Summary
Olfactory sensory neurons express just one out of a possible ~1000 odorant receptor genes, reflecting an exquisite mode of gene regulation. In one model, once an odorant receptor is chosen for expression, other receptor genes are suppressed by a negative feedback mechanism, ensuring a stable functional identity of the sensory neuron for the lifetime of the cell. The signal transduction mechanism subserving odorant receptor gene silencing remains obscure, however. Here we demonstrate in the zebrafish that odorant receptor gene silencing is dependent on receptor activity. Moreover, we show that signaling through G protein βγ subunits is both necessary and sufficient to suppress the expression of odorant receptor genes, and likely acts through histone methylation to maintain the silenced odorant receptor genes in transcriptionally inactive heterochromatin. These results provide new insights linking receptor activity with the epigenetic mechanisms responsible for ensuring the expression of one odorant receptor per olfactory sensory neuron.
Introduction
Sensory systems receive and process external stimuli to convey information about the organism’s environment. Primary sensory neurons – the nervous system’s initial points of contact with the sensory world – are tuned to respond to different types of stimuli (light, touch, sound, etc.) or to a subset of stimuli within a given modality. The receptive field properties of a given primary sensory neuron – and therefore the overall logic of sensory processing – are determined by the particular receptors expressed by the cell. In the vertebrate olfactory system, the identification and discrimination of an odorant’s molecular identity from myriad chemical structures in odor space begins with the activation of odorant receptors expressed by olfactory sensory neurons in the nose. A large multigene family of olfactory-specific G protein-coupled receptors (GPCRs) initially identified in the rat (Buck and Axel, 1991) constitutes what is now referred to as the OR family of odorant receptors (Mombaerts, 2004). Each olfactory sensory neuron expresses a single OR allele, which defines the receptive field properties of the cell by virtue of the receptor’s ligand tuning properties (Chess et al., 1994; DeMaria and Ngai, 2010; Lewcock and Reed, 2004; Serizawa et al., 2003). Olfactory sensory neurons expressing the same OR in turn converge upon spatially invariant glomeruli in the olfactory bulb, the site of the first synaptic relay in olfactory sensory processing (Mombaerts et al., 1996; Ressler et al., 1994; Vassar et al., 1994). Thus, activation of specific odorant receptors by an odorant elicits a characteristic pattern of activity in the olfactory bulb.
The highly regulated expression of OR genes according to the “one receptor, one neuron” rule defines the functional identity of the sensory neuron by determining the odorants to which the cell responds. ORs also play a role in the precise targeting of the olfactory sensory neurons’ axons in the olfactory bulb (Imai et al., 2006; Mombaerts et al., 1996; Sakano, 2010; Serizawa et al., 2006; Wang et al., 1998), which underlies the anatomical basis of the olfactory sensory map. How is the expression of one OR gene initially established and maintained in each neuron to safeguard the cell’s identity and ensure its appropriate innervation in the olfactory bulb? The complexity of the regulatory mechanisms governing OR gene expression is daunting, considering the large size of the OR gene family, which ranges from 50~150 genes in fish to >1000 genes in rodents (Alioto and Ngai, 2005; Mombaerts, 2004; Niimura and Nei, 2005; Zhang and Firestein, 2002). Previous studies have shown that individual sensory neurons can in rare instances sequentially express multiple OR genes, with such gene switching events occurring more frequently when the initial OR gene expressed by the cell is a pseudogene (Lewcock and Reed, 2004; Serizawa et al., 2003; Shykind et al., 2004). These observations support a model involving a negative feedback loop in which a functional OR, once selected, silences the expression of all other OR genes in the genome (Lewcock and Reed, 2004; Serizawa et al., 2003; Serizawa et al., 2004; Shykind, 2005; Shykind et al., 2004). In this manner, OR gene silencing prevents gene switching and ensures the stable expression of a single OR in each olfactory sensory neuron. Recent studies have revealed the importance of epigenetic regulation of OR gene expression by repressive histone modifications, which maintain all but the actively transcribed OR gene in transcriptionally inactive heterochromatin (Magklara et al., 2011). The intracellular signaling mechanisms connecting OR-dependent events and OR gene silencing remain largely unknown, however.
In the present study, we examine the role of receptor-mediated activity in OR gene regulation. Using pharmacologic and genetic approaches in the zebrafish, we demonstrate a pivotal role of heterotrimeric G protein βγ subunits in OR gene silencing. Manipulations that enhance receptor-mediated activity, in particular through direct activation of Gβγ signaling, decrease the number of cells expressing a given OR gene. Conversely, inhibition of Gβγ signaling deregulates OR gene expression, causing an increase in the number of sensory neurons expressing a specific OR and the aberrant expression of multiple ORs per cell. We further show that perturbations of histone 3 lysine 9 (H3K9) methylation states characteristic of transcriptionally inactive chromatin lead to perturbations of OR gene expression similar to those caused by perturbations of Gβγ signaling. The effects of blocking both Gβγ signaling and H3K9 methylation simultaneously are not additive, suggesting that these two processes function in the same pathway to negatively regulate OR gene expression. Finally, genome-wide RNA transcript profiling by deep sequencing (RNA-Seq) of olfactory sensory neurons reveals that the expression of histone modifying enzymes involved in the regulation of repressive histone methylation marks is influenced by Gβγ activity, providing a possible mechanistic link between receptor activity at the plasma membrane and chromatin structure in the nucleus. Our results establish a role of G protein-mediated receptor activity in the negative feedback loop that ensures the singularity of OR gene expression and the maintenance of olfactory sensory neuron identity.
Results
OR activity-dependent suppression of OR gene expression
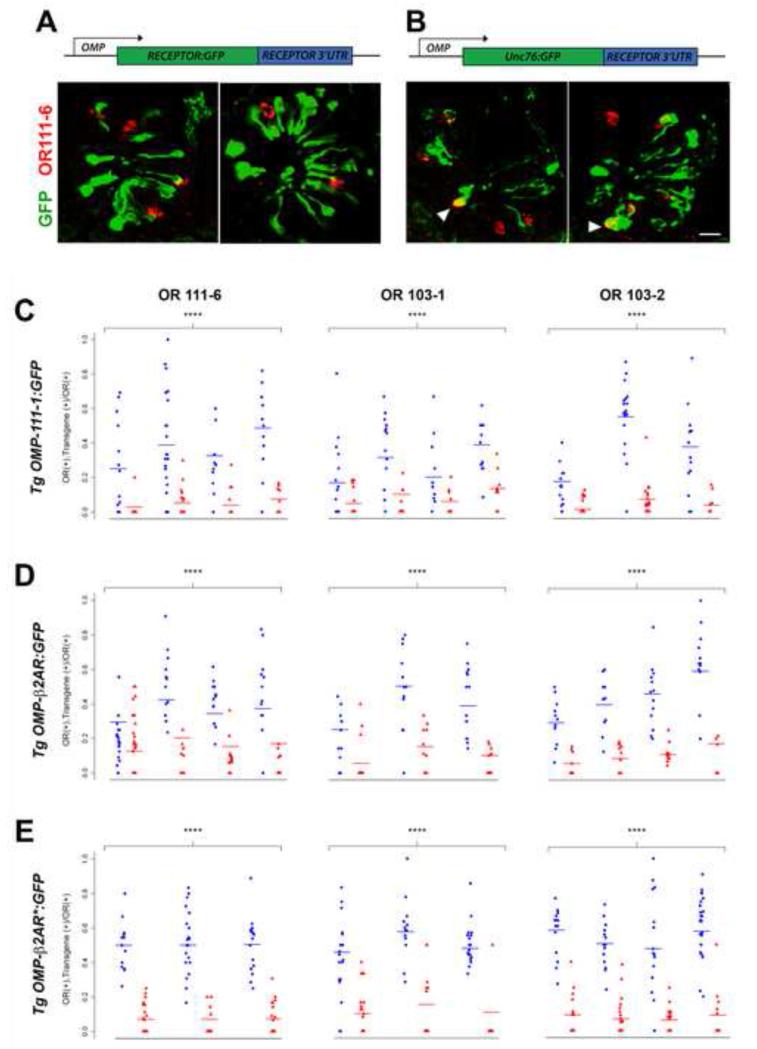
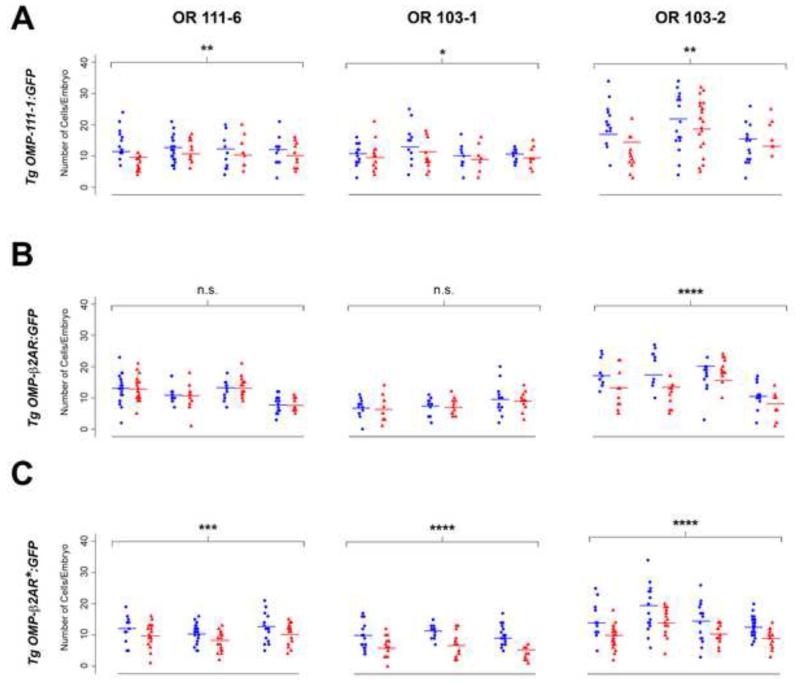
To explore the mechanisms underlying OR-dependent gene silencing, we developed an approach that allows for the transient expression of transgenes in the olfactory system of zebrafish embryos. Transgene constructs incorporating the zebrafish olfactory marker protein (OMP) promoter (Celik et al., 2002) were used to drive the widespread expression of genes in olfactory sensory neurons (see Experimental Procedures for details). OMP and OMP promoter-driven transgenes are expressed in the olfactory placode starting at ~24 hours post-fertilization (hpf), coinciding with the initial appearance of mature olfactory sensory neurons and labeling cells that innervate glomeruli in the olfactory bulb (Celik et al., 2002). Expression of OMP and OMP transgenes is therefore restricted to mature cells (and possibly also late stage maturing cells) in the zebrafish olfactory sensory neuron lineage. We first asked whether expression of an OR transgene under the control of the OMP promoter could suppress the expression of endogenous OR genes. Zebrafish embryos injected at the one-cell stage with OMP transgenes containing either a zebrafish OR:GFP fusion (OMP-OR111-1:GFP) or GFP control (OMP-unc76:GFP) exhibited widespread expression of the transgene in olfactory sensory neurons when assayed at 3 days post-fertilization (dpf) (Figure 1A, B). Using such transiently transgenic fish, we determined whether forced expression of an ectopic OR gene in olfactory sensory neurons influences the number of cells that express endogenous OR genes. In these experiments, we performed RNA in situ hybridizations for 3 different OR genes (OR111-6, a representative from the same subfamily as the transgene, and OR103-1 and OR103-2, two genes from a different subfamily (Alioto and Ngai, 2005)) on 3 dpf embryos and quantitated the number of cells expressing each receptor. As shown in Figure 2A and Table 1, expression of the OMP-OR111-1:GFP transgene had a modest but significant effect on endogenous OR gene expression, with a 10-15% reduction in the number of cells expressing all OR genes assayed, as compared to the GFP control (p-values = 0.05-0.002 from Poisson regression; see Experimental Procedures).
