Abstract
G × E in psychiatry may explain why environmental risk factors have big impact in some individuals but not in others, and conversely why relatives that are genetically at risk for disease do not all develop disease. Here we discuss two novel methods that use an aggregate genome-wide measure of genetic risk to detect G × E and estimate its effect in the population using data currently available and data we anticipate will be available in the near future. The first method exploits summary statistics from large-scale genome-wide association studies ignorant of the environmental conditions and detects G × E in an out-of-sample risk-profiling framework. The second method relies on larger samples and is based on a mixed linear model framework. It estimates variance explained directly from single nucleotide polymorphisms and environmental measures. Both methods have great potential to improve public health interventions focusing on risk-based screening that is informed by both genetic and environmental risk factors.
Key words: Gene–environment interaction, polygenic, psychiatric genetics
Introduction
The importance of gene-by-environment interaction (G × E) in psychiatry has intuitive appeal and is often discussed. However, the dearth of data to investigate G × E means that empirical evidence is modest. Here we consider how the era of genome-wide genotyping opens new approaches for G × E research. G × E in disease aetiology implies environmental factors control disease outcome conditional on genetic predisposition, and conversely, genetic factors control disease outcome conditional on environmental exposure. It explains why genetic and environmental factors can have a big impact in some individuals but not in others. G × E research aims to identify non-marginal genetic and environmental effects, that is, effects of a specific genetic (or environmental) risk factor that does not act in the population as a whole when averaging over all other variables but only act conditional on an environmental (or genetic) variable.
Interpretation of G × E research requires careful consideration of the hypotheses tested, first recognising that presence of statistical interaction depends on – and may be induced by – the scale of measurement. In the past 15 years, the majority of G × E researches have used a molecular genetic approach focusing on candidate genes and specific environmental risk factors. Researchers hypothesised environmental factors controlled disease outcome only if a single genetic mutation was present (or vice versa, a genetic mutation to control disease outcome only under certain environmental circumstances). For complex genetic diseases including psychiatric disorders, the effect of a specific genetic variant is usually very small effect and hence the prior probability that a specific genetic variant interacts with a specific environmental factor is also very small. Therefore, the majority of published G × E studies have suffered from lack of replication, low-power, a publication bias towards positive results and major methodological concerns (for a critical review see Duncan & Keller, 2011). Interpretation of candidate gene G × E research is further complicated by the wide diversity of methods and reporting standards used, precluding meta-analytical evaluation of G × E evidence (Modinos et al. 2013). However, lack of power and replication was not limited to G × E studies but was also inherent in the decade of candidate gene association studies which were not powered to detect the small effect sizes that we now know operate in human diseases. Significant findings were almost never replicated and candidate genes that were selected based on their potential involvement in candidate biological pathways (e.g., neurotransmitter systems) have generally not shown robust association with psychiatric disease suggesting that current understanding of the biological basis of psychiatric disease is lacking.
The era of genome-wide association studies (GWAS) promised new hope but early studies were also underpowered (Manolio et al. 2009). More recent statistical analyses of aggregate single nucleotide polymorphism (SNP) effects, however, have shown that genetic variation in complex psychiatric disorders is polygenic in nature. Odds ratio of the individual variants generally range from 1.1 to 1.4; consequently, very large sample sizes are required to estimate those SNP effects with high precision. Recent efforts to increase sample sizes are now starting to pay off; the latest mega-analyses of GWAS data for schizophrenia has identified >100 genome-wide significant associations (2014), together explaining 7% of the variation in out of sample prediction. Other psychiatric diseases are expected to follow if GWAS sample sizes continue to increase.
Comparison of effect sizes of individual genetic risk variants with environmental risk factors shows that much larger risk can be attributed to individual environmental factors. For example, recent meta-analyses of the association between schizophrenia and urbanicity and migrant status revealed a pooled odds ratio of 2.39 for urbanicity (Vassos et al. 2012) and odds ratios of 2.7 and 4.5, for first and second generation migrants to European countries, respectively (Dealberto, 2010).
Taking these lessons forward to novel G × E research where we hypothesise that exposure to environmental risk factor increases the risk of disease only in those that are genetically susceptible, we will have to redefine being genetically susceptible in our study design. A single mutation in a candidate gene is unlikely to have a big impact in the population, by taking the aggregate effect of all mutations; however, we can differentiate people being at high genetic risk for disease. A polygenic architecture for psychiatric disorders of many weakly contributing variants means that genetic effects interact on the scale of disease (i.e., affected v. not affected) but act more additively on a susceptibility to disease scale (i.e., liability scale) (Zammit et al. 2010). On the disease scale, environmental variables are expected to combine interactively with genetic variants (combined), but more unknown is whether the genetic variants and environmental factors interact on the underlying scale where genetic effects combine more additively.
