Abstract
In order to understand the link between the genetic background of patients and wound clinical outcomes, it is critical to have a reliable method to assess the phenotypic characteristics of healed wounds. In this study, we present a novel imaging method that provides reproducible, sensitive and unbiased assessments of post-surgical scarring. We used this approach to investigate the possibility that genetic variants in orofacial clefting genes are associated with suboptimal healing. Red-green-blue (RGB) digital images of post-surgical scars of 68 patients, following unilateral cleft lip repair, were captured using the 3dMD image system. Morphometric and colorimetric data of repaired regions of the philtrum and upper lip were acquired using ImageJ software and the unaffected contralateral regions were used as patient-specific controls. Repeatability of the method was high with interclass correlation coefficient score > 0.8. This method detected a very significant difference in all three colors, and for all patients, between the scarred and the contralateral unaffected philtrum (P ranging from 1.20−05 to 1.95−14). Physicians’ clinical outcome ratings from the same images showed high inter-observer variability (overall Pearson coefficient = 0.49) as well as low correlation with digital image analysis results. Finally, we identified genetic variants in TGFB3 and ARHGAP29 associated with suboptimal healing outcome.
Keywords: wound healing, cleft palate, genes, genotype-phenotype, Tgfb3, Arhgap29, scar, imaging, digital
Introduction
Wound healing is a complex event requiring the coordination of many biological processes to achieve proper tissue repair (1). Wound healing can be conceptually divided into three overlapping phases: the inflammatory phase, the proliferation phase and the maturation phase (1). Alterations in one of these biological steps will affect other events in the wound healing process. In addition to reducing quality-of-life, poor wound healing results in huge financial impact. It is estimated that the annual health care costs related to acute and chronic non-healing wounds exceed $25 billion (2).
Acute and chronic wounds are complex medical problems. Current diagnosis relies primarily on clinical assessment of the wound over time, including scarring, pain, exudates, and edema in conjunction with the history of the patient. Prognosis for the wound outcome should be a critical part of clinical management, but despite advanced research in the field of wound healing, tools to predict healing outcomes or identify individuals at risk are still lacking. A pilot study of 18 Caucasians reported the possible contribution of copy number variations in HLA-DRB5 in the disease pathogenesis of keloids (3). A larger genome-wide association study reported the identification of four loci for increased risk for keloids in Japanese population (4). Recently, genetic variation in neuronal nitric oxide synthase associated protein was shown to be associated with an increased incidence of lower extremity amputation and diabetic peripheral neuropathy (5). Although targeted approaches have identified a few potential biomarkers (e.g., MMP-2, MMP-9, elastase) that predict chronic wound outcomes, no progress beyond clinically-based prognosis has been made (6).
In addition to these unique conditions, patients in the general population do not heal equally, suggesting that genetic and/or environmental factors contribute to tissue repair. For example, recent empiric evidence suggests a genetic contribution to predisposition to surgical site infection (7). Identifying genetic variants that contribute to wound healing likely could be readily translated into improving clinical decision making and practice to improve wound healing. Perhaps the most immediate application for such information would be in its use as a predictive tool for wound healing outcomes as such information may help to counsel the patient and guide clinical management.
Understanding the genetic basis of wound healing among affected individuals requires accurate characterization of the wound healing phenotype as evidence by mature scars. As of now, this phenotypic characterization relies primarily on visual inspection and rating using established analog scales (8, 9). This can be very challenging and subjective, as these studies rely on evaluations by one or more observers, and thus can vary greatly between raters. Therefore, there is a need to develop a standardized, objective and quantitative method to accurately evaluate healing, including scaring, which can become a standard tool in the evaluation of surgical outcomes. Among the different parameters characterizing a scar, the color of the skin is an important component that is largely determined by the distribution of blood vessels and pigmentation. Computerized image analysis of skin colors using a pixel averaging procedure had been proposed as a simple cost-effective and non-invasive method to evaluate scar color (10, 11).
Herein we describe a new image analysis method to identify color differences between the scarred area and unaffected areas within the same individual, and that the amplitude of the difference is correlated with the wound healing outcome. A collection of digital images of patients with surgically repaired cleft lip and palate enrolled in the Iowa Oral Cleft Study was analyzed to systematically obtain color profiles of scars and unaffected facial areas. Our study establishes a new reliable and objective method to characterize the phenotype of labial wounds in individuals with oral clefts and demonstrate an association with genetic variants in candidate genes related to craniofacial morphology.
Material and Methods
Patient population
All work involving human subjects was reviewed and approved by the University of Iowa Institutional Review Board (IRB) according to the Declaration of Helsinki. Informed consent from all participants was obtained prior to the study. Patients seeking care from the The University of Iowa Craniofacial clinic and affected individuals identified through the Iowa Registry of Congenital and Inherited Disorders (IRCID) that participated in the Iowa Oral Cleft Study were included in the present study. Among all enrolled individuals with oral clefts, sixty-eight had surgically repaired unilateral cleft lip and palate, as well as facial images and DNA available. Their demographics are presented in Table 1. The mean age was 10.2 years and 60% were males. Among the 68 participants, 41% had cleft lip only, and 59% had cleft lip and palate. All participants were non-syndromic cases. Specific information about the labial surgery was not recorded for all participants. Because labial repair commonly occurs at 3-6 months of age, and the youngest participant was 2 years old, the acquisition of the photograph would have occurred at least a year after the survey for all participants. At the time of image capture, participants were in an indoor environment under controlled climate and lighting conditions and had not eaten or engaged in strenuous physical activity for at least one hour prior to photography.
Table 1.
