Abstract
Background
New methods are needed to improve health behaviors such as adherence to colorectal cancer (CRC) screening. There is increasing availability of personalized genetic information to inform medical decisions. It is not known if such information motivates behavioral change.
Objective
To determine, in average risk persons, if individualized gene-environment risk assessment about CRC susceptibility improves adherence to screening.
Design
Two-arm, randomized, controlled trial
Setting
Four medical school affiliated primary care practices
Patients
783 patients at average risk for CRC, but not adherent with screening at study entry
Intervention
Patients were randomized to usual care or to receipt of Gene Environmental Risk Assessment (GERA), which assessed Methylene Tetrahydrofolate Reductase (MTHFR) polymorphisms and serum folate level. Based on pre-specified polymorphism/folate level combinations, GERA participants were told they were at either “elevated” or at “average” risk for CRC.
Measurements
The primary outcome was receipt of CRC screening within 6 months of study entry.
Results
CRC screening rates were not statistically significantly different between usual care (35.7%) and GERA (33.1%) arms overall. After adjustment for baseline participant factors, the odds ratio (OR) for screening completion for GERA vs usual care was 0.88 (95% CI 0.64 - 1.22). Within the GERA arm, there was no significant difference in screening rates between GERA average risk (38.1%) and GERA elevated risk (26.9%) groups. Odds ratios for elevated vs. average risk remained non-significant after adjustment for covariates (OR=0.75, 95% CI 0.39 - 1.42).
Limitations
Only one personalized, gene-environment interaction and only one health behavior, colorectal cancer screening, were assessed.
Conclusion
In average risk persons, there was no positive association between CRC screening uptake and feedback of a single personalized gene-environment risk assessment (GERA). Additional studies will be required to assess whether other approaches to providing GERA affect screening utilization differently. These findings raise concern about the effectiveness of moderately predictive genetic risk assessment to promote favorable healthcare behavior.
Funding
National Institutes of Health (USA)
Introduction
Genetic testing for cancer susceptibility is offered most frequently for single gene disorders. A mutation in the adenomatous polyposis coli (APC) gene confers a colorectal cancer (CRC) risk of approximately 90%.1 Identification of this mutation carries substantial clinical impact. However, most cancers do not arise from single mutations. Instead, susceptibility results from the modest contributions of many genes, usually with environmental interaction. Identification of these genes and their modifiers is an active area of research.2
Gene-environment risk assessment (GERA) for common diseases is anticipated to become a common component of healthcare.3,4 In this project, we assessed polymorphic variants of Methylene TetraHydroFolate Reductase (MTHFR), an important regulator of cellular folate metabolism. Specific MTHFR polymorphisms are associated with variable susceptibility to colon cancer.5, 6, 7, 8 Epidemiological data also supports a link between serum folate level and CRC risk.9 Assessment of the gene (MTHFR) and environment interaction (serum folate) is attractive because data already exist that allows for CRC risk stratification among “average risk” persons. Risk elevation for CRC associated with specific polymorphisms in combination with low folate is roughly 1.5-2.0× baseline in US Whites and African Americans.8
Unlike genetic testing for high-risk individuals such as in familial adenomatous polyposis, little is known about the impact on health behaviors or the psychological effects of providing average risk individuals with moderately predictive genetic information. 4,10,11,12,13 Understanding the benefits and harms of such testing in average risk individuals is important as similar testing is marketed directly to the public in an unsupervised fashion.14
We conducted a prospective, randomized controlled trial assessing the impact of GERA feedback on CRC screening among average risk individuals who were not adherent to screening recommendations at study entry. CRC prevention is an optimal area to study the emerging role of moderately predictive genetic testing. CRC is the second leading cause of cancer death in the US.15 Screening can reduce disease incidence and mortality.16
Our primary hypothesis was that the provision of personalized GERA information would improve CRC screening uptake compared to usual care. Further, we anticipated that participants characterized as “elevated” risk by GERA would screen more than those at “average” risk. Although MTHFR and folate testing provided personalized risk information, the intent of this project was not to evaluate the specific, predictive value of this combination for colorectal neoplasia.
Methods
Design Overview
We conducted a randomized, controlled trial to compare CRC screening utilization in the GERA arm to the usual care (control) arm, and also to detect differences in screening proportion between the GERA average and elevated risk groups.12 To increase the statistical power for comparisons within the GERA arm, participants were randomized 2:1 to GERA or to usual care, respectively. Participants provided baseline, three week and six month survey data, and agreed to medical record review to ascertain CRC screening status. GERA arm subjects also participated in GERA counseling and disclosure sessions, and had blood drawn for genotyping and blood-based folate assessment.
Setting and Participants
Participants in this institutional review board approved trial were recruited from four Internal Medicine and Family Medicine practices affiliated with Thomas Jefferson University. We searched scheduling, billing and medical records databases applying filters matching eligibility requirements which included: (a) men and women 50-79y; (b) average risk for CRC defined as: asymptomatic, without a personal history of colorectal adenomas or cancer, inflammatory bowel disease, or a family history of familial adenomatous polyposis (FAP), hereditary non-polyposis colorectal cancer (HNPCC) or CRC in a first-degree relative; (c) non-adherence with CRC screening guidelines at study initiation defined as none of the following: home stool blood test in the last 12 months; barium enema or flexible sigmoidoscopy in the last 5 years, or colonoscopy in the last 10 years; (d) no antibiotics or anti-folate medications (sulfasalazine or methotrexate) within the past 6 weeks; (e) English speaking.