Figure 1. Expression of OMP-receptor transgenes in zebrafish embryos.
Zebrafish were injected with OMP-OR111-1:GFP (A) or OMP-unc76:GFP (B) at the one-cell stage and harvested at 3 dpf to simultaneously localize endogenous OR111-6 expression by RNA in situ hybridization (cells labeled in red) and GFP expression by immunohistochemistry (cells labeled in green). Arrowheads in (B) indicate olfactory sensory neurons co-expressing OR111-6 with GFP. Bar = 20 μm. Quantitation of the fraction of OR111-6-, OR103-1-, and OR103-2-expressing neurons positive for GFP transgene expression is shown for OMP-OR111-1:GFP (C), OMP-β2AR:GFP (D), and OMP-β2AR*:GFP (E). Plots show the proportion of double-positive cells (number of green and red cells/number of red cells) per embryo injected with the indicated transgene construct (red triangles) or OMP-unc76:GFP control (blue circles). Fitted cell proportions (horizontal bars) and p-values for the test of transgene effects were obtained from generalized linear models with a binomial distribution (see Methods). **** p < 10−5. See also Figure S1.
Figure 2. Suppression of receptor transgenes in embryonic zebrafish olfactory sensory neurons.
The number of cells expressing endogenous odorant receptors OR111-6 (left column), OR103-1 (middle column) or OR103-2 (right column) was determined by RNA in situ hybridization on 3 dpf zebrafish embryos previously injected at the one-cell stage with the following receptor-containing transgene construct: OMP-OR111-1:GFP (A); OMP-β2AR:GFP (B); OMP- β2AR*:GFP (C). Plots show the number of receptor-positive cells per embryo injected with the indicated transgene construct (red triangles) or OMP-unc76:GFP control (blue circles); data were derived from the same experiments presented in Figure 1. Each pair of treatment and control plots represents an independent experiment for a particular transgene and probe; within an experiment, 15-20 embryos were analyzed for each treatment and control condition. Fitted cell counts (horizontal bars) and p-values for the test of transgene effects were obtained from generalized linear models with a Poisson distribution (see Experimental Procedures). Results from the generalized linear model are summarized in Table 1. * p = 0.05; ** p ≤ 0.003; *** p < 10−3; **** p < 10−5; n.s., not significant (p > 0.05).
Table 1. Quantitation of OR gene expression under conditions affecting GPCR expression or Gβγ signaling.
| Treatment |
OR (+) cells Estimated fold-change (treatment/control) |
p-value | Number of experiments |
|
|---|---|---|---|---|
|
| ||||
|
OMP-OR111-
1:GFP |
OR111- 6 |
0.84 | 0.003 | 4 |
| OR103- 1 |
0.88 | 0.05 | 3 | |
| OR103- 2 |
0.85 | 0.002 | 3 | |
|
| ||||
| OMP-β2AR:GFP | OR111-6 | 0.99 | 0.8 | 4 |
| OR103- 1 |
0.94 | 0.5 | 3 | |
| OR103- 2 |
0.77 | < 10−5 | 4 | |
|
| ||||
| OMP-β2AR*:GFP | OR111-6 | 0.80 | < 10−3 | 3 |
| OR103- 1 |
0.59 | < 10−5 | 3 | |
| OR103- 2 |
0.71 | < 10−5 | 4 | |
|
| ||||
| Gallein | OR111-1 | 1.5 | < 10−5 | 4 |
| OR111-6 | 1.6 | < 10−5 | 3 | |
| OR103- 2 |
1.6 | < 10−5 | 3 | |
|
| ||||
| OMP-GRKct | OR111-1 | 1.9 | < 10−5 | 3 |
| OR111-6 | 1.7 | < 10−5 | 3 | |
| OR103- 2 |
1.6 | < 10−5 | 3 | |
|
| ||||
|
OMP-Gβ1 +
OMP-G γ 1 − |
OR111-1 | 0.69 | < 10−5 | 3 |
| OR111-6 | 0.62 | < 10−5 | 3 | |
| OR103- 2 |
0.47 | < 10−5 | 3 | |
Embryos subjected to the indicated treatments (together with their respective controls) were analyzed by RNA in situ hybridization using probes for OR111-6, OR103-1, or OR103-2. The ratios of OR-positive cells in treatment vs. control embryos were estimated using generalized linear models with Poisson distribution (see Experimental Procedures). The p-values measure the statistical significance of transgene or drug effects under the Poisson regression models; for each condition (treatment or control) and OR probe, 3 to 4 independent experiments were performed, each typically comprising 15-20 embryos.
Mouse olfactory sensory neurons expressing a β2-adrenergic receptor (β2AR) in place of an endogenous OR appear to function normally in terms of mutually exclusive expression with ORs and axon targeting (Feinstein et al., 2004). We were therefore interested in whether the human β2AR would behave similarly to ORs in our transgenic assays. Zebrafish embryos were injected with an OMP-β2AR:GFP transgene or OMP-unc76:GFP control, and the number of cells expressing endogenous OR genes was determined (Figure 2B and Table 1). No statistically significant effect of the β2AR:GFP transgene on OR111-6 or OR103-1 expression was observed. However, we did observe a modest yet highly significant decrease in the number of cells expressing OR103-2 (~25% decrease, p < 10−5). Thus, OR- or β2AR-encoding transgenes can – to a limited and variable extent – suppress the expression of endogenous OR genes. Interestingly, OMP transgenes containing either OR or β2AR receptor coding sequences were co-expressed with endogenous ORs in ~10-fold fewer cells than the control OMP-unc76:GFP transgene (p < 10−5 from a generalized linear model with a binomial distribution; Figure 1C-E), indicating that expression of endogenous ORs and transgenic receptors is largely mutually exclusive. In addition, as previously demonstrated in the mouse (Nguyen et al., 2007), OMP transgenes containing OR coding sequences are suppressed in the olfactory sensory neuron environment: when OMP-GFP transgene constructs were co-injected together with an OMP-mCherry plasmid, the ratio of cells expressing GFP vs. mCherry was ~3-fold lower when the OMP-GFP transgene also encoded an OR (Figure S1). These observations suggest that the relatively modest effect of receptor misexpression on endogenous OR expression is at least in part due to the reduced number of cells that express receptor-containing transgenes.
We hypothesized that activity of the encoded receptor is necessary to silence other OR gene loci. The β2AR, whose structure and function have been intensively studied (Rosenbaum et al., 2009), provides a means of exploring the role of receptor activity in OR gene silencing. Specifically, a conserved aspartate-arginine-tyrosine (DRY) tripeptide motif at the cytoplasmic base of the receptor’s third transmembrane domain modulates the activity of β2AR and related GPCRs (Rosenbaum et al., 2009). Substitution of the aspartate residue in the DRY motif to asparagine (NRY mutant) renders β2AR constitutively active (Rasmussen et al., 1999). When the constitutively active NRY mutant (β2AR*) was expressed under the control of the OMP promoter, we observed a significant reduction in cells expressing endogenous OR genes (Figure 2C and Table 1); OR111-6-positive cells were reduced by 20% (p < 10−3), whereas the numbers of cells expressing OR103-1 and OR103-2 were reduced by 30-40% (p < 10− 5). These results suggest that GPCR activity can suppress the expression of endogenous OR genes by olfactory sensory neurons.
