Abstract
Pearl millet (Pennisetum glaucum) is an important cereal of traditional farming systems that has the natural ability to withstand various abiotic stresses. The present study aims at the identification and validation of major differentially expressed genes in response to drought stress in P. glaucum by Suppression Subtractive Hybridization (SSH) analysis. Twenty-two days old seedlings of P. glaucum cultivar PPMI741 were subjected to drought stress by treatment of 30 % Polyethylene glycol for different time periods 30 min (T1), 2 h (T2), 4 h (T3), 8 h (T4), 16 h (T5), 24 h (T6) and 48 h (T7) respectively, monitored by examining the RWC of seedlings. Total RNA was isolated to construct drought responsive subtractive cDNA library through SSH, sequenced to identify the differentially expressed genes in response to drought stress and validated by qRT-PCR.745 ESTs were assembled into a collection of 299 unigenes having 52 contigs and 247 singletons. All 745 ESTs were submitted to ENA-EMBL databases (Accession no. HG516611- HG517355). After analysis, 10 differentially expressed genes were validated namely Abscisic stress ripening protein, Ascorbate peroxidase, Inosine-5′-monophosphate dehydrogenase, Putative beta-1, 3-glucanase, Glyoxalase, Rab7, Aspartic proteinase Oryzasin, DnaJ—like protein and Calmodulin—like protein by qRT-PCR. The identified ESTs reveal a major portion of the stress responsive transcriptome that may prove to be a vent to unravel molecular basis underlying tolerance of pearl millet (Pennisetum glaucum) to drought stress. These genes could be utilized for transgenic breeding or transferred to crop plants through marker assisted selection for the development of better drought resistant cultivars having enhanced adaptability to survive harsh environmental conditions.
Electronic supplementary material
The online version of this article (doi:10.1007/s12298-015-0287-1) contains supplementary material, which is available to authorized users.
Keywords: Drought stress, ESTs, Pearl millet, qRT-PCR, Stress responsive genes
Introduction
Water deficiency is an important abiotic factor which restricts the crop productivity in the semi-arid tropics. Along with this, the altering climatic conditions are also expected to contribute towards drought stresses with enhanced severity in the near future. Consequently, sustainable and equitable worldwide food security is mainly dependent on the development of crop plants with better adaptation to water-limited environments (Kholova et al. 2014).
The decrease in rainfall due to world-wide climatic shifts has been predicted to reduce crop yield in semi-arid areas. The reduction of crop yield currently affects approximately 3.6 billion ha (25 % of upland in the world) in semi-arid and arid areas (UNEP report 1991). In these areas, desertification and population growth will exacerbate food shortage. Pearl millet [Penniseteum glaucum (L.R.Br)] belongs to the family Poaceae and is the most widely grown type of millet, which has strong development of underground organs and tends to have efficient adaptive mechanisms to cope with drought (Bezançon et al. 2009). India is the largest producer of pearl millet in Asia, both in terms of area (about 9 million ha) and production (8.3 million tons) with an average productivity of 930 kg/ha during the past 3 years (ICAR-AICPMIP report 2014). Drought is one of the main environmental constraints to agricultural productivity worldwide. Many efforts have been made to elucidate the mechanisms of drought tolerance in plants through molecular and genomics approaches, and a number of genes that respond to drought stress at the transcriptional level have been reported (Guo et al. 2009). Drought limits the agricultural production by preventing the crop plants from expressing their full genetic potential (Mitra 2001). Drought is actually a meteorological event which implies the absence of rainfall for a period of time, long enough to cause moisture depletion in soil and plant tissue (Fatima et al. 2014) but it is hard to exactly define drought because it has different meaning in different areas. In a very arid region there has to be really long period of no precipitation to be considered as drought. On the other hand in tropical areas a period of six days with no rain at all can be considered as a drought event.
Several techniques have been utilized for identifying differentially expressed genes (Liang and Pardee 1992; Vos et al. 1995; Hubank and Schatz 1994). Among the various methods for differential transcriptome analysis, Suppression Subtractive Hybridization (SSH) proves to be an efficient approach to isolate and identify cDNAs of differentially expressed genes in the absence of sequence information (Mishra et al. 2007; Almeida et al. 2013; Ding et al. 2014; Khan et al. 2014). In this technique differentially expressed genes can be normalized and enriched over 1000-fold in a single round of hybridization (Diatchenko et al. 1996). This would substantially increase the chances for identification of rare transcripts involved in drought stress. The concept of using cDNAs as a route to expedite gene discovery was first demonstrated in the early 1980s (Putney et al. 1983). Later gene discovery in most plants was done primarily by sample sequencing of expressed sequence tags (ESTs) (Lim et al. 1996; Covitz et al. 1998).
SSH has successfully been used to identify genes responsive to various biotic and abiotic stresses in various plant species such as Saccharum (Watt 2003); Festuca (Zhang et al. 2005); Pennisetum (Mishra et al. 2007); Agrostis (Xu et al. 2009); Triticum (Chauhan et al. 2011). Thus in the current study we focus on identifying, validating and establishing the putative functions of the major differentially expressed genes in P. glaucum during drought stress response using SSH, real-time quantitative qRT-PCR and homology searches.
Materials and methods
Seed material and stress treatment
Seeds of Penniseteum glaucum cultivar PPMI741 (a drought tolerant parental line) were procured from the Pearl Millet Breeding Unit, Indian Agricultural Research Institute, New Delhi, India. Seeds, washed thoroughly with distilled water and germinated in duplicates in autoclaved pots (15 cm diameter and 8 in. depth) containing autoclaved Soilrite™ at 33 °C with 16 h light/8 h dark photoperiod at the National Phytotron Facility, IARI, New Delhi.
Drought stress was induced by the treatment of 30 % Polyethylene glycol. One of the two sets of 22 days old seedlings were exposed to 30 % PEG 6000 (Polyethylene glycol) and 1 mM MES for different time periods 30 min (T1), 2 h (T2), 4 h (T3), 8 h (T4), 16 h (T5), 24 h (T6) and 48 h (T7) respectively to stimulate drought stress at −1.25 Mpa (osmotic potential). The other set of seedlings was maintained as control.
Relative water content (RWC)
Another set of drought stressed seedlings were used for determination of relative water content. Relative water content test of seedling subjected to drought stress was measured as per the procedure of Barrs and Weatherley (1962). The seedling samples were weighed to obtain fresh weight (FW) and were immediately hydrated by soaking in double distilled water in a closed Petri dish for 4 h under normal room light and temperature for turgidity. Thereafter the samples were taken out and the surface was wiped using tissue paper and immediately weighed to obtain turgid weight (TW). Samples were then packed in butter paper and dried at 80 °C for 24 h in and dry weight was measured (DW). Relative water content of the samples was calculated as per the following formula:
The seedlings were then cleansed thoroughly with RNAse OUT™ and flash frozen immediately using liquid nitrogen and stored at −80 °C for RNA isolation so as to preserve the stage specific transcript.
