Abstract
Background
Diabetes mellitus is the most common chronic endocrine disorder, affecting an estimated population of 382 million people worldwide. It is associated with microvascular and macrovascular complications, including diabetic nephropathy (DN); primary cause of end-stage renal disease. Different inflammatory and angiogenic molecules in various pathways are important modulators in the pathogenesis and progression of diabetic nephropathy. Differential disease risk in DN may be partly attributable to genetic susceptibility. In this meta-analysis, we aimed to determine which of the previously investigated genetic variants in these pathways are significantly associated with the development of DN and to examine the functional role of these genes.
Methods
A systematic search was conducted to collect and analyze all studies published till June 2013; that investigated the association between genetic variants involved in inflammatory cytokines and angiogenesis and diabetic nephropathy. Genetic variants associated with DN were selected and analyzed by using Comprehensive Meta Analysis software. Pathway analysis of the genes with variants showing significant positive association with DN was performed using Genomatix Genome Analyzer (Genomatix, Munich, Germany).
Results
After the inclusion and exclusion criteria for this analysis, 34 studies were included in this meta-analysis. 11 genetic variants showed significant positive association with DN in a random-effects meta-analysis. These included genetic variants within or near VEGFA, CCR5, CCL2, IL-1, MMP9, EPO, IL-8, ADIPOQ and IL-10. rs1800871 (T) genetic variant in IL-10 showed protective effect for DN. Most of these eleven genetic variants were involved in GPCR signaling and receptor binding pathways whereas four were involved in chronic kidney failure. rs833061 [OR 2.08 (95% CI 1.63-2.66)] in the VEGFA gene and rs3917887 [OR 2.04 (95% CI 1.64-2.54)] in the CCL2 gene showed the most significant association with the risk of diabetic nephropathy.
Conclusions
Our results indicate that 11 genetic variants within or near VEGFA, CCR5, CCL2, IL-1, MMP9, EPO, IL-8, ADIPOQ and IL-10 showed significant positive association with diabetic nephropathy. Gene Ontology or pathway analysis showed that these genes may contribute to the pathophysiology of DN. The functional relevance of the variants and their pathways can lead to increased biological insights and development of new therapeutic targets.
Electronic supplementary material
The online version of this article (doi:10.1186/s12881-014-0103-8) contains supplementary material, which is available to authorized users.
Keywords: Diabetic nephropathy, Inflammatory cytokines, Angiogenesis, Genetic variants, Meta-analysis, Pathways, SNP
Background
Diabetes mellitus (DM) is a set of metabolic disorders characterized by hyperglycemia resulting from defects in insulin secretion and/or action. The prevalence of the disease, which is becoming a major world-wide health problem, is increasing rapidly [1]. In 2013, 382 million cases of DM worldwide were estimated, and that number is expected to increase to 592 million cases in 2035 [1]. Diabetes mellitus is associated with microvascular and macrovascular complications, including diabetic nephropathy (DN), a primary cause of end-stage renal disease (ESRD) [2]. Due to the global increase in prevalence of diabetes there has been a concomitant rise in the number of patients with diabetic nephropathy (DN) indicating a prevalence of 30-40% of the patients with type 1 (T1DM) and type 2 diabetes (T2DM) being affected [3].
Mostly, individuals with long durations of diabetes and poor glycemic control develop progressive diabetic nephropathy. However, some patients appear to be at increased risk while others remain relatively protected [4]. Genetic predisposition plays an important role in the risk to developing diabetic nephropathy though the incidence and severity is affected by the extent of control of the abnormal metabolic state associated with diabetes mellitus [5].
In recent years, there has been an increased understanding of the genetic and molecular basis of development and progression of diabetic nephropathy. Although diabetic nephropathy is traditionally considered a nonimmune disease, recent findings indicate a significant role of immune-mediated inflammatory processes in the pathophysiology of diabetic nephropathy. These include the up-regulation of inflammatory mediators, higher urinary levels of monocyte chemoattractant protein-1 and increasing macrophage influx with the progression of diabetic nephropathy [6–8]. In streptozotocin (STZ) diabetic rats, an increase in glomeruli capillaries area was observed in comparison to normal age-matched controls, implying a pronounced angiogenic reaction to the diabetic condition [9]. Higher concentration of angiogenic markers, vascular endothelial growth factor (VEGF) and transforming growth factor β (TGF β) in plasma and urine of diabetic nephropathy patients, emphasize the role of angiogenesis [10,11]. Chen and Ziyadeh, [12] reported attenuation of diabetic albuminaria by blockade of VEGF signaling pathway. Further, a recent study showed anti-angiogenic and anti-inflammatory DNA vaccination ameliorates the progression of glomerular pathology in an animal model of diabetic nephropathy [13]. As inflammation and angiogenesis play a crucial role in the pathomechanism of diabetic nephropathy, many studies have investigated the association of genetic polymorphism in these pathway genes with the risk of diabetic nephropathy. However, there is a lack of consistency among the reported studies due to small sample size, limited power and sparseness of data. Therefore, the aim of this meta-analysis is to analyze all published studies that investigated the association of genetic variants involved in inflammatory cytokines and angiogenesis with diabetic nephropathy and to assess their role in different biochemical pathways.
Methods
Literature search strategy
The articles relevant to this study were searched from PubMed, Embase and Cochrane Library in June, 2013. Different possible variations and combinations of the following search terms were used: ‘diabetes mellitus’, ‘diabetic nephropathy’, ‘ESRD’, ‘inflammatory cytokines’, ‘angiogenesis’, ‘genetic variants’, ‘polymorphism’, ‘SNPs’ and ‘gene’. Additional search query was used by combining the names of the specific genes and genetic variants with the term ‘diabetic nephropathy’. The reference list of each relevant publication was also examined to identify additional studies appropriate for inclusion in the meta-analysis. In the initial search no filter for language preference was used.
Inclusion and exclusion criteria
Only those studies were included in which the cases had diabetes mellitus with macroalbuminuria, overt proteinuria, ESRD due to diabetic nephropathy or diabetic nephropathy identified by biopsy and controls had diabetic mellitus with normoalbuminaria after > 10 years of diabetes duration. Cases with macroalbuminaria and/or overt proteinuria and/or ESRD were merged together for comparison with controls. The selected studies should provide detailed genotypic data of the genes in the inflammatory and angiogenic pathways. Only articles in English language were included. The cases and controls in the studies included were matched by duration of diabetes and/or age. Lack of information related to the distribution of genotypes or alleles within the case and control groups is one of our exclusion criteria. Studies in which the controls were non-diabetic and/or there was comparison of different stages of diabetic nephropathy were excluded. Review and meta-analysis articles were excluded.
Data extraction and Statistical analysis
Following information was extracted from each selected study like PMID No, first author’s surname, journal & year of publication, sample size, ethnicity, methodology, type and duration of diabetes, criteria for diabetic nephropathy and frequency of alleles.
For each SNP, the allelic data from different studies was pooled and minor allele frequencies were calculated and compared between cases and controls. Data were entered into a database and a statistical analysis was performed using SPSS (version 17.0; SPSS, Chicago, IL). The pooled odds ratio was used to estimate the association between the genetic variants and diabetic nephropathy in this meta-analysis. The Odds Ratio (OR), at allele level and statistically significant P-values were validated for all the studies and recalculated for some of them, in order to remove any adjustments made within each study in cases where two different groups merged together, such as in the case of macroalbuminaria and ESRD groups. A p-value <0.05 was considered to be statistically significant. Deviation from Hardy–Weinberg equilibrium (HWE) was tested using the De Finetti program (http://ihg.gsf.de/cgi-bin/hw/hwa1.pl) for all the studies. The percentage of variation across studies due to heterogeneity rather than chance was estimated via Higgins I2 statistics using Comprehensive Meta Analysis Version 2.0 software (Biostat, Englewood NJ, 2005). I2 is estimated by the ratio (Q-df)/Q, where Q is the Cochran’s Q statistic and df is degrees of freedom. I2 lies between 0 and 100% with values over 50% indicating high heterogeneity [14]. The random-effects model was performed using Comprehensive Meta Analysis Version 2.0 software. The random effects model assumes that there is a different underlying effect size for each study. Random effects model takes into account the diversity between the studies and is preferred in majority of genetic association studies. Random effects model is generally used in the presence or anticipation of any heterogeneity between studies [15]. The sub-group analysis was performed for diabetes mellitus type (type 1 or type 2), diabetic nephropathy stage (established diabetic nephropathy or advanced diabetic nephropathy) and ethnicity (European or Asian origin). The genes showing association with diabetic nephropathy were used as input core data for Genomatix Pathway Analysis (Genomatix, Munich, Germany).