Gβγ signaling is necessary and sufficient for suppression of OR gene expression
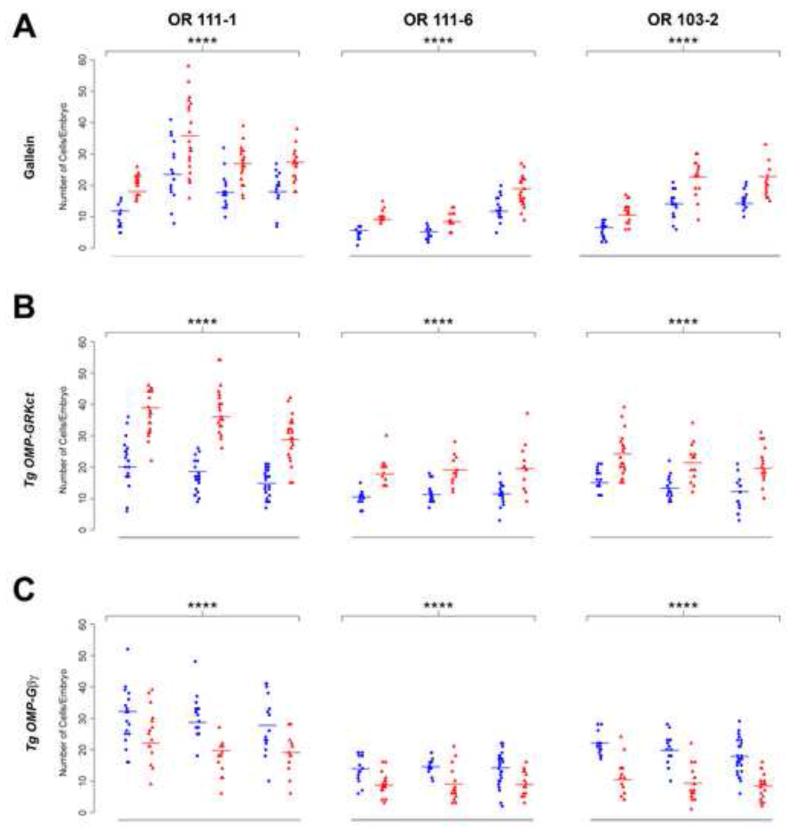
Through what intracellular signaling pathway does receptor activity suppress OR gene expression? Odor-evoked signaling is mediated through receptor-dependent activation of Gαolf (Belluscio et al., 1998), a Gαs isoform enriched in mature olfactory sensory neurons (Jones et al., 1990), ultimately leading to membrane depolarization due to the increased synthesis of cAMP by type III adenylyl cyclase and opening of cyclic nucleotide-gated cation channels (DeMaria and Ngai, 2010). Considering that the ORs and β2AR can couple through either Gαs or Gαolf (Kajiya et al., 2001), Gαs/Gαolf signaling would seem to be the most likely pathway for subserving OR gene silencing. However, previous studies have shown that a constitutively active Gαs mutant expressed in lieu of an intact OR fails to suppress the expression of other OR genes (Imai et al., 2006). Consistent with these observations, ectopic expression of constitutively active Gαs in zebrafish olfactory sensory neurons did not reduce the number of cells expressing endogenous OR genes (Figure S2). We therefore wished to determine whether G protein βγ subunits – which would be released from G protein αβγ heterotrimers upon receptor-mediated activation but not by ectopic expression of constitutively active Gα subunits – are required for OR gene silencing. This issue was addressed with four independent experimental approaches (Figure 3 and Figure S2). First, we blocked Gβγ activity globally by exposing embryonic zebrafish to gallein, a small molecule inhibitor of Gβγ (Bonacci et al., 2006). Fish were treated with drug starting at 16-20 hours post-fertilization (hpf), a developmental stage that precedes the first onset of OR gene expression by 4-8 hours (Barth et al., 1996). Compared to untreated fish, gallein treatment resulted in a 1.5- to 1.6-fold increase in the number of olfactory sensory neurons expressing OR111-1, OR111-6, or OR103-2 (p < 10−5; Figure 3A and Table 1). To address the possibility that inhibition of G protein signaling alters the absolute number of mature olfactory sensory neurons by delaying or inhibiting neuronal maturation, we quantitated the number of mature olfactory sensory neurons in stably transgenic OMP-Gal4;UASGCaMP1.6 fish in which a UAS-GCaMP reporter is driven by an OMP-Gal4 driver (Figure S2). We found indistinguishable numbers of transgene-positive mature olfactory sensory neurons (detected using an anti-GFP antibody) in control and drug-treated fish; patterns of innervation of the olfactory bulb by olfactory sensory neurons in drug-treated embryos were qualitatively normal (Figure S2). We also assessed the number of cells expressing phospho-histone H3 (a marker of cells in mitosis) and activated caspase (a marker of apoptotic cells) in the olfactory placodes of embryos treated with gallein; no significant difference was observed for either marker (Figure S2). These control experiments indicate that gallein treatment (and other drug treatments used in this study – see below) does not cause a major shift in the dynamics of proliferation, maturation or survival in the olfactory sensory neuron lineage.
Figure 3. OR gene expression is negatively regulated by Gβγ signaling.
The effects of altering G protein βγ signaling were quantitated in 3 dpf zebrafish embryos treated with 100 μM gallein (A), or previously injected with OMP-GRKct (B) or OMP-Gβ1 + OMP-Gγ13 transgene constructs (C). The number of cells expressing endogenous odorant receptors OR111-6 (left column), OR103-1 (middle column) or OR103-2 (right column) was determined by RNA in situ hybridization. Plots show the number of receptor-positive cells per embryo in each treatment (red triangles) or corresponding control condition (blue circles): vehicle control for gallein treatment, OMP-unc76:GFP for transgene injections. Experimental design and generalized linear model-based analysis are described in Experimental Procedures. Fitted cell counts are indicated by horizontal bars and results from the generalized linear model are summarized in Table 1. Inhibition of Gβγ signaling by gallein treatment or olfactory-specific GRKct expression results in a highly significant increase in the number of cells expressing an individual OR, whereas activation of Gβγ signaling by over-expression of Gβ1 and Gγ13 in olfactory sensory neurons results in a highly significant reduction (**** p < 10−5 for all treatments). See also Figure S2.
In a second approach, we inhibited Gβγ activity specifically in mature olfactory sensory neurons by expressing under the control of the OMP promoter a peptide comprising the C-terminal 195 amino acids of G protein receptor kinase 2 (GRKct). GRKct functions as a dominant-negative inhibitor by binding to Gβγ and preventing its interaction with downstream effectors (Koch et al., 1994). In zebrafish embryos injected with an OMP-GRKct transgene, endogenous OR genes were expressed in 1.6- to 1.9-fold more cells than in control embryos (p < 10−5; Figure 3B and Table 1). Expression of an OMP transgene encoding an N-terminal peptide of receptor for activated C kinase 1 (RACKnt) – which, like GRKct binds to Gβγ subunits and prevents their interaction with downstream effectors (Chen et al., 2004; Chen et al., 2008) – similarly caused a significant increase in the number of cells expressing endogenous OR (Figure S2). Thus, both pharmacologic and genetic inhibition of Gβγ activity result in an increase in the number of olfactory sensory neurons expressing a given OR gene. Moreover, the similar magnitude of the effects observed with gallein treatment and OMP transgenes encoding Gβγ inhibitory peptides indicates that the perturbations in OR expression are restricted mainly to mature olfactory sensory neurons.
As a complementary approach to probe the role of Gβγ signaling in OR gene regulation, we pursued a gain-of-function strategy in which zebrafish Gβγ subunits were ectopically expressed in olfactory sensory neurons using the OMP promoter. Over-expression of Gβγ can result in constitutive Gβγ signaling by exceeding the cell’s pool of available (GDP-bound) Gα (Faure et al., 1994). We therefore asked whether ectopic expression of Gβ1 and Gγ13 – the Gβγ isoforms that are enriched in olfactory sensory neurons (Kerr et al., 2008) – would negatively regulate OR gene expression. Consistent with the results of inhibiting Gβγ with gallein or GRKct, co-injection of OMP-Gβ1 and OMP-Gγ13 transgenes resulted in a 1.4- to 2-fold decrease in the number of cells expressing endogenous OR genes (p < 10−5; Figure 3C and Table 1). Together these observations indicate that Gβγ signaling is both necessary and sufficient to suppress OR gene expression.
Inhibition of Gβγ signaling results in aberrant OR co-expression
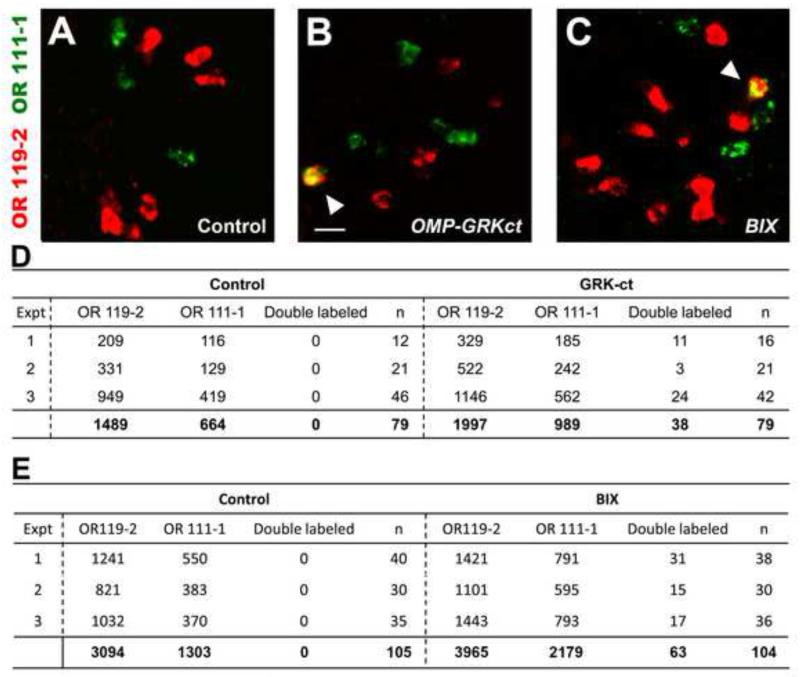
The increase in the number of cells expressing a particular OR in gallein-treated and OMP-GRKct- and OMP-RACKnt-injected embryos (Figure 3, Table 1 and Figure S2) suggests that olfactory sensory neurons express multiple ORs when Gβγ signaling is inhibited. To test this idea directly, we asked whether inhibition of Gβγ signaling by expression of GRKct would lead to the aberrant co-expression of multiple OR genes by individual cells. Zebrafish embryos were injected either with OMP-GRKct or control OMP-unc76:GFP transgenes, harvested at 3dpf, and subjected to double-label RNA in situ hybridization using probes for two endogenous ORs, OR111-1 and OR119-2 (Figure 4). Since individual olfactory sensory neurons typically express just a single OR gene, we expected that co-localization of two OR probes would not be observed in embryos in which Gβγ signaling was unperturbed. Consistent with this expectation, out of a total of 2153 cells positive for either OR, cells co-expressing both receptors were entirely absent in control embryos (Figure 4A, D). In striking contrast, OR111-1 and OR119-2 co-localized in 38 out of 2948 cells (1.3%) positive for either receptor alone in embryos expressing the OMP-GRKct transgene (Figure 4B, D). Thus, Gβγ signaling is required to prevent co-expression of multiple OR genes by an individual olfactory sensory neuron.
Figure 4. Inhibition of Gβγ signaling or H3K9 methylation results in aberrant co-expression of multiple OR genes.
