Abstract
OBJECTIVE
Copy number variations (CNVs) in the CYP2D6 gene contribute to interindividual variation in drug metabolism. Since the most common duplicated allele in Asian populations is the nonfunctional CYP2D6*36 allele, the goal of this study was to identify CNV assays that can differentiate between multiple copies of the CYP2D6*36 allele and multiple copies of other CYP2D6 alleles.
METHODS
We determined CYP2D6 gene copy numbers in 32 subjects with known CYP2D6 CNVs from the Coriell Japanese-Chinese panel using four qRT-PCR assays. These assays target different regions of the CYP2D6 gene: 5′-flanking region (5′flank), intron 2 (Int2), intron 6 (Int6), and exon 9 (Ex9). The specific target site of the Ex9 assay was verified by sequencing the PCR amplicon.
RESULTS
Three of the CYP2D6 CNV assays (5′-flank, Int2, and Int6) estimated CYP2D6 copy numbers that were concordant for all 32 subjects. However, the Ex9 assay was concordant in only 10 of 32 samples. The 10 concordant samples did not contain any CYP2D6*36 alleles and the 22 discordant samples contained at least one CYP2D6*36 allele. Also, the Ex9 assay accurately quantified all of the non-CYP2D6*36 alleles in all samples. Ex9 amplicon sequencing indicated that it targets a region of CYP2D6 exon 9 that undergoes partial gene-conversion in the CYP2D6*36 allele.
CONCLUSION
CYP2D6 Ex9 CNV assay can be used to determine the copy number of non-CYP2D6*36 alleles. Selective amplification of non-CYP2D6*36 sequence by the Ex9 assay should be useful in determining the number of functional copies of CYP2D6 in Asian populations.
Keywords: Cytochrome P450 2D6, copy number variation, genotyping
INTRODUCTION
Copy number variation (CNV) involving deletion or multiplication of DNA segments is a primary source of variation in the human genome (1). Recent studies have suggested an increasing role for CNVs in many diseases including cancer, developmental diseases, mental illness, autoimmune diseases, and infectious diseases (2, 3). A number of genes involved in drug metabolism also exhibit copy number polymorphisms -- UGT2B17, UGT2B28 (4), SULTA1 (5), GSTT1, GSTM1 (6) and CYP2D6 (7, 8).
Cytochrome P450 2D6 (CYP2D6) is involved in the metabolism of ~20–25% of commonly prescribed drugs (9). Its activity is affected by environmental and genetic factors that lead to large interindividual variability in drug metabolism (10, 11). Genetically, CYP2D6 is highly polymorphic; variants include single nucleotide polymorphisms (SNPs), insertions, deletions, as well as, copy number variations resulting from CYP2D6 gene deletion or multiplication. Not all CNVs are functionally similar; CNVs in CYP2D6 gene can cause either reduced metabolism (gene deletions) or increased metabolism (>2 functional copies). The frequencies of CYP2D6 CNVs vary considerably between ethnic populations (7, 8). For example, the frequency of the CYP2D6 whole gene deletion (CYP2D6*5) ranges from 1–7% in all the three major ethnic populations (7) and the frequency of CYP2D6 gene multiplication is as high as 45% in Asians (12). The genetics of CYP2D6 are further complicated by the variety of CYP2D6 alleles that exist as multiple copies. While the functional CYP2D6*1 and *2 duplications are frequent among all ethnicities (7, 8), the nonfunctional CYP2D6*4 and *36 allele duplications are more common among Caucasians (7) and Asians (12, 13), respectively. These studies emphasize that the number of copies of a gene detected by the CNV assays cannot reasonably be used to estimate the phenotypic consequence without identification of the sequence of each copy.
The currently available genome-wide association studies (GWAS) arrays have relatively few probes covering the CYP2D6 locus, and thus are insensitive for accurately identifying allele specific CYP2D6 CNVs. The determination of CYP2D6 gene copy number is also complicated by the presence of highly homologous pseudogenes, CYP2D8 and CYP2D7 and by the presence of CYP2D6-CYP2D7 fusion genes like CYP2D6*36, which includes the 5′ end of the CYP2D6 gene fused with the 3′end of the CYP2D7 gene due to gene-recombination. Furthermore, because of the large sizes of CYP2D6 gene deletion and duplications, relatively difficult techniques including long-template PCR and Southern blotting are commonly used for copy number estimation (14). Recently, several qRT-PCR assays have been reported (12, 14, 15). Commercial qRT-PCR assays have also become available; however, since the exact positions of the target sequences are not provided for these assays, their ability to distinguish between specific alleles and fusion genes must be determined experimentally. Since our preliminary studies showed discrepancies between some of the qRT-PCR assays, the objective of this study was to evaluate the ability of several of these assays to amplify several different CYP2D6 alleles. The results of our study suggest that assay selection is important for the accurate determination of CYP2D6 gene copy number.
