Abstract
Background and Objectives
Resistance to macrolide can be mediated by erm and msrA genes in Staphylococcus aureus. There are the evidences that show erm genes may be causative agent of inducible or constitutive resistance. The aim of this study was to investigate the incidence of inducible clindamycin resistance and determine the most frequency of erm and msrA genes among S. aureus isolates.
Materials and Methods
In this study a total of 124 non duplicated clinical isolates of S. aureus were tested with disk diffusion method. All isolates were tested by PCR for mecA, ermA, ermB, ermC and msrA genes.
Results
According to PCR results, 48.4% had mecA gene and 51.6% were mecA negative. By phenotypic D-test method, 32.3% revealed inducible resistance and recorded as D and D+. Sensitive and constitutive phenotypes were found in 54.8% and 12.9% of isolates respectively. Inducible clindamycin resistance was more prevalent in MRSA (29%) than MSSA isolates (2.4%). Among studied erm genes, the most frequency genes were ermA and ermC with 41.1% and 17.7% respectively. Three isolates of them had D phenotype, while the PCR results of erm genes were negative. All isolates were negative for ermB or msrA genes.
Conclusion
Since S. aureus isolates with inducible resistance may mutate and change to constitutive resistance, to prevent treatment failure, we suggest that inducible resistance test be performed on erythromycin resistant/clindamycin sensitive isolates.
Keywords: D- test, Inducible clindamycin resistance, Staphylococcus aureus
INTRODUCTION
In the past decade Staphylococcus aureus especially methicillin-resistant strains were known as the most important pathogens that were frequently isolated, and caused serious and life threatening clinical infections such as nosocomial and community-acquired infections (1-3). Vancomycin and teicoplanin are commonly used to treat the infections with methicillin-resistant S. aureus (MRSA)(1), however, recently isolation of S. aureus with decrease susceptibility or resistance to glycopeptides (4) caused encourage of physicians to prescribe of other alternative treatments such as Macrolide - Lincosamide - Streptogramin (MLS) (5). Macrolide (erythromycin), lincosamide (clindamycin), and streptogramin (quinupristin-dalfopristin) antimicrobial agents (collectively MLSB agents) have been used to treat staphylococcal infections (6).
However, among MLS because of pharmacokinetics properties such as good oral absorption and excellent tissue penetration, clindamycin is the most used antibiotic, but excessive use of MLS in the treatment of infections, has been led to increase of resistance to these antibiotics (5). Resistance to MLS antibiotics among staphylococci can be occurred by various mechanisms, including: I- an active efflux encoded by msrA gene (cause resistance to macrolids and type B streptogramins, and not to clindamycin) (6), II- Enzymatic inactivation of antibiotic (7) and III- ribosomal target modification that is the major mechanism of resistance (8) and affects macrolides, lincosamides, and type B streptogramins (MLSB resistance)(6, 9). In staphylococci, the four genes, ermA, ermB, ermC and ermF, are frequently involved in resistance to MLS (10). The expression of MLSB resistance can be inducible or constitutive and is not related to the type of the erm genes (8). S. aureus isolates with constitutive resistance in vitro, demonstrate resistance to both erythromycin and clindamycin whereas S. aureus isolates that harbor inducible resistance are resistant to erythromycin but appear susceptible to clindamycin (iMLSB) (11). Although, after contact to clindamycin in vivo, they may mutate and produce constitutive resistance that becoming resistant to all MLS antibiotics (12) and may cause treatment failure (13-14). In addition, isolates with msrA-mediated efflux pump also have the same phenotype and are resistant to erythromycin and sensitive to clindamycin, however they cannot produce constitutive resistance during treatment (14).
Lack of identity of inducible clindamycin resistance leads to false laboratory reports and could lead to clinical failure when clindamycin is used therapeutically and cause treatment problems (6, 15). On the other hand, labeling of staphylococci as clindamycin resistant, while they are only resistance to erythromycin, could stop prescription of clindamycin, in cases that infections have occurred by truly clindamycin-susceptible staphylococcal isolates (6, 16). A simple laboratory test (as titled D-zone test) can differentiate between staphylococci that have inducible erm genes-mediated resistance and those which have efflux pump-mediated resistance (14).
