Abstract
Exosomes are 40–100 nm nano-sized vesicles that are released from many cell types into the extracellular space. Such vesicles are widely distributed in various body fluids. Recently, mRNAs and microRNAs (miRNAs) have been identified in exosomes, which can be taken up by neighboring or distant cells and subsequently modulate recipient cells. This suggests an active sorting mechanism of exosomal miRNAs, since the miRNA profiles of exosomes may differ from those of the parent cells. Exosomal miRNAs play an important role in disease progression, and can stimulate angiogenesis and facilitate metastasis in cancers. In this review, we will introduce the origin and the trafficking of exosomes between cells, display current research on the sorting mechanism of exosomal miRNAs, and briefly describe how exosomes and their miRNAs function in recipient cells. Finally, we will discuss the potential applications of these miRNA-containing vesicles in clinical settings.
Keywords: Exosome, Extracellular microRNA, Circulating microRNA, Sorting, Cell-to-cell communication
Introduction
Exosomes, membrane-bound vesicles of 40–100 nm in diameter, are present in almost all biological fluids [1], [2], [3]. They are released from most cell types into the extracellular space after fusion with the plasma membrane [1], [2], [3]. Lipids and proteins are the main components of exosome membranes, which are enriched with lipid rafts [1], [2], [3]. In addition to the proteins, various nucleic acids have recently been identified in the exosomal lumen, including mRNAs, microRNAs (miRNAs), and other non-coding RNAs (ncRNAs) [4]. These exosomal RNAs can be taken up by neighboring cells or distant cells when exosomes circulate, and they subsequently modulate recipient cells. The discovery of their function in genetic exchange between cells has brought increasing attention to exosomes.
MicroRNAs are a class of 17–24 nt small, noncoding RNAs, which mediate post-transcriptional gene silencing by binding to the 3′-untranslated region (UTR) or open reading frame (ORF) region of target mRNAs [5]. The involvement of miRNAs in many biological activities has been well documented, including cell proliferation, cell differentiation, cell migration, disease initiation, and disease progression [6], [7], [8], [9], [10]. Accumulating evidence has shown that miRNAs can stably exist in body fluids, including saliva [11], [12], urine [13], breast milk [14], and blood [11], [15], [16]. In addition to being packed into exosomes or microvesicles, extracellular miRNAs can be loaded into high-density lipoprotein (HDL) [17], [18], or bound by AGO2 protein outside of vesicles [16]. All these three modes of action protect miRNAs from degradation and guarantee their stability. Given the transportability of vesicles, the role of miRNAs in exosomes is gaining increasing attention. Conveying information via circulating vesicles is deemed to be the third way of intercellular communication that is as essential as the cell-to-cell contact-dependent signaling and signaling via transfer of soluble molecules [19], [20].
Formation and secretion of exosomes require enzymes [21], [22] and ATP [23], and the miRNA and mRNA profiles of exosomes differ from those of the parent cells [24]. Therefore, cells may possess an active selecting mechanism for exosomes and their cargos. Besides, functions of the transferred exosomal molecular constituents in the recipient cells are under investigation. Hereby, this review will concisely introduce the origin and trafficking of exosomes and discuss the sorting mechanism and function of exosomal miRNAs.
Formation and secretion of exosomes
Exosomes were first discovered by Pan and Johnstone in 1983 [25]. They reported that the release of transferrin receptors into the extracellular space during the maturation of sheep reticulocytes was associated with a type of small vesicle [23], [25]. In 1989, Johnstone defined such functional vesicles as exosomes [26]. To date, a series of extracellular vesicles have been described [27]. However, in the last three decades, no unified terminology for extracellular vesicles has been presented. The definition for such extracellular vesicles named as microvesicles, exosomes, and microparticles remains confusing among different reports [28], [29], [30]. Now, according to the way of vesicular secretion from cells, extracellular vesicles can be grouped into two general classes. One of these classes is known as microvesicles, which are directly shed from the cell membrane. The other is known as exosomes, which are released by exocytosis when multivesicular bodies (MVBs) fuse with the plasma membrane [31]. Here, we mainly focus on the second group of vesicles, i.e., exosomes.
Exosomes can be revealed using transmission microscopy, possessing a cup-shaped morphology after negative staining [1], [2], [3]. These vesicles can be concentrated in the 1.10–1.21 g/ml section of a sucrose density gradient [1], [2], [3]. They can also be identified by the presence of proteins common to most exosomes, such as the tetraspanin proteins CD63, CD9, and CD81 [1], [2], [3].
As mentioned above, exosomes are originally formed by endocytosis. First, the cell membrane is internalized to produce endosomes. Subsequently, many small vesicles are formed inside the endosome by invaginating parts of the endosome membranes. Such endosomes are called MVBs. Finally, the MVBs fuse with the cell membrane and release the intraluminal endosomal vesicles into the extracellular space to become exosomes [32].
The regulatory molecules involved in the release of exosomes were identified by Ostrowski and colleagues, who observed that Rab27a and Rab27b were associated with exosome secretion. Knockdown of Rab27 or their effectors, SYTL4 and EXPH5, could inhibit secretion of exosomes in HeLa cells [33]. Moreover, Yu et al. discovered that both the tumor repressor protein p53 and its downstream effector TSAP6 could enhance exosome production [34]. Baietti et al. found that syndecan-syntenin interacted directly with ALIX protein via Leu-Tyr-Pro-X(n)-Leu motif to support the intraluminal budding of endosomal membranes, which is an important step in exosome formation [35]. All of these studies indicate that a set of molecules act as a regulatory network and are responsible for the formation and secretion of exosomes in parent cells.
The trafficking of exosomes
Exosomes present in body fluids play an important role in exchanging information between cells. In general, there are three mechanisms of interaction between exosomes and their recipient cells. First, the transmembrane proteins of exosomes directly interact with the signaling receptors of target cells [36]. Second, the exosomes fuse with the plasma membrane of recipient cells and deliver their content into the cytosol [37]. Third, the exosomes are internalized into the recipient cells and have two fates. In one case, some engulfed exosomes may merge into endosomes and undergo transcytosis, which will move exosomes across the recipient cells and release them into neighboring cells. In the other case, endosomes fused from engulfed exosomes will mature into lysosomes and undergo degradation [37], [38]. Some recent studies have reported the factors influencing internalization of exosomes in recipient cells. Koumangoye et al. observed that disruption of exosomal lipid rafts resulted in the inhibition of internalization of exosomes and that annexins, which are related to cell adhesion and growth, were essential for the uptake of exosomes in the breast carcinoma cell line BT-549 [39]. Escrevente et al. described a decrease in exosome uptake after the ovarian carcinoma cell line SKOV3 and its derived exosomes were treated with protease K, which indicated that the proteins mediating exosome internalization are presented on the surface of both the cells and the exosomes [40]. However, the detailed mechanism of exosome internalization is still not well understood.