Potential participants were mailed a letter outlining the study (see Supplement, “GERA Study Description”), a pamphlet reviewing standard CRC screening options, a consent form and information describing how to opt out of the study. We telephoned individuals who had not opted out, confirmed eligibility, obtained verbal consent and administered a baseline survey containing demographics, past medical and multivitamin (MVI) use history. Participants also responded to previously described questionnaires regarding CRC screening knowledge and the possible role of genes and diet in cancer development.17 Finally, they completed the Impact of Event Scale (IES), a well validated assessment tool for psychological distress.18
Randomization and Intervention
Study arm allocation was implemented by a secure, web-based application using randomization tables generated prior to the start of study.
GERA Intervention Arm
Participants randomized to GERA met with specially trained study nurses rather than genetic counselors. This research design reflected an effort to study feasible approaches to genetic test disclosure that could be incorporated in primary care settings, as there appear to be an inadequate number of genetic counselors to meet health service needs even in high risk settings.19 Study nurses underwent training by genetic counselors prior to study initiation as well as regular monitoring of disclosure sessions to ensure process uniformity.
Employing a standardized, study pamphlet describing GERA, the nurse and intervention arm participants together reviewed the purpose of the gene-environment risk assessment, the potential range of results from genotyping and folate measurement and how results would be disclosed. A basic description of genes and their importance in cancer was provided. An average lifetime risk of CRC of 1 in 20 was described. Participants were told their GERA results would be either “elevated” or “average”. An “elevated” result should not be viewed as a guarantee of disease. It was described as only one potential risk factor for CRC, suggesting modestly increased risk compared to a similarly aged person. Conversely, an “average” risk result was not assurance of protection against CRC now or in the future, it simply indicated the absence of this risk factor. The study nurse emphasized that GERA was a method to stratify CRC risk, but was not intended to be a substitute for screening.
Venipuncture was performed to assess serum folate and MTHFR polymorphism for participants randomized to the GERA arm. Folate analysis was conducted in a single commercial laboratory employing a standard 125I RIA kit with controls. All genotyping was performed at Fox Chase's Genomics Facility. After DNA extraction, separate PCR amplifications of exons 4 and 7 of the MTHFR gene were completed, followed by pyrosequencing specific for codons 677 and 1298.
Characterization of GERA-associated CRC risk was based on specific combinations of age specific serum folate and Methylene TetraHydroFolate Reductase (MTHFR) polymorphism (Supplemental Table 1). MTHFR is an important regulator of cellular folate metabolism. Separately, either MTHFR polymorphic variant or serum folate level is associated with variable susceptibility to CRC. 5,6,9,20,21 The majority of compound genotype combinations carry a modestly elevated CRC risk in the setting of low folate, compared to carriers of these same genotypes with normal serum folate level or to persons with less common combinations (TT/AA and CC/CC) regardless of folate level. 8,22,23
“Low” folate status was conventionally assigned to any individual in the 25th percentile or below for age-based population norms, according to US National Health and Nutrition Examination Survey (NHANES) data at study commencement.24
Within two weeks of blood collection, GERA results were disclosed and explained to every participant first by telephone and then by mail (see Supplement, “Results Mailing”). In the disclosure session, the same points about multiple potential risk factors (GERA being only one) were emphasized as was the importance of screening regardless of GERA result.
Usual care (control) Arm
Usual care participants completed the same baseline questionnaire as GERA participants; however they did not meet with a study nurse, discuss GERA, participate in decision counseling or provide a blood sample.
Regardless of study arm or risk, all participants were encouraged to undergo screening.
Outcomes and Follow-up
Control and intervention participants subsequently completed similar procedures including:
Receipt of a two-card fecal immunochemical test kit with instructions to minimize the impact of variable access to CRC screening.
Three week and six month post enrollment questionnaires were administered by telephone. These questionnaires asked about CRC screening utilization and repeated the IES assessment.
As part of the post-visit three-week and six month telephone questionnaires, participants randomized to GERA were also asked about receipt of counseling and GERA results. Participants who recalled receiving GERA results were given the Multidimensional Inventory of Cancer Risk Assessment (MICRA) which measures psychological reactions specific to genetic testing.25 MICRA includes three subscales, including distress, uncertainty, and positive experiences (where a higher score reflects fewer positive experiences).
The primary study outcome of CRC screening within 6 months of enrollment was calculated based upon electronic and manual medical chart review. Electronic reviews were conducted first, searching for completion date of any screening test (stool blood test, sigmoidoscopy, barium enema or colonoscopy). If no electronic entry was found, paper charts were reviewed. Chart abstractors were blinded to study arm allocation.
Statistical Analysis
A target sample size of N=1760 was selected to provide 80% power to detect a clinically relevant 10% absolute difference between the proportion of participants screened in the “elevated” compared to the “average” risk groups of the GERA arm. This sample size also provided 99% power to detect a 10% absolute difference in the screening proportions between the usual care and GERA arms. As planned, the study's data and safety monitoring board (DSMB) performed periodic interim analyses to monitor the impact of the GERA intervention. When it was recognized that the effect of the study intervention was substantially smaller than hypothesized, the DSMB recommended stopping recruitment because further enrollment was extremely unlikely to yield statistically significant differences for increased screening in the intervention arm.
Potential confounders associated with CRC screening identified from a priori literature reviews were compared across study arms using Chi-square tests and Wilcoxon rank sum tests for categorical and continuous variables, respectively. Factors examined included age, gender, race, marital status, education, regular multi-vitamin use, impact of events scale, CRC screening knowledge scale, and genetics and diet knowledge scale. IES results are dichotomized as higher stress (score ≥26 vs. ≤25) based on standard convention.18
The primary efficacy analysis used univariate logistic regression to compare the proportions of patients completing CRC screening within six months of enrollment between study arms. Statistical significance was assessed by the Wald Chi-square test. Following intention-to-treat methodology, all enrolled study participants were included in the primary outcome analysis. Participants without chart audit information (n=6) were included in the analysis as non-adherent. Within the GERA arm, the association of GERA risk (elevated vs. average) with CRC screening was evaluated in a similar way. Participants randomized to the GERA arm who did not receive GERA risk determination were excluded from this analysis (n=77, due to lab error, blood not drawn, or no receipt of GERA counseling). To explore whether the study's results would be affected by a substantial imbalance in the proportion of participants with unknown GERA risk across risk groups, we performed sensitivity analyses in the GERA arm. We considered two extremes: all participants with unknown risk were (1) allocated to the average risk group, or (2) designated as elevated risk.
