Abstract
Purpose
Spliced variant forms of androgen receptor (AR-Vs) have been identified recently in castration-resistant prostate cancer (CRPC) cell lines and clinical samples. We identified the cistrome and gene signature of AR-Vs in CRPC cell lines and determined the clinical significance of AR-Vs regulated genes.
Materials and Methods
The CRPC cell line 22Rv1, which expresses both full length androgen receptor (AR-FL) and AR-Vs endogenously, was used as the research model. We established 22Rv1-ARFL−/ARVs+ and 22Rv1- ARFL−/ARVs− through RNA interference. ChIP-seq and microarray techniques were used to identify cistrome and gene expression profiles of AR-Vs in the absence of androgen.
Results
AR-Vs binding sites were identified in 22Rv1-ARFL−/ARVs+. A set of genes was found to be regulated uniquely by AR-Vs, but not by full-length AR in the absence of androgen. Integrated analysis revealed that some genes are modulated by AR-Vs directly. Unsupervised clustering analysis showed that the AR-Vs gene signature can differentiate not only benign from malignant prostate tissue, but also localized prostate cancer from metastatic CRPC specimens. Some genes that were modulated uniquely by AR-Vs also correlated with histological grade and biochemical failure.
Conclusions
We conclude that AR-Vs can bind to DNA independently of full-length AR in the absence of androgen and modulate a unique set of genes that is not regulated by AR-FL. The AR-Vs gene signature correlates with disease progression, and distinguishes between primary cancer and CRPC specimens, as well as benign and malignant prostate specimens.
Keywords: Prostatic Neoplasms, Castration-Resistant, Receptors, Androgen, Alternative Splicing, Cistrome, Gene signature
Introduction
Prostate cancer (PCa) is the most frequently diagnosed non-cutaneous malignancy in men and the second leading cause of male cancer-related mortality in the United States. Annually, there are approximately 233,000 newly diagnosed cases and 29,480 deaths1. For men with locally advanced or metastatic disease, androgen deprivation therapy (ADT) that disrupts androgen receptor (AR) signaling is the accepted initial treatment. In most patients PCa regresses upon treatment; however, relapse inevitably occurs, which ultimately progresses to castration-resistant prostate cancer (CRPC)2. However, AR signaling remains active in CRPC, suggesting that the CRPC phenotype may arise as a result of failure to suppress recurrent AR activity3.
In CRPC, AR undergoes alternative splicing with cryptic exons giving rise to truncated, constitutively active AR-Vs. All ARs have similar structural backbones, containing the N-terminal transactivation domain (TAD) and DNA binding domain (DBD). However, AR-Vs lack the ligand-binding domain (LBD) compared to the full length AR (AR-FL). Conventional ADT focuses on androgen-dependent activation of AR by either androgen ablation or antiandrogens that impede binding of endogenous ligand. Therefore, constitutively active, AR-Vs that lack the LBD are a compelling mechanism for CRPC cells to circumvent ADT. To date, over 20 different AR-Vs have been identified at the mRNA level in cell lines and CRPC tissue samples4–10. However, mechanisms by which these AR-Vs modulate gene expression in the absence of androgen remains unclear. Clarifying this mechanism is critical to understand the disease progression in CRPC patients who have failed ADT.
To characterize the AR-Vs cistrome, we used chromatin immunoprecipitation coupled with next-generation sequencing (ChIP-seq) in CRPC 22Rv1 cells under androgen-depleted conditions. The identified binding sites of AR-Vs were found to be independent of AR-FL as well as androgens, and can modulate a unique set of genes identified through a microarray-based approach. Based on the gene expression profiles, a unique gene signature for AR-Vs was identified. The clinical significance of the AR-Vs gene signature was determined with PCa specimens from the clinic.
Materials and Methods
Cell culture and transfection
22Rv1 cells were obtained from ATCC and maintained at 37°C and 5% CO2 in RPMI containing 10% Fetal Bovine Serum (FBS) and 1% antibiotic/antimycotic (Invitrogen, Life Technologies). Transient transfections were performed by electroporation (350V, 10ms. BTX, Harvard Apparatus). Cells (5×106) were incubated in 500 µL of RPMI 1640 containing 10% CSS (Charcoal Stripped Serum) with 20µg of shRNA or 750nmol siRNA for 15 minutes at room temperature. Overnight recovery was carried out in RPMI 1640 containing 10% CSS. Media was replenished with RPMI 1640 (10% CSS), and cells were harvested at 72hrs post-transfection.
AR knockdown
We produced shRNA suppression vectors in plasmid pFRT-H1P (Invitrogen) to target AR exon 1 (shAR Ex1 target sequence CAAGGGAGGTTACACCAAA) or exon 8 (shAR Ex8 target sequence TCAGTTCACTTTTGACCTGCTAATCA), which specifically knock down expression of all AR isoforms or full-length AR, respectively. We also purchased siRNA targeting AR exon 1 (siAR Ex1, Dharmacon D-003400-02 target sequence CAAGGGAGGUUACACCAAA) and siRNA targeting AR exon 7 (siAR Ex7, Dharmacon D-003400-01 target sequence GGAACUCGAUCGUAUCAUU).
Real-time RT-PCR
RNA was isolated with Trizol, and cDNA was prepared using Superscript III First-Strand Synthesis System (Both Invitrogen). Two-step Real-time PCR was performed with SYBR Green PCR Master Mix (Applied Biosystems, Life Technologies) on ABI PRISM 7700 SDS. Primer sequences are described in Supplementary Table 1.
Western blotting
Cells were washed twice in PBS and lysed with RIPA buffer (50mM Tris-HCl pH 7.4, 150mM NaCl, 2mM EDTA, 1% NP-40, 0.1% SDS). Equal amounts of protein were seperated on 4–12% Bis-Tris gels (Invitrogen) in 1× MOPS running buffer and transferred to nitrocellulose. Western blots were probed with AR (N-20, Santa Cruz Biotechnology), β-actin (#4967L, Cell Signaling), and α-tubulin (#2144, Cell Signaling). Membranes were immersed in chemiluminescence reagents (SuperSignal West Dura Chemiluminescent Substrate) and exposed to HyBlot CL autoradiography film for signal detection.