Demographics of the studied population
| Category | N (Freq %) |
|---|---|
| Age: | |
| Mean | 10.24 |
| Range | 2-44 |
| Gender: | |
| Male | 41 (60.29) |
| Female | 27 (39.71) |
| Cleft Type | |
| Cleft Lip Only (CLO) | 28 (41.18) |
| Cleft Lip and Palate (CLP) | 40 (58.82) |
| Race | |
| Caucasian | 63 (92.65) |
| Black/ African American | 1 (1.47) |
| Other | 4 (5.88) |
| Ethnicity | |
| Hispanic | 4 (5.88) |
| Non-Hispanic | 63 (92.65) |
| Unknown | 1 (1.47) |
Image capture and data collection
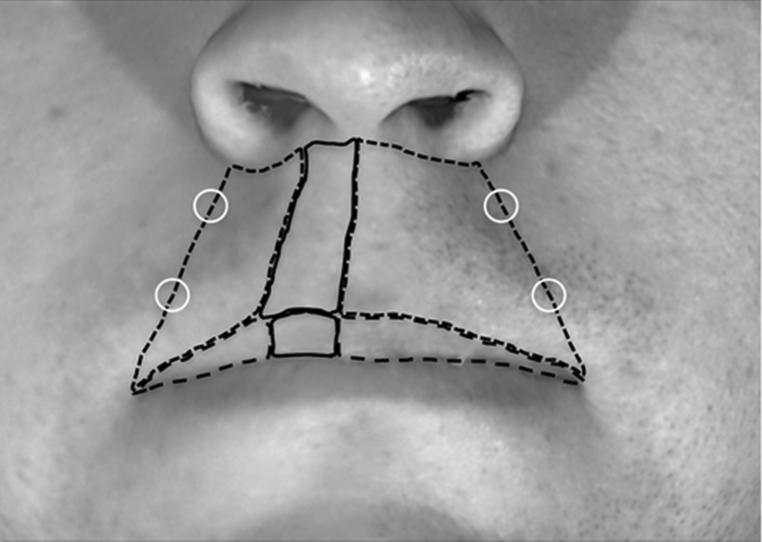
Three-dimensional images of our study population were captured using the 3dMD imaging system (Atlanta, GA, USA). The camera was calibrated using a black and white scale and pictures were taken under regular office lighting. Using the 3dMD image analysis manipulation program, each 3D photograph was configured to the same axial positions based on landmark locations, so that the midline of the pupil was at the same horizontal level as the tragus of the ear. Once the uniform position of each image was obtained, the inter-commissure width of the mouth was measured using 3dMD software tools, and recorded for future calibration of the ImageJ software. The uniformly positioned images were exported from 3dMD as two-dimensional image files and calibrated for size. Windows tablet PCs were specifically used to allow for an accurate method of hand tracing (by means of a stylus) critical regions of each image within the ImageJ program. Using this system, we obtained size and color information from the key facial areas as illustrated in Figure 1. In the few images where mirroring the wound outline to the contralateral unaffected region of the philtrum or lip was not possible, an outline of comparable size and similar shape that avoided confounding regions (such as regions of shadowing or outlines that wouldn't fit the opposite side due to asymmetries) was used. Upon tracing the selected facial regions on the philtrum and lip, the area was measured using ImageJ software. Red, green and blue (RGB) color histogram profiles were also collected from every traced region of the face. Mean values for each color of each region was recorded and used for further analysis.
Figure 1.
Landmarks for the digital image analysis. Area of the philtrum (blue lines), area of the lip (green lines) and affected area in both the philtrum and the lip region (white lines) were measured using ImageJ. Color histogram for these areas as well as four control points (white circle) were obtained with ImageJ.
Reliability of the digital assessment method
To determine the reliability of our method, 15 images were randomly selected to undergo two rounds of complete tracing and data extraction by a single investigator completed within a week from each other. Wound and control regions were traced according to the above protocol with area and color measurements being collected during each round. To measure the similarity of the data between rounds, Intraclass Correlation Coefficients were calculated using SPSS [SPSS Inc., Chicago, Ill, (12)] for the inter-commissure width, area and color values, followed by Student t-test analysis calculated in NCSS (13).
Objective visual scar assessment
Four physicians (two dermatologists, one pediatric otolaryngologist, one facial plastic surgeon) independently and subjectively graded the scars of all the patients. The clinicians rated the same photographs that were used for the digital imaging analysis. They graded the scar using a visual numerical score (0-5, with 5 being an imperceptible scar) similar to the one previously described (14) including the following criteria: redness, scar width, color, texture and overall appearance.
Genotyping analysis
We genotyped each individual for 17 single nucleotide polymorphisms (SNPs) at seven loci previously associated with orofacial cleft - ABCA4/ARHGAP29 (15), IRF6 (15, 16), MSX1 (17), 8q24 (15), FOXE1 (18), TGFB3 (17), and MAFB (15) (for details, see Table 2). Genotyping was completed using TaqMan SNP Genotyping Assays (Applied BioSystems/Life Technologies, Forser City, CA) and detected using the Applied BioSystems Prism 7900HT.
Table 2.
Summary of genes tested for association with scar phenotype
| Gene | SNPs tested | Expression in the face | Role in clefting | Expression in the skin | Role in scar/wound healing |
|---|---|---|---|---|---|
| ABCA4-ARHGAP29 | rs560426 | In the oral epithelium, medial edge epithelium of murine embryos (36) | Genetic variants associated with NSCLP (15, 36) | Predominantly in the epidermis (36) | ND |
| Interferon Regulatory Factor 6 IRF6 | rs2235371, rs642961 | In the oral epithelium, medial edge epithelium of murine embryos (37, 38) | Mutations cause VWS and PPS (34), genetic variants associated with NSCLP (16, 39) Knockout mice have orofacial anomalies (37, 40) | Predominantly in suprabasal layers of the epidermis (37) | Patients with VWS have increased likelihood of worse surgical cleft outcome (35) |
| Muscle Segment Homeobox Gene of Drosophila MSX1 | rs4075, rs12532 | Restricted to the anterior of the first upper molar site in palatal mesenchyme (41) | Genetic variants associated with NSCLP (17) Knockout mice have cleft palate (42) |
Predominantly in epidermal structures in fetal skin (43) | ND |
| 8q24 | rs987525 | ND | Genetic variants associated with NSCLP (15, 44) | ND | ND |
| Forkhead/wingled-helix domain transcription factor FOXE1 | rs4460498, rs3758249, rs14443434 | In oral epithelium of murine embryos (45) | Genetic variants associated with NSCLP (18, 46) | In the epidermis and hair follicles (47) | ND |
| Transforming growth factor beta 3 TGFB3 | rs1012861 | In the medial edge epithelium (48) | Genetic variants associated with NSCLP (17) Knockout mice have cleft palate (49) |
In all layers of the epidermis (50) | Plays a role in scarless wound healing (29, 30) |
| V-maf musculoaponeurotic fibrosarcoma oncogene homolog B MAFB | rs11696257 | In the oral epithelium of murine embryos (15) | Genetic variants associated with NSCLP (15) | In differentiated keratinocytes and hair follicles (51, 52) | ND |
NSCLP, non-syndromic cleft lip and palate
VWS, Van der Woude Syndrome
PPS, Popliteal Pterygium Syndrome
ND, not determined
Statistical analysis
Statistical analysis was performed with appropriate test for each study as indicated in tables or figure legend. A level of P < 0.05 was considered statistically significant. Given the exploratory nature of our work and the small sample size, we did not compensate for multiple comparisons. Distribution of the data was verified and means were used for normal distribution, while a Wilcoxon rank test of medians was used in the cases where the distribution was not normal.
Results
Establishment of digital parameters and repeatability
To perform digital analysis of facial images, we defined multiple areas within the lip and the philtrum (Fig 1). On the lip, the surgical scar constituted the affected area, whereas the contralateral identical area represented the non-affected area (or control). The same distinction between surgical scar (affected area) and contralateral area (non-affected control) was applied to the philtrum region of the face. In addition, four unaffected areas (two on each side) were artificially selected at the border of the philtrum spanning a line between the ala of the nose and the labial commissure (Fig 1).
