Abstract
Studying evolution in the laboratory provides a means of understanding the processes, dynamics and outcomes of adaptive evolution in precisely controlled and readily replicated conditions. The advantages of experimental evolution are maximized when selection is well defined, which enables linking genotype, phenotype and fitness. One means of maintaining a defined selection is continuous culturing: chemostats enable the study of adaptive evolution in nutrient-limited environments in which growth is sub-maximal, whereas cells in turbidostats evolve in nutrient abundance that allows maximal growth. Although the experimental effort required for continuous culturing is considerable relative to the experimental simplicity of serial batch culture, the opposite is true of the environments they produce: continuous culturing results in simplified and constant conditions whereas serial batch cultures are complex and dynamic. The comparative simplicity of the selective environment that is unique to continuous culturing provides an ideal experimental system for addressing key questions in adaptive evolution.
Introduction
Experimental evolution with microbes commenced at least 130 years ago with the work of Darwin's contemporary, Reverend W. H. Dallinger [1]. However, for many years progress in experimental evolution was limited by the inability to comprehensively characterize the genetic variation associated with adaptive evolution. The advent of genomic technologies solved this problem, first through the use of DNA microarrays to identify nucleotide [2,3] and structural [4] variation, and subsequently with the application of quantitative high throughput DNA sequencing [5-9]. Whole genome sequencing of both individual lineages and entire populations is no longer a roadblock to progress, and has rapidly become a routine experimental method that has transformed the field of experimental evolution. These technological advances mean that many long-standing questions in evolutionary biology can now be addressed with unprecedented detail, precision and rigor.
The dawn of a new era in experimental evolution warrants revisiting the major goals of the research program of experimental evolution. These goals have been discussed in recent publications [10,11], including those accompanying this article, and can be summarized as follows: 1) understanding the molecular basis of adaptation at the functional and mechanistic level, 2) understanding the consequences of adaptive mutations on organismal phenotypes and physiology, 3) defining the predictability and repeatability of adaptive evolution, 4) mapping the distribution of fitness effects of mutations, 5) determining how parameters such as population size and strength of selection affect adaptation, and 6) identifying the parameters that affect the dynamics of adaptive evolution.
In general (but not exclusively [12,13]), experimental microbial evolution entails selection of de novo mutations that arise in an initially genetically clonal population. Thus, experimental evolution in microbes differs from experimental evolution in animals such as worms [14], flies [15] and mice [16], which typically entails selection on standing (pre-existing) genetic variation by founding populations with genetically heterogeneous individuals. When undertaking experimental evolution with microbes, the ease of maintaining large populations (108-1010 individuals) with short generation times (20-360 minutes) that typically have small genome sizes (106-107 bases) with typical mutation rates of 10−7-10−9 substitutions/bp/generation means that mutation supply is extremely high. In many experimental evolution scenarios it is reasonable to assume that on average every possible one base substitution in a microbial genome is introduced into the population each generation. Thus, selection has ample diversity on which to act.
Technically, experimental evolution with microbes entails selection over prolonged periods of culturing in laboratory conditions. This can be achieved by simply passaging cells in culture flasks (i.e. batch cultures) using the method of serial transfer. For the practiced experimentalist there are few microbiology techniques that are simpler than transferring a sample from one population to inoculate a new culture containing fresh medium and thus initiate a new round of population growth. Moreover, the method of serial dilution of batch cultures is readily amenable to parallelization using microtiter plates and robotic liquid handling, which enable the simultaneous analysis of hundreds of populations [9,17].
Alternatively, long-term selection can be performed using methods of continuous culturing including chemostats and turbidostats. In contrast to serial transfer of batch cultures, long term selection using continuous culture can be logistically challenging and less amenable to large-scale multiplexing, leading to the reasonable question: “why bother?” The goal of our article is to argue that the answer to this question lies in the great utility of maintaining a continuous, and invariant, selection during experimental evolution. Continuous culturing, using chemostats or turbidostats, provides the only means of ensuring a sustained and invariant selective pressure, a feature that greatly simplifies the goal of connecting adaptive genotypes with their phenotypic consequences and explaining why they result in increased fitness. As a result, continuous culturing is ideally suited to addressing some of the central goals of experimental evolution.
The principle of the chemostat
The principle of the chemostat differs in several respects from batch culture [18]. In a chemostat, fresh medium is continuously added to the growing culture at a defined rate and at the same rate culture is removed. Eventually, the culture reaches a steady-state in which the cells grow continuously at a constant rate and the growth rate of the population is equal to the rate at which it is diluted [19,20]. Through the process of continuous dilution a growing population of cells can be maintained in a chemostat indefinitely. An essential requirement of the chemostat is the use of a defined medium in which a single nutrient is present at a growth limiting concentration [21]. A nutrient is said to be limiting in the chemostat when its concentration dictates the steady-state cell density, such that increasing the concentration of the limiting nutrient results in a proportional increase in the steady-state cell density. In the steady-state condition the concentration of the growth-limiting nutrient is typically in the low micromolar range. Thus, cells in a chemostat grow continuously in a chemically defined environment where all nutrients but one are present in excess. This environment is most similar to a batch culture just prior to nutrient exhaustion and has been described as placing the cells in an environment in which they are “poor, not starving” [22] or “hungry” [23]. The low concentration of the growth-limiting nutrient defines the selection imposed on cells. A variety of growth-limiting nutrients can be used, so long as they are essential for growth of the organism. Typically, these are sources of carbon, nitrogen, phosphorous or sulfur, though non-essential nutrients can also be made essential by the use of appropriate auxotrophic mutants that are defective in a biosynthetic pathway. Increases in fitness in the chemostat environment are typically achieved by improved capabilities in the acquisition or utilization of the growth-limiting nutrient.