Double label RNA in situ hybridizations were performed to identify olfactory sensory neurons expressing either OR111-1 (green) or OR119-2 (red) in 3 dpf zebrafish embryos previously injected with OMP-unc76:GFP (panel A), OMP-GRKct (panel B), or treated with BIX01294. Note the lack of OR co-expression in the OMP-unc76:GFP control, which contrasts with the occurrence of cells co-expressing OR111-1 and OR119-2 in OMP-GRKct-injected and BIX-treated fish (arrowheads). Bar in (B) = 20 μm. The numbers of cells positive for either or both receptor, from 3 independent experiments for either OMP-GRKct-injected or BIX-treated fish, are tabulated in panels C and D (n = number of fish scored).
Zebrafish OR genes reside in transcriptionally inactive chromatin
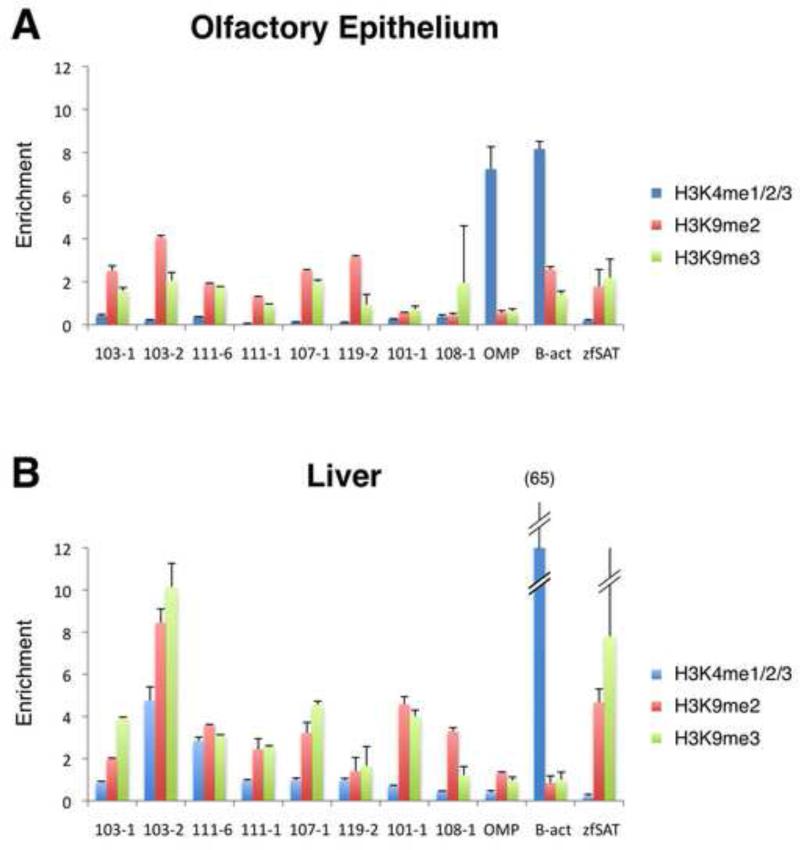
What are the downstream events that mediate Gβγ suppression of OR gene expression? Recent studies in the mouse have demonstrated that OR gene loci reside in inactive heterochromatin characterized by trimethyl histone H3 lysine 9 (H3K9me3) and other inhibitory histone methylation marks (Magklara et al., 2011). These observations have led to a model in which all OR genes are repressed in olfactory neuron progenitors, such that expression of a given OR involves selective protection or derepression of that gene by demethylation of H3K9me3. We hypothesized that Gβγ suppresses OR gene expression by interacting with the pathways regulating histone lysine methylation. To address this hypothesis, we first asked whether inhibitory histone methylation marks are enriched in chromatin associated with OR genes in the zebrafish, as they are in the mouse. Since only a small fraction of zebrafish olfactory sensory neurons in the adult express a given OR gene (~1-2%; (Barth et al., 1996)), when assaying the total population of cells in the olfactory epithelium we expect that chromatin associated with OR genes will show an overall enrichment of inhibitory histone methylation marks relative to chromatin modifications associated with actively transcribed genes. Chromatin immunoprecipitation (ChIP) was performed on native chromatin from whole olfactory epithelium or liver (in which ORs are not expressed) from adult zebrafish using antibodies specific for the inhibitory histone methylation marks H3K9me2 and H3K9me3, and mono-, di- and tri-methyl histone 3 lysine 4 (H3K4me1/2/3), which is associated with actively transcribed genes. Following immunoprecipitation, quantitative PCR (qPCR) was performed for eight OR genes, OMP and β-actin as controls for genes actively transcribed in the olfactory epithelium (β-actin only in liver), and satellite DNA (Ekker et al., 1992) as a control for H3K9me3-containing heterochromatin. As shown in Figure 5, H3K9me2 and H3K9me3 are enriched in chromatin associated with the eight OR genes tested, generally at levels comparable to satellite DNA and greater than observed for OMP. H3K9me2 and H3K9me3 are enriched relative to H3K4me1/2/3 for OR genes and satellite DNA in both olfactory epithelium and liver; for the OR genes, the enrichment of these repressive marks relative to H3K4me1/2/3 is in most cases greater in olfactory epithelium than in liver. In contrast, chromatin associated with actively transcribed OMP and β-actin in olfactory epithelium and β-actin in liver demonstrate comparatively greater enrichment of H3K4me1/2/3 relative to H3K9me2 and H3K9me3.
Figure 5. Zebrafish OR genes are associated with chromatin enriched in repressive histone methylation marks.
Chromatin immunoprecipitation followed by quantitative PCR (ChIP-qPCR) was performed on lysates prepared from whole olfactory epithelium (A) or liver (B) from adult zebrafish. Immunoprecipitations were carried out using antibodies directed against H3K4me1/2/3 (associated with transcriptionally active genes), or H3K9me2 and H3K9me3 (associated with transcriptionally inactive chromatin). qPCR was performed on the immunoprecipitated samples using primers for the eight indicated OR genes, OMP, β-actin (B-act) and satellite sequence (zfSAT); enrichment relative to input DNA is expressed as 2−ΔCt, where Ct = the PCR cycle number at which detection crossed threshold in the qPCR reaction and ΔCt = (CtChiP – Ctinput). Data are representative of 3 independent experiments. Note the enrichment of H3K9me2 and H3K9me3 repressive marks relative to H3k4me1/2/3 for OR genes in both tissues, particularly in the olfactory epithelium. In contrast, H3K4me1/2/3 shows greater enrichment relative to H3K9me2 and H3K9me3 for transcriptionally active genes (OMP and β-actin in olfactory epithelium and β-actin in liver). Error bars represent standard errors of the means.
It is unclear whether the enrichment of both H3K9me2 and H3K9me3 in chromatin associated with OR genes in olfactory epithelium reflects the heterogeneity of cells represented in the tissue preparation (which contains not only non-neuronal cells in the sensory mucosa but also cells from the non-sensory portion of the tissue) or heterogeneity in H3K9 methylation within the olfactory sensory neuron lineage itself. Nonetheless, based on this representative sampling of eight OR genes we conclude that zebrafish OR genes reside in transcriptionally inactive chromatin characterized by the inhibitory histone methylation marks H3K9me2 and H3K9me3.
H3K9 methylation suppresses OR gene expression and is required to enforce the “one receptor, one neuron” rule
Although OR genes are localized to heterochromatin characterized by repressive H3K9me3 marks (Magklara et al., 2011) (Figure 5), it remains to be determined whether H3K9 demethylation (or inhibition of H3K9 methylation) is sufficient to release ORs from transcriptional repression. Having confirmed that repressive histone methylation marks are enriched in chromatin associated with zebrafish OR genes, we therefore asked whether inhibition of H3K9 methylation would affect OR gene expression. H3K9 can be methylated to H3K9me2 by G9a histone methyltransferase and/or G9a-like protein (GLP), and then to H3K9me3 by the addition of a third and final methyl group catalyzed by SETDB1/2 (Black et al., 2012). Thus, the conversion of H3K9 to H3K9me2 and H3K9me3 – both inhibitory histone methylation marks – can be blocked by inhibiting G9a/GLP histone methyltransferase activity. Embryonic zebrafish were treated with BIX01294 (BIX), a small molecule inhibitor of G9a and GLP (Kubicek et al., 2007), and scored for the number of cells expressing representative OR genes. BIX-treated embryos showed a 1.5-1.6-fold greater number of cells expressing OR111-1 and OR103-2 as compared to untreated control fish (Figure 6A and Table 2); similar results were obtained with another G9a/GLP inhibitor, UNC0638 ((Vedadi et al., 2011); Figure S3). Trimethylated H3K9 can be demethylated by the lysine-specific demethylase Lsd1 (Black et al., 2012).
Figure 6. Perturbations in OR gene expression caused by inhibition of Gβγ signaling and H3K9 methylation.