Results
The initial literature search yielded 751 citations with 111 involving inflammatory cytokines and angiogenesis related to diabetic nephropathy in humans. 25 research articles representing 34 different studies were selected after exclusion of studies which included progression of nephropathy, meta-analysis, reviews etc. The selected 34 studies contained 55 genetic variants in 18 genes of inflammatory cytokines and angiogenesis which were associated with diabetic nephropathy. Table 1 includes the details and references of all the studies used in this meta-analysis. The detailed information of the studied SNPs and the corresponding pooled odds ratios and p-values are presented in Additional file 1a, b, c, d and e.
Table 1.
Details of the genes and studies in this meta-analysis study
| Gene | SNP | Article | No. of studies | Population | Case definition | T1D/T2D | OR a |
|---|---|---|---|---|---|---|---|
| CCR5 (Chemokine Receptor 5) | rs7637813 | Pettigrew et al, [16] | 1 | Irish | EDNb | 1 | 1.173(0.932-1.478) |
| 10577983 | Pettigrew et al, [16] | 1 | 1.163(0.935-1.447) | ||||
| rs2227010 | Pettigrew et al, [16] | 1 | 1.0(0.805-1.248) | ||||
| rs17765882 | Pettigrew et al, [16] | 1 | 1.501(0.980-2.298) | ||||
| rs2734648 | Tregouet et al, [17] | 3 | Danish | EDN | 1 | 0.969(0.788-1.191) | |
| Tregouet et al, [17] | Finnish | EDN | 1 | 1.186(0.947-1.486) | |||
| Tregouet et al, [17] | French | EDN | 1 | 1.082(0.853-1.373) | |||
| Del 32/rs333 | Ahluwalia et al, [18] | 4 | North Indian | EDN | 2 | 2.58 (1.98–3.37) | |
| Ahluwalia et al, [18] | South Indian | EDN | 2 | 0.88 (0.51–1.5) | |||
| Pettigrew et al, [16] | Irish | EDN | 1 | 1.242(0.859-1.794) | |||
| Mlynarski et al, [19] | American | ADNc | 1 | 0.959(0 .652 1.410) | |||
| 59029 G.A/rs1799987 | Pettigrew et al, [16] | 10 | Irish | EDN | 1 | 1.017(0.818-1.263) | |
| Ahluwalia et al, [18] | North Indian | EDN | 2 | 2.22 (1.71–2.87) | |||
| Ahluwalia et al, [18] | South Indian | EDN | 2 | 2.17 (1.43–3.29) | |||
| Buraczynska et al, [20] | Polish | EDN | 2 | 1.83(1.43-2.34) | |||
| Mlynarski et al, [19] | American | ADN | 1 | 0.904(0.664- 1.23) | |||
| Nakajima et al, [21] | Japanese | EDN | 2 | 1.179(0.855-1.627) | |||
| Tregouet et al, [17] | Danish | EDN | 1 | 1.112(0.918-1.348) | |||
| Tregouet et al, [17] | Finnish | EDN | 1 | 1.152(0.938-1.415) | |||
| Tregouet et al, [17] | French | EDN | 1 | 1.063(0.845-1.336) | |||
| Prasad et al, [22] | Indian | ADN | 2 | 1.379(1.047-1.818) | |||
| ADIPOQ (Adiponectin, C1Q And Collagen Domain Containing) | rs266729 | Zhang et al, [23] | 2 | European | EDN + ADN | 1 | 1.120(0.929-1.35) |
| Wu et al, [24] | Taiwanese | EDN | 2 | 1.39(1.0-1.9) | |||
| rs17300539 | Vionnet et al, [25] | 4 | Danish | EDN and Retd | 1 | 1.348(0.965-1.884) | |
| Vionnet et al, [25] | Finnish | EDN and Ret | 1 | 1(0.570-1.754) | |||
| Vionnet et al, [25] | French | EDN and Ret | 1 | 1.472(0.998-2.171) | |||
| Prior et al, [26] | British | EDN and Ret | 1 | 1.924 (1.024-3.617) | |||
| IL8 (Interleukin-8) | rs4073 | Ahluwalia et al, [18] | 2 | North Indian | EDN | 2 | 1.44 (1.1–1.88) |
| Ahluwalia et al, [18] | South Indian | EDN | 2 | 1.5 (0.96–2.33) | |||
| CCL2 (Chemokine ligand 2) | rs1024611 | Ahluwalia et al, [18] | 2 | North Indian | EDN | 2 | 1.04 (0.8–1.36) |
| Joo et al, [27] | Korean | ADN | 2 | 0.91(0.65-1.2) | |||
| rs3917887 | Ahluwalia et al, [18] | 2 | North Indian | EDN | 2 | 2.03 (1.57–2.63) | |
| Ahluwalia et al, [18] | South Indian | EDN | 2 | 1.7 (1.12–2.57) | |||
| MMP9 (Matrix Metallopeptidase 9) | rs17576 | Ahluwalia et al, [18] | 2 | North Indian | EDN | 2 | 1.81 (1.40–2.34) |
| Ahluwalia et al, [18] | South Indian | EDN | 2 | 2.19 (2.45–3.31) | |||
| IL-10 (Interleukin-10) | –592C/A/rs1800872 | Arababadi et al, [28] | 2 | Iranian | EDN | 2 | 1.484(0.944-2.331) |
| Ezzidi et al, [2]/ Mtiraoui et al, [29] | Tunisian | EDN | 2 | 0.915(0.751-1.114) | |||
| –819C/T/rs1800871 | Ezzidi et al, [2]/ Mtiraoui et al, [29] | 1 | 0.777(0.631-0.958) | ||||
| –1082G/A/rs1800896 | Ezzidi et al, [2]/ Mtiraoui et al, [29] | 1 | 0.841(0.694-1.018) | ||||
| IL-1 (Interleukin-1) | IL1A | Loughrey et al, [30] | 1 | Caucasian | EDN | 1 | 0.66 (0.42-1.04) |
| IL1B | Loughrey et al, [30] | 1 | 1.971(1.221-3.182) | ||||
| IL1RI | Loughrey et al, [30] | 1 | 0.94(0.618-1.429) | ||||
| VEGFA (Vascular endothelial growth factor A) | - 2549 I/D/rs35569394 | Yang et al, [31] | 2 | British | EDN and Ret | 1 | 1.619(1.038-2.525) |
| Buraczynska et al, [32] | Polish | EDN | 2 | 1.09(0.77-1.54) | |||
| +405/rs2010963 | Buraczynska et al, [32] | 2 | Polish | EDN | 2 | 0.949(0.658-1.369) | |
| McKnight et al, [33] | Irish | EDN and Ret | 1 | 0.88 (0.69-1.12) | |||
| -1499C > T/rs833061 | McKnight et al, [33] | 2 | Irish | EDN and Ret | 1 | 2.08(1.63-2.66) | |
| -2578C > A/rs699947 | McKnight et al, [33] | 2 | Irish | EDN and Ret | 1 | 0.92(0.72-1.17) | |
| rs2146323 | Tregouet et al, [17] | 3 | Danish | EDN | 1 | 0.899(0.732-1.105) | |
| Tregouet et al, [17] | Finnish | EDN | 1 | 0.914(0.739-1.131) | |||
| Tregouet et al, [17] | French | EDN | 1 | 0.854(0.667-1.095) | |||
| rs3024997 | Tregouet et al, [17] | 3 | Danish | EDN | 1 | 0.967(0.786-1.191) | |
| Tregouet et al, [17] | Finnish | EDN | 1 | 0.986(0.773-1.257) | |||
| Tregouet et al, [17] | French | EDN | 1 | 1.124(0.879-1.438) | |||
| rs3025000 | Tregouet et al, [17] | 3 | Danish | EDN | 1 | 0.962(0.781-1.186) | |
| Tregouet et al, [17] | Finnish | EDN | 1 | 0.974(0.765-1.240) | |||
| Tregouet et al, [17] | French | EDN | 1 | 1.087(0.843-1.402) | |||
| 936C/T/rs3025039 | Kim et al, [34] | 2 | korean | EDN + ADN | 2 | 1.632(0.984-2.708) | |
| VEGFB (Vascular endothelial growth factor B) | rs12366035 | Tregouet et al, [17] | 3 | Danish | EDN | 1 | 0.784(0.633-0.971) |
| Tregouet et al, [17] | Finnish | EDN | 1 | 0.92(0.727-1.163) | |||
| Tregouet et al, [17] | French | EDN | 1 | 1.144(0.887-1.474) | |||
| VEGFC (Vascular endothelial growth factor C) | rs585706 | Tregouet et al, [17] | 3 | Danish | EDN | 1 | 0.8(0.562-1.139) |
| Tregouet et al, [17] | Finnish | EDN | 1 | 1.318(0.917-1.895) | |||
| Tregouet et al, [17] | French | EDN | 1 | 1.271(0.882-1.832) | |||
| IL-6 (Interleukin-6) | rs2069827 | Ng et al, [35] | 1 | Caucasian | EDN + ADN | 2 | 0.65(0.365-1.159) |
| rs1800796 | Ng et al, [35] | 0.935(0.588-1.487) | |||||
| rs1800795 | Ng et al, [35] | 0.829(0.623-1.104) | |||||
