Abstract
Biomarker is the measurable change associated with a physiological or pathophysiological process. Unlike blood which has mechanisms to keep the internal environment homeostatic, urine is more likely to reflect changes of the body. As a result, urine is likely to be a better biomarker source than blood. However, since the urinary proteome is affected by many factors, including diuretics, careful evaluation of those effects is necessary if urinary proteomics is used for biomarker discovery. Here, we evaluated the effects of three commonly-used diuretics (furosemide, F; hydrochlorothiazide, H; and spirolactone, S) on the urinary proteome in rats. Urine samples were collected before and after intragastric administration of diuretics at therapeutic doses and the proteomes were analyzed using label-free liquid chromatography–tandem mass spectrometry (LC–MS/MS). Based on the criteria of P ⩽ 0.05, a fold change ⩾2, a spectral count ⩾5, and false positive rate (FDR) ⩽1%, 14 proteins (seven for F, five for H, and two for S) were identified by Progenesis LC–MS. The human orthologs of most of these 14 proteins are stable in the healthy human urinary proteome, and ten of them are reported as disease biomarkers. Thus, our results suggest that the effects of diuretics deserve more attention in future urinary protein biomarker studies. Moreover, the distinct effects of diuretics on the urinary proteome may provide clues to the mechanisms of diuretics.
Keywords: Urinary proteome, Diuretics, Biomarkers
Introduction
Biomarker is the measurable change associated with a physiological or pathophysiological process. Unlike blood is homeostatic, urine is more likely to reflect changes of the body. In other words, urine is likely to be a better biomarker source than blood [1]. Saving urinary protein samples on the membrane can facilitate storage of samples in large numbers and help to speed up the biomarker research in urine proteome [2]. Furthermore, compared to plasma, urine can be collected continuously and noninvasively. Second, the urinary proteome directly reflects the conditions of the urinary system. Third, it can also reflect the physiological status of the whole human body [3]. These advantages make the urinary proteome a suitable source for disease biomarker discovery.
To date, many urinary biomarkers have been reported in a variety of diseases [3], such as various chronic and acute renal injuries [4], bladder cancer [5], prostate cancer [6] and coronary artery disease [7]. However, studies focusing on the urinary protein biomarker discovery still face certain challenges. A major issue is that the urinary proteomic pattern of an individual may be affected by multiple factors, such as gender, age, diet [8], medication, daily activities, exercises [9,10], smoking [11], stress, menstrual cycle and other physiological variations. Environmental factors including temperature and humidity may also affect the urinary proteome. Therefore, these factors should be taken into consideration in the urinary biomarker research.
Effects of some factors, such as gender, age, daily activity and environmental conditions, have been investigated previously [12–14]. However, effects of some other factors, especially medication, are difficult to examine, since the regular therapeutic process of patients should not be disturbed during urine collection. Therefore, influences of medications on the urinary proteome should be taken into account during data analysis and interpretation.
Diuretics are among the most commonly used medications. They are used to induce negative fluid and sodium balances in a variety of clinical situations, including hypertension, heart failure, renal failure, nephritic syndrome, and cirrhosis [15]. However, it remains unclear whether and how diuretics affect the urinary proteome, which hampers the urinary biomarker discovery for those diseases.
In this study, we examined the effects of furosemide, hydrochlorothiazide, and spirolactone on the urinary proteome using label-free quantitative proteomics [16]. These drugs represent thiazide diuretics, loop diuretics, and potassium-sparing diuretics, respectively, which are the three types of commonly used diuretics with different modes of action [17]. The rat urine samples were collected before and after the diuretics were administered, digested using the filter aided proteome preparation (FASP) method [16], and analyzed with a high-speed TripleTOF 5600 system. Progenesis LC–MS was used to quantify the urinary proteins.
Results and discussion
The effects of diuretics on rat urine volumes
In order to evaluate the direct effects of diuretics on rats, urine samples were collected before and 1, 3, or 5 days after the diuretics were administered intragastrically. As shown in Table S1, the rat urinary volumes increased significantly (∼2–3 folds, P < 0.05) after the administration of furosemide (F) and hydrochlorothiazide (H), especially within the first 8 h after lavage. This period is the effective time of the diuretics. However, there is no significant increase in urine output (P > 0.05), after the rats were administered with spirolactone (S), probably due to the fact that spirolactone is not an efficient diuretic on its own and usually is applied in combination with other diuretics.