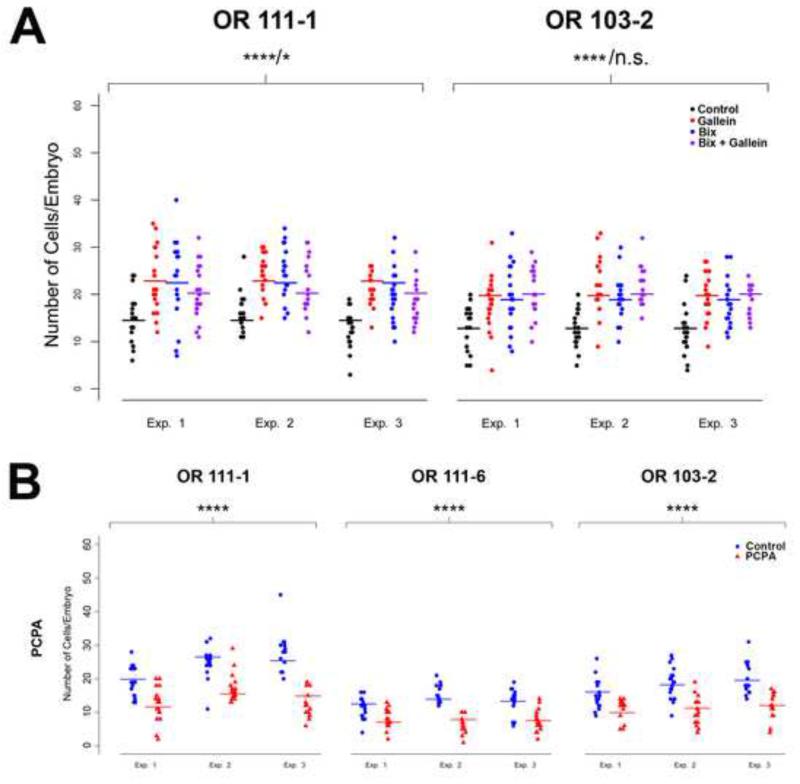
(A) The effects of inhibiting G protein βγ signaling and H3K9 methylation individually or in combination were quantitated in 3 dpf zebrafish embryos treated with 100 μM gallein, 20 μM BIX01294, or 100 μM gallein plus 20 μM BIX01294. The number of cells expressing endogenous odorant receptors OR111-1 (left) or OR103-2 (right) was determined by RNA in situ hybridization. Plots show the number of receptor-positive cells per embryo in control fish (black) or fish treated with gallein (red), BIX (blue), gallein + BIX (purple). Three independent experiments were performed, as indicated. Experimental design and GLM-based analysis are as described in Figure 2 and Experimental Procedures. Fitted cell counts are indicated by horizontal bars and results from the GLM are summarized in Table 2. Inhibition of Gβγ signaling by gallein treatment or H3K9 methylation by BIX results in similar increases in the number of cells expressing an individual OR (**** p < 10−5 for all drug treatments vs. control). Treatment with gallein + BIX resulted in a slight but significant decrease in the number of cells expressing OR111-1 compared to the average number of OR111-1 (+) cells from gallein- or BIX-treated embryos (* p = 0.0012); no significant difference in the number of OR103-2 (+) cells was found between fish treated with gallein + BIX vs. the average number from gallein- or BIX-treated fish (n.s., p = 0.30). (B) The effect of inhibiting histone demethylation was quantitated in 3 dpf zebrafish embryos treated with 75 μM trans-2-phenylcyclopropylamine (PCPA), an inhibitor of the histone demethylase Lsd1. The number of cells expressing endogenous odorant receptors OR111-1 (left column), OR111-6 (center column) or OR103-2 (right column) was determined by RNA in situ hybridization. Plots show the number of receptor-positive cells per embryo in control fish (blue circles) or fish treated with PCPA (red triangles). Three independent experiments were performed, as indicated. Fitted cell counts are indicated by horizontal bars and results from the GLM are summarized in Table 2. (**** p < 10−5). See also Figures S2 and S3.
Table 2. Quantitation of OR gene expression under conditions affecting Gβγ signaling and H3K9 methylation.
| Comparison |
OR (+) cells Estimated fold-change |
p-value | Number of experiments |
|
|---|---|---|---|---|
|
| ||||
| BIX vs. Control | OR111-1 | 1.6 | < 10−5 | 3 |
| OR103- 2 |
1.5 | < 10−5 | 3 | |
|
| ||||
| Gallein vs. Control | OR111-1 | 1.6 | < 10−5 | 3 |
| OR103- 2 |
1.5 | < 10−5 | 3 | |
|
| ||||
|
BIX+Gallein vs.
Control |
OR111-1 | 1.4 | < 10−5 | 3 |
| OR103- 2 |
1.6 | < 10−5 | 3 | |
|
| ||||
| Gallein vs. BIX | OR111-1 | 1.0 | 0.7 | 3 |
| OR103- 2 |
1.0 | 0.3 | 3 | |
|
| ||||
|
BIX+Gallein vs.
Average of BIX & Gallein |
OR111-1 | 0.89 | 0.001 | 3 |
| OR103- 2 |
1.0 | 0.3 | 3 | |
|
| ||||
| PCPA vs. Control | OR111-1 | 0.59 | < 10−5 | 3 |
| OR111-6 | 0.56 | < 10−5 | 3 | |
| OR103- 2 |
0.62 | < 10−5 | 3 | |
Embryos subjected to the indicated treatments were analyzed by RNA in situ hybridization using probes for OR111-1, OR111-6 or OR103-2. The ratio of OR-positive cells in each comparison was estimated using a generalized linear model with Poisson distribution (see Experimental Procedures). The p-values measure the statistical significance of comparisons under the Poisson regression models; three experiments were performed for each condition and OR probe, each typically comprising 15-20 embryos.
Accordingly, treatment of embryos with the Lsd1 inhibitor trans-2-phenylcyclopropylamine (PCPA) (Jie et al., 2009; Lee et al., 2006) caused a 1.6- to 1.8-fold decrease in the number of OR-positive cells (Figure 6B and Table 2). Together these observations demonstrate that H3K9 demethylation is both necessary and sufficient to activate OR gene expression and further identify the specific involvement of the histone modifying enzymes G9a/GLP and Lsd1 in this process (see also (Lyons et al., 2013)).
Does negative regulation of OR gene expression by H3K9 methylation reflect a role of this epigenetic modification in ensuring the expression of just one OR gene per olfactory sensory neuron? To address this question, zebrafish embryos were treated with BIX and then subjected to double-label RNA in situ hybridization with probes for OR111-1 and OR119-2. As we observed in gallein-treated fish, out of 6144 cells positive for either OR in BIX-treated embryos, 63 (~1%) showed co-expression of both receptor genes. In contrast, zero out of 4397 receptor-positive cells exhibited co-expression of the two receptors in control embryos (Figure 4A, C, E). These results provide functional evidence that methylation of H3K9 by G9a/GLP is required to restrict each olfactory sensory neuron to express a single OR.
Gβγ signaling and histone methylation operate in the same pathway to silence OR gene expression
Inhibition of G9a/GLP results in changes in OR gene expression similar to those observed with manipulations that inhibit Gβγ signaling: in both cases there is an increase in the number of cells expressing a given OR and the appearance of cells aberrantly expressing multiple ORs. We therefore wondered whether the silencing of OR gene expression by Gβγ signaling is mediated by H3K9 methylation. To test this hypothesis, we treated zebrafish embryos with gallein (Gβγ inhibitor) and BIX (G9a/GLP histone methyltransferase inhibitor) either alone or in combination. We reasoned that if Gβγ and G9a/GLP suppress OR expression by acting within the same pathway, treatment with both drugs simultaneously should result in an increase in OR-positive cells no greater than the increase observed with either drug alone. On the other hand, if Gβγ and G9a/GLP operate via parallel or independent pathways impinging on OR gene regulation, we would expect an additive effect of inhibiting both Gβγ or G9a/GLP. As shown in Figure 6A and Table 2, embryos treated simultaneously with gallein and BIX showed a 1.4- to 1.6-fold increase in the numbers of cells expressing OR111-1 or 103-2 (p < 10−5), similar to the effect of gallein or BIX treatment alone. The number of cells expressing OR103-2 in embryos treated with BIX plus gallein was indistinguishable from the number found in fish treated with either drug individually (p = 0.3). Curiously, there was a slight (~10%) but significant decrease in cells expressing OR111-1 in fish treated with both drugs compared to BIX- or gallein-treated fish (p = 0.001). This latter observation indicates a mild negative interaction between these two drug treatments that may reflect toxicity associated with globally blocking both pathways simultaneously. Nonetheless, it is noteworthy that the effect of blocking Gβγ signaling and H3K9 methylation was not additive, consistent with the hypothesis that Gβγ-dependent OR gene silencing is mediated by repressive histone methylation.
Gβγ signaling regulates expression of G9a histone methyltransferase
The results presented thus far suggest that receptor-activated Gβγ signaling limits each olfactory sensory neuron to expressing a single OR gene by promoting methylation of histone residues associated with transcriptionally inactive heterochromatin. To gain a global view of how Gβγ signaling affects gene expression, we performed RNA transcript profiling by deep sequencing (RNA-Seq) on olfactory sensory neurons purified by fluorescence activated cell sorting (FACS) from embryonic TgOMP-Gal4;UAS-GCaMP zebrafish. Pairs of RNA samples from purified cells isolated from gallein-treated and control fish were sequenced, and data from three independent experiments were normalized and analyzed to identify genes showing significant changes in expression in cells from gallein-treated fish relative to controls. From an analysis that incorporated multiple hypothesis testing to control the false discovery rate (Benjamini and Hochberg, 1995), approximately 5,000 out of ~21,000 mapped genes were found to exhibit differential expression between treatment and control with a false discovery rate (FDR) of less than 0.05 (Table S1). Read counts for all but 4 of the ~150 annotated zebrafish OR genes (Alioto and Ngai, 2005) were too low to allow reliable quantitation of their expression levels (data not shown).
In light of our results implicating H3K9 methylation as a mediator of Gβγ-dependent OR silencing, we focused on 101 genes encoding proteins involved in histone methylation (Black et al., 2012; Kouzarides, 2007) (Table S2); 21 genes showed significant differences in expression (adjusted p-value < 0.05) between gallein and untreated cells. Consistent with our pharmacologic analysis of H3K9 methylation, expression of ehmt2 – which encodes G9a histone methyltransferase – was down-regulated 2.2-fold in OMP-positive cells from gallein-treated fish compared to controls (adjusted p-value = 0.03; Table 3). Similarly, the lysine demethylase KDM6B, which demethylates H3K27me3 – another hallmark of transcriptionally inactive chromatin (Black et al., 2012; Kouzarides, 2007) – to H3K27me1, was up-regulated 1.7-fold in gallein-treated fish (adjusted p-value < 10−8; Table 3). Thus, inhibition of Gβγ signaling results in changes in expression of histone modifying enzymes that may underlie a decrease in repressive histone methylation marks.
