Abstract
Enhanced vascular arginase activity can impair endothelium-dependent vasorelaxation by decreasing L-arginine availability to endothelial nitric oxide (NO) synthase, thereby reducing NO production and uncoupling NOS function. Elevated angiotensin II (Ang II) is a key component of endothelial dysfunction in many cardiovascular diseases and has been linked to elevated arginase activity. In this study we explored the signaling pathway leading to increased arginase expression/activity in response to Ang II in bovine aortic endothelial cells (BAEC). Our previous studies indicate involvement of p38 mitogen activated protein kinase (MAPK) in Ang II-induced arginase upregulation and reduced NO production. In this study, we further investigated the Ang II-transcriptional regulation of arginase 1 in endothelial cells. Our results indicate the involvement of ATF-2 transcription factor of the AP1 family in arginase 1 upregulation and in limiting NO production. Using small interfering RNA (siRNA) targeting ATF-2, we showed that this transcription factor is required for Ang II-induced arginase 1 gene upregulation and increased arginase 1 expression and activity, leading to reduced NO production. Electrophoretic mobility shift assay and chromatin immunoprecipitation assay further confirmed the involvement of ATF-2. Moreover, our data indicate that p38 MAPK phosphorylates ATF-2 in response to Ang II. Collectively, our results indicate that Ang II increases endothelial arginase activity/expression through a p38 MAPK/ATF-2 pathway leading to reduced endothelial NO production. These signaling steps might be therapeutic targets for preventing vascular endothelial dysfunction associated with elevated arginase activity/expression.
Keywords: Arginase, nitric oxide (NO), transcription factors, ATF-2, p38 MAPK
1. INTRODUCTION
Arginase is the hydrolytic enzyme responsible for conversion of L-arginine into urea and L-ornithine (Demougeot et al., 2005; Yang et al., 2006). There are two distinct isoforms of arginase; arginase 1, largely found in the liver as a component of the urea cycle and arginase 2, which is predominant in kidney. Both isoforms, however, have been found in vascular tissue (Bachetti et al., 2004; Berkowitz et al., 2003).
Elevated arginase activity has been associated with cardiovascular pathologies such as hypertension, diabetes, atherosclerosis, ischemic reperfusion injury, erectile dysfunction and sickle cell anemia (Bagnost et al., 2008; Bivalacqua et al., 2007; Jeyabalan et al., 2008; Morris et al., 2005; Romero et al., 2008). In these conditions, elevation of arginase has been shown to mediate vascular dysfunction through limiting nitric oxide (NO) production or availability. Arginase can reciprocally regulate NO production in endothelial cells by competing with nitric oxide synthase (NOS) for the substrate L-arginine (Bagnost et al., 2008; Berkowitz et al., 2003; Romero et al., 2008).
Angiotensin II (Ang II) actions in endothelial cells are mostly associated with endothelial NOS (eNOS) dysfunction and uncoupling, which lead to decreased levels of NO and increased superoxide production (Satoh et al., 2008).
Elevated arginase activity also has been associated with systemic hypertension. Inhibition of arginase has been reported to decrease blood pressure and improve vascular function of resistance vessels in adult hypertensive rats (Bagnost et al., 2008; Demougeot et al., 2005). These findings thus suggest a central role for arginase in diseases in which vascular dysfunction is linked to elevated levels of Ang II.
However, it should be emphasized that global inhibition of arginase in the body may be dangerous. Arginase 1 is a crucial enzyme in the urea cycle for disposal of harmful ammonia and complete knock-out of the arginase 1 gene in mice is lethal by 2 weeks of age because of hyper-ammonemia (Iyer et al., 2002). Use of arginase inhibitors carries the risk of reducing its function to very low levels. Hence, identifying the steps - e.g. signaling proteins or transcription factors - that directly enhance arginase in endothelial cells may be very beneficial targets in specifically limiting vascular arginase activity without unwanted global side effects.
We have reported a role for arginase 1 upregulation in vascular dysfunction in a model of Ang II-induced vascular endothelial dysfunction and hypertension (Shatanawi et al., 2011). We have also shown that Ang II elevates arginase through a RhoA/Rho kinase (ROCK) and p38 mitogen activated protein kinase (MAPK) pathway. Others have reported that Ang II elevates arginase 1 levels in isolated rat periglomerular vessels (Hultstrom et al., 2009). Additionally, high glucose and reactive oxygen species increase arginase activity in bovine coronary endothelial cells via a RhoA/ROCK mechanism (Chandra et al., 2012; Romero et al., 2008).
