Abstract
Publicly-accessible resources have promoted the advance of scientific discovery. The era of genomics and big data has brought the need for collaboration and data sharing in order to make effective use of this new knowledge. Here, we describe the web resources for cancer genomics research and rate them on the basis of the diversity of cancer types, sample size, omics data comprehensiveness, and user experience. The resources reviewed include data repository and analysis tools; and we hope such introduction will promote the awareness and facilitate the usage of these resources in the cancer research community.
Keywords: Cancer, Genomics, Data integration, Resource, Collaboration
Introduction
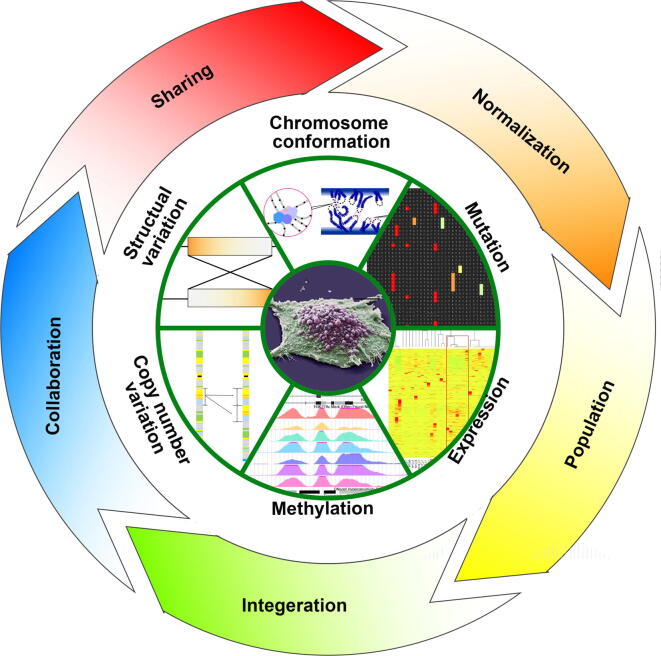
There has been accumulating evidence over the last two to three decades supporting that cancer is a disease of the genome. Previous studies have followed a one-by-one approach to examine the molecular mechanisms of cancer, although this approach is one-sided and inefficient. With the development of high-throughput sequencing technologies, recent years have witnessed a great data explosion and systematic study of the cancer genome. For the first time, data were made available for the complete genome sequences including point mutations and structural alternations for a large number of cancer types, enabling the differentiation of cancer subtypes in an unprecedented global view. However, the effective use of the massive amounts of cancer genome data remains a challenge due to the limitations of computational methodologies and insufficient collaboration and sharing (Figure 1). In this paper, we describe several popular and effective web-based cancer genomics data repositories, along with tools and resources (Table 1) to manage and analyze these data. We have rated the resources based on data comprehensiveness and ease-of-use according to our own experience.
Figure 1.
The future of cancer research In the era of big data, one of the major challenges is to make full use of multi-dimensional data from heterogeneous sources, including different omics data and a variety of medical data from bedside. The success in the battle against cancer will largely depend on population-sized information from both genomic and clinical resources, advanced algorithms for data mining, open and sharing circumstances.
Table 1.