The principle of the turbidostat
A turbidostat is analogous to a chemostat in that the culture is continuously diluted by the addition of fresh medium. However, in contrast to a chemostat, the goal of a turbidostat is to avoid cells ever experiencing nutrient limitation. This is achieved by continuous addition of fresh medium to the growing culture to maintain a specific cell density. As with a chemostat, the culture is continuously diluted by the addition of medium and the removal of an equivalent volume culture. However, in the case of a turbidostat, all nutrients are present in excess and the dilution rate is set near the maximal growth rate of the cells. In practice this is achieved by constant monitoring of cell density and automated addition of media when the density exceeds the specified value. The resulting steady-state environment is most similar to a batch culture during the mid-log exponential phase of growth, when growth rate is maximal, and nutrients are in abundant supply.
Unlike a chemostat, the growth rate of cells in a turbidostat is determined by intrinsic properties of the cell. As the turbidostat environment is never nutrient poor, the ability of cells to grow is not constrained by nutrient abundance. Instead, the limits to growth are inherent properties of the cell that determine how rapidly it can replicate. Factors that likely limit the rate a cell can reproduce itself when resources are abundant include the rate of nutrient uptake and the rate of macromolecular and organelle biogenesis, as well as complex molecular processes such as DNA replication, transcription and translation. In principle, increases in fitness in the turbidostat might result from enhancements in any of these processes. Variants on turbidostats include devices in which the ability to grow maximally is constrained by an environmental agent, for example by adding growth inhibitors such as high ethanol or antibiotics [24,25].
Distinction from serial transfer in batch cultures
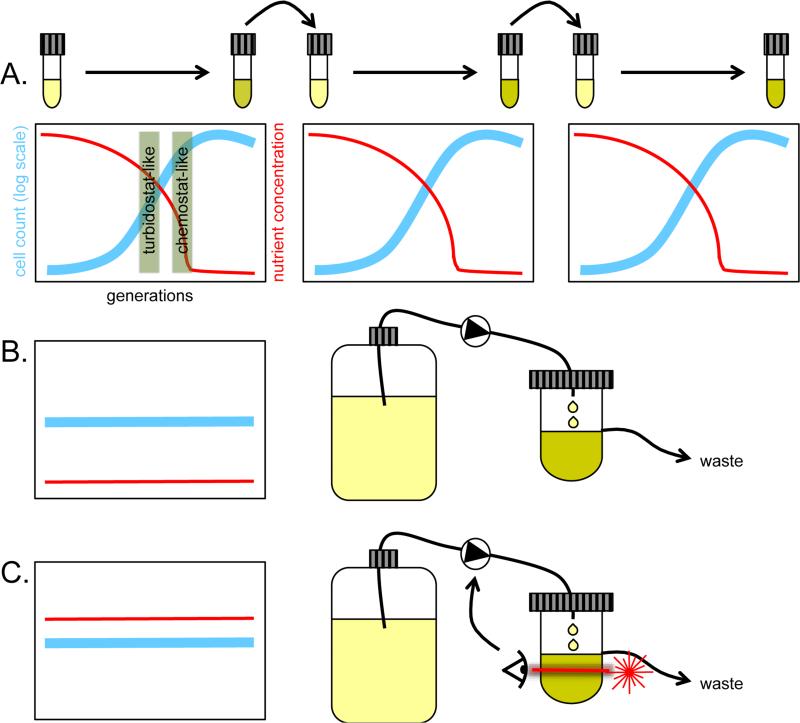
Despite the very different selective pressures that operate in the chemostat compared with a turbidostat, both methods share the principle of continuous culturing and therefore a continuous selection. A comparable constancy of selective pressure is not possible using serial dilution of batch cultures, even when great care is taken to transfer from exponentially growing cultures prior to the onset of stationary phase [26,27]. Regardless of whether an undefined medium is used, in which the environmental factor that determines the population size at the end of each growth phase is unknown, or a defined medium in which the nutrient that is first exhausted and therefore determines the final population size is pre-determined, dramatically fluctuating levels of nutrient abundance are characteristic of experimental evolution using serial transfer (Figure 1). In fact, a batch culture of cells experiences both a turbidostat-like and chemostat-like environment during each growth cycle in addition to experiencing near or complete starvation, depending on the period length of the transfer cycle.
Figure 1.
Continuous versus discontinuous modes of selection used in microbial experimental evolution. A. Serially diluted cultures experience variations in nutrient level and cell density over each growth cycle, including turbidostat-like and chemostat-like phases. B. Chemostat cultures grow at a set dilution rate and experience constant nutrient limitation akin to that seen in batch cultures just before nutrient exhaustion. C. Turbidostats can grow cultures at their maximal growth rate by tuning dilution rate based on culture density, generating constant population size and selection pressure.
It is certainly true that the repeated cycles of feast and famine in a serial transfer experiment impose a strong selection on cells. However, at this point we do not understand which phase of the growth cycle is the predominant selective force in a serial passage regime, and the relative importance of factors, such as nutritional abundance, intracellular processes and excreted products, is likely to change over the course of a single passage during a serial dilution evolution experiment. As a result, increased fitness in batch culture may result from decreased duration in lag phase (the time taken to reinitiate growth upon encountering fresh medium), increased growth rate during the growth phase or a decreased probability to enter a quiescent, non-reproductive, state upon nutrient depletion [28,29]. It is quite plausible that alleles that improve fitness in each of these growth phases are antagonistically pleiotropic with respect to each other. Thus, allele frequencies may fluctuate throughout each serial passage or different lineages may specialize in optimizing one or more of each of the phases of batch culture growth. Continuous culturing provides a means of avoiding this complexity. The constancy of selection in a chemostat or turbidostat enables the selection to be precisely defined and indefinitely maintained providing considerable advantages for addressing the following key questions using experimental evolution.