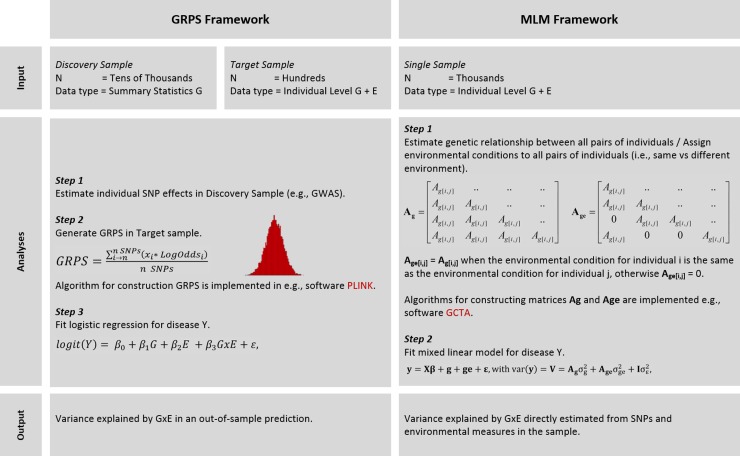
Here we describe two methods (see Fig. 1 for a schematic of the methods) that utilise the aggregate effect of genetic mutations to study potential G × E in a more powerful and reliable way compared with the candidate-gene approach.
Fig. 1.
Summary of genetic risk profiling framework and mixed linear model framework for detecting and estimating G × E. GRPS, genetic risk profile score; MLM, mixed linear model; G, genetic condition; E, environmental condition; G × E, gene–environment interaction; Ag, genetic relationship matrix; Age, gene–environment relationship matrix; MLM, framework can also be applied in a bivariate setting in which the two traits represent the two environments; environmental conditions can be binary, ordinary and continues.
G × E in a risk-profiling framework
Genomic risk profile scores (GRPS) can be used as proxy estimate of genetic effects in a G × E study in which individuals have both genome-wide genotypes and measures for environmental risk factors. GRPS are quantitative scores calculated for each individual and are an estimate of an individuals' genetic risk for disease. GRPS were first applied to schizophrenia GWAS data where they provided evidence for a substantial polygenic component to the risk of schizophrenia involving many loci of very small individual effect (Purcell et al. 2009).
GRPS are calculated using estimates of genetic effect sizes derived from an independent GWAS ‘discovery sample’. It is important that no individuals (or their close relatives) from the GRPS sample (or ‘target sample’) are included in the discovery sample. GRPS are constructed in two steps: firstly, individual effect sizes of risk alleles (e.g., beta from linear regression, odds ratio from logistic regression or best linear unbiased prediction (BLUP) from linear-mixed models (Yang et al. 2014)) are estimated in the discovery sample. Secondly, for each individual within the target sample a GRPS is computed by taking the number of risk alleles an individual possesses weighted by the effect size of that allele from the discovery sample, averaged over the number of loci included in the GRPS.
The GRPS and the environmental variable of interest can now be included as individual terms as well as a product term in a risk-profiling framework. Fitting nested (increasingly more restricted) models allows testing for significance of the GRPS, the environmental factor of interest, and their interaction effect. For a disease trait (binary Y), the full logistic regression equation will be:
where G is the multi-locus GRPS, E is the environmental moderator variable and G × E is the interaction term between G and E. Under this model significant genetic and environmental effects already imply G × E on the disease (Y) scale, but a significant G × E term implies G × E on the underlying liability to disease. In this framework, multiple GRPS and multiple environmental factors can be included in the analyses to allow for hypothesis-driven identification of G × E effects. For example, GRPS can be based on SNPs that are selected based on functional annotation or physical position in the genome.
In the context of schizophrenia, the latest mega analysis published by the Psychiatric Genomics Consortium (PGC) (2014) (their Figure 3) shows that if individuals in independent target samples are ranked on GRPS then the odds of disease in the 10th decile is 7–20-fold (varying between samples) greater than the odds of disease in the first decile.
A G × E application may consider, for example, neonatal vitamin D level (McGrath et al. 2010b) as an environmental risk factor for schizophrenia that is hypothesised to interact with the GRPS. In this application, the G × E analysis could be extended by considering multiple GRPS and hence multiple genetic and G × E terms based on biological function, e.g., one based on SNPs in calcium ion channel genes (e.g., Purcell et al. 2014) and one based on the remaining SNPs.