The function of exosomes
Exosomes can be released from many cell types, such as blood cells, endothelial cells, immunocytes, platelets, and smooth muscle cells [41], [42], [43]. It is believed that exosomes can regulate the bioactivities of recipient cells by the transportation of lipids, proteins, and nucleic acids while circulating in the extracellular space. Several reports have shown that exosomes play important roles in immune response, tumor progression, and neurodegenerative disorders. Esther et al. reported that activated T cells could recruit dendritic cell (DC)-derived exosomes that contain major histocompatibility complex (MHC) class II to down-regulate the immune response during interaction of T cells and DCs [44]. Exosomes derived from platelets that were treated with thrombin and collagen stimulated proliferation and increased chemoinvasion in the lung adenocarcinoma cell line A549 [45]. Exosomes derived from SGC7901 promoted the proliferation of SGC7901 and another gastric cancer cell line, BGC823 [46]. In addition, CD147-positive exosomes derived from epithelial ovarian cancer cells promoted angiogenesis in endothelial cells in vitro [47]. Interestingly, Webber et al. incubated exosomes derived from a mesothelioma cell line, a prostate cancer cell line, a bladder cancer cell line, a colorectal cancer cell line, and a breast cancer cell line with primary fibroblasts in vitro, and found that fibroblasts could be transformed into myofibroblasts [48]. A similar phenomenon was also observed by Cho et al., who described that tumor-derived exosomes converted mesenchymal stem cells within the stroma of the tumor tissue into cancer-associated myofibroblasts [49]. Although the function of exosomes has been documented in the aforementioned studies, it remains an open question which specific class of molecules contained in exosomes influences the recipient cells.
The sorting mechanism for exosomal miRNAs
As described above, a wide variety of molecules are contained in exosomes, including proteins, lipids, DNAs, mRNAs, and miRNAs, which are recorded in the ExoCarta database [50]. Among these molecules, miRNAs have attracted most attention, due to their regulatory roles in gene expression. Goldie et al. demonstrated that, among small RNAs, the proportion of miRNA is higher in exosomes than in their parent cells [51]. As some profiling studies have shown, miRNAs are not randomly incorporated into exosomes. Guduric-Fuchs et al. analyzed miRNA expression levels in a variety of cell lines and their respective derived exosomes, and found that a subset of miRNAs (e.g., miR-150, miR-142-3p, and miR-451) preferentially enter exosomes [52]. Similarly, Ohshima et al. compared the expression levels of let-7 miRNA family members in exosomes derived from the gastric cancer cell line AZ-P7a with those from other cancer cell lines, including the lung cancer cell line SBC-3/DMS-35/NCI-H69, the colorectal cancer cell line SW480/SW620, and the stomach cancer cell line AZ-521. As a result, they found that members of the let-7 miRNA family are abundant in exosomes derived from AZ-P7a, but are less abundant in exosomes derived from other cancer cells [53]. Moreover, some reports have shown that exosomal miRNA expression levels are altered under different physiological conditions. The level of miR-21 was lower in exosomes from the serum of healthy donors than those glioblastoma patients [29]. Levels of let-7f, miR-20b, and miR-30e-3p were lower in vesicles from the plasma of non-small-cell lung carcinoma patients than normal controls [30]. Different levels of eight exosomal miRNAs, including miR-21 and miR141, were also found between benign tumors and ovarian cancers [54]. All these studies show that parent cells possess a sorting mechanism that guides specific intracellular miRNAs to enter exosomes.
According to previous studies, there exists a class of miRNAs that are preferentially sorted into exosomes, such as miR-320 and miR-150. Members of the miR-320 family are widely distributed in exosomes derived from normal tissue and tumors [29], [41], [52], [55], [56]. miR-150 is highly expressed in exosomes derived from the HEK293T cell line, peripheral blood of tumor patients, colony-stimulating factor 1 (CSF-1)-induced bone marrow-derived macrophages, and the serum of colon cancer patients [52], [54], [55], [57], [58]. In addition, some miRNAs, miR-451 for example, are highly expressed in exosomes derived from normal cells, such as the HMC-1 cell line, the HEK293T cell line, primary T lymphocytes, and Epstein–Barr virus-transformed lymphoblastoid B-cells [52], [59], [60], [61]. Other miRNAs, such as miR-214 and miR-155, are enriched in exosomes derived from tumor cell lines or peripheral blood from cancer patients [54], [58], [62].