In order to provide confidence in our measurements, repeatability measurements for 15 patients one week apart were performed. The intraclass correlation coefficient (ICC) ranges between 0 and 1, in which 0 implies null repeatability and 1 indicates perfect repeatability. We considered values > 0.8 indicators of excellent repeatability, and values between 0.7 and 0.8 acceptable. As shown in Table 3, our method demonstrated excellent repeatability from ICC scores > 0.8 for the vast majority of the repeated measurements (29/34 measurements, or 85.3%), acceptable repeatability > 0.7 for a few measurements (3/34 measurements, or 8.8%), and only the contralateral unaffected area of the lip did not meet our acceptable criteria (ICC = 0.640).
Table 3.
Repeated measures of 15 images are not significantly different
| Interclass Correlation Coefficients for scar color | Interclass Correlation Coefficients for scar size | |||
|---|---|---|---|---|
| Green | Blue | Red | ||
| Philtrum | ||||
| Scar area | 0.704 | 0.803 | 0.860 | 0.934 |
| Total unaffected area | 0.963 | 0.980 | 0.994 | ND |
| Contralateral unaffected area | 0.925 | 0.935 | 0.901 | 0.798 |
| Lip | ||||
| Scar area | 0.879 | 0.908 | 0.930 | 0.884 |
| Total unaffected area | 0.961 | 0.977 | 0.943 | ND |
| Contralateral unaffected area | 0.946 | 0.952 | 0.928 | 0.640 |
| Random controls | ||||
| Control 1 | 0.985 | 0.988 | 0.988 | NA |
| Control 2 | 0.944 | 0.936 | 0.772 | NA |
| Control 3 | 0.976 | 0.974 | 0.984 | NA |
| Control 4 | 0.946 | 0.959 | 0.941 | NA |
ND, Not determined; NA, Not applicable
Interclass correlation coefficients were complemented with a one-sample t-test of the mean difference between round 1 and round 2. This more conservative statistical analysis shows that some, but not all the data was statistically significant (Table 4). In cases where the sample demonstrated a non-normal distribution, a Wilcoxon Signed Rank test of medians was used instead. Together, these data demonstrate acceptable to excellent repeatability in our measurements insuring high quality in further analyses.
Table 4.
Repeatability of the digital scar measurements (Student t-test)
| P value after Student t-test for color | P value after Student t-test for scar size | |||
|---|---|---|---|---|
| Green | Blue | Red | ||
| Philtrum | ||||
| scar area | 0.293 | 0.293 | 0.268 | 0.284 |
| total unaffected area | 0.670 | 0.798 | 0.595 | ND |
| contralateral unaffected area | 0.039 | 0.083 | 0.042 | 0.458 |
| Lip | ||||
| scar area | 0.589 | 0.842 | 0.977 | 0.119 |
| total unaffected area | 0.754 | 0.894 | 0.754 | ND |
| contralateral unaffected area | 0.037 | 0.032 | 0.251 | 0.908 |
| Random controls | ||||
| control 1 | 0.222 | 0.222 | 0.147 | NA |
| control 2 | 0.292 | 0.316 | 0.349 | NA |
| control 3 | 0.023 | 0.016 | 0.159 | NA |
| control 4 | 0.234 | 0.268 | 0.377 | NA |
ND: not determined; NA: not applicable;
The color profile of several unaffected skin areas is similar
As part of the validation of our novel digital imaging analysis technique, we tested whether the color profile of our control points (control points 1 to 4 and the contralateral unaffected area of the philtrum) were correlated. We used ImageJ to obtain the red, green and blue color intensity for each area and calculated the Pearson correlation coefficients (r) between these measurements (Table 5). A value of 1 indicates a perfect linear equation between two data points as shown in the 2×2 matrix between identical areas (Table 5, diagonal). Our data show that the color profiles of the four philtrum controls were significantly correlated with each other (P < 0.001). Furthermore, the color of the contralateral unaffected area was significantly correlated to the color of controls 1, 3 and 4. Overall, these data support the validity of our method to adequately capture the color profile of unaffected areas and indicates similar color profiles between all these areas, allowing us to use them interchangeably.
Table 5.
Color of unaffected contralateral and random control areas is significantly similar
| Contralateral Unaffected | Control 1 | Control 2 | Control 3 | Control 4 | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Green | Blue | Red | Green | Blue | Red | Green | Blue | Red | Green | Blue | Red | Green | Blue | Red | ||
| Contralateral Unaffected | ra | 1 | 1 | 1 | 0.343 | 0.405 | 0.459 | 0.025NS | 0.126NS | 0.210NS | 0.393 | 0.418 | 0.516 | 0.219NS | 0.325 | 0.335 |
| Control 1 | r | 0.343 | 0.405 | 0.459 | 1 | 1 | 1 | 0.694 | 0.748 | 0.633 | 0.832 | 0.860 | 0.837 | 0.686 | 0.758 | 0.605 |
| Control 2 | r | 0.025NS | 0.126NS | 0.210NS | 0.694 | 0.748 | 0.633 | 1 | 1 | 1 | 0.569 | 0.651 | 0.521 | 0.763 | 0.779 | 0.764 |
| Control 3 | r | 0.393 | 0.418 | 0.516 | 0.832 | 0.860 | 0.837 | 0.569 | 0.651 | 0.521 | 1 | 1 | 1 | 0.757 | 0.814 | 0.703 |
| Control 4 | r | 0.219NS | 0.325 | 0.335 | 0.686 | 0.758 | 0.605 | 0.763 | 0.779 | 0.764 | 0.757 | 0.814 | 0.703 | 1 | 1 | 1 |
Pearson correlation coefficient
non-significant
The color profile of the affected skin is significantly different than unaffected skin
The parameters that contribute to the visual appearance of the scar include texture and color. Therefore, we hypothesized that the color of the affected lip and philtrum is different than the respective unaffected skin. As described above, we used the contralateral area of the lip and philtrum as controls since they are the most relevant unaffected regions. Using ImageJ to obtain the red, green and blue mean color values for each participant, our data indicate that the values followed a normal distribution among the population (data not shown). Using Student t-test analysis, we found a statistically significant color difference between the mean color of the affected philtrum area and the contralateral unaffected philtrum control (Table 6, P ranging from 1.20 × 10−05 to 1.95 × 10−14). For the lip region, only the red color was significantly different in affected and unaffected area (P = 1.2 × 10−03), while no statistical difference was observed for the green and blue colors (Table 6).
Table 6.
Color of the affected and unaffected skin is significantly different
Mean color value of affected and unaffected were substracted.
P value after Student t-test on the mean color difference.
