Abstract
Optogenetic systems enable precise spatial and temporal control of cell behavior. We engineered a light-activated CRISPR/Cas9 effector (LACE) system that induces transcription of endogenous genes in the presence of blue light. This was accomplished by fusing the light-inducible heterodimerizing proteins CRY2 and CIB1 to a transactivation domain and the catalytically inactive dCas9, respectively. The versatile LACE system can be easily directed to new DNA sequences for the dynamic regulation of endogenous genes.
Genome engineering technologies have enabled activation or repression of endogenous genes in mammalian cells using synthetic transcription factors that can be targeted to almost any DNA sequence1–12. Most recently, the CRISPR/Cas9 system of Streptococcus pyogenes was reengineered to function in mammalian cells as a transcription factor4–12. Unlike previous systems that utilize programmable zinc-finger proteins (ZFPs)1 or transcription activator-like effectors (TALEs)2–4 to target specific sites within the genome, the CRISPR/Cas9 system is directed to a DNA sequence by a single guide RNA (gRNA).13 Thus targeting the CRISPR/Cas9 system to new DNA sequences only requires alteration of the short gRNA sequence and does not involve reengineering of a protein DNA-binding domain. The simplicity of this system has enabled its rapid development as a tool for many diverse applications in biology.
Several optogenetic transcriptional control systems have been developed for dynamically regulating gene expression with light.4, 14–20 Most of these systems use light-inducible heterodimerizing proteins from plants.21 For instance, fusion of one heterodimerizing protein to a ZFP18 or TALE4 and fusion of its binding partner to a transcriptional activation domain, such as VP64, enables light-dependent recruitment of the activation domain to the DNA sequence targeted by the ZFP or TALE. These systems enable control over expression of any gene in a reversible, tunable, and spatially defined manner. However, the reengineering of the ZFP or TALE DNA-binding domain to target new sequences can be laborious and require specialized expertise. This is particularly a concern for gene activation with systems that must target several sequences in a gene promoter to synergistically achieve robust activation.22, 23
To address these limitations, we adapted the CRISPR/Cas9 activator system for optogenetic control. First, as was done previously with TALEs,4 we fused the light-inducible heterodimerizering proteins CRY2 and CIB1 from Arabidopsis thaliana to the VP64 transactivation domain and either the N- or C- terminus of dCas9, the catalytically inactive form of Cas9 (D10A, H840A)13 (Supplementary Results, Supplementary Fig. 1). We used the N-terminal fragment of CIB1 (CIBN) and either the full-length CRY2 (CRY2FL) or a truncated CRY2 (CRY2PHR) that has higher levels of activity.16 The plasmids encoding one of the two VP64 fusions (CRY2FL-VP64 or CRY2PHR-VP64) and one of the two dCas9 fusions (dCas9-CIBN or CIBN-dCas9) were co-transfected with four gRNAs targeting the human IL1RN promoter7 into HEK293T cells. Four gRNAs were used together based on the previous observation that multiple gRNAs are necessary to robustly activate gene expression with constitutive dCas9-VP64 transcription factors.6–12 We expected that the gRNAs would direct the binding of dCas9 to the IL1RN promoter in both the light and dark, but VP64 would co-localize with dCas9 via CRY2-CIBN interactions and induce transcription only in the presence of blue light (Fig. 1a, Supplementary Fig. 1). All of the fusion proteins were expressed well as determined by western blot for the FLAG epitope tag (Supplementary Figs. 2–3). Beginning at 24 hours post-transfection, transfected cells were incubated in the dark or under blue light for three days and expression of the IL1RN gene was determined by qRT-PCR. The combination of CRY2FL with dCas9-CIBN or CIBN-dCas9 showed detectable levels of gene activation above negligible background levels of IL1RN expression in the dark (p<0.05) (Fig. 1b). However, the levels of activation were orders of magnitude lower than the constitutively active dCas9-VP64 fusion (Fig. 1b),7 which is typical of systems in which a direct fusion protein is divided into inducible dimerizing fragments.
Figure 1.