SDS–PAGE analysis of the urine samples
As a first step of the sample analysis, the urine samples collected on different days were separated by SDS–PAGE. As shown in Figure 1A, the protein patterns of the urine samples in the H group changed only modestly among those obtained before and 1, 3 and 5 days after the diuretic administration. However, for the F and S groups, there were some significant changes among samples obtained at different time points, especially those on Day 3 after gavage for the F group (Figure 1B) and Day 1 for the S group (Figure 1C). Therefore, normal urine samples, Day 3 for the F and H group and Day 1 for the S group were further analyzed using 1D-LC–MS/MS.
Figure 1.
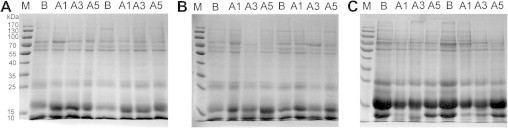
SDS–PAGE of the urine samples from rats treated with different diuretics Urine protein samples were separated by SDS–PAGE and stained using Commassie brilliant blue for the hydrochlorothiazide group (H, A), the furosemide group (F, B) and the spirolactone group (S, C). M, markers; B, normal rat urine samples; A1, A3 and A5, urine samples obtained 1, 3, 5 days after the diuretics were administered.
The changes of the rat urine proteome after diuretic administration
To investigate the changes of the urine proteome after diuretic administration, a total of 18 LC–MS/MS runs of urine samples from three different rats in each diuretic group were analyzed. The 18 datasets were analyzed using Progenesis LC–MS and Mascot Daemon. The false discovery rate (FDR) was adjusted to be less than 1%. As a result, we identified 331, 302, and 325 proteins in the F, S and H group, respectively (Tables S2–S4). The identified peptides are listed in Table S5. All the Supplementary materials can be found in the urinary protein biomarker database [3] (http://122.70.220.102/biomarker).
The coefficients of variation (CVs) for each of the three levels of sample variation—before gavage, after gavage, and between these two conditions—were calculated. As shown in Figure 2, the CV values of the samples after gavage were slightly higher than those before gavage (median CV values: 0.25 vs. 0.34 for F group; 0.35 vs. 0.39 for S group and 0.28 vs. 0.31 for H group), probably because rats respond differentially to the diuretics. In contrast, the CV values of the samples for between before and after gavage and for after gavage (median CV of F group is 0.45; median CV of S group is 0.55) are significantly higher (P < 0.05), suggesting that furosemide and spirolactone can change the urine proteome. However, the CV values of H-Between (median CV is 0.33) were not changed significantly, indicating that hydrochlorothiazide has no discernable effects on the rat urine proteome at this dosage.
Figure 2.
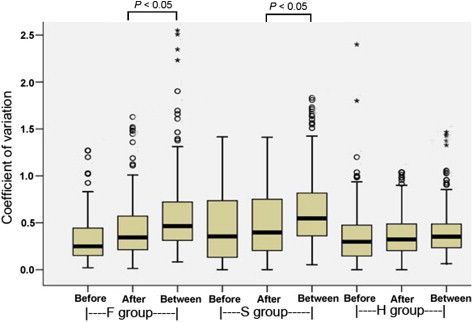
The CV values for each of the three levels of sample variation The CV values of proteins identified in each group before diuretic administration, after and between these two states were calculated using SPSS 13.0. Before indicates the CV values of urine samples before diuretic administration in the F, S and H groups, respectively; after indicates the CV values of urine samples after diuretic administration in each group; between indicates the CV values of urine samples between before and after diuretic administration in each group. (n = 3; in F and S groups, P < 0.05).