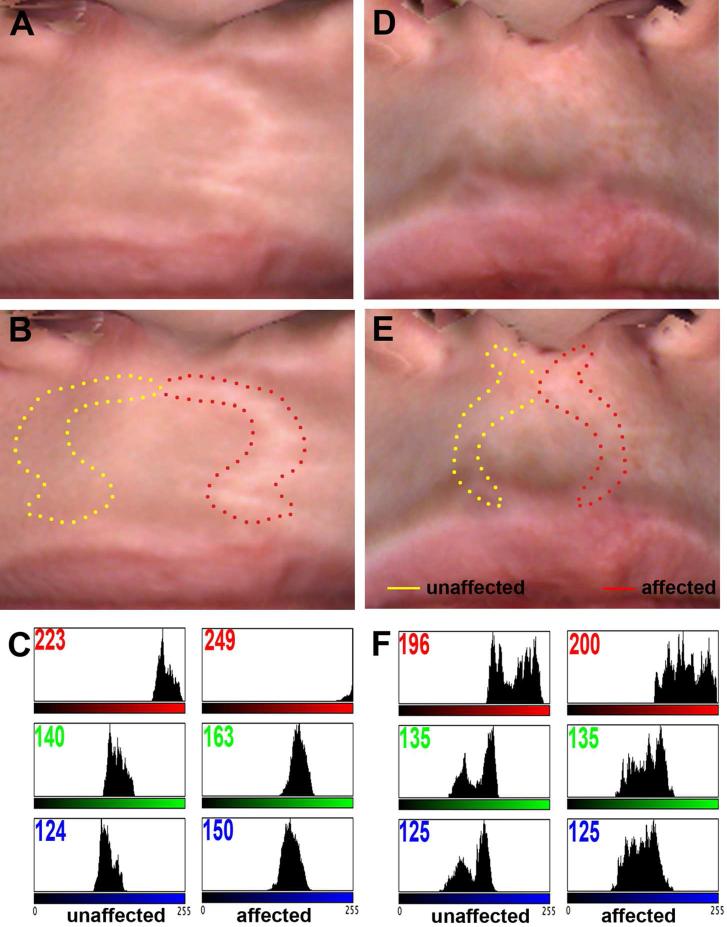
The significance of this finding is illustrated in Figure 2. The color profile of the scar of two participants with cleft lip and palate were analyzed with ImageJ. A much greater difference in mean color was observed when the scar was highly visible (Fig 2 a-c) compared to a minor difference for an imperceptible scar (Fig 2 d-f). Overall, these data demonstrate our ability to digitally quantify the color of a scar and derive a quantitative phenotype of wound healing.
Figure 2.
Digital image analysis of scar after cleft lip surgical repair. Digital macrophotographs of two patients with poor (A) and excellent (D) healing outcome. The affected (dotted red lines) and unaffected (dotted yellow lines) contralateral areas were traced for each patient (B, E). Color histograms of each area were generated using ImageJ software (C, F). The number in the top left corner of each profile corresponds to the mean color value. Note the difference in intensity in the three red, green and blue color components between unaffected and affected area in the poor healer (C), compared to the absence of difference in the good healer (F).
Clinical raters are moderately correlated amongst each other as well as with the data from the digital analysis
To fully validate our digital assessment of the scar, we asked four independent physicians to use a traditional visual analog scale to grade the surgical outcome of each patient. Five different criteria were used to characterize the scar: redness, scar width, color difference between the wound and the unaffected skin, texture and overall appearance. The Pearson correlation coefficient indicated that the ratings of the clinicians were significantly correlated for all but the between the wound and the unaffected skin criteria (Table 7). Our hypothesis was for a strong level of agreement between raters (correlation coefficient > 0.8). However, our data show a moderate level of correlation, with a Pearson correlation coefficient average of 0.495. These data indicates that clinicians only agree about half of the time and support the need for a more objective measure of scar characterization.
Table 7.
Visual raters are poorly correlated
| Part I: Redness | ||||
|---|---|---|---|---|
| Rater 1 | Rater 2 | Rater 3 | Rater 4 | |
| Rater 1 | 1a | 0.589 | 0.569 | 0.528 |
| Rater 2 | 0.589 | 1 | 0.434 | 0.649 |
| Rater 3 | 0.569 | 0.434 | 1 | 0.646 |
| Rater 4 | 0.528 | 0.649 | 0.646 | 1 |
| Part II: Scar width | ||||
|---|---|---|---|---|
| Rater 1 | Rater 2 | Rater 3 | Rater 4 | |
| Rater 1 | 1a | 0.510 | 0.508 | 0.449 |
| Rater 2 | 0.510 | 1 | 0.475 | 0.364 |
| Rater 3 | 0.508 | 0.475 | 1 | 0.610 |
| Rater 4 | 0.449 | 0.364 | 0.610 | 1 |
| Part III: Color difference between scar and unaffected skin | ||||
|---|---|---|---|---|
| Rater 1 | Rater 2 | Rater 3 | Rater 4 | |
| Rater 1 | 1a | 0.207NS | 0.273 | 0.277 |
| Rater 2 | 0.207NS | 1 | 0.468 | 0.359 |
| Rater 3 | 0.273NS | 0.468 | 1 | 0.470 |
| Rater 4 | 0.277 | 0.359 | 0.470 | 1 |
| Part IV: Texture | ||||
|---|---|---|---|---|
| Rater 1 | Rater 2 | Rater 3 | Rater 4 | |
| Rater 1 | 1a | 0.496 | 0.447 | 0.624 |
| Rater 2 | 0.496 | 1 | 0.415 | 0.400 |
| Rater 3 | 0.447 | 0.415 | 1 | 0.571 |
| Rater 4 | 0.624 | 0.400 | 0.571 | 1 |
| Part V: Overall cosmesis | ||||
|---|---|---|---|---|
| Rater 1 | Rater 2 | Rater 3 | Rater 4 | |
| Rater 1 | 1a | 0.624 | 0.613 | 0.513 |
| Rater 2 | 0.624 | 1 | 0.442 | 0.273 |
| Rater 3 | 0.613 | 0.442 | 1 | 0.563 |
| Rater 4 | 0.513 | 0.273 | 0.563 | 1 |
| Part VI: Sum of all the scores | ||||
|---|---|---|---|---|
| Rater 1 | Rater 2 | Rater 3 | Rater 4 | |
| Rater 1 | 1a | 0.637 | 0.603 | 0.561 |
| Rater 2 | 0.637 | 1 | 0.515 | 0.482 |
| Rater 3 | 0.603 | 0.515 | 1 | 0.667 |
| Rater 4 | 0.561 | 0.482 | 0.667 | 1 |
Pearson correlation coefficient
non-significant
Despite this fairly wide variation amongst raters, we wondered if their scores would correlate with the digital analysis parameters. Pearson correlation and regression analysis of the average of the clinicians’ total score showed statistical significance with the average of the wound area in the philtrum (and the percentage of the wound area over the entire philtrum area) only, yet the correlation coefficient was low (0.300 and 0.302, P = 0.02, Table 8). Similar correlations were obtained with overall appearance, the scar width, the color difference and the texture. Interestingly, the overall redness in the lip was poorly correlated (correlation coefficient 0.277, P = 0.02) with the percentage of the affected lip (Table 8). No correlation was identified between any of the surgeons's criteria and the digital difference in color profiles between affected and unaffected area (Table 8). Together, these results indicate a weak correlation between the digital analysis of the wound and the analog scoring of the same image by physicians, promoting the use of digital analysis as a more sensitive and reliable tool compared to analog rating by individuals.