Light-inducible, RNA-guided activation of endogenous human genes by the LACE system. (a) When expressed by cells, CIBN-dCas9-CIBN localizes to the DNA sequence targeted by the gRNA. In the presence of blue light CRY2 undergoes a conformational change that enables heterodimerization with CIBN, which causes translocation of CRY2FL-VP64 to the targeted DNA sequence and transcriptional activation of the downstream gene. (b) Activation of IL1RN using four targeting gRNAs, either CIBN-dCas9, dCas9-CIBN, or CIBN-dCas9-CIBN, and either CRY2FL-VP64 or CRY2PHR-VP64 in HEK293T cells. Red bars = dark-incubated cells; blue bars = cells illuminated with pulsing blue light (1s pulses at 0.067 Hz). Conditions not marked by the same letter are significantly different (p<0.05) as determined by ANOVA and Tukey’s test. NT = not tested. Data shows combined replicates from two independent experiments (n=4 ± s.d.). (c) Activation of multiple endogenous gene targets was achieved using different groups of gRNAs targeted to HBG1/2, IL1RN, or ASCL1 in HEK293T cells. Activation levels in illuminated cells that contained the LACE system were statistically similar to cells that expressed dCas9-VP64 and the same gRNAs when HBG1/2 or IL1RN were targeted. Conditions not marked by the same letter are significantly different (p<0.01) as determined by global ANOVA and Tukey’s test. NT = not tested. Data shows combined replicates from two independent experiments (n=4 ± s.d.).
In order to improve the robustness of the system, we fused CIBN to both the N- and C- terminus of dCas9 (Fig. 1a, Supplementary Fig. 1). This was motivated by our previous observation that increasing the number of VP64 domains localized to the target promoter sequence synergistically enhances gene activation.22, 23 Surprisingly, the fusion of CIBN to both dCas9 termini led to increases in IL1RN gene activation in response to blue light that were 10- to 100-fold greater than generated by dCas9 fusions to a single CIBN domain (p<0.0001) (Fig. 1b). Importantly, levels of background IL1RN expression in the dark were similar to untransfected controls (p=0.99). Furthermore, light-induced gene activation by CIBN-dCas9-CIBN and CRY2FL-VP64 was equal to gene activation by the constitutive dCas9-VP64 fusion, demonstrating efficient light-induced recruitment of VP64 to the target promoter.
To demonstrate the versatility of the system, we activated multiple endogenous gene targets by delivering the LACE system and a group of four gRNAs that target the human IL1RN, HBG1/2, or ASCL1 promoters7 to HEK293T cells (Fig. 1c). Illuminated cells in which IL1RN or HBG1/2 was targeted demonstrated significantly greater mRNA levels in the light compared to the dark (p<0.0001 and 0.005, respectively) and equivalent activation levels to dCas9-VP64 (p=0.17 and 0.35, respectively). Significant light-dependent activation was also observed when the ASCL1 locus was targeted with the LACE system (p<0.0001). However, in this case mRNA levels were not activated to the same extent as cells that received dCas9-VP64 and the same four ASCL1-targeting gRNAs (Fig. 1c). In all instances, transfected cells incubated in the dark maintained levels of the targeted gene that did not significantly differ from mock-transfected cells (p = 0.11, 0.34, and 0.07 for IL1RN, HBG1/2, and ASCL1, respectively), demonstrating negligible background in the “off” state in unstimulated conditions for four days post-transfection.
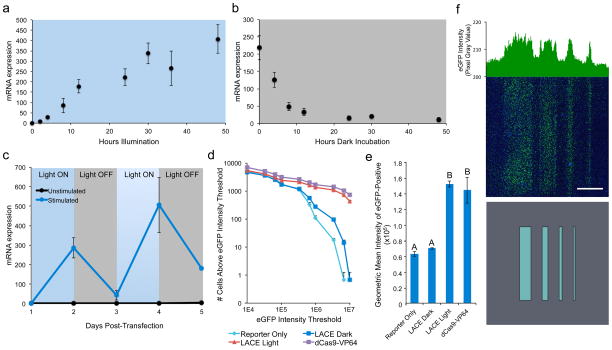
One benefit of the LACE system is that gene activation is reversible through simple removal of illumination (Fig. 2a–c). This contrasts with systems regulated by small molecules, such as the tetracycline-inducible system, that require physical removal of the inducer. HEK293T cells transiently transfected with the LACE system and four IL1RN-targeting gRNAs exhibited a significant 11-fold increase in IL1RN mRNA levels within only 2 hours and a maximum increase of 400-fold within ~30 hours (Fig. 2a). To demonstrate the reversibility of the LACE system, we incubated cells in the light for 24 hours followed by 48 hours in the dark (Fig. 2b). IL1RN mRNA levels were measured at various time points post-illumination and compared to original baseline levels. Within the first 4 hours of dark incubation, the IL1RN mRNA level decreased from ~220-fold to 125-fold over pre-illuminated cells, and within 24 hours of dark incubation the mRNA level decreased ~93% to 15-fold.
