471-2148-10-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carattoli A. Plasmids and the spread of resistance. Int J Med Microbiol. 2013;303:298–304. doi: 10.1016/j.ijmm.2013.02.001. [DOI] [PubMed] [Google Scholar]
- 7.Huang H., Yang Z.L., Wu X.M., Wang Y., Liu Y.J., Luo H. Complete genome sequence of Acinetobacter baumannii MDR-TJ and insights into its mechanism of antibiotic resistance. J Antimicrob Chemother. 2012;67:2825–2832. doi: 10.1093/jac/dks327. [DOI] [PubMed] [Google Scholar]
- 8.Wajid B., Serpedin E. Review of general algorithmic features for genome assemblers for next generation sequencers. Genomics Proteomics Bioinformatics. 2012;10:58–73. doi: 10.1016/j.gpb.2012.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jiang S.C., Paul J.H. Significance of lysogeny in the marine environment studies with isolates and a model of lysogenic phage production. Microb Ecol. 1998;35:235–243. doi: 10.1007/s002489900079. [DOI] [PubMed] [Google Scholar]
- 10.Chattoraj D.K. Control of plasmid DNA replication by iterons: no longer paradoxical. Mol Microbiol. 2000;37:467–476. doi: 10.1046/j.1365-2958.2000.01986.x. [DOI] [PubMed] [Google Scholar]
- 11.Kidgell C., Pickard D., Wain J., James K., Diem Nga L.T., Diep T.S. Characterization and distribution of a cryptic Salmonella typhi plasmid pHCM2. Plasmid. 2002;47:159–171. doi: 10.1016/s0147-619x(02)00013-6. [DOI] [PubMed] [Google Scholar]
- 12.Dreher T.W. Viral tRNAs and tRNA-like structures. Wiley Interdiscip Rev RNA. 2010;1:402–414. doi: 10.1002/wrna.42. [DOI] [PubMed] [Google Scholar]
- 13.Bailly-Bechet M., Vergassola M., Rocha E. Causes for the intriguing presence of tRNAs in phages. Genome Res. 2007;17:1486–1495. doi: 10.1101/gr.6649807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Antonenka U., Nölting C., Heesemann J., Rakin A. Horizontal transfer of Yersinia high-pathogenicity island by the conjugative RP4 attB target-presenting shuttle plasmid. Mol Microbiol. 2005;57:727–734. doi: 10.1111/j.1365-2958.2005.04722.x. [DOI] [PubMed] [Google Scholar]
- 15.Kim M., Kim S., Ryu S. Complete genome sequence of bacteriophage SSU5 specific for Salmonella enterica serovar Typhimurium rough strains. J Virol. 2012;86:10894. doi: 10.1128/JVI.01796-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gao F., Zhang C.T. GC-Profile: a web-based tool for visualizing and analyzing the variation of GC content in genomic sequences. Nucleic Acids Res. 2006;34:W686–W691. doi: 10.1093/nar/gkl040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Prentice M.B., James K.D., Parkhill J., Baker S.G., Stevens K., Simmonds M.N. Yersinia pestis pFra shows biovar-specific differences and recent common ancestry with a Salmonella enterica serovar Typhi plasmid. J Bacteriol. 2001;183:2586–2594. doi: 10.1128/JB.183.8.2586-2594.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Antonenka U., Nölting C., Heesemann J., Rakin A. Independent acquisition of site-specific recombination factors by asn tRNA gene-targeting genomic islands. Int J Med Microbiol. 2006;296:341–352. doi: 10.1016/j.ijmm.2006.02.021. [DOI] [PubMed] [Google Scholar]
- 19.Wright M.S., Haft D.H., Harkins D.M., Perez F., Hujer K.M., Bajaksouzian S. New insights into dissemination and variation of the health care-associated pathogen Acinetobacter baumannii from genomic analysis. MBio. 2014;5:e00963-13. doi: 10.1128/mBio.00963-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Garcillán-Barcia M.P., Francia M.V., de la Cruz F. The diversity of conjugative relaxases and its application in plasmid classification. FEMS Microbiol Rev. 2009;33:657–687. doi: 10.1111/j.1574-6976.2009.00168.x. [DOI] [PubMed] [Google Scholar]
- 21.Sorensen S.J., Bailey M., Hansen L.H., Kroer N., Wuertz S. Studying plasmid horizontal transfer in situ: a critical review. Nat Rev Microbiol. 2005;3:700–710. doi: 10.1038/nrmicro1232. [DOI] [PubMed] [Google Scholar]
- 22.Markowitz V.M., Mavromatis K., Ivanova N.N., Chen I.M., Chu K., Kyrpides N.C. IMG ER: a system for microbial genome annotation expert review and curation. Bioinformatics. 2009;25:2271–2278. doi: 10.1093/bioinformatics/btp393. [DOI] [PubMed] [Google Scholar]
- 23.Guo F.B., Zhang C.T. ZCURVE_V: a new self-training system for recognizing protein-coding genes in viral and phage genomes. BMC Bioinformatics. 2006;7:9. doi: 10.1186/1471-2105-7-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lowe T.M., Eddy S.R. TRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 1997;25:955–964. doi: 10.1093/nar/25.5.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gao F., Zhang C.T. Ori-Finder: a web-based system for finding oriCs in unannotated bacterial genomes. BMC Bioinformatics. 2008;9:79. doi: 10.1186/1471-2105-9-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Grant J.R., Stothard P. The CGView Server: a comparative genomics tool for circular genomes. Nucleic Acids Res. 2008;36:W181–W184. doi: 10.1093/nar/gkn179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Leekitcharoenphon P., Kaas R.S., Thomsen M.C.F., Friis C., Rasmussen S., Aarestrup F.M. SnpTree—a web-server to identify and construct SNP trees from whole genome sequence data. BMC Genomics. 2012;13:S6. doi: 10.1186/1471-2164-13-S7-S6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
GenBank accession number of genome sequences analyzed in this study