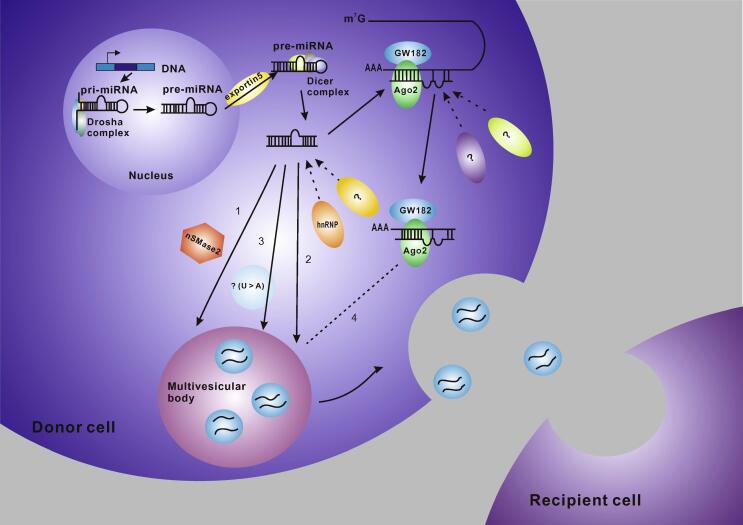
Based on current research, there are four potential modes for sorting of miRNAs into exosomes, although the underlying mechanisms remain largely unclear. These include: 1) The neural sphingomyelinase 2 (nSMase2)-dependent pathway. nSMase2 is the first molecule reported to be related to miRNA secretion into exosomes. Kosaka et al. found that overexpression of nSMase2 increased the number of exosomal miRNAs, and conversely inhibition of nSMase2 expression reduced the number of exosomal miRNAs [22]. 2) The miRNA motif and sumoylated heterogeneous nuclear ribonucleoproteins (hnRNPs)-dependent pathway. Villarroya-Beltri et al. discovered that sumoylated hnRNPA2B1 could recognize the GGAG motif in the 3′ portion of miRNA sequences and cause specific miRNAs to be packed into exosomes [59]. Similarly, another two hnRNP family proteins, hnRNPA1 and hnRNPC, can also bind to exosomal miRNAs, suggesting that they might be candidates for miRNA sorting as well. However, no binding motifs have been identified yet [59]. 3) The 3′-end of the miRNA sequence-dependent pathway. Koppers-Lalic et al. discovered that the 3′ ends of uridylated endogenous miRNAs were mainly presented in exosomes derived from B cells or urine, whereas the 3′ ends of adenylated endogenous miRNAs were mainly presented in B cells [60]. The above two selection modes commonly indicate that the 3′ portion or the 3′ end of the miRNA sequence contains a critical sorting signal. 4) The miRNA induced silencing complex (miRISC)-related pathway. It is well known that mature miRNAs can interact with assembly proteins to form a complex called miRISC. The main components of miRISC include miRNA, miRNA-repressible mRNA, GW182, and AGO2. The AGO2 protein in humans, which prefers to bind to U or A at the 5′ end of miRNAs, plays an important role in mediating mRNA:miRNA formation and the consequent translational repression or degradation of the mRNA molecule [63]. Recent studies recognized a possible correlation between AGO2 and exosomal miRNA sorting. In exosomal protein analyses, AGO2 has sometimes been identified by using mass spectrometry (MS) or Western blotting [51], [64]. Guduric-Fuchs et al. discovered that knockout of AGO2 could decrease the types or abundance of the preferentially-exported miRNAs, such as miR-451, miR-150, and miR-142-3p, in HEK293T-derived exosomes [52]. Other evidence has also supported a relationship between miRISC and exosomal miRNA sorting. First, the main components of miRISC were found to be co-localized with MVBs [65]. Second, blockage of the turnover of MVBs into lysosomes could lead to the over-accumulation of miRISCs, whereas blockage of the formation of MVBs resulted in the loss of miRISCs [66]. Third, the changes in miRNA-repressible targets levels that occur in response to cell activation may cause miRNA sorting to exosomes, partially by differentially engaging them at the sites of miRNA activity (miRISCs) and exosome biogenesis (MVBs) [55]. In summary, specific sequences present in certain miRNAs may guide their incorporation into exosomes, whereas some enzymes or other proteins may control sorting of exosomal miRNAs as well, in a miRNA sequence-independent fashion (Figure 1).
Figure 1.
The sorting mechanism for exosomal microRNAs
In animals, microRNA (miRNA) genes are transcribed into primary miRNAs (pri-miRNAs), and processed by the Drosha complex to form precursor miRNAs (pre-miRNAs), which are exported into the cytoplasm by the exportin5 complex. The pre-miRNAs undergo digestion by the Dicer complex to become mature miRNAs. Mature miRNAs are sorted into exosomes via four potential modes: (1) nSMase2-dependent pathway; (2) miRNA motif and sumoylated hnRNPs-dependent pathway; The sumoylated hnRNP family protein recognizes the GGAG motif in the 3′ portion of the miRNA sequence and guides specific miRNAs to be packed into exosomes. (3) 3′ miRNA sequence-dependent pathway; miRNAs that are preferentially sorted into exosomes have more poly(U) than poly(A) at the 3′ end. (4) The miRISC-related pathway. miRISCs co-localize with the sites of exosome biogenesis (multivesicular bodies) and their components, such as AGO2 protein and miRNA-targeted mRNA, are correlated with sorting of miRNAs into exosomes.
The function of exosomal miRNAs
Since Valadi et al. [24] described that miRNAs could be transferred between cells via exosomes, more similar observations have been reported [67], [68], [69]. The miRNAs in cell-released exosomes can circulate with the associated vehicles to reach neighboring cells and distant cells. After being delivered into acceptor cells, exosomal miRNAs play functional roles. Although it is difficult to completely exclude the effects of other exosomal cargos on recipient cells, miRNAs are considered the key functional elements. The functions of exosomal miRNAs can be generally classified into two types. One is the conventional function, i.e., miRNAs perform negative regulation and confer characteristic changes in the expression levels of target genes. For example, exosomal miR-105 released from the breast cancer cell lines MCF-10A and MDA-MB-231 reduced ZO-1 gene expression in endothelial cells and promoted metastases to the lung and brain [70]. Exosomal miR-214, derived from the human microvascular endothelial cell line HMEC-1, stimulated migration and angiogenesis in neighboring HMEC-1 cells [71]. Exosomal miR-92a, derived from K562 cells, significantly reduced the expression of integrin α5 in the human umbilical vein endothelial (HUVEC) cells and enhanced endothelial cell migration and tube formation [72]. The other one is a novel function that has been identified in some miRNAs when they are studied as exosomal miRNAs rather than intracellular miRNAs. Exosomal miR-21 and miR-29a, in addition to the classic role of targeting mRNA, were first discovered to have the capacity to act as ligands that bind to toll-like receptors (TLRs) and activate immune cells [73]. This study uncovered an entirely new function of miRNAs. To further understand this novel function of miRNAs, more investigations are worthwhile.