METHODS
Samples
We used 17 Japanese and 15 Chinese (JCH) genomic DNA samples from the HapMap human diversity panel (Coriell Institute for Medical Research, Camden, NJ, USA) for our copy number assays. We have previously reported the CYP2D6 gene copy number for these samples (12).
Copy number assays
To assess the CYP2D6 gene copy number, we used four assays: two commercial quantitative TaqMan Copy Number Assays (assay ids: Hs00010001_cn (Ex9) and Hs04502391_cn (Int6); Applied Biosystems, Forest City, CA, USA) and two assays (5′-flank and Int2) described by Hosono et al., (12). All four assays were performed along with an internal control - RNaseP TaqMan Copy Number Reference Assay (assay id: 4403326; Applied Biosystems, Forest City, CA, USA). All assays were performed in quadruplicates in a StepOne Plus PCR instrument (Applied Biosystems, Forest City, CA, USA). The assays were performed with TaqMan Genotyping PCR Master Mix (Applied Biosystems, Forest City, CA, USA) using 20 ng of genomic DNA in a 20 μL reaction. The cycling conditions were: 95°C for 10 min for initial denaturation and enzyme activation, followed by 40 cycles each of 95°C for 15 s and 60°C for 1 min. Relative quantification (RQ) was performed using CopyCaller Software (Applied Biosystems, Forest City, CA, USA), following the comparative ΔΔCT method. NA18529 (2 copies) and NA18968 (3 copies) DNA samples were used as controls based on our previous results (12). The data is reported as the estimated number of CYP2D6 copies, with the error bars representing the estimated copy numbers range. As a quality control measure, we repeated all four assays in 10% of the samples. All repeated assays yielded concordant results.
Cloning and Sequencing
To determine the target site of the Ex9 assay, we cloned the PCR product using the pCR8/GW/TOPO TA Cloning Kit (Invitrogen, Carlsbad, CA, USA). The clones were sequenced at the Indiana University DNA Sequencing Core Facility.
RESULTS
We estimated CYP2D6 gene copy number in 32 genomic DNA samples from the Coriell Japanese-Chinese Repository using four different qRT-PCR assays – two assays that target intron 6 and exon 9 (Int6 and Ex9; Applied Biosystems, Forest City, CA, USA) and two assays that target the 5′-flanking region and intron 2 (Int2 and 5′-flank; (12)). The estimated CYP2D6 gene copy numbers from three of the assays, 5′-flank, Int2 and Int6, were the same in all the samples (Figure 1). However, the Ex9 assay was concordant in only 10 (31%) of the 32 samples. Based on our previous results (12), all the 22 samples that were discordant contained at least one CYP2D6*36 allele. Since the CYP2D6*36 allele contains a gene conversion in exon 9, we cloned and sequenced the PCR amplicon from the Ex9 assay. The amplicon sequence corresponds to nucleotides 5735-5795 (61 bp) of GenBank accession # M33388.1 (Figure 2). This region is a part of the CYP2D6*36 allele that contains the CYP2D7 sequence due to partial gene recombination. The PCR amplicon sequence was that of CYP2D6, and not CYP2D7.
Figure 1. Comparison of estimated CYP2D6 gene copy numbers using four copy number assays.
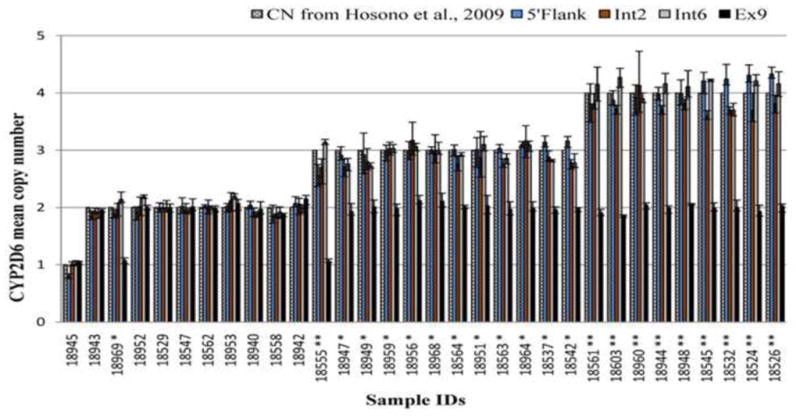
CYP2D6 copy numbers (y-axis) were estimated using CNV assays that target the 5′flanking region (blue), intron 2 (brown), intron 6 (gray), and exon 9 (black). Also included are the known copy numbers (cross hatched) from Hosono et al., (12). The sample identification numbers (x-axis) are those from the Coriell JCH diversity panel (each of the numbers are preceded by an NA). The number of * next to each sample indicates the number of CYP2D6*36 alleles present based on data from Hosono et al., (12). Each sample was assayed in quadruplicate for each of the four assays. The data presented are the mean CYP2D6 copy number with the error bars representing the range of copy numbers estimated for each assay.