1. What is the molecular basis of adaptation?
Determining the molecular basis of adaptation is critical for advancing understanding in evolutionary biology [30]. By understanding the mechanistic basis of adaptation we can begin to explain why particular outcomes of adaptive evolution are favored over other possibilities. For example, adaptation in some selective environments may entail alteration of a single biochemical pathway or protein complex whereas in other selective environments there may exist a plethora of catalytic and regulatory pathways that are potential targets for adaptive mutation. These contrasting scenarios will profoundly impact the extent to which the outcomes of repeated adaptive evolution converge at the phenotypic and genotypic level. Moreover, evolutionary dynamics within adapting populations will be shaped by the diversity of possible solutions to selective “problems.” Understanding the relationship between the selection that operates in a particular environment and the functional basis of adaptation at the molecular level is essential to predicting the tempo, trajectories and outcomes of evolutionary change.
In natural populations, few notable examples exist in which the molecular basis of adaptive evolution has been resolved to causative variants of known functional effects [31,32]. Studies of adaptation in the wild require identification of putative adaptive phenotypes and methods to map the quantitative trait loci (QTL) that often underlie these phenotypes. Such studies can provide insight into how genetic networks evolve in response to a particular selective pressure in those rare cases where it is possible to infer exactly what factor(s) drove selection of the QTL in the natural environment. Such studies provide insight into the relationship between genotype and fitness. However, individual examples of adaptation in natural populations make it difficult to derive general principles regarding this causal relationship. The goal of identifying the functional basis of adaptation is ideally suited to laboratory experimental evolution in which the type and strength of selection, the genetic background and the evolutionary parameters can be pre-defined and controlled. Through the combination of whole genome sequencing, genetics and strain reconstruction, the identification and quantification of fitness effects associated with adaptive mutations can be achieved with a high level of rigor.
A key requirement for achieving the goal of determining the functional basis of adaptation is understanding, and controlling, the selection experienced by the population. Knowledge of the selection facilitates connecting the identified molecular variation with its functional causality. For example, the low nutrient concentration in a chemostat represents the primary selection [33]. Studies to date suggest that the primary means by which fitness is increased in a chemostat is through improved nutrient import [7,33-37]. This explains the recurrent selection for copy number variants in nutrient transporter genes [7,35-38] as well as repeated selection of adaptive mutations in regulators of nutrient transporters [7,38].
By contrast, the heterogeneous and dynamic environment of a batch culture makes it difficult to identify the selection experienced by cells in this environment. Although the spectra of mutations associated with adaptation in serial dilution are now easily identified [9], it is often difficult to explain which of these mutations are beneficial and why. Even if one tries to impose a defined stress, such as temperature, in conjunction with serial dilution, the adaptive mutations that are identified are often difficult to interpret and explain [39]. The lack of a clearly defined, experimentally-controlled selection creates a considerable challenge for experimental evolution using serial dilution for identifying the functional basis of increased fitness. By contrast, the constancy of selection in a chemostat or turbidostat simplifies the goal of connecting increased fitness with its molecular basis.
2. What are the phenotypic consequences of adaptive mutations?
In experimental evolution, the overarching objective is to select and characterize lineages with increased fitness. This is defined as a growth advantage in the relevant environment compared to the ancestral strain. Technical advances in DNA sequencing now make it possible to identify every mutation in a high-fitness lineage; however, that is only the first step in understanding the functional basis of adaptation. A persistent challenge for understanding the forces that drive adaptive evolution is connecting genotype to phenotype and explaining how the resultant phenotypic changes impact fitness. Meeting this challenge requires careful analysis of the phenotypic consequences of adaptive mutations where those phenotypes can vary from the transport kinetics of a nutrient transporter to the binding affinity of a transcription factor. An understanding of the phenotypes that are associated with increased fitness provides an explanation for why a particular class of mutations is beneficial, and why certain of these mutations may be deleterious in alternative environments (i.e., why they are antagonistically pleiotropic).
When microbes are continuously cultured in a chemostat, their fitness is frequently increased by improvements in the import and utilization of the growth-limiting nutrient. Enhanced nutrient transport capabilities are directly causative of increased fitness, and can be estimated via uptake assays that use radiolabeled nutrients. Dramatic improvements in this function have repeatedly been observed in chemostat-evolved lineages [34,40].
Another route to increased fitness in the chemostat is the inhibition of stress responses that are activated in response to nutrient deprivation [41,42]. In wildtype cells, robust stress responses are critical for surviving in environments that fluctuate in nutrient abundance. In response to starvation, many microbes exit a proliferative state and initiate a quiescent state in which they exhibit increased stress resistance. Quiescence enables long-term survival until conditions improve; however, initiation of this state in a chemostat confers a strong disadvantage as these cells will no longer contribute to future generations in the continuously diluted population. Thus, loss of a robust stress response is an expected and repeatedly observed outcome of chemostat selection [8,42,43].
The chemostat is also uniquely suited to studying the global molecular phenotypes associated with adaptation including changes in the transcriptome and proteome. A number of studies have demonstrated that a large fraction of yeast [44-46] and bacterial [47] genes change in their expression with changes in growth rate. In a batch culture or a turbidostat, adaptation is nearly always accompanied by increases in growth rate. As a result, distinguishing specific gene expression changes that directly result from adaptive mutations versus gene expression changes that are correlated with growth rate changes represents a significant challenge. The chemostat makes it possible to assay the transcriptome or proteome of both evolved and ancestral populations at the same growth rate, allowing controlled analyses of these high dimensional phenotypes [36,48,49].