Notably, current functional studies of exosomal miRNAs have some limitations. First, diverse methods are used for exosome isolation. Exosomes can be enriched from cell culture media by ultracentrifugation, density gradient separation, immunoaffinity capture, size exclusion chromatography, and ExoQuick™ Precipitation (System Biosciences, USA). Use of different exosome purification strategies could slightly affect exosomal contents, including proteins and miRNAs [74], [75], [76]. Second, the large number of variable miRNAs carried by exosomes may regulate many different signaling pathways, and will generate integral effects on recipient cells. Therefore, it is difficult to gain a thorough understanding of the functions of exosomal miRNAs. According to studies of miRNA sorting mechanisms, certain miRNAs may be classified by portions of their sequences, and the functions of each group may be elucidated separately. Third, it is difficult to identify exosomal miRNAs in a single exosome or to measure the amount of a given miRNA carried by an exosome when it is present in low abundance. Chevillet et al. quantified the number of exosomes by a NanoSight instrument (Malvern, UK) and the number of miRNA molecules in an exosome collection using a real-time PCR-based absolute quantification method. They found that, on average, most exosomes did not harbor many copies of miRNA molecule [77]. According to this study, accumulation of exosomal miRNAs in recipient cells is necessary for miRNA-based communication. More sophisticated techniques and methods need to be developed to enrich the subpopulation of miRNA-rich exosomes, and functionally sufficient quantities of exosomal miRNAs need to be determined.
Applications of exosomes and exosomal miRNAs
Exosomal miRNAs can stably exist in the blood, urine, and other body fluids of patients, and exosomes can reflect their tissue or cell of origin by the presence of specific surface proteins [1], [2], [3]. Furthermore, the amount and composition of exosomal miRNAs differ between patients with disease and healthy individuals. Thus, exosomal miRNAs show potential for use as noninvasive biomarkers to indicate disease states. Several previous studies have profiled exosomal miRNAs in different samples. It is of note that some exosomal miRNAs can be used to aid in clinical diagnosis (Table 1) [29], [30], [54], [62]. For example, a set of exosomal miRNAs, including let-7a, miR-1229, miR-1246, miR-150, miR-21, miR-223, and miR-23a, can be used as the diagnostic biomarker of colorectal cancer [57]. Another set, miR-1290 and miR-375, can be used as the prognostic marker in castration-resistant prostate cancer [56].
Table 1.
Exosomal microRNAs capable of distinguishing different pathological conditions in patients
| Sample description | Isolation strategy | Quantification method | Findings | Ref. |
|---|---|---|---|---|
| Tumor cells from glioblastoma patients at passage 1–15; serum from glioblastoma patients and controls | Ultracentrifugation | Quantitative PCR | 11 miRNAs (miR-15b, miR-16, miR-196, miR-21, miR-26a, miR-27a, miR-92, miR-93, miR-320, miR-20, and let-7a) were known to be abundant in gliomas, able to be detected in their derived microvesicles; the level of exosomal miR-21 was elevated in serum microvesicles compared with controls | [29] |
| Plasma from NSCLC patients (n = 28 for test, n = 78 for validation); plasma from controls (n = 20 for test, n = 48 for validation) | Immunobead (EpCAM) | Quantitative PCR | The levels of exosomal let-7f and/or miR-30e-3p in NSCLC patients can distinguish patients with resectable tumors from those with non-resectable tumors | [30] |
| Serum from malignant tumor patients (n = 50); serum from benign tumor patients (n = 10); serum from controls (n = 10). | Immunobead (EpCAM) and ultracentrifugation | Microarray | The levels of 8 exosomal miRNAs (miR-21, miR141, miR-200a, miR-200b, miR-200c, miR-203, miR-205, and miR-214) from malignant tumor are significantly distinct from those observed in benign tumor; exosomal miRNAs could not be detected in normal controls | [54] |
| Plasma from lung adenocarcinoma (n = 27); plasma from control (n = 9). | Size exclusion chromatography and immunobead (EpCAM) | Microarray | The levels of 12 exosomal miRNAs (miR-17-3p, miR-21, miR-106a, miR-146, miR155, miR-191, miR-192, miR-203, miR-205, miR-210, miR-212, and miR-214) are significantly different between patients and controls | [62] |
Note: NSCLC, non-small-cell lung carcinoma; EpCAM, epithelial cell adhesion molecule.
Besides the endogenous miRNAs, exogenous miRNAs can also be sorted into exosomes, which has been experimentally confirmed by Pegtel et al. [78] and Meckes et al. [79], who observed that human tumor viruses can exploit exosomes as delivery vectors to transfer their exogenous miRNAs to other non-infected cells [78], [79]. Hence, exogenous small RNAs have also been transferred by exosomes by mimicking the molecular mechanism of endogenous miRNAs transportation.
RNA interference (RNAi) has been applied to gene therapy [80], [81]. The findings on the employment of exosomes by the exogenous miRNAs suggest that combination of exosomes with RNAi technology is a promising method for gene therapy and this idea has been supported by several lines of evidence. For instance, Wahlgren et al. used plasma exosomes as gene delivery platforms to transfer exogenous siRNAs to monocytes and lymphocytes, which resulted in the silencing of the target MAPK gene [82]. In addition, Shtam et al. introduced exogenous siRNA into exosomes derived from HeLa cells, and used these transfected exosomes to knock down the target gene RAD51 in the recipient cells [83]. Moreover, the effect of exosome-siRNA gene silencing has also been validated in a mouse model [84]. It is therefore possible to use exosomes to modulate target genes for therapeutic purposes, but a great deal of additional research will be required to develop these therapies for clinical use.
Perspective
Although exosomes were first identified in the 1980s, studies on exosomes have been increasing remarkably during the last five years, especially following the discovery of functional mRNAs and miRNAs in exosomes. Exosomes play a key role in the process of cell-to-cell communication and influence the phenotype of recipient cells. However, exosome study is still in its infancy. People may want to know whether other non-coding RNAs such as long non coding RNAs could be present in exosomes, and whether they get involved in target gene regulation in recipient cells. With the discovery that exosomal miRNAs can function as ligands, a new field in exosome study has been opened up. It remains controversial whether the mechanism for packing of bioactive molecules into exosomes and secreting them into the extracellular space is an active or a passive process. More investigations on this matter will be warranted. Nonetheless, the most exciting but challenging application will be to utilize exosomes and their cargo as a clinical tool to diagnose and monitor disease, perhaps even for gene therapy, but much work remains to achieve this goal.
Competing interests
The authors declared that there are no competing interests.
Acknowledgments
This work was supported by the Projects of International Cooperation and Exchanges from the National Natural Science Foundation of China (Grant No. 31161120358), the National Basic Research Program from the Ministry of Science and Technology of China (973 program; Grant Nos. 20111CB510106 and 2015CB910603), the Open Project of State Key Laboratory of Biomembrane and Membrane Biotechnology, and the Scientific Research Foundation for Returned Scholars from the Ministry of Education of China. ML was supported by National Natural Science Foundation of China (Grant No. 31400741).