The aim of present study was to determine the incidence of inducible clindamycin resistance and investigate the prevalence of ermA, ermB, ermC, and msrA genes among the clinical isolates of S. aureus.
MATERIALS AND METHODS
Isolation and identification of bacteria
During of one year period 124 clinical isolates of S. aureus were collected from three teaching hospitals affiliated to Ahvaz Jundishapur University of Medical Sciences, Ahvaz, Iran. The bacteria which were consecutively isolated from patients in various wards and different specimens such as: catheter, blood, wound, discharge, abscess, burn, and so on, were transported to Microbiology Laboratory in School of Medicine and were confirmed by standard microbiology tests including: Gram staining, catalase, slide and tube coagulase, mannitol fermentation and production of DNase enzyme (17).
Antibiotic susceptibility testing
Antimicrobial susceptibility of the isolates was determined by using Kirby-Bauer disk diffusion method according to Clinical and Laboratory Standard Institute (CLSI) guidelines. Briefly a 0.5 McFarland suspension of bacteria were prepared and inoculated on Mueller-Hinton’s agar plates (Merck, Germany). The tested antimicrobial agents were penicillin (10U), oxacillin (1μg), cefoxitin (30μg), gentamicin (10μg), Trimetoprim-sulfametoxazol (1.25/23.75μg), azithromycin (15μg), imipenem (10μg), meropenem (10μg), ciprofloxacin (5μg) and rifampin (5μg). The minimal inhibitory concentrations (MICs) of vancomycin were determined by E-Test (Bio Mérieux) according to CLSI guidelines. S. aureus ATCC 25923 and S. aureus ATCC 29213 were included as standard strains and quality control for disk diffusion and MIC tests; respectively (CLSI, 2007).
Disk approximation test with erythromycin and clindamycin (D-Zone test)
Inducible clindamycin resistance, was determined using disk approximation test with erythromycin and clindamycin (D-zone test) as recommended by CLSI (CLSI, 2007). Briefly, 0.5 McFarland suspensions were prepared with organisms from an overnight growth and then inoculated and spread over the surface on Mueller-Hinton’s agar plates (Merck, Germany). One erythromycin disk (15 μg) and one clindamycin disk (2 μg) (MAST, Group Ltd, Merseyside, UK) were placed on the inoculated plates in a distance of 15 mm from each other. Plates were incubated at 35°C and read after 18 h. Inducible clindamycin resistance was confirmed by forming of a flattening shape of the clindamycin inhibition zone (D shape) around the erythromycin disk which indicated erythromycin had induced clindamycin resistance. Furthermore, the staphylococcal isolates were grouped to different phenotypes according to a study as previously described (14). These phenotypes were: S phenotype (sensitive to both erythromycin and clindamycin), R phenotype (constitutive resistance and were resistant to both erythromycin and clindamycin), D phenotype (resistant to erythromycin and clindamycin zone like D) and D+ (resistant to erythromycin and D shape zone for clindamycin with small colonies growing within the D zone)(14).
DNA extraction
DNA was extracted from S. aureus isolates by boiling method (2). Bacteria were inoculated on Mueller-Hinton’s agar plate overnight at 37°C. After this time, one to five colonies were suspended in 100 μl of TE buffer (10 mMTris, 1 mM EDTA, pH 7.8) and boiled for 10 minutes at 100°C. After centrifugation, bacterial suspensions at 9000 × g for 30second at 4°C, the supernatant was collected and used as DNA template for PCR reaction (2).
Identification of mecA gene
For identification of mecA gene in methicillin-resistant isolates, which were screened by resistance to oxacillin and/or cefoxitin disks, polymerase chain reaction (PCR) was performed using by primer pair mecA F 5’-GTAGAAATGACTGAACGTCCGATGA-3’ and mecA R 5’- CCAATTCCACATTGTTTCGGTCTAA-3’ that amplified a 310-bp Product (Tiwari and Sen, 2006). PCR condition in a Mastercycler (Eppendorf, Germany) were as follows: 94°C for 4 min, 35 cycle of 94°C for 1 min, 62°C for 1 min, 72°C for 45-s and final extension 5 min in 72°C. PCR products were electrophoresed on a 1% agarose gel and visualized under ultraviolet illumination. S. aureus ATCC 29213 and S. aureus ATCC 33591 were used as mecA negative and mecA positive control strains respectively.
