Abstract
Phosphate and sulfate are essential macro-elements for plant growth and development, and deficiencies in these mineral elements alter many metabolic functions. Nutritional constraints are not restricted to macro-elements. Essential metals such as zinc and iron have their homeostasis strictly genetically controlled, and deficiency or excess of these micro-elements can generate major physiological disorders, also impacting plant growth and development. Phosphate and sulfate on one hand, and zinc and iron on the other hand, are known to interact. These interactions have been partly described at the molecular and physiological levels, and are reviewed here. Furthermore the two macro-elements phosphate and sulfate not only interact between themselves but also influence zinc and iron nutrition. These intricated nutritional cross-talks are presented. The responses of plants to phosphorus, sulfur, zinc, or iron deficiencies have been widely studied considering each element separately, and some molecular actors of these regulations have been characterized in detail. Although some scarce reports have started to examine the interaction of these mineral elements two by two, a more complex analysis of the interactions and cross-talks between the signaling pathways integrating the homeostasis of these various elements is still lacking. However, a MYB-like transcription factor, PHOSPHATE STARVATION RESPONSE 1, emerges as a common regulator of phosphate, sulfate, zinc, and iron homeostasis, and its role as a potential general integrator for the control of mineral nutrition is discussed.
Keywords: PHR1, mineral homeostasis, phosphate, zinc, iron, sulfate, crosstalks, integration
Introduction
Among environmental constraints, mineral nutrition plays a key role for plant growth and development. Variations in soil nutrient composition and availability are the rule and plants have evolved mechanisms to cope with conditions ranging from extreme deficiency to toxicity due to excess. Plant breeding was oriented these last 50 years to provide crops to modern agriculture with high intrinsic growth rates and yields, under the condition that mineral nutrition was not limiting. Such condition was obtained by the massive use of fertilizers, in particular considering nitrogen (N), phosphorous (P), and potassium (K) (López-Arredondo et al., 2013). The future of agriculture will undoubtedly require to use so far uncultivated lands, some of them exhibiting unfavorable soil mineral composition, and to reduce the use of fertilizers in order to promote sustainable practices. In such a context of lower input into the environment, new cultivated plant genotypes will need to be selected in a way improving their mineral use efficiency. Reaching such a goal would be facilitated by a knowledge-based approach rooted in the understanding of how plants sense and signal changes in the availability of nutrients (López-Arredondo et al., 2013).
A wealth of knowledge was obtained these last 20 years on the physiological and morphological adaptation of plants in response to variations in availability of a given mineral element (Maathuis, 2009; Krouk et al., 2011; Gruber et al., 2013; Briat et al., 2014; Giehl and von Wirén, 2014). Genes encoding proteins involved in uptake, translocation, assimilation, and storage of macro and micro-elements have been characterized and the regulation of their expression in response to mineral status has started to be elucidated (Schachtman and Shin, 2007; Giehl et al., 2009; Gojon et al., 2009; Liu et al., 2009; Pilon et al., 2009; Hindt and Guerinot, 2012; Vigani et al., 2013). More recently multi-level studies integrating transcriptome to metabolome and to enzyme activities data enabled to begin to understand how plants reprogram various metabolic pathways in response to removal and/or resupply of mineral nutrients. It gives an insight into how plants integrate metabolism adaptation to mineral nutrition deficiency to growth (Amtmann and Armengaud, 2009; Kellermeier et al., 2014). However, it is well known that interactions between nutrients for uptake can cause imbalances if one of them is deficient or in excess (Marschner, 1995; Rouached et al., 2010). Multi-level interactions between the various mineral elements need therefore to be studied in order to understand how the different sensing and signaling pathways activated in response to changes in availability of one element are coordinately integrated with the ones of other elements.
In such a context, the principal aim of this paper is to review interactions between phosphorus (P) and sulfur (S) on one hand, and between zinc (Zn) and iron (Fe) on the other hand. In addition, phosphate (Pi) and sulfate (SO4) not only interact between themselves but also influence Zn and Fe nutrition, and these intricated nutritional cross-talks are presented, pointing out the emerging role of transcription factors (TFs) belonging to the MYB family.
The MYB Family of Transcription Factors and its Role in Abiotic Stress Responses
Due to their sessile nature plants must face and adapt to a variety of biotic (e.g., bacteria, fungi, etc.) and abiotic (e.g., cold, drought, etc.) stresses throughout their life cycle. As a consequence plants have evolved molecular mechanisms allowing a tight control of their growth and development. This process is complex and dynamic, and requires the coordinated expression of several thousands of genes.
Transcription factors are sequence-specific DNA binding proteins that play a key role in the control of genes expression by acting as transcriptional activators, repressors or both. TFs possess a modular structure that is characterized by two key domains, a DNA-binding domain (DBD) and a transcriptional regulatory domain. TFs have been categorized into various families on the basis of some specific amino acid signatures mostly present in their DBD (Charoensawan et al., 2010).
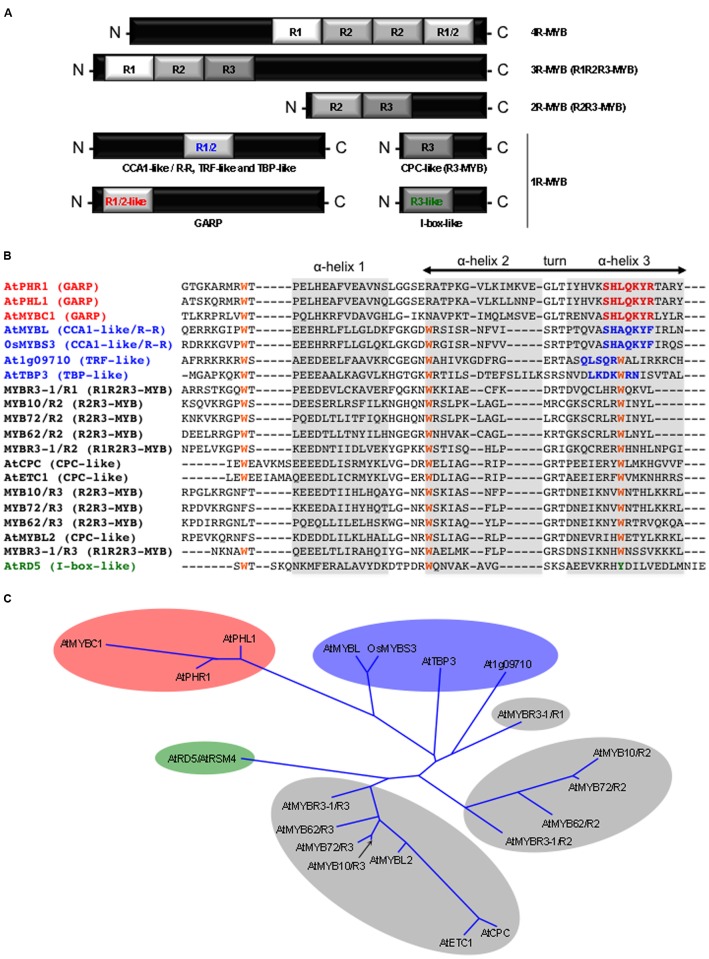
Among the various classes of TFs found in plants, the MYB family is one of the largest and most diverse (Riechmann et al., 2000; Dubos et al., 2010). MYB proteins are characterized by their DBD (Figure 1), or MYB domain. It is composed of different numbers (from 1 to 4) of imperfect repeats (R) of approximately 50 amino acids (Lipsick, 1996). Each repeat forms a helix-turn-helix (HTH) structure containing three evenly spaced tryptophan residues. These residues form a hydrophobic core playing a key role in the sequence-specific binding to DNA. The MYB gene family is divided into various groups according to the number and the type of repeat(s) found in their DBD (Stracke et al., 2001; Du et al., 2013).
FIGURE 1.
The plant MYB protein family. (A) Schematic representation of the different MYB protein classes based on the number (from 1 to 4) and the type (i.e., R1, R2, R3, R1/2, R1/2-like, and R3-like) of adjacent MYB repeat (R). It must be noted that the position of the R1/2 MYB repeat alongside the polypeptide chain of the proteins may vary between the different 1R-MYB subclasses that contain this domain (i.e., CCA1-like/R-R, TRF-like, and TBP-like). (B) Sequence alignment of MYB repeats from different classes of MYB proteins. Conserved tryptophan (W) residues are highlighted in orange. Specific amino acid signatures of the GARP, the CCA1like/R-R, TRF-like, and TBP-like, and the I-box-like 1R-MYB classes are highlighted in red, blue, and green, respectively. α-Helices are shaded in gray. The α-helix 3 is the recognition helix that makes direct contacts with DNA nitrogen bases (i.e., interacting with cis-regulatory sequences). Protein sequences were downloaded from TAIR (except for OsMYBS3, GenBank: accession AAN63154). (C) Phylogenetic relationship (unrooted tree, neighbor joining method) between the different classes of MYB repeats. The corresponding protein name is given at the extremity of each branch of the tree. The GARP, the CCA1like/R-R, TRF-like, and TBP-like, and the I-box-like repeats are shaded in red, blue and green, respectively. R1, R2, and R3 repeats are shaded in gray. (Adapted from Stracke et al., 2001, Dubos et al., 2010, and Du et al., 2013).
To date, no biological role directly related to plant responses to abiotic stresses has been clearly reported for the 3R- and 4R-MYB proteins. 3R-MYBs are found in all eukaryotic cells where they participate to the control of the cell cycle. In contrast, the role of the plant specific 4R-MYBs remains elusive.
The R2R3-MYB class (two repeats) is the largest group of MYB-proteins exclusively found in plant species. For instance, out of the 196 MYB genes found in the Arabidopsis thaliana genome, 126 encode R2R3-MYB proteins (Dubos et al., 2010). R2R3-MYBs are specifically involved in the transcriptional control of plant-specific processes, including plant responses to various abiotic stresses, such as cold or drought. This TF sub-family has been extensively studied in Arabidopsis allowing to determine the biological role played by more than half of its members (Dubos et al., 2010). For example, AtMYB14 and AtMYB15 are involved in the plant response to cold stress (Agarwal et al., 2006; Chen et al., 2013). AtMYB60 and AtMYB96 act through the ABA signaling cascade to modulate plant response to drought, by controlling stomatal movement, and root growth and cuticular wax deposition, respectively (Cominelli et al., 2005; Seo et al., 2009, 2011). In contrast, AtMYB2 and AtMYB44/AtMYBR1 regulate the expression of their target genes in response to drought in an ABA-dependent manner (Abe et al., 2003; Jaradat et al., 2013). AtMYB88 and AtMYB124/FLP paralogs are key regulators of stomata differentiation. They have recently been shown to be involved in sensing and/or transducing salt stress (and most probably other abiotic stresses; Xie et al., 2010). AtMYB20 and AtMYB73 are also involved in salt stress tolerance, whereas AtMYB108/BOS1 displays a less specific role as it is required in the response to both pathogens and abiotic stresses, including drought, salinity, and oxidative stress (Mengiste et al., 2003; Cui et al., 2013; Kim et al., 2013).
Last, the single MYB repeat proteins forms a heterogeneous group that gather genes that can be classified into five major subgroups: the CPC-like, the CCA1-like/R-R, the I- box-like, the TRF-like, the TBP-like, and the GARP (Du et al., 2013). To date, only 1R-MYBs belonging to the CPC-like (also called R3-MYB), and CCA1-like/R-R subgroups, have been associated to plant abiotic stress responses. The Arabidopsis AtMYBL2 is a CPC-like MYB protein whose activity is decreased under high light stress, and which acts as a negative regulator of anthocyanin biosynthesis (which provide a natural sunscreen for plants), and (Dubos et al., 2008). Amongt the CCA1-like/R-R group of MYB proteins, OsMYBS3 has been shown to be involved in cold tolerance in rice (Oryza sativa), whereas AtMYBL plays a role in the Arabidopsis response to salt stress (Su et al., 2010; Zhang et al., 2011). AtMYBC1 that belongs to the GARP subgroup was found to be a negative regulator of freezing tolerance in Arabidopsis (Zhai et al., 2010).
Nutrients availability is also an important environmental factor which modulates plant growth and development, and therefore crop productivity. Consequently, deficiencies in nutrient supplies are abiotic stresses against which plants have evolved signaling cascades aiming to improve nutrient acquisition and homeostasis. Similarly to the above-mentioned abiotic stresses, MYB proteins have also been found to be involved in the plant response to nutrient deficiencies. For example, two homologous R2R3-MYB proteins (namely AtMYB10 and AtMYB72) have been shown to play a key role in improving growth under Fe-limiting conditions (van de Mortel et al., 2008; Palmer et al., 2013; Zamioudis et al., 2014). However, most of the MYB proteins identified so far as involved in the regulation of mineral nutrition were associated with Pi starvation. For example AtMYB62 participates in the response to Pi shortage (Devaiah et al., 2009), whereas AtETC1 (ENHANCER OF TRY AND CPC 1) is a CPC-like MYB TF acting to modulate root hair density under Pi limited conditions (Savage et al., 2013).