The effects of different diuretics on the urinary proteome
Using the label-free quantification by the Progenesis LC–MS software, we identified seven (five up-regulated and two down-regulated) (Table 1), five (one up-regulated and four down-regulated) (Table 2) and two (one up-regulated and one down-regulated) proteins with significantly changed expression in all three rats in the F, S and H groups, respectively, according to the criteria: P ⩽ 0.05, a fold change ⩾2 and a spectral count ⩾5. Five of the seven proteins in the F group and all of the five proteins in the S group have been reported to be disease biomarkers. For example, haptoglobin is a candidate biomarker for patients with bladder cancers, acute kidney injury or diabetic nephropathy. The two significantly changed proteins in H group include beta-microseminoprotein and EGF-containing fibulin-like extracellular matrix protein 1. However, neither of them has been reported as biomarkers previously. Therefore, hydrochlorothiazide appears to have a lower impact on urine proteome than furosemide and spirolactone at the dosages tested. Interestingly, no significantly changed proteins are shared by any two groups, indicating the distinct effects of the diuretics on the urinary proteome.
Table 1.
Urinary proteins significantly changed after furosemide administration
| Accession No. | Protein name |
Fold change |
Candidate biomarkers | Refs. | ||
|---|---|---|---|---|---|---|
| Rat 1 | Rat 2 | Rat 3 | ||||
| P02781 | Prostatic steroid-binding protein C2 | 8.2↑ | 6.3↑ | 4.3↑ | No | |
| P07647 | Submandibular glandular kallikrein-9 | 3.5↑ | 6.2↑ | 5.2↑ | Yes | [18] |
| P02782 | Prostatic steroid-binding protein C1 | 7.6↑ | 5.7↑ | 5.6↑ | No | |
| P02780 | Secretoglobin family 2A member 2 | 9.6↑ | 5.0↑ | 6.2↑ | Yes | [18] |
| P22283 | Cystatin-related protein 2 | 4.7↑ | 3.7↑ | 4.3↑ | Yes | [18] |
| P08721 | Osteopontin | 7.3↓ | 7.4↓ | 5.9↓ | Yes | [19–22] |
| Q01177 | Plasminogen | 2.1↓ | 2.1↓ | 3.0↓ | Yes | [23] |
Table 2.
Urinary proteins significantly changed after spirolactone administration
| Accession No. | Protein name |
Fold change |
Candidate biomarkers | Refs. | ||
|---|---|---|---|---|---|---|
| Rat 1 | Rat 2 | Rat 3 | ||||
| P06866 | Haptoglobin | 5.0↑ | 2.1↑ | 2.2↑ | Yes | [24–29] |
| P81828 | Urinary protein 2 | 3.6↓ | 3.3↓ | 3.9↓ | Yes | [18] |
| P81827 | Urinary protein 1 | 7.3↓ | 4.3↓ | 4.4↓ | Yes | [18,30] |
| P10960 | Sulfated glycoprotein 1 | 4.0↓ | 3.1↓ | 2.4↓ | Yes | [18] |
| Q09030 | Trefoil factor 2 | 8.5↓ | 4.7↓ | 4.2↓ | Yes | [31] |
Human orthologs of the rat proteins significantly affected by the diuretics
We next evaluated the relevance of our findings to the human disease biomarkers. It is typically assumed that orthologs (co-orthologs) retain similar functions between species [32,33]. We thus transformed the significantly changed proteins with diuretic administration in rats to human orthologs. Based on the 122.R_norvegicus.orthologues database and Ensembl Compare database [34], eight of the 14 rat urinary proteins have human orthologs (Table 3). By comparing the proteins with the human core urinary proteome, we further found that seven human orthologs are relatively stable proteins in the normal human urinary proteome [35,36]. Therefore, such proteins could serve as potential urinary biomarkers, since significant qualitative or quantitative changes of these stable proteins may suggest some pathophysiological conditions [36].
Table 3.