Handled by William CS Cho
Footnotes
Peer review under responsibility of Beijing Institute of Genomics, Chinese Academy of Sciences and Genetics Society of China.
References
- 1.Simons M., Raposo G. Exosomes – vesicular carriers for intercellular communication. Curr Opin Cell Biol. 2009;21:575–581. doi: 10.1016/j.ceb.2009.03.007. [DOI] [PubMed] [Google Scholar]
- 2.Mathivanan S., Ji H., Simpson R.J. Exosomes: extracellular organelles important in intercellular communication. J Proteomics. 2010;73:1907–1920. doi: 10.1016/j.jprot.2010.06.006. [DOI] [PubMed] [Google Scholar]
- 3.Gross J.C., Chaudhary V., Bartscherer K., Boutros M. Active Wnt proteins are secreted on exosomes. Nat Cell Biol. 2012;14:1036–1045. doi: 10.1038/ncb2574. [DOI] [PubMed] [Google Scholar]
- 4.Sato-Kuwabara Y., Melo S.A., Soares F.A., Calin G.A. The fusion of two worlds: non-coding RNAs and extracellular vesicles – diagnostic and therapeutic implications (Review) Int J Oncol. 2015;46:17–27. doi: 10.3892/ijo.2014.2712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bartel D.P. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 6.Png K.J., Halberg N., Yoshida M., Tavazoie S.F. A microRNA regulon that mediates endothelial recruitment and metastasis by cancer cells. Nature. 2012;481:190–194. doi: 10.1038/nature10661. [DOI] [PubMed] [Google Scholar]
- 7.Gee H.E., Camps C., Buffa F.M., Colella S., Sheldon H., Gleadle J.M., et al. MicroRNA-10b and breast cancer metastasis. Nature. 2008;455:E8–E9. doi: 10.1038/nature07362. [DOI] [PubMed] [Google Scholar]
- 8.Tay Y., Zhang J., Thomson A.M., Lim B., Rigoutsos I. MicroRNAs to Nanog, Oct4 and Sox2 coding regions modulate embryonic stem cell differentiation. Nature. 2008;455:1124–1128. doi: 10.1038/nature07299. [DOI] [PubMed] [Google Scholar]
- 9.Kota J., Chivukula R.R., O’Donnell K.A., Wentzel E.A., Montgomery C.L., Hwang H., et al. Therapeutic delivery of miR-26a inhibits cancer cell proliferation and induces tumor-specific apoptosis. Cell. 2009;137:1005–1017. [Google Scholar]
- 10.Ma L., Teruya-Feldstein J., Weinberg R.A. Tumour invasion and metastasis initiated by microRNA-10b in breast cancer. Nature. 2007;449:682–688. doi: 10.1038/nature06174. [DOI] [PubMed] [Google Scholar]
- 11.Gallo A., Tandon M., Alevizos I., Illei G.G. The majority of microRNAs detectable in serum and saliva is concentrated in exosomes. PLoS One. 2012;7:e30679. doi: 10.1371/journal.pone.0030679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Michael A., Bajracharya S.D., Yuen P.S., Zhou H., Star R.A., Illei G.G., et al. Exosomes from human saliva as a source of microRNA biomarkers. Oral Dis. 2010;16:34–38. doi: 10.1111/j.1601-0825.2009.01604.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lv L.L., Cao Y., Liu D., Xu M., Liu H., Tang R.N., et al. Isolation and quantification of microRNAs from urinary exosomes/microvesicles for biomarker discovery. Int J Biol Sci. 2013;9:1021–1031. doi: 10.7150/ijbs.6100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhou Q., Li M., Wang X., Li Q., Wang T., Zhu Q., et al. Immune-related microRNAs are abundant in breast milk exosomes. Int J Biol Sci. 2012;8:118–123. doi: 10.7150/ijbs.8.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hu Z., Chen X., Zhao Y., Tian T., Jin G., Shu Y., et al. Serum microRNA signatures identified in a genome-wide serum microRNA expression profiling predict survival of non-small-cell lung cancer. J Clin Oncol. 2010;28:1721–1726. doi: 10.1200/JCO.2009.24.9342. [DOI] [PubMed] [Google Scholar]
- 16.Arroyo J.D., Chevillet J.R., Kroh E.M., Ruf I.K., Pritchard C.C., Gibson D.F., et al. Argonaute2 complexes carry a population of circulating microRNAs independent of vesicles in human plasma. Proc Natl Acad Sci U S A. 2011;108:5003–5008. doi: 10.1073/pnas.1019055108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tabet F., Vickers K.C., Cuesta Torres L.F., Wiese C.B., Shoucri B.M., Lambert G., et al. HDL-transferred microRNA-223 regulates ICAM-1 expression in endothelial cells. Nat Commun. 2014;5:3292. doi: 10.1038/ncomms4292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vickers K.C., Palmisano B.T., Shoucri B.M., Shamburek R.D., Remaley A.T. MicroRNAs are transported in plasma and delivered to recipient cells by high-density lipoproteins. Nat Cell Biol. 2011;13:423–433. doi: 10.1038/ncb2210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kuroiwa T., Lee E.G., Danning C.L., Illei G.G., McInnes I.B., Boumpas D.T. CD40 ligand-activated human monocytes amplify glomerular inflammatory responses through soluble and cell-to-cell contact-dependent mechanisms. J Immunol. 1999;163:2168–2175. [PubMed] [Google Scholar]
- 20.Harvey S., Martinez-Moreno C.G., Luna M., Aramburo C. Autocrine/paracrine roles of extrapituitary growth hormone and prolactin in health and disease: an overview. Gen Comp Endocrinol. 2014 doi: 10.1016/j.ygcen.2014.11.004. [DOI] [PubMed] [Google Scholar]
- 21.Kosaka N., Iguchi H., Yoshioka Y., Takeshita F., Matsuki Y., Ochiya T. Secretory mechanisms and intercellular transfer of microRNAs in living cells. J Biol Chem. 2010;285:17442–17452. doi: 10.1074/jbc.M110.107821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kosaka N., Iguchi H., Hagiwara K., Yoshioka Y., Takeshita F., Ochiya T. Neutral sphingomyelinase 2 (nSMase2)-dependent exosomal transfer of angiogenic microRNAs regulate cancer cell metastasis. J Biol Chem. 2013;288:10849–10859. doi: 10.1074/jbc.M112.446831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pan B.T., Johnstone R. Selective externalization of the transferrin receptor by sheep reticulocytes in vitro. Response to ligands and inhibitors of endocytosis. J Biol Chem. 1984;259:9776–9782. [PubMed] [Google Scholar]