PHOSPHATE STARVATION RESPONSE 1 (PHR1) and PHR1-Like 1, (PHL1) are two homologous GARP MYB proteins that play a critical role in the adaptation of plant to Pi (inorganic phosphate) deficiency. Originally, PHR1 was first identified as a regulator of Pi nutrition in Chlamydomonas reinhardtii, and named PSR1. It is required for the transcriptional induction of Pi-deficiency responsive genes in this green algae (Shimogawara et al., 1999; Wykoff et al., 1999). A genetic screen enabled to identify a PSR1 ortholog in Arabidopsis, AtPHR1 (Rubio et al., 2001). Several years later a redundant gene to AtPHR1 was characterized and named AtPHL1 (Bustos et al., 2010). These two TFs are able to interact, and they recognize the same cis element located in the promoter sequence of their target genes. A consensus sequence of this element, called PHR1 Binding Site (P1BS), has been defined as 5′-GNATATNC-3′ (Rubio et al., 2001; Bustos et al., 2010; Figure 2). Transcriptome analysis revealed that most of the genes induced in response to Pi deficiency lost this ability in phr1 phl1 double mutant plants. Furthermore, the frequency of occurrence of the P1BS element is much higher in the promoter region of genes induced in response to Pi deficiency than in others (Bustos et al., 2010). Interestingly, two Arabidopsis PHR1 orthologs, OsPHR1, and OsPHR2, have been characterized in rice (Zhou et al., 2008), and functional P1BS cis-elements have been reported in barley (Schünmann et al., 2004a,b). These data indicate that the regulatory pathway involving PHR1 to activate the expression of some genes in response to Pi starvation is likely to be conserved between monocotyledonous and dicotyledonous plants. It is noteworthy that a recent study has pointed out that the expression of AtFer1, a key gene involved in Fe homeostasis, was transcriptionally regulated by AtPHR1 and AtPHL1, providing a direct molecular link between Fe and Pi homeostasis (Bournier et al., 2013, and see below the “PHR1 involvement in Pi and Fe homeostasis interactions” section).
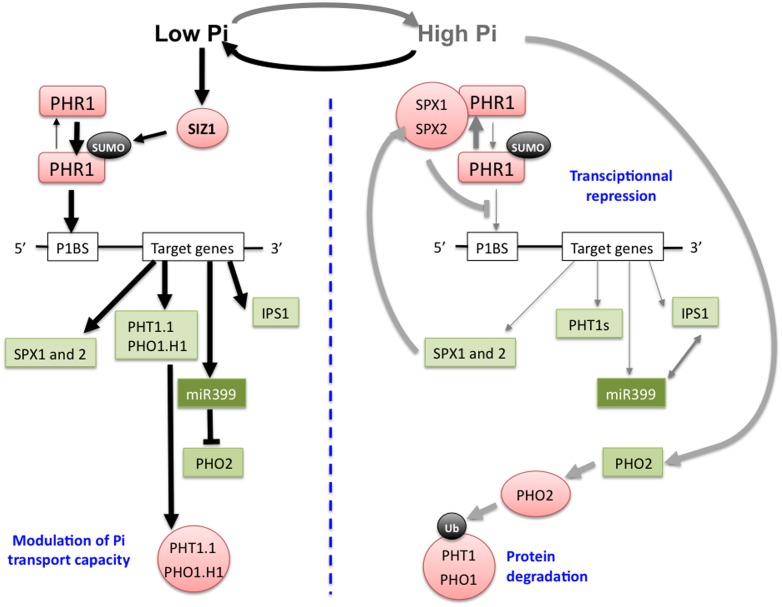
FIGURE 2.
Schematic representation of the regulatory pathways required for plant adaptation to Pi deficiency. Under low Pi nutrition conditions (left) the transcriptional activation of a set of genes necessary for Pi uptake by the roots (PHT1, PHO1), occurs through binding of the transcription factor (TF) PHOSPHATE STARVATION RESPONSE 1(PHR1) to its cis-target present in the promoter region of these genes. Under low Pi conditions, PHR1 is sumoylated by SIZ1, and this post-translational modification is likely important for PHR1 activity because Pi-deficient regulated genes are no more induced in siz1 mutant under this condition (Miura et al., 2005), although the mechanism of this regulation is unknown. Post-transcriptional regulators of Pi transporter proteins (PHT1.1, PHO1.H1) are also transcriptionally up-regulated through PHR1 activity under Pi-deficiency. Among them the miRNA miR399 negatively regulates the ubiquitin E2 conjugase PHO2 responsible of the ubiquitination of PHT1 and PHO1 proteins in order to target them for proteasome degradation. miR399-dependent inhibition of PHO2 can be titrated under high Pi through RNA mimicry via its appariement to IPS1, a non-coding RNA positively regulated by PHR1 under Pi deficiency. Under high Pi nutrition conditions (right) PHR1 target genes are transcriptionally repressed and PHO2 expression is activated promoting Pi transporters degradation. This transcriptional repression under these conditions is mediated through Pi sensing of nuclear SPX proteins which interact with PHR1 via their SPX domain in Pi-dependent manner in order to inhibit PHR1 binding to its P1BS cis-acting sequence found in the promoter region of Pi responsive genes. Green: transcripts, red: proteins, black: post-translational modifications, arrows thickness is proportional to the strength of the considered flux.
PHR1 and the Control of Pi Nutrition
P is an essential macronutrient required by living organisms. It is found in essential biological molecules including nucleic acids, ATP (a major energy carrier) and phospholipids. P is intimately linked with energy metabolism, the production of numerous metabolic intermediates, and the post-translational modification of proteins (a key parameter in signal transduction cascades; Poirier and Bucher, 2002). Its deficiency has deleterious effects on plant growth and development, evidenced by a strong decrease in shoot growth, and by the accumulation of anthocyanins. Plants acquire P by their roots as inorganic Pi. Pi concentration is heterogeneous in the soil, and often very low at the root/soil interface (Poirier and Bucher, 2002). Root architecture responds to Pi deficiency by inhibiting primary root growth and by increasing lateral root density. This is an adaptive strategy to explore more soil volume, resulting in an increased Pi uptake capacity of the plants. After having crossed the plasma membrane of epidermal and cortical root cells, Pi is distributed throughout the plant under the control of a cascade of Pi transporters belonging to the PHT and PHO1 families. For an extensive review of the Pi transporter gene family, readers are referred to Poirier and Bucher (2002), and to Nussaume et al. (2011). Plant adaptation to Pi deficiency involves wide changes in gene expression. It implicates several TFs. Few of them have been identified including WRK75, ZAT6, bHLH32, MYB62, and PHR1 (Yi et al., 2005; Chen et al., 2007; Devaiah et al., 2007a,b, 2009; Svistoonoff et al., 2007; Ticconi et al., 2009). So far, the major regulations of the expression of numerous Pi deficiency-induced genes was attributed to PHR1, via the PHR1-PHO2–miRNA399 pathway described below (Figure 2). In addition to this pathway, plants respond to Pi deficiency through pathways involving phytohormones and various metabolites (Buchner et al., 2004). Among phytohormones, cytokinins suppress the up-regulation of several genes in response to Pi deficiency, including Pi uptake transporters. It requires the cytokinin receptor CYTOKININ RESPONSE 1/WOODEN LEG/ARABIDOPSIS HISTIDINE KINASE 4 (CRE1/WOL/AHK4) pathway (Maruyama-Nakashita et al., 2004; Franco-Zorrilla et al., 2005). Among the metabolites, carbohydrates are involved in the Pi deficiency response. The expression of Pi transporters is sensitive to the carbon status of the plant, either upstream or downstream of the hexokinase (HXK) activity in glycolysis (Lejay et al., 2008).
At a molecular level Pi deficiency is regulated both at the transcriptional and post-transcriptional levels. The major actors coordinating these various regulations are PHR1 and PHL1, two TFs likely conserved amongst flowering plants.
Indeed, sensing of the Pi status of the plant is likely conserved between mono- and dicotyledonous plants, as recently reported (Wang et al., 2014a). Expression of AtPHR1 and of its rice orthologous gene OsPHR2, are not responsive to Pi, raising the question of how plants sense the intra-cellular variations of Pi concentrations. In this context, Wang et al. (2014a) recently reported that rice OsSPX1 and 2, which are nuclear proteins whose expression is itself activated by OsPHR2 under low Pi conditions, interact with OsPHR2 by their SPX domain in a Pi-dependent manner. This interaction results in the inhibition of OsPHR2 binding on its cis-acting P1BS sequence (Figure 2). Therefore, this mechanism constitutes a very efficient transcriptional regulatory feedback loop to fine tune the PHR1 dependent expression of Pi responsive genes, according to the intracellular fluctuations of Pi concentrations.
Among the genes transcriptionally regulated by PHR1/PHL1 in response to Pi deficiency are the PHT1 genes (Figure 2). They encode plasma membrane high-affinity H+/Pi co-transporters (Okumura et al., 1998; Mudge et al., 2002), preferentially expressed in the root epidermal or cortical cells, and therefore directly involved in Pi acquisition (Karthikeyan et al., 2002; Mudge et al., 2002). Once Pi has entered the root, it is loaded into the xylem sap for translocation to the shoots via PHO1, which is specifically expressed in the pericycle (Hamburger et al., 2002). PHT1 and PHO1 transporters are also post-translationally regulated during their intracellular trafficking to the plasma membrane, and the C terminus phosphorylation of PHT1 proteins retains them at the ER upon Pi refilling (Bayle et al., 2011). During the post-ER trafficking, the ubiquitin E2 conjugase PHO2 modulates the ubiquitination status of PHT1 and PHO1 transporters in order to control their rate of degradation by the proteasome. PHO2 expression is itself post-transcriptionally repressed by miR399, a small non-coding RNA up-regulated by Pi deficiency at the transcriptional level through PHR1/PHL1 activity (Fujii et al., 2005; Aung et al., 2006; Bari et al., 2006; Chiou et al., 2006; Liu et al., 2014; Figure 2). Post-translational control of Pi loading into the xylem via the degradation of PHO1 under Pi-sufficient conditions has also been reported to be mediated by PHO2 (Liu et al., 2012).
Phosphate Starvation Response 1 is itself regulated post-translationally in response to Pi deficiency through the action of SIZ1, a sumo E3 ligase (Figure 2). PHR1 can be sumoylated in vitro by SIZ1, and Pi-deficient regulated genes are no more induced in siz1 mutant in response to Pi deficiency (Miura et al., 2005). This post-translational regulation of PHR1 could modify its activity, in order to modulate the transcriptional activation of its target genes, among which the IPS1 gene (Rubio et al., 2001). IPS1 encodes a non-coding RNA whose sequence is in part complementary of the microRNA miR399, enabling post-transcriptional regulation via RNA mimicry (Liu et al., 2014). IPS1-miR399 matching would therefore lead to the inhibition of the miR399 RNA activity (Franco-Zorrilla et al., 2007) known to target PHO2 (Figure 2). It could explain the increased expression of PHT1-8 and PHT1-9 in pho2 mutant plants (Bari et al., 2006), and the decreased expression of PHO2 and of the PHT1-8 and PHT1-9 Pi transporter genes in miR399 over-expressing transgenic lines (Aung et al., 2006). The involvement of PHT1-8 and PHT1-9 in Pi uptake (Remy et al., 2012) and/or in Pi root-to-shoot translocation (Lapis-Gaza et al., 2014) has been reported. More recently an iTRAQ based quantitative membrane proteomic approach was used to search for components downstream of PHO2. It enables to show that PHO2 mediates the degradation of PHT1 proteins by interacting in the post-endoplasmic reticulum compartments where ubiquitination of endomembrane-localized PHT1-1 occurs (Huang et al., 2013). Finally the systemic effect of miR399 was evidenced by grafting experiments. It demonstrated the transport of this small RNA from leaves to root within the phloem sap, explaining why specific overexpression of the miR399 in leaves led to decrease the expression of PHO2 in roots (Pant et al., 2008).
Pi, S, and Their Biological Interactions
Sulfur is an essential element for plant growth and development. The major source of S for plants is inorganic sulfate (SO4; Leustek et al., 2000). In the cell, S is involved in different functions and aspects of plant metabolism. It is found both in reduced (amino acids, peptides, and proteins, lipoic acid, iron–sulfur clusters), and in oxidized (polysaccharides, lipids, and sulfonate group modifying proteins) forms (Kopriva, 2006). Thus S deficiency has profound effects on plant physiology. When challenged by SO4 deficiency, shoot growth is affected, resulting in a decrease in the total biomass. Modulation of root system architecture has also been observed in response to S limitation (Osmont et al., 2007). The primary root continues to grow, but the lateral roots form closer to the root tip, with an increased density. During the last two decades, a comprehensive view of SO4 transport in Arabidopsis and other plant species has emerged. Numerous “SULTR” genes encoding SO4 transporters for uptake as well as for inter-organs and subcellular distribution have been functionally characterized (Takahashi, 2010). The first key step for SO4 uptake in Arabidopsis is mainly carried out by the two high-affinity SO4 transporters SULTR1;1 and SULTR1;2 (Rouached et al., 2009). The possibility that SULTR1;2 may function as a sensor of S status or as a component of a S sensory mechanism has recently been proposed (Zhang et al., 2014).
Some key regulatory molecular mechanisms and components involved in the regulation of SO4 transport have been discovered, among which a regulatory pathway requiring miRNAs, including miR395 (Kawashima et al., 2009). This microRNA contributes to the regulation of SO4 root-to-shoot transfer involving the SULTR2;1 gene. The transcript abundance of this gene is strongly decreased under deficient S conditions (Takahashi et al., 1997, 2000). It can be explained by the increased level of miR395 under such conditions (Kawashima et al., 2009), leading to a strictly restricted expression of the SULTR2;1 gene in the xylem (Kawashima et al., 2009). miR395 acts downstream of the TF SULFUR LIMITATION 1 (SLIM1), also known as ETHYLENE-INSENSITIVE3-LIKE 3 (EIL3; Maruyama-Nakashita et al., 2006; Kawashima et al., 2009).