Figure 2.
Dynamic spatial and temporal transcriptional control using the LACE system. (a) HEK293T cells expressing the LACE system and four IL1RN-targeted gRNAs were illuminated with pulsing blue light for 52 hours (two independent experiments, n=4 ± s.d.). (b) HEK293T cells treated as in (a) were illuminated with pulsing blue light for 24 hours and then incubated in the dark starting at t=0 h (two independent experiments, n=6 ± s.d.). (c) HEK293T cells transfected with the LACE system and four IL1RN-targeted gRNAs were cycled between 24 hours of illumination and 24 hours of dark incubation for 4 days starting the day after transfection (one of two representative experiments, n=2 ± s.d.). (d) HEK293T cells were co-transfected with the LACE system or dCas9-VP64, an eGFP reporter, and a gRNA that targeted eight identical DNA sites upstream of eGFP and a minimal CMV promoter. Cells that received the LACE system were either illuminated or incubated in the dark for 24 hours, and eGFP expression was then quantified by flow cytometry. The number of eGFP-positive cells is presented as a function of the eGFP intensity threshold (one of two representative experiments, n=3 ± s.d.). (e) The geometric mean of fluorescence intensity of eGFP-positive cells in samples from (d) is shown. Letters indicate significant differences (p<0.05) as determined by global ANOVA and Tukey’s test. (f) Modified cells illuminated through a photomask of arbitrary pattern resulted in a corresponding pattern of eGFP-expressing cells. Top pattern was created using a photomask with rectangular slits of width 2, 1, 0.5, and 0.3 mm. Scale bar = 2 mm.
The LACE system also enables dynamic control of the expression of endogenous genes (Fig. 2c). Illumination of cells containing the LACE system targeted to IL1RN exhibited ~300-fold increase in mRNA levels compared to mock-transfected cells after 24 hours. Removal of illumination decreased mRNA levels to 44-fold over control after 24 hours of dark incubation. Then, re-illumination increased mRNA levels to ~500-fold over control within 24 hours. This activated level was again reduced by removing illumination, resulting in a decrease to ~180-fold after 24 hours of incubation in the dark. Non-illuminated cells maintained mRNA levels that did not significantly differ from mock-transfected cells until the fourth (p=0.02) and fifth (p=0.03) days post-transfection, at which time mRNA levels were only 3- to 4-fold higher than mock-transfected cells.
Spatially controlled activation of gene expression was achieved in cells co-transfected with the LACE system, a reporter vector containing eight repeats of a gRNA target sequence upstream of a minimal CMV promoter and the eGFP gene, and the expression plasmid for the corresponding gRNA (Fig. 2d–f, Supplementary Fig. 4). Cells transfected with LACE and incubated in the dark did not show a significant difference in eGFP levels compared to control cells transfected with empty plasmid (Fig. 2e). Cells containing the LACE system and gRNA exhibited significantly brighter eGFP fluorescence intensity when illuminated compared to when incubated in the dark (Supplementary Fig. 4). Activation of the eGFP reporter in cells transfected with the gRNA and LACE constructs, the gRNA and dCas9-VP64 expression plasmid, or an empty plasmid as a negative control was quantified by flow cytometry after 24 hours of illumination or incubation in the dark. The magnitude of the difference between cells in the light and in the dark was dependent on the threshold applied to the eGFP fluorescence intensity (Fig. 2d). The geometric mean intensities of eGFP-positive cells were statistically equivalent in illuminated cells containing the LACE system and in cells containing dCas9-VP64 (p=0.47). These two conditions were also statistically higher than both dark-incubated cells containing the LACE system and cells that only received the reporter construct (p<0.05) (Fig. 2e). When cells containing the LACE system, eGFP reporter, and gRNA were illuminated through a photomask containing an arbitrary pattern with features 0.3 to 2 mm wide, we observed a corresponding pattern of eGFP-positive cells (Fig. 2f).