Univariate logistic regression was used to assess the association of potential baseline confounders with CRC screening. Participants with missing values for a baseline variable were excluded from analyses involving that variable. The same approach was used to evaluate the relationships between baseline factors and the proportions of GERA arm patients defined as “elevated” risk.
Multivariable logistic models were fit to the data to assess the effect of the primary intervention on CRC screening after adjusting for potential confounders. Covariates in these models included study arm allocation and all a priori defined potential confounders. The effect is reported as an adjusted odds ratio and 95% two-sided confidence interval. A second multivariable model was fit to data from GERA arm participants only to evaluate the impact of GERA risk on CRC screening adherence after adjusting for these same potential confounders.
Propensity score-based analyses were conducted to further assess the effect of GERA risk on screening after controlling for confounding. The details of these analyses are described in the Appendix.
Univariate logistic models were used to explore the associations of CRC screening status and GERA risk with MICRA scores among GERA arm participants. All tests were two-sided using a 5% type 1 error, and analyses were performed using SAS version 9.3 and Stata version 12.
Results
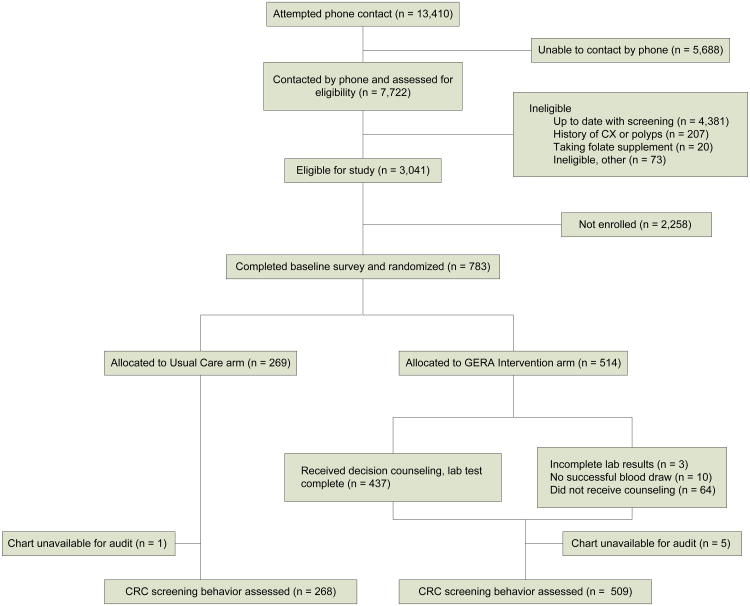
A total of 783 participants were randomized and completed the baseline survey (Figure 1). Baseline demographic, multivitamin use, psychometric and knowledge characteristics are presented in Table 1. Nearly half of the participants reported being married, and 56% indicated their race as white. Most had at least some college education. Across study arms, there were no significant differences in any of these parameters. At baseline, multivitamin use, IES score and assessments of knowledge regarding CRC screening and genes and diet were similar in participants randomized to either arm.
Figure 1. Study Flowchart.
Table 1. Baseline characteristics by study arm.
| Usual Care (n=269) | GERA (n=514) | |||
|---|---|---|---|---|
| N | % | N | % | |
| Age | ||||
| 50-59 years | 181 | 67.3 | 365 | 71.0 |
| 60-69 years | 65 | 24.2 | 107 | 20.8 |
| 70-79 years | 23 | 8.6 | 42 | 8.2 |
| Gender | ||||
| Female | 155 | 57.6 | 304 | 59.1 |
| Male | 114 | 42.4 | 210 | 40.9 |
| Race | ||||
| White | 142 | 52.8 | 294 | 57.2 |
| Black or African American | 112 | 41.6 | 202 | 39.3 |
| Other | 15 | 5.6 | 18 | 3.5 |
| Marital status† | ||||
| Married | 132 | 49.1 | 248 | 48.3 |
| Single/Divorced/Widowed | 137 | 50.9 | 265 | 51.7 |
| Education† | ||||
| Less than high school | 28 | 10.5 | 36 | 7.0 |
| High school graduate | 64 | 23.9 | 123 | 23.9 |
| Some college | 64 | 23.9 | 150 | 29.2 |
| College graduate | 112 | 41.8 | 205 | 39.9 |
| Regular Multivitamin Use | ||||
| No | 151 | 56.1 | 263 | 51.2 |
| Yes | 118 | 43.9 | 251 | 48.8 |
| Impact of Event Scale† | ||||
| 0-25 | 244 | 91.0 | 461 | 89.9 |
| 26+ | 24 | 9.0 | 52 | 10.1 |
| Knowledge Questionnaires | Median | (Q1, Q3) | Median | (Q1,Q3) |
| CRC screening knowledge | 7 | (5,8) | 7 | (5,8) |
| Genetics & diet knowledge | 5 | (3,6) | 5 | (6,7) |
Missing values excluded, n=1 for Marital status, n=1 for Education, and n=2 for Impact of Events Scale
None of the baseline characteristics differed by study arm (all P>0.05, Pearson Chi-square tests for categorical variables or Wilcoxon rank sum tests for continuous variables)
The knowledge questionnaire scores represent the number of correct responses (10 items in each questionnaire), where a higher score indicates a greater level of CRC screening knowledge or diet-genetics interaction knowledge. Elevated Impact of Event Scale scores indicate higher levels of stress.