Table 3. Expression of selected genes in olfactory sensory neurons isolated from gallein-treated and control zebrafish embryos.
| Gene Symbol |
log2 Fold-Change (gallein/control) |
Adjusted p- value |
Fold-Change (gallein/control) |
|
|---|---|---|---|---|
|
Histone modifying
enzymes |
||||
| G9a histone H3K9 methyltransferase |
ehmt2 | −1.14 | 0.03 | 0.45 |
| Kdm6 histone H3K27 demethylase |
kdm6b | 0.76 | 5 × 10−9 | 1.7 |
|
Olfactory sensory
neuron markers |
||||
| Type III adenylyl cyclase |
adcy3b | −7.83 | 10−32 | 0.0044 |
| Olfactory marker protein |
ompb | −1.63 | 10−6 | 0.32 |
| G proteins | ||||
| Gαs | gnas | −0.28 | 0.1 | -- |
| Gαolf | gnal | −1.03 | 0.2 | -- |
| Gβ1 | gnb1a | −0.68 | 0.1 | -- |
| Gγ13 | gng13b | −2.08 | 0.5 | -- |
| Gγ8 | gng8 | −1.77 | 0.8 | -- |
| Arrestins | ||||
| β-arrestin1 | arrb1 | −1.79 | 0.4 | -- |
| β-arrestin2 | arrb2a | −0.23 | 0.5 | -- |
| β-arrestin2 | arrb2b | 0.46 | 0.1 | -- |
|
Phospholipase C
isoforms |
||||
| PLC-β1 | plcb1 | −2.40 | 0.1 | -- |
| PLC-β2 | plcb2 | 0.39 | 0.9 | -- |
| PLC-β3 | plcb3 | −0.84 | 0.5 | -- |
RNA-Seq was performed on olfactory sensory neurons purified from gallein-treated and control zebrafish embryos. Data are presented for selected transcripts. Linear fold-change values are not shown in cases where adjusted p-value ≥ 0.1.
Lyons et al. recently demonstrated that final maturation of olfactory sensory neurons, as embodied by expression of OMP and type III adenylyl cyclase (Adcy3) – the adenylyl cyclase isoform responsible for generating cAMP in response to OR-mediated Gαolf activation (Wong et al., 2000) – is dependent on OR expression (Lyons et al., 2013). One question to arise from these observations is how the OR influences expression of these two genes, which are hallmarks of olfactory sensory neuron terminal differentiation. Interestingly, by RNA-Seq we found that OMP and Adcy3 are significantly down-regulated in olfactory sensory neurons isolated from gallein-treated fish as compared to controls (OMP: 3-fold reduction, adjusted p-value = 10−6; Adcy3: > 200-fold reduction, adjusted p-value = 10−32; Table 3). In contrast, no significant differences in expression of other signaling molecules, including Gαs, Gαolf, Gβ1, Gγ13, Gγ8, β-arrestins and phospholipase C β (PLC-β) isoforms, were observed between olfactory sensory neurons isolated from gallein-treated vs. control embryos (Table 3).
Discussion
The initial choice and subsequent maintenance of the single OR gene expressed by an olfactory sensory neuron are critical determinants of the cell’s functional identity. Several independent studies have demonstrated a role of the OR itself in maintaining the singularity of OR expression (Lewcock and Reed, 2004; Serizawa et al., 2003; Shykind et al., 2004) and have led to models in which functional OR protein participates in a negative feedback loop that silences the expression of other OR genes once an OR gene is initially chosen (Serizawa et al., 2004; Shykind, 2005). Recent studies have implicated epigenetic mechanisms in the regulation of OR gene expression (Clowney et al., 2012; Lyons et al., 2013; Magklara et al., 2011). How the OR itself participates in OR gene silencing and interfaces with such epigenetic mechanisms has until now remained elusive, however. The results presented here suggest that receptor signaling mediated by G protein βγ subunits is a critical link that ties together the requirement for a functional receptor with downstream epigenetic events that suppress OR gene expression.
Role of G protein βγ signaling in OR gene silencing
We have identified Gβγ subunits as the proximate downstream effector in the signal transduction cascade underlying OR-mediated gene silencing. Our interpretation is at odds with an alternative view, which contends that OR gene silencing occurs through a G protein-independent mechanism (Imai and Sakano, 2008). This latter conclusion is based on two key observations. First, expression in olfactory sensory neurons of a constitutively active Gαs mutant – which is expected to bypass the requirement for a functioning receptor – failed to prevent expression of OR genes (Imai et al., 2006). This observation alone does not address the potential role of Gβγ in silencing, however, as constitutively active Gα would not be expected to alter the levels of free Gβγ subunits in the cell. Second, OR co-expression was repressed by transgenic expression of a mutant OR in which the conserved DRY receptor activation motif was mutated to RDY (Imai et al., 2006) (see also (Nguyen et al., 2007)). Although olfactory sensory neurons expressing the RDY mutant receptor exhibited no detectable responses to the receptor’s cognate odorant, it is possible that the mutant receptor retains a level of intrinsic activity in the unliganded state sufficient to repress expression from other OR gene loci. Indeed, residual intrinsic activity is observed in a β2AR receptor containing the DRY −> RDY mutation (Nakashima et al., 2013). Whatever the case, the present results constitute strong evidence for the participation of Gβγ subunits in the regulation of OR gene expression.
Roles of Gβγ signaling and histone methylation in repressing OR gene switching
Using genetic and pharmacologic perturbations, we found that Gβγ signaling is both necessary and sufficient for repressing OR gene expression. Moreover, inhibition of Gβγ signaling or methylation of H3K9 leads to the aberrant co-localization of multiple ORs per cell. The experimentally induced co-expression of multiple ORs by an individual olfactory sensory neurons may reflect the sequential switching of expression from one OR gene to another. In this scenario, perdurance of mRNA from the initially transcribed gene could account for the presence of both transcripts within the same cell, after the first gene is no longer transcribed. The ~24 h interval between the initiation of robust OR expression at ~2 dpf (Barth et al., 1997) and our experimental endpoint at 3 dpf could allow the detection of mRNA transcribed from the first gene following a single switching event. Considering that the zebrafish genome encodes ~150 OR genes (Alioto and Ngai, 2005; Niimura and Nei, 2005), the incidence of double-positive cells in zebrafish embryos (~1%) in which Gβγ signaling or H3K9 methylation is down-regulated suggests that each olfactory sensory neuron expresses on average two OR genes at random under our experimental conditions. The frequency with which we detect the expression of two receptors per cell under conditions of reduced Gβγ signaling or H3K9 methylation in the zebrafish is consistent with the frequency of OR pseudogene −> gene switching observed in the mouse (0.1%) (Shykind et al., 2004), considering that there are 10-fold fewer OR genes in the fish genome compared to the mouse (~150 vs. ~1400). Our data suggest that Gβγ signaling and H3K9 methylation are required as part of a fail-safe mechanism to repress OR gene switching following the initiation of OR gene expression.
OR-dependent OR gene silencing acts through Gβγ signaling and histone methylation
By inhibiting Gβγ signaling and H3K9 methylation either together or individually, we provide evidence suggesting that these two processes function within the same pathway to repress OR gene expression. Consistent with these results, the expression of genes encoding enzymes involved in regulating repressive histone methylation marks are altered by perturbations in Gβγ signaling. Recent studies in the mouse have shown that OR genes become associated with heterochromatin containing the repressive histone methylation mark H3K9me3 prior to the expression of OR genes in olfactory progenitor cells (Magklara et al., 2011). These observations suggest a model in which all OR genes are initially repressed, with receptor activation involving a process in which the chosen OR gene is de-repressed by demethylation of H3K9 by the histone demethylase Lsd1 (Lyons et al., 2013; Magklara et al., 2011).
Can demethylation of H3K9 by itself affect OR gene transcription? The results from our experiments using G9a/GLP and Lsd1 inhibitors demonstrate that H3K9 demethylation is both necessary and sufficient to release OR genes from transcriptional repression. Importantly, our findings further reveal that repression by H3K9 methylation is required to prevent the expression of multiple ORs per cell and thus plays a critical role in enforcing the one receptor, one neuron rule of OR gene expression.
We propose that, following the initial selection and expression of an OR gene, the intrinsic activity of the expressed receptor – reflecting the receptor’s equilibrium between inactive and active states in the absence of bound agonist (Rosenbaum et al., 2009) – leads to the release of active Gα and Gβγ subunits from inactive Gαβγ heterotrimers. Consistent with this hypothesis, a constitutively active β2AR mutant more effectively suppresses OR expression than the wild type receptor (this study). A recent study has shown that intrinsic activity of unliganded OR influences the targeting of olfactory sensory axons to along the anterior-posterior axis of the olfactory bulb by regulating the expression of axon guidance cue receptors (Nakashima et al., 2013). Thus, intrinsic OR activity appears to play an important role in multiple gene regulatory networks governing olfactory sensory neuron development.
Whereas Gα is required for odor-evoked signal transduction (Belluscio et al., 1998) and influences axon targeting via cyclic AMP-dependent pathways (Imai et al., 2006; Nakashima et al., 2013; Serizawa et al., 2006), activated Gβγ maintains the cell’s functional identity by inhibiting transcription of other OR genes – and possibly also by stabilizing the expression of the selected OR gene – through an as yet unknown downstream effector(s) that ultimately interacts with chromatin modifying enzymes, including histone methyltransferases. Thus, there is a bifurcation in odorant receptor-mediated G protein signaling, with Gα driving primary signal transduction events and Gβγ subserving the negative feedback regulation of odorant receptor gene expression. Other GPCRs may also initiate this Gβγ-dependent gene regulatory cascade, for example in immature olfactory sensory neurons in which OR genes are not yet expressed. Consistent with this idea, repressive H3K9me3 marks are associated with OR genes in immature olfactory neuron progenitors prior to the onset of OR gene expression (Magklara et al., 2011). Down-regulation of the H3K9 demethylase Lsd1 in mature olfactory sensory neurons may also play a role in maintaining repressive histone methylation marks in chromatin associated with OR genes (Lyons et al., 2013). In this regard it is intriguing that Gβγ signaling appears to positively regulate the expression of Adcy3 (this study), which in turn negatively regulates Lsd1 expression (Lyons et al., 2013). In light of our data showing that expression of the H3K9-specific G9a histone methyltransferase is dependent on Gβγ signaling, it seems that receptor-mediated G protein activity may affect chromatin structure by targeting multiple enzymes involved in regulating histone methylation dynamics. In addition, a recent study in the mouse indicates that OR expression activates the unfolded protein response, which in turn activates the expression of Adcy3 and leads to the stabilization of OR gene choice by down-regulation of Lsd1 (Dalton et al., 2013). The unfolded protein response is thought to be used as an initial checkpoint in OR protein expression; once this pathway is down-regulated (via negative feedback), other pathways such as G protein signaling may then be required to silence other OR gene loci for the lifetime of the cell. Thus, OR expression may impinge on multiple pathways that act either sequentially or in parallel to ensure the expression of just one receptor per cell.