In contrast to the straightforward connection between phenotype and fitness in chemostat environments, the complexity and dynamics of the batch culture regime make it difficult to know which phenotypes are relevant to fitness. Increased fitness in batch cultures may result from increased growth rates during the rapid growth phase (i.e. “log phase”), the ability to eke out extra cell divisions once nutrients become scarce (i.e. “stationary phase”), or the ability to reinitiate growth more rapidly (i.e. “lag phase”) [28,29]. However, in general, fitness in batch cultures is typically estimated as the aggregate of these different growth phases. Thus, in most cases it remains unexplained why batch evolved lineages have increased fitness and which phenotypes are adaptive.
3. How reproducible is adaptive evolution?
The extent to which evolutionary outcomes are reproducible is one of the most central and enduring questions in evolutionary biology. If adaptive evolution is reproducible then it follows that the outcome of selection in a particular environment should be predictable at the phenotypic and genotypic level, subject to population size and mutation supply. Of critical importance for addressing this central question is knowledge of the type and strength of selection operating in an environment of an adapting population, as this will dictate the target size for adaptive mutations.
For some selections there may be several adaptive solutions while for other selections there may be a very limited set of solutions. Although the primary selection in a chemostat is nutrient limitation, there are a range of selective conditions that can be explored by varying the identity and concentration of the limiting nutrient. Indeed, whereas the response to selection in a sulfur-limited chemostat at the phenotypic and genotypic level is highly repeatable, the response to selection in glucose or phosphate limited chemostats appears to be more variable [36]. Thus, the reproducibility of adaptive evolution is likely to vary depending on the selection even in the comparatively simple selective environment of a chemostat.
Addressing the question of the reproducibility of adaptive evolution is significantly more difficult if the selection is not well defined. As the selection is less well-defined in batch culture regimes, and is likely to vary between experimentalists depending on the precise details of the experimental regime (e.g. the frequency of transfers, the duration of the starvation phase, whether the culture is well aerated), it is likely that this approach will encounter significant challenges in addressing the central question of the reproducibility of adaptive evolution.
4. What is the distribution of fitness effects of new mutations?
Determining the distribution of fitness effects attributable to new mutations is closely related to the question of the reproducibility of adaptive evolution. If the distribution of fitness effects is highly skewed, with a small number of mutations having large fitness effects, a limited number of solutions are expected to be observed. By contrast, if there are many possible mutations with similar fitness effects, and the distribution of fitness effects has less variance, then the outcome of adaptive evolution is likely to be more variable.
Defining the distribution of fitness effects must be conditioned on the environment in which fitness is measured. An analogy may be found with drug resistance. For some drugs, such as canavanine, drug resistance is conferred by inactivation of a single locus (CAN1) in the S. cerevisiae genome by loss of function mutations. Thus the distribution of fitness effects in a canavanine-containing environment is extremely skewed: there is a single class of mutations with high fitness, all of which result in a non-functional CAN1 gene. By contrast, the drug nystatin targets ergosterol biosynthesis, and thus mutations in at least four genes that are required for ergosterol biosynthesis can result in nystatin resistance [50]. Moreover, these mutations can result in a range of resistances. Thus the distribution of fitness effects in a nystatin-containing environment is much broader and more continuously distributed. By analogy, for some selective environments there may be a small number of possible beneficial mutations that result in similar fitness, whereas in other environments there may be tens to thousands of possible beneficial mutations associated with a range of fitness effects.
Knowledge of the selection that operates in the environment is central to anticipating and interpreting such differences. Precise control of the environment in a chemostat enables systematic exploration of the distribution of fitness effects and how they vary in different selective environments. By using different nutrient selections, it is feasible to explore how this distribution differs as a function of selection. Undertaking such studies using batch culture one is faced with a number of challenges, notably that fitness effects are typically integrated over the entirety of a growth curve, blurring any fitness differences specific to a particular phase of growth.
5. How does population size and the strength of selection affect the outcome of adaptive evolution?
Experimental evolution is ideally suited to systematically studying how variation in specific parameters such as population size, mutation supply and the strength of selection impact the outcome of adaptive evolution. In a chemostat, it is straightforward to vary population size simply by varying the concentration of limiting nutrient in the feed medium [33]. Given a constant mutation rate, varying the population size while maintaining the identical steady-state nutrient concentration enables a rigorous assessment of the effect of variation in mutation supply rate on the rate, dynamics and outcome of adaptive evolution. Alternatively mutation supply can be altered via the use of mutator strains, and studies in chemostat cultures have been key to measuring the importance of mutation supply [51].
Similarly, the strength of selection can be systematically varied by controlling dilution rate and hence residual nutrient concentration. As the dilution rate is increased the residual nutrient concentration increases thereby altering the strength of selection in a continuous and controlled manner [33]. A more extreme example is seen with glucose limitation in yeast, where at a critical dilution rate, central carbon metabolism becomes dominated by fermentation versus the mixed respirofermentative growth experienced at lower growth rates. The fitness effects of some adaptive mutations differ as the dilution rate is altered [52]. Thus, chemostats are ideally suited to studying the response to distinct, but related, selections.
One downside of using a chemostat is that cultures may experience different degrees of selection as they adapt. For example, residual nutrient concentrations generally decline over time in an evolving culture. The turbidostat and other feedback-controlled continuous culture platforms provide an alternate approach that allows the selection to continually be tuned and maintained at a constant strength. As the culture adapts, the selection pressure is ramped up to keep pace. To describe just one implementation, the “morbidostat” ramps drug concentration as a bacterial culture evolves increased drug resistance [25].
6. What are the dynamics and constraints of adaptive evolution?
Determining the dynamics of adaptive evolution has been a longstanding goal that has recently been reinvigorated by the ability to follow adaptive mutations in real time using deep sequencing [7-9]. The absence of bottlenecks in a chemostat means that every genotype has an equal opportunity of contributing to the next generation as a function of its associated fitness without the chance of being randomly purged from the populations by bottlenecking events that are characteristic of serial transfer experiments.