Abbreviations: CRC = Colorectal Cancer; Q1, Q3 = First and third quartile value (25th and 75th percentiles)
Table 2 demonstrates that CRC screening at 6 months was similar for GERA and usual care arms. In both groups, approximately 1/3 of participants completed screening during the follow-up period. The unadjusted odds ratio estimate for CRC screening for GERA versus usual care was 0.89 (95% CI 0.65 - 1.21).
Table 2. CRC screening at six months by study arm.
| Study Arm | ||||
|---|---|---|---|---|
|
| ||||
| All | Usual Care | GERA | GERA vs. Usual Care | |
| N (%) | N (%) | N (%) | Odds Ratio (95% CI) | |
|
|
|
|||
| CRC screening status | ||||
| Had screening | 266 (34.0) | 96 (35.7) | 170 (33.1) | 0.89 (0.65-1.21) |
| No screening† | 517 (66.0) | 173 (64.3) | 344 (66.9) | |
There were six participants missing data for the chart review (GERA arm n=5, Usual care arm n=1); following the intention-to-treat approach, these participants are included in the results as ‘no screening’.
Percent with CRC screening did by not differ by study arm (P>0.05, Wald Chi-square test).
Abbreviations: CI = Confidence interval; CRC = Colorectal Cancer; UC = Usual Care
Although the covariates were well-balanced in the two arms, we conducted additional analyses to assess the impact of potential confounders on the association between screening and study arm. We first determined if any participant factors were associated with greater screening adherence at 6 months (Supplemental Table 2). We then used multivariable logistic regression to evaluate the effect of study arm on screening after adjusting for a priori defined baseline patient characteristics. The adjusted odds ratio estimate was 0.88 (95% CI 0.64 - 1.22) (Table 3). Statistically significant participant factors related to CRC screening in the multivariable analysis included older age (70-79 vs. 50-59 yrs, OR=2.27, 95% CI 1.32 - 3.90; 60-69 vs. 50-59 yrs, OR=1.29, 95% CI 0.89 -1.86), higher baseline psychological stress score (OR=0.49, 95% CI 0.27 - 0.90), and greater knowledge about CRC screening (OR=1.12, 95% CI 1.01 - 1.24).
Table 3. CRC screening at six months and study arm, adjusted for covariates using multivariable logistic regression.
| Adjusted Odds Ratio estimates | |||
|---|---|---|---|
|
|
|||
| Odds Ratio | 95% CI | P value* | |
| Study Arm | NS | ||
| GERA vs Usual Care | 0.88 | 0.64-1.22 | |
| Age Category | 0.010 | ||
| 60-69 years vs 50-59 years | 1.29 | 0.89-1.86 | |
| 70-79 years vs 50-59 years | 2.27 | 1.32-3.90 | |
| Gender | NS | ||
| Male vs Female | 1.12 | 0.82-1.53 | |
| Race | NS | ||
| Black/AA vs White | 0.88 | 0.61-1.25 | |
| Other vs White | 0.87 | 0.40-1.88 | |
| Marital status† | NS | ||
| Single/Divorced/Widowed vs Married | 0.86 | 0.63-1.18 | |
| Education† | NS | ||
| <HS vs HS graduate | 2.23 | 1.20-4.16 | |
| Some college vs HS graduate | 1.13 | 0.72-1.76 | |
| College graduate vs HS graduate | 1.19 | 0.77-1.83 | |
| Regular Multivitamin Use | NS | ||
| Yes vs No | 1.32 | 0.97-1.80 | |
| Impact of Events Scale† | 0.021 | ||
| 26+ vs 0-25 | 0.49 | 0.27-0.90 | |
|
| |||
| CRC screening knowledge | 1.12 | 1.01-1.24 | 0.035 |
| Genetics & diet knowledge | 1.03 | 0.95-1.13 | NS |
p-value from Wald Chi Square test
Missing values excluded from multivariable analyses, n=4 (n=1 for Marital status, n=1 for Education, and n=2 for Impact of Events Scale)
Abbreviations: CI = Confidence interval for odds ratio; CRC = Colorectal Cancer; NS = Not significant (P>0.05)
Surprisingly, among GERA recipients, “average” risk participants had higher screening rates than “elevated” risk participants, although the odds ratio estimate of 0.60 (95% CI 0.33 - 1.07) was not significant (Table 4). Of note, 85% (437/514) of GERA participants were considered in this analysis. The remaining 77 were considered to be unknown risk as described (Figure 1). Because no difference in screening uptake was seen between risk groups, we investigated if any baseline factors may have confounded our results among participants randomized to the GERA arm (Table 5). Black participants were significantly more likely than white participants to be at “elevated” risk presumably because of well described allele distribution frequency differences across race26 (OR=5.92, 95% CI 3.26 - 10.74). Also expected was the finding that regular MVI was significantly inversely associated with “elevated” risk status (OR=0.24, 95% CI 0.13 - 0.44), as regular MVI users may have higher serum folate levels. Other significant differences included generally lower education levels in the “elevated” risk group (college graduate vs. high school graduate OR=0.36, 95% CI 0.18 - 0.71) as well as lesser baseline knowledge about CRC screening (OR = 0.78, 95% CI 0.68 - 0.88) and genetics and diet knowledge (OR=0.80, 95% CI 0.70 -0.90). To adjust for potential confounding, we fit a multivariable logistic model including all covariates to assess the effect of elevated vs. average risk designation on CRC screening. The adjusted odds ratio estimate was 0.75 (95% CI 0.39 - 1.42). The results of this analysis did not alter the conclusion that GERA risk classification had no significant effect on screening behavior (Supplemental Table 3).