| rs2069837 | Ng et al, [35] | 0.865(0.54-1.384) | |||||
| rs2069840 | Ng et al, [35] | 0.916(0.686-1.225) | |||||
| rs2069861 | Ng et al, [35] | 1.505(0.882-2.569) | |||||
| TGF-B1 (Transforming growth factor beta 1) | rs1800470 | Jahromi et al, [36] | 8 | Caucasian | EDN | 1 | 1.06(0.64-1.7) |
| Ahluwalia et al, [18] | North Indian | EDN | 2 | 1.1 (0.83–1.44) | |||
| Ng et al, [37] | Caucasian | EDN + ADN | 1 | 0.923(0.722–1.180) | |||
| McKnight et al, [33] | Irish | EDN | 1 | 1.2(0.7-2.1) | |||
| Tregouet et al, [17] | Danish | EDN | 1 | 0.979(0.799-1.199) | |||
| Tregouet et al, [17] | Finnish | EDN | 1 | 1.068(0.852-1.338) | |||
| Tregouet et al, [17] | French | EDN | 1 | 1.034(0.816-1.311) | |||
| Salgado et al, [38] | Mexican | EDN + ADN | 2 | 1.30(0.99-1.71) | |||
| Tyr81His/rs111033611 | Ahluwalia et al, [18] | 1 | North Indian | EDN | 2 | 1.14 (0.53–2.46) | |
| 915 G > C/rs1800471 | Ng et al, [37] | 3 | Caucasian | EDN + ADN | 2 | 1.022(0.627–1.665) | |
| McKnight et al, [33] | Irish | EDN | 1 | 1.0(0.66-1.5) | |||
| Salgado et al, [38] | Mexican | EDN + ADN | 2 | 4.17(1.40-12.3) | |||
| -800 A > Grs1800468 | Ng et al, [37] | 4 | Caucasian | EDN + ADN | 2 | 1.178(0.762–1.821) | |
| McKnight et al, [33] | Irish | EDN | 1 | 0.75(0.45-1.2) | |||
| Prasad et al, [22] | Indian | ADN | 2 | 1.04(0.816-1.325) | |||
| Salgado et al, [38] | Mexican | EDN + ADN | 2 | 0.98(0.46-2.09) | |||
| rs1800469 | Ng et al, [37] | 3 | Caucasian | EDN + ADN | 2 | 1.088(0.841–1.407) | |
| McKnight et al, [33] | Irish | EDN | 1 | 0.94(0.58-1.5) | |||
| Prasad et al, [22] | Indian | ADN | 2 | 0.738(0.50-1.091) | |||
| rs1800472 | Ng et al, [37] | 1 | Caucasian | EDN + ADN | 2 | 1.168(0.639–2.135) | |
| rs2241717 | Tregouet et al, [17] | 3 | Danish | EDN | 1 | 0.957(0.787-1.164) | |
| Tregouet et al, [17] | Finnish | EDN | 1 | 1.068(0.864-1.322) | |||
| Tregouet et al, [17] | French | EDN | 1 | 0.917(0.729-1.155) | |||
| rs8179181 | Tregouet et al, [17] | 3 | Danish | EDN | 1 | 1.193(0.957-1.488) | |
| Tregouet et al, [17] | Finnish | EDN | 1 | 1.022(0.799-1.308) | |||
| Tregouet et al, [17] | French | EDN | 1 | 0.944(0.722-1.234) | |||
| TGF-BR1 (TGF beta receptor 1) | rs1571589 | Tregouet et al, [17] | 3 | Danish | EDN | 1 | 0.951(0.746-1.213) |
| Tregouet et al, [17] | Finnish | EDN | 1 | 1.111(0.836-1.476) | |||
| Tregouet et al, [17] | French | EDN | 1 | 1.018(0.766-1.352) | |||
| rs928180 | Tregouet et al, [17] | 3 | Danish | EDN | 1 | 1.362(0.988-1.877) | |
| Tregouet et al, [17] | Finnish | EDN | 1 | 0.92(0.67-1.263) | |||
| Tregouet et al, [17] | French | EDN | 1 | 0.901(0.608-1.334) | |||
| TGF-BR2 (TGF beta receptor 2) | 747C > G/rs11466531 | McKnight et al, [33] | 1 | Irish | EDN | 1 | 1.37(1.0-1.89) |
| 1149G > A/ss50394788 | McKnight et al, [33] | 1 | Irish | EDN | 1 | 2.432(0.81-7.303) | |
| CCR2 (chemokine receptor type 2) | G46295A/rs1799864 | Joo et al, [27] | korean | ADN | 2 | 0.91(0.65-1.2) | |
| rs1799865 | Prasad et al, [22] | 1 | Indian | ADN | 2 | 1.551(0.901-2.671) | |
| RANTES/CCL5 (Chemokine ligand 5) | C-28G/rs2280788 | Joo et al, [27] | 2 | korean | ADN | 2 | 0.97(0.65-1.4) |
| Nakajima et al, [21] | Japanese | EDN | 2 | 1.532(1.004-2.338) | |||
| ss161639200 | Pettigrew et al, [16] | 1 | irish | EDN | 1 | 1.140(0.821-1.584) | |
| rs9898132 | Pettigrew et al, [16] | 1 | Irish | EDN | 1 | 1.092(0.746-1.599) | |
| G-403A/rs2107538 | Joo et al, [27] | 3 | Korean | ADN | 2 | 0.8(0.59-1.0) | |
| Pettigrew et al, [16] | Irish | EDN | 1 | 1.085(0.81-1.452) | |||
| Nakajima et al, [21] | Japanese | EDN | 2 | 0.88(0.632-1.225) | |||
| EPO (Erythropoietin) | rs1617640 | Tong et al, [39] | 3 | American | EDN + ADN | 2 | 1.446(1.145-1.826) |
| Tong et al, [39] | 1 | 1.535(1.320-1.787) | |||||
| Tong et al, [39] | 1 | 1.382(1.050-1.820) | |||||
| TNFα (Tumor necrosis factor) | -308/rs1800629 | Prasad et al, [22] | 1 | Indian | ADN | 2 | 1.383(0.775-2.468) |
aOR, Odds ratio (95% CI); bEDN, established diabetic nephropathy; cADN, advanced diabetic nephropathy; dRet, retinopathy.
This meta- analysis included 55 SNPs in 18 genes of inflammatory cytokines and angiogenesis which were associated with diabetic nephropathy.
Of the 55 genetic variants involved in the inflammatory cytokines and angiogenesis, 11 genetic variants in or near 9 genes were significantly associated with diabetic nephropathy after random-effects meta-analysis. Figure 1 is a Forest plot representation of the significantly associated genetic variants with diabetic nephropathy- VEGFA, CCR5 (Chemokine Receptor 5), CCL2 (Chemokine ligand 2), IL-1 (Interleukin-1), MMP9 (Matrix Metallopeptidase 9), EPO (Erythropoietin), IL-8 (Interleukin-8), ADIPOQ (Adiponectin, C1Q And Collagen Domain Containing) and IL-10 (Interleukin-1). The odds ratios of the significant associations with diabetic nephropathy were between the range of 1.24 to 2.08 for increased risk effect. After meta-analysis, 2 genetic variants- rs833061 in the VEGFA gene and rs3917887 in the CCL2 gene showed the most significant association with risk of diabetic nephropathy. Genetic variant rs833061 was analyzed in 2 Irish studies resulting in a pooled odds ratio of 2.08 (95% CI 1.63-2.66). For rs3917887, the pooled OR was 2.04 (95% CI 1.64-2.54) obtained from 2 Indian studies. rs833061 showed significant association in type 2 diabetes mellitus whereas rs3917887 was studied in type 1 diabetes mellitus. Genetic variant rs1799987 in CCR5 was the most studied SNP with ten studies of which six were of caucasian and four Asian. Five studies involved type 1 diabetes mellitus while the other five included type 2 diabetes mellitus resulting in a pooled odds ratio of 1.29 (95% CI 1.20-1.38). Figures 2 and 3 show the association of variants with diabetic nephropathy among sub-groups. The significant association between rs1799987 and diabetic nephropathy was reproduced in type 2 diabetes mellitus, Asian, Caucasian and established diabetic nephropathy subgroups. This association was not significant in the type 1 diabetes mellitus and advanced diabetic nephropathy subgroups. Other significantly associated genetic variant, rs333 (Del32) in the CCR5 gene had a pooled odds ratio of 1.24 (95% CI 1.08-1.43) from four studies. Among these four studies, two Asian studies involved type 2 diabetes mellitus whereas two Caucasian studies were for type 1 diabetes mellitus. The association was reproduced in the subgroup analysis (Figure 2).