This lack of population bottlenecks makes the chemostat an ideal platform for studying population dynamics and interactions. Early observations of neutral mutation dynamics in the chemostat seemed to indicate evolution of microbial populations could be best explained by serial clonal selective sweeps [53,54]. However, a more nuanced view has since emerged, where multiple subpopulations can coexist and compete among one another, either due to clonal reinforcement effects such as cross-feeding interactions [55,56] or clonal interference between genetically and/or phenotypically similar or distinct subpopulations [8,43,57-61]. Such interactions have even been engineered to generate novel community-level behaviors [62].
Antagonistic pleiotropy is also likely to shape the outcome of adaptive evolution. As a chemostat environment is constant there is limited scope for antagonistic pleiotropy to affect the dynamics and outcome of selection as the environment remains constant and alleles are not subject to selection in alternate environments. By contrast, the different phases of growth in a batch culture may mean that some alleles are beneficial in one phase of the growth cycle but deleterious in another. For example, constitutive expression of an allele that increases transport of nutrients when they are scarce may confer a selective advantage during stationary phase but may confer a fitness cost during the rapid growth phase. Thus, antagonistic pleiotropy may significantly affect the outcome of adaptive evolution in batch culture conditions, but have less effect in continuous culturing selections.
Caveats regarding the use of continuous culture
Despite the many advantages of continuous culturing systems they present unique challenges that may partially explain their limited adoption by researchers undertaking experimental evolution. First, chemostats and turbidostats present a number of practical difficulties, including a reputation for being difficult to build and maintain. However, the accessibility of commercial fermenters is increasing [21] as are simpler home-built devices [63-65]. Another practical challenge is the evolution of wall growth and clumps, both adaptations to the device geometry as opposed to the experimenter-imposed selection pressure (note that evolution of this trait is not limited to continuous culture, as it also arises in certain serial batch transfer implementations [66]). In addition to being undesirable “off target” adaptations that violate the key assumptions about a single dominant selection pressure, these phenotypes can interfere with the plumbing of the device, and with subsequent microbiology experiments, limiting the length of time evolution experiments can be maintained. Treatment of surfaces and/or cycling to new chambers [67] presents one work-around to limit wall growth. Strain engineering will likely be required to truly address such problems; doing so is becoming an increasingly appealing possibility as more of the mutations that underlie these phenotypes are characterized [66,68]. Another violation of the assumption of constancy is the metabolic cycle, synchronized changes in gene expression and metabolic state that occur with certain yeast strains in particular glucose-limited low dilution rate chemostats [69]. The metabolic cycle can be eliminated or desynchronized by addition of ethanol and glycerol or growing at higher growth rates. Finally, multiplexing on the scale of batch culture experiments is difficult. While arrays of >1000 microfluidic chemostats have broken through this barrier [70], the population sizes and run time horizon of these devices make them more suitable for multiplexed phenotyping than for evolution experiments.
It may be argued that the selection that operates in a continuous culture is too simple. Long term, constant selection is not likely to be the dominant experience of most natural populations (though nutrient limitation is no doubt a common occurrence). By their very design, chemostats are not well-suited to dynamic environments, although it is possible to vary them by switching media feeds, injecting reagents, changing temperature, and other perturbations [12]. However, we would argue that this simplicity is exactly the point: by modeling adaptive evolution using the simplest possible selective regimen, we greatly increase our chances of ever being able to understand and explain it completely. Notably, continuous culture is an excellent context for modeling, particularly since the steady state assumptions of most metabolic models are actually met [71].
Conclusion
Experimental evolution using continuous culturing stands on the bedrock of precisely defined selection. When combined with an organism that has a rich genetic tool kit there is the potential to make rapid progress towards answering longstanding evolutionary questions. We believe that one can draw an important parallel between experimental evolution and genetic screens. In undertaking a genetic screen the aim is to identify those molecular components that are most directly related to the cell biological process of interest. The informativeness of a genetic screen is a function of the careful definition of the selection. Thus, the art of designing genetic screens lies in defining selections that are directly relevant to the molecular processes that are of interest. A poorly defined selection yields mutants that are remotely related to the process of interest. We believe that a similar level of careful consideration and control of the selection is critical to advances in experimental evolution. The chemostat and turbidostat provides an unparalleled level of control making them indispensible tools for realizing the goals of the experimental evolution research program.
