Abstract
In this short review, we have presented a brief overview on major web resources relevant to stem cell research. To facilitate more efficient use of these resources, we have provided a preliminary rating based on our own user experience of the overall quality for each resource. We plan to update the information on an annual basis.
Keywords: Reprogramming, Direct conversion, Physical interaction, Regulatory interaction, Network
Introduction
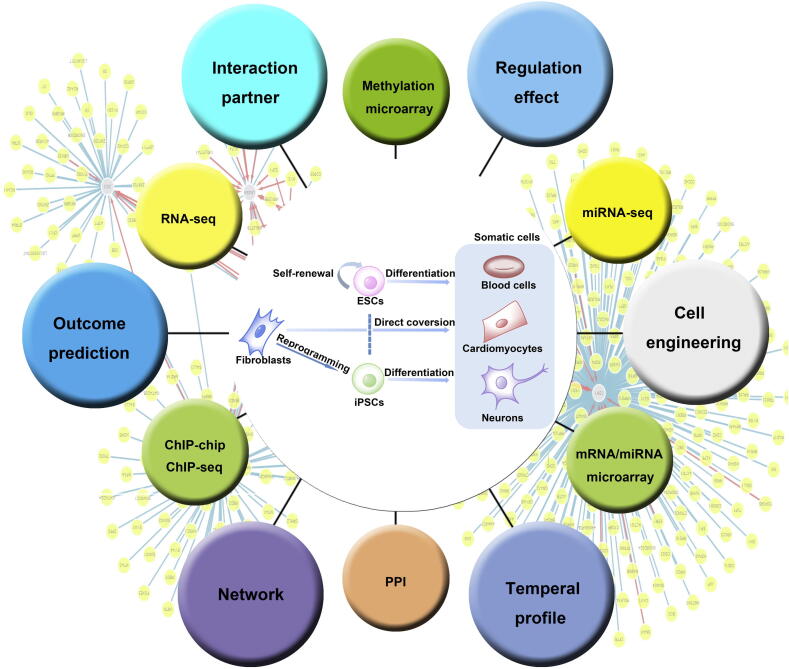
Stem cell research is at the frontier of regenerative medicine [1], [2], [3]. To avoid the ethical issues related to the use of embryonic stem cell (ESC) or somatic cell nuclear transfer (SCNT) technology, induced pluripotent stem cell (iPSC) technology has been developed and matured in recent years [4], [5]. Fibroblast and other types of terminally-differentiated cells can be reprogrammed into iPSCs using defined factors. iPSCs can be further differentiated into various tissues using tissue-specific inducing factors [6]. Differentiated cells can also be directly converted to other types of differentiated cells (also termed “trans-differentiation”) [7]. To foster the fast development in this field, several databases and web servers have been established in the past few years (Figure 1). Relevant literature and high-throughput experimental data have been curated. Available data analyses range from identification of physical interactions and regulatory partners to enrichment analysis and network construction. Here, we provide a brief overview of these web resources. Based on our own user experience of the overall quality of the resources, we have provided a preliminary rating for those resources (Table 1). The rating is mainly based on: (1) how many types of data have been included? (2) how many samples or high-throughput experiments have been included? (3) what kind of online data analysis is available? (4) is the web interface user friendly? and most importantly, (5) can we gain any novel insight by using the web tool?
Figure 1.
Integration of high-throughput data in stem cell research
In order to achieve better understanding on reprogramming, direct conversion, self-renewal, and other processes in stem cell biology, genome-wide profiling have been conducted at the end points and sometimes during those processes. Various types of high-throughput data have been collected and integrated in over a dozen of specialized web resources. The relationship among critical genes can be visualized in a variety of ways. Shown on the background is a network generated by StemCellNet.
Table 1.