Table 4. CRC screening at six months by GERA risk group.
| Within GERA arm * | ||||
|---|---|---|---|---|
|
|
||||
| Average risk | Elevated risk | Elevated risk vs. Average risk | ||
| N (%) | N (%) | N (%) | Odds Ratio (95% CI) | |
|
|
|
|||
| CRC screening status | ||||
| Had screening | 159 (34.0) | 141 (38.1) | 18 (26.9) | 0.60 (0.33-1.07) |
| No screening | 278 (66.0) | 229 (61.9) | 49 (73.1) | |
GERA arm participants with unknown GERA risk were excluded, n=77
Screening rates did by not differ within GERA arm (P>0.05, Wald Chi-square test).
Abbreviations: CI = Confidence interval; CRC = Colorectal Cancer;
Table 5. Baseline characteristics by GERA risk group.
| GERA arm* | Within GERA arm | ||||||||
|---|---|---|---|---|---|---|---|---|---|
|
| |||||||||
| Average Risk | Elevated Risk | Elevated risk vs Average Risk | |||||||
|
|
|
||||||||
| N | % | N | % | N | % | Odds Ratio | 95% CI | P value** | |
| Age Category | NS | ||||||||
| 50-59 years | 306 | 70.1 | 256 | 69.2 | 50 | 74.7 | 1.00 | Reference | |
| 60-69 years | 95 | 21.7 | 81 | 21.9 | 14 | 20.9 | 0.89 | 0.47-1.68 | |
| 70-79 years | 36 | 8.2 | 33 | 8.9 | 3 | 4.5 | 0.47 | 0.14-1.58 | |
| Gender | NS | ||||||||
| Female | 257 | 58.8 | 218 | 58.9 | 39 | 58.2 | 1.00 | Reference | |
| Male | 180 | 41.2 | 152 | 41.1 | 28 | 41.8 | 1.03 | 0.61-1.74 | |
| Race | <0.001 | ||||||||
| White | 258 | 59.0 | 241 | 65.1 | 17 | 25.4 | 1.00 | Reference | |
| Black or African American | 163 | 37.3 | 115 | 31.1 | 48 | 71.6 | 5.92 | 3.26-10.74 | |
| Other | 16 | 3.7 | 14 | 3.8 | 2 | 3.0 | 2.03 | 0.43-9.65 | |
| Marital status† | NS | ||||||||
| Married | 213 | 48.9 | 182 | 49.3 | 31 | 46.3 | 1.00 | Reference | |
| Single/Divorced/Widowed | 223 | 51.1 | 187 | 50.7 | 36 | 53.7 | 1.13 | 0.67-1.91 | |
| Education | 0.006 | ||||||||
| Less than high school | 27 | 6.2 | 19 | 5.1 | 8 | 11.9 | 1.44 | 0.55-3.72 | |
| High school graduate | 97 | 22.2 | 75 | 20.3 | 22 | 32.8 | 1.00 | Reference | |
| Some college | 133 | 30.4 | 113 | 30.5 | 20 | 29.9 | 0.60 | 0.31-1.18 | |
| College graduate | 180 | 41.2 | 163 | 44.1 | 17 | 25.4 | 0.36 | 0.18-0.71 | |
| Regular MV use | <0.001 | ||||||||
| No | 219 | 50.1 | 167 | 45.1 | 52 | 77.6 | 1.00 | Reference | |
| Yes | 218 | 49.9 | 203 | 54.9 | 15 | 22.4 | 0.24 | 0.13-0.44 | |
| Impact of Events Scale† | NS | ||||||||
| 0-25 | 392 | 89.9 | 335 | 90.8 | 57 | 85.1 | 1.00 | Reference | |
| 26+ | 44 | 10.1 | 34 | 9.2 | 10 | 14.9 | 1.73 | 0.81-3.69 | |
|
| |||||||||
| Knowledge Questionnaires | Median | (Q1, Q3) | Median | (Q1, Q3) | Median | (Q1, Q3) | Odds Ratio | 95% CI | P value** |
| CRC screening knowledge | 7 | (5,8) | 7 | (6,8) | 6 | (5,7) | 0.78 | 0.68-0.88 | <0.001 |
| Genetics & diet knowledge | 5 | (4,7) | 5 | (4,7) | 4 | (2,6) | 0.80 | 0.70-0.90 | <0.001 |
Missing value excluded, n=1 for Marital status, n=1 for IES
n=77 Participants with unknown GERA risk excluded
p-value from Wald Chi Square test
Abbreviations: CI = Confidence interval for odds ratio; CRC = Colorectal Cancer; NS = Not significant (P>0.05); Q1, Q3 = First and third quartile value (25th and 75th percentiles);
We performed additional analyses to control for potential confounders in the association between GERA risk group and CRC screening using propensity score based methods (Appendix 1). In a propensity score matched analysis with 58 pairs, the adjusted odds ratio estimate was 0.71 (95% CI 0.34-1.48) (Supplemental Table 4).
Because we had excluded GERA “risk unknown” participants (n=77) from the comparison of elevated and average risk, we performed sensitivity analyses (Supplemental Table 5). None of the analyses indicated that exclusion of unknown risk participants affected the conclusion that screening rates in the GERA elevated group were not significantly higher.
Finally, because of the inverse relationship between higher baseline IES score and subsequent screening (Supplemental Table 2), we examined the specific association between concern about genetic testing results (measured via the MICRA questionnaire) and screening uptake. Two hundred sixty four of the 283 GERA arm participants who indicated receiving risk results completed the MICRA questionnaire. Total MICRA scores as well as all subscales were significantly higher for those designated at “elevated” versus “average” risk (Table 6, top panel). However, MICRA scores were not significantly associated with screening uptake (bottom panel).