Given the importance of endothelial arginase in causing eNOS dysfunction, and the link of arginase with vascular diseases associated with elevated levels of Ang II, we sought to define the transcriptional regulation of arginase 1 in response to Ang II in relation to arginase activity and NO production. Transcriptional regulation of arginase in response to thrombin in endothelial cells has been demonstrated to occur through activator protein-1 (AP-1) consensus site (Zhu et al., 2009). More specifically, two transcription factors that bind to that site were identified - activating transcription factor-2 (ATF-2) and c-Jun. Our work examined the involvement of these two transcription factors in arginase 1 elevation in endothelial cells in response to Ang II and their impact on vascular disease through limiting NO.
2. MATERIALS AND METHODS
2.1 Cell Culture and Treatments
In all cell experiments, bovine aortic endothelial cells (BAECs) were utilized. Proliferating BAECs were purchased from Cell Applications, San Diego, CA. Cells were cultured in Endothelial Growth Medium (Cell Applications, San Diego, CA) and maintained in a humidified atmosphere at 37°C and 5% CO2. Before starting experiments, cells were adapted to grow in M199 supplemented with 50 µM L-arginine (Invitrogen, Carlsbad, CA) for 72 h to match the normal plasma L-arginine concentration which ranges from 40 to 100 µM (Romero et al., 2006). In addition, the medium was supplemented with 10% FBS (Catalog # SH30396, hyClone, GE Healthcare Life Sciences South Logan, Utah), 1% penicillin/streptomycin, and 1% L-glutamine. When cells reached 80% confluency, they then were serum starved overnight in M199 supplemented with 50 µM L-arginine, 1% L-glutamine, 1% penicillin/streptomycin and 0.2% FBS. The p38 MAPK inhibitor, SB-202190 (2 µM) (EMD biosciences, San Diego, CA), was used in some experiments and added 2 h before the addition of angiotensin II (0.1 µM, for different time points) (Sigma Aldrich, St. Louis, MO). All experiments were performed with cells from passage 3–7.
2.2 Luciferase Activity
Luciferase constructs used were generously provided by Prof. Sydney M. Morris, Jr. of the University of Pittsburgh. The constructs were transformed in E Coli competent cells (Novablue) then amplified and extracted using EndoFree ® plasmid purification kit (Qiagen, Valencia, CA).
BAECS were co-transfected with one of three arginase 1 luciferase constructs and a Renilla luciferase gene (Promega) as an internal control using Lipofectin reagent (Invitrogen, Carlsbad, CA) according to the manufacturer’s instructions. After 48 h of transfection, cells were treated with Ang II. In another set of experiments the cells also received co-transfection with siRNA for ATF-2 and c-Jun or non-targeting sc-RNA. All treatments were performed in triplicate.
Firefly luciferase activity was measured for reporter expression according to the instructions provided in the Dual-Luciferase® Reporter Assay System (Promega, San Luis Obispo, CA). Transfection efficiency was corrected by co-transfection with a plasmid containing the Renilla luciferase gene (Promega). Both Firefly and Renilla luciferase activity were measured within the same sample of cell lysate sequentially using one reaction tube. All chemiluminescence readings were obtained using a microplate luminometer (POLARStar OPTIMA, BMG Labtech). Briefly, 20 µL of cell lysate were added to a microplate well followed by adding 100 µL of luciferase substrate solution (as provided in the kit). The firefly luciferase activity (AFL) was measured immediately using 15 s as total reading time. The same well then received 100 µL of Stop & Glo reagent (contains substance to quench the enzymatic activity of Firefly and a substrate for Renilla luciferase). Renilla luciferase activity (ARL) was measured immediately using 15 s as total reading time. The corrected activity (AFL/ARL) was used to compare groups.
2.3 Arginase Activity
Arginase activity was measured using a colorimetric determination of urea production from L-arginine as described previously (Corraliza et al., 1994). Cells were in Tris buffer (50 mMTris-HCI, 0.1 mM EDTA and EGTA, pH 7.5) containing protease inhibitors (Catalog # P8340, Sigma, St. Louis, MO). These mixtures were subjected to three freeze-thaw cycles and then centrifuged for 10 m at 20,000 g. The supernatants were used for arginase activity assay.