Plants have evolved tightly controlled mechanisms allowing the coordination of the S transport and homeostasis with photosynthesis and the carbon status, in a similar manner to Pi transport system (Lejay et al., 2008). In addition to these mechanistic similarities in the regulation of the Pi and SO4 transport systems, plants have developed coordinated and tightly controlled mechanisms to maintain intracellular homeostasis of both elements in response to their external availability. It has been reported that plant cells operate a rapid replacement of sulfolipids by phospholipids under S deficiency, and the replacement of phospholipids by sulfolipids during Pi deficiency (Essigmann et al., 1998; Hartel et al., 1998; Yu et al., 2002; Sugimoto et al., 2007). Such a metabolic switch attests the P/S nutritional interdependency. Interestingly, two genes necessary for the replacement of phospholipids by sulfolipids in Pi-deficient plants, SQD1 and SQD2, contain a PHR1 binding sequence (P1BS) in their promoter, and are up-regulated by Pi deficiency in a PHR1-dependant manner (Franco-Zorrilla et al., 2004; Stefanovic et al., 2007). The accumulation of SO4 and Pi was affected in Arabidopsis lines characterized by a very low inositol-6-phosphate (phytic acid, PA) content (Belgaroui et al., 2014). The expression of genes involved in the SO4 and Pi transport or signaling was altered in these low PA mutants. PA emerged thus as a component of the co-regulation of SO4 and Pi homeostasis. Hsieh et al. (2009) reported that the increased accumulation of miR395, known to be up-regulated by SO4 starvation, is suppressed in Pi-deficient plants. It could be therefore a mean to increase the S translocation from root to shoot by SULTR2;1, enhancing thus the sulfolipid biosynthesis in replacement of phospholipids under Pi deficiency. Evidences for co-regulation of Pi and S signaling pathways are starting to emerge from recent data. Rouached et al. (2011a) reported that the SO4 concentration increases in roots and decreases in shoots of Arabidopsis Pi-deficient plants. These results indicate an adaptive regulation of the SO4 transport process upon Pi deficiency in plants, and particularly the inter-organ SO4 distribution. Interestingly, PHR1 is required for this process to take place, likely through its positive regulatory role on SULTR1;3 expression (Rouached et al., 2011a). PHR1 has also a negative effect on the expression of SULTR2;1 and SULTR3;4 in response to Pi deficiency (Rouached et al., 2011a). Noteworthy, such a function is also preserved in Chlamydomonas reinhardtii via the PHR1 ortholog PSR1 (Moseley et al., 2009). An integrative model for the regulation of the expression of genes involved in intracellular and inter-organ SO4 transport under Pi deficiency in Arabidopsis has been proposed, in which PHR1 plays an integrative role (Rouached, 2011). Considered together, these data reveals an unsuspected level of complexity and interconnection in the regulation of SO4 and Pi homeostasis in plants.
Fe, Zn, and Their Biological Interactions
Zn is an essential microelement for cell life. It is the only metal represented in all six classes of enzymes: oxidoreductases, transferases, hydrolases, lyases, isomerases, and ligases (Coleman, 1998). It also plays a structural role in regulatory proteins (Berg and Shi, 1996). Despite its essentiality, high concentrations of Zn can be toxic for the cell, and cause oxidative stress, a decrease in accumulation of ATP, disintegration of cell organelles, and biogenesis of vacuoles (Sresty and Madhava Rao, 1999; Xu et al., 2013).
Zn deficient plants exhibit deformed and chlorotic leaves, and interveinal necrosis, leading to decrease biomass production. Transport across the plasma membrane is achieved by transporters belonging to the ZIP (ZRT, IRT-like protein) family (Mäser et al., 2001) such as AtIRT1 (Vert et al., 2002; Barberon et al., 2011). A passive Zn influx can also occur through the depolarization-activated non-selective cation channel (DA-NSCC; Piñeros and Kochian, 2003), or by the voltage-independent NSCCs (VI-NSCCs; Demidchik and Maathuis, 2007). Zn loading into the xylem sap involves plasma membrane transporters which are members of the P1B-ATPases family, namely AtHMA2, and AtHMA4 (Hussain et al., 2004; Verret et al., 2004). For further information on the Zn transport in plants, readers are referred to Sinclair and Krämer (2012). So far, how plants sense and transmit the signal of Zn deficiency remains poorly understood.
Fe, as Zn, is essential. It is also potentially toxic when in excess because of its reactivity with oxygen which catalyzes the formation of free radicals able to oxidize organic molecules, ultimately leading to cell death (Briat et al., 2010b). Fe is a key element to ensure the electron flow through the PSII-b6f/Rieske-PSI complex. It is therefore essential for CO2 fixation by the photosynthesis process. Indeed it has been well documented that Fe is a limiting factor for biomass production, as well as for the quality of plant products (Briat et al., 2014). Fe enters the plant via the roots, from where it is distributed within the plant. According to the plant family considered – i.e., graminacea plants versus other plants – two mechanisms prevail for mining Fe from the soil solution. They involve, respectively, chelation of the ferric iron (Fe3+) by small organic molecules, among which methionine derivatives constitute the mugineic acid (MAs) family, or its reduction in its ferrous form (Fe2+) prior to transport across the plasma membrane of root epidermal cells (Curie et al., 2009; Morrissey and Guerinot, 2009; Conte and Walker, 2011). This transport is achieved by transmembrane proteins exhibiting transporter activities, such as YS1 responsible of the transport of Fe3+-MAs complexes in graminacea, or IRT1 responsible of the root uptake of Fe2+ by non-graminacea plants (Curie et al., 2001; Vert et al., 2002). Many other transporters, as well as soluble proteins, responsible for Fe long distance distribution, sub-cellular compartmentation, and storage have also been characterized this last decade (Morrissey and Guerinot, 2009; Briat et al., 2010a; Kobayashi and Nishizawa, 2012). The expression of these genes encoding proteins responsible of Fe homeostasis at the whole plant level is precisely regulated through integrated pathways modulated by the sensing of the Fe status of the plant. These controls mainly operate at the transcriptional level via a netwotk of TFs, most of them belonging to the bHLH family (Ivanov et al., 2012; Kobayashi and Nishizawa, 2012; Hindt and Guerinot, 2012), but also at the post-transcriptional level (Ravet et al., 2012). How plants sense their Fe status is now the new frontier in this field of research. It could be achieved, at least in part, by some TFs and/or by Hemerythrin motif-containing Really interesting new gene and Zinc-finger proteins (HRZs)/BRUTUS (BTS), that were recently identified both in rice and Arabidopsis, respectively (Kobayashi and Nishizawa, 2014).
Fe deficiency in Arabidopsis leads to the activation of expression of IRT1, the primary transporter responsible of root Fe uptake. IRT1 has a weak substrate specificity and contributes therefore to the accumulation of a broad range of divalent transition metals including Zn (Vert et al., 2002; Arrivault et al., 2006; Haydon et al., 2012). Conversely, excess Zn causes physiological Fe deficiency. Early studies reported an absence of IRT1 protein in Arabidopsis roots from plants grown under Zn excess conditions (Connolly et al., 2002), suggesting that the known post-translational regulation of IRT1 protein levels through ubiquitin-mediated proteasomal degradation (Kerkeb et al., 2008; Barberon et al., 2011) might predominate under this condition. However, it was reported more recently that IRT1 protein in Arabidopsis grown under excess Zn increased in abundance, comparatively to plants grown under standard Zn nutrition conditions (Fukao et al., 2011; Shanmugam et al., 2011). IRT1 could therefore be a major contributor to Zn deficiency.
MTP3 (a member of the Cation Diffusion Facilitator family), HMA3 (belonging to the P1B-type ATPase family), and ZIF1 (a member of the Major Facilitator Superfamily transporters) are vacuolar membrane proteins required for Zn tolerance, and encoded by genes which are transcriptionaly activated in response to excess Zn or Fe deficiency (Becher et al., 2003; Arrivault et al., 2006; van de Mortel et al., 2006; Haydon and Cobbett, 2007; Haydon et al., 2012). Indeed MTP3 and HMA3 gene expression is decreased in the fit mutant. This mutant lacks a bHLH TF that activates root Fe deficiency responses, in particular the transcriptional up-regulation of the Fe(III)-chelate reductase FRO2 and of the Fe2+ transporter IRT1 (Colangelo and Guerinot, 2006). Interestingly, transcriptional regulation of the ZIF1 gene under Fe deficiency is independent of Fe deficiency-induced Zn accumulation (Arrivault et al., 2006; Haydon et al., 2012). The bHLH TF POPEYE, controlling a set of genes expressed in the stele and important for the control of Fe homeostasis was reported to directly interact with the ZIF1 promoter and to repress its transcription, although there is a concurrent net increase in ZIF1 transcript levels, under Fe deficiency (Long et al., 2010).
These results evidence the complexity of the cross-talks between the pathways at work to regulate Fe deficiency and Zn excess in order to establish an integrated response, and the necessity of additional work in the future to decipher them.
The two macro-elements Pi and S on one hand, and the two metals Zn and Fe on the other hand, do not only interact between themselves as reported above. Indeed Pi and S status of the plant can also influence Zn and Fe nutrition, and these aspects will now be reviewed below.
S and Fe Homeostasis Interactions
From a biochemical point of view, Fe and S are known to interact for the building of Fe–S clusters, which are a major sink for Fe, and known to be essential for photosynthesis, respiration, and many cellular enzymatic reactions (Couturier et al., 2013). However, it is only recently that Fe and S interactions have been documented at physiological and molecular levels, although the cross-talks between the pathways regulating the integrated homeostasis of these two elements remain to be deciphered. It is well established that leaf Fe concentration decreases in S-deficient tomato, comparatively to S-sufficient control plants. This is consistent with the observation that the expression level of the LeFRO1 gene encoding a root Fe3+-chelate reductase, the activity of this reductase, and the reduction based of 59Fe uptake, are decreased in response to S starvation (Zuchi et al., 2009). In agreement with these observations a strong repression of expression of the Arabidopsis thaliana IRT1 Fe2+ transporter in response to S deficiency was recently reported (Forieri et al., 2013). Reciprocally, it has been observed that Fe starvation modifies S uptake and assimilation. At a genome wide level, data mining of transcriptomes from Fe deficient Arabidopsis plants revealed a cluster of S-metabolism related genes (including genes encoding plasma membrane and tonoplast S transporters, and enzymes of the S assimilation pathway such as adenosine phosphosulfate reductase) co-expressed with Fe-deficient genes (Schuler et al., 2011). Also, the high affinity S transporter SULTR1;1 mRNA abundance is 2.5-fold lower in absence of Fe (Forieri et al., 2013). However, a contrasted report was recently published concerning this last point (Paolacci et al., 2014). It indicated that the expression of most of the group 2 and 4 S transporters were up-regulated in Fe starved tomato plants. It turned out to be also the case of the SIST1;1 and SIST1;2 tomato high affinity transporters of the group 1, to which belongs the SULTR1;1 transporter from Arabidopsis. Resolution of these discrepancies concerning the impact of Fe deficiency on the regulation of expression of high affinity SO4 transporters clearly requires further work.
The interaction between Fe and S metabolisms has not only been studied in plants acquiring Fe from the soil through a reduction-based strategy as it occurs in tomato (Solanum lycopersicum) or Arabidopsis. It has also been investigated with graminaceous plants. Synthesis of Fe3+-chelators of the MAs family (Kobayashi and Nishizawa, 2012) and their release in the rhizosphere were reduced in barley (Hordeum vulgare L.) plants grown under S starvation (Kuwajima and Kawai, 1997; Astolfi et al., 2006). However, transcript abundance of the YS1 gene encoding the Fe3+-MAs transporter (Curie et al., 2001) increased in response to Fe deficiency at the same level, whatever the S nutrition conditions imposed (Astolfi et al., 2012). The modifications of S metabolism occurring in response to Fe deficiency have also been investigated in durum wheat (Triticum turgidum L.; Ciaffi et al., 2013). These authors have shown that Fe deficiency under S sufficient nutrition conditions led to a S deficiency response at a molecular level. This response was characterized by an increase in abundance of some of the transcripts encoding enzymes of the S assimilation pathway, including APS reductase, ATP sulfurylase, sulfite reductase, and serine acetyltransferase. Furthermore, the activity of the corresponding enzymes was also found to be increased by Fe shortage. However, changes in mRNA abundance of some other genes of the S assimilation pathway, and the activity of the corresponding enzymes, were observed to be uncoupled in their response to Fe or S deprivation. In addition, the expression of the wheat SULTR1;3 high affinity SO4 transporter was significantly increased in roots and shoots in response to both S or Fe shortage, with the highest expression level observed under Fe deficiency conditions. In contrast, the expression of the SULTR1.1 transporter gene, mainly expressed in roots, was strongly induced in response to S deficiency, but unaffected by Fe deficiency (Ciaffi et al., 2013).
In conclusion, the interactions between Fe and S metabolisms are attested both in graminaceous and non-graminaceous plants. These interactions have started to be documented at a molecular level, reporting that Fe deficiency modifies the expression of genes involved in S transport and assimilation, and vice-versa. Nevertheless, the characterization of these interactions is still in its infancy, and more work is needed to understand the complexity of the integration of the various pathways involved. Of particular interest would be the study of a possible role of the synthesis of Fe–S cluster, and of their relative abundance in response to various nutritional stress, as driving forces of the Fe–S interactions (Couturier et al., 2013; Forieri et al., 2013).