Table 6. MICRA score at 3 week post-baseline by GERA risk group and CRC screening within 6 months.
| MICRA score (3 weeks)† by GERA risk group | |||||
|---|---|---|---|---|---|
|
| |||||
| Median (Q1,Q3) | Elevated vs. Average Risk | ||||
|
| |||||
| Average risk, N=224 | Elevated risk, N=40 | Odds Ratio | 95% CI | P value* | |
| MICRA total score | 12 (6.5, 19) | 20.5 (12.5,28.5) | 1.11 | 1.07-1.16 | <0.001 |
| MICRA subscale | |||||
| Distress | 0 (0,0) | 1 (0,5.5) | 1.28 | 1.15-1.42 | <0.001 |
| Uncertainty | 0 (0,3) | 2 (0,10) | 1.11 | 1.05-1.17 | <0.001 |
| Positive Experiences | 6 (2,12) | 9.5 (6,16) | 1.06 | 1.01-1.11 | 0.028 |
|
| |||||
| MICRA score (3 weeks) by CRC screening within six months | |||||
|
| |||||
| Median (Q1, Q3) | CRC Screening vs None | ||||
|
| |||||
| No CRC Screening, N=167 | Had CRC Screening, N=97 | Odds Ratio | 95% CI | P value* | |
|
| |||||
| MICRA total score | 14 (8,21) | 13 (6,10) | 0.98 | 0.95-1.00 | NS |
| Micra subscale | |||||
| Distress | 0 (0,1) | 0 (0,0) | 0.98 | 0.91-1.07 | NS |
| Uncertainty | 0 (0,3) | 0 (0,4) | 0.98 | 0.93-1.03 | NS |
| Positive Experiences | 8 (3,14) | 6 (2,12) | 0.98 | 0.94-1.02 | NS |
Sample size includes 264 GERA risk patients who completed the initial MICRA survey at 3 weeks (survey is after GERA test results were given).
p-value from Wald Chi-square test
Abbreviations: CI = Confidence interval for odds ratio; CRC = Colorectal Cancer; NS = Not significant (P>0.05); Q1, Q3 = First and third quartile value (25th and 75th percentiles)
Discussion
This study examined the effect of combined gene (MTHFR genotype) and environment (serum folate level) assessment, or GERA, on subsequent CRC screening in average risk individuals not currently adherent with screening. We found no significant difference in screening uptake at 6 months between those randomized to GERA or usual care arms, or between GERA participants identified at “elevated” versus “average” risk.
A dominant claim in the media is that enhanced knowledge of individual genetic make-up will promote a healthier lifestyle27,28 and several companies market gene testing services directly to patients. Personalized, gene-based risk assessment could provide an attractive potential motivator to improve an array of health behaviors ranging from cancer screening to smoking cessation.29 However, most relevant research about the impact of genetic testing on health behavior decision-making has focused on persons with high risk, single gene mutations. If personalized, genetic information proves to be an effective motivator, its greatest benefit will come when testing expands to populations at average risk for common disease.
Previous studies in this broader arena have not emphasized specific disease risk, instead they tend to concentrate on broader “lifestyle” issues of diet, exercise and smoking cessation.4 Those studies evaluating genetic feedback on cancer screening have had small sample sizes and focus exclusively on populations at substantially elevated risk for diseases like hereditary breast and ovarian cancer (BRCA1/2)29 or CRC (FAP and HNPCC).12 Overall, there is little evidence that moderately predictive, personalized information effectively promotes positive behavior change in average risk settings.4,30 Our findings that the GERA intervention failed to improve CRC screening participation are consistent with the small published literature. For example, Bloss et al. reported there was no significant change in screening test utilization in an uncontrolled, convenience sample of persons who purchased an online genomics test.31
There is concern that moderately predictive genetic testing may have a negative psychological impact, either through increased alarm about newly identified risk, or the fear about misinterpretation of imprecise information.13 Although we found some increase in distress related to GERA receipt, this effect did not appear related to screening rates (Table 6). In addition, a lack of familiarity with genomics on the part of patients (and providers) has been cited as a concern limiting the impact of genetic testing.28 We did see a consistent increase over time in knowledge about CRC screening, and the role of genetic risk (data not shown), suggesting the educational component of the intervention was successful. However, neither baseline knowledge, nor increasing post intervention knowledge was associated with screening uptake.
In a recent report by the National Science Foundation, the most important issue identified for social science researchers was how to motivate people to change their behavior.32 The current study addresses an important question: does an intervention built on participant specific gene-environment risk assessment improve utilization of a clinically effective risk reduction strategy, CRC screening? As utilized in this study, personalized GERA is ineffective in this role. Potential explanations worthy of future study include the possibility that individuals with more behavioral risk factors may focus on genetic, not behavioral explanations as causes for potential illness.33 As a result, they are unable or unwilling to alter behavior to reduce their risk. Similarly, some have speculated that genetic risk is perceived as immutable, and hence those persons who carry such a predisposition believe their fate is sealed.29 Provision of FIT cards was intended to minimize screening barriers for motivated individuals. However, additional efforts to facilitate screening utilization may have produced different results. Alternatively, despite consistent reminders about the value of accepted screening methods, some participants may have believed that GERA testing itself was a form of screening or that folate supplementation alone was adequate protection against CRC. Finally, perhaps the most likely explanation is that the perceived risk associated with moderately predictive testing is simply not great enough to motivate behavior change, particularly in persons not oriented towards healthy behaviors like cancer screening.27
This study had several limitations. At present, for common diseases with a polygenic basis, nearly any gene or gene plus environment assessment can identify only modest predisposition to disease. Such testing is not intended to be deterministic.28 We studied only one combination of gene and environmental interaction. Other combinations or other forms of personalized information could be more effective motivational tools. We also studied only one risk reduction behavior, CRC screening. A wide range of behavioral interventions have been tried with uneven effect to increase such screening. Hence, generalizability to other behavioral targets remains an open question. Finally, a better understanding of the impact of the perceived risk associated with GERA results might explain why our intervention was ineffective.