Algorithms for the construction of GRPSs have been implemented in the software PLINK (http://pngu.mgh.harvard.edu/~purcell/plink/; option – score) (Purcell et al. 2007). The prediction accuracy of the constructed GRPS and the G × E effects can be analysed in a statistical software package of choice.
G × E in a mixed linear model framework
The estimation of variance explained by aggregate SNP effects, sometimes called SNP heritability (see the Introduction section) is based on a mixed linear model framework. In this framework, genetic variance is estimated from genetic similarity among pairs of individuals who are not related in the classical sense. The basic idea behind this method is that for a polygenic trait, pairs of individuals who show higher genetic similarity also show higher resemblance at the trait level. Genetic similarity for each pair of individuals (defined in the model as sharing 0, 1 or 2 alleles at each locus) is measured from all the SNPs and the aggregated SNP effects are treated as random variables in a mixed linear model (Yang et al. 2010, 2011). The model can be augmented with G × E effects. Similarly to the SNP effects, the G × E effects are included as random effects in the model. In matrix notation, the linear mixed model including a G × E term can be written as:
where y is an n × 1 vector containing the phenotypes (e.g., 1, affected v. 0, unaffected), g, ge and ε are vectors of length n of the aggregate effects of all the SNPs from all of the individuals, the genotype–environment interaction effects from all of the individuals, and the residual effects, respectively. The variance of y is the sum of the genetic variance, the interaction variance and the error variance. For example, when the environmental condition is binary, Ag is the genetic relationship matrix (GRM) estimated from all SNPs and elements in Age = Ag for all pairs of individuals sharing the same environment and elements in Age = 0 for the pairs of individuals in different environments.
The estimate of the aggregate SNP effects (g) reflects the genetic variation that is captured by common SNPs and the estimate of the G × E effects (ge) reflects the proportion of the variance attributable to G × E. Through application of restricted maximum likelihood estimation (REML), the proportion of genetic and/or G × E variance to the total variance can be estimated. Significance of the parameters can be tested by likelihood ratio tests comparing the likelihood under the full and reduced models.
When the E variable is binary, G × E can also be investigated using a bivariate mixed model with the two traits representing the two environments (Falconer & Latyszewski, 1952). Under this more general framework the SNP-heritabilities are not forced to be the same under the two conditions, an additional degree of freedom is however included in the model. A genetic correlation across the environments that is significantly less than one implies existence of G × E. However, when the SNP-heritabilities of the trait in the two environments differ a genetic correlation across environments that is equal to one does not necessary imply absence of G × E; in this scenario it is however most likely that the G × E term reflects a scale effect (Lynch & Walsh, 1998). To detect possible scale effects, data could be transformed prior to analysis; any variance that can be removed through transformation of the data can be labelled as a scale effect. Interactions reflecting scale effects can however not always be removed or even reduced by a transformation of scale (Falconer & Mackay, 1996).
For disease traits, interpretation of the estimates of the SNP-heritability of the two traits (i.e., heritability of the disease in the two environments) is problematic, exacerbated by potentially different lifetime disease prevalences under the two environmental conditions, which may be unknown or difficult to estimate. We advise to use the bivariate approach to test for significance of the G × E term, rather than interpreting the estimates of SNP-heritability in the two environments, since the correlation is not affected by the ascertainment imposed on the disease in the two environments. A magnitude of the interaction variance can be estimated in the univariate mixed linear model. Analysis under both the univariate and bivariate G × E frameworks is recommended to gain further insight into the estimated interaction.
Application of the mixed model method requires data sets in which all individuals are measured for genome-wide genotypes and the environmental risk factor. In large data sets the genetic and G × E terms can be partitioned by fitting multiple GRMs based on specific notations such as functional pathways, similar to fitting multiple GRPS × E interaction terms in the risk profiling framework.
Estimation of genetic variance and G × E variance in a linear mixed model has been implemented in the software GCTA (http://www.complextraitgenomics.com/software/gcta/) (Yang et al. 2011).
Comparison of the two methods
The method of choice, risk-profiling or mixed linear model, primarily depends on the data available to researchers. The mixed linear model method requires large samples measured for both individual level genetic and environmental measures (based on power considerations (e.g., Dudbridge, 2013; Visscher et al. 2014), we estimate a sample size of at least 5000 individuals). Because individual studies are generally too small, researchers are likely to combine data from several studies into one study. The gain in sample size, however, often comes with a loss in coherence of both genetic and environmental measures. Recently developed methods in statistical genetics allow harmonisation of the genetic data, e.g., through imputation of SNP data to a common reference panel. Harmonisation of environmental measures across studies, however, needs more thoughtful discussion in the field, and new data collection efforts should aim for harmonisation with other studies (e.g., PhenX Toolkit; Hamilton & Tabitha, 2014).