PHR1 Involvement in Pi and Fe Homeostasis Interactions
Clear physiological links have been established between Fe and Pi (Hirsch et al., 2006; Ward et al., 2008). The complexation of Fe by Pi in soils leads to the formation of precipitates, decreasing the availability of these two elements for plants. As a consequence, the high affinity root Fe2+ uptake system, which is induced by Fe deficiency, is also activated under Pi excess conditions (Ward et al., 2008). Conversely, Pi starvation promotes metal accumulation in plants, mainly aluminum, and Fe (Misson et al., 2005; Hirsch et al., 2006; Ward et al., 2008). From a developmental point of view, it is also well documented that a decrease of primary root growth under Pi deficiency is partly due to Fe toxicity (Ward et al., 2008; Ticconi et al., 2009). Once taken up by the roots, Pi is translocated to the shoots, and Fe can interact with Pi inside roots leading to a reduced Pi translocation to the shoots (Cumbus et al., 1977; Mathan and Amberger, 1977). The same kind of interaction has been described in leaves in which high Pi content favors chlorosis, even if the leaf Fe level is sufficient (Dekock et al., 1978). Finally, in seeds Fe is stored in vacuoles under the form of globoids composed in part of phytate (inositol hexakisphosphate = IP6)-Fe complexes (Lanquar et al., 2005). These observations indicate that Pi is a very efficient Fe chelator. As a consequence, changes in Pi homeostasis will strongly influence Fe availability.
At the molecular level, transcriptome analysis of Pi deficient plants revealed an increase in abundance of transcripts from Fe excess responsive genes, and reciprocally a decrease in abundance of transcripts from Fe deficiency responsive genes (Misson et al., 2005; Müller et al., 2007; Thibaud et al., 2010). In this context, the induction of expression of the AtFER1 gene, encoding the Fe storage protein ferritin, in response to Pi deficiency was interpreted by some authors as a consequence of an increase in available Fe under such conditions (Hirsch et al., 2006). Indeed this interpretation is likely wrong since the abundance of AtFER1 mRNA in response to Pi starvation was recently shown to be mediated by PHR1 and PHL1, through their binding to a P1BS cis-element found in the AtFER1 promoter, independently of the plant Fe nutrition conditions (Bournier et al., 2013). Furthermore, the Fe-dependent IDRS cis-acting element present in the AtFER1 proximal promoter (Petit et al., 2001a) is not required for Pi-deficient induction of AtFER1 expression (Bournier et al., 2013). Moreover, AtFER3 and AtFER4 ferritins genes, lacking P1BS cis-element in their promoter and known to be induced by Fe excess (Petit et al., 2001b), have their expression unchanged by Pi starvation (Bournier et al., 2013). Finally, induction of AtFER1 expression in response to Fe excess is not altered in phr1 (Bournier et al., 2013) nor in phr1xphl1 (Bustos et al., 2010) mutant plants.
At a cellular level, Fe distribution around the vessels was abnormal in phr1x phl1 double mutant (Bournier et al., 2013), suggesting that PHR1 and PHL1 may link Fe and Pi homeostasis not only by regulating AtFER1 gene expression, but also by having a broader regulatory function of Fe metabolism. Indeed, genome wide analysis of Pi starved wild-type (Misson et al., 2005; Müller et al., 2007; Thibaud et al., 2010) and double phr1xphl1 mutant (Bustos et al., 2010) plants revealed other Fe homeostasis related genes such as NAS3 (NICOTIANAMINE SYNTHASE 3) and YSL8. These genes were induced upon Pi starvation in wild type, and exhibit a decreased induction in the double mutant plants. Moreover, Fe deficiency responsive genes, including FRO3, IRT1, IRT2, NAS1, and FRO6 were repressed upon Pi starvation in wild type and miss-regulated in the phr1xphl1 double mutant plants. These observations strengthen a global role of PHR1 and PHL1 in the overall control of Fe homeostasis that is, by this mean, integrated to the Pi status of the plant.
PHR1 Involvement in Pi and Zn Interactions
Pi and Zn homeostasis in plants are known to strongly interact (Loneragan et al., 1982; Cakmak and Marschner, 1986; Huang et al., 2000; Zhu et al., 2001; Misson et al., 2005; Bouain et al., 2014a; Khan et al., 2014). Nevertheless, the molecular bases of the Pi–Zn interactions remain so far poorly understood. Long-term Pi deprivation leads to Zn over-accumulation (Misson et al., 2005), and inversely Zn starvation appears to cause an over-accumulation of Pi (Loneragan et al., 1982; Huang et al., 2000; Bouain et al., 2014a; Khan et al., 2014). Transcriptome data from roots of Pi- or Zn-deficient Arabidopsis plants (Hammond et al., 2003; Wintz et al., 2003; Wu et al., 2003; Misson et al., 2005; van de Mortel et al., 2006; Müller et al., 2007; Bustos et al., 2010; Rouached et al., 2011b; Woo et al., 2012), could induce the expression of many Pi-related genes (van de Mortel et al., 2006), and that Pi starvation modifies the expression of genes involved in the maintenance of metal homeostasis such as Zn and Fe (Misson et al., 2005; Bustos et al., 2010). Considered together, this transcriptome data reinforce the evidence of a cross-talk between Pi and Zn signaling pathways in planta. Cross-analysis of these expression data showed that Pi and Zn deficiencies often act in anopposite manner on these sets of genes. Many genes induce by Zn deficiency are repressed by Pi deficiency (Bustos et al., 2010; Bouain et al., 2014b). Interestingly, it was observed that most of these genes were further repressed in the phr1 mutant genetic background (Bustos et al., 2010; Bouain et al., 2014a). This observation further supports the implication of PHR1 in the regulation of expression of Pi-related genes under Zn limitation in Arabidopsis (Bouain et al., 2014b). It agrees with a report by Khan et al. (2014) showing the involvement of AtPHR1 in the cross-talk between Pi and Zn homeostasis. As aforementioned, PHR1 was already known as a major regulator of Pi deficiency signaling through its involvement in the PHR1-miR399-PHO2 regulatory pathway (Bari et al., 2006). However, this regulatory pathway is not involved in the over-accumulation of Pi in the shoot in response to Zn deficiency (Khan et al., 2014). The Zn-responsive signaling pathway in which PHR1 is involved thus remains to be elucidated. Khan et al. (2014) also identified two additional genes that are involved in the control of Pi accumulation in response to Zn deficiency, namely PHO1 and its homolog PHO1;H3. PHO1 is a well-characterized root-to-shoot Pi transporter (Hamburger et al., 2002). It is likely one of the final targets of the Zn-deficiency signaling pathway. Since its level of expression is unchanged in response to Zn deficiency, one favorite hypothesis is that its activity would be regulated through a protein–protein interaction, considering that a similar mechanism involving PHO1 and PHO2 has already been reported (Liu et al., 2012). The PHO1;H3 gene appeared to be specifically and strongly induced by Zn deficiency (Khan et al., 2014). PHO1;H3 constitutes thus an interesting entry point to study Pi–Zn crosstalk and the regulation of Pi transport under Zn limitation. A first model illustrating Pi–Zn crosstalk in plant has been proposed (Bouain et al., 2014b; Kisko et al., 2014). In the future, this model would benefit from an in depth characterization of the molecular mechanisms underlying the effect of Pi starvation on Zn nutrition, together with the effects of Zn deficiency on Pi nutrition. The identified and characterized key genes and mechanisms acting in the coordination of Pi and Zn transport and signaling in plants could help to point to specific mutants or genetic variants that could be used in breeding programs. Taken together, this knowledge should have important consequences for both basic and applied research in agronomy and should be valuable to plant biologists, agronomists and breeders.
PHR1 as an Integrator of Multiple Nutrition Signals and Beyond
PHR1 was initially described as a major transcriptional regulator of Pi homeostasis. It activates the transcription of Pi deficiency responsive genes encoding Pi transporters (Rubio et al., 2001), as well as regulatory RNA (Fujii et al., 2005) and proteins involved in the post-transcriptional and post-translational regulation of these transporters, respectively (Aung et al., 2006; Bari et al., 2006; Liu et al., 2012; Figure 2). More recently, few reports indicated that PHR1 regulated also the expression of genes required for S, Fe, and Zn transport and homoeostasis (Rouached et al., 2011a; Bournier et al., 2013; Khan et al., 2014), linking the metabolism of these nutriments to Pi nutrition (Figure 3). PHR1 is therefore the first molecular link common to various pathways controlling mineral nutrition of both macro- and micro-elements. It can be extended to some aspects of the control of plant response to water stress, for which AtPHL1 activity is required (Elfving et al., 2011). As mentioned above, in response to Pi starvation PHR1 controls the transcription of post-transcriptional and post-translational regulators such as miR399, IPS1, and PHO2 (Figure 2). Whether or not these PHR1-dependent regulators play a role in the regulation of S, Fe, or Zn metabolisms is unknown, and would deserve to be investigated. Furthermore, the requirement of PHR1 has been documented in the case of two by two interactions of only some of the elements considered: Pi and S, Pi and Fe, Pi and Zn but to our knowledge no reports mentioned a direct role of PHR1 in the Fe–S or Fe–Zn interactions.
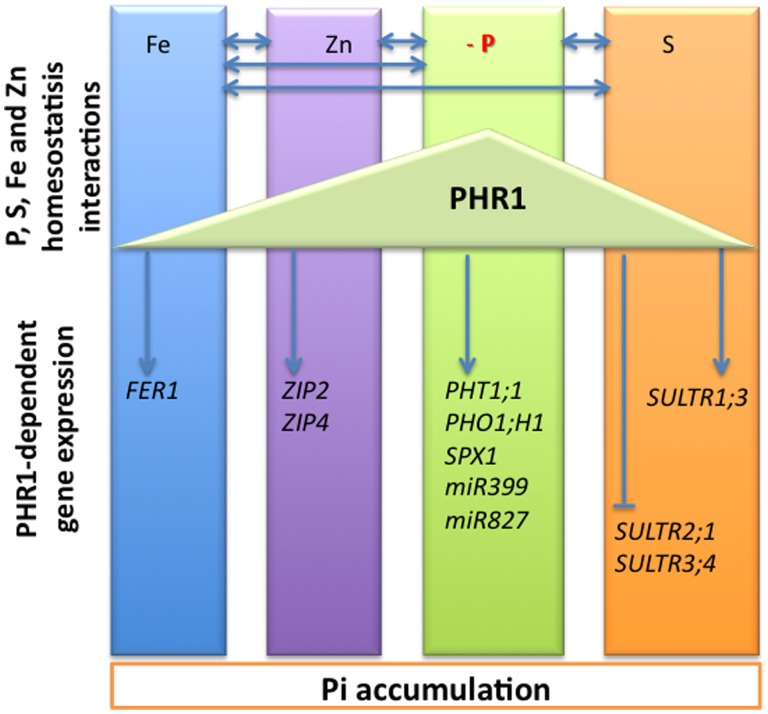
FIGURE 3.
Schematic representation of the macro- and micronutrients homeostasis crosstalks. The interactions between phosphorus (P), sulfur (S), iron (Fe), and zinc (Zn) homeostasis are indicated by left right arrows. At a molecular level, the PHR1 transcription factor was initially identified as a key regulator of the expression of phosphate starvation induced (PSI) genes, including phosphate transporters PHT1;1, PHO1;H 1, and genes involved in phosphate deficiency sensing and signaling including SPX1, miR399, and miR827. PHR1 appeared also as a regulator of the expression of genes involved in sulfate transport including the sulfate transporters SULTR1;3, SULTR2;1, and SULTR3;4. The arrowheads and flat ended lines indicate the positive and negative effects of PHR1, respectively. The transcriptional regulation of some genes involved in maintaining Fe and Zn homeostasis has also been shown to be PHR1-dependent; it includes the FER1 gene encoding the Fe storage protein ferritin, and the ZIP2 and ZIP4 genes encoding zinc transporters.
PHR1 has been the most studied regulator of Pi deficiency response, but it is known that other regulators are involved. Among them TFs including WRKY75 (Devaiah et al., 2007a), ZAT6 (Devaiah et al., 2007b), MYB62 (Devaiah et al., 2009), PTF1 (Yi et al., 2005), bHLH32 (Chen et al., 2007), and WRKY45 (Wang et al., 2014b) have been reported. Furthermore, not only additional TFs have to be considered. In addition to the well-characterized role of miR399 in the regulation of Pi homeostasis in Arabidopsis, other miRNAs (miRNA778, miRNA827, and miRNA2111) were reported to be specifically and strongly induced in response to Pi starvation (Fujii et al., 2005; Hsieh et al., 2009; Pant et al., 2009). The role of these miRNA in the potential cross-talks between pathways regulating homeostasis of various mineral nutriments has already been suggested for miRNA82, involved in the cross-talk between Pi-limitation and nitrate-limitation signaling pathways affecting anthocyanin synthesis (Pant et al., 2009; Liu et al., 2014). Finally, chromatin modification could also be concerned through H2A.Z histone deposition via the nuclear actin-related protein ARP6 in order to activate the expression of many genes related to the Pi deficiency response (Smith et al., 2010).
Integration of pathways controlling two by two nutriment homeostasis has started to be documented. However a survey of transcriptome data reveals that the role played by PHR1 and PHL1 in these interactions could be wider (Figure 4). Indeed PHR1 and PHL1 control transcript accumulation of key genes of Fe homeostasis as well as genes whose expression is directly dependent on S or Pi availability. In consequence, a major challenge in the future will be to consider mineral nutrition as a system, and to develop tools enabling to model integrative gene networks that will take into account the availability of a maximum of macro-and micro-elements, and their interactions, at a given time.