Major web resources for stem cell research
| Name | Link | Main features | Rating | Refs. |
|---|---|---|---|---|
| CellNet | http://cellnet.hms.harvard.edu/ | Cell type classification; gene regulatory networks; refinement of factors for cell engineering | ★★★★★ | [8] |
| LifeMap | http://discovery.lifemapsc.com/ | Differentiation, development, and regenerative medicine; graphical display of embryonic development ontology tree | ★★★★★ | [9] |
| ESCAPE | http://www.maayanlab.net/ESCAPE/ | Multiple data types for human and mouse ESCs; network construction, enrichment analysis, lineage prediction | ★★★★★ | [10] |
| StemCellNet | http://stemcellnet.sysbiolab.eu/ | Network with physical interaction and regulation; interactive visualization of the network online | ★★★★★ | [11] |
| HSC-explorer | http://mips.helmholtz-muenchen.de/HSC/ | Early stage of hematopoiesis; interactive graphical display with many functionalities | ★★★★★ | [12] |
| SyStemCell | http://lifecenter.sgst.cn/SyStemCell/ | Clear indication of up or down regulation; co-localization analysis for discovery of novel correlation | ★★★★☆ | [13] |
| CORTECON | http://cortecon.neuralsci.org/ | NGS data from in vitro cortical development; gene, cluster, disease, KEGG pathway, and GO term | ★★★★☆ | [14] |
| SCDE | http://discovery.hsci.harvard.edu/ | Tissue and cancer stem cells; curation on experiments; enrichment analysis; code sharing | ★★★★☆ | [15] |
| StemBase | http://www.stembase.ca/?path=/ | Detailed curation of experiment information; correlation and mutual information analysis | ★★★★☆ | [16] |
| CODEX | http://codex.stemcells.cam.ac.uk/ | NGS data for ESCs and haematopoietic cells | ★★★☆☆ | [17] |
| ESCD | http://biit.cs.ut.ee/escd/ | ESCs, embryonic carcinoma cells; search by GO terms | ★★☆☆☆ | [18] |
Note: Our rating is mainly based on the number of data types included; the number of samples or high-throughput experiments included, the kinds of online data analysis available, whether the web interface is user-friendly, and most importantly, whether users can gain any novel insight by using the web tool.
CellNet
Among the available web resources, CellNet is the most practical tool for somatic cell reprogramming and direct conversion [8]. Analyses on the gene regulatory network (GRN) have been conducted on 20 mouse cell lines or tissue types and 16 human cell lines or tissue types, and several characteristic GRN modules have been identified for each cell line or tissue type. The main aim of CellNet is to facilitate cell engineering, not limited to stem cell biology. User-uploaded gene expression profiles are compared with the benchmark profiles, and three types of analysis results can be obtained. The first is cell and tissue type classification, basically indicating how close the engineered cell is to any of the benchmark cells or tissues. The second is the GRN status, i.e., the evaluation of the establishment of the characteristic GRN modules for intended target cell or tissue. The third is the network influence score. For each of the critical transcriptional regulators of the intended target cell or tissue, the distance to the expected expression level will be calculated and the top 50 down-regulated regulators will be highlighted. Overall, CellNet provides a practical guide to fill the gap between the engineered cell and the intended target. Although CellNet is not specifically designed for stem cell research, this unique application on cell engineering is the main reason we gave it a 5-star rating.
LifeMap
LifeMap contains a large collection of the literature and gene expression data relevant to stem cell differentiation, embryonic development and regenerative medicine [9]. Information is available for cell types including ESCs, iPSCs, embryonic progenitor cells, adult stem cells, primary cells, and fully-differentiated somatic cells from human and mouse. Retrievable information include gene expression, signaling pathways, cell types, developmental stages, anatomical compartments, differentiation protocols, diseases, cell therapies, and literature references. Illustrative and interactive images are provided for better user experience. LifeMap is more like an encyclopedia for embryonic development and regenerative medicine. The main highlights include comprehensive curation of both literature and gene expression information, interactive graphical interface of the full development tree, and unique information on regenerative medicine. Registration is required for the access of the full features.
ESCAPE
The Embryonic Stem Cell Atlas from Pluripotency Evidence (ESCAPE) database is developed based on gene sets from published experiments on human and mouse ESCs [10]. The curated data types include chromatin immunoprecipitation (ChIP) data for protein−DNA interaction, regulatory information from loss-of-function and gain-of-function (Logof) experiments, protein–protein interaction (PPI) using key factors as baits, miRNA−target interactions from popular miRNA websites, potential key regulators from RNAi experiments, ESC- or differentiating ESC- specific proteins, histone modifications, miRNA expression, and time-course expression. In addition to the retrieval of the collected information, these gene sets can also be used to construct interaction and regulatory networks, conduct enrichment analysis for user-supplied gene lists, and predict one of the four lineages during ESC differentiation, the latter being a unique feature among the available web tools described in this article. The network is built upon the input gene list, curated ChIP, PPI, and Logof data.