Table 8.
Correlation analysis between visual and digital scar assessment
| Clinicians's average score | ||||||
|---|---|---|---|---|---|---|
| Redness | Scar width | Color difference | Texture | Overall appearance | Total | |
| Philtrum | ||||||
| green | −0.155a,NS | 0.028NS | 0.248NS | −0.111NS | 0.035NS | 0.022NS |
| blue | −0.067NS | 0.041NS | 0.247NS | −0.099NS | 0.055NS | 0.052NS |
| red | −0.009NS | 0.014NS | 0.258NS | −0.072NS | 0.085NS | 0.075NS |
| affected area | 0.029NS | 0.338** | 0.334** | 0.225NS | 0.250* | 0.300* |
| percentage of affected area | 0.029NS | 0.349* | 0.363** | 0.180NS | 0.257* | 0.302* |
| Lip | ||||||
| green | −0.142NS | 0.048NS | 0.044NS | 0.089NS | 0.065NS | 0.032NS |
| blue | −0.102NS | 0.030NS | 0.322NS | 0.067NS | 0.055NS | 0.084NS |
| red | −0.035NS | 0.013NS | 0.158NS | 0.066NS | 0.109NS | 0.083NS |
| affected area | 0.201NS | 0.010NS | −0.020NS | 0.111NS | 0.130NS | 0.103NS |
| percentage of affected area | 0.277* | 0.077NS | 0.103NS | 0.153NS | 0.215NS | 0.202NS |
Pearson correlation coefficient
non significant after Pearson correlation analysis
significant after Pearson correlation analysis (P < 0.05)
significant after Pearson correlation analysis (P < 0.01)
Genetic variants associated with cleft lip wound repair outcome
In order to determine whether genetic variants contribute to the appearance of the surgical outcome of lip repair, we regressed the mean color difference between the affected and unaffected areas within the same individual, as well as the percentage of philtrum and lip covered by wounded area, on 17 SNPs at seven chromosomal loci, one SNP at a time, using ordinary least squares (OLS). Our data showed significant correlation between rs1012861, a genetic variant upstream of TGFB3, and the red color of the philtrum (P = 0.025, Table 9) and the percentage of the wound area in the lip (P = 0.023, Table 9). Significant association was also observed with the percentage of wound area in the philtrum and a genetic variant rs560426 within the ABCA4/ARHGAP29 region (P = 0.031, Table 9). A marginal correlation was observed between the same SNP and the blue color of the lip (P = 0.059, Table 9). No correlation was identified with IRF6, FOXE1, MAFB, MSX1, and the chromosomal region 8q24 (Table 9 and data not shown).
Table 9.
Regression analysis for candidate genes
| ABCA4/ARHGAP29 rs560426 chr1: 94553438 | IRF6 rs642961 chr1: 209989270 | TGFB3 rs1012861 chr14: 76378579 | |
|---|---|---|---|
| Philtrum | |||
| green | 0.566a | 0.251 | 0.107 |
| blue | 0.728 | 0.319 | 0.099 |
| red | 0.861 | 0.278 | 0.025 |
| percentage of scar | 0.031 | 0.764 | 0.094 |
| Lip | |||
| green | 0.089 | 0.420 | 0.103 |
| blue | 0.059 | 0.538 | 0.118 |
| red | 0.143 | 0.881 | 0.081 |
| percentage of scar | 0.601 | 0.931 | 0.023 |
Data are P values after regression analysis. P < 0.05 are considered significant.
Discussion
There is an unmet need for a tool to identify risk factors for suboptimal wound healing and subsequently, prominent scars in order to be able to screen at risk individuals and design targeted clinical practices to address these risks. The present study establishes a new objective, reliable, and non-invasive digital imaging method to characterize the phenotype of surgical scars. Moreover, for the first time, single nucleotide polymorphisms in TGFB3 and in ABCA4/ARHGAP29 were identified as genetic variants associated with objective measures of scar characteristics.
Cutaneous scarring is the final outcome of the wound healing process, and has the potential to affect individuals physiologically and psychologically. Surgery is the method of choice to correct cleft lip and palate, unavoidably leaving scars with the potential to affect both speech and facial aesthetics (19, 20). Typically, scars can be defined by size, texture, and color. We utilized digital images acquired using a three-dimensional software package and the ImageJ software to develop a novel method to characterize the scar of patients with unilateral scarring in the lip. This method presents numerous advantages over existing tools [for review (21)].
First, it is easy to use. It only requires a digital image and ImageJ software that is freely accessible online (http://rsb.info.nih.gov/ij/). The use of computerized color was initially reported for assessing burn scar hypertrophy (22) and expanded to other wounds and cutaneous diseases (23-26), yet had not been reported for surgical wounds. Although our camera was internally calibrated, it does not require elaborated adjustments as with other systems (24) nor does it require calibration with an external color scale like the Macbeth Color Checker (GretagMacbeth, Windsor, NY). In fact, our method employs the patient's own unaffected skin from the same image as a control, and our data demonstrate that the contralateral unaffected area and four random areas around the philtrum can be interchangeably used as controls (Table 5). This opens the possibility to extend our patient population to individuals with bilateral clefting, and design further studies for any wounded individuals as long as unaffected skin is present in the digital image. Overall, it presents the major advantage of being internally controlled for each individual and is independent of illumination or other image settings.
Second, it is non-invasive. Other non-invasive color assessment include reflectance spectrophotometers , tristimulus colorimetric instruments and narrow-band simple reflectance meters. Although they have been used for the characterization of wounds and skin diseases, they are expensive, sometimes too big to be placed on certain body parts, and require direct application on the scar, leading to a change in color by simple application pressure of the device.
Third, our method is highly reliable and generates continuous quantitative traits, allowing greater variation and more precise and powerful analysis. This is in contrast to the vast majority of rating scales like the Vancouver Burn Scar Scale (8), that are ordinal and filled by clinicians. If length and width can be objectively measured, color assessment requires clinical and personal judgments. Even if one can visually discern variations in the appearance of different skin, it is difficult to quantify such differences based on a person's assessment. This is illustrated by our own data (Table 7) showing moderate correlation amongst four independent raters, particularly in characterizing the color difference between the scar and the unaffected skin (r = 0.2, Table 7). Our digital analysis, however, identified significant differences in all three color channels between the color of the scar and the unaffected skin (Table 6). The total score of the clinicians’ rating correlated significantly with the size of the scar only, pointing to the predominance of this criterion in a subjective overall characterization of a scar. It further emphasizes the higher sensitivity of our method, giving us more power to perform analysis of scar outcome. From a practical perspective, we believe this method appears to have sufficient consistency and uniformity to provide refined and standardized scar assessment. Ultimately, this may help provide a much needed tool to assess wound healing that eliminates many of the historical challenges such as logistical difficulties in arranging face-to-face interactions for scar assessment and high inter-rater variability.