Acknowledgements
We thank the editors, Frank Rosenzweig and Gavin Sherlock, for helpful comments on the manuscript. MD was supported by grants from the National Science Foundation (1120425) and the National Institute of General Medical Sciences (R01 GM094306 and P41 GM103533) from the National Institutes of Health. MD is a Rita Allen Foundation Scholar and a Fellow in the Genetic Networks program at the Canadian Institute for Advanced Research. DG was supported by the National Institutes of Health (GM107466), the National Science Foundation (MCB-1244219), and a Dupont Young Professor award.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Bell G. Selection. second Oxford Univ Press; 2008. [Google Scholar]
- 2.Gresham D, Ruderfer DM, Pratt SC, Schacherer J, Dunham MJ, Botstein D, et al. Genome-wide detection of polymorphisms at nucleotide resolution with a single DNA microarray. Science. 2006;311:1932–1936. doi: 10.1126/science.1123726. [DOI] [PubMed] [Google Scholar]
- 3.Herring CD, Raghunathan A, Honisch C, Patel T, Applebee MK, Joyce AR, et al. Comparative genome sequencing of Escherichia coli allows observation of bacterial evolution on a laboratory timescale. Nat Genet. 2006;38:1406–1412. doi: 10.1038/ng1906. [DOI] [PubMed] [Google Scholar]
- 4.Dunham MJ, Badrane H, Ferea T, Adams J, Brown PO, Rosenzweig F, et al. Characteristic genome rearrangements in experimental evolution of Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 2002;99:16144–16149. doi: 10.1073/pnas.242624799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Araya CL, Payen C, Dunham MJ, Fields S. Whole-genome sequencing of a laboratory-evolved yeast strain. BMC Genomics. 2010;11:88. doi: 10.1186/1471-2164-11-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barrick JE, Yu DS, Yoon SH, Jeong H, Oh TK, Schneider D, et al. Genome evolution and adaptation in a long-term experiment with Escherichia coli. Nature. 2009;461:1243–1247. doi: 10.1038/nature08480. [DOI] [PubMed] [Google Scholar]
- 7.Hong J, Gresham D. Molecular specificity, convergence and constraint shape adaptive evolution in nutrient-poor environments. PLoS Genet. 2014;10:e1004041. doi: 10.1371/journal.pgen.1004041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kvitek DJ, Sherlock G. Whole Genome, Whole Population Sequencing Reveals That Loss of Signaling Networks Is the Major Adaptive Strategy in a Constant Environment. PLoS Genet. 2013;9:e1003972. doi: 10.1371/journal.pgen.1003972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lang GI, Rice DP, Hickman MJ, Sodergren E, Weinstock GM, Botstein D, et al. Pervasive genetic hitchhiking and clonal interference in forty evolving yeast populations. Nature. 2013:1–6. doi: 10.1038/nature12344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Elena SF, Lenski RE. Evolution experiments with microorganisms: the dynamics and genetic bases of adaptation. Nat Rev Genet. 2003;4:457–469. doi: 10.1038/nrg1088. [DOI] [PubMed] [Google Scholar]
- 11.Desai MM. Statistical questions in experimental evolution. J. Stat. Mech. 2013;2013:P01003. [Google Scholar]
- 12.Piotrowski JS, Nagarajan S, Kroll E, Stanbery A, Chiotti KE, Kruckeberg AL, et al. Different selective pressures lead to different genomic outcomes as newly-formed hybrid yeasts evolve. BMC Evol Biol. 2012;12:46. doi: 10.1186/1471-2148-12-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Burke MK, Liti G, Long AD. Standing genetic variation drives repeatable experimental evolution in outcrossing populations of Saccharomyces cerevisiae. Mol Biol Evol. 2014 doi: 10.1093/molbev/msu256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Teotónio H, Chelo IM, Bradić M, Rose MR, Long AD. Experimental evolution reveals natural selection on standing genetic variation. Nat Genet. 2009;41:251–257. doi: 10.1038/ng.289. [DOI] [PubMed] [Google Scholar]
- 15.Turner TL, Miller PM. Investigating Natural Variation in Drosophila Courtship Song by the Evolve and Resequence Approach. Genetics. 2012;191:633–642. doi: 10.1534/genetics.112.139337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Swallow JG, Koteja P, Carter PA, Garland T. Artificial selection for increased wheel-running activity in house mice results in decreased body mass at maturity. J. Exp. Biol. 1999;202:2513–2520. doi: 10.1242/jeb.202.18.2513. [DOI] [PubMed] [Google Scholar]
- 17.Lang GI, Botstein D, Desai MM. Genetic variation and the fate of beneficial mutations in asexual populations. Genetics. 2011;188:647–661. doi: 10.1534/genetics.111.128942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kubitschek HE. Introduction to research with continuous cultures. Prentice Hall; 1970. [Google Scholar]
- 19.Novick A, Szilard L. Description of the chemostat. Science. 1950;112:715–716. doi: 10.1126/science.112.2920.715. [DOI] [PubMed] [Google Scholar]
- 20.Monod J. La technique de culture continue: Théorie et applications. Annales De l'Institut Pasteur; Paris: 1950. pp. 390–410. [Google Scholar]
- 21.Ziv N, Brandt NJ, Gresham D. The use of chemostats in microbial systems biology. Journal of Visualized Experiments : JoVE. 2013 doi: 10.3791/50168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Saldanha AJ, Brauer MJ, Botstein D. Nutritional homeostasis in batch and steady-state culture of yeast. Mol Biol Cell. 2004;15:4089–4104. doi: 10.1091/mbc.E04-04-0306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ferenci T. Hungry bacteria – definition and properties of a nutritional state Ferenci - 2008 - Environmental Microbiology - Wiley Online Library. Environ. Microbiol. 2001 doi: 10.1046/j.1462-2920.2001.00238.x. [DOI] [PubMed] [Google Scholar]