- 24.Valadi H., Ekstrom K., Bossios A., Sjostrand M., Lee J.J., Lotvall J.O. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol. 2007;9:654–659. doi: 10.1038/ncb1596. [DOI] [PubMed] [Google Scholar]
- 25.Pan B.T., Johnstone R.M. Fate of the transferrin receptor during maturation of sheep reticulocytes in vitro: selective externalization of the receptor. Cell. 1983;33:967–978. doi: 10.1016/0092-8674(83)90040-5. [DOI] [PubMed] [Google Scholar]
- 26.Johnstone R.M., Bianchini A., Teng K. Reticulocyte maturation and exosome release: transferrin receptor containing exosomes shows multiple plasma membrane functions. Blood. 1989;74:1844–1851. [PubMed] [Google Scholar]
- 27.Cocucci E., Racchetti G., Meldolesi J. Shedding microvesicles: artefacts no more. Trends Cell Biol. 2009;19:43–51. doi: 10.1016/j.tcb.2008.11.003. [DOI] [PubMed] [Google Scholar]
- 28.Hunter M.P., Ismail N., Zhang X., Aguda B.D., Lee E.J., Yu L., et al. Detection of microRNA expression in human peripheral blood microvesicles. PLoS One. 2008;3:e3694. doi: 10.1371/journal.pone.0003694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Skog J., Wurdinger T., van Rijn S., Meijer D.H., Gainche L., Sena-Esteves M., et al. Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers. Nat Cell Biol. 2008;10:1470–1476. doi: 10.1038/ncb1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Silva J., Garcia V., Zaballos A., Provencio M., Lombardia L., Almonacid L., et al. Vesicle-related microRNAs in plasma of nonsmall cell lung cancer patients and correlation with survival. Eur Respir J. 2011;37:617–623. doi: 10.1183/09031936.00029610. [DOI] [PubMed] [Google Scholar]
- 31.Heijnen H.F., Schiel A.E., Fijnheer R., Geuze H.J., Sixma J.J. Activated platelets release two types of membrane vesicles: microvesicles by surface shedding and exosomes derived from exocytosis of multivesicular bodies and alpha-granules. Blood. 1999;94:3791–3799. [PubMed] [Google Scholar]
- 32.Gruenberg J., van der Goot F.G. Mechanisms of pathogen entry through the endosomal compartments. Nat Rev Mol Cell Biol. 2006;7:495–504. doi: 10.1038/nrm1959. [DOI] [PubMed] [Google Scholar]
- 33.Ostrowski M., Carmo N.B., Krumeich S., Fanget I., Raposo G., Savina A., et al. Rab27a and Rab27b control different steps of the exosome secretion pathway. Nat Cell Biol. 2010;12 doi: 10.1038/ncb2000. 19–30; sup pp 1–13. [DOI] [PubMed] [Google Scholar]
- 34.Yu X., Harris S.L., Levine A.J. The regulation of exosome secretion: a novel function of the p53 protein. Cancer Res. 2006;66:4795–4801. doi: 10.1158/0008-5472.CAN-05-4579. [DOI] [PubMed] [Google Scholar]
- 35.Baietti M.F., Zhang Z., Mortier E., Melchior A., Degeest G., Geeraerts A., et al. Syndecan-syntenin-ALIX regulates the biogenesis of exosomes. Nat Cell Biol. 2012;14:677–685. doi: 10.1038/ncb2502. [DOI] [PubMed] [Google Scholar]
- 36.Munich S., Sobo-Vujanovic A., Buchser W.J., Beer-Stolz D., Vujanovic N.L. Dendritic cell exosomes directly kill tumor cells and activate natural killer cells via TNF superfamily ligands. Oncoimmunology. 2012;1:1074–1083. doi: 10.4161/onci.20897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mulcahy L.A., Pink R.C., Carter D.R. Routes and mechanisms of extracellular vesicle uptake. J Extracell Vesicles. 2014 doi: 10.3402/jev.v3.24641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tian T., Zhu Y.L., Hu F.H., Wang Y.Y., Huang N.P., Xiao Z.D. Dynamics of exosome internalization and trafficking. J Cell Physiol. 2013;228:1487–1495. doi: 10.1002/jcp.24304. [DOI] [PubMed] [Google Scholar]
- 39.Koumangoye R.B., Sakwe A.M., Goodwin J.S., Patel T., Ochieng J. Detachment of breast tumor cells induces rapid secretion of exosomes which subsequently mediate cellular adhesion and spreading. PLoS One. 2011;6:e24234. doi: 10.1371/journal.pone.0024234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Escrevente C., Keller S., Altevogt P., Costa J. Interaction and uptake of exosomes by ovarian cancer cells. BMC Cancer. 2011;11:108. doi: 10.1186/1471-2407-11-108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liao J., Liu R., Yin L., Pu Y. Expression profiling of exosomal miRNAs derived from human esophageal cancer cells by Solexa high-throughput sequencing. Int J Mol Sci. 2014;15:15530–15551. doi: 10.3390/ijms150915530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Saunderson S.C., Dunn A.C., Crocker P.R., McLellan A.D. CD169 mediates the capture of exosomes in spleen and lymph node. Blood. 2014;123:208–216. doi: 10.1182/blood-2013-03-489732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wieckowski E., Whiteside T.L. Human tumor-derived vs dendritic cell-derived exosomes have distinct biologic roles and molecular profiles. Immunol Res. 2006;36:247–254. doi: 10.1385/IR:36:1:247. [DOI] [PubMed] [Google Scholar]
- 44.Nolte-‘t Hoen E.N., Buschow S.I., Anderton S.M., Stoorvogel W., Wauben M.H. Activated T cells recruit exosomes secreted by dendritic cells via LFA-1. Blood. 2009;113:1977–1981. doi: 10.1182/blood-2008-08-174094. [DOI] [PubMed] [Google Scholar]