In contrast, the risk-profiling method requires individual level genetic and environmental measures in only the target sample with the GRPS constructed based on GWAS summary statistics from a larger independent discovery sample that is (most likely) ignorant of the environmental conditions. Since the dearth of data sets informative for both genetic and environmental measures has been a limiting factor in G × E research, the risk-profiling framework is likely to be more widely applied and allows statistical power to be leveraged from larger samples not measured for the environment. In the risk-profiling framework, identification of true interaction effects largely depends on the prediction accuracy of the genetic and environmental factors. Prediction accuracy of GRPS is driven by precision with which the individual SNP effects are estimated in the ‘discovery sample’ in which large sample size generates higher precision. Combined efforts in the psychiatric genetics community (PGC) have achieved 34 241 schizophrenia cases, and 45 604 controls in 2014 (2014) resulting in high precision of the estimation of individual SNP effects and consequently large prediction accuracy of the GRPS. Precision of the environmental measures largely depends on the measure of interest. By nature, some environmental variables are measured with greater precision (e.g., migrant status) than others (e.g., age of first cannabis use). Large discovery sample sizes and well-defined environmental variables in the target samples will increase precision and prediction accuracy in the risk-profiling framework. Interpretation of results must consider the likely representation of unmeasured environmental risk factors in the discovery sample.
In both frameworks, the genetic architecture underlying the disease is a determinant in the statistical power of a study, this factor is however beyond our control. For example, larger sample sizes are required when common SNPs explain less variance (i.e., SNP heritability is low), which can be due to, for example, common SNPs being not in sufficient linkage disequilibrium (LD) with the causal variants or total heritability being low. In the risk-profiling framework, prediction efficacy (e.g., the amount of variance that can be explained by the GRPS) depends on the sample size of the discovery sample whereas the ability to detect variance explained that is significantly larger than zero depends on the size of the target sample. The same applies to variance explained by G × E. When sufficient samples are available and the researcher can apply both methods, the mixed linear model framework is to be preferred. It estimates the variance explained directly from individual-level genotype data, accounting for the correlation structure between the SNPs.
Multi-locus G × E success
G × E studies that estimate genetic risk from genome-wide genotypes are in their early days since there are few data sets of sufficient size informative for both genetic and environmental factors. Much larger sample sizes, however, are expected to become available in the coming years allowing application of both frameworks to a wide variety of psychiatric diseases, either with direct measures of genetic risk and environment or through proxy-measures of both entities.
Using genetic summary statistics on alcohol problems in young adults from the Avon Longitudinal Study of Parents and Children (ALSPAC, n = 4304 individuals), Salvatore et al. (2014) show an association between the derived GRPS and alcohol problems in adolescents in an independent population based Finnish sample (FinnTwinn12, n = 1162). They also demonstrated interaction between the GRPS and two environmental factors: parental knowledge and peer deviance. Genetic factors related to alcohol problems were more pronounced under conditions of low parental knowledge and high peer deviance.
When environmental measures are not available in a case-control sample, association between genetic factors underlying the disease and the potential environmental moderator can be studied in samples with healthy individuals. Power et al. (2014) used GRPS for schizophrenia risk (Ripke et al. 2013), to explore the genetic relationship between schizophrenia and cannabis use in a population sample in which <1% would be expected to have lifetime schizophrenia. Cannabis use is well established to be much higher among schizophrenic patients compared with the general population, causality and its direction, however, is still under debate (e.g., Ferdinand et al. 2005; Green et al. 2005; McGrath et al. 2010a; Kuepper et al. 2011). GRPS for schizophrenia were associated with cannabis use (ever v. never as well as quantity of use) in a sample of 2082 healthy individuals. This result does not exclude the possibility of a causal relationship but shows that at least part of the association between schizophrenia and cannabis use may be due to a shared genetic aetiology.
An approach to study G × E when measures on environmental risk factors are not directly available is to use epigenetic markers as proxies for the environment. Epigenetic markers associated with for example smoking behaviour (Shenker et al. 2013; Zeilinger et al. 2013) can be included as proxy-environmental moderators in the model both as a main effect and in an interaction term with genetic risk.
An example of a G × E study in a linear mixed model framework is a bivariate analysis of schizophrenia in which the two traits represent two different populations: European and African descent (de Candia et al. 2013). The genetic correlation derived from SNP similarity within and between populations was estimated at 0.66 (s.e. = 0.23) and was significantly different from zero but not from one. The results were not suggestive of G × E interaction and suggested that many schizophrenia risk alleles are shared across ethnic groups.