ChIP and ChIP-qPCR
ChIP experiments were performed as described previously (http://younglab.wi.mit.edu/hESRegulation/Young_Protocol.doc). Briefly, chromatin was crosslinked for 10 min at room temperature with 1% formaldehyde added directly to cell culture medium. Crosslinked chromatin was sonicated, diluted, and immunoprecipitated with Protein G-plus Agarose beads (Calbiochem) pre-bound with AR antibody (N-20, Santa Cruz) at 4°C overnight. The precipitated protein-DNA complexes were eluted, and crosslinking was reversed at 65°C for 16 hr. DNA fragments were purified and analyzed by real-time PCR. The primers used in real-time PCR are listed in Supplementary Table 1.
ChIP-seq and data analysis
ChIP-seq libraries were prepared using methods described previously11, and high throughput sequencing was performed using the Illumina HiSeq2000 platforms at the Mayo Genome Core Facility. Raw reads were checked for sequencing quality and mapped to human reference genome (hg19/GRCh37) using bowtie212. Uniquely mapped reads were used for downstream peak calling. MACS213 was used to identify AR binding sites with p-value cutoff 1E-5. Raw and processed data have been deposited into the NCBI Gene Expression Omnibus with accession number GSE58478 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?token=ehsxaswytfennul&acc=GSE58478).
Microarray analysis
RNA was isolated and quality checked on an Agilent 2100 Bioanalyzer (Agilent Technologies). Biotin labeled cDNA was generated and hybridized to the Illumina HumanHT-12 v4 Expression BeadChip arrays platform (Illumina, San Diego, CA) in the Advanced Genomic Technology Center Gene Expression Core at Mayo Clinic. Differentially expressed genes were detected using a moderated t-test implemented in the R package Limma14. Multiple testing was adjusted by Benjamini and Hochberg’s false discovery rate (FDR)15.
Patient Microarray data analysis (UMN-Mayo Partnership)
Gene expression data were collected from PCa patient tissues in a project funded by the Minnesota Partnership for Biotechnology and Medical Genomics collaborative venture with the Mayo Clinic and the University of Minnesota. Surgical specimens were obtained with written patient consent from the Mayo Clinic/NIH Specialized Program of Research Excellence (SPORE) in Prostate Cancer tumor bank with institutional review board approval (#1937 - 00). Data were collected and analyzed from tissues (frozen at −80°C) that included normal prostates as well as varying Gleason patterns of aggressive and non-aggressive PCa. Approximately 20,000 cells were laser capture–microdissected (LCM) from each of the benign and malignant frozen prostate tissues. The mRNA extracted from these cells was amplified linearly and analyzed on Affymetrix U133 plus 2.0 microarray chips16. Ninety nine laser capture microdissected prostate specimens, consisting of normal epithelium (n=17), benign prostatic hyperplasia (BPH) (n=10), prostatic intraepithelial neoplasia (PIN) (n=4), prostate cancers of Gleason Pattern (GP) 3 (n=31), GP4 (n=20), GP5 (n=10) and lymph node (LN) metastasis (n=7) from 81 individual treatment-naive patients were included in this profiling study.
Patient Microarray data (Harvard database in Gene Expression Omnibus)
Data from a Harvard database17 publicly available on the Gene Expression Omnibus (GEO) database (accession number GSE32269) was analyzed for correlations of gene expression with progression to CRPC. This data base is comprised of data from 120 snap-frozen CT-guided bone marrow biopsies from patients with castration-resistant prostate cancer. Of these, 29 independent biopsies containing mostly tumor were identified as CRPC group through careful microscopic examination, and 4 samples containing no tumor were identified as normal bone marrow. Twenty two primary tumors samples were isolated by LCM of frozen biopsies from hormone-naive patients. Affymetrix platform HG-U133A arrays were used for this analysis.