The LACE system provides a straightforward and robust optogenetic method to regulate the expression of endogenous genes using the CRISPR/dCas9 system with spatial and temporal control. When co-transfected with gRNAs into mammalian cells that are stimulated with blue light, LACE produces a high level of transcriptional activation that is, in some cases, comparable to those observed with dCas9-VP64. In addition, at endogenous loci, there is minimal and often insignificant background activity in dark-incubated cells that co-express the LACE system and gRNAs. Compared to previous systems based on TALEs4 and ZFPs18, the optogenetic CRISPR/dCas9 system affords greater flexibility and simplicity in targeting any endogenous gene. Thus LACE expands optogenetic control of gene expression to nearly any regulatory element in the genome, in addition to the regulation of transgene constructs. LACE also enables the potential for multiplex gene regulation with combinations of gRNAs.9, 12 We believe this tool will prove useful for studies in basic science, synthetic biology, and tissue engineering that require precise spatial and/or temporal control of cellular behavior.24 For example, LACE may be used to study the stability of feedback loops or epigenetic marks induced by dynamic patterns of gene activation, cell-cell interactions in specific geometric patterns, or morphogenesis of multiple tissues types.25
Online Methods
Plasmid Construction
CRY2FL-VP64, CIBN-dCas9, dCas9-CIBN, and CIBN-dCas9-CIBN were cloned using Gibson Assembly. CRY2FL was PCR-amplified and assembled into a pcDNA3.1 vector containing dCas9-VP647 that was digested with SacII and BstEII to remove the dCas9 sequence. dCas9-CIBN was cloned by digesting pcDNA3.1-dCas9-VP64 with XbaI and AscI and using Gibson Assembly to insert the PCR-amplified CIBN gene to create pcDNA3.1-dCas9-CIBN. CIBN-dCas9-CIBN was generated by digesting pcDNA3.1-dCas9-CIBN with ClaI and inserting the PCR-amplified CIBN gene to create pcDNA3.1-CIBN-dCas9-CIBN. pcDNA3.1-dCas9-CIBN was generated by digesting pcDNA3.1-CIBN-dCas9-CIBN with AflII and AscI and using Gibson Assembly to insert the fragment 5′-GAAACGCAAAGTTGGGCGCGCCtaaCTTAAGTTTAAACCGCTGATCA-3′, which was created by annealing complementary oligonucleotides (IDTDNA) and contains a stop sequence (lowercase letters) flanked by sequences that are homologous to the digested vector backbone (uppercase letters). CRY2(PHR)-VP64 was acquired from Addgene4. CRY2FL and CIBN were provided by Chandra Tucker.16 The eGFP reporter construct was described previously18 (Addgene ID 60718) and contains 8 repeats of the sequence 5′-AAAGGTCGAGAAACTGCAAA-3′ upstream of a minimal CMV promoter and the eGFP gene. The promoter and eGFP transgene sequence is provided in the Supplementary Information. The gRNA that targeted these repeat sequences was cloned by annealing two oligonucleotides (IDTDNA): 5′-caccgAAAGGTCGAGAAACTGCAAA-3′ and 5′-aaacTTTGCAGTTTCTCGACCTTTc-3′, phosphorylating the resulting double-stranded DNA product using T4 DNA Ligase (NEB), and ligating into a BbsI-digested gRNA plasmid backbone as described previously7. The design and sequences of all other gRNAs and the corresponding gRNA expression plasmids were described previously.7
All new plasmids described in this study are available from Addgene (plasmid ID 60551-4, 60718-9). All new protein sequences and the sequence of the promoter driving the eGFP reporter are provided in Supplementary Figures 5 and 6.