Figure 1.
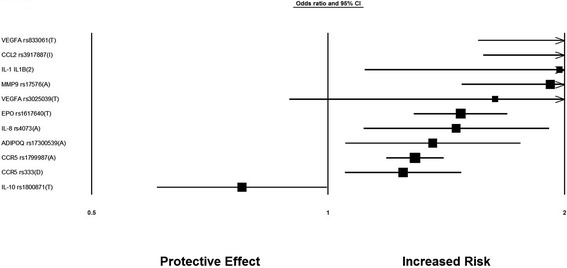
Forest Plot for genetic variants involved in inflammatory cytokines and angiogenesis and significantly associated with diabetic nephropathy after meta-analysis.
Figure 2.
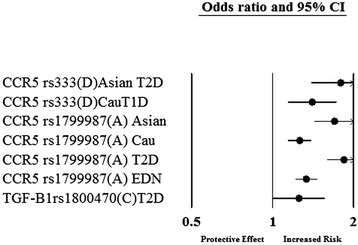
Genetic variants significantly associated with diabetic nephropathy after meta-analysis in a subgroup; T1D- type 1 diabetes mellitus, T2D- type 2 diabetes mellitus, Cau-Caucasian origin, EDN- established diabetic nephropathy.
Figure 3.
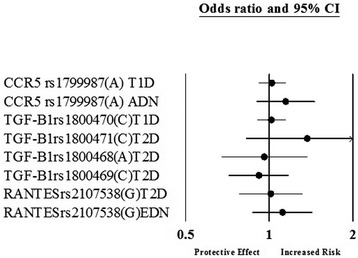
Genetic variants not significantly associated with diabetic nephropathy after meta-analysis in a subgroup; T1D- type 1 diabetes mellitus, T2D- type 2 diabetes mellitus, EDN- established diabetic nephropathy, ADN- advanced diabetic nephropathy.
The genetic variant rs17300539 in ADIPOQ gene was studied in four european studies, all involving type 1 diabetes mellitus. rs17300539 was significantly associated with diabetic nephropathy with a pooled odds ratio of 1.35 (95% CI 1.09-1.68). rs4073 SNP in IL-8 gene had a pooled OR of 1.45 (95% CI 1.16-1.82) from two Indian studies involving type 2 diabetes mellitus. In the same Indian studies, another genetic variant rs17576 in the MMP9 gene was significantly associated with diabetic nephropathy with a pooled OR of 1.91 (95% CI 1.54-2.38). The genetic variant rs3025039 in the VEGF gene was found to be significantly associated with diabetic nephropathy with a pooled OR of 1.63 (95% CI 0.98-2.70). Two studies in the Korean population investigated rs3025039 in type 2 diabetic patients. The genetic variant rs1617640 in the EPO gene was investigated in three American studies in which one study included type 2 diabetes mellitus whereas other two were for type 1 diabetes mellitus. After meta-analysis, the pooled OR of 1.47 (95% CI 1.31-1.65) showed a significant association with diabetic nephropathy. There was only one study in type 1 diabetes Caucasian population for the SNP IL1B in the IL-1 gene. IL1B showed a significant association with risk of diabetic nephropathy having an OR of 1.97 (95% CI 1.22-3.18).
In this meta-analysis, there was only one SNP which had a protective effect against diabetic nephropathy. Genetic variant rs1800871 in the IL-10 gene was studied in a Tunisian study and had an OR of 0.77 (95% CI 0.63-0.98). In our meta-analysis, there were 44 genetic variants in the genes involved in inflammatory cytokines and angiogenesis that were not found to be significantly associated with DN (Figure 4). It is important to mention that among the 44 SNPs, only 22 were investigated in two or more than two studies while the others had a single study each. Genetic variant rs1800470 showed a significant association with diabetic nephropathy in the type 2 diabetes mellitus subgroup with an OR of 1.25 (95% CI 1.04-1.51) (Figure 2).
Figure 4.
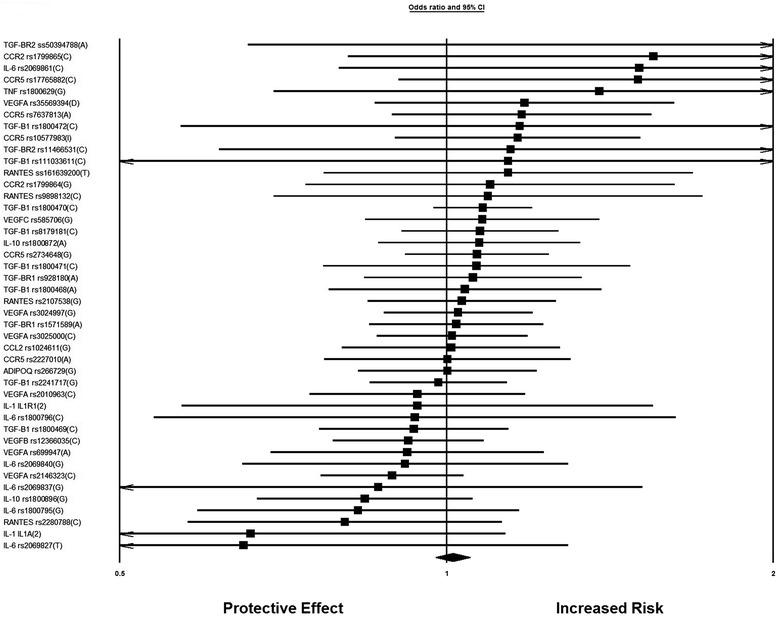
Forest Plot for genetic variants involved in inflammatory cytokines and angiogenesis and not significantly associated with diabetic nephropathy after meta-analysis.
For this meta-analysis, genes showing significant association with diabetic nephropathy were analyzed to assess their contribution in different functional pathways. G protein coupled-receptors (GPCRs) or seven-transmemebrane receptors are the most prevailing class of signaling transduction molecules in humans. In the GPCR signaling pathways, inflammatory cytokine and angiogenic genes positively associated with diabetic nephropathy encode proteins present on the extra-cellular membrane. Among the nine genes showing significant association with diabetic nephropathy in our study- IL10, VEGFA, EPO, IL1 and IL8 were a part of GPCR signaling pathway (Figure 5). In the molecular function based pathway, VEGFA, EPO, IL1, IL8, IL10, ADIPOQ and CCL2 were involved in receptor binding. EPO and ADIPOQ are inhibited by TNF (Tumor necrosis factor) , whereas, other genes are activated by TNF. VEGFA is activated by SMAD3 (Mothers against decapentaplegic homolog 3) present in the intra-cellular region (Figure 6). Disease pathway analysis showed IL6, EPO, ADIPOQ and CCR2 to be involved in chronic kidney failure. The hierarchical layout shows that IL6, EPO, ADIPOQ and CCR2 function by influencing angiotensin I-converting enzyme (ACE) gene expression. ACE further interacts with G-protein beta3-subunit (GNB3) and this interaction is also associated with hypertension in many populations (Figure 7).
Figure 5.
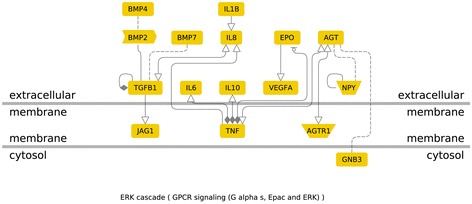
Genes involved in GPCR signaling Pathway: (G protein coupled-receptors are seven-transmemebrane receptors involved in signal transduction in humans).
Figure 6.
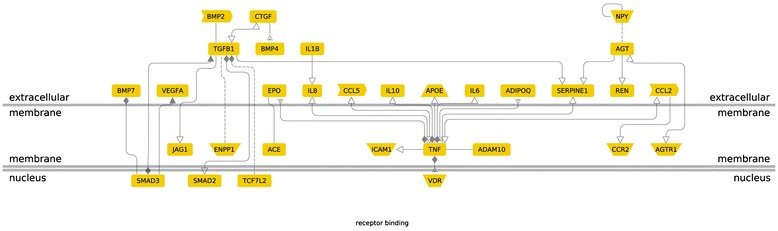
Genes involved in receptor binding pathway.
Figure 7.
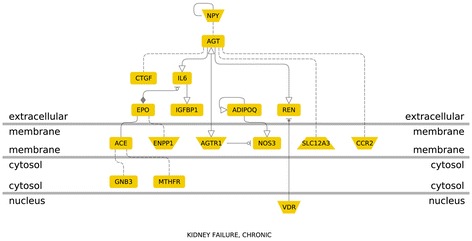
Genes involved in chronic kidney failure (Disease Pathway).