Human orthologs of rat proteins significantly changed after diuretic administration
| Rat potein ID | Rat protein name | Human protein ID | Human protein name | Human core urinary proteome |
|---|---|---|---|---|
| Q01177 | Plasminogen | P00747a | Plasminogen | Yes |
| Q09030 | Trefoil factor 2 | Q03403a | Trefoil factor 2 | Yes |
| P08721 | Osteopontin | P10451a | Osteopontin | Yes |
| O35568 | EGF-containing fibulin-like extracellular matrix protein 1 | Q12805a | EGF-containing fibulin-like extracellular matrix protein 1 | Yes |
| P10960 | Sulfated glycoprotein 1 | P07602a | Sulfated glycoprotein 1 | No |
| P06866 | Haptoglobin | P00738a | Haptoglobin | Yes |
| P02781 | Prostatic steroid-binding protein C2 | P11684b | Secretoglobin family 1A member 1 | Yes |
| P07647 | Submandibular glandular kallikrein-9 | P06870b | Kallikrein-1 | Yes |
Note:a Protein present in the 122.R_norvegicus.orthologues database; b Proteins present in the Ensembl Compare database.
Conclusions
In this manuscript, the effects of three diuretics on the urinary proteome were examined in rats. We have shown for the first time through a proteomic approach that some candidate urinary biomarkers may be affected by diuretics, suggesting that the effects of diuretics should be carefully evaluated in the future urinary protein biomarker studies. The results obtained here could help minimize the interference of diuretics with biomarker discovery using the urinary proteomics. In addition, the studies on these significantly changed proteins may provide insights into mechanisms of diuretics as well as renal clearance of proteins. However, some limitations of this study should be noted. First, the findings in this study need to be verified in humans. Second, it would be ideal to validate our results in studies with a higher statistical power. Furthermore, the effects of doses and durations of diuretics on the urinary proteome should also be evaluated in the future. In addition, other commonly used medications, such as glucocorticoids and angiotensin-converting enzyme inhibitors (ACEIs), may likewise affect the urinary proteome and should also be further studied.
Materials and methods
Animals and ethics statement
Pathogen-free male Sprague-Dawley rats (150–160 g) were purchased from the Institute of Laboratory Animal Science, Chinese Academy of Medical Science (Beijing, China). They were given a standard laboratory diet and free access to tap water. The rats were maintained in a room with controlled temperature (22 ± 1 °C) and humidity (65%–70%) and a 12:12 h light:dark cycle. The study was performed after the rats had been allowed to acclimate for 1 week. This study was approved by the Institute of Basic Medical Sciences Animal Ethics Committee, Peking Union Medical College (Animal Welfare Assurance Number: A5518). All rats received humane care in compliance with the institutional animal care guidelines approved by the Institutional Animal Care and Use Committee of the Peking Union Medical College.
Intragastric administration of diuretics and urine collection
Urine samples from 15 rats were collected after each rat was given 1 ml saline by intragastric administration for 24 h using metabolic cages, and these samples were used as controls. Then, the rats were randomly divided into three groups with five rats in each group. Each group of rats was given either 20 mg/(kg·d) of furosemide, 20 mg/(kg·d) of spirolactone, or 25 mg/(kg·d) of hydrochlorothiazide, respectively. The dosing volumes of diuretics were adjusted to 1 ml. All rats were given diuretics by intragastric administration for 5 days, and the rat urine samples were collected on 1, 3 and 5 days after diuretic administration as described above. The samples were acidified immediately with hydrochloric acid and then cooled to 4 °C to prevent bacterial growth and proteolysis. No rats died during the experiments.
Acetone precipitation
All the urine samples were centrifuged at 5000 × g for 30 min, and the pellets were removed. The supernatants were precipitated with 75% v/v acetone for 12 h followed by centrifugation at 12 000 × g for 30 min. After removing supernatant, the pellets were thoroughly air-dried, resuspended in the lysis buffer (8 M urea, 2 M thiourea, 50 mM Tris and 25 mM DTT) and subjected to protein quantitation by the Bradford assay.
SDS–PAGE analysis
For each sample, 30 μg of proteins were dissolved in the PAGE sample buffer (50 mM Tris–HCl, pH 6.8, 50 mM DTT, 0.5% SDS and 10% glycerol) and incubated at 97 °C for 5 min. The proteins were then resolved by SDS–PAGE. After electrophoresis, the gels were stained using Coomassie brilliant blue.