Amplification of erm and msrA determinants by PCR
DNA amplification was performed using specific primers for detection of erm and msrA genes. Oligonucleotide primers used for PCR were as follows: ermA/F:5’– TATCTTATCGTTGAGAAGGGATT–3’, ermA/R: 5’ – CTACACTTGGCTTAGGATGAAA–3’ which amplified a 139 bp, ermB/F: 5’ – CTATCTGATTGTTGAAGAAGGATT–3’, ermB/R: 5’ –TTTACTCTTGGTTTAGGATGAAA–3’ which amplified a 142 bp, ermC/F:5’– CTTGTTGATCACGATAATTTCC −3’, ermC/R 5’ –ATCTTTTAGCAAACCCGTATTC −3’ which amplified a 190 bp and msrA/ F: 5’ –TCCAATCATTGCACAAAATC −3’, msrA/ R: 5’ –AATTCCCTCTATTTGGTGGT −3’ which amplified a 163 bp amplicon (10). PCR reactions were adjusted according to conditions described in previous study with some modifications (10). Each reaction was carried out in a final volume of 25μl with 1X PCR buffer, 0.2U Taq polymerase, 2mM MgCl2, 200μM of dNTP, and 0.4μM of each primer. Amplification conditions were as follows: Initial denaturation, 95°C for 3 min; 35 cycles of 95°C for 30 s, various annealing temperatures (58°C for ermC, 62.8°C for ermA, 59°C for ermB and 55°C for msrA) for 30 s and 72°C for 45 s and final extension at 72°C for 7min. PCR products were analyzed by separating on 1.5% agarose gel electrophoresis, then were stained with ethidium bromide solution and finally visualized in gel documentation system (10). One S. aureus isolate with ermA and another one with ermC were sequenced and used as positive control for identification of these genes. We also used another native isolate as positive control for msrA gene. Furthermore, a reaction containing all materials except DNA was used as negative control. Distilled water was used instead of DNA in negative control reaction.
Statistical analyses
The results were analyzed using the SPSS for windows software version 19. Fisher’s exact test or chi-square, as appropriate, was used to compare frequencies. P –value of ≤0.017 was considered as statistically significant.
RESULTS
In this study a total of 124 S. aureus isolates, which were collected from different hospital wards were examined. The frequencies of S. aureus isolated from different clinical samples are shown in Table 1. The results of antimicrobial sensitivity test showed that all isolates were susceptible to vancomycin (100% susceptible) and the majority of them were resistant to penicillin (96.8%). The results of antibiotic susceptibility testing for other antibiotics are shown in the Table 2.
Table 1.
Frequency of S. aureus isolates in various clinical specimens and different phenotypes
| Phenotypes | ||||||
|---|---|---|---|---|---|---|
| Specimen | D (%) | D+(%) | S (%) | R (%) | Negative (%) | Number (%) |
| Burn | 17.7 | 0 | 7.3 | 1.6 | 0 | 33 (26.6) |
| Wound | 4.8 | 0 | 16.1 | 4 | 0 | 31 (25) |
| Blood culture | 3.2 | 0 | 8.1 | 2.4 | 0 | 17 (13.7) |
| Catheter | 4 | 0 | 7.3 | 0.8 | 0 | 15 (12.1) |
| Discharge | 0 | 0 | 6.5 | 1.6 | 0 | 10 (8.1) |
| Trachea | 0.8 | 0 | 3.2 | 0.8 | 0 | 6 (4.8) |
| Urine culture | 0 | 0 | 2.4 | 0 | 0 | 3 (2.4) |
| Corneal lesion | 0 | 0 | 1.6 | 0.8 | 0 | 3 (2.4) |
| Abscess | 0 | 0 | 0.8 | 0.8 | 0 | 2 (1.6) |
| Nasal swab | 0.8 | 0 | 0.8 | 0 | 0 | 2 (1.6) |
| Nail infection | 0 | 0.8 | 0 | 0 | 0 | 1 (0.8) |
| Pleural effusion | 0 | 0 | 0.8 | 0 | 0 | 1 (0.8) |
| Total | 31.5 | 0.8 | 54.8 | 12.9 | 0 | 124 (100) |
D: Resistant to erythromycin and clindamycin zone like D, D+: Resistant to erythromycin and D shape zone for clindamycin with small colonies growing within the D zone, S: Sensitive, R: Resistant, Negative: Resistant to erythromycin and susceptible to clindamycin and lack of D shape zone
Table 2.