Isolation of total RNA
For each sample including control and corresponding stress exposed Pearl millet cultivar PPMI741 seedlings 100 mg of leaf tissues were ground in pre-chilled mortar and pestle into a fine powder in liquid nitrogen, and total RNA was isolated using the method proposed by Chomczynski and Sacchi (1987). The isolated RNA was dissolved in RNase-free water and quantified by spectrophotometry using Nanodrop (Thermo Scientific, USA) and gel electrophoresis with formaldehyde agarose gel. It was stored at −80 °C till further processing.
Preparation of double-stranded cDNA driver from control plant
Double-stranded cDNA was prepared from 1.5 μg of poly (A) + control RNA (driver population) and stressed RNA (tester population) using the PCR-Select ™ cDNA Subtraction kit (Clonetech, USA). The cDNA from various durations of drought stress (30 min, 2, 4, 8, 12, 24 and 48 h) were pooled so as to include various stress responsive genes expressed at various durations. Control cDNA from corresponding control plants were also pooled and forward subtracted library was constructed using PCR select cDNA subtraction kit.
Construction of drought responsive subtractive cDNA library through SSH
The cDNAs enriched for differentially expressed genes, obtained after the secondary PCR following RsaI restriction digestion, adapter ligation and hybridization, were ligated into a pGEM®-T Easy vector (Promega, USA). Electrocompetent E. coli cells (NEB 10 beta, New England Biolabs, MA, USA) were transformed with the ligated product using an electroporator (Eppendorf Multiorator 30672, USA) using standard electroporation protocol. The transformed cells were spread on LA-Ampicillin (100 μg/ml) plates supplemented with of 100 μl of IPTG (0.1 M) and 10 μl X-Gal (100 mg/ml) and incubated at 37 °C for overnight. White colonies were randomly picked and colony PCR was carried out. False positives and colony mixture were also removed via the colony PCR analysis. The confirmed recombinant colonies were stored at −80 ° C, in 96-well format flat bottom cryo-storage plates containing LB medium supplemented with 20 % glycerol.
Sequencing and data analysis
Positive clones from the subtracted cDNA library having insert size of more than 500 bp were sequenced by single pass sequencing using the vector specific sequencing primer T7 by an automated DNA sequencer (sequencing were performed at Macrogen Inc., South Korea, through Sequencher Tech Pvt. Ltd, Ahmedabad, India). The obtained raw sequence reads were screened, edited manually to trim vector/adaptor sequences using VecScreen tool (www.ncbi.nlm.nih.gov/Vecscreen) and EditSeq of DNASTAR™ Navigator Suite. The edited ESTs which had more than 100 bp in length were assembled and clustered into contigs and singletons using SeqMan Pro program of the DNASTAR™ Navigator Suite.
Database searches
BLASTALL (NCBI) is used for batch execution of BLASTX (Altschul et al. 1990) to search for sequence similarity between processed DNA-sequence outputs and public non-redundant amino acid databases. The unigenes obtained were functionally categorized under biological process, cellular component and molecular function by Rice Genome Annotation Project (RGAP).
Validation of differentially expressed genes by qRT PCR
Validation of genes was performed by qRT-PCR using Light Cycler® 480 system (Roche, Germany) equipped with a 96 well plates system and KAPA SYBR® FAST Master Mix reagent (KAPA Biosystems, USA). Tissues from three biological samples of each treatment were pooled. The qRT-PCR experiments were performed with three technical replicates with 10 μl containing 4 μl of cDNA (diluted according to initial concentration so as to contain 150 ng cDNA), 1.0 μl of forward and reverse primers, and 5 μl of KAPA SYBR® FAST qPCR Master Mix according to the manufacturer’s instructions.
The following thermal cycling profile was used for all qRT-PCR (Table 1). All quantifications were normalized to the Pg -Actin gene (used as housekeeping gene and amplified in the same conditions) as per 2−ΔΔCT method (Livak and Schmittgen 2001).
Table 1.
Programme for qRT-PCR validation of differentially expressed genes of p. glaucum
| PCR stages | Sub stage | Temperature | Time | Cycles |
|---|---|---|---|---|
| Pre incubation | 95 °C | 3 min | 1 | |
| Amplification | Denaturation | 95 °C | 10 s | 30 |
| Annealing | 60 °C | 20 s | ||
| Extension | 72 °C | 10 s | ||
| Mating curve | 95 °C | 5 s | 1 | |
| 60 °C | 1 min | |||
| 97 °C | Continuous | |||
| 40 °C | Hold |
Result and discussion
Relative water content
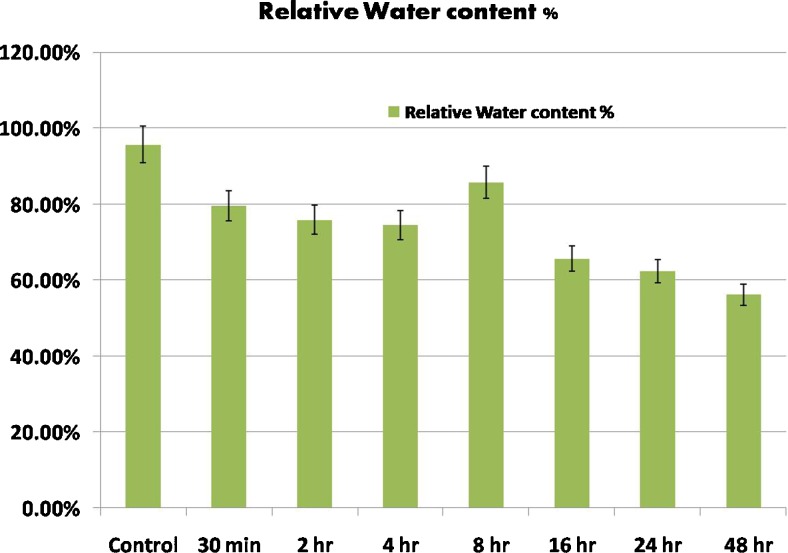
The relative water content (RWC) of drought treated seedlings decreased with the increased duration of drought stress. RWC was calculated as 79.63 % in 30 min, 75.89 % in 2 h, 74.53 % in 4 h. However it increased to 85.86 % in 8 h, which decreased further to 65.74, 62.37 and 56.20 % respectively in 16, 24 and 48 h (Figs. 1 and 2).
Fig. 1.