Curiously, conditional knockout of Gγ13 in mouse olfactory sensory neurons results in dramatic down-regulation of Gαolf, Gβ1 and the guanine nucleotide exchange factor RIC8B, as well as mislocalization of type III adenylyl cyclase to the sensory neuron soma (Li et al., 2013). In contrast, Gαolf and Gβ1 expression is unaffected by inhibition of Gβγ signaling in zebrafish olfactory sensory neurons (this study). Perturbations in Gβγ signaling therefore appear to affect the expression of multiple components of the olfactory signal transduction cascade in both fish and mammals, although these effects may exhibit species-specific differences. Considering that fish possess ~10-fold fewer OR genes than mammals, it is possible that mammals evolved additional mechanisms to restrict the expression of one OR per cell from a highly expanded OR gene repertoire. Whatever the case, the identified role of Gβγ signaling in OR gene silencing now allows an informed search for the downstream interaction partners of G protein βγ subunits (Dupre et al., 2009) that function to ensure the singularity of OR gene expression by an individual olfactory sensory neuron. Such partners in turn would interact with the genetic and epigenetic network that regulates histone methylation and perhaps other aspects of chromatin structure associated with OR gene loci. It is interesting to note that interaction of Gβγ with RACKnt – which we found to inhibit OR gene silencing when ectopically expressed in olfactory sensory neurons – blocks Gβγ activation of PLC-β, type II adenylyl cyclase and PI3 kinase, but not MAP kinase (Chen et al., 2004; Chen et al., 2008). With these clues in mind, the present study lays the groundwork for a directed search for the molecules bridging G protein signaling and OR gene regulation in olfactory sensory neurons.
Experimental Procedures
Transgenes and zebrafish
Plasmids containing the OMP promoter were used to drive widespread expression of transgene sequences in olfactory sensory neurons. Zebrafish embryos were co-injected at the one-cell stage with plasmid DNA (80ng/μl) and Tol2 messenger RNA (25ng/μl) to facilitate robust transient transgenic expression. For drug treatments, zebrafish embryos were treated at 16-20 hpf with 100 μM gallein 20 μM BIX, 20 μM UNC0638, 75 μM PCPA, or a control solution of EM containing DMSO to match the amount used to deliver drug in the corresponding experimental treatment (typically 0.2%). At 3 dpf, embryos were sorted for GFP fluorescence (in the case of transgenic fish), fixed in 4% paraformaldehyde, and processed for RNA in situ hybridizations and immunohistochemistry.
RNA in situ hybridization and immunohistochemistry
Whole mount fluorescent RNA in situ hybridization on 3dpf embryos using FITC-tyramide detection was performed as described (Welten et al., 2006). Co-localization of endogenous OR gene expression with transgene expression was detected via immunohistochemical labeling of GFP with Alexa568 detection in conjunction with FITC-tyramide-based RNA in situ hybridization. Simultaneous detection of mRNAs encoded by two OR genes was carried out using FITC-/Cy3-tyramide two-color RNA in situ hybridization.
Imaging and statistical analysis
Fixed embryos were embedded in 1.2% low melting point agarose gel and imaged head-on by confocal microscopy. Image stacks were analyzed using NIH ImageJ and Adobe Photoshop. Generalized linear models (GLM) were used to analyze the number of cells expressing endogenous ORs and to test for differences between treatment and control conditions (McCullagh and Nelder, 1989).
ChIP-qPCR
Olfactory epithelia and liver of 3-6 month old zebrafish adults were dissected and native chromatin was prepared essentially as described (Magklara et al., 2011). Immunoprecipitated DNA was purified using a MinElute PCR Purification Kit (Qiagen) and amplified using the WGA4 Whole Genome Amplification kit (SIGMA). Duplicate or triplicate aliquot of each amplified reaction was then subjected to quantitative PCR. Enrichment over input is expressed as 2−ΔCt, where Ct = the PCR cycle number at which detection crossed threshold in the qPCR reaction and ΔCt = (CtChiP – Ctinput).
RNA-Seq of FACS-purified olfactory sensory neurons
For fluorescent labeling and FACS purification of olfactory sensory neurons, a stable OMP-Gal4 transgenic zebrafish line transgenic line was generated using a plasmid construct containing 1.4 kb OMP 5′ promoter sequence (Celik et al., 2002) inserted upstream of the Gal4 transactivator and crossed with UAS-GCaMP1.6 transgenic fish (Del Bene et al., 2010). OMP-Gal4;UAS-GCaMP1.6 transgenic zebrafish embryos were collected at 16-20 hpf and treated with 100 μM gallein or DMSO (control). Heads of 5 dpf embryos were dissected and dissociated with trypsin and collagenase and sorted for GFP fluorescence; cells were collected in 1ml of Trizol and stored at −80°C. Three pairs of FACS-purified gallein-treated and control cells were analyzed by RNA-Seq. Differential expression (DE) analysis was carried out within the framework of generalized linear models (GLM) as implemented in the Bioconductor R package edgeR (Robinson et al., 2010). A likelihood ratio test of differential expression between treated and control libraries identified 5,094 differentially expressed genes at a false discovery rate (Benjamini and Hochberg, 1995) < 0.05. See Table S1 for the list of DE genes. RNA-Seq data have been deposited in the NCBI Gene Expression Omnibus and are accessible through GEO Series accession number GSEXXXXX (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSEXXXXX).
Detailed information for all methods can be found in Supplemental Experimental Procedures.
Supplementary Material
Acknowledgments
This work was supported by grants to J.N. from the National Institute on Deafness and Other Communication Disorders. T.F. was also supported by a training grant from the National Human Genome Research Institute (T32 HG00047); S.R.W. was the recipient of a Graduate Research Fellowship from the National Science Foundation. We thank the members of the Ngai lab, past and present, for their invaluable advice and suggestions over the course of this study.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alioto TS, Ngai J. The odorant receptor repertoire of teleost fish. BMC Genomics. 2005;6:173. doi: 10.1186/1471-2164-6-173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barth AL, Dugas JC, Ngai J. Noncoordinate expression of odorant receptor genes tightly linked in the zebrafish genome. Neuron. 1997;19:359–369. doi: 10.1016/s0896-6273(00)80945-9. [DOI] [PubMed] [Google Scholar]
- Barth AL, Justice NJ, Ngai J. Asynchronous onset of odorant receptor expression in the developing zebrafish olfactory system. Neuron. 1996;16:23–34. doi: 10.1016/s0896-6273(00)80020-3. [DOI] [PubMed] [Google Scholar]
- Belluscio L, Gold GH, Nemes A, Axel R. Mice deficient in Golf are anosmic. Neuron. 1998;20:69–81. doi: 10.1016/s0896-6273(00)80435-3. [DOI] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J Roy Stat Soc Series B (Methodological) 1995;57:289–300. [Google Scholar]
- Black JC, Van Rechem C, Whetstine JR. Histone lysine methylation dynamics: establishment, regulation, and biological impact. Mol Cell. 2012;48:491–507. doi: 10.1016/j.molcel.2012.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonacci TM, Mathews JL, Yuan C, Lehmann DM, Malik S, Wu D, Font JL, Bidlack JM, Smrcka AV. Differential targeting of Gbetagamma-subunit signaling with small molecules. Science. 2006;312:443–446. doi: 10.1126/science.1120378. [DOI] [PubMed] [Google Scholar]
- Buck L, Axel R. A novel multigene family may encode odorant receptors: a molecular basis for odor recognition. Cell. 1991;65:175–187. doi: 10.1016/0092-8674(91)90418-x. [DOI] [PubMed] [Google Scholar]
- Celik A, Fuss SH, Korsching SI. Selective targeting of zebrafish olfactory receptor neurons by the endogenous OMP promoter. Eur J Neurosci. 2002;15:798–806. doi: 10.1046/j.1460-9568.2002.01913.x. [DOI] [PubMed] [Google Scholar]
- Chen S, Dell EJ, Lin F, Sai J, Hamm HE. RACK1 regulates specific functions of Gbetagamma. J Biol Chem. 2004;279:17861–17868. doi: 10.1074/jbc.M313727200. [DOI] [PubMed] [Google Scholar]
- Chen S, Lin F, Shin ME, Wang F, Shen L, Hamm HE. RACK1 regulates directional cell migration by acting on G betagamma at the interface with its effectors PLC beta and PI3K gamma. Mol Biol Cell. 2008;19:3909–3922. doi: 10.1091/mbc.E08-04-0433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chess A, Simon I, Cedar H, Axel R. Allelic inactivation regulates olfactory receptor gene expression. Cell. 1994;78:823–834. doi: 10.1016/s0092-8674(94)90562-2. [DOI] [PubMed] [Google Scholar]
- Clowney EJ, LeGros MA, Mosley CP, Clowney FG, Markenskoff-Papadimitriou EC, Myllys M, Barnea G, Larabell CA, Lomvardas S. Nuclear aggregation of olfactory receptor genes governs their monogenic expression. Cell. 2012;151:724–737. doi: 10.1016/j.cell.2012.09.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalton RP, Lyons DB, Lomvardas S. Co-opting the unfolded protein response to elicit olfactory receptor feedback. Cell. 2013;155:321–332. doi: 10.1016/j.cell.2013.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del Bene F, Wyart C, Robles E, Tran A, Looger L, Scott EK, Isacoff EY, Baier H. Filtering of visual information in the tectum by an identified neural circuit. Science. 2010;330:669–673. doi: 10.1126/science.1192949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeMaria S, Ngai J. The cell biology of smell. J Cell Biol. 2010;191:443–452. doi: 10.1083/jcb.201008163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupre DJ, Robitaille M, Rebois RV, Hebert TE. The role of Gbetagamma subunits in the organization, assembly, and function of GPCR signaling complexes. Annu Rev Pharmacol Toxicol. 2009;49:31–56. doi: 10.1146/annurev-pharmtox-061008-103038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekker M, Fritz A, Westerfield M. Identification of two families of satellite-like repetitive DNA sequences from the zebrafish (Brachydanio rerio) Genomics. 1992;13:1169–1173. doi: 10.1016/0888-7543(92)90033-o. [DOI] [PubMed] [Google Scholar]