The results of antibiogram test for S. aureus isolates
| Antibiotic | Sensitive (%) | Intermediate (%) | Resistant (%) |
|---|---|---|---|
| Penicillin | 4 (3.2) | - | 120 (96.8) |
| Oxacillin | 69 (55.6) | - | 55 (44.4) |
| Cefoxitin | 64(51.6) | - | 60(48.4) |
| Gentamicin | 71 (57.3) | 2 (1.6) | 51 (41.1) |
| Trimetoprim-sulfametoxazole | 84 (67.7) | - | 40 (32.3) |
| Azithromycin | 66 (53.2) | - | 58 (46.8) |
| Vancomycin | 124 (100) | - | - |
| Imipenem | 103 (83.1) | 2 (1.6) | 19 (15.3) |
| Meropenem | 122 (98.4) | - | 2(1.6) |
| Ciprofloxacin | 68(54.8) | 3(2.4) | 53(42.7) |
| Rifampin | 100 (80.6) | 13 (10.5) | 11 (8.9) |
| Clindamycin | 66(53.2) | - | 58(46.8) |
| Erythromycin | 66(53.2) | 1(0.8) | 57(46) |
As mentioned above, the results of vancomycin E-test showed that all of staphylococcal isolates were sensitive to vancomycin and their MIC to this antibiotic was in range 0.5μg/ml to 2μg/ml, with MIC 50 = 1μg/ml and MIC 90 = 1.5μg/ml. Based on the results of D-Zone test, different phenotypes of S. aureus including S phenotype (54.8%), R phenotype (12.9%), D phenotype (31.5%) and D+(0.8%) were observed (Fig. 1). The prevalence of different phenotype among each specimen is shown in Table 1.
Fig. 1.
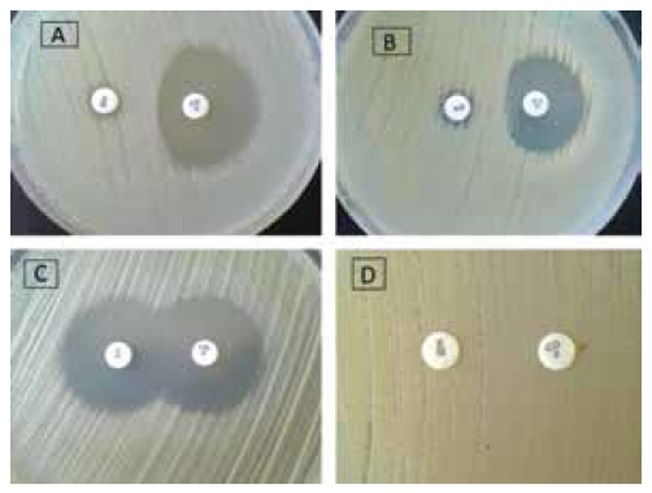
Four phenotypes observed in the study. A: D phenotype, B: D+ phenotype, C: S phenotype, D: R phenotype. (E: erythromycin 15μg, CD: clindamycin 2 μg)
The electrophoresis results of PCR products showed that 48.4% and 51.6% of isolates were positive and negative for mecA gene, respectively. The rate of inducible clindamycin resistance in methicillin-resistant isolates was higher than in MSSA isolates (P-value< 0.001). The rate of D, D+, S and R phenotypes among MRSA isolates were 29%, 0%,6.5% and 12.9% respectively. Among MSSA isolates 2.4%, 0.8%, 48.4% and 0% had D, D+, S and R phenotypes respectively. According to PCR results, 41.1% isolates had ermA (Fig. 2) but 17.7% contained ermC (Fig. 3). Twenty isolates (16.1%) were positive for both ermA and ermC. Three isolates had D phenotype while the results of erm genes PCR were negative. All isolates were negative for ermB and msrA genes. The result of PCR for erm genes according to sensitivity to methicillin is shown in Table 3.