Conclusion
The goal of G × E research now and in the near future is the identification of novel genetic pathways that do not have marginal effects and the discovery of environmental risk factors that affect only a subpopulation of genetically susceptible individuals. Increasing sample sizes in psychiatric genetics research are starting to show that genetic risk predictors could have utility for stratification of individuals into high- and low-risk groups for developing disease (PGC–SCZ, 2014). Augmenting these genetic risk predictors with environmental moderators should increase prediction accuracy. Diagnostic use of multi-locus genetic risk predictors is a long-term goal that might come closer once informed by environmental predictors. Real progress in G × E research requires concerted effort of collection of informative data sets.
Acknowledgements
None.
Financial Support
AV is supported by an NHMRC John Cade Fellowship awarded to John McGrath (1056929). NW is supported by NHMRC 613602 and 1047956.
Statement of Interest
The authors declare no conflict of interest.
Ethical Standards
None.
References
- Ripke Stephan, Neale Benjamin M, Corvin Aiden, Walters James TR, Farh Kai-How, Holmans Peter A, Lee Phil, Bulik-Sullivan Brendan, Collier David A, Huang Hailiang, Pers Tune H, Agartz Ingrid, Agerbo Esben, Albus Margot, Alexander Madeline, Amin Farooq, Bacanu Silviu A, Begemann Martin, Belliveau Richard A Jr, Bene Judit, Bergen Sarah E, Bevilacqua Elizabeth, Bigdeli Tim B, Black Donald W, Bruggeman Richard, Buccola Nancy G, Buckner Randy L, Byerley William, Cahn Wiepke, Cai Guiqing, Campion Dominique, Cantor Rita M, Carr Vaughan J, Carrera Noa, Catts Stanley V, Chambert Kimberly D, Chan Raymond CK, Chen Ronald YL, Chen Eric YH, Cheng Wei, Cheung Eric FC, Chong Siow Ann, Cloninger C Robert, Cohen David, Cohen Nadine, Cormican Paul, Craddock Nick, Crowley James J, Curtis David, Davidson Michael, Davis Kenneth L, Degenhardt Franziska, Del Favero Jurgen, Demontis Ditte, Dikeos Dimitris, Dinan Timothy, Djurovic Srdjan, Donohoe Gary, Drapeau Elodie, Duan Jubao, Dudbridge Frank, Durmishi Naser, Eichhammer Peter, Eriksson Johan, Escott-Price Valentina, Essioux Laurent, Fanous Ayman H, Farrell Martilias S, Frank Josef, Franke Lude, Freedman Robert, Freimer Nelson B, Friedl Marion, Friedman Joseph I, Fromer Menachem, Genovese Giulio, Georgieva Lyudmila, Giegling Ina, Giusti-Rodríguez Paola, Godard Stephanie, Goldstein Jacqueline I, Golimbet Vera, Gopal Srihari, Gratten Jacob, Haan de Lieuwe, Hammer Christian, Hamshere Marian L, Hansen Mark, Hansen Thomas, Haroutunian Vahram, Hartmann Annette M, Henskens Frans A, Herms Stefan, Hirschhorn Joel N, Hoffmann Per, Hofman Andrea, Hollegaard Mads V, Hougaard David M, Ikeda Masashi, Joa Inge, Julià Antonio, Kahn René S, Kalaydjieva Luba, Karachanak-Yankova Sena, Karjalainen Juha, Kavanagh David, Keller Matthew C, Kennedy James L, Khrunin Andrey, Kim Yunjung, Klovins Janis, Knowles James A, Konte Bettina, Kucinskas Vaidutis, Kucinskiene Zita Ausrele, Kuzelova-Ptackova Hana, Kähler Anna K, Laurent Claudine, Keong Jimmy Lee Chee, Lee S Hong, Legge Sophie E, Lerer Bernard, Li Miaoxin, Li Tao, Liang Kung-Yee, Lieberman Jeffrey, Limborska Svetlana, Loughland Carmel M, Lubinski Jan, Lönnqvist Jouko, Macek Milan Jr, Magnusson Patrik KE, Maher Brion S, Maier Wolfgang, Mallet Jacques, Marsal Sara, Mattheisen Manuel, Mattingsdal Morten, McCarley Robert W, McDonald Colm, McIntosh Andrew M, Meier Sandra, Meijer Carin J, Melegh Bela, Melle Ingrid, Mesholam-Gately Raquelle I, Metspalu Andres, Michie Patricia T, Milani Lili, Milanova Vihra, Mokrab Younes, Morris Derek W, Mors Ole, Murphy Kieran C, Murray Robin M, Myin-Germeys Inez, Müller-Myhsok Bertram, Nelis Mari, Nenadic Igor, Nertney Deborah A, Nestadt Gerald, Nicodemus Kristin K, Nikitina-Zake Liene, Nisenbaum Laura, Nordin Annelie, O'Callaghan