Figure 2. Sequence alignment of the Ex9 assay amplicon with the CYP2D6, CYP2D7and CYP2D8 exon 9 region.

A portion of the exon 9 sequence from CYP2D6*1, CYP2D6*36, and the corresponding regions from CYP2D7 and CYP2D8 pseudogenes are shown. This corresponds to nucleotides 5700-5819 (120 bp) of M33388.1. The sequence of the Ex9 assay amplicon is highlighted in gray and the position of the assay described by Schaeffeler et al., (15) is underlined. The lower case letters indicate differences from the CYP2D6 gene. * indicates DNA sequence conservation across all four genes.
DISCUSSION
The results from our studies indicate the importance of selecting the appropriate assay when determining the CYP2D6 gene copy number. Multiple assays exist, and they may serve different purposes. Three of the four assays that we tested estimated the same CYP2D6 gene copy numbers in all the samples, presumably all copies of CYP2D6 (*1, *2, *36 and *10) irrespective of the allele involved in the CNV. These copy number estimations were concordant with our previous results (12). In contrast, the CYP2D6 gene copy number estimated by one of the commercial assays (Ex9) was discordant with the other three assays. The Ex9 assay detected all of the alleles, except the CYP2D6*36 allele. The lack of detection of CYP2D6*36 is likely because the reverse primer of the assay lies in a region of CYP2D6 exon 9 that has a different sequence in the CYP2D6*36 allele due to gene-conversion to CYP2D7. Although the exact primer sequences and locations of the amplicon are not provided for the Ex9 assay, by cloning and resequencing, we determined the sequence of the target region. The Ex9 assay target sequence appears to be located within the sequence targeted by the assay described by Schaeffeler et al., (15), but the primers are not the same. Therefore, the Ex9 assay would not be useful in detecting the total number of CYP2D6 alleles in Asian populations where the CYP2D6*36 allele is common. However, since the CYP2D6*36 allele is a nonfunctional allele, the Ex9 assay should give a better estimation of the number of functional copies, particularly in Asian population. An exception would be other rare nonfunctional alleles, such as CYP2D6*4 which is present in <1% frequency in Asians; this allele would also be detected by the Ex9 assay. When combined with one of the other copy number assays, the Ex9 assay can be indirectly used to identify the number of CY2D6*36 alleles. Since the current method for CYP2D6*10 genotyping is the presence of the SNP, 100T, a mutation which is also present in CYP2D6*36, Ex9 copy number assay should help to distinguish between *10 and *36 alleles. In populations where the CYP2D6*36 alleles are quite rare, such as the Caucasian and African American populations, all four assays studied are likely to produce similar results. Our results also indicate that all these assays should also be useful in detecting the CYP2D6*5 (gene deletion) in samples that have only one copy due to a whole gene deletion in CYP2D6*5. In conclusion, these results indicate that it is critical that copy number assays are designed to target appropriate regions of the CYP2D6 gene. Not all copy number variants are the same and interpretable data may not be obtained if CNV assays for any gene are not appropriately targeted.
Acknowledgments
SOURCES OF SUPPORT: This work was supported by grants from the NIH-Pharmacogenetics Research Network (5U01GM061373, D.A.F.), the National Center for Research Resources (K24RR020815, D.A.F.), NIH-NIGMS (1R01GM088076, T.C.S.), the Indiana University Cancer Center, the US Department of Defense Predoctoral Fellowship (BC083078, A.R.) and the Ministry of Education, Culture, Sports, Science and Technology of Japan (N.H., M.K., and Y.N.).
Footnotes
DISCLAIMER: None.