Fig. 2.
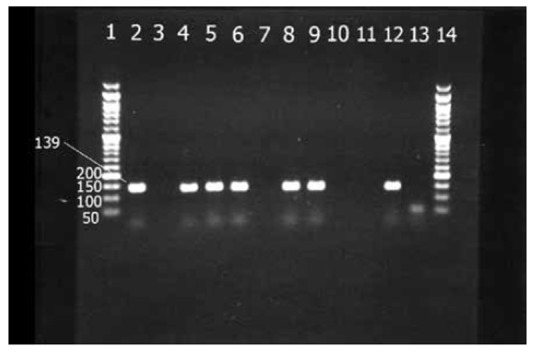
Electerophoresis results of ermA gene. Lanes 1& 14: 50 bp DNA ladder, Lanes 2,4 – 6 and 8-9 isolates with ermA gene in 139 bp, Lanes 3, 7, 10-11 isolates with negative ermA gene. Lanes 12 & 13, Positive and negative controls respectively.
Fig. 3.

Electerophoresis results of ermC gene. Lanes1 and 9: 50 bp DNA ladder, Lanes 2 and 5: isolates with ermC gene in 190 bp, Lanes 3, 4 and 6: isolates with negative ermC gene. Lanes 7 & 8 positive and negative controls respectively.
Table 3.
Results of erm genes PCR according sensitivity to methicillin for the S. aureus isolates
| Genotypee | Results | Sensitivity to methicilin | ||
|---|---|---|---|---|
| MRSA (%) | MSSA (%) | |||
| PCR results | ermA | positive | 39.5 | 1.6 |
| negative | 2.4 | 1.6 | ||
| ermC | positive | 16.9 | 0.8 | |
| negative | 25 | 2.4 | ||
| ermB | positive | 0 | 0 | |
| negative | 0 | 0 | ||
| msrA | positive | 0 | 0 | |
| negative | 0 | 0 | ||
DISCUSSION
For microbiology laboratories there is important to correctly recognize and report an S. aureus isolate, which is truly clindamycin susceptible when it’s erythromycin resistant, and clindamycin susceptible. This true result may depend obtained by using a simple disk diffusion, described as D –zone test, because of this test can exclude inducible clindamycin resistance (18). Prevalence of S. aureus isolates with inducible resistance can be depending on geographic region, patient’s age, species of bacteria, sample origin and source of the strains like community or nosocomial. Prevalence of inducible rate is also different from a hospital to another hospital and even among patients (10, 19-20).
The results of our study have shown that incidence of inducible clindamycin resistance was 32.3% among all isolates. Rahbar et al. in Iran showed that 10.8% of S. aureus isolates had iMLSB(21). Jethwani et al. in India showed that 43% of S. aureus isolates were iMLSB (22). Dizbay et al., in Turkey, reported that 90% erythromycin resistant, clindamycin sensitive S. aureus showed inducible clindamycin resistance (23).
As mentioned, the rate of inducible resistance may vary depending on the resistance bacteria to methicillin (21). In our study iMLSB was most prevalent in MRSA (29%) compared to MSSA (3.2%) isolates (P-value < 0.001). The prevalence of iMLSB resistance in MRSA has been previously iMLSB reported as highly variable, from12.3% to 35.9%, in different parts of the world (7, 11, 15, 19, 21, 24-25). Similar to MRSA, the prevalence rate of iMLSB variable among MSSA isolates. In present study 3.2% of MSSA had iMLSB phenotype. This rate has been reported variously from different countries. Some of these reports showed iMLSB rates from 4% to 68% (7, 11, 15, 19, 21, 25).