The greatest variations in color between the affected and unaffected areas were observed in the red channel. Using two-dimensional images extracted from photographs acquired with a Di3D stereo photogrammetry system, Ayoub et al reported an elevated redness in scar tissue from individuals with unilateral orofacial clefts and increased luminosity in scars from individuals with unilateral cleft lip but not cleft lip and palate (27). These data are in agreement with our findings, yet we did not evaluate luminosity and did not distinguish between cleft lip and cleft lip and palate. It would be interesting to consider these two cleft types separately in the future, as it becomes increasingly evident that these two phenotypes are developmentally distinct (28).
Phenotypic variations in healing indicate that part of our genetic make-up could contribute to wound healing outcomes. One of the major challenges in performing genotype-phenotype association studies is using phenotypic measures with limited variation. We believe that our novel digital analysis tool generates a well-characterized scar phenotype that can capture detailed variation. Using this method for phenotypic assessment, we identified single nucleotide polymorphisms in TGFB3 and ABCA4/ARHGAP29 to be associated with scars with colors that differed the most from unaffected skin. These data confirm a previously known role for TGFB3 in scarless wound healing (29, 30) and its requirement for proper wound healing in vivo (31). The other polymorphism is in an intronic region of ABCA4 that is believed to regulate ARHGAP29 (32). ARHGAP29 belongs to the family of guanine activating protein regulating the level of active RhoA-GTPase, which is essential for keratinocyte adhesion and migration (33). Although no role for ARHGAP29 in wound healing has been described, we speculate it could affect levels of active RhoA to regulate cellular migration. ARHGAP29 interacts genetically with Interferon Regulatory Factor 6 (IRF6) (32), a transcription factor that when mutated, leads to two clefting syndromes (Van der Woude syndrome and Popliteal Pterygium syndrome) (34). We showed that patients with VWS were more likely to have wound healing complications following surgical cleft repair compared to patients with a non-syndromic cleft (35). We did not detect an association between genetic variants in IRF6 and poor surgical outcomes. This could be due to the low number of informative individuals (none for one of the SNPs) or an association between IRF6 and wound healing with other genetic variants that we have not tested for yet.
There are a few limitations to our study. First, we investigated a population of individuals with clefts who could have a unique genetic background and have been exposed to particular environment constituting confounding factors. The fact that we identified genetic variants in TGFB3 and ARHGAP29 may be related to their clefts rather than wound healing. Moreover, other cleft genes did not show association with poor healing. Therefore, we believe that these two genes play a critical role in tissue repair. As TGFB3 and ARHGAP29 are associated with orofacial clefts (17, 32), one could speculate that embryologic development of the palate and tissue repair share a common gene regulatory network involving at least TGFB3, ARHGAP29 and IRF6. Second, our study was a pilot and should be expanded and replicated in another sample of individuals with oral clefts and for other surgical procedures. Furthermore, it was exploratory and was not designed to find the causal genetic variant. Future studies should include genome wide association studies to evaluate risk variant genome wide, as previously described for keloid scars (4), instead of the candidate-gene approach.
In conclusion, our study is the first step towards standardized, reliable wound healing evaluation and the potential to predict wound repair outcome following surgery or trauma. This is of high clinical significance to patients undergoing surgical procedures with the potential of designing interventions tailored to an individual's genomic and genetic profile.
Acknowledgements
The authors wish to thank Jeff Murray for his constant support and the patients and their family for their participation. The authors wish to acknowledge the technical assistance of Dr. Brian Swick and Dr. Jill Lightfoot for scoring images, and Chika Takeuchi for genotyping. The original images and DNA samples were collected with funds from the Center for Disease Control and Prevention (CDC) grant 1R01DD000295. Partial support for this project was also provided by AR055313 and DE08559 (MD). Brian Smith was supported by T32 DEO14678-09. The contents of this work are the sole responsibility of the authors and do not necessarily represent the official views of the CDC and NIH.
Footnotes
None of the authors have a conflict of interest.
Literature cited
- 1.Baum CL, Arpey CJ. Normal cutaneous wound healing: clinical correlations with cellular and molecular events. Dermatol Surg. 2005;31(6):674–86. doi: 10.1111/j.1524-4725.2005.31612. [DOI] [PubMed] [Google Scholar]
- 2.Sen CK, Gordillo GM, Roy S, Kirsner R, Lambert L, Hunt TK, et al. Human skin wounds: a major and snowballing threat to public health and the economy. Wound Repair Regen. 2009;17(6):763–71. doi: 10.1111/j.1524-475X.2009.00543.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shih B, Bayat A. Comparative genomic hybridisation analysis of keloid tissue in Caucasians suggests possible involvement of HLA-DRB5 in disease pathogenesis. Arch Dermatol Res. 2012;304(3):241–9. doi: 10.1007/s00403-011-1182-4. [DOI] [PubMed] [Google Scholar]
- 4.Nakashima M, Chung S, Takahashi A, Kamatani N, Kawaguchi T, Tsunoda T, et al. A genome-wide association study identifies four susceptibility loci for keloid in the Japanese population. Nat Genet. 2010;42(9):768–71. doi: 10.1038/ng.645. [DOI] [PubMed] [Google Scholar]
- 5.Margolis DJ, Gupta J, Thom SR, Townsend RR, Kanetsky PA, Hoffstad O, et al. Diabetes, lower extremity amputation, loss of protective sensation, and neuronal nitric oxide synthase associated protein in the chronic renal insufficiency cohort study. Wound Repair Regen. 2013;21(1):17–24. doi: 10.1111/j.1524-475X.2012.00866.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Broadbent J, Walsh T, Upton Z. Proteomics in chronic wound research: potentials in healing and health. Proteomics Clin Appl. 2010;4(2):204–14. doi: 10.1002/prca.200900152. [DOI] [PubMed] [Google Scholar]