Results
AR-Vs bind to DNA independently of androgen treatment and full-length AR
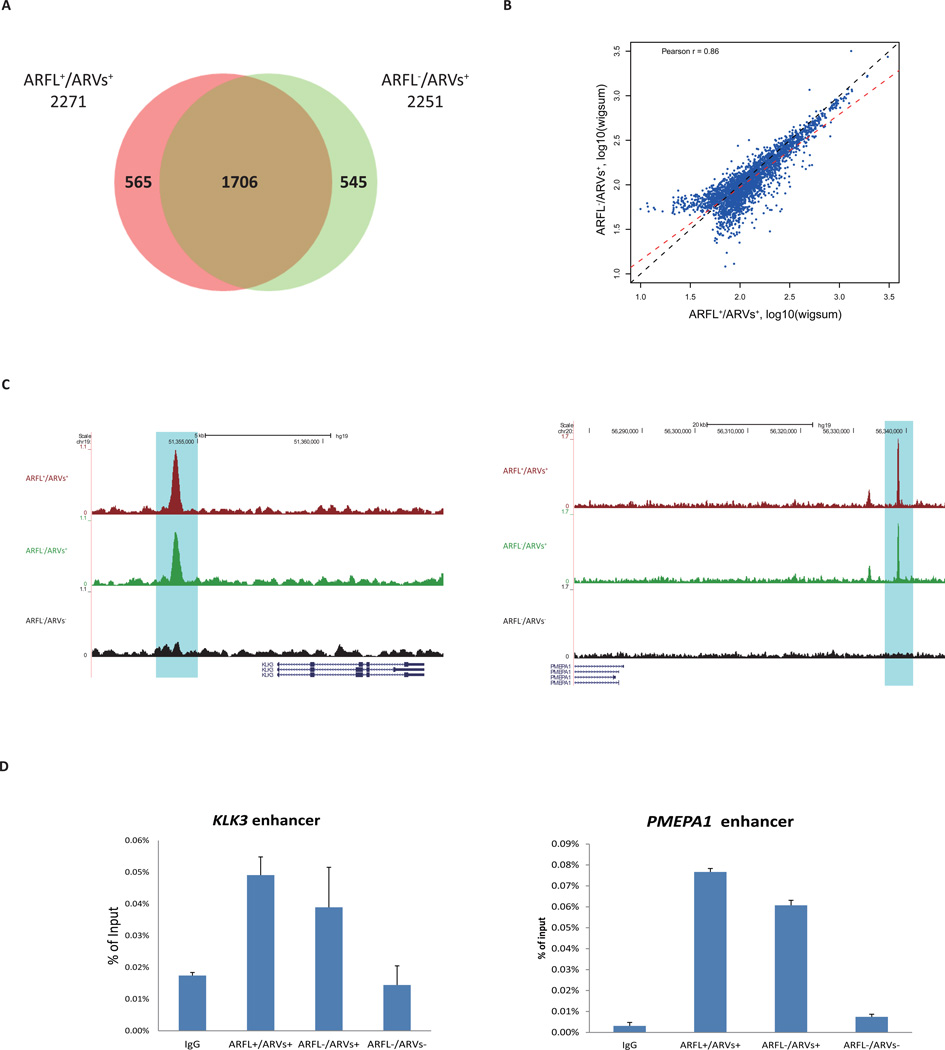
To interrogate the cistrome, CRPC 22Rv1 cells were utilized since they express both AR-FL and AR-Vs endogenously. Cells were transfected with non-target siRNA (ARFL+/ARVs+), siAR Ex7 to knockdown AR-FL (ARFL−/ARVs+), or siAR Ex1 to knockdown all AR (ARFL−/ARVs−) (Fig. S1A). ChIP-seq experiments were performed on each of these three groups with two biological replicates. Very few AR binding sites were identified in ARFL−/ARVs− control cells, suggesting that the knockdown had eliminated nearly all AR-DNA interaction, which was consistent with protein levels as determined by Western blots (Fig. S1B). Qualitative analysis showed that the majority (~75%) of identified AR binding sites in ARFL+/ARVs+ and ARFL−/ARVs+ overlapped (Fig. 1A). Comparing the cistromes quantitatively showed that AR binding sites in ARFL+/ARVs+ and ARFL−/ARVs+ are similar (Fig. 1B). These data suggest that AR-FL has minimal effect on AR-Vs binding to DNA in the absence of androgens. Peak intensity at the enhancer regions of KLK3 and PMEPA1 in ARFL+/ARVs+ and ARFL−/ARVs+ were comparable, while no peak was detected in ARFL−/ARVs− (Fig. 1C). ChIP-qPCR for KLK3 enhancer and PMEPA1 enhancer validated these findings (Fig. 1D). These data suggest that AR-Vs binding in 22Rv1 cells is independent of androgen and full-length AR.
Figure 1. ChIP-seq data analyses.
(A) Qualitative comparison of AR cistrome in ARFL+/ARVs+ group and ARFL−/ARVs+ group with Venn diagram.
(B) Quantitative comparison of AR binding in ARFL+/ARVs+ and ARFL−/ARVs+. The MACS called peaks from two groups were consolidated. Each dot represents a binding site. The signal intensities (reads coverage) of peaks were measured by wigsum. Black dashed line indicates diagonal line, red dashed line indicates linear regression line.
(C) Examples of AR-binding events around KLK3 and PMEPA1 genes loci in ARFL+/ARVs+, ARFL−/ARVs+, and ARFL−/ARVs− groups.
(D) ChIP-qPCR validation for KLK3 enhancer and PMEPA1 enhancer.
AR-Vs regulate a distinct set of genes independently of full-length AR
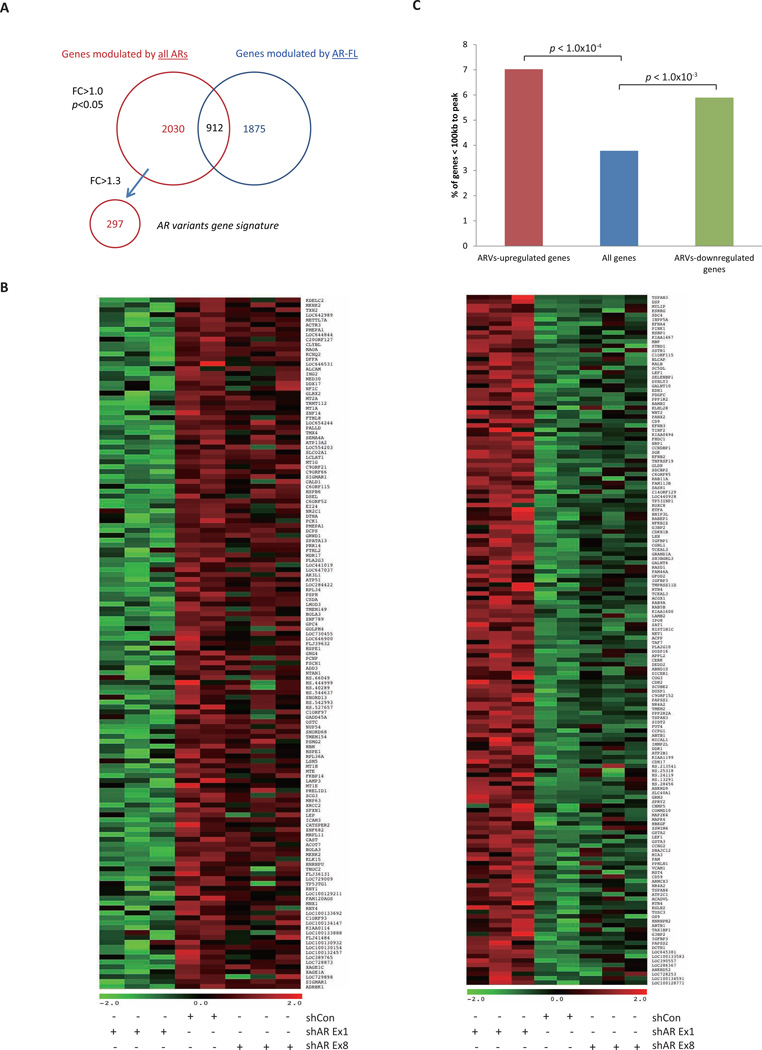
To identify AR-Vs regulated genes, 22Rv1 cells were transfected with empty vector shCon (ARFL+/ARVs+), shAR Ex1 (ARFL−/ARVs−), and shAR Ex8 (ARFL−/ARVs+) (Fig. S1A). Gene expression profiles were determined. Western blots for protein were used to validate the efficiency of knocking down the AR isoforms (Fig. S1C). Pair-wise comparisons identified 2,942 probes that were differentially expressed between shAR Ex1 and shCon, which should be regulated by both AR-FL and AR-Vs, and 2,787 probes that were differentially expressed between shAR Ex8 and shCon, which should be regulated by AR-FL (fold change>1.0; P<0.05). Subtracting 912 probes common to both sets left 2,030 probes that were uniquely regulated by AR-Vs (Fig. 2A). Two hundred ninety seven probes were uniquely upregulated (n=141) or downregulated (n=156) by AR-Vs at least 1.3 fold, in an AR-FL-independent manner in the absence of androgen (Fig. 2B; Supplementary Tables S2; P<0.05). Validation by real-time RT-PCR showed that AR-Vs regulate a subset of genes independently of AR-FL (Fig. S2).