References
- 1.Sebat J, Lakshmi B, Troge J, Alexander J, Young J, Lundin P, et al. Large-scale copy number polymorphism in the human genome. Science. 2004 Jul 23;305(5683):525–8. doi: 10.1126/science.1098918. [DOI] [PubMed] [Google Scholar]
- 2.Shrestha S, Tang J, Kaslow RA. Gene copy number: learning to count past two. Nat Med. 2009 Oct;15(10):1127–9. doi: 10.1038/nm1009-1127. [DOI] [PubMed] [Google Scholar]
- 3.Wain LV, Armour JA, Tobin MD. Genomic copy number variation, human health, and disease. Lancet. 2009 Jul 25;374(9686):340–50. doi: 10.1016/S0140-6736(09)60249-X. [DOI] [PubMed] [Google Scholar]
- 4.McCarroll SA, Hadnott TN, Perry GH, Sabeti PC, Zody MC, Barrett JC, et al. Common deletion polymorphisms in the human genome. Nat Genet. 2006 Jan;38(1):86–92. doi: 10.1038/ng1696. [DOI] [PubMed] [Google Scholar]
- 5.Hebbring SJ, Adjei AA, Baer JL, Jenkins GD, Zhang J, Cunningham JM, et al. Human SULT1A1 gene: copy number differences and functional implications. Hum Mol Genet. 2007 Mar 1;16(5):463–70. doi: 10.1093/hmg/ddl468. [DOI] [PubMed] [Google Scholar]
- 6.Bolt HM, Thier R. Relevance of the deletion polymorphisms of the glutathione S-transferases GSTT1 and GSTM1 in pharmacology and toxicology. Curr Drug Metab. 2006 Aug;7(6):613–28. doi: 10.2174/138920006778017786. [DOI] [PubMed] [Google Scholar]
- 7.Bradford LD. CYP2D6 allele frequency in European Caucasians, Asians, Africans and their descendants. Pharmacogenomics. 2002 Mar;3(2):229–43. doi: 10.1517/14622416.3.2.229. [DOI] [PubMed] [Google Scholar]
- 8.Sistonen J, Sajantila A, Lao O, Corander J, Barbujani G, Fuselli S. CYP2D6 worldwide genetic variation shows high frequency of altered activity variants and no continental structure. Pharmacogenet Genomics. 2007 Feb;17(2):93–101. doi: 10.1097/01.fpc.0000239974.69464.f2. [DOI] [PubMed] [Google Scholar]
- 9.Ingelman-Sundberg M. Pharmacogenetics of cytochrome P450 and its applications in drug therapy: the past, present and future. Trends Pharmacol Sci. 2004 Apr;25(4):193–200. doi: 10.1016/j.tips.2004.02.007. [DOI] [PubMed] [Google Scholar]
- 10.Ingelman-Sundberg M. Genetic and environmental causes for interindividual variability in drug pharmacokinetics. International Congress Series; 2001 August; 2001. pp. 175–86. [Google Scholar]
- 11.Eichelbaum M, Ingelman-Sundberg M, Evans WE. Pharmacogenomics and individualized drug therapy. Annu Rev Med. 2006;57:119–37. doi: 10.1146/annurev.med.56.082103.104724. [DOI] [PubMed] [Google Scholar]
- 12.Hosono N, Kato M, Kiyotani K, Mushiroda T, Takata S, Sato H, et al. CYP2D6 genotyping for functional-gene dosage analysis by allele copy number detection. Clin Chem. 2009 Aug;55(8):1546–54. doi: 10.1373/clinchem.2009.123620. [DOI] [PubMed] [Google Scholar]
- 13.Johansson I, Oscarson M, Yue QY, Bertilsson L, Sjoqvist F, Ingelman-Sundberg M. Genetic analysis of the Chinese cytochrome P4502D locus: characterization of variant CYP2D6 genes present in subjects with diminished capacity for debrisoquine hydroxylation. Mol Pharmacol. 1994 Sep;46(3):452–9. [PubMed] [Google Scholar]
- 14.Meijerman I, Sanderson LM, Smits PH, Beijnen JH, Schellens JH. Pharmacogenetic screening of the gene deletion and duplications of CYP2D6. Drug Metab Rev. 2007;39(1):45–60. doi: 10.1080/03602530600952206. [DOI] [PubMed] [Google Scholar]
- 15.Schaeffeler E, Schwab M, Eichelbaum M, Zanger UM. CYP2D6 genotyping strategy based on gene copy number determination by TaqMan real-time PCR. Hum Mutat. 2003 Dec;22(6):476–85. doi: 10.1002/humu.10280. [DOI] [PubMed] [Google Scholar]