Cell Culture, Transfections, and Gene Expression
Cells were maintained in DMEM supplemented with 10% FBS and 1% penicillin/streptomycin. For transfections, 1.8×105 cells were plated per well of a 24-well plate and transfected with Lipofectamine 2000 (Invitrogen) the next day according to manufacturer’s instructions. Each well was transfected with 400, 235, and 165 ng of gRNA expression plasmid, CIBN-dCas9-CIBN plasmid, and CRY2-VP64 plasmid, respectively. For activation of the LACE system, cells were illuminated beginning 24 hours after transfection. For quantitative real-time PCR, cells were illuminated for 3 days, and then mRNA was purified using Qiagen RNeasy spin prep columns. cDNA was synthesized using the SuperScript® VILO cDNA Synthesis Kit (Life Technologies). Relative levels of cDNA were detected using Quanta PerfeCta® SYBR® Green FastMix® (Quanta Biosciences) and CFX96 Real-Time PCR Detection System (Bio-Rad). Raw data was normalized to GAPDH levels and cells transfected with an empty plasmid control using the ΔΔCt method. Primer sequences and standard curves have been previously described.7 For imaging of eGFP fluorescence, cells were illuminated 24 hours after transfection for 24 hours and then fixed with 4% paraformaldehyde. eGFP and DAPI images were exposed for 21 ms and 163 ms, respectively through a 10x objective. Patterned images were transfected using Lipofectamine 3000 (Invitrogen) with 735, 515, 250, and 1000 ng of CIBN-dCas9-CIBN, CRY2FL-VP64, eGFP reporter plasmid, and gRNA plasmid, respectively. Images were acquired using the scan and stitch feature of the Zeiss Axio Observer Z1 fluorescence microscope (10x magnification). Contrast and brightness were adjusted equally for all images using FIJI ImageJ 1.48r. Flow cytometry experiments were performed with a BD Accuri C6 flow cytometer after 24 hours of illumination or incubation in the dark.
Illumination
Cells were illuminated using a custom-built 4×6 LED array26 (~16 mW/cm2), measured using a Newport Power Meter held directly above LEDs, Model No. 1931-C). Six blue Rebel LEDs (450 nm, 48 Lumens @ 700 mA, Philips Lumileds) pre-mounted on a 10 mm square base (Luxeon Star LEDs, Ontario, Canada) were connected in series, and four of these LED series were connected in parallel and mounted on a printed circuit board (Radio Shack). The LED array was powered using an Arduino Uno microcontroller programmed to pulse illumination for 1 second at 0.067 Hz. Current was regulated using a BuckPuck AC Driver (700 mA, Luxeon Star LEDs). For all experiments except patterning, the LED array was held 8″ above the incubator shelf with illumination facing downward such that cells were illuminated from the top of the dish. For patterning experiments cells were plated in 35 mm dishes (StemCell Technologies) that were set atop of the upward-facing LED array. A photomask made by a 3D printer was placed between the upward-facing LED array and the bottom of the 35 mm culture dish. Plates containing cells incubated in the dark were wrapped in aluminum foil.
Western Blot
Proteins were expressed from LACE plasmids transfected into human HEK293T cells. Cells were lysed in RIPA buffer (Sigma R0278). Lysates were mixed with loading buffer and boiled for 5 min; equal volumes of protein were run in a NuPAGE Novex 4–12% Bis-Tris polyacrylamide gel and transferred to a nitrocellulose membrane. Non-specific antibody binding was blocked with 5% non-fat milk in TBS-T (50 mM Tris, 150 mM NaCl, and 0.1% Tween-20) for 30 min at RT. Membrane was then incubated with an anti-FLAG primary antibody (Sigma F7425, 1:1000) in 5% milk in TBS-T overnight at 4°C and then an anti-rabbit/HRP conjugated secondary antibody (Sigma A6154, 1:5000) in 5% milk in TBS-T for 1 hr at RT. Membrane was visualized using the Immun-Star™ WesternC™ Chemiluminescence Kit (Bio-Rad, 170-5070) according to manufacturer’s dirctions, and image was captured using a ChemiDoc XRS+ System and processed using ImageLab software (Bio-Rad). After image capture, the membrane was stripped of the primary and secondary antibodies using Restore PLUS Western blot stripping buffer (Thermo Scientific, 46430) for 10 minutes before blocking in 5% milk in TBS-T for 30 min at RT. Membrane was then probed with an anti-GAPDH antibody (Cell Signaling 2118L, 1:5000) in 5% milk in TBS-T for 1 hr at RT and then the anti-rabbit/HRP conjugated secondary antibody for 1 hr at RT.