FASP cleanup and overnight digestion
FASP was performed using NANOSPE 10 K OMEGA centrifugal devices (PALL life sciences, NY, Washington, USA) following previously described procedures [37]. Briefly, 100 μg of urinary proteins samples were mixed with 0.2 ml of 8 M urea in 0.1 M Tris–HCl, pH 8.5, loaded onto the membrane filter and centrifuged at 14,000 × g for 35 min. Then the samples were reduced and alkylated. Finally, sequencing grade modified trypsin was added at a protein-to-enzyme ratio of 50:1, followed by incubation overnight at 37 °C. The digested peptides were eluted from the filters twice using 0.1 ml of 50 mM ammonium bicarbonate and then desalted by solid-phase extraction using the Oasis HLB Extraction Cartridge (Waters, Milford, MA, USA), dried in a SpeedVac, resuspended with 20 μl of 0.1% formic acid and stored at −80 °C until use.
LC–MS/MS
Urine samples from three rats in each group were analyzed using an AB SCIEX (Framingham, MA, USA) Triple-TOF 5600 mass spectrometer. Each sample was analyzed once. Briefly, the tryptic peptides (2 μg in each sample) were analyzed using a RP C18 capillary LC column (100 μm × 150 mm, 3 μm) from Michrom Bioresources (Michrom BioResources, Westford, MA, USA). The eluted gradient was 5%–30% buffer B (0.1% formic acid, 99.9% acetonitrile; flow rate, 0.5 μl/min) for 100 min. The MS data were acquired by Triple-TOF MS using an ion spray voltage of 3 kV, curtain gas of 20 psi, nebulizer gas of 30 psi, and an interface heater temperature of 150 °C. The precursors were acquired in 500 ms ranging from 350 to 1250 Da, and the product ion scans were acquired in 50 ms ranging from 250 to 1800 Da. A rolling collision energy setting was used. A total of 30 product ion scans were collected if exceeding a threshold of 125 counts per second (counts/s) and with a +2 to +5 charge-state for each cycle.
Database searching and protein identification
The Mascot Daemon software (version 2.4.0, Matrix Science, London) was used to search the MS/MS data against the SwissProt_rat database (release 2012_07; taxonomy: Rattus; containing 7787 sequences). The carbamidomethylation of cysteines was set as a fixed modification. The oxidation of methionine and protein N-terminal acetylation were set as variable modifications. The specificity of trypsin digestion was set for cleavage after K or R, and two missed trypsin cleavage sites were allowed. The mass tolerances in MS and MS/MS were both set to 0.05 Da. After the Mascot search, the significance threshold and ion score cut-off were set to 0.05 using MudPIT protein scoring. FDR was adjusted to less than 1% when the search result was exported.
Label-free quantification
For label-free quantification, the acquired raw data files corresponding to the different samples were imported into the Progenesis LC–MS software version 4.1 (Nonlinear Dynamics, Newcastle upon Tyne, UK) for feature detection, alignment, and quantification. All sample features were aligned according to retention times by automatic alignment to maximally overlay all the two-dimensional (m/z and retention time) feature maps. Then, the single-charged peptides and the peptides with charge states higher than three were excluded from the analysis. After alignment, the samples were divided into the appropriate groups: furosemide before (normal urine samples) and after (urine samples collected after gavage); hydrochlorothiazide before and after; and spirolactone before and after. Urine samples before and after gavage from the same rats were internal control. The peak lists generated by the Progenesis LC–MS software were used for protein identification as described above and then re-imported into the software. For quantification, only unique peptides were included, and the total cumulative abundance was calculated by summing the individual abundance of all peptides assigned to each protein [38].
Statistical analysis
Percentages of variances were calculated from the median CV, which is the standard deviation divided by the mean of a measurement. P values were generated by t-test.
Authors’ contributions
XL and YG designed the experiment; XL performed the animal experiments, prepared the samples and analyzed the data; MZ and ML performed the LC–MS/MS analysis; LJ analyzed the data. XL wrote the manuscript; all authors read and approved the final manuscript.
Competing interests
The authors have declared that there are no competing interests.
Acknowledgements
This work was supported by the National Basic Research Program of China (Grant Nos. 2012CB517606 and 2013CB530805), 111 Project (Grant No. B08007), National Natural Science Foundation of China (Grant No. 31200614), Beijing Natural Science Foundation (Grant No. 5132028) and Key Basic Research Program of the Ministry of Science and Technology of China (Grant No. 2013FY114100).