- 45.Janowska-Wieczorek A., Wysoczynski M., Kijowski J., Marquez-Curtis L., Machalinski B., Ratajczak J., et al. Microvesicles derived from activated platelets induce metastasis and angiogenesis in lung cancer. Int J Cancer. 2005;113:752–760. doi: 10.1002/ijc.20657. [DOI] [PubMed] [Google Scholar]
- 46.Qu J.L., Qu X.J., Zhao M.F., Teng Y.E., Zhang Y., Hou K.Z., et al. Gastric cancer exosomes promote tumour cell proliferation through PI3K/Akt and MAPK/ERK activation. Dig Liver Dis. 2009;41:875–880. doi: 10.1016/j.dld.2009.04.006. [DOI] [PubMed] [Google Scholar]
- 47.Millimaggi D., Mari M., D’Ascenzo S., Carosa E., Jannini E.A., Zucker S., et al. Tumor vesicle-associated CD147 modulates the angiogenic capability of endothelial cells. Neoplasia. 2007;9:349–357. doi: 10.1593/neo.07133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Webber J., Steadman R., Mason M.D., Tabi Z., Clayton A. Cancer exosomes trigger fibroblast to myofibroblast differentiation. Cancer Res. 2010;70:9621–9630. doi: 10.1158/0008-5472.CAN-10-1722. [DOI] [PubMed] [Google Scholar]
- 49.Cho J.A., Park H., Lim E.H., Lee K.W. Exosomes from breast cancer cells can convert adipose tissue-derived mesenchymal stem cells into myofibroblast-like cells. Int J Oncol. 2012;40:130–138. doi: 10.3892/ijo.2011.1193. [DOI] [PubMed] [Google Scholar]
- 50.Simpson R.J., Kalra H., Mathivanan S. ExoCarta as a resource for exosomal research. J Extracell Vesicles. 2012 doi: 10.3402/jev.v1i0.18374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Goldie B.J., Dun M.D., Lin M., Smith N.D., Verrills N.M., Dayas C.V., et al. Activity-associated miRNA are packaged in Map1b-enriched exosomes released from depolarized neurons. Nucleic Acids Res. 2014;42:9195–9208. doi: 10.1093/nar/gku594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Guduric-Fuchs J., O’Connor A., Camp B., O’Neill C.L., Medina R.J., Simpson D.A. Selective extracellular vesicle-mediated export of an overlapping set of microRNAs from multiple cell types. BMC Genomics. 2012;13:357. doi: 10.1186/1471-2164-13-357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ohshima K., Inoue K., Fujiwara A., Hatakeyama K., Kanto K., Watanabe Y., et al. Let-7 microRNA family is selectively secreted into the extracellular environment via exosomes in a metastatic gastric cancer cell line. PLoS One. 2010;5:e13247. doi: 10.1371/journal.pone.0013247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Taylor D.D., Gercel-Taylor C. MicroRNA signatures of tumor-derived exosomes as diagnostic biomarkers of ovarian cancer. Gynecol Oncol. 2008;110:13–21. doi: 10.1016/j.ygyno.2008.04.033. [DOI] [PubMed] [Google Scholar]
- 55.Squadrito M.L., Baer C., Burdet F., Maderna C., Gilfillan G.D., Lyle R., et al. Endogenous RNAs modulate microRNA sorting to exosomes and transfer to acceptor cells. Cell Rep. 2014;8:1432–1446. doi: 10.1016/j.celrep.2014.07.035. [DOI] [PubMed] [Google Scholar]
- 56.Huang X., Yuan T., Tschannen M., Sun Z., Jacob H., Du M., et al. Characterization of human plasma-derived exosomal RNAs by deep sequencing. BMC Genomics. 2013;14:319. doi: 10.1186/1471-2164-14-319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ogata-Kawata H., Izumiya M., Kurioka D., Honma Y., Yamada Y., Furuta K., et al. Circulating exosomal microRNAs as biomarkers of colon cancer. PLoS One. 2014;9:e92921. doi: 10.1371/journal.pone.0092921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rana S., Malinowska K., Zoller M. Exosomal tumor microRNA modulates premetastatic organ cells. Neoplasia. 2013;15:281–295. doi: 10.1593/neo.122010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Villarroya-Beltri C., Gutierrez-Vazquez C., Sanchez-Cabo F., Perez-Hernandez D., Vazquez J., Martin-Cofreces N., et al. Sumoylated hnRNPA2B1 controls the sorting of miRNAs into exosomes through binding to specific motifs. Nat Commun. 2013;4:2980. doi: 10.1038/ncomms3980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Koppers-Lalic D., Hackenberg M., Bijnsdorp I.V., van Eijndhoven M.A., Sadek P., Sie D., et al. Nontemplated nucleotide additions distinguish the small RNA composition in cells from exosomes. Cell Rep. 2014;8:1649–1658. doi: 10.1016/j.celrep.2014.08.027. [DOI] [PubMed] [Google Scholar]
- 61.Ekstrom K., Valadi H., Sjostrand M., Malmhall C., Bossios A., Eldh M., et al. Characterization of mRNA and microRNA in human mast cell-derived exosomes and their transfer to other mast cells and blood CD34 progenitor cells. J Extracell Vesicles. 2012 doi: 10.3402/jev.v1i0.18389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rabinowits G., Gercel-Taylor C., Day J.M., Taylor D.D., Kloecker G.H. Exosomal microRNA: a diagnostic marker for lung cancer. Clin Lung Cancer. 2009;10:42–46. doi: 10.3816/CLC.2009.n.006. [DOI] [PubMed] [Google Scholar]
- 63.Frank F., Sonenberg N., Nagar B. Structural basis for 5′-nucleotide base-specific recognition of guide RNA by human AGO2. Nature. 2010;465:818–822. doi: 10.1038/nature09039. [DOI] [PubMed] [Google Scholar]
- 64.Melo S.A., Sugimoto H., O’Connell J.T., Kato N., Villanueva A., Vidal A., et al. Cancer exosomes perform cell-independent microRNA biogenesis and promote tumorigenesis. Cancer Cell. 2014;26:707–721. doi: 10.1016/j.ccell.2014.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gibbings D.J., Ciaudo C., Erhardt M., Voinnet O. Multivesicular bodies associate with components of miRNA effector complexes and modulate miRNA activity. Nat Cell Biol. 2009;11:1143–1149. doi: 10.1038/ncb1929. [DOI] [PubMed] [Google Scholar]