Major web resources for cancer genomics research
| Name | Link | Main features | Rating | Refs. |
|---|---|---|---|---|
| CGHub | https://cghub.ucsc.edu/ | Comprehensive data repository; huge data size | ★★★★★ | [1] |
| EGA | https://www.ebi.ac.uk/ega/ | Comprehensive data repository; huge data size | ★★★★★ | |
| COSMIC | http://cancer.sanger.ac.uk | Largest somatic mutation database; genome sequencing paper curation | ★★★★★ | [2] |
| CPRG | http://www.broadinstitute.org/software/cprg | Interface for cancer program resources | ★★★★☆ | |
| GDAC | http://gdac.broadinstitute.org/ | Data analysis; automatic pipelines; user-friendly reports | ★★★★☆ | [3] |
| SNP500Cancer | http://snp500cancer.nci.nih.gov | Sequence and genotype verification of SNPs | ★★★★☆ | [4] |
| canEvolve | www.canevolve.org/ | Comprehensive analysis of tumor profile; Data from 90 studies involving more than 10,000 patients | ★★★★☆ | [5] |
| MethyCancer | http://methycancer.psych.ac.cn | Relationship among DNA methylation, gene expression and cancer | ★★★★☆ | [6] |
| SomamiR | http://compbio.uthsc.edu/SomamiR/ | Correlation between somatic mutation and microRNA; genome-wide displaying | ★★★★☆ | [7] |
| cBioPortal | http://www.cbioportal.org/public-portal/ | Graphical summaries; gene alteration; processed data; visualization | ★★★★★ | [8] |
| UCSC Cancer Genomics Browser | https://genome-cancer.soe.ucsc.edu/ | Clinical information; gene expression; copy number variation; visualization | ★★★★☆ | [9] |
| CGWB | https://cgwb.nci.nih.gov/ | Visualization; gene mutation and variation; automated analysis pipeline | ★★★★☆ | [10] |
| GDSC | http://www.cancerrxgene.org | Drug sensitivity information; drug response information | ★★★★☆ | [11] |
| canSAR | https://cansar.icr.ac.uk/ | Multidisciplinary information; drug discovery | ★★★★☆ | [12] |
| NONCODE | http://www.noncode.org/ | ncRNAs; lncRNAs; up-to-date and comprehensive resource | ★★★★☆ | [13], [14] |
Note: Rating is given on the basis of the diversity of cancer types, sample size, omics data comprehensiveness, and user experience.
Cancer Genomics Hub
The Cancer Genomics Hub (CGHub) is a central repository for the genomic information generated through three different programs at the National Cancer Institute (NCI) of the United States, namely, The Cancer Genome Atlas (TCGA), the Cancer Cell Line Encyclopedia (CCLE), and the Therapeutically Applicable Research to Generate Effective Treatments (TARGET) projects [1]. CGHub is hosted at the University of California, Santa Cruz (UCSC), with controlled data access to ensure patient privacy. CGHub holds nearly 1.9 PB of data, covering 42 cancer types and normal controls (https://cghub.ucsc.edu/summary_stats.html). Till Dec 2014, there have been more than 10,000 samples from TCGA alone (https://tcga-data.nci.nih.gov/tcga/tcgaHome2.jsp) and more than 500 papers have been published by researchers from the TCGA Research Network and those who used TCGA data in their work (http://cancergenome.nih.gov/publications). The launch of CGHub will promote the sharing of cancer data, collaboration between cancer researchers, and potentially facilitate the personalized medicine.
European Genome-phenome Archive
The European Genome-phenome Archive (EGA) is a data center for all types of sequencing and genotyping experiments. Almost 58% of all studies in EGA are related to cancer, including data generated by the International Cancer Genome Consortium (ICGC). Since its founding in 2008, ICGC has produced terabytes of data from about 12,232 donors and 50 cancer projects (https://dcc.icgc.org/repository/release_17). Somatic variant data are openly accessible at the ICGC Data Portal (https://dcc.icgc.org/repository), whereas raw sequence data, germline mutations, and clinical data are held at EGA with controlled access.
Catalogue Of Somatic Mutations In Cancer
The Catalogue Of Somatic Mutations In Cancer (COSMIC) is the largest database of somatic mutations and their effects on human cancer [2]. The database is curated manually from published literature, allowing very precise definitions of disease types and patient details. The database is updated every 2 months and has thus far integrated 15,047 whole cancer genomes from 1,058,292 samples, including 2,710,499 coding mutations, 10,567 gene fusions, 61,232 genomic rearrangements, 702,652 copy number variations (CNVs), and 118,886,698 abnormal expression variants. Data can be queried by key words and downloaded by registered users. COSMIC has also stored curated, large-scale systematic screens and whole-genome shotgun sequencing papers for reference. The huge, manually-curated, and regularly-updated dataset of the COSMIC database makes it an invaluable resource for cancer studies.
Cancer Program Resource Gateway
The Broad Institute is one of the most famous academic centers for cancer study. Its Cancer Program aims to investigate the fundamental mechanisms of cancer and research from discovery to clinical applications. The Program releases many datasets and tools for scientific research, which are held at the Broad Cancer Program Resource Gateway (CPRG). We will describe Broad’s Genome Data Analysis Center (GDAC), one of these resources, as an example.