Footnotes
Peer review under responsibility of Beijing Institute of Genomics, Chinese Academy of Sciences and Genetics Society of China.
Supplementary material
The urine volumes of rats before and after diuretic administration
Proteins identified in furosemide group
Proteins identified in spirolactone group
Proteins identified in hydrochlorothiazide group
Peptides identified in furosemide, spirolactone and hydrochlorothiazide groups
References
- 1.Gao Y. Urine—an untapped goldmine for biomarker discovery? Sci China Life Sci. 2013;56:1145–1146. doi: 10.1007/s11427-013-4574-1. [DOI] [PubMed] [Google Scholar]
- 2.Jia L., Liu X., Liu L., Li M., Gao Y. Urimem, a membrane that can store urinary proteins simply and economically, makes the large-scale storage of clinical samples possible. Sci China Life Sci. 2013 doi: 10.1007/s11427-013-4582-1. [DOI] [PubMed] [Google Scholar]
- 3.Shao C, Li M, Li X, Wei L, Zhu L, Yang F, et al. A tool for biomarker discovery in the urinary proteome: a manually curated human and animal urine protein biomarker database. Mol Cell Proteomics 2011;10:M111.010975. [DOI] [PMC free article] [PubMed]
- 4.Rosner M.H. Urinary biomarkers for the detection of renal injury. Adv Clin Chem. 2009;49:73–97. doi: 10.1016/s0065-2423(09)49004-8. [DOI] [PubMed] [Google Scholar]
- 5.Vrooman O.P., Witjes J.A. Urinary markers in bladder cancer. Eur Urol. 2008;53:909–916. doi: 10.1016/j.eururo.2007.12.006. [DOI] [PubMed] [Google Scholar]
- 6.Montagut C., Albanell J., Bellmunt J. Prostate cancer. Multidisciplinary approach: a key to success. Crit Rev Oncol Hematol. 2008;68:S32–36. doi: 10.1016/j.critrevonc.2008.07.009. [DOI] [PubMed] [Google Scholar]
- 7.Zimmerli L.U., Schiffer E., Zürbig P., Good D.M., Kellmann M., Mouls L. Urinary proteomic biomarkers in coronary artery disease. Mol Cell Proteomics. 2008;7:290–298. doi: 10.1074/mcp.M700394-MCP200. [DOI] [PubMed] [Google Scholar]
- 8.Mullen W., Gonzalez J., Siwy J., Franke J., Sattar N., Mullan A. A pilot study on the effect of short-term consumption of a polyphenol rich drink on biomarkers of coronary artery disease defined by urinary proteomics. J Agric Food Chem. 2011;59:12850–12857. doi: 10.1021/jf203369r. [DOI] [PubMed] [Google Scholar]
- 9.Kohler M., Franz S., Regeniter A., Ikonen A., Walpurgis K., Thomas A. Comparison of the urinary protein patterns of athletes by 2D-gel electrophoresis and mass spectrometry-a pilot study. Drug Test Anal. 2009;1:382–386. doi: 10.1002/dta.80. [DOI] [PubMed] [Google Scholar]
- 10.Kohler M., Walpurgis K., Thomas A., de Maree M., Mester J., Schänzer W. Effects of endurance exercise on the urinary proteome analyzed by 2-D PAGE and Orbitrap MS. Proteomics Clin Appl. 2010;4:568–576. doi: 10.1002/prca.200900209. [DOI] [PubMed] [Google Scholar]
- 11.Airoldi L., Magagnotti C., Iannuzzi A.R., Marelli C., Bagnati R., Pastorelli R. Effects of cigarette smoking on the human urinary proteome. Biochem Biophys Res Commun. 2009;381:397–402. doi: 10.1016/j.bbrc.2009.02.055. [DOI] [PubMed] [Google Scholar]
- 12.Li Y., Fu C., Zhou X., Xiao Z., Zhu X., Jin M. Urine interleukin-18 and cystatin-C as biomarkers of acute kidney injury in critically ill neonates. Pediatr Nephrol. 2012;27:851–860. doi: 10.1007/s00467-011-2072-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Doi K., Katagiri D., Negishi K., Hasegawa S., Hamasaki Y., Fujita T. Mild elevation of urinary biomarkers in prerenal acute kidney injury. Kidney Int. 2012;82:1114–1120. doi: 10.1038/ki.2012.266. [DOI] [PubMed] [Google Scholar]