Eadbhard, O'Dushlaine Colm, O'Neill F. Anthony, Oh Sang-Yun, Olincy Ann, Olsen Line, Van Os Jim, Psychosis Endophenotypes International Consortium, Pantelis Christos, Papadimitriou George N, Papiol Sergi, Parkhomenko Elena, Pato Michele T, Paunio Tiina, Pejovic-Milovancevic Milica, Perkins Diana O, Pietiläinen Olli, Pimm Jonathan, Pocklington Andrew J, Powell John, Price Alkes, Pulver Ann E, Purcell Shaun M, Quested Digby, Rasmussen Henrik B, Reichenberg Abraham, Reimers Mark A, Richards Alexander L, Roffman Joshua L, Roussos Panos, Ruderfer Douglas M, Salomaa Veikko, Sanders Alan R, Schall Ulrich, Schubert Christian R, Schulze Thomas G, Schwab Sibylle G, Scolnick Edward M, Scott Rodney J, Seidman Larry J, Shi Jianxin, Sigurdsson Engilbert, Silagadze Teimuraz, Silverman Jeremy M, Sim Kang, Slominsky Petr, Smoller Jordan W, So Hon-Cheong, Spencer ChrisCA, Stahl Eli A, Stefansson Hreinn, Steinberg Stacy, Stogmann Elisabeth, Straub Richard E, Strengman Eric, Strohmaier Jana, Stroup T Scott, Subramaniam Mythily, Suvisaari Jaana, Svrakic Dragan M, Szatkiewicz Jin P, Söderman Erik, Thirumalai Srinivas, Toncheva Draga, Tosato Sarah, Veijola Juha, Waddington John, Walsh Dermot, Wang Dai, Wang Qiang, Webb Bradley T, Weiser Mark, Wildenauer Dieter B, Williams Nigel M, Williams Stephanie, Witt Stephanie H, Wolen Aaron R, Wong Emily HM, Wormley Brandon K, Xi Hualin Simon, Zai Clement C, Zheng Xuebin, Zimprich Fritz, Wray Naomi R, Stefansson Kari, Visscher Peter M, Wellcome Trust Case-Control Consortium, Adolfsson Rolf, Andreassen Ole A, Blackwood Douglas HR, Bramon Elvira, Buxbaum Joseph D, Børglum Anders D, Cichon Sven, Darvasi Ariel, Domenici Enrico, Ehrenreich Hannelore, Esko Tõnu, Gejman Pablo V, Gill Michael, Gurling Hugh, Hultman Christina M, Iwata Nakao, Jablensky Assen V, Jönsson Erik G, Kendler Kenneth S, Kirov George, Knight Jo, Lencz Todd, Levinson Douglas F, Li Qingqin S, Liu Jianjun, Malhotra Anil K, McCarroll Steven A, McQuillin Andrew, Moran Jennifer L, Mortensen Preben B, Mowry Bryan J, Nöthen Markus M, Ophoff Roel A, Owen Michael J, Palotie Aarno, Pato Carlos N, Petryshen Tracey L, Posthuma Danielle, Rietschel Marcella, Riley Brien P, Rujescu Dan, Sham Pak C, Sklar Pamela, Clair David St, Weinberger Daniel R, Wendland Jens R, Werge Thomas, Daly Mark J, Sullivan Patrick F & O'Donovan Michael C (2014). Biological insights from 108 schizophrenia-associated genetic loci. Nature 511, 421–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dealberto MJ (2010). Ethnic origin and increased risk for schizophrenia in immigrants to countries of recent and longstanding immigration. Acta Psychiatrica Scandinavica 121, 325–339. [DOI] [PubMed] [Google Scholar]
- de Candia TR, Lee SH, Yang J, Browning BL, Gejman PV, Levinson DF, Mowry BJ, Hewitt JK, Goddard ME, O'Donovan MC, Purcell SM, Posthuma D, Visscher PM, Wray NR and Keller MC (2013). Additive genetic variation in schizophrenia risk is shared by populations of African and European descent. American Journal of Human Genetics 93, 463–470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudbridge F (2013). Power and predictive accuracy of polygenic risk scores. PLoS Genetics 9, e1003348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan LE, Keller MC (2011). A critical review of the first 10 years of candidate gene-by-environment interaction research in psychiatry. American Journal of Psychiatry 168, 1041–1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falconer DF, Mackay TFC (1996). Introduction to Quantitative Genetics. Longman: Essex. [Google Scholar]
- Falconer DS, Latyszewski M (1952). The environment in relation to selection for size in mice. Journal of Genetics 51, 67–80. [Google Scholar]
- Ferdinand RF, Sondeijker F, van der Ende J, Selten JP, Huizink A and Verhulst FC (2005). Cannabis use predicts future psychotic symptoms, and vice versa. Addiction 100, 612–618. [DOI] [PubMed] [Google Scholar]