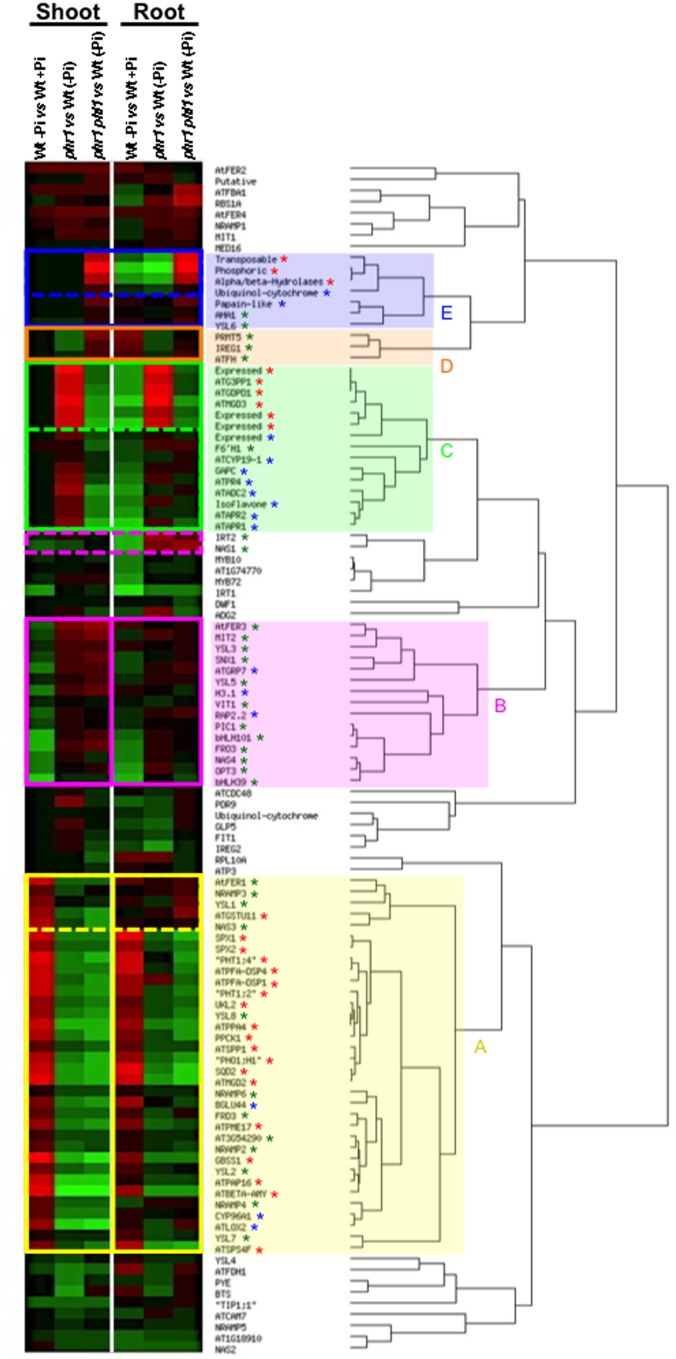
FIGURE 4.
A wider role for PHR1 and PHL1 in the regulation of plant mineral nutrition. Transcript abundance values of genes involved in iron (Fe) homeostasis or that respond to phosphate (Pi) or sulfur (S) deficiency in wild type or phr1 and phr1 phl1 mutants, in the presence or absence of Pi, were selected from microarrays data (Bustos et al., 2010). These data were hierarchically clustered using EPCLUST with the default parameters (http://www.bioinf.ebc.ee/EP/EP/EPCLUST/). (A yellow square): Genes whose mRNA abundance is positively dependent (at least) on PHR1 activity in shoots and roots, or in shoots only (dashed lines); (B pink square): Genes whose mRNA abundance is negatively dependent (at least) on PHR1 activity in shoots and roots or in roots only (dashed lines); (C green square): revealed that Zn deficiency Genes whose mRNA abundance is positively dependent on PHR1 activity but negatively dependent on PHL1 activity both in shoots and roots or in shoots only (dashed lines). (D orange square): Genes whose mRNA abundance is negatively dependent on PHR1 activity but positively dependent on PHL1 activity both in shoots and roots. (E blue square): Genes whose mRNA abundance is negatively dependent on both PHR1 and PHL1 activities in shoots and roots or in shoots only (dashed lines). Red star, Pi deficiency response; blue star, S deficiency response; green star, Fe homeostasis.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2015.00290/abstract
References
- Abe H., Urao T., Ito T., Seki M., Shinozaki K., Yamaguchi-Shinozaki K. (2003). Arabidopsis AtMYC2 (bHLH) and AtMYB2 (MYB) function as transcriptional activators in abscisic acid signaling. Plant Cell 15 63–78 10.1105/tpc.006130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agarwal M., Hao Y., Kapoor A., Dong C. H., Fujii H., Zheng X., et al. (2006). A R2R3 type MYB transcription factor is involved in the cold regulation of CBF genes and in acquired freezing tolerance. J. Biol. Chem. 281 37636–37645 10.1074/jbc.M605895200 [DOI] [PubMed] [Google Scholar]
- Amtmann A., Armengaud P. (2009). Effects of N, P, K and S on metabolism: new knowledge gained from multi-level analysis. Curr. Opin. Plant Biol. 12 275–283 10.1016/j.pbi.2009.04.014 [DOI] [PubMed] [Google Scholar]
- Arrivault S., Senger T., Krämer U. (2006). The Arabidopsis metal tolerance protein AtMTP3 maintains metal homeostasis by mediating Zn exclusion from the shoot under Fe deficiency and Zn oversupply. Plant J. 46 861–879 10.1111/j.1365-313X.2006.02746.x [DOI] [PubMed] [Google Scholar]
- Astolfi S., Cesco S., Zuchi S., Neumann G., Roemheld V. (2006). Sulphur starvation reduces phytosiderophores release by Fe-deficient barley plants. Soil Sci. Plant Nutr. 52 80–85 10.1111/j.1747-0765.2006.00010.x [DOI] [Google Scholar]
- Astolfi S., Zuchi S., Neumann G., Cesco S., Sanità di Toppi L., Pinton R. (2012). Response of barley plants to Fe defciency and Cd contamination as affected by S starvation. J. Exp. Bot. 63 1241–1250 10.1093/jxb/err344 [DOI] [PubMed] [Google Scholar]
- Aung K., Lin S. I., Wu C. C., Huang Y. T., Su C. L., Chiou T. J. (2006). pho2 a phosphate overaccumulator, is caused by a nonsense mutation in a microRNA399 target gene. Plant Physiol. 141 1000–1011 10.1104/pp.106.078063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barberon M., Zelazny E., Robert S., Conejero G., Curie C., Friml J., et al. (2011). Monoubiquitin-dependent endocytosis of the iron-regulated transporter 1 (IRT1) transporter controls iron uptake in plants. Proc. Natl. Acad. Sci. U.S.A. 108 450–458 10.1073/pnas.1100659108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bari R., Datt Pant B., Stitt M., Scheible W. R. (2006). PHO2 microRNA399 and PHR1 define a phosphate-signaling pathway in plants. Plant Physiol. 141 988–999 10.1104/pp.106.079707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayle V., Arrighi J. F., Creff A., Nespoulous C., Vialaret J., Rossignol M., et al. (2011). Arabidopsis thaliana high-affinity phosphate transporters exhibit multiple levels of posttranslational regulation. Plant Cell 23 1523–1535 10.1105/tpc.110.081067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becher M., Talke I. N., Krall L., Krämer U. (2003). Cross-species microarray transcript pro- filing reveals high constitutive expression of metal homeostasis genes in shoots of the zinc hyperaccumulator Arabidopsis halleri. Plant J. 37 251–268 10.1046/j.1365-313X.2003.01959.x [DOI] [PubMed] [Google Scholar]
- Belgaroui N., Zaidi I., Farhat A., Chouayekh H., Bouain N., Chay S., et al. (2014). Over-expression of the bacterial phytase US417 in Arabidopsis reduces the concentration of phytic acid and reveals its involvement in the regulation of sulfate and phosphate homeostasis and signaling. Plant Cell Physiol. 55 1912–1924 10.1093/pcp/pcu122 [DOI] [PubMed] [Google Scholar]
- Berg J. M., Shi Y. (1996). The galvanization of biology: a growing appreciation for the roles of zinc. Science 271 1081–1085 10.1126/science.271.5252.1081 [DOI] [PubMed] [Google Scholar]
- Bouain N., Kisko M., Rouached A., Dauzat M., Lacombe B., Belgaroui N., et al. (2014a). Phosphate/zinc interaction analysis in two lettuce varieties reveals contrasting effects on biomass, photosynthesis, and dynamics of Pi transport. Biomed. Res. Int. 2014 548254 10.1155/2014/548254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouain N., Shahzad Z., Rouached A., Khan G. A., Berthomieu P., Abdelly C., et al. (2014b). Phosphate and zinc transport and signalling in plants: toward a better understanding of their homeostasis interaction. J. Exp. Bot. 65 5725–5741 10.1093/jxb/eru314 [DOI] [PubMed] [Google Scholar]
- Bournier M., Tissot N., Mari S., Boucherez J., Lacombe E., Briat J. F., et al. (2013). Arabidopsis ferritin 1 (AtFer1) gene regulation by the phosphate starvation response 1 (AtPHR1) transcription factor reveals a direct molecular link between iron and phosphate homeostasis. J. Biol. Chem. 288 22670–22680 10.1074/jbc.M113.482281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briat J. F., Dubos C., Gaymard F. (2014). Iron nutrition, biomass production, and plant product quality. Trends Plant Sci. 20 33–40 10.1016/j.tplants.2014.07.005 [DOI] [PubMed] [Google Scholar]
- Briat J. F., Duc C., Ravet K., Gaymard F. (2010a). Ferritins and iron storage in plants. Biochim. Biophys. Acta 1800 806–814 10.1016/j.bbagen.2009.12.003 [DOI] [PubMed] [Google Scholar]
- Briat J. F., Ravet K., Arnaud N., Duc C., Boucherez J., Touraine B., et al. (2010b). New insights into ferritin synthesis and function highlight a link between iron homeostasis and oxidative stress in plants. Ann. Bot. 105 811–822 10.1093/aob/mcp128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchner P., Prosser I., Hawkesford M. J. (2004). Phylogeny and expression of paralogous and orthologous sulphate transporter genes in diploid and hexaploid wheats. Genome 47 526–534 10.1139/g04-011 [DOI] [PubMed] [Google Scholar]
- Bustos R., Castrillo G., Linhares F., Puga M. I., Rubio V., Pérez-Pérez J., et al. (2010). A central regulatory system largely controls transcriptional activation and repression responses to phosphate starvation in Arabidopsis. PLoS Genet. 6:e1001102 10.1371/journal.pgen.1001102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cakmak I., Marschner H. (1986). Mechanism of phosphorus induced zinc deficiency in cotton. I. Zinc deficiency-enhanced uptake rate of phosphorus. Physiol. Plant. 68 483–490 10.1111/j.1399-3054.1986.tb03386.x [DOI] [Google Scholar]
- Charoensawan V., Wilson D., Teichmann S. A. (2010). Lineage-specific expansion of DNA-binding transcription factor families. Trends Plant Sci. 26 388–393 10.1016/j.tig.2010.06.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Chen Z., Kang J., Kang D., Gu H., Qin G. (2013). AtMYB14 regulates cold tolerance in Arabidopsis. Plant Mol. Biol. Rep. 31 87–97 10.1007/s11105-012-0481-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z.-H., Nimmo G. A., Jenkins G. I., Nimmo H. G. (2007). BHLH32 modulates several biochemical and morphological processes that respond to Pi starvation in Arabidopsis. Biochem. J. 405 191–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiou T. J., Aung K., Lin S. I., Wu C. C., Chiang S. F., Su C. L. (2006). Regulation of phosphate homeostasis by MicroRNA in Arabidopsis. Plant Cell 18 412–421 10.1105/tpc.105.038943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciaffi M., Paolacci A. R., Celletti S., Catarcione G., Kopriva S., Astolfi S. (2013). Transcriptional and physiological changes in the S assimilation pathway due to single or combined S and Fe deprivation in durum wheat (Triticum durum L.) seedlings. J. Exp. Bot. 64 1663–1675 10.1093/jxb/ert027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colangelo E. P., Guerinot M. L. (2006). Put the metal to the petal: metal uptake and transport throughout plants, Curr. Opin. Plant Biol. 9 322–330 10.1016/j.pbi.2006.03.015 [DOI] [PubMed] [Google Scholar]
- Coleman J. E. (1998). Zinc enzymes. Curr. Opin. Chem. Biol. 2 222–234 10.1016/S1367-5931(98)80064-1 [DOI] [PubMed] [Google Scholar]
- Cominelli E., Galbiati M., Vavasseur A., Conti L., Sala T., Vuylsteke M., et al. (2005). A guard-cell-specific MYB transcription factor regulates stomatal movements and plant drought tolerance. Curr. Biol. 15 1196–1200 10.1016/j.cub.2005.05.048 [DOI] [PubMed] [Google Scholar]
- Connolly E. L., Fett J. P., Guerinot M. L. (2002). Expression of the IRT1 metal transporter is controlled by metals at the levels of transcript and protein accumulation. Plant Cell 14 1347–1357 10.1105/tpc.001263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conte S. S., Walker E. L. (2011). Transporters contributing to iron trafficking in plants. Mol. Plant 4 464–476 10.1093/mp/ssr015 [DOI] [PubMed] [Google Scholar]
- Couturier J., Touraine B., Briat J. F., Gaymard F., Rouhier N. (2013). The iron-sulfur cluster assembly machineries in plants: current knowledge and open questions. Front. Plant Sci. 4:259 10.3389/fpls.2013.00259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui M. H., Yoo K. S., Hyoung S., Nguyen H. T., Kim Y. Y., Kim H. J., et al. (2013). An Arabidopsis R2R3-MYB transcription factor, AtMYB20 negatively regulates type 2C serine/threonine protein phosphatases to enhance salt tolerance. FEBS Lett. 587 1773–1778 10.1016/j.febslet.2013.04.028 [DOI] [PubMed] [Google Scholar]
- Cumbus I. P., Hornsey D. J., Robinson L. W. (1977). The influence of phosphorus, zinc and manganese on absorption and translocation of iron in watercress. Plant Soil 48 651–660 10.1007/BF00145775 [DOI] [Google Scholar]
- Curie C., Cassin G., Couch D., Divol F., Higuchi K., Le Jean M., et al. (2009). Metal movement within the plant : contribution of nicotianamine and yellow stripe 1-like transporters. Ann. Bot. 103 1–11 10.1093/aob/mcn207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curie C., Panaviene Z., Loulergue C., Dellaporta S. L., Briat J. F., Walker E. L. (2001). Maize yellow stripe1 encodes a membrane protein directly involved in Fe (III) uptake. Nature 409 346–349 10.1038/35053080 [DOI] [PubMed] [Google Scholar]
- Dekock P. C., Hall A., Inkson R. H. E. (1978). Active iron in plant leaves. Ann. Bot. 43 737–740. [Google Scholar]
- Demidchik V., Maathuis F. J. M. (2007). Physiological roles of nonselective cation channels in plants: from salt stress to signalling and development. New Phytol. 175 387–404 10.1111/j.1469-8137.2007.02128.x [DOI] [PubMed] [Google Scholar]