In summary, this large, randomized trial found no effect on CRC screening rates in an average risk population exposed to personalized genetic and environmental risk information. Further study would be required to assess whether other gene-enviromental risk information and different means of presenting individualized results for common diseases like CRC will spur more healthy behaviors to reduce risk. Genetic and molecular testing to predict response to specific therapeutic options has an increasing role in healthcare delivery, however the potential for similar testing to motivate behavioral change is less clear.
Supplementary Material
Acknowledgments
The authors gratefully acknowledge the invaluable contributions of James Cocroft, Anett Petrich, Heidi Swan, Thomas Wolf and members of the Fox Chase Cancer Center Genomics Facility.
Supported by NIH grant R01CA112230 (DSW) and P30 CA006927
Appendix: Propensity score methods for covariate adjustment
We used propensity score analysis to adjust for differences in patient characteristics between the GERA elevated and average risk groups. All a priori selected confounders were included in a logistic regression model to estimate a propensity score (Rosenbaum and Rubin, 1983) for each participant, defined as the probability of being in the GERA elevated risk group given the covariates. Participants were excluded from the analysis due to missing values for covariates (n=2) or insufficient overlap of propensity scores between the two risk groups (n=25). We conducted a 1:1 matched analysis, where GERA elevated risk participants were paired to GERA average risk participants using the propensity score. Matches were chosen using a SAS macro (Parsons, 2004), and were considered successful if the difference in propensity scores was less than 0.02. Matches were found for 58 of 67 elevated risk participants. Fisher's Exact and Wilcoxon rank-sum tests were used to test for balanced covariate distributions across paired samples (Supplemental Table 6). Conditional logistic regression was then used to estimate the adjusted odds ratio and 95% confidence interval for the impact of GERA risk on CRC screening.
Rosenbaum, Paul R., and Donald B. Rubin. “The central role of the propensity score in observational studies for causal effects.” Biometrika 70.1 (1983): 41-55.
Parsons LS, “Performing a 1:N Case-Control Match on Propensity Score”. Proceedings of the Twenty-ninth annual SAS users group international conference, Cary, NC: SAS Institute Inc, 2004.
Footnotes
Clinicaltrials.gov Registration NCT0087360
References
- 1.Levin B, Lieberman DA, McFarland B, et al. Screening and surveillance for the early detection of colorectal cancer and adenomatous polyps, 2008: a joint guideline from the American Cancer Society, the US Multi-Society Task Force on Colorectal Cancer, and the American College of Radiology. Gastroenterology. 2008;134:1570–95. doi: 10.1053/j.gastro.2008.02.002. [DOI] [PubMed] [Google Scholar]
- 2.Ghazarian AA, Simonds NI, Bennett K, et al. A review of NCI's extramural grant portfolio: identifying opportunities for future research in genes and environment in cancer. Cancer Epidemiology, Biomarkers & Prevention. 2013;22:501–7. doi: 10.1158/1055-9965.EPI-13-0156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Green ED, Guyer MS. Charting a course for genomic medicine from base pairs to bedside. Nature. 2011;470:204–13. doi: 10.1038/nature09764. [DOI] [PubMed] [Google Scholar]
- 4.McBride CM, Bryan AD, Bray MS, Swan GE, Green ED. Health behavior change: can genomics improve behavioral adherence? American Journal of Public Health. 2012;102:401–5. doi: 10.2105/AJPH.2011.300513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kim YI. Folate and cancer prevention: a new medical application of folate beyond hyperhomocysteinemia and neural tube defects. Nutrition Reviews. 1999;57:314–21. doi: 10.1111/j.1753-4887.1999.tb06905.x. [DOI] [PubMed] [Google Scholar]
- 6.Ma J, Stampfer MJ, Giovannucci E, et al. Methylenetetrahydrofolate reductase polymorphism, dietary interactions, and risk of colorectal cancer. Cancer Research. 1997;57:1098–102. [PubMed] [Google Scholar]
- 7.Ryan BM, Weir DG. Relevance of folate metabolism in the pathogenesis of colorectal cancer. The Journal of Laboratory and Clinical Medicine. 2001;138:164–76. doi: 10.1067/mlc.2001.117161. [DOI] [PubMed] [Google Scholar]
- 8.Keku T, Millikan R, Worley K, et al. 5,10-Methylenetetrahydrofolate reductase codon 677 and 1298 polymorphisms and colon cancer in African Americans and whites. Cancer Epidemiology, Biomarkers & Prevention. 2002;11:1611–21. [PubMed] [Google Scholar]
- 9.Giovannucci E, Stampfer MJ, Colditz GA, et al. Multivitamin use, folate, and colon cancer in women in the Nurses' Health Study. Annals of Internal Medicine. 1998;129:517–24. doi: 10.7326/0003-4819-129-7-199810010-00002. [DOI] [PubMed] [Google Scholar]
- 10.Scheuner MT, Sieverding P, Shekelle PG. Delivery of genomic medicine for common chronic adult diseases: a systematic review. JAMA. 2008;299:1320–34. doi: 10.1001/jama.299.11.1320. [DOI] [PubMed] [Google Scholar]
- 11.Heshka JT, Palleschi C, Howley H, Wilson B, Wells PS. A systematic review of perceived risks, psychological and behavioral impacts of genetic testing. Genetics in Medicine. 2008;10:19–32. doi: 10.1097/GIM.0b013e31815f524f. [DOI] [PubMed] [Google Scholar]