StemCellNet
StemCellNet is mainly a network tool for stem cell biology [11]. The datasets supporting the network construction include physical protein interactions with key regulators, transcriptional regulatory interactions from ChIP binding experiments, generic physical and regulatory interactions from public resources, and stemness gene sets from the literature. The constructed network can be visualized online or downloaded (as exemplified in Figure 1). The online network display can be refined according to several options. The node size can be adjusted based on the number of appearances of the specific gene in the stemness datasets. Users can also evaluate the importance of the nodes based on the number of key stemness neighbors. In addition, analysis on the significance of enrichment can be performed on the network for each of the stemness gene sets. The network can also be annotated by incorporating user-uploaded gene expression profiles. Trimming of the network can be achieved by applying one or several of the filters. The network functionality in StemCellNet is the best among the web tools reviewed in this article.
HSC-explorer
HSC-explorer is a curated database for hematopoietic stem cells (HSCs) [12]. This database is focused on the early stage of hematopoiesis. At the time of the writing of this manuscript, over 7000 experimentally-validated interactions have been collected from 217 publications. Detailed data statistics is shown on the homepage. Search results can be displayed as both tables and graphical networks. The interactions are carefully curated with links to the original publications when necessary. The graphical network is user-friendly with a variety of functionalities. The heterogeneous network nodes include gene/protein, SNP, CpG site, drug, pathway, disease, organism, and environment, among others. The types of directional interactions include increasing, decreasing and affecting the expression, quantity, activity, etc. of one entity by the other. Detailed information can be displayed on mouseover at the nodes or edges. In addition to the retrieval of directly-collected information, several topics with special interest in hematopoiesis have been curated. Overall, this database is a good resource for researchers interested in hematopoiesis.
SyStemCell
SyStemCell collected 285 stem cell related publications at the initial release [13]. The majority of the data is on human and mouse, although a small amount of data is on rat and rhesus macaque. The data types include mRNA expression, protein expression, DNA methylation and hydromethylation, histone modification, miRNA information, and transcription factor (TF) regulation. The search results are displayed as increase, detected, and decrease with different colors. Annotations include information from gene ontology (GO), BioCarta, the NCBI BioSystems database, and the database of Differentially Expressed Proteins in human Cancer (dbDEPC). Other functionalities include data browsing and co-localization analysis. The co-localization analysis can be used to discover novel correlation among the selected features. The last release of SyStemCell was on Feb 10, 2012. Therefore, data in the past three years may not be available at this website.
CORTECON
CORTECON is a neural stem cell (NSC)-specific resource and a repository for gene expression in the in vitro developing human cortex [14]. The web tool is mainly based on one high-throughput sequencing study by the authors themselves. The temporal expression data can be retrieved by gene, disease, KEGG pathway, or GO term. Every gene belongs to one of the clusters according to the temporal expression profile. But a gene may be associated with several diseases or multiple stages of cortical development. In general, the relationship among gene cluster, disease, KEGG pathway, GO term, and development stage seems to be many-to-many. Since this is a single study-based web tool, interpretation of the search results shall be cautioned.
SCDE
The Stem Cell Discovery Engine (SCDE) is mainly focused on resources for cancer stem cells [15]. Over 53 relevant datasets (1098 assays) have been curated in the database, including samples from blood, intestine, and brain, almost all from human and mouse. User-specified gene lists can be compared against the curated datasets. They can also be compared against molecular signatures in GeneSigDB, MSigDB, and WikiPathway. SCDE has recently evolved into two components, Stem Cell Commons and Galaxy, although both appear to be in the process of further development. The Galaxy is mainly devoted to data analysis mentioned above. The Stem Cell Commons (http://stemcellcommons.org/) is being developed into an integrated platform, including browse, search, analysis, visualization, and code sharing. Users can also upload data to the Stem Cell Commons. The main goals are to promote discovery and reproducibility in stem cell research.
StemBase
StemBase has curated 62 experiments and 217 samples from mouse, human, and rat [16]. The database can be searched in simple and advanced modes. A portion of the expression information can be retrieved by specifying several fields. The retrieved information can be annotated by GO terms and relevant publications. An additional feature in StemBase is the correlation and mutual information of expression among the specified genes or probes. The expression of each probe can also be viewed on the UCSC genome browser, which seems to be a unique feature. StemBase was originally designed in 2007 without any major update. Therefore, most of the data collected are not so up-to-date.