Our results showed that S phenotype rate with 48.4% was the most prevalent among MSSA isolates, while, its rate was 6.5% among MRSA (P-value < 0.001). Similar to D phenotype, the prevalence of S phenotype is very variable among MSSA and MRSA in different countries. The prevalence rate of S phenotype in MSSA has been reported 14% to 90.9% and among MRSA isolates from 0% to 26.3% (7, 15, 21-22).
We detected constitutive resistance (12.9%) only in MRSA, but it was not found in MSSA isolates. This type of resistance has been reported from 8 to 64.6% in MRSA and 1.6 to 13% in MSSA isolates in different parts of the world (11, 15, 21-23, 26).
The results of PCR in our study showed that only ermA with 41.1% and ermC with 17.7% were found among studied isolates. No ermB or msrA was detected in this study. Westh et al. in Denmark showed that among S. aureus strains isolated from 1959 to 1988, ermA and ermC were responsible for 98% resistance to erythromycin (27). Cetin et al. in Turkey found that 62% and 17% of S. aureus isolates were positive for ermA & ermC genes respectively (26). Saderi et al. in Tehran reported 60.3% and 54.8% of genes belonged to ermA and ermC respectively in S. aureus strains (28). We have not found any msrA gene, although different rates of msrA genes among S. aureus isolates had been reported (10, 26). Lina et al. showed that msrA was more prevalent in coagulase-negative staphylococci (29). Prevalence of ermB is low and few studies reported this gene in S. aureus. Coutinho et al. reported that between 45 isolates of S. aureus only, 1 isolate had ermB(20), while in Aktas et al. report, this rate was 8.3% (10). Lina et al. showed only one isolate with ermB among 144 isolates of S. aureus (29). Cetin et al. reported the same as present study, found no ermB gene in 47 S. aureus isolates (26). We detected that 16.1% of isolates had both erm A & erm C. Some of the studies also found both erm A and erm C among S. aureus isolates (10, 26).
In our study three S. aureus isolates showed inducible clindamycin resistance (D phenotype), while the results of erm genes PCR were negative. Similar findings have been previously reported. Aktas et al. found that 16.6% of S. aureus isolates were PCR negative (10). Saderi et al. in Tehran studied S. aureus strains for ermA & ermC and reported that 33.3% of strains were negative for both genes (28). Other phenotypes including Hazy D (HD) or Negative (Neg) that previously described (14), were not found in our study. In the present study, all the erythromycin resistant isolates and clindamycin susceptible showed inducible resistance and no negative phenotype was identified among them. In conclusion, we recommend that microbiology laboratories in hospitals perform the D test for any S. aureus isolate that is resistance to erythromycin and sensitive to clindamycin.
Acknowledgments
This study was jointly funded by Vice-Chancellor for research affairs and Infectious Disease & Tropical Research Center of Ahvaz Jundishapur University of Medical Sciences (Project No. 88114). We appreciate the Center for Developing Clinical Research for the consultation and statistical analysis. Thanks also should be extended to the people of the Research Consultation Center (RCC), for their technical support.