Variation in Relative Water Content (RWC) of drought stressed plants by PEG 6000 from different time duration (A-Control, B-30 min, C-2 h, D- 4 h, E- 8 h, F- 16 h, G- 24 h and H-48 h)
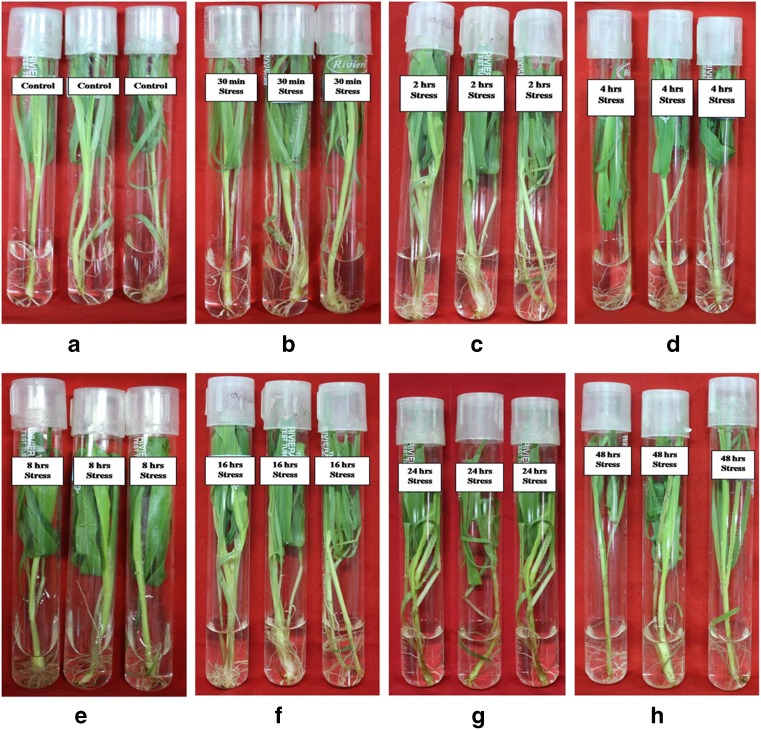
Fig. 2.
Expression analysis of whole seedling of P. glaucum under drought stress condition. The relative water content (RWC) of drought stressed plants by PEG 6000 from different time duration (a Control, b 30 min, c 2 h, d 4 h, e 8 h, f 16 h, g 24 h and h 48 h)
Isolation of total RNA from leave tissues of P. glaucum seedling
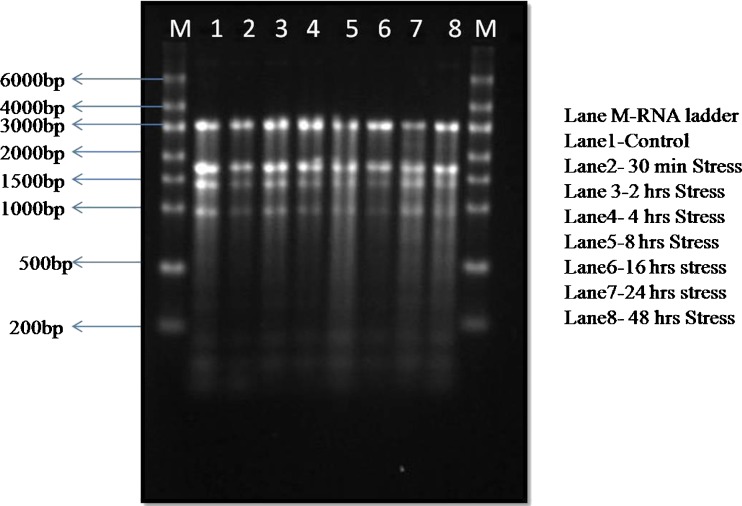
The integrity of total RNA was examined by electrophoresis. RNA samples were run on denaturing 1.2 % formaldehyde agarose gel stained with ethidium bromide (10 mg/ml) (Fig. 3). The presence of two bright bands corresponding to ribosomal 28S and 18S rRNA with a ratio of intensities of ~2:1 confirmed the integrity of RNA. The OD ratio of 260/280 nm of the total RNA isolated from each treatment ranged from 2.06 to 2.13 and the concentrations measured by the Nano Drop ND-1000 spectrophotometer quantifications ranged from 765.0 to 2638.0 (ng/μl) (Table 2).
Fig. 3.
Agarose gel electrophoresis showing RNA extraction by Qiazol method of P. glaucum (whole seedling) treated by drought stress
Table 2.
Quantification of total RNA isolated from control and experiment seedlings at different time periods of drought stress
| S.N. | Treatment | OD ratio 260/280 nm | OD ratio 260/230 nm | Concentration (ng/μl) |
|---|---|---|---|---|
| 1 | Control | 2.09 | 1.89 | 2638.0 ng/μl |
| 2 | 30 min | 2.10 | 2.10 | 1483.4 ng/μl |
| 3 | 2 h | 2.12 | 2.32 | 1456.9 ng/μl |
| 4 | 4 h | 2.07 | 2.21 | 2820.8 ng/μl |
| 5 | 8 h | 2.07 | 2.11 | 1501.8 ng/μl |
| 6 | 16 h | 2.10 | 2.21 | 1709.9 ng/μl |
| 7 | 24 h | 2.13 | 2.09 | 1655.3 ng/μl |
| 8 | 48 h | 2.06 | 1.97 | 765.9 ng/μl |
Construction of subtractive cDNA libraries to identify drought stress responsive genes
RNA of 2.0 μg concentration was used to synthesize cDNA in a long distance polymerase chain reaction (LD-PCR) and observed on 1.2 % agarose EtBr gel in 1X TAE buffer. The bands with variable size were visible with absolute background smear showing very high concentration of amplified product (Fig. 4). The amplified cDNA was digested using RsaI restriction enzyme and amplified using primary and secondary PCR to obtain the secondary hybridized product which was then ligated into pGEMT easy vector and transformed into E. coli. A total of 2400 clones were obtained based on screening by selection of recombinant clones by alpha complementation and visualized as blue-white colonies on the LA plate with (amp+Xgal+IPTG). Out of these 2400 colonies, 1344 clones (four 96 well-format plates) showing the insert sizes more than 250 bp were identified using colony PCR using the primer Sp6: 5′TATTTAGGTGACACTATA G and T7 primer: 5′TAATACGACTCACTATAGG primer. A total of 745 good ESTs were obtained from single pass sequencing and analysis of the obtained sequences. The EST library thus, constructed contained transcripts responsive to early heat stress (15 min) to transcripts responsive to late heat stress (48 h). Using the DNA STAR version 8 software, the 745 ESTs yielded 299 unigenes which were assembled into 52 contigs and 247 singletons. (Supplementary Table 1). All 745 ESTs were submitted to ENA-EMBL databases (Accession no. HG516611-HG517355).