Publication Bias
Thirty six genetic variants were lacking a good number of studies (i.e. less than three studies), thus it was not possible to calculate the publication bias. Of the other nineteen studies, six SNP studies (ADIPOQ rs17300539; RANTES rs2107538; TGF-B1 rs1800468; TGF-B1 rs2241717; VEGFA rs3024997; VEGFA rs3025000) did not account for publication bias as evident from the symmetric funnel plot and thirteen did show evidence of publication bias (Additional file 2).
Discussion
This meta-analysis, including 55 genetic variants in 18 genes from 34 published studies, explored the association between the genetic variants within the genes involved in inflammatory cytokines and angiogenesis pathways and diabetic nephropathy. The results showed that 11 genetic variants were significantly associated with diabetic nephropathy.
Cytokines act as pleiotropic polypeptides regulating inflammatory and immune responses through actions on cells. The relationships between inflammation and the development and progression of diabetic nephropathy involve complex molecular networks and processes. Many studies have shown a strong association of circulating inflammatory markers and proinflammatory cytokines with the risk of developing of diabetic complications [40]. Evidence from studies where immunosuppressive strategies reduce renal macrophage accumulation and attenuate the development of diabetic nephropathy, support the role of inflammation in diabetic complications [40].
Angiogenesis, the formation of new blood vessels from pre-existing vasculature, has been implicated in the genesis of diverse diabetic complications including diabetic nephropathy. Several studies have reported an increase in potent stimulators of angiogenesis in diabetic nephropathy [41,42]. In addition, the therapeutic efficacies of anti–angiogenic strategies have further demonstrated the involvement of angiogenesis in the progression of diabetic nephropathy [13,43]. In this context, genetics of inflammatory cytokines and angiogenesis is important to analyze the involved genes and their role in susceptibility to diabetic nephropathy.
In this meta-analysis, variants within or near VEGFA (two variants), CCR5 (two variants), CCL2, IL-1, MMP9, EPO, IL-8, ADIPOQ and IL-10 were significantly associated with diabetic nephropathy. Three genetic variants (two in the VEGFA and one in the EPO gene) belonged to angiogenesis pathway whereas the other variants were included in inflammatory cytokines. These results support a role of the inflammatory cytokines and angiogenesis pathways in the pathogenesis of diabetic nephropathy.
It has been reported that there is an increased VEGF expression in patients with diabetic nephropathy and antibodies against VEGF in the early stages of experimental diabetes can ameliorate the renal dysfunction [31]. In this meta-analysis, two genetic variants in VEGFA gene were significantly associated with diabetic nephropathy. Both genetic variants were analysed in studies with moderate sample size. and more studies are needed to establish true effect sizes in such cases [44]. T allele of rs833061 genetic variant in VEGFA gene was found to increase the risk of diabetic nephropathy whereas Mooyaart et al. [44] in their study considered C allele of rs833061 as having a protective effect.
CCL2, also known as Monocyte chemo-attractant protein-1(MCP-1), is the strongest known chemo-tactic factor for monocytes and is upregulated in diabetic nephropathy [18]. In this meta-analysis, the insertion at CCL2 rs3917887 showed two fold higher risk of diabetic nephropathy as compared to controls. Chinoy et al. have reported the association CCL2 rs3917887 with development of inflammatory myopathies [45].
CCR5 is a β-chemokine receptor involved in a migration of monocytes, NK cells and some T-cells to the inflammation site [20]. CCR5 rs1799987 was the most studied genetic variant in inflammatory cytokines. This SNP is reported to increase the protein level by increasing the transcriptional activity of the CCR5 gene [46]. Also, for genetic variant rs1799987 in CCR5 gene A allele was the risk factor for diabetic nephropathy in our study whereas Mooyaart et al. [44] considered G allele of rs1799987 as a protective allele. The 32 –bp deletion in CCR5 rs333 changes the open reading frame of the gene resulting in a trunacated protein [47]. This polymorphism showing a significant association with DN in this meta-analysis, is shown to promote renal fibrosis instead of normal tissue repair [48].
ADIPOQ encodes adiponectin, a hormone exclusively secreted by the adipose tissue [23]. A allele of rs17300539 in the promoter of ADIPOQ is a risk factor for DN. It has been suggested that this genetic variation may contribute to an increased risk of developing nephropathy partly through the increase in adiponectin levels [25].
IL-8 is a member of the CXC chemokine family and is associated with a variety of proinflammatory activities [49]. rs4073 lies in the regulatory region and increases the protein level. An increase in the urinary excretion of IL-8 in DN patients has been reported [18]. In their study, Skov et al., showed that IL-8 is an antibody therapeutic target in inflammatory diseases [49].
The pro-inflammatory cytokine interleukin 1 (IL-1) stimulates kidney mesangial cell proliferation and extracellular matrix expansion, contributing to pathogenesis of DN [30]. In this meta-analysis, there is only one study showing allele IL1B*2 associated with a nearly two fold higher risk of diabetic nephropathy. Additional studies are required to establish the reported risk factor.
MMP9 rs17576 lies in the substrate binding region and leads to over-accumulation of extracellular matrix, leading to renal damage [18]. Rysz et al. reported an increase in MMP-9 in diabetic nephropathy when compared with diabetes with normal renal function [50].
EPO is a potent angiogenic factor involved in diabetic micro-vascular complications [39]. Garcia et al. reported EPO increased the rate of renal damage [51]. T allele of genetic variant rs1617640 in EPO gene was a risk allele for diabetic nephropathy in this meta-analysis. This finding was in agreement with Williams et al. [52] who in their study showed T allele of rs1617640 as a risk factor for diabetic nephropathy. On the contrary, Mooyaart et al. [44] considered T allele to have a protective effect for diabetic nephropathy.
Interleukin-10 (IL-10) fulfils the criteria for an anti-inflammatory and immunosuppressive cytokine [53]. Wong et al. reported a correlation between IL-10 levels and extent of renal damage in diabetic nephropathy [54]. There were two studies investigating the protective effect of T allele of rs1800871 for diabetic nephropathy. Both the studies were done in Tunisian population by the same authors with apparently the same study group (Table 1- ref. 2, 29). Since the methodology and results were identical for the two studies, we considered them as a single study to avoid overestimation of effects.
An important factor that bears on the interpretation of meta-analysis of the genetics of diabetic nephropathy is the adequacy of phenotype definition. This problem is reflected in studies showing that most patients with type 1 diabetes categorized initially as having microalbuminaria, undergo regression to normoalbuminaria [52]. To avoid this problem in our meta-analysis, we only included studies where diabetic nephropathy was defined by macroalbuminaria or ESRD. As with all meta-analysis, one major limitation of this study is publication bias. Only published data in journals was included in this study which may lead to ignoring the negative or non-significant association studies. Heterogeneity between the studies can affect the interpretation of results. One of the potential reason for heterogeneity is the winner’s curse, which appears as an upward bias in the estimated effect of a newly identified allele on disease risk when the study design lacks sufficient statistical power. . The winner’s curse manifests mostly in genome‐wide association (GWA) studies in which 300 000–1 000 000 single‐nucleotide polymorphisms are tested [55]. All the studies included in this meta-analysis were candidate gene based. Further, to minimize the overestimation caused by the winner’s curse, effect sizes and minor allele frequencies were calculated from pooled estimates from the initial reporting study and other subsequent studies. Random effects model was performed to account for any possible heterogeneity.
In this study, enrichment analysis of the genes significantly associated with diabetic nephropathy, generated three functional pathways- GPCR signaling, receptor binding and chronic kidney failure. GPCRs transduce extracellular signals inside the cell through activation of heterotrimeric G proteins and/or via other G protein-independent signaling pathways. Agonist-induced GPCR phosphorylation by G protein-coupled receptor kinases (GRKs) are involved in a number of important systems related to the development and progression of Diabetic nephropathy [56]. Presence of these genes in the functional pathways supports their significant association with diabetic nephropathy as concluded in this meta-analysis. However, further experiments are required to elucidate the exact role of these genes in the pathogenesis of diabetic nephropathy.
Considering a worldwide increase in diabetes, there is an urgent need to provide patients protection from the development and progression of diabetic nephropathy. Identification of genes involved in the primary mechanisms contributing to onset and progression of diabetic nephropathy is important for developing new therapeutic interventions. Inflammatory cytokines and angiogenic factors play a diverse role in the pathogenesis of diabetic nephropathy. The recognition of the genes/genetic variants as having a significant association with risk of diabetic nephropathy will provide new therapeutic targets. Furthermore, this will also help to identify patients with increased risk of diabetic nephropathy.
Conclusions
This study identified 11 genetic variants in or near 9 genes -VEGFA, CCR5, CCL2, IL-1, MMP9, EPO, IL-8, ADIPOQ and IL-10, significantly associated with diabetic nephropathy. Further studies are required to better understand their functional relevance in the pathogenesis of diabetic nephropathy. These genetic variants within the inflammatory cytokines and angiogensis pathways might be good candidates for identifying genetic predisposition to DN as well as revealing the pathogenesis of diabetic nephropathy.