References
- 1.Guerin F, Buu-Hoi A, Mainardi JL, Kac G, Colardelle N, Vaupre S, et al. Outbreak of methicillin-resistant Staphylococcus aureus with reduced susceptibility to glycopeptides in a Parisian hospital. J Clin Microbiol. 2000;38:2985–2988. doi: 10.1128/jcm.38.8.2985-2988.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nunes EL, dos Santos KR, Mondino PJ, Bastos Mdo C, Giambiagi-deMarval M. Detection of ileS-2 gene encoding mupirocin resistance in methicillin-resistant Staphylococcus aureus by multiplex PCR. Diagn Microbiol Infect Dis. 1999;34:77–81. doi: 10.1016/s0732-8893(99)00021-8. [DOI] [PubMed] [Google Scholar]
- 3.Soge OO, Meschke JS, No DB, Roberts MC. Characterization of methicillin-resistant Staphylococcus aureus and methicillin-resistant coagulase-negative Staphylococcus spp. isolated from US West Coast public marine beaches. J Antimicrob Chemother. 2009;64:1148–55. doi: 10.1093/jac/dkp368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tiwari HK, Sen MR. Emergence of vancomycin resistant Staphylococcus aureus (VRSA) from a tertiary care hospital from northern part of India. BMC Infect Dis. 2006;6:156. doi: 10.1186/1471-2334-6-156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Prabhu K, Rao S, Rao V. Inducible clindamycin resistance in Staphylococcus aureus isolated from clinical samples. J Lab Physicians. 2011;3:25–27. doi: 10.4103/0974-2727.78558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zelazny AM, Ferraro MJ, Glennen A, Hindler JF, Mann LM, Munro S, et al. Selection of strains for quality assessment of the disk induction method for detection of inducible clindamycin resistance in Staphylococci: a CLSI collaborative study. J Clin Microbiol. 2005;43:2613–2615. doi: 10.1128/JCM.43.6.2613-2615.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yilmaz G, Aydin K, Iskender S, Caylan R, Koksal I. Detection and prevalence of inducible clindamycin resistance in staphylococci. J Med Microbiol. 2007;56(Pt 3):342–345. doi: 10.1099/jmm.0.46761-0. [DOI] [PubMed] [Google Scholar]
- 8.Wondrack L, Massa M, Yang BV, Sutcliffe J. Clinical strain of Staphylococcus aureus inactivates and causes efflux of macrolides. Antimicrob Agents Chemother. 1996;40:992–998. doi: 10.1128/aac.40.4.992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Prunier AL, Malbruny B, Tande D, Picard B, Leclercq R. Clinical isolates of Staphylococcus aureus with ribosomal mutations conferring resistance to macrolides. Antimicrob Agents Chemother. 2002;46:3054–6305. doi: 10.1128/AAC.46.9.3054-3056.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Aktas Z, Aridogan A, Kayacan CB, Aydin D. Resistance to macrolide, lincosamide and streptogramin antibiotics in staphylococci isolated in Istanbul, Turkey. J Microbiol. 2007;45:286–290. [PubMed] [Google Scholar]
- 11.Gupta V, Datta P, Rani H, Chander J. Inducible clindamycin resistance in Staphylococcus aureus: a study from North India. J Postgrad Med. 2009;55:176–179. doi: 10.4103/0022-3859.57393. [DOI] [PubMed] [Google Scholar]
- 12.Navaneeth BV. A preliminary in vitro study on inducible and constitutive clindamycin resistance in Staphylococcus aureus from a South Indian tertiary care hospital. Int J Infect Dis. 2006;10:184–185. doi: 10.1016/j.ijid.2005.03.007. [DOI] [PubMed] [Google Scholar]
- 13.Siberry GK, Tekle T, Carroll K, Dick J. Failure of clindamycin treatment of methicillin-resistant Staphylococcus aureus expressing inducible clindamycin resistance in vitro. Clin Infect Dis. 2003;1(37):1257–1260. doi: 10.1086/377501. [DOI] [PubMed] [Google Scholar]
- 14.Steward CD, Raney PM, Morrell AK, Williams PP, McDougal LK, Jevitt L, et al. Testing for induction of clindamycin resistance in erythromycin-resistant isolates of Staphylococcus aureus. J Clin Microbiol. 2005;43:1716–1721. doi: 10.1128/JCM.43.4.1716-1721.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chelae S, Laohaprertthisarn V, Phengmak M, Kongmuang U, Kalnauwakul S. Detection of inducible clindamycin resistance in staphylococci by disk diffusion induction test. J Med Assoc Thai. 2009 Jul;92:947–951. [PubMed] [Google Scholar]