Fig. 4.
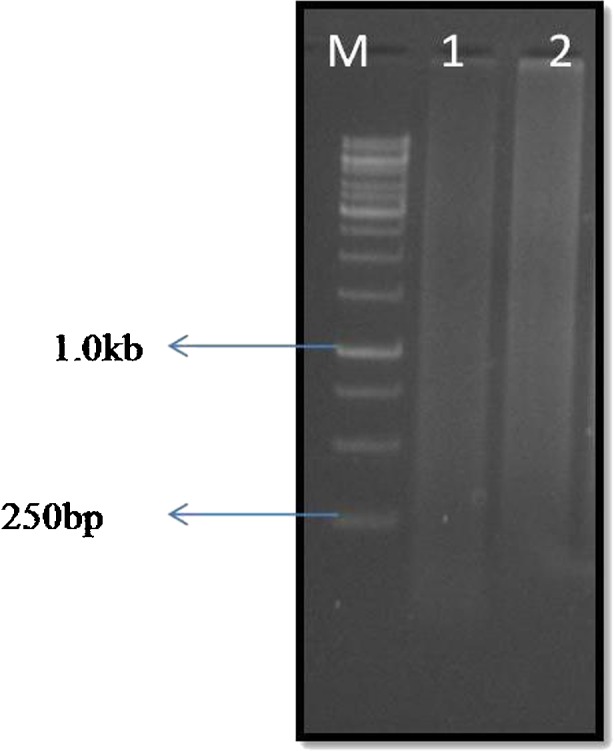
Agarose gel electrophoresis showing LD- PCR cycle optimization. Lane M: 1.0 kb DNA ladder; Lane 1: control; Lane 2: Final optimization at 24th cycle (Plant sample: P. glaucum, drought stress by PEG 6000: 30 min to 48 h)
Identification and in silico characterization of drought stress responsive ESTs in P. glaucum
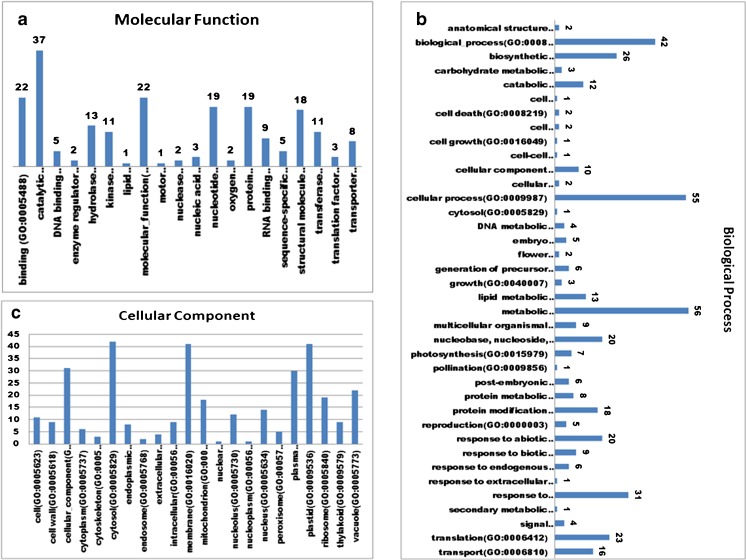
A total of 745 ESTs were identified in which 16 % were classified under the “Response to Abiotic/Biotic stimulus (GO: 0009628)/ (GO: 0009607). BLASTX was performed against non-redundant protein databases NCBI (www.ncbi.nlm.nih.gov) and Rice Genome Annotation Project (RGAP) to determine putative functions of these unigenes. The products of many of the unigenes predicted by BLASTX were homologous to proteins involved in protection against stress damage. A total of 16 % drought stress modulated genes were categorized using the GO (Gene Ontology) IDs available through RGAP BLASTx Locus IDs. These locus IDs were used to assign GO terms fewer than three main categories viz. biological process, cellular component and molecular function (Fig. 5). On the basis of molecular and biological functions, the gene products were categorized into transcription factors, kinase activity, cellular homeostasis during environmental stresses, cell-cell signaling, signal transduction. The result suggests presence of complex network of stress signaling processes (Table 3).
Fig. 5.
Functional categorization of annotated unigenes of P. glaucum SSH library. a Molecular function b Biological process and c Cellular component
Table 3.
Details of some important ESTs obtained in response to drought in the forward SSH library and reverse library their BLASTX analysis
| EST Id | Locus ID | Gene product |
|---|---|---|
| Forward SSH library | ||
| >Contig_6 | LOC_Os01g57962.1 | Photosystem I P700 chlorophyll a apoprotein A2, |
| >Contig_12 | LOC_Os11g06720.1 | Abscisic stress ripening protein |
| >Contig_23 | LOC_Os08g43560.1 | Ascorbate Peroxidase |
| >Contig_11 | LOC_Os11g48090.2 | Inosine-5′-monophosphate dehydrogenase |
| >Contig_53 | LOC_Os09g33500.2 | Putative beta-1, 3-glucanase |
| >Contig_44 | LOC_Os02g13640.1 | Glyoxalase |
| >Contig_15 | LOC_Os03g17870.1 | Metallothionein, putative, expressed |
| >Contig_26 | LOC_Os10g38206.1 | NADPH-dependent oxidoreductase, putative, |
| >Contig_33 | LOC_Os11g48040.1 | Mitochondrial carrier protein, putative, expressed |
| >Contig_47 | LOC_Os12g05860.1 | Cupin domain containing protein, expressed |
| >Contig_60 | LOC_Os02g13840.1 | Citrate synthase, putative, expressed |
| >Contig_50 | LOC_Os05g47980.1 | ATP synthase, putative, expressed |
| >Contig_35 | LOC_Os04g16856.1 | Chloroplast 30S ribosomal protein S7, putative, |
| >Contig_36 | LOC_Os09g33500.2 | Transketolase, putative, expressed |
| >Contig_51 | LOC_Os01g55470.1 | Transposon protein, putative |
| >Contig_45 | LOC_Os05g23740.1 | DnaK family protein, putative, expressed |
| >Contig_58 | LOC_Os11g11390.1 | Ribosomal protein, putative, expressed |
| >Contig_43 | LOC_Os01g17170.1 | Magnesium-protoporphyrin IX monomethylestercyclase, chloroplast precursor, putative, expressed |
| >Contig_45 | LOC_Os08g39140.2 | Hsp70 proteins |
| >Contig_37 | LOC_Os05g49200.1 | Aspartic proteinase oryzasin-1 precursor, putative, expressed |
| >Contig_28 | LOC_Os06g46770.1 | Ubiquitin family protein, putative, expressed |
| >Contig_35 | LOC_Os04g16856.1 | Chloroplast 30S ribosomal protein |
| >Contig_18 | LOC_Os06g46149.2 | Serine/arginine rich (SR) |
| Reverse SSH library | ||
| >Contig_11 | LOC_Os07g34589.3 | Translation initiation factor SUI1, putative, expressed |
| >Contig_16 | LOC_Os05g23610.1 | Protein phosphatase inhibitor 2 containing protein, expressed |
| >Contig_20 | LOC_Os07g38110.1 | tic20, putative, expressed |
| >Contig_21 | LOC_Os01g16240.1 | OsCam1-3—Calmodulin, expressed |
| >Contig_19 | LOC_Os05g33380.1 | Fructose-bisphospatealdolaseisozyme, putative, expressed |
| >Contig_13 | LOC_Os01g57966.1 | Photosystem I assembly protein ycf3, putative, expressed |
| >Contig_14 | LOC_Os03g17000.2 | NAD dependent epimerase/dehydratase family domain containing protein, expressed |
Validation of differentially expressed genes by qRT PCR
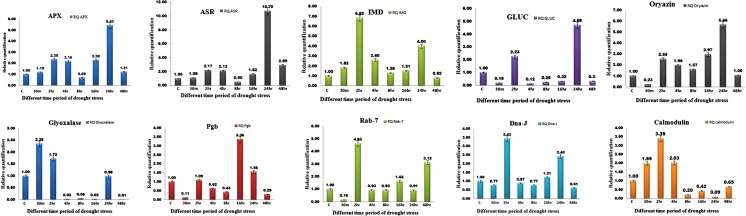