- 66.Lee Y.S., Pressman S., Andress A.P., Kim K., White J.L., Cassidy J.J., et al. Silencing by small RNAs is linked to endosomal trafficking. Nat Cell Biol. 2009;11:1150–1156. doi: 10.1038/ncb1930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kogure T., Lin W.L., Yan I.K., Braconi C., Patel T. Intercellular nanovesicle-mediated microRNA transfer: a mechanism of environmental modulation of hepatocellular cancer cell growth. Hepatology. 2011;54:1237–1248. doi: 10.1002/hep.24504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Chiba M., Kimura M., Asari S. Exosomes secreted from human colorectal cancer cell lines contain mRNAs, microRNAs and natural antisense RNAs, that can transfer into the human hepatoma HepG2 and lung cancer A549 cell lines. Oncol Rep. 2012;28:1551–1558. doi: 10.3892/or.2012.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Montecalvo A., Larregina A.T., Shufesky W.J., Stolz D.B., Sullivan M.L., Karlsson J.M., et al. Mechanism of transfer of functional microRNAs between mouse dendritic cells via exosomes. Blood. 2012;119:756–766. doi: 10.1182/blood-2011-02-338004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zhou W., Fong M.Y., Min Y., Somlo G., Liu L., Palomares M.R., et al. Cancer-secreted miR-105 destroys vascular endothelial barriers to promote metastasis. Cancer Cell. 2014;25:501–515. doi: 10.1016/j.ccr.2014.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.van Balkom B.W., de Jong O.G., Smits M., Brummelman J., den Ouden K., de Bree P.M., et al. Endothelial cells require miR-214 to secrete exosomes that suppress senescence and induce angiogenesis in human and mouse endothelial cells. Blood. 2013;121:S1–S15. doi: 10.1182/blood-2013-02-478925. [DOI] [PubMed] [Google Scholar]
- 72.Umezu T., Ohyashiki K., Kuroda M., Ohyashiki J.H. Leukemia cell to endothelial cell communication via exosomal miRNAs. Oncogene. 2013;32:2747–2755. doi: 10.1038/onc.2012.295. [DOI] [PubMed] [Google Scholar]
- 73.Fabbri M., Paone A., Calore F., Galli R., Gaudio E., Santhanam R., et al. MicroRNAs bind to toll-like receptors to induce prometastatic inflammatory response. Proc Natl Acad Sci U S A. 2012;109:E2110–E2116. doi: 10.1073/pnas.1209414109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Rekker K., Saare M., Roost A.M., Kubo A.L., Zarovni N., Chiesi A., et al. Comparison of serum exosome isolation methods for microRNA profiling. Clin Biochem. 2014;47:135–138. doi: 10.1016/j.clinbiochem.2013.10.020. [DOI] [PubMed] [Google Scholar]
- 75.Tauro B.J., Greening D.W., Mathias R.A., Ji H., Mathivanan S., Scott A.M., et al. Comparison of ultracentrifugation, density gradient separation, and immunoaffinity capture methods for isolating human colon cancer cell line LIM1863-derived exosomes. Methods. 2012;56:293–304. doi: 10.1016/j.ymeth.2012.01.002. [DOI] [PubMed] [Google Scholar]
- 76.Taylor D.D., Zacharias W., Gercel-Taylor C. Exosome isolation for proteomic analyses and RNA profiling. Methods Mol Biol. 2011;728:235–246. doi: 10.1007/978-1-61779-068-3_15. [DOI] [PubMed] [Google Scholar]
- 77.Chevillet J.R., Kang Q., Ruf I.K., Briggs H.A., Vojtech L.N., Hughes S.M., et al. Quantitative and stoichiometric analysis of the microRNA content of exosomes. Proc Natl Acad Sci U S A. 2014;111:14888–14893. doi: 10.1073/pnas.1408301111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Pegtel D.M., Cosmopoulos K., Thorley-Lawson D.A., van Eijndhoven M.A., Hopmans E.S., Lindenberg J.L., et al. Functional delivery of viral miRNAs via exosomes. Proc Natl Acad Sci U S A. 2010;107:6328–6333. doi: 10.1073/pnas.0914843107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Meckes D.G., Jr., Shair K.H., Marquitz A.R., Kung C.P., Edwards R.H., Raab-Traub N. Human tumor virus utilizes exosomes for intercellular communication. Proc Natl Acad Sci U S A. 2010;107:20370–20375. doi: 10.1073/pnas.1014194107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ryther R.C., Flynt A.S., Phillips J.A., 3rd, Patton J.G. SiRNA therapeutics: big potential from small RNAs. Gene Ther. 2005;12:5–11. doi: 10.1038/sj.gt.3302356. [DOI] [PubMed] [Google Scholar]
- 81.Koutsilieri E., Rethwilm A., Scheller C. The therapeutic potential of siRNA in gene therapy of neurodegenerative disorders. J Neural Transm Suppl. 2007;72:43–49. doi: 10.1007/978-3-211-73574-9_7. [DOI] [PubMed] [Google Scholar]
- 82.Wahlgren J., De L.K.T., Brisslert M., Vaziri Sani F., Telemo E., Sunnerhagen P., et al. Plasma exosomes can deliver exogenous short interfering RNA to monocytes and lymphocytes. Nucleic Acids Res. 2012;40:e130. doi: 10.1093/nar/gks463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Shtam T.A., Kovalev R.A., Varfolomeeva E.Y., Makarov E.M., Kil Y.V., Filatov M.V. Exosomes are natural carriers of exogenous siRNA to human cells in vitro. Cell Commun Signal. 2013;11:88. doi: 10.1186/1478-811X-11-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Pan Q., Ramakrishnaiah V., Henry S., Fouraschen S., de Ruiter P.E., Kwekkeboom J., et al. Hepatic cell-to-cell transmission of small silencing RNA can extend the therapeutic reach of RNA interference (RNAi) Gut. 2012;61:1330–1339. doi: 10.1136/gutjnl-2011-300449. [DOI] [PubMed] [Google Scholar]