Acknowledgements
This study was supported and funded by Strategic Center for Diabetes Research, King Saud University, Saudi Arabia.
Abbreviation
- DM
Diabetes mellitus
- T1DM
Type 1 Diabetes mellitus
- T2DM
Type 2 diabetes mellitus
- DN
Diabetic Nephropathy
- SNP
Single nucleotide Polymorphism
- DNA
Deoxyribonucleic acid
- bp
Base pair
- OR
Odds ratio
- CI
Confidence interval
- HWE
Hardy–Weinberg equilibrium
- ESRD
End stage renal disease
- NK Cells
Natural killer cells
- STZ
Streptozotocin
- VEGFA
Vascular endothelial growth factor A
- CCR5
Chemokine (C-C motif) receptor type 5
- CCL2
Chemokine ligand 2
- IL1
Interleukin-1
- MMP9
Matrix metallopeptidase 9
- EPO
Erythropoietin
- IL8
Interleukin 8
- ADIPOQ
Adiponectin C1Q and collagen domain containing
- IL10
Interleukin 10
- GRKs
G protein-coupled receptor kinases
- GPCRs
G protein-coupled receptors
- IL1B
Interleukin-1 beta
- IL6
Interleukin 6
- MCP1
Monocyte chemoattractant protein-1
- BMP4
Bone morphogenetic protein 4
- BMP2
Bone morphogenetic protein 2
- BMP7
Bone morphogenetic protein 7
- TGFB1
Transforming growth factor beta 1
- JAG1
jagged 1 protein
- SMAD2
Mothers against decapentaplegic homolog 2
- SMAD3
Mothers against decapentaplegic homolog 3
- ENPP1
Ectonucleotide pyrophosphatase/phosphodiesterase family member 1
- TCF7L2
transcription factor 7-like 2
- ACE
angiotensin I converting enzyme
- APOE
Apolipoprotein E
- SERPINEI
Serpin peptidase Inhibitor, Clade E (Nexin, Plasminogen activator Inhibitor Type 1), Member 1
- ICAM1
Intercellular Adhesion Molecule 1
- TNF
Tumor necrosis factor
- ADAM10
A Disintegrin and metalloproteinase domain-containing protein 10
- VDR
Vitamin D (1,25- Dihydroxyvitamin D3) Receptor
- NPY
Neuropeptide Y
- GNB3
Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-3
- REN
Rennin
- CCL5
Chemokine ligand 5
- CCR2
Chemokine (C-C motif) receptor 2
- AGT
Angiotensinogen
- AGTR1
Angiotensin II receptor
- CTGF
Connective tissue growth factor
- MTHFR
Methylenetetrahydrofolate reductase
- NOS3
Nitric oxide synthase 3
- SLC12A3
Solute carrier family 12 (Sodium/Chloride Transporter), member 3
Additional files
a: Details of SNPs included in the meta-analysis, for genes CCR5, ADIPOQ, IL-8 and CCL2. b: Details of SNPs included in the meta-analysis, for genes IL-10, MMP9 and IL-1. c: Details of SNPs included in the meta-analysis, for genes VEGFA, VEGFB and VEGFC. d: Details of SNPs included in the meta-analysis, for genes IL-6, CCR2, EPO, TNFα and CCL5. e: Details of SNPs included in the meta-analysis, for genes TGF-B1, TGF-BR1 and TGF-BR2.
Funnel plots of nineteen SNPs with more than two studies. a: Funnel plot of ADIPOQ rs17300539. b: Funnel plot of CCR5 rs333. c: Funnel plot of CCR5 rs2734648. d: Funnel plot of EPO rs1617640. e: Funnel plot of CCR5 rs1799987. f: Funnel plot of RANTES rs2107538. g: Funnel plot of TGF-B1 rs1800468. h: Funnel plot of TGF-B1 rs1800469. i: Funnel plot of TGF-B1 rs2241717. j: Funnel plot of TGF-B1 rs8179181. k: Funnel plot of TGF-BR1 rs928180. l: Funnel plot of TGF-BR1 rs1571589. m: Funnel plot of TGF-B1 rs1800471. n: Funnel plot of TGF-B1 rs1800470. o: Funnel plot of VEGFA rs3024997. p: Funnel plot of VEGFA rs3025000. q: Funnel plot of VEGFA rs2146323. r: Funnel plot of VEGFB rs12366035. s: Funnel plot of VEGFC rs585706.
Footnotes
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
NN carried out the literature search, the analysis and interpretation of data and drafted the manuscript. KS conceived of the study, and participated in its design and coordination and helped to draft the manuscript. SAQ and DAN participated in the literature search and have been involved in drafting the manuscript. All authors have read and approved the final version of the manuscript.
Contributor Information
Nyla Nazir, Email: npeerzada@ksu.edu.sa.
Khalid Siddiqui, Email: ksiddiqui@ksu.edu.sa.
Sara Al-Qasim, Email: sqassim@dsrcenter.org.
Dhekra Al-Naqeb, Email: dalnaqeb@dsrcenter.org.
References
- 1.International Diabetes Federation: IDF Diabetes Atlas. 6th edition. Brussels, Belgium: International Diabetes Federation; 2013. [http://www.idf.org/diabetesatlas]
- 2.Ezzidi I, Mtiraoui N, Kacem M, Mallat SG, Mohamed MB, Chaieb M, Mahjoub T, Almawi WY. Interleukin-10–592C/A, −819C/T and –1082A/G promoter variants affect the susceptibility to nephropathy in Tunisian type 2 diabetes (T2DM) patients. Clin Endocrinol. 2009;70:401–407. doi: 10.1111/j.1365-2265.2008.03337.x. [DOI] [PubMed] [Google Scholar]
- 3.Zhang D, Efendic S, Brismar K, Gu HF. Effects of MCF2L2, ADIPOQ and SOX2 genetic polymorphisms on the development of nephropathy in type 1 Diabetes Mellitus. BMC Med Genet. 2010;11:116. doi: 10.1186/1471-2350-11-116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Freedman BI, Bostrom M, Daeihagh P, Bowden DW. Genetic Factors in Diabetic Nephropathy. Clin J Am Soc Nephrol. 2007;2:1306–131. doi: 10.2215/CJN.02560607. [DOI] [PubMed] [Google Scholar]
- 5.Blech I, Katzenellenbogen M, Katzenellenbogen A, Wainstein J, Rubinstein A, Harman-Boehm I, Cohen J, Pollin TI, Glaser B. Predicting diabetic nephropathy using a multifactorial genetic model. PLoS One. 2011;6(4):e18743. doi: 10.1371/journal.pone.0018743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xu ZG, Lanting L, Vaziri ND, Li Z, Sepassi L, Rodriguez-Iturbe B, Natarajan R. Upregulation of angiotensin II type I receptor, inflammatory mediators, and enzymes of arachidonate metabolism in obese Zucker rat kidney: reversal by angiotensin II type 1 receptor blockade. Circulation. 2005;111:1962–1969. doi: 10.1161/01.CIR.0000161831.07637.63. [DOI] [PubMed] [Google Scholar]
- 7.Tashiro K, Koyanagi I, Saitoh A, Shimizu A, Shike T, Ishiguro C, Koizumi M, Funabiki K, Horikoshi S, Shirato I, Tomino Y. Urinary levels of monocyte chemoattractant protein-1 (MCP-1) and inter-leukin-8 (IL-8), and renal injuries in patients with type 2 diabetic nephropathy. J Clin Lab Anal. 2002;16:1–4. doi: 10.1002/jcla.2057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lewis A, Steadman R, Manley P, Craig K, de la Motte C, Hascall V, Phillips AO. Diabetic nephropathy, inflammation, hyaluronan and interstitial fibrosis, Histol. Histopathol. 2008;23:731–739. doi: 10.14670/HH-23.731. [DOI] [PubMed] [Google Scholar]
- 9.Nyengaard JR, Rasch R. The impact of experimental diabetes mellitus in rats on glomerular capillary number and sizes. Diabetologia. 1993;36:189–94. doi: 10.1007/BF00399948. [DOI] [PubMed] [Google Scholar]
- 10.Sharma K, Ziyadeh FN. Hyperglycemia and diabetic kidney disease. The case for transforming growth factor-beta as a key mediator. Diabetes. 1995;44:1139–46. doi: 10.2337/diab.44.10.1139. [DOI] [PubMed] [Google Scholar]
- 11.Kim NH, Kim KB, Kim DL, Kim SG, Choi KM, Baik SH, Choi DS, Kang YS, Han SY, Han KH, Ji YH, Cha DR. Plasma and urinary vascular endothelial growth factor and diabetic nephropathy in type 2 diabetes mellitus. Diabet Med. 2004;21:545–551. doi: 10.1111/j.1464-5491.2004.01200.x. [DOI] [PubMed] [Google Scholar]