The quantitative up-regulation of the selected genes for their expression in response to drought stress clearly showed that the subtractive cDNA libraries constructed in this study were substantially enriched for stress responsive genes. Differential expression analysis of genes encoding Inosine-5′-monophosphate dehydrogenase, Putative beta-1, 3-glucanase, DnaJ-like protein, Calmodulin-like protein and Rab7 were up-regulated from early time showing the maximum expression of 6.82, 4.69, 3.43, 3.38 and 4.62 fold respectively, after 2 h of stress treatment. On the other hand ASR, APX and Aspartic proteinase oryzasin gene exhibited over expression to the level of 10.7, 5.42 and 5.65 fold respectively after 24 h of continuous dehydration. The expression profiling of Glyoxalase and Ubiquitin-Conjugating enzyme E2-7 genes suggested higher early expression level of 2.34 fold after 30 min drought stress. The expression remained stable up to 16 h that again reached to the peak value of 3.36 fold afterwards displaying late expression (Fig. 6).
Fig. 6.
RT-qPCR analysis of ten drought responsive genes identified in a SSH library of P. glaucum cultivar PPMI741. Relative levels of expression of Abscisic stress ripening protein (Asr), Ascorbate peroxidase (Apx), Inosine-5′-monophosphate dehydrogenase(IMD), Putative beta-1, 3-glucanase (GLUC), Glyoxalase, Rab7, Aspartic proteinase Oryzasin, DnaJ-like protein (Dna-J), Calmodulin-like protein, Ubiquitin-Conjugating enzyme E2-7 (Pgb) in RNA from 22 days old seedlings were determined after exposing seedlings to 30 % PEG 6000 for 30 min to 48 h
Discussion
Subtractive suppression hybridization is a powerful technique that enables researchers to compare two populations of mRNA and obtain clones of genes that are expressed in one population but not in the other (Almeida et al. 2013; Ding et al. 2014; Khan et al. 2014). In the present study, drought stress responsive subtractive cDNA library was constructed from 22 days old pearl millet seedlings [P. glaucum cv. PPMI741] subjected to drought stress at room temperature by 30 % PEG 6000 for different time period (30 min, 2, 4, 8, 16, 24 and 48 h). Plants with high capacity for water retention can better survive drought stress. During 0–48 h of 30 % PEG600 treatment, P. glaucum PPMI741 samples were observed to have high relative water content (RWC) which indicates its drought tolerance ability.
Based on the above observation, P. glaucum PPMI741 seedlings were selected to be used to create a subtractive cDNA library. Further, a total of 745 ESTs, found in the drought responsive subtracted cDNA library, were submitted to ENA-EMBL databases accession no. HG516611-HG517355. These EST sequences from the subtracted cDNA library are rich sources of drought stress-related genes that can help in understanding the molecular basis of drought tolerance in pearl millet by revealing a major part of the stress-responsive transcriptome (Mishra et al. 2007). More than 25 % of the ESTs generated were classified into response to abiotic stimulus category. Five percent of the ESTs in the subtracted cDNA library showed no homology to any protein in the database. These uncharacterized ESTs provide new candidate genes for investigation to elucidate their role in drought stress.
Validation of differential expression of ten selected genes in P. glaucum at various durations (30 min, 2, 4, 8, 16, 24 and 24 h) viz. Abscisic stress ripening protein (EST ID: Contig_12_ Pg _FSSH), Ascorbate peroxidase (EST ID: Contig_23_Pg_FSSH), Glyoxalase (EST ID: Contig_24_Pg_FSSH), Rab7(EST ID: Singlet_223_Pg_FSSH), Aspartic proteinase Oryzasin (EST ID: Contig_37_Pg_FSSH-P), Ubiquitin-Conjugating enzyme E2-7 (EST ID: Contig_27_ Pg_FSSH), DnaJ-like protein (EST ID:4F_E10_Pg_FSSH), Calmodulin-like protein (EST ID: Singlet_204_Pg_FSSH), Putative beta-1,3-glucanase (EST ID: Contig_53_Pg_FSSH) and Inosine-5′-monophosphate dehydrogenase (EST ID: Contig_11_Pg_FSSH) (Fig. 6) by qRT-PCR analysis corroborated their significant role in drought stress management. Genes like Inosine-5′-monophosphate dehydrogenase, Putative beta-1, 3-glucanase, DnaJ-like protein, Calmodulin-like protein and Rab7 were up regulated from early time having a maximum expression of 6.82, 4.69, 3.43, 3.38 and 4.62 fold respectively after 2 h of stress treatment.
Proteins containing Inosine-5′-monophosphate dehydrogenase (IMPDH) domain (PF00478) along with Cystathionine binding synthase (CBS) domain has been classified in this subgroup. The upregulation of these genes in response to drought stress indicates that CBS domain containing proteins may have an important role to play in plants tolerance to drought, salt and osmotic stress conditions by trying to maintain a balance in the generation and removal of reactive oxygen species (ROS) such as hydrogen peroxide (H2O2) (Finkel and Holbrook 2000; Allen and Tresini 2000). In present research, Inosine-5′-monophosphate dehydrogenase (ESTS ID: Contig_11-FSSH-Pg) showed higher expression at 2 and 24 h of drought stress (30 % PEG6000) in whole seedling of P. glaucum which in coherence with the previous findings.