- Green B, Young R and Kavanagh D (2005). Cannabis use and misuse prevalence among people with psychosis. British Journal of Psychiatry: the Journal of Mental Science 187, 306–313. [DOI] [PubMed] [Google Scholar]
- Hamilton C, Tabitha P (2014). PhenX. Available from: https://http://www.phenxtoolkit.org/
- Kuepper R, van Os J., Lieb R, Wittchen HU, Hofler M and Henquet C (2011). Continued cannabis use and risk of incidence and persistence of psychotic symptoms: 10 year follow-up cohort study. BMJ 342, d738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch M, Walsh B. (1998). Genetics and Analysis of Quantitative Traits. Sinauer Associates, Inc: Sunderland, Massachusetts, USA. [Google Scholar]
- Manolio TA, Collins FS, Cox NJ, Goldstein DB, Hindorff LA, Hunter DJ, McCarthy MI, Ramos EM, Cardon LR, Chakravarti A, Cho JH, Guttmacher AE, Kong A, Kruglyak L, Mardis E, Rotimi CN, Slatkin M, Valle D, Whittemore AS, Boehnke M, Clark AG, Eichler EE, Gibson G, Haines JL, Mackay TF, McCarroll SA and Visscher PM (2009). Finding the missing heritability of complex diseases. Nature 461, 747–753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGrath J, Welham J, Scott J, Varghese D, Degenhardt L, Hayatbakhsh MR, Alati R, Williams GM, Bor W and Najman JM (2010a). Association between cannabis use and psychosis-related outcomes using sibling pair analysis in a cohort of young adults. Archives of General Psychiatry 67, 440–447. [DOI] [PubMed] [Google Scholar]
- McGrath JJ, Eyles DW, Pedersen CB, Anderson C, Ko P, Burne TH, Norgaard-Pedersen B, Hougaard DM and Mortensen PB (2010b). Neonatal vitamin D status and risk of schizophrenia: a population-based case-control study. Archives of General Psychiatry 67, 889–894. [DOI] [PubMed] [Google Scholar]
- Modinos G, Iyegbe C, Prata D, Rivera M, Kempton MJ, Valmaggia LR, Sham PC, van Os J. and McGuire P (2013). Molecular genetic gene–environment studies using candidate genes in schizophrenia: a systematic review. Schizophrenia Research 150, 356–365. [DOI] [PubMed] [Google Scholar]
- Power RA, Verweij KJ, Zuhair M, Montgomery GW, Henders AK, Heath AC, Madden PA, Medland SE, Wray NR and Martin NG (2014). Genetic predisposition to schizophrenia associated with increased use of cannabis. Molecular Psychiatry, doi:10.1038/mp.2014.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ and Sham PC (2007). PLINK: a tool set for whole-genome association and population-based linkage analyses. American Journal of Human Genetics 81, 559–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell SM, Wray NR, Stone JL, Visscher PM, O'Donovan MC, Sullivan PF and Sklar P (2009). Common polygenic variation contributes to risk of schizophrenia and bipolar disorder. Nature 460, 748–752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell SM, Moran JL, Fromer M, Ruderfer D, Solovieff N, Roussos P, O'Dushlaine C, Chambert K, Bergen SE, Kahler A, Duncan L, Stahl E, Genovese G, Fernandez E, Collins MO, Komiyama NH, Choudhary JS, Magnusson PK, Banks E, Shakir K, Garimella K, Fennell T, DePristo M, Grant SG, Haggarty SJ, Gabriel S, Scolnick EM, Lander ES, Hultman CM, Sullivan PF, McCarroll SA and Sklar P (2014). A polygenic burden of rare disruptive mutations in schizophrenia. Nature 506, 185–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ripke S, O'Dushlaine C, Chambert K, Moran JL, Kahler AK, Akterin S, Bergen SE, Collins AL, Crowley JJ, Fromer M, Kim Y, Lee SH, Magnusson PK, Sanchez N, Stahl EA, Williams S, Wray NR, Xia K, Bettella F, Borglum AD, Bulik-Sullivan BK, Cormican P, Craddock N, de Leeuw C, Durmishi N, Gill M, Golimbet V, Hamshere ML, Holmans P, Hougaard DM, Kendler KS, Lin K, Morris DW, Mors O, Mortensen PB, Neale BM, O'Neill FA, Owen MJ, Milovancevic MP, Posthuma D, Powell J, Richards AL, Riley BP, Ruderfer D, Rujescu D, Sigurdsson E, Silagadze T, Smit