CODEX
CODEX is devoted to next-generation sequencing (NGS) experiments including ChIP-seq, RNA-seq, and DNase-seq [17]. The datasets are divided by species (human and mouse data). The regulatory information derived from the datasets can also be retrieved. The CODEX server consists of three sections, i.e., HAEMCODE for haematopoietic cells, ESCODE for embryonic stem cells, and CODEX for all cell types. Due to the limited NGS data available for stem cell-related experiments, CODEX is of limited use at the present time.
ESCD
The Embryonic Stem Cell Database (ESCD) has mainly collected datasets on key transcription factor binding, RNAi knockdown, and protein overexpression experiments [18]. Data from both human and mouse samples have been included. In addition to ESCs, data for embryonic carcinoma cells have also been included. ESCD can be queried by gene IDs and GO terms. The major weakness of ESCD is the limited data types and datasets covered.
Other resources
Several other resources are available on the web. StemCellDB (http://stemcells.nih.gov/research/nihresearch/scunit/Pages/Default.aspx) is established by the NIH Stem Cell Unit with an aim for direct comparison of human ESC lines, adult stem cells, and iPSCs [19]. PluriNetWork (http://www.ibima.med.uni-rostock.de/IBIMA/PluriNetWork/) has curated 274 pluripotency genes in mouse with 574 interactions (the current data statistics) [20]. The network can be downloaded for further exploration. FunGenES was originally designed for mouse ESC differentiation [21]. However, the web server is no longer active. Additionally, large amount of data is available from some worldwide collaboration projects with broad scope, including ENCODE (http://genome.ucsc.edu/ENCODE/), TCGA (https://icgc.org/), and Roadmap Epigenomics (http://www.roadmapepigenomics.org/). However, a portion of the data from these projects has already been curated in some of the web tools described above.
Concluding remarks
It is an ongoing effort to develop efficient tools for the better understanding of reprogramming, differentiation, and trans-differentiation. Some of the web resources are continuously updated or upgraded. We shall point out that a good portion of the web resources have not been well maintained since the initial publication. New tools will surely emerge in the future. The continuous effort on web maintenance should be carefully considered when developing new web tools. We ourselves are also in the process of developing an integrated web server for stem cell research. Mere collection of public data will be far from sufficient in the future. A major effort should be focused on enhancing our fundamental understanding of the mechanism regarding the maintenance of pluripotency and gaining precise control of the reprogramming, differentiation, and direct conversion.
Competing interests
The authors declare that there are no conflicts of interest.
Acknowledgements
This work was supported by the grants from the National Basic Research Program of China (973 Program; Grant No. 2014CB964901) and the National High-tech R&D Program of China (863 Program; Grant No. 2015AA020100) awarded to HL by the Ministry of Science and Technology of China.
Handled by Xiangdong Fang
Footnotes
Peer review under responsibility of Beijing Institute of Genomics, Chinese Academy of Sciences and Genetics Society of China.