Figure 2. Microarray data analyses.
(A) A Venn diagram indicates the number of probe sets differentially expressed in common or unique between shRNA conditions identified by microarray.
(B) Heat map representing differential expression of AR-Vs unique upregulated genes (141 probe sets; left) and downregulated genes (156 probe sets; right) between shRNA conditions (fold change > 1.3; P<0.05). Each row on the heatmap represents a probe set; each column represents an individual sample. Gene expression on each probe set was standardized to the mean of samples where red color is higher than the mean and green color is lower than the mean.
(C) Percentage of AR-Vs upregulated, downregulated and all genes within 100kb of AR-Vs bindings.
To determine whether the identified AR-Vs binding sites are associated with expression of AR target genes, we analyzed the correlation between ChIP-seq data and expression data. A higher percentage of genes that were upregulated or downregulated by AR-Vs was found within 100kb of the AR-Vs binding sites compared to the distribution of genome-wide genes (P<0.05; Fig. 2C). This suggests that in the absence of androgen, AR-Vs bind to genomic regions, and exert functional changes in the expression of specific target genes.
AR-Vs regulated gene correlate with castration-resistance prostate cancer
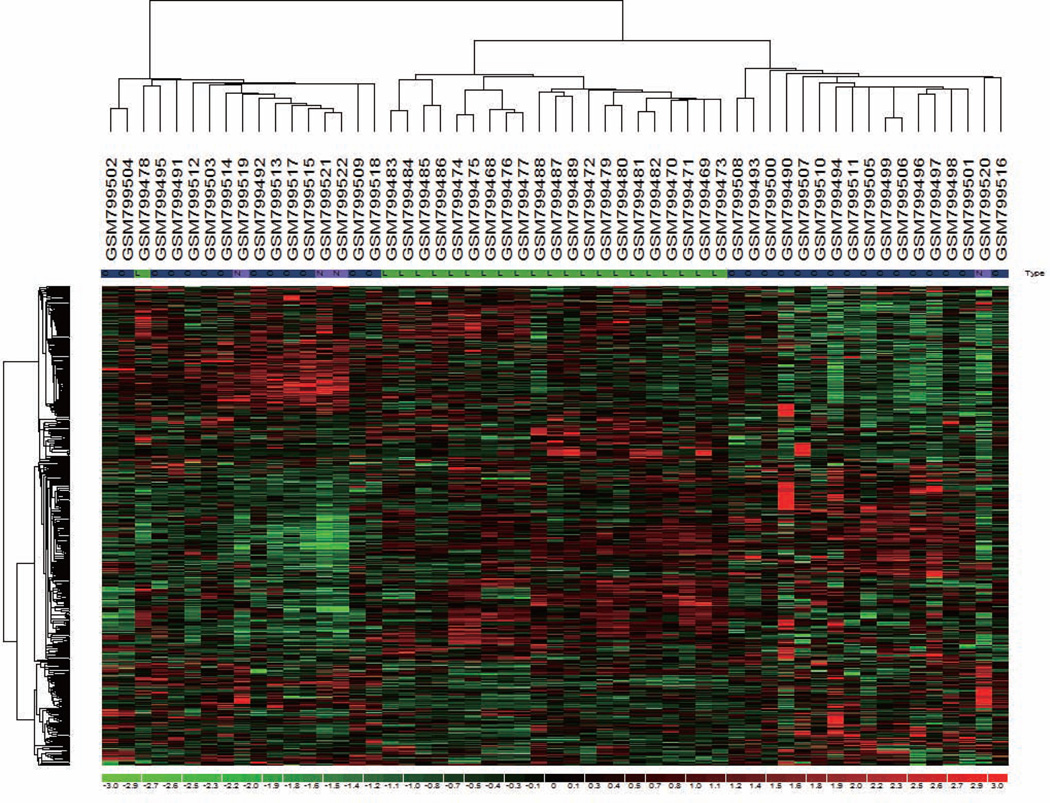
To establish the clinical significance of AR-Vs-regulated genes identified in 22Rv1 cells, we chose 297 probes as the AR-Vs gene signature for further analysis (Fig. 2A; Supplementary Tables S2; P<0.05). The expression of this signature was analyzed in primary PCa and CRPC specimens from the Harvard database17 (GSE32269). Unsupervised clustering of 55 specimens generated three major clusters, which strikingly differentiated localized prostate cancer (LPC, primary PCa) from CRPC specimens (Fig. 3). One hundred eighty probes (135 genes) were identified that were differentially expressed between these two groups (Supplementary Table S3; p<0.05). Among these genes, KIAA0494, CDH2, WNT2, NFIC, AK4, CSDA, TUSC3, PMEPA1, LAMB2, MIA3 were determined to be AR-Vs primary target genes based on integrated analysis of ChIP-seq and the gene expression profile. These data suggest that the AR-Vs-gene signature can distinguish primary PCa from CRPC specimens and may correlate with the progression of CRPC.
Figure 3.
Unsupervised clustering of localized prostate cancer (LPC) and castration-resistant prostate cancer (CRPC) by AR-Vs gene signature. Unsupervised clustering using 297 probe sets (259 genes) AR-Vs gene signature separates LPC and CRPC. Tissue-type designation N, L and C refers to normal, localized prostate cancer and castration-resistant prostate cancer, respectively.