Broad’s GDAC
It is important but generally time-consuming or sometimes even impossible for most labs to analyze terabytes of sequence data. However, the Firehose system from Broad’s GDAC is changing the status quo. The GDAC systematically analyzes data from TCGA pilot and extends to other diseases as well. Firehose now assembles ∼40 terabytes of TCGA data and reliably executes more than 6000 pipelines per month. GDAC obtains and processes the TCGA data every 2 weeks, and makes them available afterward [3]. Firehose contains series of standardized pipelines for genomic analysis and the computing environment is accessible to the public so that people can install and run their own tools for data analysis. Taking advantage of the powerful computational environment at Broad, Firehose provides continuously-updated data and results in different tiers, including version-stamped standardized datasets, analysis results, and biologist-friendly reports.
SNP500Cancer
The SNP500Cancer database, a part of the Cancer Genome Anatomy Project (CGAP) (http://cgap.nci.nih.gov/Tools), is a repository for sequence and genotype verification of single nucleotide polymorphisms (SNPs) in cancer and other complex diseases [4]. The main aim of the SNP500Cancer project is to re-sequence reference samples to find known or novel SNPs for molecular epidemiology studies in cancer. The database provides sequence information for anonymized control DNA samples and can be queried by gene, gene ontology pathway, chromosome, the SNP Database (dbSNP) ID, or SNP500Cancer SNP ID. SNP500Cancer is an insightful resource for researchers to select SNPs for further analysis due to its high confidence, nonetheless its data volume is limited.
canEvolve
With the rapid development of biological technologies, genome-wide tumor profiling has grown drastically in scale and availability. canEvolve fulfills the need for data integration and interpretation. canEvolve contains integrated data from 90 studies involving more than 10,000 patients. Data analysis can be performed at different levels: (1) primary analysis like mRNA, microRNA (miRNA) and protein expression, genome variations and protein–protein interactions; (2) integrative analysis of gene and miRNA expression, gene expression and CNVs, and gene set enrichment analysis; (3) network analysis; and (4) survival analysis. All the aforementioned data can be queried and visualized in table or graph format. canEvolve is a comprehensive functional genomics platform and is fully accessible to the public [5].
MethyCancer
DNA methylation plays a crucial role in the development of cancer and is associated with oncogene activation, chromosomal instability, and tumor suppressor gene silencing. MethyCancer is designed to interpret the relationship between DNA methylation, gene expression, and cancer. MethyCancer houses data on (1) DNA methylation, (2) CNV and cancer information, (3) CpG Island clones, and (4) the correlations between these datasets. The database can be easily searched with the user-friendly MethyView display [6].
SomamiR
miRNA sequence variation is contributable to a variety of cancers. SomamiR is a database created to investigate the association of somatic and germline mutations with miRNA function in cancer. The mutation data are collected from the Gene Expression Omnibus (GEO) and the Pediatric Cancer Genome Project (PCGP), whereas the miRNA data are from miRBase release 18. There are many ways to search SomamiR for somatic mutations that affect miRNA target sites, genome-wide association studies (GWAS) in cancer, candidate gene associations in cancer, KEGG pathways, and mutation information in a genome browser. SomamiR also contains a set of experimentally-validated somatic and germline mutations that disrupt miRNA function associated with cancer. The integration of mutation data in SomamiR will enable scientists to more accurately predict whether mutations would affect miRNA binding and consequently functional regulation [7].
cBioPortal
The cBioPortal for Cancer Genomics is an accessible portal for researchers to explore, visualize, and analyze multidimensional cancer genomics data [8]. The portal contains datasets from many published cancer studies including CCLE and TCGA. It is worthy of note that cBioPortal processes original molecular profiling data from cancer tissues and cell lines into smaller datasets. Researchers can interactively explore patterns of genetic alterations across samples in a single study, compare gene alteration frequencies across multiple studies, or summarize all relevant genomic variations in an individual tumor sample through the cBioPortal web query interface. It also supports biological pathway exploration, survival analysis, and data downloading service.