- 12.McBride CM, Koehly LM, Sanderson SC, Kaphingst KA. The behavioral response to personalized genetic information: will genetic risk profiles motivate individuals and families to choose more healthful behaviors? Annual Review of Public Health. 2010;31:89–103. doi: 10.1146/annurev.publhealth.012809.103532. [DOI] [PubMed] [Google Scholar]
- 13.Ransohoff DF, Khoury MJ. Personal genomics: information can be harmful. European Journal of Clinical Investigation. 2010;40:64–8. doi: 10.1111/j.1365-2362.2009.02232.x. [DOI] [PubMed] [Google Scholar]
- 14.Bloss CS, Wineinger NE, Darst BF, Schork NJ, Topol EJ. Impact of direct-to-consumer genomic testing at long term follow-up. Journal of Medical Genetics. 2013;50:393–400. doi: 10.1136/jmedgenet-2012-101207. [DOI] [PubMed] [Google Scholar]
- 15.Jemal A, Simard EP, Dorell C, et al. Annual Report to the Nation on the Status of Cancer, 1975-2009, featuring the burden and trends in human papillomavirus(HPV)-associated cancers and HPV vaccination coverage levels. Journal of the National Cancer Institute. 2013;105:175–201. doi: 10.1093/jnci/djs491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Qaseem A, Denberg TD, Hopkins RH, Jr, et al. Screening for colorectal cancer: a guidance statement from the American College of Physicians. Annals of Internal Medicine. 2012;156:378–86. doi: 10.7326/0003-4819-156-5-201203060-00010. [DOI] [PubMed] [Google Scholar]
- 17.Myers RE, Manne SL, Wilfond B, et al. A randomized trial of genetic and environmental risk assessment (GERA) for colorectal cancer risk in primary care: trial design and baseline findings. Contemporary Clinical Trials. 2011;32:25–31. doi: 10.1016/j.cct.2010.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Horowitz M, Wilner N, Alvarez W. Impact of event scale: a measure of subjective stress. Psychosom Med. 1979;41:209–18. doi: 10.1097/00006842-197905000-00004. [DOI] [PubMed] [Google Scholar]
- 19.Wilfond BS, Fost N. The cystic fibrosis gene: medical and social implications for heterozygote detection. JAMA. 1990;263:2777–83. [PubMed] [Google Scholar]
- 20.Konings EJ, Goldbohm RA, Brants HA, Saris WH, van den Brandt PA. Intake of dietary folate vitamers and risk of colorectal carcinoma: results from The Netherlands Cohort Study. Cancer. 2002;95:1421–33. doi: 10.1002/cncr.10866. [DOI] [PubMed] [Google Scholar]
- 21.Houlston RS, Tomlinson IP. Polymorphisms and colorectal tumor risk. Gastroenterology. 2001;121:282–301. doi: 10.1053/gast.2001.26265. [DOI] [PubMed] [Google Scholar]
- 22.Pufulete M, Al-Ghnaniem R, Leather AJ, et al. Folate status, genomic DNA hypomethylation, and risk of colorectal adenoma and cancer: a case control study. Gastroenterology. 2003;124:1240–8. doi: 10.1016/s0016-5085(03)00279-8. [DOI] [PubMed] [Google Scholar]
- 23.Ulvik A, Evensen ET, Lien EA, et al. Smoking, folate and methylenetetrahydrofolate reductase status as interactive determinants of adenomatous and hyperplastic polyps of colorectum. American Journal of Medical Genetics. 2001;101:246–54. doi: 10.1002/ajmg.1370. [DOI] [PubMed] [Google Scholar]
- 24.McDowell MA, Lacher DA, Pfeiffer CM, et al. Blood Folate Levels: The Latest NHANES Results. Vol. 6. Hyattsville, MD: National Center for Health Statistics; May, 2008. 2008. [PubMed] [Google Scholar]
- 25.Cella D, Hughes C, Peterman A, et al. A brief assessment of concerns associated with genetic testing for cancer: the Multidimensional Impact of Cancer Risk Assessment (MICRA) questionnaire. Health Psychology. 2002;21:564–72. [PubMed] [Google Scholar]
- 26.Botto LD, Yang Q. 5,10-Methylenetetrahydrofolate reductase gene variants and congenital anomalies: a HuGE review. American Journal of Epidemiology. 2000;151:862–77. doi: 10.1093/oxfordjournals.aje.a010290. [DOI] [PubMed] [Google Scholar]
- 27.Smith RA, Duffy SW, Tabar L. Breast cancer screening: the evolving evidence. Oncology. 2012;26:471–5. 9–81, 85–6. [PubMed] [Google Scholar]
- 28.Evans JP, Meslin EM, Marteau TM, Caulfield T. Genomics. Deflating the genomic bubble. Science. 2011;331:861–2. doi: 10.1126/science.1198039. [DOI] [PubMed] [Google Scholar]
- 29.Marteau TM, French DP, Griffin SJ, et al. Effects of communicating DNA-based disease risk estimates on risk-reducing behaviours. The Cochrane database of systematic reviews. 2010:CD007275. doi: 10.1002/14651858.CD007275.pub2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Reid RJ, McBride CM, Alford SH, et al. Association between health-service use and multiplex genetic testing. Genetics in Medicine. 2012;14:852–9. doi: 10.1038/gim.2012.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bloss CS, Schork NJ, Topol EJ. Effect of direct-to-consumer genomewide profiling to assess disease risk. The New England Journal of Medicine. 2011;364:524–34. doi: 10.1056/NEJMoa1011893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Giles J. Social science lines up its biggest challenges. Nature. 2011;470:18–9. doi: 10.1038/470018a. [DOI] [PubMed] [Google Scholar]
- 33.O'Neill SC, McBride CM, Alford SH, Kaphingst KA. Preferences for genetic and behavioral health information: the impact of risk factors and disease attributions. Annals of Behavioral Medicine. 2010;40:127–37. doi: 10.1007/s12160-010-9197-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.