AR-Vs regulated gene correlate with prostate cancer disease progression
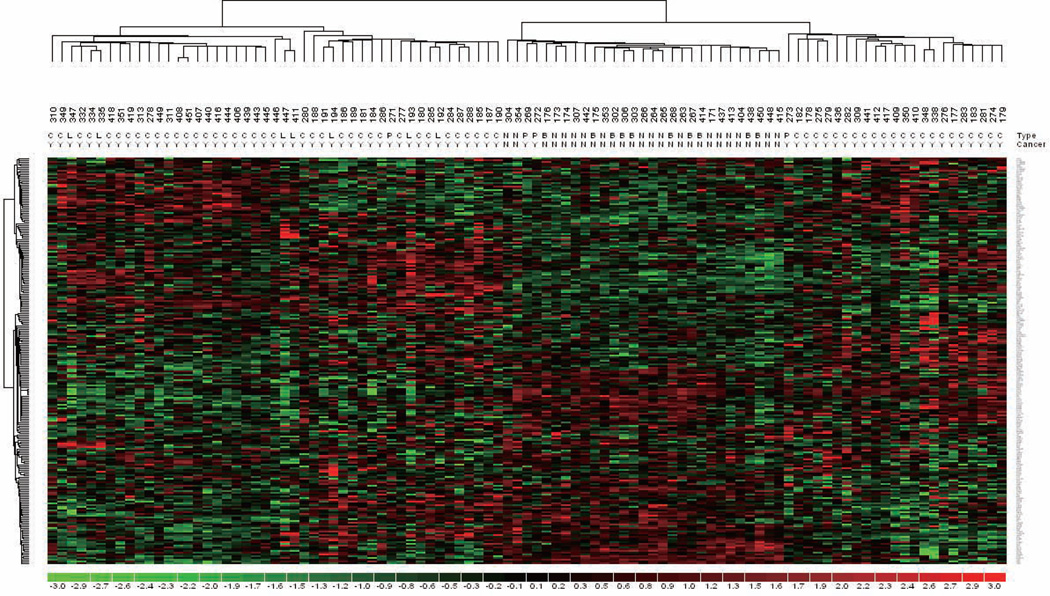
We next investigated whether the gene signature correlates with PCa disease progression. Expression data of the 297 probes was analyzed against mRNA expression data sets derived from the UMN-Mayo Partnership prostate specimens. Unsupervised clustering of 99 specimens by the AR-Vs gene signature generated three major clusters, which strikingly differentiated non-cancerous (normal and BPH) from pre-cancer/cancerous (PIN, primary cancer and metastasis) prostate specimens (Fig. 4). Two hundred thirty probe sets (141 genes) were differentially expressed (Supplementary Table S4; p<0.05). Among these genes, FHDC1, LAMB2, MAOA, KLK15, COMMD10, LXN, TP53TG1, SPATA13, PMEPA1, SLCO2A1, KIAA0494, TUSC3 and AK4 are potential primary target genes of AR-Vs in 22Rv1 cells. These results suggest that the AR-Vsgene signature can distinguish benign and malignant prostate specimens.
Figure 4.
Unsupervised clustering of prostate cancer specimens by AR-Vs gene signature. Unsupervised clustering using 297 probe sets (231 genes) AR-Vs gene signature separates benign and malignant prostate specimens. Tissue-type designation N, B, P, C, L refers to normal epithelium, benign prostatic hyperplasia, prostatic intraepithelial neoplasia, localized prostate cancer and lymph node metastasis, respectively. Malignant specimens are marked as “Y” and benign samples are marked as “N”.
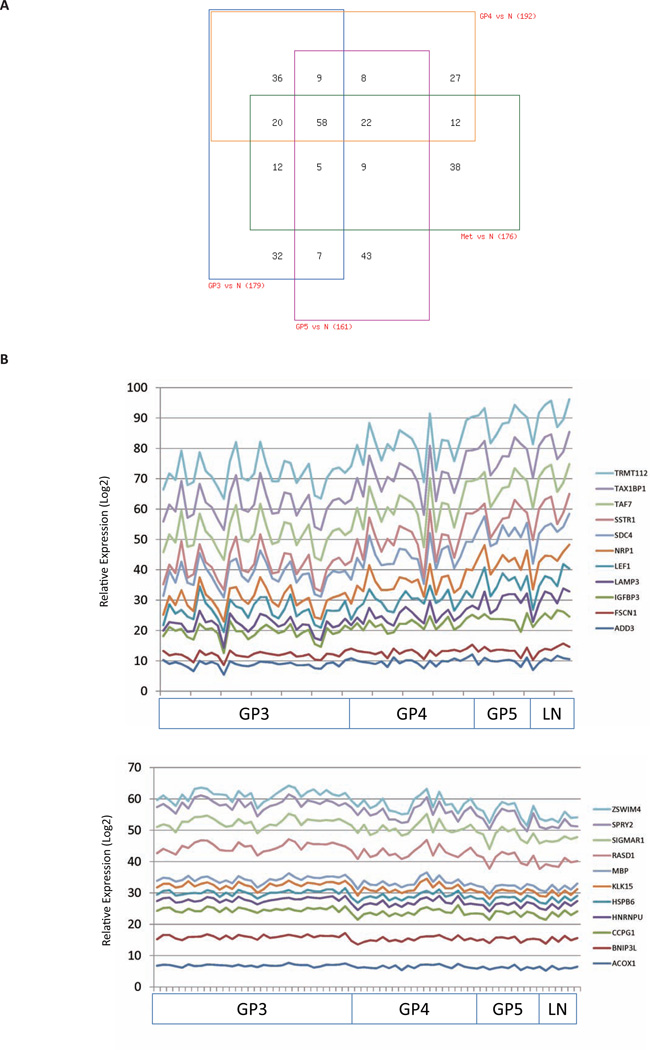
Pair-wise comparisons of normal epithelium and Gleason pattern 3 (GP3), GP4, GP5 and lymph node (LN) specimens showed differential expression of 179, 192, 161 and 176 probes respectively (Fig. 5A; Supplementary Table S5; p<0.05). Expression of a core of 58 probes was found to be consistently altered in GP3, GP4, GP5 and LN specimens compared with normal prostate epithelium. Further analysis showed that 165 probes (112 genes) were associated significantly, either positively (n=85) or negatively (n=80), with primary Gleason patterns and LN metastasis (Fig. 5B; Supplementary Table S6; p<0.05). KLK15, KIAA0494, NFKBIZ, CDH2, CSDA, FHDC1, AK4, MAOA, TUSC3 were identified as AR-Vs direct target genes associated with this progression. These results indicate that AR-Vs-regulated genes correlate with histological grade and metastatic status.
Figure 5. AR-Vs gene signature correlates with prostate cancer disease progression.
(A) Differentially expressed AR-Vs regulated probe sets in normal epithelium, localized cancer, and metastatic lymph node (LN) metastases. Pairwise comparisons were made between Gleason Pattern (GP)3, GP4, GP5, and LN metastases and normal epithelium, respectively, using the limma package implemented in R to identify differentially expressed probe sets (one-way ANOVA; p<0.05). Expression patterns of probe sets that are common or unique in GP3, GP4, GP5, and LN metastatic specimens are summarized in a 4-way Venn diagram. A detailed description of probe set tissue distribution is listed in Supplementary Table S5.