- 7.Lee JP, Hopf HW, Cannon-Albright LA. Empiric evidence for a genetic contribution to predisposition to surgical site infection. Wound Repair Regen. 2013;21(2):211–5. doi: 10.1111/wrr.12024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Baryza MJ, Baryza GA. The Vancouver Scar Scale: an administration tool and its interrater reliability. The Journal of burn care V rehabilitation. 1995;16(5):535–8. doi: 10.1097/00004630-199509000-00013. [DOI] [PubMed] [Google Scholar]
- 9.Draaijers LJ, Tempelman FR, Botman YA, Tuinebreijer WE, Middelkoop E, Kreis RW, et al. The patient and observer scar assessment scale: a reliable and feasible tool for scar evaluation. Plast Reconstr Surg. 2004;113(7):1960–5. doi: 10.1097/01.prs.0000122207.28773.56. discussion 6-7. [DOI] [PubMed] [Google Scholar]
- 10.Karamfilov T, Weichold S, Karte K, Vilser W, Wollina U. Remittance spectroscopy mapping of human skin in vivo. Skin Res Technol. 1999;5:49–52. doi: 10.1111/j.1600-0846.1996.tb00073.x. [DOI] [PubMed] [Google Scholar]
- 11.Takiwaki H, Shirai S, Watanabe Y, Nakagawa K, Arase S. A rudimentary system for automatic discrimination among basic skin lesions on the basis of color analysis of video images. J Am Acad Dermatol. 1995;32(4):600–4. doi: 10.1016/0190-9622(95)90344-5. [DOI] [PubMed] [Google Scholar]
- 12.Shrout PE, Fleiss JL. Intraclass correlations: uses in assessing rater reliability. Psychological bulletin. 1979;86(2):420–8. doi: 10.1037//0033-2909.86.2.420. [DOI] [PubMed] [Google Scholar]
- 13.Hintze J. NCSS 2007. NCSS, LLC; Kaysville, Utah: 2007. [Google Scholar]
- 14.Sniezek PJ, Walling HW, DeBloom JR, 3rd, Messingham MJ, VanBeek MJ, Kreiter CD, et al. A randomized controlled trial of high-viscosity 2-octyl cyanoacrylate tissue adhesive versus sutures in repairing facial wounds following Mohs micrographic surgery. Dermatol Surg. 2007;33(8):966–71. doi: 10.1111/j.1524-4725.2007.33199.x. [DOI] [PubMed] [Google Scholar]
- 15.Beaty TH, Murray JC, Marazita ML, Munger RG, Ruczinski I, Hetmanski JB, et al. A genome-wide association study of cleft lip with and without cleft palate identifies risk variants near MAFB and ABCA4. Nat Genet. 2010;42(6):525–31. doi: 10.1038/ng.580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rahimov F, Marazita ML, Visel A, Cooper ME, Hitchler MJ, Rubini M, et al. Disruption of an AP-2alpha binding site in an IRF6 enhancer is associated with cleft lip. Nat Genet. 2008;40(11):1341–7. doi: 10.1038/ng.242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lidral AC, Romitti PA, Basart AM, Doetschman T, Leysens NJ, Daack-Hirsch S, et al. Association of MSX1 and TGFB3 with nonsyndromic clefting in humans. Am J Hum Genet. 1998;63(2):557–68. doi: 10.1086/301956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Moreno LM, Mansilla MA, Bullard SA, Cooper ME, Busch TD, Machida J, et al. FOXE1 association with both isolated cleft lip with or without cleft palate, and isolated cleft palate. Hum Mol Genet. 2009;18(24):4879–96. doi: 10.1093/hmg/ddp444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kummer AW, Clark SL, Redle EE, Thomsen LL, Billmire DA. Current practice in assessing and reporting speech outcomes of cleft palate and velopharyngeal surgery: a survey of cleft palate/craniofacial professionals. Cleft Palate Craniofac J. 2012;49(2):146–52. doi: 10.1597/10-285. [DOI] [PubMed] [Google Scholar]
- 20.Christofides E, Potgieter A, Chait L. A long term subjective and objective assessment of the scar in unilateral cleft lip repairs using the Millard technique without revisional surgery. J Plast Reconstr Aesthet Surg. 2006;59(4):380–6. doi: 10.1016/j.bjps.2005.04.037. [DOI] [PubMed] [Google Scholar]
- 21.Perry DM, McGrouther DA, Bayat A. Current tools for noninvasive objective assessment of skin scars. Plast Reconstr Surg. 2010;126(3):912–23. doi: 10.1097/PRS.0b013e3181e6046b. [DOI] [PubMed] [Google Scholar]
- 22.Davey RB, Sprod RT, Neild TO. Computerised colour: a technique for the assessment of burn scar hypertrophy. A preliminary report. Burns. 1999;25(3):207–13. doi: 10.1016/s0305-4179(98)00137-5. [DOI] [PubMed] [Google Scholar]
- 23.Kaartinen IS, Valisuo PO, Alander JT, Kuokkanen HO. Objective scar assessment—a new method using standardized digital imaging and spectral modelling. Burns. 2011;37(1):74–81. doi: 10.1016/j.burns.2010.03.008. [DOI] [PubMed] [Google Scholar]
- 24.Yamamoto T, Takiwaki H, Arase S, Ohshima H. Derivation and clinical application of special imaging by means of digital cameras and Image J freeware for quantification of erythema and pigmentation. Skin Res Technol. 2008;14(1):26–34. doi: 10.1111/j.1600-0846.2007.00256.x. [DOI] [PubMed] [Google Scholar]
- 25.Setaro M, Sparavigna A. Quantification of erythema using digital camera and computer-based colour image analysis: a multicentre study. Skin Res Technol. 2002;8(2):84–8. doi: 10.1034/j.1600-0846.2002.00328.x. [DOI] [PubMed] [Google Scholar]
- 26.Whitmer K, Barford B, Turner M, Sullivan D, Sommers M. Digital image analysis of facial erythema over time in persons with varied skin pigmentation. Skin Res Technol. 2011 doi: 10.1111/j.1600-0846.2011.00505.x. [DOI] [PubMed] [Google Scholar]
- 27.Ayoub A, Bell A, Simmons D, Bowman A, Brown D, Lo TW, et al. 3D Assessment of Lip Scarring and Residual Dysmorphology Following Surgical Repair of Cleft Lip and Palate: A Preliminary Study. Cleft Palate Craniofac J. 2011;48(4):379–87. doi: 10.1597/10-057. [DOI] [PubMed] [Google Scholar]
- 28.Harville EW, Wilcox AJ, Lie RT, Vindenes H, Abyholm F. Cleft lip and palate versus cleft lip only: are they distinct defects? Am J Epidemiol. 2005;162(5):448–53. doi: 10.1093/aje/kwi214. [DOI] [PubMed] [Google Scholar]
- 29.Ferguson MW, Duncan J, Bond J, Bush J, Durani P, So K, et al. Prophylactic administration of avotermin for improvement of skin scarring: three double-blind, placebo-controlled, phase I/II studies. Lancet. 2009;373(9671):1264–74. doi: 10.1016/S0140-6736(09)60322-6. [DOI] [PubMed] [Google Scholar]