- Faure M, Voyno-Yasenetskaya TA, Bourne HR. cAMP and beta gamma subunits of heterotrimeric G proteins stimulate the mitogen-activated protein kinase pathway in COS-7 cells. J Biol Chem. 1994;269:7851–7854. [PubMed] [Google Scholar]
- Feinstein P, Bozza T, Rodriguez I, Vassalli A, Mombaerts P. Axon guidance of mouse olfactory sensory neurons by odorant receptors and the beta2 adrenergic receptor. Cell. 2004;117:833–846. doi: 10.1016/j.cell.2004.05.013. [DOI] [PubMed] [Google Scholar]
- Imai T, Sakano H. Odorant receptor-mediated signaling in the mouse. Curr Opin Neurobiol. 2008;18:251–260. doi: 10.1016/j.conb.2008.07.009. [DOI] [PubMed] [Google Scholar]
- Imai T, Suzuki M, Sakano H. Odorant receptor-derived cAMP signals direct axonal targeting. Science. 2006;314:657–661. doi: 10.1126/science.1131794. [DOI] [PubMed] [Google Scholar]
- Jie Z, Li T, Jia-Yun H, Qiu J, Ping-Yao Z, Houyan S. Trans-2-phenylcyclopropylamine induces nerve cells apoptosis in zebrafish mediated by depression of LSD1 activity. Brain Res Bull. 2009;80:79–84. doi: 10.1016/j.brainresbull.2009.04.013. [DOI] [PubMed] [Google Scholar]
- Jones DT, Masters SB, Bourne HR, Reed RR. Biochemical characterization of three stimulatory GTP-binding proteins: the large and small forms of Gs and the olfactory-specific G-protein, Golf. J Biol Chem. 1990;265:2671–2676. [PubMed] [Google Scholar]
- Kajiya K, Inaki K, Tanaka M, Haga T, Kataoka H, Touhara K. Molecular bases of odor discrimination: Reconstitution of olfactory receptors that recognize overlapping sets of odorants. J Neurosci. 2001;21:6018–6025. doi: 10.1523/JNEUROSCI.21-16-06018.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerr DS, Von Dannecker LE, Davalos M, Michaloski JS, Malnic B. Ric-8B interacts with G alpha olf and G gamma 13 and co-localizes with G alpha olf, G beta 1 and G gamma 13 in the cilia of olfactory sensory neurons. Mol Cell Neurosci. 2008;38:341–348. doi: 10.1016/j.mcn.2008.03.006. [DOI] [PubMed] [Google Scholar]
- Koch WJ, Hawes BE, Allen LF, Lefkowitz RJ. Direct evidence that Gi-coupled receptor stimulation of mitogen-activated protein kinase is mediated by G beta gamma activation of p21ras. Proc Natl Acad Sci U S A. 1994;91:12706–12710. doi: 10.1073/pnas.91.26.12706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- Kubicek S, O’Sullivan RJ, August EM, Hickey ER, Zhang Q, Teodoro ML, Rea S, Mechtler K, Kowalski JA, Homon CA, et al. Reversal of H3K9me2 by a small-molecule inhibitor for the G9a histone methyltransferase. Mol Cell. 2007;25:473–481. doi: 10.1016/j.molcel.2007.01.017. [DOI] [PubMed] [Google Scholar]
- Lee MG, Wynder C, Schmidt DM, McCafferty DG, Shiekhattar R. Histone H3 lysine 4 demethylation is a target of nonselective antidepressive medications. Chem Biol. 2006;13:563–567. doi: 10.1016/j.chembiol.2006.05.004. [DOI] [PubMed] [Google Scholar]
- Lewcock JW, Reed RR. A feedback mechanism regulates monoallelic odorant receptor expression. Proc Natl Acad Sci USA. 2004;101:1069–1074. doi: 10.1073/pnas.0307986100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F, Ponissery-Saidu S, Yee KK, Wang H, Chen ML, Iguchi N, Zhang G, Jiang P, Reisert J, Huang L. Heterotrimeric G protein subunit Ggamma13 is critical to olfaction. J Neurosci. 2013;33:7975–7984. doi: 10.1523/JNEUROSCI.5563-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyons DB, Allen WE, Goh T, Tsai L, Barnea G, Lomvardas S. An epigenetic trap stabilizes singular olfactory receptor expression. Cell. 2013;154:325–336. doi: 10.1016/j.cell.2013.06.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magklara A, Yen A, Colquitt BM, Clowney EJ, Allen W, Markenscoff-Papadimitriou E, Evans ZA, Kheradpour P, Mountoufaris G, Carey C, et al. An epigenetic signature for monoallelic olfactory receptor expression. Cell. 2011;145:555–570. doi: 10.1016/j.cell.2011.03.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCullagh P, Nelder JA. Generalized Linear Models. 2nd edn Chapman & Hall/CRC Press; 1989. [Google Scholar]
- Mombaerts P. Genes and ligands for odorant, vomeronasal and taste receptors. Nat Rev Neurosci. 2004;5:263–278. doi: 10.1038/nrn1365. [DOI] [PubMed] [Google Scholar]
- Mombaerts P, Wang F, Dulac C, Chao SK, Nemes A, Mendelsohn M, Edmondson J, Axel R. Visualizing an olfactory sensory map. Cell. 1996;87:675–686. doi: 10.1016/s0092-8674(00)81387-2. [DOI] [PubMed] [Google Scholar]
- Nakashima A, Takeuchi H, Imai T, Saito H, Kiyonari H, Abe T, Chen M, Weinstein LS, Yu CR, Storm DR, et al. Agonist-independent GPCR activity regulates anterior-posterior targeting of olfactory sensory neurons. Cell. 2013;154:1314–1325. doi: 10.1016/j.cell.2013.08.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen MQ, Zhou Z, Marks CA, Ryba NJ, Belluscio L. Prominent roles for odorant receptor coding sequences in allelic exclusion. Cell. 2007;131:1009–1017. doi: 10.1016/j.cell.2007.10.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niimura Y, Nei M. Evolutionary dynamics of olfactory receptor genes in fishes and tetrapods. Proc Natl Acad Sci U S A. 2005;102:6039–6044. doi: 10.1073/pnas.0501922102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen SG, Jensen AD, Liapakis G, Ghanouni P, Javitch JA, Gether U. Mutation of a highly conserved aspartic acid in the beta2 adrenergic receptor: constitutive activation, structural instability, and conformational rearrangement of transmembrane segment 6. Mol Pharmacol. 1999;56:175–184. doi: 10.1124/mol.56.1.175. [DOI] [PubMed] [Google Scholar]
- Ressler KJ, Sullivan SL, Buck LB. Information coding in the olfactory system: evidence for a stereotyped and highly organized epitope map in the olfactory bulb. Cell. 1994;79:1245–1255. doi: 10.1016/0092-8674(94)90015-9. [DOI] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenbaum DM, Rasmussen SG, Kobilka BK. The structure and function of G-protein-coupled receptors. Nature. 2009;459:356–363. doi: 10.1038/nature08144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakano H. Neural map formation in the mouse olfactory system. Neuron. 2010;67:530–542. doi: 10.1016/j.neuron.2010.07.003. [DOI] [PubMed] [Google Scholar]
- Serizawa S, Miyamichi K, Nakatani H, Suzuki M, Saito M, Yoshihara Y, Sakano H. Negative feedback regulation ensures the one receptor-one olfactory neuron rule in mouse. Science. 2003;302:2088–2094. doi: 10.1126/science.1089122. [DOI] [PubMed] [Google Scholar]
- Serizawa S, Miyamichi K, Sakano H. One neuron-one receptor rule in the mouse olfactory system. Trends Genet. 2004;20:648–653. doi: 10.1016/j.tig.2004.09.006. [DOI] [PubMed] [Google Scholar]
- Serizawa S, Miyamichi K, Takeuchi H, Yamagishi Y, Suzuki M, Sakano H. A neuronal identity code for the odorant receptor-specific and activity-dependent axon sorting. Cell. 2006;127:1057–1069. doi: 10.1016/j.cell.2006.10.031. [DOI] [PubMed] [Google Scholar]
- Shykind BM. Regulation of odorant receptors: one allele at a time. Hum Mol Genet. 2005;14:R33–39. doi: 10.1093/hmg/ddi105. Spec No 1. [DOI] [PubMed] [Google Scholar]
- Shykind BM, Rohani SC, O’Donnell S, Nemes A, Mendelsohn M, Sun Y, Axel R, Barnea G. Gene switching and the stability of odorant receptor gene choice. Cell. 2004;117:801–815. doi: 10.1016/j.cell.2004.05.015. [DOI] [PubMed] [Google Scholar]
- Vassar R, Chao SK, Sitcheran R, Nunez JM, Vosshall LB, Axel R. Topographic organization of sensory projections to the olfactory bulb. Cell. 1994;79:981–991. doi: 10.1016/0092-8674(94)90029-9. [DOI] [PubMed] [Google Scholar]
- Vedadi M, Barsyte-Lovejoy D, Liu F, Rival-Gervier S, Allali-Hassani A, Labrie V, Wigle TJ, Dimaggio PA, Wasney GA, Siarheyeva A, et al. A chemical probe selectively inhibits G9a and GLP methyltransferase activity in cells. Nat Chem Biol. 2011;7:566–574. doi: 10.1038/nchembio.599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F, Nemes A, Mendelsohn M, Axel R. Odorant receptors govern the formation of a precise topographic map. Cell. 1998;93:47–60. doi: 10.1016/s0092-8674(00)81145-9. [DOI] [PubMed] [Google Scholar]
- Welten MC, de Haan SB, van den Boogert N, Noordermeer JN, Lamers GE, Spaink HP, Meijer AH, Verbeek FJ. ZebraFISH: fluorescent in situ hybridization protocol and three-dimensional imaging of gene expression patterns. Zebrafish. 2006;3:465–476. doi: 10.1089/zeb.2006.3.465. [DOI] [PubMed] [Google Scholar]
- Wong ST, Trinh K, Hacker B, Chan GC, Lowe G, Gaggar A, Xia Z, Gold GH, Storm DR. Disruption of the type III adenylyl cyclase gene leads to peripheral and behavioral anosmia in transgenic mice. Neuron. 2000;27:487–497. doi: 10.1016/s0896-6273(00)00060-x. [DOI] [PubMed] [Google Scholar]
- Zhang X, Firestein S. The olfactory receptor gene superfamily of the mouse. Nat Neurosci. 2002;5:124–133. doi: 10.1038/nn800. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.