- Devaiah B. N., Karthikeyan A. S., Raghothama K. G. (2007a). WRKY75 transcription factor is a modulator of phosphate acquisition and root development in Arabidopsis. Plant Physiol. 143 1789–1801 10.1104/pp.106.093971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devaiah B. N., Nagarajan V. K., Raghothama K. G. (2007b). Phosphate homeostasis and root development in Arabidopsis are synchronized by the zinc finger transcription factor ZAT6. Plant Physiol. 145 147–159 10.1104/pp.107.101691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devaiah B. N., Madhuvanthi R., Karthikeyan A. S., Raghothama K. G. (2009). Phosphate starvation responses and gibberellic acid biosynthesis are regulated by the MYB62 transcription factor in Arabidopsis. Mol. Plant 2 43–58 10.1093/mp/ssn081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du H., Wang Y. B., Xie Y., Liang Z., Jiang S. J., Zhang S. S., et al. (2013). Genome-wide identification and evolutionary and expression analyses of MYB-related genes in land plants. DNA Res. 20 437–448 10.1093/dnares/dst021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubos C., Le Gourrierec J., Baudry A., Huep G., Lanet E., Debeaujon I., et al. (2008). MYBL2 is a new regulator of flavonoid biosynthesis in Arabidopsis thaliana. Plant J. 55 940–953 10.1111/j.1365-313X.2008.03564.x [DOI] [PubMed] [Google Scholar]
- Dubos C., Stracke R., Grotewold E., Weisshaar B., Martin C., Lepiniec L. (2010). MYB transcription factors in Arabidopsis. Trends Plant Sci. 15 573–581 10.1016/j.tplants.2010.06.005 [DOI] [PubMed] [Google Scholar]
- Elfving N., Davoine C., Benlloch R., Blomberg J., Brännström K., Müller D., et al. (2011). The Arabidopsis thaliana Med25 mediator subunit integrates environmental cues to control plant development. Proc. Natl. Acad. Sci. U.S.A. 108 8245–8250 10.1073/pnas.1002981108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Essigmann B., Guler S., Narang R. A., Linke D., Benning C. (1998). Phosphate availability affects the thylakoid lipid composition and the expression of SQD1 a gene required for sulfolipid biosynthesis in Arabidopsis thaliana. Proc. Natl. Acad. Sci. U.S.A. 95 1950–1955 10.1073/pnas.95.4.1950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forieri I., Wirtz M., Hell R. (2013). Toward new perspectives on the interaction of iron and sulfur metabolism in plants. Front. Plant Sci. 4:357 10.3389/fpls.2013.00357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franco-Zorrilla J. M., Gonzalez E., Bustos R., Linhares F., Leyva A., Paz-Ares J. (2004). The transcriptional control of plant responses to phosphate limitation. J. Exp. Bot. 55 285–293 10.1093/jxb/erh009 [DOI] [PubMed] [Google Scholar]
- Franco-Zorrilla J. M., Martín A. C., Leyva A., Paz-Ares J. (2005). Interaction between phosphate-starvation, sugar, and cytokinin signaling in Arabidopsis and the roles of cytokinin receptors CRE1/AHK4 and AHK3. Plant Physiol. 138 847–857 10.1104/pp.105.060517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franco-Zorrilla J. M., Valli A., Todesco M., Mateos I., Puga M. I., Rubio-Somoza I., et al. (2007). Target mimicry provides a new mechanism for regulation of microRNA activity. Nat. Genet. 39 1033–1037 10.1038/ng2079 [DOI] [PubMed] [Google Scholar]
- Fujii H., Chiou T. J., Lin S. I., Aung K., Zhu J. K. (2005). A miRNA involved in phosphate-starvation response in Arabidopsis. Curr. Biol. 15 2038–2043 10.1016/j.cub.2005.10.016 [DOI] [PubMed] [Google Scholar]
- Fukao Y., Ferjani A., Tomioka R., Nagasaki N., Kurata R., Nishimori Y., et al. (2011) iTRAQ analysis reveals mechanisms of growth defects due to excess zinc in Arabidopsis Plant Physiol. 155 1893–1907 10.1104/pp.110.169730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giehl R. F., Meda A. R., von Wirén N. (2009). Moving up, down, and everywhere: signaling of micronutrients in plants. Curr. Opin. Plant Biol. 12 320–327 10.1016/j.pbi.2009.04.006 [DOI] [PubMed] [Google Scholar]
- Giehl R. F., von Wirén N. (2014). Root nutrient foraging. Plant Physiol. 166 509–517 10.1104/pp.114.245225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gojon A., Nacry P., Davidian J. C. (2009). Root uptake regulation: a central process for NPS homeostasis in plants. Curr. Opin. Plant Biol. 12 328–338 10.1016/j.pbi.2009.04.015 [DOI] [PubMed] [Google Scholar]
- Gruber B. D., Giehl R. F., Friedel S., von Wirén N. (2013). Plasticity of the Arabidopsis root system under nutrient deficiencies. Plant Physiol. 163 161–179 10.1104/pp.113.218453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamburger D., Rezzonico E., MacDonald-Comber Petétot J., Somerville C., Poirier Y. (2002). Identification and characterization of the Arabidopsis PHO1 gene involved in phosphate loading to the xylem. Plant Cell 14 889–902 10.1105/tpc.000745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond J. P., Bennett M. J., Bowen H. C., Broadley M. R., Eastwood D. C., May S. T., et al. (2003). Changes in gene expression in Arabidopsis shoots during phosphate starvation and the potential for developing smart plants. Plant Physiol. 132 578–586 10.1104/pp.103.020941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartel H., Essigmann B., Lokstein H., Hoffmann-Benning S., Peters-Kottig M., Benning C. (1998). The phospholipid-deficient pho1 mutant of Arabidopsis thaliana is affected in the organization, but not in the light acclimation, of the thylakoid membrane. Biochim. Biophys. Acta 1415 205–218 10.1016/S0005-2736(98)00197-7 [DOI] [PubMed] [Google Scholar]
- Haydon M. J., Cobbett C. S. (2007). A novel major facilitator superfamily protein at the tonoplast influences zinc tolerance and accumulation in Arabidopsis. Plant Physiol. 143 1705–1719 10.1104/pp.106.092015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haydon M. J., Kawachi M., Wirtz M., Stefan H., Hell R., Krämer U. (2012). Vacuolar nicotianamine has critical and distinct roles under iron deficiency and for zinc sequestration in Arabidopsis. Plant Cell 24 724–737 10.1105/tpc.111.095042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hindt M. N., Guerinot M. L. (2012). Getting a sense for signals: regulation of the plant iron deficiency response. Biochim. Biophys. Acta 1823 1521–1530 10.1016/j.bbamcr.2012.03.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirsch J., Marin E., Floriani M., Chiarenza S., Richaud P., Nussaume L., et al. (2006). Phosphate deficiency promotes modification of iron distribution in Arabidopsis plants. Biochimie 88 1767–1771 10.1016/j.biochi.2006.05.007 [DOI] [PubMed] [Google Scholar]
- Hsieh L. C., Lin S. I., Shih A. C., Chen J. W., Lin W. Y., Tseng C. Y., et al. (2009). Uncovering small RNA-mediated responses to phosphate deficiency in Arabidopsis by deep sequencing. Plant Physiol. 151 2120–2132 10.1104/pp.109.147280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C., Barker S. J., Langridge P., Smith F. W., Graham R. D. (2000). Zinc deficiency up-regulates expression of high-affinity phosphate transporter genes in both phosphate-sufficient and -deficient barley roots. Plant Physiol. 124 415–422 10.1104/pp.124.1.415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang T. K., Han C. L., Lin S. I., Chen Y. J., Tsai Y. C., Chen Y. R., et al. (2013). Identification of downstream components of ubiquitin-conjugating enzyme PHOSPHATE2 by quantitative membrane proteomics in Arabidopsis roots. Plant Cell 25 4044–4060 10.1105/tpc.113.115998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussain D., Haydon M. J., Wang Y., Wong E., Sherson S. M., Young J., et al. (2004). P-type ATPase heavy metal transporters with roles in essential zinc homeostasis in Arabidopsis. Plant Cell 16 1327–1339 10.1105/tpc.020487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov R., Brumbarova T., Bauer P. (2012). Fitting into the harsh reality: regulation of iron-deficiency responses in dicotyledonous plants. Mol. Plant. 5 27–42 10.1093/mp/ssr065 [DOI] [PubMed] [Google Scholar]
- Jaradat M. R., Feurtado J. A., Huang D., Lu Y., Cutler A. J. (2013). Multiple roles of the transcription factor AtMYBR1/AtMYB44 in ABA signaling, stress responses, and leaf senescence. BMC Plant Biol. 13:192 10.1186/1471-2229-13-192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karthikeyan A. S., Varadarajan D. K., Mukatira U. T., D’Urzo M. P., Damsz B., Raghothama K. G. (2002). Regulated expression of Arabidopsis phosphate transporters. Plant Physiol. 130 221–233 10.1104/pp.020007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawashima C. G., Yoshimoto N., Maruyama-Nakashita A., Tsuchiya Y. N., Saito K., Takahashi H., et al. (2009). Sulphur starvation induces the expression of microRNA-395 and one of its target genes but in different cell types. Plant J. 57 313–321 10.1111/j.1365-313X.2008.03690.x [DOI] [PubMed] [Google Scholar]
- Kellermeier F., Armengaud P., Seditas T. J., Danku J., Salt D. E., Amtmann A. (2014). Analysis of the root system architecture of Arabidopsis provides a quantitative readout of crosstalk between nutritional signals. Plant Cell 26 1480–1496 10.1105/tpc.113.122101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerkeb L., Mukherjee I., Chatterjee I., Lahner B., Salt D. E., Connolly E. L. (2008). Iron-induced turnover of the Arabidopsis IRON-regulated transporter1 metal transporter requires lysine residues. Plant Physiol. 146 1964–1973 10.1104/pp.107.113282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan G. A., Bouraine S., Wege S., Li Y., de Carbonnel M., Berthomieu P., et al. (2014). Coordination between zinc and phosphate homeostasis involves the transcription factor PHR1 the phosphate exporter PHO1 and its homologue PHO1;H3 in Arabidopsis. J. Exp. Bot. 65 871–884 10.1093/jxb/ert444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J. H., Nguyen N. H., Jeong C. Y., Nguyen N. T., Hong S. W., Lee H. (2013). Loss of the R2R3 MYB, AtMyb73 causes hyper-induction of the SOS1 and SOS3 genes in response to high salinity in Arabidopsis. J. Plant Physiol. 170 1461–1465 10.1016/j.jplph.2013.05.011 [DOI] [PubMed] [Google Scholar]
- Kisko M., Bouain N., Rouached A., Choudhary S. P., Rouached H. (2014). Molecular mechanisms of phosphate and zinc signaling crosstalk in plants: phosphate and zinc loading into root xylem in Arabidopsis. Environ. Exp. Bot. 114 57–64 10.1016/j.envexpbot.2014.05.013 [DOI] [Google Scholar]
- Kobayashi T., Nishizawa N. K. (2012). Iron uptake, translocation, and regulation in higher plants. Annu. Rev. Plant Biol. 63 131–152 10.1146/annurev-arplant-042811-105522 [DOI] [PubMed] [Google Scholar]
- Kobayashi T., Nishizawa N. K. (2014). Iron sensors and signals in response to iron deficiency. Plant Sci. 224 36–43 10.1016/j.plantsci.2014 [DOI] [PubMed] [Google Scholar]
- Kopriva S. (2006). Regulation of sulfate assimilation in Arabidopsis and beyond. Ann. Bot. 97 479–495 10.1093/aob/mcl006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krouk G., Ruffel S., Gutiérrez R. A., Gojon A., Crawford N. M., Coruzzi G. M., et al. (2011). A framework integrating plant growth with hormones and nutrients. Trends Plant Sci. 16 178–182 10.1016/j.tplants.2011.02.004 [DOI] [PubMed] [Google Scholar]
- Kuwajima K., Kawai S. (1997). “Relationship between sulfur metabolism and biosynthesis of phytosiderophores in barley roots,” in Plant Nutrition for Sustainable Food Production and Environnment. Developments in Plant and Soil Sciences Vol. 78 eds Ando, T., Fujita, K., Mae, T., Matsumoto, H., Mori, S., Sekiya J. (Dordrech: Kluwer Academic Publishers; ) 285–286. [Google Scholar]