- 14.Jin J., Ku Y.H., Kim Y., Kim Y., Kim K., Lee J.Y. Differential proteome profiling using iTRAQ in microalbuminuric and normoalbuminuric type 2 diabetic patients. Exp Diabetes Res. 2012;2012:168602. doi: 10.1155/2012/168602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Reddy P., Mooradian A.D. Diuretics: an update on the pharmacology and clinical uses. Am J Ther. 2009;16:74–85. doi: 10.1097/MJT.0b013e31818d3f67. [DOI] [PubMed] [Google Scholar]
- 16.Wisniewski J.R., Zougman A., Nagaraj N., Mann M. Universal sample preparation method for proteome analysis. Nat Methods. 2009;6:359–362. doi: 10.1038/nmeth.1322. [DOI] [PubMed] [Google Scholar]
- 17.Wile D. Diuretics: a review. Ann Clin Biochem. 2012;49:419–431. doi: 10.1258/acb.2011.011281. [DOI] [PubMed] [Google Scholar]
- 18.Wang Y., Chen Y., Zhang Y., Wu S., Ma S., Hu S. Differential ConA-enriched urinary proteome in rat experimental glomerular diseases. Biochem Biophys Res Commun. 2008;371:385–390. doi: 10.1016/j.bbrc.2008.04.082. [DOI] [PubMed] [Google Scholar]
- 19.Ozer J.S., Dieterle F., Troth S., Perentes E., Cordier A., Verdes P. A panel of urinary biomarkers to monitor reversibility of renal injury and a serum marker with improved potential to assess renal function. Nat Biotechnol. 2010;28:486–494. doi: 10.1038/nbt.1627. [DOI] [PubMed] [Google Scholar]
- 20.Hoffmann D., Fuchs T.C., Henzler T., Matheis K.A., Herget T., Dekant W. Evaluation of a urinary kidney biomarker panel in rat models of acute and subchronic nephrotoxicity. Toxicology. 2010;277:49–58. doi: 10.1016/j.tox.2010.08.013. [DOI] [PubMed] [Google Scholar]
- 21.Rouse R.L., Zhang J., Stewart S.R., Rosenzweig B.A., Espandiari P., Sadrieh N.K. Comparative profile of commercially available urinary biomarkers in preclinical drug-induced kidney injury and recovery in rats. Kidney Int. 2011;79:1186–1197. doi: 10.1038/ki.2010.463. [DOI] [PubMed] [Google Scholar]
- 22.Fuchs T.C., Frick K., Emde B., Czasch S., von Landenberg F., Hewitt P. Evaluation of novel acute urinary rat kidney toxicity biomarker for subacute toxicity studies in preclinical trials. Toxicol Pathol. 2012;40:1031–1048. doi: 10.1177/0192623312444618. [DOI] [PubMed] [Google Scholar]
- 23.Kentsis A., Lin Y.Y., Kurek K., Calicchio M., Wang Y.Y., Monigatti F. Discovery and validation of urine markers of acute pediatric appendicitis using high-accuracy mass spectrometry. Ann Emerg Med. 2010;55 doi: 10.1016/j.annemergmed.2009.04.020. 62–70.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Malard V., Gaillard J.C., Berenguer F., Sage N., Quemeneur E. Urine proteomic profiling of uranium nephrotoxicity. Biochim Biophys Acta. 2009;1794:882–891. doi: 10.1016/j.bbapap.2009.01.010. [DOI] [PubMed] [Google Scholar]
- 25.Li H., Li C., Wu H., Zhang T., Wang J., Wang S. Identification of Apo-A1 as a biomarker for early diagnosis of bladder transitional cell carcinoma. Proteome Sci. 2011;9:21. doi: 10.1186/1477-5956-9-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zager R.A., Vijayan A., Johnson A.C. Proximal tubule haptoglobin gene activation is an integral component of the acute kidney injury “stress response”. Am J Physiol Renal Physiol. 2012;303:F139–148. doi: 10.1152/ajprenal.00168.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Riaz S., Alam S.S., Srai S.K., Skinner V., Riaz A., Akhtar M.W. Proteomic identification of human urinary biomarkers in diabetes mellitus type 2. Diabetes Technol Ther. 2010;12:979–988. doi: 10.1089/dia.2010.0078. [DOI] [PubMed] [Google Scholar]