Plant beta-1,3-glucanase (ESTS ID: Contig_53-FSSH-Pg) one of the typical PR proteins, can catalyze the hydrolysis of β-1,3-glucans, which are a major component of the cell wall of most fungi while little has been found in higher plants so far (Stone and Clarke 1992; Leubner-Metzger and Meins 1999).
Moreover, DNAJ-like protein are mostly used in model plants and crops and do not always maintain stable expression levels among different tissues, experimental conditions and species (Zhong et al. 2011 and Zhu et al. 2013). Systematic validations of reference genes have mainly focused on models and important crop species such as Arabidopsis (Czechowski et al. 2005), rice (Jain et al. 2006), wheat (Paolacci et al. 2009), barley (Janska et al. 2013). DNAJ were the most stable genes in PEG treatment. It is known to be up regulated in heat stress but down regulate in salt stress at different time duration in Ammopipathus mangolicus (Yan et al. 2014). In the present study, DNAJ-like protein was up regulated gradually at 4, 8, 16 and 24 h but it was observed to be highly expressed at 2 h in drought stress (30 % PEG treatment).
Expression profiling of Calmodulin-like protein gene (ESTS ID: Singlet_204-FSSH-Pg) suggests the up-regulation up to 3.38 fold and down-regulation up to level of 0.087 fold at 2 and 24 h respectively of treatment times. Previous studies have shown that Calmodulin-like protein plays positive role in plant stress tolerance (Liu et al. 1998) and shown to have enhanced salt and drought tolerance in transgenic rice plants (Mallikarjuna et al. 2011). However, its down regulation at 2 h shows that calmodulin-like protein gene might be involved in late drought stress response in P. glaucum.
The role of Rab7 in abiotic stress tolerance, such as salinity and drought has been validated and earlier studies have shown over expression of AtRab7, PgRab7 and PjRab7 in A. thaliana and tobacco under conditions of NaCl stress (Mazel and Levine 2002). The Rab7 proteins are important component of the vesicle trafficking system in all eukaryotes (Zerial and McBride 2001). The role of AtRab7 protein in eukaryotes seems to be associated with the late endocytosis, where it functions in the fusion of late endosomes to lysosomes or vacuoles.
On the other hand ASR, Apx and Aspartic proteinase Oryzasin genes were up-regulated at later hours showing 10.7, 5.42 and 5.65 fold up regulation respectively at 24 h. Glyoxalase and Ubiquitin-Conjugating enzyme E2-7 early and late expressed 2.34 and 3.36 fold respectively 30 min, 16 h compared to the control Pg- actin of drought stress. Similar results were obtained in previous studies.
The Asr gene family (Abscisic acid, Stress and Ripening), classified as a new group of Late Embryogenesis Abundant (LEA) (Caramelo and Iusem 2009; Battaglia et al. 2008), has been reported to be induced under water stress (Chang et al. 1998) and involved in adaptation to dry climates (Frankel et al. 2003). It is also reported to be involved in abscisic acid signaling and has been used to develop transgenic Arabidopsis with enhance drought and salt tolerance gene was up-regulated during drought and salt stress in transgenic Arabidopsis (Yang et al. 2005). Our investigation also showed up-regulation of Asr gene after 24 h drought stress.
Some genes were found to be up-regulated at later hours of stress induction which were down regulated at initial or extreme late phases of investigation. For example, ascorbate peroxidase (ESTS ID: Contig_23-FSSH-Pg) gene was found to be up regulated in P. glaucum up to 5.42 fold at 24 h after drought treatment but after 4 h and 48 h decreased indicating that the gene has transient expression. In stressful environmental conditions, there is an enhanced production of reactive oxygen species (ROS) in plants that causes significant damage to cells. Antioxidant defenses which can detoxify ROS are present in plants. A major hydrogen peroxide detoxifying enzyme is ascorbate peroxidase that catalyses conversion of H2O2 into water, using ascorbate as a specific electron donor (Caverzan et al. 2012). Modulated expression of ascorbate peroxidase in our library indicated the Pennisetum response to drought stress.
Aspartic Proteinase Oryzasin gene (ESTS ID: Contig_37-FSSH-Pg) classified in aspartic proteinases and metalloproteinase’s are present at later stages expressed in salt, heat drought stress in rice, barley and Coffea arabica (Dominguez and Cejudo 1996). The said gene is expressed under drought stress in our investigation with up regulated expression at 2 to 24 h of drought stress (30 % PEG6000) and down regulated at 30 min and 48 h indicating that the gene is not responding during early stress conditions and late stress conditions.
Another unigene with its expression profile validated by qRT-PCR is Glyoxalasegene (ESTS ID: Contig_24-FSSH-Pg) that showed higher expression level 30 min and 2 h. After 2 h this gene down regulated in drought stress. Analysis of expression patterns using Gene investigator (Zimmermann et al. 2004) showed that GLX2-1 transcript levels are elevated during several abiotic stresses including anoxia, hypoxia, drought, light, osmotic and salt. In earlier studies exposure of the plants to salt stress had led to increase in GLX2-1 transcript levels of 15 fold increases in 24 h (Devanathan et al. 2014) which confirms that GLX2-1 is induced during abiotic stress and suggests that it may have a role in stress tolerance.
Conclusion
Differentially expressed genes in important crop plants provide a sturdy foundation for the development of drought resistant varieties of economically important crops that can impart sustainable and equitable global food security. The current study focuses on identifying the major differentially expressed genes in P. glaucum during drought stress response using SSH and validating them by qRT-PCR analysis. The putative functions of these P. glaucum ESTs were established based on the homology searches and the presumed biological implications of the products of these differentially expressed genes in relation to the complex networks of stress-adaptive processes in Pennisetum are also discerned. The study will be a valuable resource in the investigations of drought tolerance, as well as other characteristics, of P. glaucum.
Electronic supplementary material
(XLSX 70 kb)
Acknowledgments
The authors are grateful to Project Director, National Research Centre on Plant Biotechnology (NRCPB) for providing necessary facilities to carry out this research. Dr. Tara Satyavati, Principal Scientist, Division of Genetics, Indian Agricultural Research Institute, New Delhi is thanked for providing Pearl millet seeds. National Phytotron Facility, Indian Agricultural Research Institute is acknowledged for providing controlled glass chambers for carrying out the experiments. The financial grant of senior research fellowship provided ICAR (Indian Council of Agricultural Research) under NICRA (National Initiative on Climate Resilient Agriculture) project is duly acknowledged.