- 12.Chen S, Ziyadeh FN. Vascular endothelial growth factor and diabetic nephropathy. Curr Diab Rep. 2008;8:470–6. doi: 10.1007/s11892-008-0081-3. [DOI] [PubMed] [Google Scholar]
- 13.Celec P, Hodosy J, Gardlík R, Behuliak M, Pálffy R, Pribula M, Jáni P, Turňa J, Sebeková K. The effects of anti-inflammatory and anti- angiogenic DNA vaccination on diabetic nephropathy in rats. Hum Gene Ther. 2012;23:158–166. doi: 10.1089/hum.2011.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Higgins JP, Thompson SG, Deeks JJ, Altman DG. Measuring inconsistency in meta-analyses. Brit Med J. 2003;327:557–560. doi: 10.1136/bmj.327.7414.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kavvoura FK, Ioannidis JPA. Methods for meta-analysis in genetic association studies: a review of their potential and pitfalls. Hum Genet. 2008;123:1–14. doi: 10.1007/s00439-007-0445-9. [DOI] [PubMed] [Google Scholar]
- 16.Pettigrew KA, McKnight AJ, Patterson CC, Kilner J, Sadlier DM, Maxwell AP. Resequencing of the CCL5 and CCR5 genes and investigation of variants for association with diabetic nephropathy. J Hum Genet. 2010;55:248–251. doi: 10.1038/jhg.2010.15. [DOI] [PubMed] [Google Scholar]
- 17.Tregouet DA, Groop PH, McGinn S, Forsblom C, Hadjadj S, Marre M, Parving HH, Tarnow L, Telgmann R, Godefroy T, Nicaud V, Rousseau R, Parkkonen M, Hoverfalt A, Gut I, Heath S, Matsuda F, Cox R, Kazeem G, Farrall M, Gauguier D, Brand-Herrmann SM, Cambien F, Lathrop M, Vionnet N. G/T substitution in intron 1 of the UNC13B gene is associated with increased risk of nephropathy in patients with type 1 diabetes. Diabetes. 2008;57:2843–2850. doi: 10.2337/db08-0073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ahluwalia TS, Khullar M, Ahuja M, Kohli HS, Bhansali A, Mohan V, Venkatesan R, Rai TS, Sud K, Singal PK. Common Variants of Inflammatory Cytokine Genes Are Associated with Risk of Nephropathy in Type 2 Diabetes among Asian Indians. PLoS One. 2009;4(4):e5168. doi: 10.1371/journal.pone.0005168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mlynarski WM, Placha GP, Wolkow PP, Bochenski JP, Warram JH, Krolewski AS. Risk of diabetic nephropathy in type 1 diabetes is associated with functional polymorphisms in RANTES receptor gene (CCR5): a sex-specific effect. Diabetes. 2005;54:3331–3335. doi: 10.2337/diabetes.54.11.3331. [DOI] [PubMed] [Google Scholar]
- 20.Buraczynska M, Zukowski P, Wacinski P, Berger-Smyka B, Dragan M, Mozul S. Chemotactic cytokine receptor 5 gene polymorphism: Relevance to microvascular complications in type 2 diabetes. Cytokine. 2012;58(2):213–217. doi: 10.1016/j.cyto.2012.01.007. [DOI] [PubMed] [Google Scholar]
- 21.Nakajima K, Tanaka Y, Nomiyama T, Ogihara T, Ikeda F, Kanno R, Iwashita N, Sakai K, Watada H, Onuma T, Kawamori R. RANTES promoter genotype is associated with diabetic nephropathy in type 2 diabetic subjects. Diabetes Care. 2003;26:892–898. doi: 10.2337/diacare.26.3.892. [DOI] [PubMed] [Google Scholar]
- 22.Prasad P, Tiwari AK, Kumar KMP, Ammini AC, Gupta A, Gupta R, Thelma BK. Association of TGFbeta1, TNFalpha, CCR2 and CCR5 gene polymorphisms in type-2 diabetes and renal insufficiency among Asian Indians. BMC Med Genet. 2007;8:20. doi: 10.1186/1471-2350-8-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang D, Ma J, Brismar K, Efendic S, Gu HF. A single nucleotide polymorphism alters the sequence of SP1 binding site in the adiponectin promoter region and is associated with diabetic nephropathy among type 1 diabetic patients in the Genetics of Kidneys in Diabetes Study. J Diabetes Complicat. 2009;23:265–272. doi: 10.1016/j.jdiacomp.2008.05.004. [DOI] [PubMed] [Google Scholar]
- 24.Wu LSH, Hsieh CH, Pei D, Hung YJ, Kuo SW, Lin E. Association and interaction analyses of genetic variants in ADIPOQ, ENPP1, GHSR, PPARγ and TCF7L2 genes for diabetic nephropathy in a Taiwanese population with type 2 diabetes. Nephrol Dial Transplant. 2009;24:3360–3366. doi: 10.1093/ndt/gfp271. [DOI] [PubMed] [Google Scholar]
- 25.Vionnet N, Tregouët D, Kazeem G, Gut I, Groop PH, Tarnow L, Parving HH, Hadjadj S, Forsblom C, Farrall M, Gauguier D, Cox R, Matsuda F, Heath S, Thevard A, Rousseau R, Cambien F, Marre M, Lathrop M. Analysis of 14 Candidate Genes for Diabetic Nephropathy on Chromosome 3q in European Populations Strongest Evidence for Association With a Variant in the Promoter Region of the Adiponectin Gene. Diabetes. 2006;55(11):3166–3174. doi: 10.2337/db06-0271. [DOI] [PubMed] [Google Scholar]
- 26.Prior SL, Javid J, Gill GV, Bain SC, Stephens JW. The adiponectin rs17300539 G > A variant and nephropathy risk. Kidney Int. 2008;74:1361. doi: 10.1038/ki.2008.474. [DOI] [PubMed] [Google Scholar]
- 27.Joo KW, Hwang YH, Kim JH, Oh KH, Kim H, Shin HD, Chung WK, Yang J, Park KS, Ahn C. MCP-1 and RANTES Polymorphisms in Korean Diabetic End-Stage Renal Disease. J Korean Med Sci. 2007;22:611–5. doi: 10.3346/jkms.2007.22.4.611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Arababadi MK, Mirzaei MR, Sajadi SMA, Hassanshahi G, Ahmadabadi BN, Salehabadi VA, Derakhshan R, Kennedy D. Interleukin (IL)-10 gene polymorphisms are associated with type 2 diabetes with and without nephropathy: A study of patients from the southest region of Iran. Inflammation. 2012;35(3):797–802. doi: 10.1007/s10753-011-9376-7. [DOI] [PubMed] [Google Scholar]
- 29.Mtiraoui N, Ezzidi I, Kacem M, Ben Hadj Mohamed M, Chaieb M, Haj Jilani AB, Mahjoub T, Almawi WY. Predictive value of interleukin-10 promoter genotypes and haplotypes in determining the susceptibility to nephropathy in type 2 diabetes patients. Diabetes Metab Res Rev. 2009;25(1):57–63. doi: 10.1002/dmrr.892. [DOI] [PubMed] [Google Scholar]
- 30.Loughrey BV, Maxwell AP, Fogarty DG, Middleton D, Harron JC, Patterson CC, Darke C, Savage DA. An interleukin 1B allele, which correlates with a high Secretor phenotype, is associated with diabetic Nephropathy. Cytokine. 1998;10(12):984–988. doi: 10.1006/cyto.1998.0388. [DOI] [PubMed] [Google Scholar]
- 31.Yang B, Cross DF, Ollerenshaw M, Millward BA, Demaine AG. Polymorphisms of the vascular endothelial growth factor and susceptibility to diabetic microvascular complications in patients with type 1 diabetes mellitus. J Diabetes Complicat. 2003;17:1–6. doi: 10.1016/S1056-8727(02)00181-2. [DOI] [PubMed] [Google Scholar]
- 32.Buraczynska M, Ksiazek P, Gaszczyk IB, Jozwiak L. Association of the VEGF gene polymorphism with diabetic retinopathy in type 2 diabetes patients. Nephrol Dial Transplant. 2007;22:827–832. doi: 10.1093/ndt/gfl641. [DOI] [PubMed] [Google Scholar]
- 33.McKnight A-J, Maxwell AP, Patterson CC, Brady HR, Savage DA. Association of VEGF-1499C[right arrow]T polymorphism with diabetic nephropathy in type 1 diabetes mellitus. J Diabetes Complicat. 2007;21:242–245. doi: 10.1016/j.jdiacomp.2006.05.005. [DOI] [PubMed] [Google Scholar]
- 34.Kim HW, Ko GJ, Kang YS, Lee MH, Song HK, Kim HK, Cha DR. Role of the VEGF 936 C/T polymorphism in diabetic microvascular complications in type 2 diabetic patients. Nephrology. 2009;14:681–688. doi: 10.1111/j.1440-1797.2009.01085.x. [DOI] [PubMed] [Google Scholar]