- 30.Ferguson MW, O'Kane S. Scar-free healing: from embryonic mechanisms to adult therapeutic intervention. Philos Trans R Soc London B Biol Sci. 2004;359(1449):839–50. doi: 10.1098/rstb.2004.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Le M, Naridze R, Morrisson J, Biggs LC, Rhea L, Schutte BC, et al. Transforming growth factor beta 3 is required for proper excisional wound repair in vivo. PLoS ONE. 2012;7(10):e48040. doi: 10.1371/journal.pone.0048040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Leslie E, Mansilla MA, Biggs LC, Schuette K, Bullard S, Cooper M, et al. Expression and mutation analyses implicate ARHGAP29 as the etiologic gene for the cleft lip with or without cleft palate locus identified by genome-wide association on chromosome 1p22. Birth Defects Res A Clin Mol Teratol. 2012 doi: 10.1002/bdra.23076. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jackson B, Peyrollier K, Pedersen E, Basse A, Karlsson R, Wang Z, et al. RhoA is dispensable for skin development, but crucial for contraction and directed migration of keratinocytes. Mol Biol Cell. 2011;22(5):593–605. doi: 10.1091/mbc.E09-10-0859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kondo S, Schutte BC, Richardson RJ, Bjork BC, Knight AS, Watanabe Y, et al. Mutations in IRF6 Van der Woude and popliteal pterygium syndromes. Nat Genet. 2002;32(2):285–9. doi: 10.1038/ng985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jones JL, Canady JW, Brookes JT, Wehby GL, L'Heureux J, Schutte BC, et al. Wound complications after cleft repair in children with Van der Woude syndrome. J Craniofac Surg. 2010;21(5):1350–3. doi: 10.1097/SCS.0b013e3181ec6aad. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Leslie EJ, Mansilla MA, Biggs LC, Schuette K, Bullard S, Cooper M, et al. Expression and mutation analyses implicate ARHGAP29 as the etiologic gene for the cleft lip with or without cleft palate locus identified by genome-wide association on chromosome 1p22. Birth Defects Res A Clin Mol Teratol. 2012;94(11):934–42. doi: 10.1002/bdra.23076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ingraham CR, Kinoshita A, Kondo S, Yang B, Sajan S, Trout KJ, et al. Abnormal skin, limb and craniofacial morphogenesis in mice deficient for interferon regulatory factor 6 (Irf6). Nat Genet. 2006;38(11):1335–40. doi: 10.1083/ng1903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Knight AS, Schutte BC, Jian R, Dixon MJ. Developmental expression analysis of the mouse and chick orthologues of IRF6: the gene mutated in Van der Woude syndrome. Dev Dyn. 2006;235:1441–7. doi: 10.1002/dvdy.20598. [DOI] [PubMed] [Google Scholar]
- 39.Zucchero TM, Cooper ME, Maher BS, Daack-Hirsch S, Nepomuceno B, Ribeiro L, et al. Interferon regulatory factor 6 (IRF6) gene variants and the risk of isolated cleft lip or palate. N Engl J Med. 2004;351(8):769–80. doi: 10.1056/NEJMoa032909. [DOI] [PubMed] [Google Scholar]
- 40.Richardson RJ, Dixon J, Malhotra S, Hardman M, Knowles L, Boot-Handford RP, et al. Irf6 is a key determinant of the keratinocyte proliferation-differentiation switch. Nat Genet. 2006 doi: 10.1038/ng1894. [DOI] [PubMed] [Google Scholar]
- 41.Zhang Z, Song Y, Zhao X, Zhang X, Fermin C, Chen Y. Rescue of cleft palate in Msx1-deficient mice by transgenic Bmp4 reveals a network of BMP and Shh signaling in the regulation of mammalian palatogenesis. Development. 2002;129(17):4135–46. doi: 10.1242/dev.129.17.4135. [DOI] [PubMed] [Google Scholar]
- 42.Satokata I, Maas R. Msx1 deficient mice exhibit cleft palate and abnormalities of craniofacial and tooth development. Nat Genet. 1994;6(4):348–56. doi: 10.1038/ng0494-348. [DOI] [PubMed] [Google Scholar]
- 43.Stelnicki EJ, Komuves LG, Holmes D, Clavin W, Harrison MR, Adzick NS, et al. The human homeobox genes MSX-1, MSX-2, and MOX-1 are differentially expressed in the dermis and epidermis in fetal and adult skin. Differentiation. 1997;62(1):33–41. doi: 10.1046/j.1432-0436.1997.6210033.x. [DOI] [PubMed] [Google Scholar]
- 44.Birnbaum S, Ludwig KU, Reutter H, Herms S, Steffens M, Rubini M, et al. Key susceptibility locus for nonsyndromic cleft lip with or without cleft palate on chromosome 8q24. Nat Genet. 2009;41(4):473–7. doi: 10.1038/ng.333. [DOI] [PubMed] [Google Scholar]
- 45.Dathan N, Parlato R, Rosica A, De Felice M, Di Lauro R. Distribution of the titf2/foxe1 gene product is consistent with an important role in the development of foregut endoderm, palate, and hair. Dev Dyn. 2002;224(4):450–6. doi: 10.1002/dvdy.10118. [DOI] [PubMed] [Google Scholar]
- 46.Vieira AR, Avila JR, Daack-Hirsch S, Dragan E, Felix TM, Rahimov F, et al. Medical sequencing of candidate genes for nonsyndromic cleft lip and palate. PLoS Genet. 2005;1(6):e64. doi: 10.1371/journal.pgen.0010064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Eichberger T, Regl G, Ikram MS, Neill GW, Philpott MP, Aberger F, et al. FOXE1, a new transcriptional target of GLI2 is expressed in human epidermis and basal cell carcinoma. J Invest Dermatol. 2004;122(5):1180–7. doi: 10.1111/j.0022-202X.2004.22505.x. [DOI] [PubMed] [Google Scholar]
- 48.Fitzpatrick DR, Denhez F, Kondaiah P, Akhurst RJ. Differential expression of TGF beta isoforms in murine palatogenesis. Development. 1990;109(3):585–95. doi: 10.1242/dev.109.3.585. [DOI] [PubMed] [Google Scholar]
- 49.Kaartinen V, Voncken JW, Shuler C, Warburton D, Bu D, Heisterkamp N, et al. Abnormal lung development and cleft palate in mice lacking TGF-b3 indicates defects of epithelial-mesenchymal interaction. Nat Genet. 1995;11:415–21. doi: 10.1038/ng1295-415. [DOI] [PubMed] [Google Scholar]
- 50.Levine JH, Moses HL, Gold LI, Nanney LB. Spatial and temporal patterns of immunoreactive transforming growth factor beta1, beta2 and beta3 during excisional wound repair. Am J Pathol. 1993;143(2):368–80. [PMC free article] [PubMed] [Google Scholar]
- 51.Miyai M, Tanaka YG, Kamitani A, Hamada M, Takahashi S, Kataoka K. c-Maf and MafB transcription factors are differentially expressed in Huxley's and Henle's layers of the inner root sheath of the hair follicle and regulate cuticle formation. J Dermatol Sci. 2010;57(3):178–82. doi: 10.1016/j.jdermsci.2009.12.011. [DOI] [PubMed] [Google Scholar]
- 52.Borrelli S, Fanoni D, Dolfini D, Alotto D, Ravo M, Grober OM, et al. C/EBPdelta gene targets in human keratinocytes. PLoS One. 2010;5(11):e13789. doi: 10.1371/journal.pone.0013789. [DOI] [PMC free article] [PubMed] [Google Scholar]