References
- Allen RG, Tresini M. Oxidative stress and gene regulation. Free Radic Biol Med. 2000;28:463–499. doi: 10.1016/S0891-5849(99)00242-7. [DOI] [PubMed] [Google Scholar]
- Almeida CMA, Donato MTS, Amaral DOJ, Lima GSA, Brito GG, Lima MMA, Correia MTS, Silva MV. Differential gene expression in sugarcane induced by salicylic acid and under water deficit conditions. Agric Sci Res. 2013;3(1):38–44. [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman D. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Barrs HD, Weatherley PE. A re-examination of the relative turgidity technique for estimating water deficit in leaves. Aust J Biol Sci. 1962;15:413–428. [Google Scholar]
- Battaglia M, Olvera-Carrillo Y, Garciarrubio A, Campos F, Covarrubias AA. The enigmatic LEA proteins and other hydrophilins. Plant Physiol. 2008;148:6–24. doi: 10.1104/pp.108.120725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bezançon G, Pham JL, Deu M, Vigouroux Y, Sagnard F, Mariac C, Kapran I, Mamadou A, Gerard B, Ndjeunga J. Changes in the diversity and geographic distribution of cultivated millet (Pennisetum glaucum [L.] R. Br.) and sorghum (Sorghum bicolor (L.) Moench) varieties in Niger between 1976 and 2003. Genet Resour Crop Evol. 2009;56:223–236. doi: 10.1007/s10722-008-9357-3. [DOI] [Google Scholar]
- Caramelo JJ, Iusem ND. When cells lose water: lessons from biophysics and molecular biology. Prog Biophys Mol Biol. 2009;99(1):1–6. doi: 10.1016/j.pbiomolbio.2008.10.001. [DOI] [PubMed] [Google Scholar]
- Caverzan A, Passaia G, Rosa SB, Rebeiro CW, Lazzarotto F, Margis-Pinheiro M. Plant responses to stresses: role of ascorbate peroxidise in antioxidant protection. Genet Mol Biol. 2012;35(4):1011–1019. doi: 10.1590/S1415-47572012000600016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang YC, Ma JF, Matsumoto H. Mechanisms of Al-induced iron chlorosis in wheat (Triticum aestivum): al-inhibited biosynthesis and secretion of phytosiderophore. Physiol Plant. 1998;102:9–15. doi: 10.1034/j.1399-3054.1998.1020102.x. [DOI] [PubMed] [Google Scholar]
- Chauhan H, Khurana N, Tayagi AK, Khurana JP, Khurana P. Characterization of high temperature stress responsive gene in bread wheat (Tritcum aestivum L.) and their various stages of development. Plant Mol Biol. 2011;75:35–51. doi: 10.1007/s11103-010-9702-8. [DOI] [PubMed] [Google Scholar]
- Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidiniumthiocyanate-phenol-chloroform extraction. Anal Biochem. 1987;162:156–159. doi: 10.1016/0003-2697(87)90021-2. [DOI] [PubMed] [Google Scholar]
- Covitz PA, Smith LS, Long SR. Expressed sequence tags from a root-hair-enriched Medicago truncatula cDNA Library. Plant Physiol. 1998;117(4):1325–1332. doi: 10.1104/pp.117.4.1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czechowski T, Stitt M, Altmann T, Udvardi M, Scheible WR. Genomewide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiol. 2005;139:5–17. doi: 10.1104/pp.105.063743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devanathan S, Erban A, Perez-Torres R, Kopka J, Christopher A, Makaroff CA. Arabidopsis thaliana Glyoxalase 2–1 is required during abiotic stress but is not essential under normal plant growth. PLoS ONE. 2014;9(4):1–12. doi: 10.1371/journal.pone.0095971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diatchenko L, Lau YF, Campbell AP, Chenchik A, Moqadam F, Huang B, Lukyanov S, Lukyanov K, Gurskaya N, Sverdlov ED, Siebert PD. Suppression subtractive hybridization: a method for generating differentially regulated or tissue-specific cDNA probes and libraries. Proc Natl Acad Sci U S A. 1996;93:6025–6030. doi: 10.1073/pnas.93.12.6025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding H, Zhang ZM, Qin FF, Dai LX, Li CJ, Ci DW, Song WW. Isolation and characterization of drought-responsive genes from peanut roots by suppression subtractive hybridization. Electron J Biotechnol. 2014;7(6):304–310. doi: 10.1016/j.ejbt.2014.09.004. [DOI] [Google Scholar]
- Dominguez F, Cejudo FJ. Characterization of the endoproteases appearing during wheat grain development. Plant Physiol. 1996;112:1211–1217. doi: 10.1104/pp.112.3.1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fatima S, Arshad M, Chaudhari SK, Ali A, Amjad MS, Kausar R (2014) Utilization of synthetics for drought tolerance in bread wheat (Triticum aestivum L.). International J Bioscience 5(1):104–112
- Finkel T, Holbrook NJ. Oxidants, oxidative stress and the biology of ageing. Nature. 2000;408:239–247. doi: 10.1038/35041687. [DOI] [PubMed] [Google Scholar]
- Frankel N, Hasson E, Iusem ND, Rossi MS. Adaptive evolution of the water stress-induced gene Asr2 in Lycopersicon species dwelling in arid habitats. Mol Biol Evol. 2003;20:1955–1962. doi: 10.1093/molbev/msg214. [DOI] [PubMed] [Google Scholar]
- Guo P, Baum M, Grando S, Ceccarelli S, Bai G, Li R, von Korff M, Varshney RK, Graner A, Valkoun J. Differentially expressed genes between drought-tolerant and drought-sensitive barley genotypes in response to drought stress during the reproductive stage. J Exp Bot. 2009;60(12):3531–3544. doi: 10.1093/jxb/erp194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubank M, Schatz DG. Identifying differences in mRNA expression by representational difference analysis of cDNA. Nucleic Acids Res. 1994;22(25):5640–5648. doi: 10.1093/nar/22.25.5640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ICAR- AICRP (All India Coordinated Research Project on Pearl Millet) (2014) Annual report 2013–2014. http://www.aicrpmip.res.in
- Jain M, Nijhawan A, Tyagi AK, Khurana JP. Validation of housekeeping genes as internal control for studying gene expression in rice by quantitative real time PCR. Biochem Biophys Res Commun. 2006;345:646–651. doi: 10.1016/j.bbrc.2006.04.140. [DOI] [PubMed] [Google Scholar]
- Janska A, Hodek J, Svoboda P, Zamecnık J, Prasil IT. The choice of reference gene set for assessing gene expression in barley (Hordeumvulgare L.) under low temperature and drought stress. Mol Genet Genomics. 2013;288:639–649. doi: 10.1007/s00438-013-0774-4. [DOI] [PubMed] [Google Scholar]