- 35.Ng DP, Nurbaya S, Ye S, Krolewski AS. An IL-6 haplotype on human chromosome 7p21 confers risk for impaired renal function in type 2 diabetes. Kidney Int. 2008;74(4):521–527. doi: 10.1038/ki.2008.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jahromi MM, Millward BA, Demaine AG. Significant correlation between association of polymorphism in codon 10 of transforming growth factor-β1 T(29) C with type 1 diabetes and patients with nephropathy disorder. J Interf Cytokine Res. 2010;30(2):59–66. doi: 10.1089/jir.2009.0026. [DOI] [PubMed] [Google Scholar]
- 37.Ng DPK, Warram JH, Krolewski AS. TGF- β1 as a genetic susceptibility locus for advanced diabetic nephropathy in type 1 diabetes mellitus: An investigation of multiple known DNA sequence variants. Am J Kidney Disease. 2003;41(1):22–28. doi: 10.1053/ajkd.2003.50011. [DOI] [PubMed] [Google Scholar]
- 38.Salgado-V A, Martinez JA, Rosas M, Garcia-Mena J, Utrera-Barillas D, Gomez-Diaz R, Pena JE, Parra EJ, Cruz M. Association of polymorphisms within the transforming growth factor-β1 gene with diabetic nephropathy and serum cholesterol and triglyceride concentrations. Nephrology. 2010;15:644–648. doi: 10.1111/j.1440-1797.2010.01302.x. [DOI] [PubMed] [Google Scholar]
- 39.Tong Z, Yang Z, Patel S, Chen H, Gibbs D, Yang X, Hau VS, Kaminoh Y, Harmon J, Pearson E, Buehler J, Chen Y, Yu B, Tinkham NH, Zabriskie NA, Zeng J, Luo L, Sun JK, Prakash M, Hamam RN, Tonna S, Constantine R, Ronquillo CC, Sadda S, Avery RL, Brand JM, London N, Anduze AL, King GL, Bernstein PS. Promoter polymorphism of the erythropoietin gene in severe diabetic eye and kidney complications. Proc Natl Acad Sci U S A. 2008;105(19):6998–7003. doi: 10.1073/pnas.0800454105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Elmarakby AA, Sullivan JC. Relationship between oxidative stress and inflammatory cytokines in diabetic nephropathy. Cardiovasc Ther. 2012;30:49–59. doi: 10.1111/j.1755-5922.2010.00218.x. [DOI] [PubMed] [Google Scholar]
- 41.Nasu T, Maeshima Y, Kinomura M, Hirokoshi-Kawahara K, Tanabe K, Sugiyama H, Sonoda H, Sato Y, Makino H. Vasohibin-1, a negative feedback regulator of angiogenesis, ameliorates renal alterations in a mouse model of diabetic nephropathy. Diabetes. 2009;58:2365–2375. doi: 10.2337/db08-1790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sonezaki K, Maezawa Y, Takemoto M, Kobayashi K, Tokuyama T, Takada-Watanabe A, Simoyama T, Sato S, Saito Y, Yokote K. Alteration of VEGF and Angiopoietins Expressions in Diabetic Glomeruli Implicated in the Development of Diabetic Nephropathy. Advanced Studies Med Sci. 2013;1:11–28. [Google Scholar]
- 43.Zent R, Pozzi A. Angiogenesis in diabetic nephropathy. Semin Nephrol. 2007;27:161–171. doi: 10.1016/j.semnephrol.2007.01.007. [DOI] [PubMed] [Google Scholar]
- 44.Mooyaart AL, Valk EJ, van Es LA, Bruijn JA, de Heer E, Freedman BI, Dekkers OM, Baelde HJ. Genetic associations in diabetic nephropathy: a meta-analysis. Diabetologia. 2011;54:544–553. doi: 10.1007/s00125-010-1996-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chinoy H, Salway F, Fertig N, Tait BD, Oddis CV, Ollier WE, Cooper RG. Monocyte chemotactic protein-1 single nucleotide polymorphisms do not confer susceptibility for the development of adult onset polymyositis/dermatomyositis in UK Caucacians. Rheumatology. 2007;46(4):604–7. doi: 10.1093/rheumatology/kel359. [DOI] [PubMed] [Google Scholar]
- 46.Narvatilova Z. Polymorphisms in CCL2 & CCL5 chemikines/chemokine receptors genes and their association with diseases. Biomed Pap Med Fac Univ Palacky Oloumuc Czech Repub. 2006;150(2):191–204. doi: 10.5507/bp.2006.028. [DOI] [PubMed] [Google Scholar]
- 47.Gonzalez E, Dhanda R, Bamshad M, Mummidi S, Geevarghese R, Catano G, Anderson SA, Walter EA, Stephan KT, Hammer MF, Mangano A, Sen L, Clark RA, Ahuja SS, Dolan MJ, Ahuja SK. Global survey of genetic variation in CCR5, RANTES and MIP-alpha:impact on the epidemiology of the HIV-1 pandemic. Proc Natl Acad Sci U S A. 2001;98:5199–5204. doi: 10.1073/pnas.091056898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vielhauer V, Anders HJ, Mack M, Cihak J, Strutz F, Stangassinger M, Luckow B, Gröne HJ, Schlöndorff D. Obstructive nephropathy in the mouse: progressive fibrosis correlates with tubulointerstitial chemokine expression and accumulation of CC chemokine receptor 2- and 5- positive leukocytes. J Am Soc Nephrol. 2001;12:1173–1187. doi: 10.1681/ASN.V1261173. [DOI] [PubMed] [Google Scholar]
- 49.Skov L, Beurskens FJ, Zachariae CO, Reitamo S, Teeling J, Satijn D, Knudsen KM, Boot EP, Hudson D, Baadsgaard O, Parren PW, van de Winkel JG. IL-8 as Antibody Therapeutic Target in Inflammatory Diseases: Reduction of Clinical Activity in Palmoplantar Pustulosis. J Immunol. 2008;181(1):669–679. doi: 10.4049/jimmunol.181.1.669. [DOI] [PubMed] [Google Scholar]
- 50.Rysz J, Banach M, Stolarek RA, Pasnik J, Cialkowska-Rysz A, Koktysz R, Piechota M, Baj Z. Serum matrix metalloproteinases MMP-2 and MMP-9 and metalloproteinase tissue inhibitors TIMP-1 and TIMP-2 in diabetic nephropathy. J Nephrol. 2007;20(4):444–52. [PubMed] [Google Scholar]
- 51.Garcia DL, Anderson S, Rennke HG, Brenner BM. Anemia lessens and its prevention with recombinant human erythropoietin worsens glomerular injury and hypertension in rats with reduced renal mass. Proc Natl Acad Sci U S A. 1988;85(16):6142–6. doi: 10.1073/pnas.85.16.6142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Williams WW, Salem RM, McKnight AJ, Sandholm N, Forsblom C, Taylor A, Guiducci C, McAteer JB, McKay GJ, Isakova T, Brennan EP, Sadlier DM, Palmer C, Soderlund J, Fagerholm E, Harjutsalo V, Lithovius R, Gordin D, Hietala K, Kyto J, Parkkonen M, Rosengard-Barlund M, Thorn L, Syreeni A, Tolonen N, Saraheimo M, Waden J, Pitkaniemi J, Sarti C, Tuomilehto J, Tryggvason K, et al. Association testing of previously reported variants in a large case–control meta-analysis of diabetic nephropathy. Diabetes. 2012;61(8):2187–2194. doi: 10.2337/db11-0751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Myśliwska J, Zorena K, Semetkowska-Jurkiewicz E, Rachoń D, Suchanek H, Myśliwski A. High levels of circulating interleukin-10 in diabetic nephropathy patients. Eur Cytokine Netw. 2005;16(2):117–22. [PubMed] [Google Scholar]
- 54.Wong CK, Ho AW, Tong PC, Yeung CY, Kong AP, Lun SW, Chan JC, Lam CW. Aberrant activation profile of cytokines and mitogen-activated protein kinases in type 2 diabetic patients with nephropathy. Clin Exp Immunol. 2007;149(1):123–31. doi: 10.1111/j.1365-2249.2007.03389.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nakaoka H, Inoue I. eLS. Chichester: John Wiley & Sons Ltd; 2010. The Winner’s Curse. [Google Scholar]
- 56.Wang FL, Tang LQ, Wei W. The connection between GRKs and various signaling pathways involved in diabetic nephropathy. Mol Biol Rep. 2012;39(7):7717–7726. doi: 10.1007/s11033-012-1608-x. [DOI] [PubMed] [Google Scholar]