AB, Stefansson H, Steinberg S, Suvisaari J, Tosato S, Verhage M, Walters JT, Levinson DF, Gejman PV, Laurent C, Mowry BJ, O'Donovan MC, Pulver AE, Schwab SG, Wildenauer DB, Dudbridge F, Shi J, Albus M, Alexander M, Campion D, Cohen D, Dikeos D, Duan J, Eichhammer P, Godard S, Hansen M, Lerer FB, Liang KY, Maier W, Mallet J, Nertney DA, Nestadt G, Norton N, Papadimitriou GN, Ribble R, Sanders AR, Silverman JM, Walsh D, Williams NM, Wormley B, Arranz MJ, Bakker S, Bender S, Bramon E, Collier D, Crespo-Facorro B, Hall J, Iyegbe C, Jablensky A, Kahn RS, Kalaydjieva L, Lawrie S, Lewis CM, Linszen DH, Mata I, McIntosh A, Murray RM, Ophoff RA, Van Os J., Walshe M, Weisbrod M, Wiersma D, Donnelly P, Barroso I, Blackwell JM, Brown MA, Casas JP, Corvin AP, Deloukas P, Duncanson A, Jankowski J, Markus HS, Mathew CG, Palmer CN, Plomin R, Rautanen A, Sawcer SJ, Trembath RC, Viswanathan AC, Wood NW, Spencer CC, Band G, Bellenguez C, Freeman C, Hellenthal G, Giannoulatou E, Pirinen M, Pearson RD, Strange A, Su Z, Vukcevic D, Langford C, Hunt SE, Edkins S, Gwilliam R, Blackburn H, Bumpstead SJ, Dronov S, Gillman M, Gray E, Hammond N, Jayakumar A, McCann OT, Liddle J, Potter SC, Ravindrarajah R, Ricketts M, Tashakkori-Ghanbaria A, Waller MJ, Weston P, Widaa S, Whittaker P, McCarthy MI, Stefansson K, Scolnick E, Purcell S, McCarroll SA, Sklar P, Hultman CM and Sullivan PF (2013). Genome-wide association analysis identifies 13 new risk loci for schizophrenia. Nature Genetics 45, 1150–1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salvatore JE, Aliev F, Edwards AC, Evans DM, Macleod J, Hickman M, Lewis G, Kendler KS, Loukola A, Korhonen T, Latvala A, Rose RJ, Kaprio J and Dick DM (2014). Polygenic scores predict alcohol problems in an independent sample and show moderation by the environment. Genes 5, 330–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shenker NS, Polidoro S, van Veldhoven K., Sacerdote C, Ricceri F, Birrell MA, Belvisi MG, Brown R, Vineis P and Flanagan JM (2013). Epigenome-wide association study in the European Prospective Investigation into Cancer and Nutrition (EPIC-Turin) identifies novel genetic loci associated with smoking. Human Molecular Genetics 22, 843–851. [DOI] [PubMed] [Google Scholar]
- Vassos E, Pedersen CB, Murray RM, Collier DA and Lewis CM (2012). Meta-analysis of the association of urbanicity with schizophrenia. Schizophrenia Bulletin 38, 1118–1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visscher PM, Hemani G, et al. (2014). Statistical power to detect genetic (co)variance of complex traits using SNP data in unrelated samples. PLoS Genetics 10, e1004269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Benyamin B, McEvoy BP, Gordon S, Henders AK, Nyholt DR, Madden PA, Heath AC, Martin NG, Montgomery GW, Goddard ME and Visscher PM (2010). Common SNPs explain a large proportion of the heritability for human height. Nature Genetics 42, 565–569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Lee SH, Goddard ME and Visscher PM (2011). GCTA: a tool for genome-wide complex trait analysis. American Journal of Human Genetics 88, 76–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Zaitlen NA, et al. (2014). Advantages and pitfalls in the application of mixed-model association methods. Nature Genetics 46, 100–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zammit S, Lewis G, Dalman C and Allebeck P (2010). Examining interactions between risk factors for psychosis. British Journal of Psychiatry: the Journal of Mental Science 197, 207–211. [DOI] [PubMed] [Google Scholar]
- Zeilinger S, Kuhnel B, Klopp N, Baurecht H, Kleinschmidt A, Gieger C, Weidinger S, Lattka E, Adamski J, Peters A, Strauch K, Waldenberger M and Illig T (2013). Tobacco smoking leads to extensive genome-wide changes in DNA methylation. PLoS ONE 8, e63812. [DOI] [PMC free article] [PubMed] [Google Scholar]