Statistical Analysis
Data is shown as mean ± standard deviation, with sample numbers indicated in figure legends. Qualitative images shown are representative of at least two experiments performed in duplicate. To combine replicate experiments in Figure 1c, data normalization was performed to account for the variation in basal gene expression between replicate experiments (see Supplementary Data). Statistical significance was determined by performing a one-way or multi-way ANOVA test. Differences with p<0.05 were considered significant. Tukey’s or Student’s pairwise t-test was used for pairwise comparisons as appropriate. Statistical analysis was performed using JMP® Pro 11. All quantitative raw data is provided in Supplementary Data.
Supplementary Material
Acknowledgments
Chandra Tucker provided CRY2 and CIBN plasmids. Paul Vosburgh designed and fabricated the 3D photomasks. This work was supported by an NIH Director’s New Innovator Award (DP2OD008586), NSF Faculty Early Career Development (CAREER) Award (CBET-1151035), NIH R01DA036865, NIH R03AR061042, NIH P30AR066527, and an American Heart Association Scientist Development Grant (10SDG3060033). L.R.P. was supported by an American Heart Association Predoctoral Fellowship and NIH Biotechnology Training Grant (T32GM008555).
Footnotes
Author Contributions
L.R.P. and C.A.G. designed experiments, analyzed the data, and wrote the manuscript. L.R.P. performed experiments.
The authors declare no competing financial interests.
References
- 1.Beerli RR, Dreier B, Barbas CF., 3rd Positive and negative regulation of endogenous genes by designed transcription factors. Proc Natl Acad Sci U S A. 2000;97:1495–1500. doi: 10.1073/pnas.040552697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhang F, Cong L, Lodato S, Kosuri S, Church GM, Arlotta P. Efficient construction of sequence-specific TAL effectors for modulating mammalian transcription. Nat Biotech. 2011;29:149–153. doi: 10.1038/nbt.1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Miller JC, Tan S, Qiao G, Barlow KA, Wang J, Xia DF, et al. A TALE nuclease architecture for efficient genome editing. Nat Biotechnol. 2011;29:143–148. doi: 10.1038/nbt.1755. [DOI] [PubMed] [Google Scholar]
- 4.Konermann S, Brigham MD, Trevino AE, Hsu PD, Heidenreich M, Cong L, et al. Optical control of mammalian endogenous transcription and epigenetic states. Nature. 2013;500:472–476. doi: 10.1038/nature12466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Qi LS, Larson MH, Gilbert LA, Doudna JA, Weissman JS, Arkin AP, et al. Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell. 2013;152:1173–1183. doi: 10.1016/j.cell.2013.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gilbert LA, Larson MH, Morsut L, Liu Z, Brar GA, Torres SE, et al. CRISPR-mediated modular RNA-guided regulation of transcription in eukaryotes. Cell. 2013;154:442–451. doi: 10.1016/j.cell.2013.06.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Perez-Pinera P, Kocak DD, Vockley CM, Adler AF, Kabadi AM, Polstein LR, et al. RNA-guided gene activation by CRISPR-Cas9-based transcription factors. Nat Methods. 2013;10:973–976. doi: 10.1038/nmeth.2600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Maeder ML, Linder SJ, Cascio VM, Fu Y, Ho QH, Joung JK. CRISPR RNA-guided activation of endogenous human genes. Nat Methods. 2013;10:977–979. doi: 10.1038/nmeth.2598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cheng AW, Wang H, Yang H, Shi L, Katz Y, Theunissen TW, et al. Multiplexed activation of endogenous genes by CRISPR-on, an RNA-guided transcriptional activator system. Cell Res. 2013;23:1163–1171. doi: 10.1038/cr.2013.