- Lanquar V., Lelievre F., Bolte S., Hames C., Alcon C., Neumann D., et al. (2005). Mobilization of vacuolar iron by AtNRAMP3 and AtNRAMP4 is essential for seed germination on low iron. EMBO J. 24 4041–4051 10.1038/sj.emboj.7600864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapis-Gaza H. R., Jost R., Finnegan P. M. (2014). Arabidopsis PHOSPHATE TRANSPORTER1 genes PHT1; 8 and PHT1; 9 are involved in root-to-shoot translocation of orthophosphate. BMC Plant Biol. 14:334 10.1186/s12870-014-0334-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lejay L., Wirth J., Pervent M., Cross J. M. F., Tillard P., Gojon A. (2008). Oxidative pentose phosphate pathway-dependent sugar sensing as a mechanism for regulation of root ion transporters by photosynthesis. Plant Physiol. 146 2036–2053 10.1104/pp.107.114710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leustek T., Martin M. N., Bick J. A., Davies J. P. (2000). Pathways and regulation of sulfur metabolism revealed through molecular and genetic studies. Ann. Rev. Plant Physiol. Plant Mol. Biol. 51 141–165 10.1146/annurev.arplant.51.1.141 [DOI] [PubMed] [Google Scholar]
- Lipsick J. S. (1996). One billion years of Myb. Oncogene 13 223–235. [PubMed] [Google Scholar]
- Liu T. Y., Chang C. Y., Chiou T. J. (2009). The long-distance signaling of mineral macronutrients. Curr. Opin. Plant Biol. 12 312–319 10.1016/j.pbi.2009.04.004 [DOI] [PubMed] [Google Scholar]
- Liu T. Y., Huang T. K., Tseng C. Y., Lai Y. S., Lin S. I., Lin W. Y., et al. (2012). PHO2-dependent degradation of PHO1 modulates phosphate homeostasis in Arabidopsis. Plant Cell 24 2168–2183 10.1105/tpc.112.096636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu T. Y., Lin W. Y., Huang T. K., Chiou T. J. (2014). MicroRNA-mediated surveillance of phosphate transporters on the move. Trends Plant Sci. 19 647–655 10.1016/j.tplants.2014.06.004 [DOI] [PubMed] [Google Scholar]
- Loneragan J. K., Grunes D. L., Welch R. M., Aduayi E. A., Tengah A., Lazar V. A., et al. (1982). Phosphorus accumulation and toxicity in leaves in relation to zinc supply. Soil Sc. Soc. Am. J. 46 345–352 10.2136/sssaj1982.03615995004600020027x [DOI] [Google Scholar]
- Long T. A., Tsukagoshi H., Busch W., Lahner B., Salt D. E., Benfey P. N. (2010). The bHLH transcription factor POPEYE regulates response to iron deficiency in Arabidopsis roots. Plant Cell 22 2219–2236 10.1105/tpc.110.074096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-Arredondo D. L., Leyva-González M. A., Alatorre-Cobos F., Herrera-Estrella L. (2013). Biotechnology of nutrient uptake and assimilation in plants. Int. J. Dev. Biol. 57 595–610 10.1387/ijdb.130268lh [DOI] [PubMed] [Google Scholar]
- Maathuis F. J. (2009). Physiological functions of mineral macronutrients. Curr. Opin. Plant Biol. 12 250–258 10.1016/j.pbi.2009.04.003 [DOI] [PubMed] [Google Scholar]
- Marschner H. (1995). Mineral Nutrition of Higher Plants. London: Academic Press [Google Scholar]
- Maruyama-Nakashita A., Nakamura Y., Tohge T., Saito K., Takahashi H. (2006). Arabidopsis SLIM1 is a central transcriptional regulator of plant sulphur response and metabolism. Plant Cell 18 3235–3251 10.1105/tpc.106.046458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruyama-Nakashita A., Nakamura Y., Yamaya T., Takahashi H. (2004). A novel regulatory pathway of sulfate uptake in Arabidopsis roots: implication of CRE1/WOL/AHK4-mediated cytokinin-dependent regulation. Plant J. 38 779–789 10.1111/j.1365-313X.2004.02079.x [DOI] [PubMed] [Google Scholar]
- Mäser P., Thomine S., Schroeder J. I., Ward J. M., Hirschi K., Sze H., et al. (2001). Phylogenetic relationships within cation transporter families of Arabidopsis. Plant Physiol. 126 1646–1667 10.1104/pp.126.4.1646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathan K. K., Amberger A. (1977). Influence of iron on the uptake of phosphorous by maize. Plant Soil 48 413–422 10.1007/BF00010097 [DOI] [Google Scholar]
- Mengiste T., Chen X., Salmeron J., Dietrich R. (2003). The BOTRYTIS SUSCEPTIBLE1 gene encodes an R2R3MYB transcription factor protein that is required for biotic and abiotic stress responses in Arabidopsis. Plant Cell 15 2551–2565 10.1105/tpc.014167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misson J., Raghothama K. G., Jain A., Jouhet J., Block M. A., Bligny R., et al. (2005). A genome-wide transcriptional analysis using Arabidopsis thaliana Affymetrix gene chips determined plant responses to phosphate deprivation. Proc. Natl. Acad. Sci. U.S.A. 102 11934–11939 10.1073/pnas.0500778102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miura K., Rus A., Sharkhuu A., Yokoi S., Karthikeyan A. S., Raghothama K. G., et al. (2005). The Arabidopsis SUMO E3 ligase SIZ1 controls phosphate deficiency responses. Proc. Natl. Acad. Sci. U.S.A. 102 7760–7765 10.1073/pnas.0500778102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrissey J., Guerinot M. L. (2009). Iron uptake and transport in plants: the good, the bad, and the ionome. Chem. Rev. 109 4553–4567 10.1021/cr900112r [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moseley J. L., Gonzalez-Ballester D., Pootakham W., Bailey S., Grossman A. R. (2009). Genetic interactions between regulators of Chlamydomonas phosphorus and sulfur deprivation responses. Genetics 181 889–905 10.1534/genetics.108.099382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mudge S. R., Rae A. L., Diatloff E., Smith F. W. (2002). Expression analysis suggests novel roles for members of the Pht1 family of phosphate transporters in Arabidopsis. Plant J 31 341–353 10.1046/j.1365-313X.2002.01356.x [DOI] [PubMed] [Google Scholar]
- Müller R., Morant M., Jarmer H., Nilsson L., Hamborg Nielsen T. H. (2007). Genome-wide analysis of the Arabidopsis leaf transcriptome reveals interaction of phosphate and sugar metabolism. Plant Physiol. 143 156–171 10.1104/pp.106.090167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nussaume L., Kanno S., Javot H., Marin E., Pochon N., Ayadi A., et al. (2011). Phosphate import in plants: focus on the PHT1 transporters. Front. Plant Sci. 2:83 10.3389/fpls.2011.00083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okumura S., Mitsukawa N., Shirano Y., Shibata D. (1998). Phosphate transporter gene family of Arabidopsis thaliana. DNA Res. 5 261–269 10.1093/dnares/5.5.261 [DOI] [PubMed] [Google Scholar]
- Osmont K. S., Sibout R., Hardtke C. S. (2007). Hidden branches: developments in root system architecture. Annu. Rev. Plant Biol. 58 93–113 10.1146/annurev.arplant.58.032806.104006 [DOI] [PubMed] [Google Scholar]
- Palmer C. M., Hindt M. N., Schmidt H., Clemens S., Guerinot M. L. (2013). MYB10 and MYB72 are required for growth under iron-limiting conditions. PLoS Genet. 9:e1003953 10.1371/journal.pgen.1003953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pant B. D., Buhtz A., Kehr J., Scheible W. R. (2008). MicroRNA399 is a long-distance signal for the regulation of plant phosphate homeostasis. Plant J. 53 731–738. 10.1111/j.1365-313X.2007.03363.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pant B. D., Musialak-Lange M., Nuc P., May P., Buhtz A., Kehr J., et al. (2009). Identification of nutrient responsive Arabidopsis and rapeseed microRNAs by comprehensive real-time polymerase chain reaction profiling and small RNA sequencing. Plant Physiol. 150 1541–1555 10.1104/pp.109.139139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paolacci A. R., Celletti S., Catarcione G., Hawkesford M. J., Astolfi S., Ciaffi M. (2014). Iron deprivation results in a rapid but not sustained increase of the expression of genes involved in iron metabolism and sulfate uptake in tomato (Solanum lycopersicum L.) seedlings. J. Integr. Plant Biol. 56 88–100 10.1111/jipb.12110 [DOI] [PubMed] [Google Scholar]
- Petit J. M., van Wuytswinkel O., Briat J. F., Lobréaux S. (2001a). Characterization of an iron-dependent regulatory sequence involved in the transcriptional control of AtFer1 and ZmFer1 plant ferritin genes by iron. J. Biol. Chem. 276 5584–5590 10.1074/jbc.M005903200 [DOI] [PubMed] [Google Scholar]
- Petit J. M., Briat J. F., Lobréaux S. (2001b). Structure and differential expression of the four members of the Arabidopsis thaliana ferritin gene family. Biochem. J. 359 575–582 10.1042/0264-6021:3590575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilon M., Cohu C. M., Ravet K., Abdel-Ghany S. E., Gaymard F. (2009). Essential transition metal homeostasis in plants. Curr. Opin. Plant Biol. 12 347–357 10.1016/j.pbi.2009.04.011 [DOI] [PubMed] [Google Scholar]
- Piñeros M., Kochian L. (2003). Differences in whole-cell and single-channel ion currents across the plasma membrane of mesophyll cells from two closely related Thlaspi species. Plant Physiol. 131 583–594 10.1104/pp.011932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poirier Y., Bucher M. (2002). Phosphate transport and homeostasis in Arabidopsis. Arabidopsis Book 1:e0024 10.1199/tab.0024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravet K., Reyt G., Arnaud N., Krouk G., Djouani el-B., Boucherez J., et al. (2012). Iron and ROS control of the DownSTream mRNA decay pathway is essential for plant fitness. EMBO J. 31 175–186 10.1038/emboj.2011.341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remy E., Cabrito T. R., Batista R. A., Teixeira M. C., Sá-Correia I., Duque P. (2012). The Pht1; 9 and Pht1; 8 transporters mediate inorganic phosphate acquisition by the Arabidopsis thaliana root during phosphorus starvation. New Phytol. 195 356–371 10.1111/j.1469-8137.2012.04167.x [DOI] [PubMed] [Google Scholar]
- Riechmann J. L., Heard J., Martin G., Reuber L., Jiang C. Z., Keddie J., et al. (2000). Arabidopsis transcription factors: genome-wide comparative analysis among eukaryotes. Science 290 2105–2110 10.1126/science.290.5499.2105 [DOI] [PubMed] [Google Scholar]
- Rouached H. (2011). Multilevel coordination of phosphate and sulfate homeostasis in plants. Plant Signal. Behav. 6 952–955 10.4161/psb.6.7.15318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouached H., Secco D., Arpat A. B. (2009). Getting the most sulfate from soil: regulation of sulfate uptake transporters in Arabidopsis. J. Plant Physiol. 166 893–902 10.1016/j.jplph.2009.02.016 [DOI] [PubMed] [Google Scholar]
- Rouached H., Secco D., Arpat B. A. (2010). Regulation of ion homeostasis in plants: current approaches and future challenges. Plant Signal. Behav. 5 501–502 10.4161/psb.11027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouached H., Secco D., Arpat A. B., Poirier Y. (2011a). The transcription factor PHR1 plays a key role in the regulation of sulfate shoot-to-root flux upon phosphate starvation in Arabidopsis. BMC Plant Biol. 11:19 10.1186/1471-2229-11-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouached H., Stefanovic A., Secco D., Bulak Arpat A., Gout E., Bligny R., et al. (2011b). Uncoupling phosphate deficiency from its major effects on growth and transcriptome via PHO1 expression in Arabidopsis. Plant J. 65 557–570 10.1111/j.1365-313X.2010.04442.x [DOI] [PubMed] [Google Scholar]
- Rubio V., Linhares F., Solano R., Martín A. C., Iglesias J., Leyva A., et al. (2001). A conserved MYB transcription factor involved in phosphate starvation signaling both in vascular plants and in unicellular algae. Genes Dev. 15 2122–2133 10.1101/gad.204401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savage N., Yang T. J., Chen C. Y., Lin K. L., Monk N. A., Schmidt W. (2013). Positional signaling and expression of ENHANCER OF TRY AND CPC1 are tuned to increase root hair density in response to phosphate deficiency in Arabidopsis thaliana. PLoS ONE 8:e75452 10.1371/journal.pone.0075452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schachtman D. P., Shin R. (2007). Nutrient sensing and signaling: NPKS. Annu. Rev. Plant Biol. 58 47–69 10.1146/annurev.arplant.58.032806.103750 [DOI] [PubMed] [Google Scholar]
- Schuler M., Keller A., Backes C., Philippar K., Lenhof H. P., Bauer P. (2011). Transcriptome analysis by GeneTrail revealed regulation of functional categories in response to alterations of iron homeostasis in Arabidopsis thaliana. BMC Plant Biol. 11:87 10.1186/1471-2229-11-87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schünmann P. H., Richardson A. E., Smith F. W., Delhaize E. (2004a). Characterization of promoter expression patterns derived from the Pht1 phosphate transporter genes of barley (Hordeum vulgare L.). J. Exp. Bot. 55 855–865 10.1093/jxb/erh103 [DOI] [PubMed] [Google Scholar]