References
- 1.Young R.A. Control of the embryonic stem cell state. Cell. 2011;144:940–954. doi: 10.1016/j.cell.2011.01.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Krupalnik V., Hanna J.H. Stem cells: the quest for the perfect reprogrammed cell. Nature. 2014;511:160–162. doi: 10.1038/nature13515. [DOI] [PubMed] [Google Scholar]
- 3.Wang H., Zhang Q., Fang X. Transcriptomics and proteomics in stem cell research. Front Med. 2014;8:433–444. doi: 10.1007/s11684-014-0336-0. [DOI] [PubMed] [Google Scholar]
- 4.Takahashi K., Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 5.Takahashi K., Tanabe K., Ohnuki M., Narita M., Ichisaka T., Tomoda K., et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 6.Hartfield E.M., Yamasaki-Mann M., Ribeiro Fernandes H.J., Vowles J., James W.S., Cowley S.A., et al. Physiological characterisation of human iPS-derived dopaminergic neurons. PLoS One. 2014;9:e87388. doi: 10.1371/journal.pone.0087388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xue Y., Ouyang K., Huang J., Zhou Y., Ouyang H., Li H., et al. Direct conversion of fibroblasts to neurons by reprogramming PTB-regulated microRNA circuits. Cell. 2013;152:82–96. doi: 10.1016/j.cell.2012.11.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cahan P., Li H., Morris S.A., Lummertz da Rocha E., Daley G.Q., Collins J.J. CellNet: network biology applied to stem cell engineering. Cell. 2014;158:903–915. doi: 10.1016/j.cell.2014.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Edgar R., Mazor Y., Rinon A., Blumenthal J., Golan Y., Buzhor E., et al. LifeMap discovery: the embryonic development, stem cells, and regenerative medicine research portal. PLoS One. 2013;8:e66629. doi: 10.1371/journal.pone.0066629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xu H., Baroukh C., Dannenfelser R., Chen E.Y., Tan C.M., Kou Y., et al. ESCAPE: database for integrating high-content published data collected from human and mouse embryonic stem cells. Database (Oxford) 2013;2013:bat045. doi: 10.1093/database/bat045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pinto J.P., Reddy Kalathur R.K., Machado R.S., Xavier J.M., Braganca J., Futschik M.E. StemCellNet: an interactive platform for network-oriented investigations in stem cell biology. Nucleic Acids Res. 2014;42:W154–W160. doi: 10.1093/nar/gku455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Montrone C., Kokkaliaris K.D., Loeffler D., Lechner M., Kastenmuller G., Schroeder T., et al. HSC-explorer: a curated database for hematopoietic stem cells. PLoS One. 2013;8:e70348. doi: 10.1371/journal.pone.0070348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yu J., Xing X., Zeng L., Sun J., Li W., Sun H., et al. SyStemCell: a database populated with multiple levels of experimental data from stem cell differentiation research. PLoS One. 2012;7:e35230. doi: 10.1371/journal.pone.0035230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.van de Leemput J., Boles N.C., Kiehl T.R., Corneo B., Lederman P., Menon V., et al. CORTECON: a temporal transcriptome analysis of in vitro human cerebral cortex development from human embryonic stem cells. Neuron. 2014;83:51–68. doi: 10.1016/j.neuron.2014.05.013. [DOI] [PubMed] [Google Scholar]
- 15.Ho Sui S.J., Begley K., Reilly D., Chapman B., McGovern R., Rocca-Sera P., et al. The Stem Cell Discovery Engine: an integrated repository and analysis system for cancer stem cell comparisons. Nucleic Acids Res. 2012;40:D984–D991. doi: 10.1093/nar/gkr1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Porter C.J., Palidwor G.A., Sandie R., Krzyzanowski P.M., Muro E.M., Perez-Iratxeta C., et al. StemBase: a resource for the analysis of stem cell gene expression data. Methods Mol Biol. 2007;407:137–148. doi: 10.1007/978-1-59745-536-7_11. [DOI] [PubMed] [Google Scholar]
- 17.Sanchez-Castillo M., Ruau D., Wilkinson A.C., Ng F.S., Hannah R., Diamanti E., et al. CODEX: a next-generation sequencing experiment database for the haematopoietic and embryonic stem cell communities. Nucleic Acids Res. 2015;43:D1117–D1123. doi: 10.1093/nar/gku895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jung M., Peterson H., Chavez L., Kahlem P., Lehrach H., Vilo J., et al. A data integration approach to mapping OCT4 gene regulatory networks operative in embryonic stem cells and embryonal carcinoma cells. PLoS One. 2010;5:e10709. doi: 10.1371/journal.pone.0010709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mallon B.S., Chenoweth J.G., Johnson K.R., Hamilton R.S., Tesar P.J., Yavatkar A.S., et al. StemCellDB: the human pluripotent stem cell database at the National Institutes of Health. Stem Cell Res. 2013;10:57–66. doi: 10.1016/j.scr.2012.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Som A., Harder C., Greber B., Siatkowski M., Paudel Y., Warsow G., et al. The PluriNetWork: an electronic representation of the network underlying pluripotency in mouse, and its applications. PLoS One. 2010;5:e15165. doi: 10.1371/journal.pone.0015165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schulz H., Kolde R., Adler P., Aksoy I., Anastassiadis K., Bader M., et al. The FunGenES database: a genomics resource for mouse embryonic stem cell differentiation. PLoS One. 2009;4:e6804. doi: 10.1371/journal.pone.0006804. [DOI] [PMC free article] [PubMed] [Google Scholar]