(B) Expression of AR-Vs gene signature correlates with Gleason pattern and metastasis. A Spearman rank linear regression model assesses the correlation of probe set expression with Gleason Pattern and presence of lymph node metastases. The y-axis represents a Log2 scale of relative probe set expression. The 11 most significantly positively correlated probe sets (p<0.05) are plotted in a stacked line graph (upper panel). The 11 most significantly negatively correlated probe sets (p<0.05) are plotted in a stacked line graph (lower panel). Probe sets that associate with disease progression are listed in Supplementary Table S6.
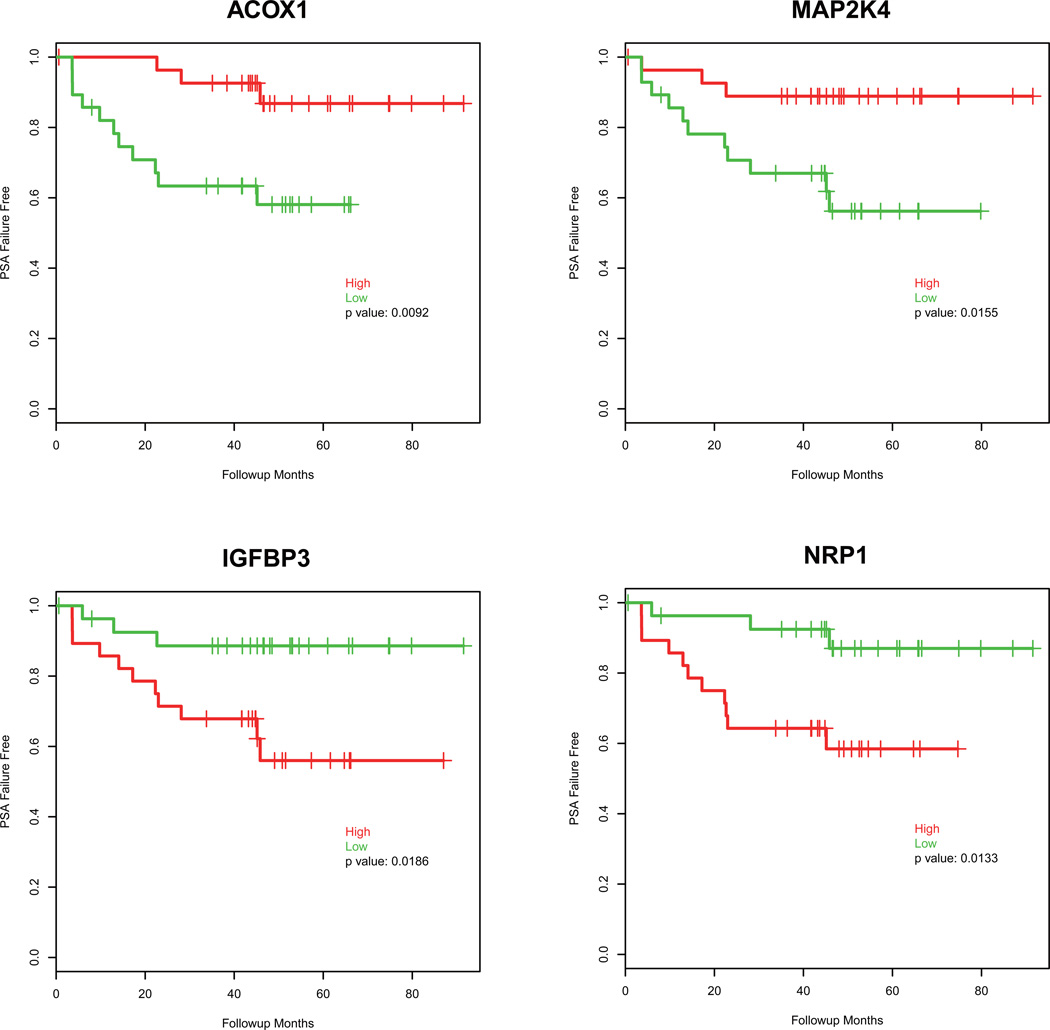
Fifty-six patients with localized prostate cancer (GP3, GP4 or GP5) at the time of surgery were followed up for an average of 42.5 ± 22.42 months, where 15 patients had biochemical recurrence (PSA failure) within that time period (Supplementary Table S7). Cox model analysis showed that expression of 34 probes was significantly associated with the time of PSA failure (Supplementary Table S8; p<0.05). To visualize the association, the Log rank test was used to assess the differences between high expression and low expression of 34 probes. The top 4 genes that were significantly associated with time to PSA failure are shown in Fig. 6. Low expression of ACOX1 and MAP2K4, and high expression of IGFBP3 and NRP1 were significantly associated with a shorter time to PSA failure (p<0.05). These results demonstrate that some ARVs-regulated genes correlate with biochemical recurrence of prostate cancer after prostatectomy.
Figure 6.
Expression of AR-Vs gene signature correlates with biochemical recurrence. Follow-up data was available for 56 patients, and univariate association was conducted for time to biochemical recurrence (p<0.05). Probe set expression of the four significantly associated genes was extracted, and the median expression was used to dichotomize patients into “high” (red) and “low” (green) for Kaplan-Meier plots. Log rank test was used to determine differences in time to biochemical recurrence.
Discussion
In this study, we show the cistrome and regulatory landscape of AR-Vs in the absence of androgen using the CRPC 22Rv1 CRPC cell line that expresses both AR-FL and AR-Vs endogenously. An AR-Vs gene signature was generated based on these data. We demonstrate that the AR-Vs gene signature can differentiate not only hormone-naive from CRPC, but also benign from malignant specimens. The correlation of the AR-Vs gene signature with histological grade, lymph node metastases and biochemical recurrence indicates that some AR-Vs regulated genes may be responsible for more aggressive disease. Further characterization of these genes may identify novel therapeutic targets in CRPC patients.