- Khan MS, Khraiwesh B, Pugalenthi G, Gupta RS, Singh J, Duttamajumder SK, Kapur R. Subtractive hybridization-mediated analysis of genes and in silico prediction of associated microRNAs under waterlogged conditions in sugarcane (Saccharum spp.) FEBS Open Bio. 2014;4:533–541. doi: 10.1016/j.fob.2014.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kholova J, Hash CT, Kakkera A, Kočová M, Vadez V. Constitutive water-conserving mechanisms are correlated with the terminal drought tolerance of pearl millet [Pennisetum glaucum (L.) R. Br.] J Exp Bot. 2014;61(2):369–377. doi: 10.1093/jxb/erp314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leubner-Metzger G, Meins F., Jr . Functions and regulation of plant b-1,3-glucanases (PR-2) In: Datta SK, Muthukrishnan S, editors. Pathogenesis-related proteins plants. Boca Raton: CRC Press; 1999. pp. 49–76. [Google Scholar]
- Liang P, Pardee AB. Differential display of eukaryotic messenger RNA by means of the polymerase chain reaction. Science. 1992;257:967–971. doi: 10.1126/science.1354393. [DOI] [PubMed] [Google Scholar]
- Lim CO, Kim HY, Kim MG, Lee SI, Chung WS, Park SH, Hwang I, Cho MJ. Expressed sequence tags of Chinese cabbage flower bud cDNA. Plant Physiol. 1996;111:577–588. doi: 10.1104/pp.111.2.577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q, Kasuga M, Sakuma Y, Abe H, Miura S, Yamaguchi-Shinozaki K, Shinozaki K. Two transcription factors, DREB1 and DREB2, with an EREBP/AP2 DNA binding domain separate two cellular signal transduction pathways in drought- and low-temperature-responsive gene expression, respectively, in Arabidopsis. Plant Cell. 1998;10:1391–1406. doi: 10.1105/tpc.10.8.1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Mallikarjuna N, Senapathy S, Jadhav DR, Saxena K, Sharma HC, Upadhyaya HD, Rathore A, Varshney R. Progress in the utilization of Cajanus platycarpus (Benth.) Maesen in pigeonpea improvement. Plant Breed. 2011;130:507–514. doi: 10.1111/j.1439-0523.2011.01870.x. [DOI] [Google Scholar]
- Mazel A, Levine A. Induction of glucosyltransferase transcription and activity during superoxide-dependent cell death in Arabidopsis plants. Plant Physiol Biochem. 2002;40:133–140. doi: 10.1016/S0981-9428(01)01351-1. [DOI] [Google Scholar]
- Mishra RN, Reddy PS, Nair S, Markandeya G, Reddy AR, Sopory SK, Reddy MK. Isolation and characterization of expressed sequence tags (ESTs) from subtracted cDNA libraries of Pennisetumglaucum seedlings. Plant Mol Biol. 2007;64:713–732. doi: 10.1007/s11103-007-9193-4. [DOI] [PubMed] [Google Scholar]
- Mitra J. Genetics and genetic improvement of drought resistance in crop plants. Curr Sci. 2001;80:758–762. [Google Scholar]
- Paolacci A, Tanzarella O, Porceddu E, Ciaffi M. Identification and validation of reference genes for quantitative RT-PCR normalization in wheat. BMC Mol Biol. 2009;10:11. doi: 10.1186/1471-2199-10-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putney SD, Herlihy WC, Schimmel P. A new troponin T and cDNA clones for 13 different muscle proteins, found by shotgun sequencing. Nature. 1983;302:718–721. doi: 10.1038/302718a0. [DOI] [PubMed] [Google Scholar]
- Stone BA, Clarke AE. Chemistry and physiology of higher plant 1,3-β-glucanase (callose) In: Stone BA, Clark AE, editors. Chemistry and biology of (1–3)-β-glucans. Melbourne: La Trobe University Press; 1992. pp. 365–429. [Google Scholar]
- Vos P, Hogers R, Bleeker M, Reijans M, Lee T, Hornes M, Frijters A, Pot J, Peleman J, Kuiper M, Zabeau M. AFLP: a new technique for DNA fingerprinting. Nucleic Acids Res. 1995;23:4407–4414. doi: 10.1093/nar/23.21.4407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watt DA. Aluminium responsive gene in sugarcane, identification and analysis of expression under oxidative stress. J Bot. 2003;54:1163–1175. doi: 10.1093/jxb/erg128. [DOI] [PubMed] [Google Scholar]
- Xu Y, Tian J, Gianfagna T, Huang B. Effects of SAG12-ipt expression on cytokinin production, growth and senescence of creeping bentgrass (Agrostisstolonifera L.) under heat stress.Plant. Growth Regul. 2009;57(3):281–291. doi: 10.1007/s10725-008-9346-8. [DOI] [Google Scholar]
- Yan X, Dong X, Zhang W, Yin H, Xiao H. Reference gene selection for quantitative real-time PCR normalization in Reaumuria soongorica. PLoS ONE. 2014;9(8):1–10. doi: 10.1371/journal.pone.0104124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C, Chen Y, Jauh GY, Wang C. A lily ASR protein involves abscisic acid signaling and confers drought and salt resistance in arabidopsis. Plant Physiol. 2005;139(2):836–846. doi: 10.1104/pp.105.065458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zerial M, McBride H. Rab proteins as membrane organizers. Nat Rev Mol Cell Biol. 2001;2:107–119. doi: 10.1038/35052055. [DOI] [PubMed] [Google Scholar]
- Zhang JH, Huang WD, Liu YP, Pan QH. Effects of the temperature acclimation pretreatment on the ultra structure of mesophyll cells in young grape plants (Vitis viniferaL.cv.Jingxiu) under cross-temperature stresses. J Integr Plant Biol. 2005;47:959. doi: 10.1111/j.1744-7909.2005.00109.x. [DOI] [Google Scholar]
- Zhong HY, Chen JW, Li CQ, Chen L, Wu JY. Selection of reliable reference genes for expression studies by reverse transcription quantitative realtime PCR in litchi under different experimental conditions. Plant Cell Rep. 2011;30:641–653. doi: 10.1007/s00299-010-0992-8. [DOI] [PubMed] [Google Scholar]
- Zhu J, Zhang L, Li W, Han S, Yang W. Reference gene selection for quantitative. PLoS ONE. 2013;8:1–10. doi: 10.1371/journal.pone.0053196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann P, Hirsch-Hoffmann M, Hennig L, Gruissem W. GENEVESTIGATOR. Arabidopsis microarray database and analysis toolbox. Plant Physiol. 2004;136:2621–2632. doi: 10.1104/pp.104.046367. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(XLSX 70 kb)