UCSC Cancer Genomics Browser
The UCSC Cancer Genomics Browser is an online analysis tool for hosting, visualizing, and analyzing information on cancer genomics and clinical research [9]. The browser provides users with measurements from a single experiment and the associated clinical information for multiple samples. Furthermore, two or more datasets can be viewed at the same time to allow comparisons of gene expression and CNV across different data and cancer types. Data downloading and clinical heatmap are also supported. The browser currently contains genome-wide experiments on 71,870 samples, most of which are from large-scale international cancer projects including TCGA, CCLE, and the Stand Up To Cancer initiative.
Cancer Genome Work Bench
The Cancer Genome Work Bench (CGWB) is an effective tool with a focus on gene expression and mutations, CNV, and methylation [10]. It provides an automated analysis pipeline for users to visualize and analyze genomic information and gene expression variations in each sample. There are two main viewers in CGWB—Heatmap viewer and Genome browser, both of which can present user-specified information such as gene expression, somatic mutations and pathway context that the mutations may be involved in, and users can toggle between these two viewers. It’s of note that CGWB has kept clinical data and individual genotypes private and permission is needed for access to these data.
Genomics of Drug Sensitivity in Cancer
The Genomics of Drug Sensitivity in Cancer (GDSC) database is the largest open knowledgebase of drug sensitivity and drug response in cancer cells [11]. The GDSC project includes experiments describing the response to almost 200 anticancer therapeutics in more than 1000 cancer cell lines. The cell line drug sensitivity data were integrated with large genomic datasets in the COSMIC database to identify molecular markers of cancer drug response. In the GDSC website, drug sensitivity can be queried by cancer gene, cancer cell line, and compound. Data retrieved are presented in a variety of graphical representations for view and downloading. The large collection of cell lines, drug sensitivity, and genomic datasets have enhanced our understanding of cancer cell genomic heterogeneity and facilitated the discovery of new patient-specific treatments.
canSAR
canSAR is an open cancer-centered cross-discipline knowledgebase that aims to facilitate translational cancer research [12]. The knowledgebase features multidisciplinary information. For instance, for a given protein or drug, researchers can obtain information on current understanding of the protein or drug, such as the expression or mutation of the protein in cancer, cellular sensitivity profiles, and specific binding proteins of the drug. A large collection of human proteome data is housed in canSAR right now. Moreover, this database also contains data and annotations for cancer, nontransformed cell lines and protein 3D structure summaries.
NONCODE
Noncoding RNAs (ncRNAs) function in a variety of cancer types. NONCODE is a regularly-updated knowledgebase of ncRNAs (except for tRNAs and rRNAs) from several species such as human and mouse [13]. NONCODE was first launched in 2005 and was reported in Science NetWatch [14]. Version 4.0 of NONCODE has 595,854 ncRNA entries, among which 210,831 (about 35%) are long ncRNAs (lncRNAs). More than 80% of the ncRNA data in NONCODE are experimentally-derived, thus providing users with a highly-credible resource. In addition to providing lncRNA expression profiles across tissues, NONCODE provides an online pipeline called iLncRNA for users to analyze customized RNA-seq data. The database also has an ID conversion function that changes RefSeq or Ensembl IDs into NONCODE IDs.
Concluding remarks
Recent advances in genome and related information technologies have accelerated the bridging of scientific research and clinical application, with the creation of publicly-accessible data repository and analytical tools. Cancer genome data from large-scale projects such as TCGA and ICGC are being used by large firms to develop new targets and biomarkers. This review provides a brief introduction to some representative web-based resources that can be divided into five categories. First, resources such as CGHub, EGA, COSMIC, and cBioPortal serve as an encyclopedia of common cancers and omics data types. Other resources offer tools for data analysis and integration, and house some analysis results for query (e.g., CPRG, GDAC, and canEvolve). The third class of tools is mainly used for visualization (e.g., UCSC cancer browser and CGWB). The fourth class includes databases that focus on inferring the association of specific biological features to cancer (e.g., MethyCancer, SomamiR, and NONCODE). Finally, databases such as GDSC and CanSAR support the application of genomics to drug discovery. Many other useful resources for cancer research were not included in this paper due to the scope limit. However, the move from bench to bedside remains a daunting task. One of the major obstacles is the heterogeneity of cancer cells and the variability in the response to anticarcinogens among patients with similar symptoms. To address this issue, personal genomics projects should be initiated in a larger population than the average sample size for the available projects. In addition, advanced computational and statistical methodologies aimed at big data management and data mining should be developed to establish the clinical relevance of cancer genomic discovery.