Chromatin immunoprecipitation combined with microarray (ChIP-chip) or next-generation sequencing (ChIP-seq) have been used to characterize the AR cistrome in androgen dependent PCa cell lines18–20, CRPC cell lines21–23 and PCa tissues24. The effects of DHT treatment on the AR cistrome have been characterized in 22Rv1 cells with ChIP-chip22 and ChIP-seq techniques23. However, these studies focused on AR binding events in the presence of androgen, and did not determine the contribution of AR-Vs to the AR binding sites in the absence of androgen. Thus, to our knowledge, this is the first report of the AR-Vs cistrome in the absence of androgen in a CRPC cell line.
Gene expression profiles that are regulated by AR-Vs have been addressed in several reports5–7, 25–27, in which specific AR-Vs were either overexpressed in PCa model systems6, 7, 26 or a specific AR-V was knocked down in a cell line that endogenously expresses multiple truncated AR-Vs5, 25. In our present study, the 22Rv1 cell model with endogenous co-expression of AR-FL and AR-Vs is more reflective of clinical PCa and thus represents an ideal cell-based model for gene profiling of AR-Vs target genes.
An important question has been whether AR-Vs regulate canonical androgen-regulated genes, or if they regulate a unique subset of genes. Studies using LNCaP cells ectopically expressing either AR-V76 or ARV567es7 demonstrated that AR-Vs can induce canonical androgen-regulated genes such as PSA (KLK3), KLK2, NKX3.1, FKBP5 and TMPRSS2 in the absence of androgens. Another study assessing androgen/AR-regulated transcription expression following AR-V7 or AR-FL knockdown in CWR-R1 cells, suggested that the AR-Vs transcriptional program represents a subset of the broader androgen/AR transcriptional program25. These data are in contrast to other studies showing some, but not all, canonical androgen-regulated genes (e.g. PSA, TMPRSS2, FKBP5) being regulated by AR-Vs in CRPC cell lines or clinical specimens5, 27. In our present study, although many identified genes are not canonical androgen-regulated genes, within the AR-Vs gene signature, we can still identify known androgen-regulated genes, such as the PMEPA1 gene. The repertoire of AR-Vs target genes is likely to be highly complex, overlapping with known androgen-regulated genes, and implicates some novel target genes. Thus, more experiments need to evaluate whether or not these signature genes are also regulated by activated AR-FL in the same cell line.
Several analyses that correlated AR-FL binding sites with gene expression profiles19, 20, 22 suggested that AR regulates not only adjacent target genes in a linear fashion, but also distal genes that may be far away from binding sites via looping structures within chromosomes28. It has been reported that AR-FL can modulate target genes directly in the absence of androgen23. The distances between binding sites and transcription start sites (TSS) can vary from 25kb to 250kb3. Thus we hypothesized that AR-Vs modulate primary target genes in a manner similar to AR-FL and chose 100kb for our analysis. The correlation between AR-Vs cistrome and regulated genes suggests that AR-Vs can bind to DNA in the absence of androgen and directly modulate target gene expression, suggesting AR-Vs may play an important role in CRPC through regulating target genes directly.
It has been suggested that AR-Vs and AR-FL can co-occupy the promoter of a canonical androgen-responsive gene, e.g.,KLK3, in a mutually-dependent manner in the absence of androgen29. This study29 used the KLK3 promoter region, whereas our study used the KLK3 enhancer region (approximately 4Kb upstream from the promoter), and our ChIP-seq data shows that the AR-Vs binding to the KLK3 promoter region is much weaker than the enhancer region. Also, in this study29, only AR-V7 binding was analyzed. Since several AR-Vs are expressed in 22Rv1 cells, including AR-V1, AR-V3, AR-V4, AR-V7 and other minor isoforms8, the cistrome we identified represents a more comprehensive analysis of AR-Vs binding. Our findings that AR-Vs bind to DNA and the cistrome independently of AR-FL, are consistent with observations that many AR-Vs are located in the nucleus in the absence of androgen30, while most AR-FL is located in the cytoplasm and needs ligand binding to translocate into the nucleus3.
Conclusions
Overall, our study identified an AR-Vs cistrome and a distinct AR-Vs-gene signature derived from an in vitro CRPC model. This signature was validated in a cohort of clinical PCa specimens. These findings have revealed the clinical relevance for a number of target genes for AR-Vs. Further analysis and characterization of these signaling networks may lead to novel drugable targets for CRPC patients.
Supplementary Material
Acknowledgements
Huihuang Yan, PhD, Division of Biomedical Statistics and Informatics, Mayo Clinic, provided preliminary ChIP-seq data processing. Kevin M. Regan, Department of Urology Research, and Yu Zhao, PhD, Department of Biochemistry and Molecular Biology, Mayo Clinic, provided technical support.
Funding: The work was supported by Prostate Cancer SPORE CA091956 and the T.J. Martell Foundation. Peter E. Lonergan was supported by a Fulbright Scholarship from the US Department of State.
Glossary
- PCa
Prostate cancer
- CRPC
castration-resistant prostate cancer
- AR
androgen receptor
- AR-FL
full-length androgen receptor
- AR-Vs
androgen receptor splice variants
- LBD
ligand binding domain
- ChIP-seq
chromatin immunoprecipitation coupled with next-generation sequencing
- ChIP-chip
chromatin immunoprecipitation coupled with microarray
- BPH
benign prostatic hyperplasia
- PIN
prostatic intraepithelial neoplasia
- GP
Gleason Pattern
- LN
lymph node
- TSS
transcription start sites
Footnotes
Publisher's Disclaimer: DISCLAIMER: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our subscribers we are providing this early version of the article. The paper will be copy edited and typeset, and proof will be reviewed before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to The Journal pertain.
Disclosure of potential conflicts of interest: No potential conflicts of interest were disclosed.