- Schünmann P. H., Richardson A. E., Vickers C. E., Delhaize E. (2004b). Promoter analysis of the barley Pht1;1 phosphate transporter gene identifies regions controlling root expression and responsiveness to phosphate deprivation. Plant Physiol. 136 4205–4214 10.1104/pp.104.045823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo P. J., Lee S. B., Suh M. C., Park M. J., Go Y. S., Park C. M. (2011). The MYB96 transcription factor regulates cuticular wax biosynthesis under drought conditions in Arabidopsis. Plant Cell 23 1138–1152 10.1105/tpc.111.083485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo P. J., Xiang F., Qiao M., Park J. Y., Lee Y. N., Kim S. G., et al. (2009). The MYB96 transcription factor mediates abscisic acid signaling during drought stress response in Arabidopsis. Plant Physiol. 151 275–289 10.1104/pp.109.144220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shanmugam V., Lo J. C., Wu C. L., Wang S. L., Lai C. C., Connolly E. L., et al. (2011). Differential expression and regulation of iron-regulated metal transporters in Arabidopsis halleri and Arabidopsis thaliana — the role in zinc tolerance. New Phytol. 190 125–137 10.1111/j.1469-8137.2010.03606.x [DOI] [PubMed] [Google Scholar]
- Shimogawara K., Wykoff D. D., Usuda H., Grossman A. R. (1999). Chlamydomonas reinhardtii mutants abnormal in their responses to phosphorus deprivation. Plant Physiol. 120 685–694 10.1104/pp.120.3.685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinclair S. A., Krämer U. (2012). The zinc homeostasis network of land plants. Biochim. Biophys. Acta 1823 1553–1567 10.1016/j.bbamcr.2012.05.016 [DOI] [PubMed] [Google Scholar]
- Smith A., Jain A., Deal R., Nagarajan V., Poling M., Raghothama K., et al. (2010). Histone H2A.Z régulâtes the expression of several classes of phosphate starvation response genes, but not as a transcriptional activator. Plant Physiol. 152 217–225 10.1104/pp.109.145532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sresty T. V. S., Madhava Rao K. V. (1999). Ultrastructural alterations in response to zinc and nickel stress in the root cells of pigeon pea. Environ. Exp. Bot. 41 3–13 10.1016/S0098-8472(98)00034-3 [DOI] [Google Scholar]
- Su C. F., Wang Y. C., Hsieh T. H., Lu C. A., Tseng T. H., Yu S. M. (2010). A novel MYBS3-dependent pathway confers cold tolerance in rice. Plant Physiol. 153 145–158 10.1104/pp.110.153015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugimoto K., Sato N., Tsuzuki M. (2007). Utilization of a chloroplast membrane sulfolipid as a major internal sulfur source for protein synthesis in the early phase of sulfur starvation in Chlamydomonas reinhardtii. FEBS Lett. 581 4519–4522 10.1016/j.febslet.2007.08.035 [DOI] [PubMed] [Google Scholar]
- Stefanovic A., Ribot C., Rouached H., Wang Y., Chong J., Belbahri L., et al. (2007). Members of the PHO1 gene family show limited functional redundancy in phosphate transfer to the shoot, and are regulated by phosphate deficiency via distinct pathways. Plant J. 50 982–994 10.1111/j.1365-313X.2007.03108.x [DOI] [PubMed] [Google Scholar]
- Stracke R., Werber M., Weisshaar B. (2001). The R2R3-MYB gene family in Arabidopsis thaliana. Curr. Opin. Plant Biol. 4 447–456 10.1016/S1369-5266(00)00199-0 [DOI] [PubMed] [Google Scholar]
- Svistoonoff S., Creff A., Reymond M., Sigoillot-Claude C., Ricaud L., Blanchet A., et al. (2007). Root tip contact with low-phosphate media reprograms plant root architecture. Nat. Genet. 39 792–796 10.1038/ng2041 [DOI] [PubMed] [Google Scholar]
- Takahashi H. (2010). Regulation of sulfate transport and assimilation in plants. Int. Rev. Cell. Mol. Bio. 281 129–159 10.1016/S1937-6448(10)81004-4 [DOI] [PubMed] [Google Scholar]
- Takahashi H., Watanabe-Takahashi A., Smith F. W., Blake-Kalff M., Hawkesford M. J., Saito K. (2000). The role of three functional sulphate transporters involved in uptake and translocation of sulphate in Arabidopsis thaliana. Plant J. 23 171–182 10.1046/j.1365-313x.2000.00768.x [DOI] [PubMed] [Google Scholar]
- Takahashi H., Yamazaki M., Sasakura N., Watanabe A., Leustek T., Engler J. A., et al. (1997). Regulation of sulfur assimilation in higher plants: a sulfate transporter induced in sulfate starved roots plays a central role in Arabidopsis thaliana. Proc. Natl Acad. Sci. U.S.A. 94 11102–11107 10.1073/pnas.94.20.11102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thibaud M. C., Arrighi J. F., Bayle V., Chiarenza S., Creff A., Bustos R., et al. (2010). Dissection of local and systemic transcriptional responses to phosphate starvation in Arabidopsis. Plant J. 64 775–789 10.1111/j.1365-313X.2010.04375.x [DOI] [PubMed] [Google Scholar]
- Ticconi C., Lucero R., Sakhonwasee S., Adamson A., Creff A., Nussaume L., et al. (2009). ER-resident proteins PDR2 and LPR1 mediate the developmental response of root meristems to phosphate availability. Proc. Natl. Acad. Sci. U.S.A. 106 14174–14179 10.1073/pnas.0901778106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van de Mortel J. E., Almar Villanueva L., Schat H., Kwekkeboom J., Coughlan S., Moerland P. D., et al. (2006). Large expression differences in genes for iron and zinc homeostasis, stress response, and lignin biosynthesis distinguish roots of Arabidopsis thaliana and the related metal hyperaccumulator Thlaspi caerulescens. Plant Physiol. 142 1127–1147 10.1104/pp.106.082073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van de Mortel J. E., Schat H., Moerland P. D., Ver Loren van Themaat E., van der Ent S., Blankestijn H., et al. (2008). Expression differences for genes involved in lignin, glutathione and sulphate metabolism in response to cadmium in Arabidopsis thaliana and the related Zn/Cd-hyperaccumulator Thlaspi caerulescens. Plant Cell Environ. 31 301–324 10.1111/j.1365-3040.2007.01764.x [DOI] [PubMed] [Google Scholar]
- Verret F., Gravot A., Auroy P., Leonhardt N., David P., Nussaume L., et al. (2004). Overexpression of AtHMA4 enhances root-to-shoot translocation of zinc and cadmium and plant metal tolerance. FEBS Lett. 576 306–312 10.1016/j.febslet.2004.09.023 [DOI] [PubMed] [Google Scholar]
- Vert G., Grotz N., Dedaldechamp F., Gaymard F., Guerinot M. L., Briat J. F., et al. (2002). IRT1 an Arabidopsis transporter essential for iron uptake from the soil and for plant growth. Plant Cell 14 1223–1233 10.1105/tpc.001388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vigani G., Zocchi G., Bashir K., Philippar K., Briat J. F. (2013). Signals from chloroplasts and mitochondria for iron homeostasis regulation. Trends Plant Sci. 18 305–311 10.1016/j.tplants.2013.01.006 [DOI] [PubMed] [Google Scholar]
- Wang Z., Ruan W., Shi J., Zhang L., Xiang D., Yang C., et al. (2014a). Rice SPX1 and SPX2 inhibit phosphate starvation responses through interacting with PHR2 in a phosphate-dependent manner. Proc. Natl. Acad. Sci. U.S.A. 111 14953–14958 10.1073/pnas.1404680111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Xu Q., Kong Y. H., Chen Y., Duan J. Y., Wu W. H., et al. (2014b). Arabidopsis WRKY45 transcription factor activates PHOSPHATE TRANSPORTER1; 1 expression in response to phosphate starvation. Plant Physiol. 164 2020–2029 10.1104/pp.113.235077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward J. T., Lahner B., Yakubova E., Salt D. E., Raghothama K. G. (2008). The effect of iron on the primary root elongation of Arabidopsis during phosphate deficiency. Plant Physiol. 147 1181–1191 10.1104/pp.108.118562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wintz H., Fox T., Wu Y. Y., Feng V., Chen W., Chang H. S., et al. (2003). Expression profiles of Arabidopsis thaliana in mineral deficiencies reveal novel transporters involved in metal homeostasis. J. Biol. Chem. 278 47644–47653 10.1074/jbc.M309338200 [DOI] [PubMed] [Google Scholar]
- Woo J., MacPherson C. R., Liu J., Wang H., Kiba T., Hannah, M. A.,et al. (2012). The response and recovery of the Arabidopsis thaliana transcriptome to phosphate starvation. BMC Plant Biol. 12:62 10.1186/1471-2229-12-62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu P., Ma L., Hou X., Wang M., Wu Y., Liu F., et al. (2003). Phosphate starvation triggers distinct alterations of genome expression in Arabidopsis roots and leaves. Plant Physiol. 132 1260–1271 10.1104/pp.103.021022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wykoff D. D., Grossman A. R., Weeks D. P., Usuda H., Shimogawara K. (1999). Psr1 a nuclear localized protein that regulates phosphorus metabolism in Chlamydomonas. Proc. Natl. Acad. Sci. U.S.A. 96 15336–15341 10.1073/pnas.96.26.15336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Z., Li D., Wang L., Sack F. D., Grotewold E. (2010). Role of the stomatal development regulators FLP/MYB88 in abiotic stress responses. Plant J. 64 731–739 10.1111/j.1365-313X.2010.04364.x [DOI] [PubMed] [Google Scholar]
- Xu Q., Chu W., Qiu H., Fu Y., Cai S., Sha S. (2013). Responses of Hydrilla verticillata (L.f.) Royle to zinc: in situ localization, subcellular distribution and physiological and ultrastructural modifications. Plant Physiol. Biochem. 69 43–48 10.1016/j.plaphy.2013.04.018 [DOI] [PubMed] [Google Scholar]
- Yi K., Wu Z., Zhou J., Du L., Guo L., Wu Y., et al. (2005). OsPTF1 a novel transcription factor involved in tolerance to phosphate starvation in rice. Plant Physiol. 138 2087–2096 10.1104/pp.105.063115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu B., Xu C., Benning C. (2002). Arabidopsis disrupted in SQD2 encoding sulfolipid synthase is impaired in phosphate-limited growth. Proc. Natl. Acad. Sci. U.S.A. 99 5732–5737 10.1073/pnas.082696499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamioudis C., Hanson J., Pieterse C. M. (2014). β -Glucosidase BGLU42 is a MYB72-dependent key regulator of rhizobacteria-induced systemic resistance and modulates iron deficiency responses in Arabidopsis roots. New Phytol. 204 368–379 10.1111/nph.12980 [DOI] [PubMed] [Google Scholar]
- Zhai H., Bai X., Zhu Y., Li Y., Cai H., Ji W., et al. (2010). A single-repeat R3-MYB transcription factor MYBC1 negatively regulates freezing tolerance in Arabidopsis. Biochem. Biophys. Res. Commun. 394 1018–1023 10.1016/j.bbrc.2010.03.114 [DOI] [PubMed] [Google Scholar]
- Zhang B., Pasini R., Dan H., Joshi N., Zhao Y., Leustek T., et al. (2014). Aberrant gene expression in the Arabidopsis SULTR1; 2 mutants suggests a possible regulatory role for this sulfate transporter in response to sulfur nutrient status. Plant J. 77 185–197 10.1111/tpj.12376 [DOI] [PubMed] [Google Scholar]
- Zhang X., Ju H. W., Chung M. S., Huang P., Ahn S. J., Kim C. S. (2011). The R-R-type MYB-like transcription factor, AtMYBL, is involved in promoting leaf senescence and modulates an abiotic stress response in Arabidopsis. Plant Cell Physiol. 52 138–148 10.1093/pcp/pcq180 [DOI] [PubMed] [Google Scholar]
- Zhou J., Jiao F., Wu Z., Li Y., Wang X., He X., et al. (2008). OsPHR2 is involved in phosphate-starvation signaling and excessive phosphate accumulation in shoots of plants. Plant Physiol. 146 1673–1686 10.1104/pp.107.111443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y. G., Smith S. E., Smith F. A. (2001). Plant growth and cation composition of two cultivars of spring wheat (Triticum aestivum L.) differing in P uptake efficiency. J. Exp. Bot. 52 1277–1282 10.1093/jexbot/52.359.1277 [DOI] [PubMed] [Google Scholar]
- Zuchi S., Cesco S., Varanini Z., Pinton R., Astolfi S. (2009). Sulphur deprivation limits Fe-deficiency responses in tomato plants. Planta 230 85–94 10.1007/s00425-009-0919-1 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.