- 24.Avrahami-Moyal L, Engelberg D, Wenger JW, Sherlock G, Braun S. Turbidostat culture of Saccharomyces cerevisiae W303-1A under selective pressure elicited by ethanol selects for mutations in SSD1 and UTH1. FEMS Yeast Res. 2012;12:521–533. doi: 10.1111/j.1567-1364.2012.00803.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Toprak E, Veres A, Michel J-B, Chait R, Hartl DL, Kishony R. Evolutionary paths to antibiotic resistance under dynamically sustained drug selection. Nat Genet. 2011;44:101–105. doi: 10.1038/ng.1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Torres EM, Dephoure N, Panneerselvam A, Tucker CM, Whittaker CA, Gygi SP, et al. Identification of aneuploidy-tolerating mutations. Cell. 2010;143:71–83. doi: 10.1016/j.cell.2010.08.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sandberg TE, Pedersen M, LaCroix RA, Ebrahim A, Bonde M, Herrgard MJ, et al. Evolution of Escherichia coli to 42 °C and Subsequent Genetic Engineering Reveals Adaptive Mechanisms and Novel Mutations. Mol Biol Evol. 2014 doi: 10.1093/molbev/msu209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lenski RE, Mongold JA, Sniegowski PD, Travisano M, Vasi F, Gerrish PJ, et al. Evolution of competitive fitness in experimental populations of E. coli: what makes one genotype a better competitor than another? Antonie Van Leeuwenhoek. 1998;73:35–47. doi: 10.1023/a:1000675521611. [DOI] [PubMed] [Google Scholar]
- 29.Rozen D, Lenski R. Long-Term Experimental Evolution in Escherichia coli. VIII. Dynamics of a Balanced Polymorphism. Am. Nat. 2000;155:24–35. doi: 10.1086/303299. [DOI] [PubMed] [Google Scholar]
- 30.Dean AM, Thornton JW. Mechanistic approaches to the study of evolution: the functional synthesis. Nat Rev Genet. 2007;8:675–688. doi: 10.1038/nrg2160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Steiner CC, Weber JN, Hoekstra HE. Adaptive variation in beach mice produced by two interacting pigmentation genes. PLoS Biol. 2007;5:e219. doi: 10.1371/journal.pbio.0050219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jones FC, Grabherr MG, Chan YF, Russell P, Mauceli E, Johnson J, et al. The genomic basis of adaptive evolution in threespine sticklebacks. Nature. 2012;484:55–61. doi: 10.1038/nature10944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gresham D, Hong J. The functional basis of adaptive evolution in chemostats. FEMS Microbiol Rev. 2014 doi: 10.1111/1574-6976.12082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Brown CJ, Todd KM, Rosenzweig RF. Multiple duplications of yeast hexose transport genes in response to selection in a glucose-limited environment. Mol Biol Evol. 1998;15:931–942. doi: 10.1093/oxfordjournals.molbev.a026009. [DOI] [PubMed] [Google Scholar]
- 35.Gresham D, Usaite R, Germann SM, Lisby M, Botstein D, Regenberg B. Adaptation to diverse nitrogen-limited environments by deletion or extrachromosomal element formation of the GAP1 locus. Proc Natl Acad Sci USA. 2010;107:18551–18556. doi: 10.1073/pnas.1014023107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gresham D, Desai MM, Tucker CM, Jenq HT, Pai DA, Ward A, et al. The repertoire and dynamics of evolutionary adaptations to controlled nutrient-limited environments in yeast. PLoS Genet. 2008;4:e1000303. doi: 10.1371/journal.pgen.1000303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sonti RV, Roth JR. Role of gene duplications in the adaptation of Salmonella typhimurium to growth on limiting carbon sources. Genetics. 1989;123:19–28. doi: 10.1093/genetics/123.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kvitek DJ, Sherlock G. Reciprocal Sign Epistasis between Frequently Experimentally Evolved Adaptive Mutations Causes a Rugged Fitness Landscape. PLoS Genet. 2011;7:e1002056. doi: 10.1371/journal.pgen.1002056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tenaillon O, Rodriguez-Verdugo A, Gaut RL, McDonald P, Bennett AF, Long AD, et al. The Molecular Diversity of Adaptive Convergence. Science. 2012;335:457–461. doi: 10.1126/science.1212986. [DOI] [PubMed] [Google Scholar]
- 40.Wang L, Spira B, Zhou Z, Feng L, Maharjan RP, Li X, et al. Divergence involving global regulatory gene mutations in an Escherichia coli population evolving under phosphate limitation. Genome Biol Evol. 2010;2:478–487. doi: 10.1093/gbe/evq035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ferenci T. Maintaining a healthy SPANC balance through regulatory and mutational adaptation. Mol Microbiol. 2005;57:1–8. doi: 10.1111/j.1365-2958.2005.04649.x. [DOI] [PubMed] [Google Scholar]
- 42.Notley-McRobb L, King T, Ferenci T. rpoS mutations and loss of general stress resistance in Escherichia coli populations as a consequence of conflict between competing stress responses. J Bacteriol. 2002;184:806–811. doi: 10.1128/JB.184.3.806-811.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Maharjan R, Seeto S, Notley-McRobb L, Ferenci T. Clonal adaptive radiation in a constant environment. Science. 2006;313:514–517. doi: 10.1126/science.1129865. [DOI] [PubMed] [Google Scholar]
- 44.Brauer MJ, Huttenhower C, Airoldi EM, Rosenstein R, Matese JC, Gresham D, et al. Coordination of growth rate, cell cycle, stress response, and metabolic activity in yeast. Mol Biol Cell. 2008;19:352–367. doi: 10.1091/mbc.E07-08-0779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Regenberg B, Grotkjaer T, Winther O, Fausbøll A, Akesson M, Bro C, et al. Growth-rate regulated genes have profound impact on interpretation of transcriptome profiling in Saccharomyces cerevisiae. Genome Biol. 2006;7:R107. doi: 10.1186/gb-2006-7-11-r107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Castrillo JI, Zeef LA, Hoyle DC, Zhang N, Hayes A, Gardner DCJ, et al. Growth control of the eukaryote cell: a systems biology study in yeast. J Biol. 2007;6:4. doi: 10.1186/jbiol54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ishii N, Nakahigashi K, Baba T, Robert M, Soga T, Kanai A, et al. Multiple high-throughput analyses monitor the response of E. coli to perturbations. Science. 2007;316:593–597. doi: 10.1126/science.1132067. [DOI] [PubMed] [Google Scholar]