References
- 1.Siegel R, Ma J, Zou Z, et al. Cancer statistics, 2014. CA Cancer J Clin. 2014;64:9. doi: 10.3322/caac.21208. [DOI] [PubMed] [Google Scholar]
- 2.Debes JD, Tindall DJ. Mechanisms of androgen-refractory prostate cancer. N Engl J Med. 2004;351:1488. doi: 10.1056/NEJMp048178. [DOI] [PubMed] [Google Scholar]
- 3.Lu J, Tindall DJ. The role of androgen receptor-regulated genes in prostate cancer progression. In: Socorro S, editor. Androgen receptors : structural biology, genetics and molecular defects. United States: NOVA SCIENCE PUBLISHERS; 2014. pp. 107–128. [Google Scholar]
- 4.Dehm SM, Schmidt LJ, Heemers HV, et al. Splicing of a novel androgen receptor exon generates a constitutively active androgen receptor that mediates prostate cancer therapy resistance. Cancer Res. 2008;68:5469. doi: 10.1158/0008-5472.CAN-08-0594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Guo Z, Yang X, Sun F, et al. A novel androgen receptor splice variant is up-regulated during prostate cancer progression and promotes androgen depletion-resistant growth. Cancer Res. 2009;69:2305. doi: 10.1158/0008-5472.CAN-08-3795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hu R, Dunn TA, Wei S, et al. Ligand-independent androgen receptor variants derived from splicing of cryptic exons signify hormone-refractory prostate cancer. Cancer Res. 2009;69:16. doi: 10.1158/0008-5472.CAN-08-2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sun S, Sprenger CC, Vessella RL, et al. Castration resistance in human prostate cancer is conferred by a frequently occurring androgen receptor splice variant. J Clin Invest. 2010;120:2715. doi: 10.1172/JCI41824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Marcias G, Erdmann E, Lapouge G, et al. Identification of novel truncated androgen receptor (AR) mutants including unreported pre-mRNA splicing variants in the 22Rv1 hormone-refractory prostate cancer (PCa) cell line. Hum Mutat. 2010;31:74. doi: 10.1002/humu.21138. [DOI] [PubMed] [Google Scholar]
- 9.Hu R, Isaacs WB, Luo J. A snapshot of the expression signature of androgen receptor splicing variants and their distinctive transcriptional activities. Prostate. 2011;71:1656. doi: 10.1002/pros.21382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Watson PA, Chen YF, Balbas MD, et al. Constitutively active androgen receptor splice variants expressed in castration-resistant prostate cancer require full-length androgen receptor. Proc Natl Acad Sci U S A. 2010;107:16759. doi: 10.1073/pnas.1012443107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Peng JC, Valouev A, Swigut T, et al. Jarid2/Jumonji coordinates control of PRC2 enzymatic activity and target gene occupancy in pluripotent cells. Cell. 2009;139:1290. doi: 10.1016/j.cell.2009.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang Y, Liu T, Meyer CA, et al. Model-based analysis of ChIP-Seq (MACS) Genome biology. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wu ZJ, Irizarry RA, Gentleman R, et al. A model-based background adjustment for oligonucleotide expression arrays. Journal of the American Statistical Association. 2004;99:909. [Google Scholar]
- 15.Benjamini Y, Hochberg Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. Journal of the Royal Statistical Society. Series B (Methodological) 1995;57:289. [Google Scholar]
- 16.Kube DM, Savci-Heijink CD, Lamblin AF, et al. Optimization of laser capture microdissection and RNA amplification for gene expression profiling of prostate cancer. BMC Mol Biol. 2007;8:25. doi: 10.1186/1471-2199-8-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cai C, Wang H, He HH, et al. ERG induces androgen receptor-mediated regulation of SOX9 in prostate cancer. J Clin Invest. 2013;123:1109. doi: 10.1172/JCI66666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang Q, Li W, Liu XS, et al. A hierarchical network of transcription factors governs androgen receptor-dependent prostate cancer growth. Mol Cell. 2007;27:380. doi: 10.1016/j.molcel.2007.05.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Massie CE, Lynch A, Ramos-Montoya A, et al. The androgen receptor fuels prostate cancer by regulating central metabolism and biosynthesis. EMBO J. 2011;30:2719. doi: 10.1038/emboj.2011.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Takayama K, Tsutsumi S, Katayama S, et al. Integration of cap analysis of gene expression and chromatin immunoprecipitation analysis on array reveals genome-wide androgen receptor signaling in prostate cancer cells. Oncogene. 2011;30:619. doi: 10.1038/onc.2010.436. [DOI] [PubMed] [Google Scholar]
- 21.Wang Q, Li W, Zhang Y, et al. Androgen receptor regulates a distinct transcription program in androgen-independent prostate cancer. Cell. 2009;138:245. doi: 10.1016/j.cell.2009.04.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen H, Libertini SJ, George M, et al. Genome-wide analysis of androgen receptor binding and gene regulation in two CWR22-derived prostate cancer cell lines. Endocr Relat Cancer. 2010;17:857. doi: 10.1677/ERC-10-0081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Decker KF, Zheng D, He Y, et al. Persistent androgen receptor-mediated transcription in castration-resistant prostate cancer under androgen-deprived conditions. Nucleic Acids Res. 2012;40:10765. doi: 10.1093/nar/gks888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sharma NL, Massie CE, Ramos-Montoya A, et al. The androgen receptor induces a distinct transcriptional program in castration-resistant prostate cancer in man. Cancer Cell. 2013;23:35. doi: 10.1016/j.ccr.2012.11.010. [DOI] [PubMed] [Google Scholar]
- 25.Li Y, Chan SC, Brand LJ, et al. Androgen receptor splice variants mediate enzalutamide resistance in castration-resistant prostate cancer cell lines. Cancer Res. 2013;73:483. doi: 10.1158/0008-5472.CAN-12-3630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hu R, Lu C, Mostaghel EA, et al. Distinct transcriptional programs mediated by the ligand-dependent full-length androgen receptor and its splice variants in castration-resistant prostate cancer. Cancer Res. 2012;72:3457. doi: 10.1158/0008-5472.CAN-11-3892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hornberg E, Ylitalo EB, Crnalic S, et al. Expression of androgen receptor splice variants in prostate cancer bone metastases is associated with castration-resistance and short survival. PLoS One. 2011;6:e19059. doi: 10.1371/journal.pone.0019059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wu D, Zhang C, Shen Y, et al. Androgen receptor-driven chromatin looping in prostate cancer. Trends Endocrinol Metab. 2011;22:474. doi: 10.1016/j.tem.2011.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cao B, Qi Y, Zhang G, et al. Androgen receptor splice variants activating the full-length receptor in mediating resistance to androgen-directed therapy. Oncotarget. 2014 doi: 10.18632/oncotarget.1802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chan SC, Li Y, Dehm SM. Androgen receptor splice variants activate androgen receptor target genes and support aberrant prostate cancer cell growth independent of canonical androgen receptor nuclear localization signal. J Biol Chem. 2012;287:19736. doi: 10.1074/jbc.M112.352930. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.