- 28.Jiang H., Guan G., Zhang R., Liu G., Cheng J., Hou X. Identification of urinary soluble E-cadherin as a novel biomarker for diabetic nephropathy. Diabetes Metab Res Rev. 2009;25:232–241. doi: 10.1002/dmrr.940. [DOI] [PubMed] [Google Scholar]
- 29.Bhensdadia N.M., Hunt K.J., Lopes-Virella M.F., Michael Tucker J., Mataria M.R., Alge J.L. Urine haptoglobin levels predict early renal functional decline in patients with type 2 diabetes. Kidney Int. 2013;83:1136–1143. doi: 10.1038/ki.2013.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cutillas P.R., Chalkley R.J., Hansen K.C., Cramer R., Norden A.G., Waterfield M.D. The urinary proteome in Fanconi syndrome implies specificity in the reabsorption of proteins by renal proximal tubule cells. Am J Physiol Renal Physiol. 2004;287:F353–364. doi: 10.1152/ajprenal.00018.2004. [DOI] [PubMed] [Google Scholar]
- 31.Lemberger S.I., Dorsch R., Hauck S.M., Amann B., Hirmer S., Hartmann K. Decrease of Trefoil factor 2 in cats with feline idiopathic cystitis. BJU Int. 2011;107:670–677. doi: 10.1111/j.1464-410X.2010.09500.x. [DOI] [PubMed] [Google Scholar]
- 32.Koonin E.V. Orthologs, paralogs, and evolutionary genomics. Annu Rev Genet. 2005;39:309–338. doi: 10.1146/annurev.genet.39.073003.114725. [DOI] [PubMed] [Google Scholar]
- 33.Remm M., Storm C.E., Sonnhammer E.L. Automatic clustering of orthologs and in-paralogs from pairwise species comparisons. J Mol Biol. 2001;314:1041–1052. doi: 10.1006/jmbi.2000.5197. [DOI] [PubMed] [Google Scholar]
- 34.Shaye D.D., Greenwald I. OrthoList: a compendium of C. elegans genes with human orthologs. PLoS One. 2011;6:e20085. doi: 10.1371/journal.pone.0020085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nagaraj N., Mann M. Quantitative analysis of the intra- and inter-individual variability of the normal urinary proteome. J Proteome Res. 2011;10:637–645. doi: 10.1021/pr100835s. [DOI] [PubMed] [Google Scholar]
- 36.Sun W., Chen Y., Li F., Zhang L., Yang R., Zhang Z. Dynamic urinary proteomic analysis reveals stable proteins to be potential biomarkers. Proteomics Clin Appl. 2009;3:370–382. doi: 10.1002/prca.200800061. [DOI] [PubMed] [Google Scholar]
- 37.Wisniewski J.R., Zougman A., Mann M. Combination of FASP and StageTip-based fractionation allows in-depth analysis of the hippocampal membrane proteome. J Proteome Res. 2009;8:5674–5678. doi: 10.1021/pr900748n. [DOI] [PubMed] [Google Scholar]
- 38.Stoop M.P., Singh V., Stingl C., Martin R., Khademi M., Olsson T. Effects of natalizumab treatment on the cerebrospinal fluid proteome of multiple sclerosis patients. J Proteome Res. 2013;12:1101–1107. doi: 10.1021/pr3012107. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The urine volumes of rats before and after diuretic administration
Proteins identified in furosemide group
Proteins identified in spirolactone group
Proteins identified in hydrochlorothiazide group
Peptides identified in furosemide, spirolactone and hydrochlorothiazide groups