- 48.Zhong S, Miller SP, Dykhuizen DE, Dean AM. Transcription, Translation, and the Evolution of Specialists and Generalists. Mol Biol Evol. 2009;26:2661–2678. doi: 10.1093/molbev/msp187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ferea TL, Botstein D, Brown PO, Rosenzweig RF. Systematic changes in gene expression patterns following adaptive evolution in yeast. Proc Natl Acad Sci USA. 1999;96:9721–9726. doi: 10.1073/pnas.96.17.9721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gerstein AC, Lo DS, Otto SP. Parallel genetic changes and nonparallel gene-environment interactions characterize the evolution of drug resistance in yeast. Genetics. 2012;192:241–252. doi: 10.1534/genetics.112.142620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chao L, Cox EC. Competition between high and low mutating strains of Escherichia coli. Evolution. 1983:125–134. doi: 10.1111/j.1558-5646.1983.tb05521.x. [DOI] [PubMed] [Google Scholar]
- 52.Maharjan R, McKenzie C, Yeung A, Ferenci T. The basis of antagonistic pleiotropy in hfq mutations that have opposite effects on fitness at slow and fast growth rates. Heredity (Edinb) 2013;110:10–18. doi: 10.1038/hdy.2012.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Novick A, Szilard L. Experiments with the Chemostat on spontaneous mutations of bacteria. Proc Natl Acad Sci USA. 1950;36:708–719. doi: 10.1073/pnas.36.12.708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Paquin C, Adams J. Frequency of fixation of adaptive mutations is higher in evolving diploid than haploid yeast populations. Nature. 1983;302:495–500. doi: 10.1038/302495a0. [DOI] [PubMed] [Google Scholar]
- 55.Treves DS, Manning S, Adams J. Repeated evolution of an acetate- crossfeeding polymorphism in long-term populations of Escherichia coli. Mol Biol Evol. 1998;15:789–797. doi: 10.1093/oxfordjournals.molbev.a025984. [DOI] [PubMed] [Google Scholar]
- 56.Kinnersley M, Wenger J, Kroll E, Adams J, Sherlock G, Rosenzweig F. Ex uno plures: clonal reinforcement drives evolution of a simple microbial community. PLoS Genet. 2014;10:e1004430. doi: 10.1371/journal.pgen.1004430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Helling RB, Vargas CN, Adams J. Evolution of Escherichia coli during growth in a constant environment. Genetics. 1987;116:349–358. doi: 10.1093/genetics/116.3.349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rosenzweig RF, Sharp RR, Treves DS, Adams J. Microbial evolution in a simple unstructured environment: genetic differentiation in Escherichia coli. Genetics. 1994;137:903–917. doi: 10.1093/genetics/137.4.903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Maharjan RP, Ferenci T, Reeves PR, Li Y, Liu B, Wang L. The multiplicity of divergence mechanisms in a single evolving population. Genome Biol. 2012;13:R41. doi: 10.1186/gb-2012-13-6-r41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kao KC, Sherlock G. Molecular characterization of clonal interference during adaptive evolution in asexual populations of Saccharomyces cerevisiae. Nat Genet. 2008;40:1499–1504. doi: 10.1038/ng.280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Payen C, Di Rienzi SC, Ong GT, Pogachar JL, Sanchez JC, Sunshine AB, et al. The dynamics of diverse segmental amplifications in populations of Saccharomyces cerevisiae adapting to strong selection. G3 (Bethesda) 2014;4:399–409. doi: 10.1534/g3.113.009365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bernstein HC, Paulson SD, Carlson RP. Synthetic Escherichia coli consortia engineered for syntrophy demonstrate enhanced biomass productivity. J Biotechnol. 2012;157:159–166. doi: 10.1016/j.jbiotec.2011.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Miller AW, Befort C, Kerr EO, Dunham MJ. Design and Use of Multiplexed Chemostat Arrays. Journal of Visualized Experiments : JoVE. 2013 doi: 10.3791/50262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Takahashi CN, Miller AW, Ekness F, Dunham MJ, Klavins E. A Low Cost, Customizable Turbidostat for Use in Synthetic Circuit Characterization. ACS Synth. Biol. 2014:140801103853005. doi: 10.1021/sb500165g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Toprak E, Veres A, Yildiz S, Pedraza JM, Chait R, Paulsson J, et al. Building a morbidostat: an automated continuous-culture device for studying bacterial drug resistance under dynamically sustained drug inhibition. Unknown. 2013;8:555–567. doi: 10.1038/nprot.nprot.2013.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Oud B, Guadalupe-Medina V, Nijkamp JF, de Ridder D, Pronk JT, van Maris AJA, et al. Genome duplication and mutations in ACE2 cause multicellular, fast-sedimenting phenotypes in evolved Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 2013;110:E4223–31. doi: 10.1073/pnas.1305949110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.de Crécy E, Metzgar D, Allen C, Pénicaud M, Lyons B, Hansen CJ, et al. Development of a novel continuous culture device for experimental evolution of bacterial populations. Appl. Microbiol. Biotechnol. 2007;77:489–496. doi: 10.1007/s00253-007-1168-5. [DOI] [PubMed] [Google Scholar]
- 68.Vidal O, Longin R, Prigent-Combaret C, Dorel C, Hooreman M, Lejeune P. Isolation of an Escherichia coli K-12 mutant strain able to form biofilms on inert surfaces: involvement of a new ompR allele that increases curli expression. J Bacteriol. 1998;180:2442–2449. doi: 10.1128/jb.180.9.2442-2449.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Tu BP, Kudlicki A, Rowicka M, McKnight SL. Logic of the yeast metabolic cycle: temporal compartmentalization of cellular processes. Science. 2005;310:1152–1158. doi: 10.1126/science.1120499. [DOI] [PubMed] [Google Scholar]
- 70.Dénervaud N, Becker J, Delgado-Gonzalo R, Damay P, Rajkumar AS, Unser M, et al. A chemostat array enables the spatio-temporal analysis of the yeast proteome. Proc Natl Acad Sci USA. 2013;110:15842–15847. doi: 10.1073/pnas.1308265110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Weaver DS, Keseler IM, Mackie A, Paulsen IT, Karp PD. A genome-scale metabolic flux model of Escherichia coli K-12 derived from the EcoCyc database. BMC Syst Biol. 2014;8:79. doi: 10.1186/1752-0509-8-79. [DOI] [PMC free article] [PubMed] [Google Scholar]