Competing interests
The authors declared that there are no competing interests.
Acknowledgements
This research was supported by the Strategic Priority Research Program of the Chinese Academy of Sciences, Stem Cell and Regenerative Medicine Research (Grant No. XDA01040405), the National High-tech R&D Program of China (863 Program, 2012AA022502) and the National “Twelfth Five-Year” Plan for Science & Technology Support of China (2013BAI01B09) awarded to XF and the National Natural Science Foundation of China (Grant No. 31471236) awarded to YL.
Handled by Qiang Tian
Footnotes
Peer review under responsibility of Beijing Institute of Genomics, Chinese Academy of Sciences and Genetics Society of China.
References
- 1.Wilks C., Cline M.S., Weiler E., Diehkans M., Craft B., Martin C., et al. The Cancer Genomics Hub (CGHub): overcoming cancer through the power of torrential data. Database (Oxford)2014;2014:bau093. doi: 10.1093/database/bau093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Forbes S.A., Beare D., Gunasekaran P., Leung K., Bindal N., Boutselakis H., et al. COSMIC: exploring the world’s knowledge of somatic mutations in human cancer. Nucleic Acids Res. 2015;43:D805–D811. doi: 10.1093/nar/gku1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Marx V. Drilling into big cancer-genome data. Nat Methods. 2013;10:293–297. doi: 10.1038/nmeth.2410. [DOI] [PubMed] [Google Scholar]
- 4.Packer B.R., Yeager M., Burdett L., Welch R., Beerman M., Qi L., et al. SNP500Cancer: a public resource for sequence validation, assay development, and frequency analysis for genetic variation in candidate genes. Nucleic Acids Res. 2006;34:D617–D621. doi: 10.1093/nar/gkj151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Samur M.K., Yan Z., Wang X., Cao Q., Munshi N.C., Li C., et al. CanEvolve: a web portal for integrative oncogenomics. PLoS One. 2013;8:e56228. doi: 10.1371/journal.pone.0056228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.He X., Chang S., Zhang J., Zhao Q., Xiang H., Kusonmano K., et al. MethyCancer: the database of human DNA methylation and cancer. Nucleic Acids Res. 2008;36:D836–D841. doi: 10.1093/nar/gkm730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bhattacharya A., Ziebarth J.D., Cui Y. SomamiR: a database for somatic mutations impacting microRNA function in cancer. Nucleic Acids Res. 2013;41:D977–D982. doi: 10.1093/nar/gks1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gao J., Aksoy B.A., Dogrusoz U., Dresdner G., Gross B., Sumer S.O., et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6:l1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Goldman M., Craft B., Swatloski T., Ellrott K., Cline M., Diekhans M., et al. The UCSC cancer genomics browser: update 2013. Nucleic Acids Res. 2013;41:D949–D954. doi: 10.1093/nar/gks1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang J., Finney R.P., Rowe W., Edmonson M., Yang S.H., Dracheva T., et al. Systematic analysis of genetic alterations in tumors using Cancer Genome WorkBench (CGWB) Genome Res. 2007;17:1111–1117. doi: 10.1101/gr.5963407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yang W., Soares J., Greninger P., Edelman E.J., Lightfoot H., Forbes S., et al. Genomics of Drug Sensitivity in Cancer (GDSC): a resource for therapeutic biomarker discovery in cancer cells. Nucleic Acids Res. 2013;41:D955–D961. doi: 10.1093/nar/gks1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bulusu K.C., Tym J.E., Coker E.A., Schierz A.C., Al-Lazikani B. CanSAR: updated cancer research and drug discovery knowledgebase. Nucleic Acids Res. 2014;42:D1040–D1047. doi: 10.1093/nar/gkt1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu C., Bai B., Skogerbo G., Cai L., Deng W., Zhang Y., et al. NONCODE: an integrated knowledge database of non-coding RNAs. Nucleic Acids Res. 2005;33:D112–D115. doi: 10.1093/nar/gki041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Netwatch Databases: decoding the noncode. Science. 2005;307:329. [Google Scholar]