- 16.Fiebelkorn KR, Crawford SA, McElmeel ML, Jorgensen JH. Practical disk diffusion method for detection of inducible clindamycin resistance in Staphylococcus aureus and coagulase-negative staphylococci. J Clin Microbiol. 2003;41:4740–744. doi: 10.1128/JCM.41.10.4740-4744.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Winn W, Allen S, Janda W, Koneman E, Procop G, Schreckenberger P, et al. Koneman’s Color Atlas and Textbook of Diagnostic microbiology. 6. Lippincott Williams & Wilkins; Philadelphia, USA: 2006. Gram-Positive Cocci Part I; pp. 623–671. [Google Scholar]
- 18.Fernandes CJ, O’Sullivan MV, Cai Y, Kong F, Zeng X, Gilbert GL, et al. Agar dilution method for detection of inducible clindamycin resistance in Staphylococcus spp. J Clin Microbiol. 2007;45:4018–4020. doi: 10.1128/JCM.01158-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Levin TP, Suh B, Axelrod P, Truant AL, Fekete T. Potential clindamycin resistance in clindamycin-susceptible, erythromycin-resistant Staphylococcus aureus: report of a clinical failure. Antimicrob Agents Chemother. 2005;49:1222–1224. doi: 10.1128/AAC.49.3.1222-1224.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Coutinho Vde L, Paiva RM, Reiter KC, de-Paris F, Barth AL, Machado AB. Distribution of erm genes and low prevalence of inducible resistance to clindamycin among staphylococci isolates. Braz J Infect Dis. 2010;14:564–568. doi: 10.1016/s1413-8670(10)70113-6. [DOI] [PubMed] [Google Scholar]
- 21.Rahbar M, Hajia M. Inducible clindamycin resistance in Staphylococcus aureus: a cross-sectional report. Pak J Biol Sci. 2007;10:189–192. doi: 10.3923/pjbs.2007.189.192. [DOI] [PubMed] [Google Scholar]
- 22.Jethwani UN MSA, Shah Latika N. Detection of inducible clindamycin resistance by an automated system in a tertiary care hospital. Afr J Microbiol Res. 2011;5:3. [Google Scholar]
- 23.Dizbay M, Gunal O, Ozkan Y, Ozcan Kanat D, Altuncekic A, Arman D. Constitutive and inducible clindamycin resistance among nosocomially acquired staphylococci. Mikrobiyol Bul. 2008;42:217–221. [PubMed] [Google Scholar]
- 24.Tashakori M, Mohseni Moghadam F, Ziasheikholeslami N, Jafarpour P, Bahsoun M, Hadavi M, et al. Staphylococcus aureus nasal carriage and patterns of antibiotic resistance in bacterial isolates from patients and staff in a dialysis center of southeast Iran. Iran J Microbiol. 2014;6:5. [PMC free article] [PubMed] [Google Scholar]
- 25.Seifi N, Kahani N, Askari E, Mahdipour S, Naderi NM. Inducible clindamycin resistance in Staphylococcus aureus isolates recovered from Mashhad, Iran. Iran J Microbiol. 2012;4:82–6. [PMC free article] [PubMed] [Google Scholar]
- 26.Cetin ES, Gunes H, Kaya S, Aridogan BC, Demirci M. Distribution of genes encoding resistance to macrolides, lincosamides and streptogramins among clinical staphylococcal isolates in a Turkish university hospital. J Microbiol Immunol Infect. 2010;43:524–529. doi: 10.1016/S1684-1182(10)60081-3. [DOI] [PubMed] [Google Scholar]
- 27.Westh H, Hougaard DM, Vuust J, Rosdahl VT. Prevalence of erm gene classes in erythromycin-resistant Staphylococcus aureus strains isolated between 1959 and 1988. Antimicrob Agents Chemother. 1995;39:369–373. doi: 10.1128/aac.39.2.369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Saderi H, Emadi B, Owlia P. Phenotypic and genotypic study of macrolide, lincosamide and streptogramin B (MLSB) resistance in clinical isolates of Staphylococcus aureus in Tehran, Iran. Med Sci Monit. 2011;17:48–53. doi: 10.12659/MSM.881386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lina G, Quaglia A, Reverdy ME, Leclercq R, Vandenesch F, Etienne J. Distribution of genes encoding resistance to macrolides, lincosamides, and streptogramins among staphylococci. Antimicrob Agents Chemother. 1999;43:1062–1066. doi: 10.1128/aac.43.5.1062. [DOI] [PMC free article] [PubMed] [Google Scholar]