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Farzadfard F, Perli SD, Lu TK. Tunable and multifunctional eukaryotic transcription factors based on CRISPR/Cas. ACS Synth Biol. 2013;2:604–613. doi: 10.1021/sb400081r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mali P, Aach J, Stranges PB, Esvelt KM, Moosburner M, Kosuri S, et al. CAS9 transcriptional activators for target specificity screening and paired nickases for cooperative genome engineering. Nat Biotechnol. 2013;31:833–838. doi: 10.1038/nbt.2675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kabadi AM, Ousterout DG, Hilton IB, Gersbach CA. Multiplex CRISPR/Cas9-based genome engineering from a single lentiviral vector. Nucleic Acids Res. 2014;42:e147. doi: 10.1093/nar/gku749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337:816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shimizu-Sato S, Huq E, Tepperman JM, Quail PH. A light-switchable gene promoter system. Nat Biotech. 2002;20:1041–1044. doi: 10.1038/nbt734. [DOI] [PubMed] [Google Scholar]
- 15.Yazawa M, Sadaghiani AM, Hsueh B, Dolmetsch RE. Induction of protein-protein interactions in live cells using light. Nat Biotechnol. 2009;27:941–945. doi: 10.1038/nbt.1569. [DOI] [PubMed] [Google Scholar]
- 16.Kennedy MJ, Hughes RM, Peteya LA, Schwartz JW, Ehlers MD, Tucker CL. Rapid blue-light-mediated induction of protein interactions in living cells. Nat Methods. 2010;7:973–975. doi: 10.1038/nmeth.1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang X, Chen X, Yang Y. Spatiotemporal control of gene expression by a light-switchable transgene system. Nat Methods. 2012;9:266–269. doi: 10.1038/nmeth.1892. [DOI] [PubMed] [Google Scholar]
- 18.Polstein LR, Gersbach CA. Light-inducible spatiotemporal control of gene activation by customizable zinc finger transcription factors. J Am Chem Soc. 2012;134:16480–16483. doi: 10.1021/ja3065667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Muller K, Engesser R, Metzger S, Schulz S, Kampf MM, Busacker M, et al. A red/far-red light-responsive bi-stable toggle switch to control gene expression in mammalian cells. Nucleic Acids Res. 2013;41:e77. doi: 10.1093/nar/gkt002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Motta-Mena LB, Reade A, Mallory MJ, Glantz S, Weiner OD, Lynch KW, et al. An optogenetic gene expression system with rapid activation and deactivation kinetics. Nat Chem Biol. 2014;10:196–202. doi: 10.1038/nchembio.1430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gautier A, Gauron C, Volovitch M, Bensimon D, Jullien L, Vriz S. How to control proteins with light in living systems. Nat Chem Biol. 2014;10:533–541. doi: 10.1038/nchembio.1534. [DOI] [PubMed] [Google Scholar]
- 22.Perez-Pinera P, Ousterout DG, Brunger JM, Farin AM, Glass KA, Guilak F, et al. Synergistic and tunable human gene activation by combinations of synthetic transcription factors. Nat Methods. 2013;10:239–242. doi: 10.1038/nmeth.2361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Maeder ML, Linder SJ, Reyon D, Angstman JF, Fu Y, Sander JD, et al. Robust, synergistic regulation of human gene expression using TALE activators. Nat Methods. 2013;10:243–245. doi: 10.1038/nmeth.2366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Olson EJ, Tabor JJ. Optogenetic characterization methods overcome key challenges in synthetic and systems biology. Nat Chem Biol. 2014;10:502–511. doi: 10.1038/nchembio.1559. [DOI] [PubMed] [Google Scholar]
- 25.Chakraborty S, Ji H, Kabadi AM, Gersbach CA, Christoforou N, Leong KW. A CRISPR/Cas9-Based System for Reprogramming Cell Lineage Specification. Stem Cell Reports. 2014;3:940–947. doi: 10.1016/j.stemcr.2014.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Polstein LR, Gersbach CA. Light-inducible gene regulation with engineered zinc finger proteins. Methods Mol Biol. 2014;1148:89–107. doi: 10.1007/978-1-4939-0470-9_7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.