In brief, 25 µL of supernatant was heated with MnCl2 (10 mM) for 10 m at 56°C to activate arginase. The mixture was then incubated with 50 µL L-arginine (0.5 M, pH 9.7) for one hour at 37°C to hydrolyze the L-arginine. The hydrolysis reaction was stopped with acid and the mixture was then heated at 100°C with 25 µL of α-isonitrosopropiophenone (9% α-ISPF in EtOH) for 45 m. The samples were kept in dark at room temperature for 10 m then absorbance was measured at 540 nm.
2.4 siRNA Transfection
BAECs were transfected with siRNA targeting ATF-2 or c-Jun (Dharmacon, Lafayette, CO) using siPORT Amine (Ambion, Austin, TX), according to the manufacturer’s instructions. Scrambled siRNA (non-targeting siRNA) served as control to validate the specificity of the siRNAs. In brief, cells were transfected with 50 nM of targeting or non-targeting siRNA for 48 h. Specific mRNA depletion was analyzed by Western blot.
2.5 Western Blot Analysis
Cells were lysed in Ripa buffer (Upstate Biotechnology, Temecula, CA) containing protease and phosphatase inhibitors (Catalog # P5726 and P0044, Sigma, St. Louis, MO). Cell lysates were centrifuged for 10 m at 20,000 g, and supernatants were collected for Western blotting analysis. Protein estimation was carried out in supernatants using protein assay kit (Bio Rad, Hercules, CA). Equal amounts of protein were loaded, separated by electrophoresis using 10% SDS-PAGE gels, and transferred into nitrocellulose membranes. The blots were blocked using 5% bovine serum albumin (Sigma, St. Louis, MO), incubated with their respective primary and secondary antibodies; anti-arginase 1 (Catalog # 610708, 1:1000, BD Biosciences, San Diego, CA), anti-phospho-ATF-2 (Catalog #9221, 1:1000), anti-total-ATF-2 (Catalog #9226 1:1000), anti-c-Jun (Catalog #9165, 1:1000) (Cell signaling, Boston, MA), anti-actin (Catalog #A2066, 1:1000, Sigma, St. Louis, MO), followed by the respective secondary antibodies. Signals were detected using chemiluminescence. To quantify the resultant blots, individual band intensities were measured (arbitrary units) and ratios of protein to actin were calculated per sample using NIH ImageJ software.
2.6 Electrophoretic Mobility Shift Assay (EMSA)
We also examined the binding of nuclear extracts of BAECs to the DNA probe of arginase 1. Nuclear extracts were prepared from vehicle versus Ang II treated BAECs using NE-PER Nuclear and Cytoplasmic Extraction Reagents (Thermo Fisher Scientific, Rockford, IL) according to the manufacturer’s protocol. Protein concentrations were determined in both nuclear and cytoplasmic extracts using a protein assay kit (Bio Rad, Hercules, CA). The sequences of arginase 1 promoter DNA probes 5' biotin labeled and encompassing the −3,157 bp AP-1 binding region were used for EMSA as reported in literature (Gray et al., 2005; Zhu et al., 2009). The sequences are as follows: wild-type AP-1 probe, CCA GTC TGA CTC TCA GAA CC and mutant AP-1 probe, CCA GTC GTC CTC TCA GAA CC. These sequences were custom synthesized by Integrated DNA Technologies (IDT, Coralville, IA). The Light Shift Chemiluminescent EMSA Kit (Catalog #20148 ,Thermo Fisher Scientific, Rockford, IL) was used to perform all EMSA experiments according to the company’s protocols. In brief, 5’ biotin end-labeled arginase 1 DNA (200 pM) was incubated with 25 µg/µl nuclear extract. The DNA -protein complexes were resolved by electrophoresis under native conditions on 6% polyacrylamide DNA retardation gels (Invitrogen, Carlsbad, CA). The DNA-protein complex was then transferred to a positively charged nylon membrane (Zeta-Probe®, Bio Rad, Hercules, CA) and crosslinked using UV lamp (λmax=254 nm). The biotin labeled DNA-protein complex was then probed with streptavidin-HRP conjugate, incubated with the substrate (peroxide), and detected with chemiluminescence.
2.7 Supershift Assay
ATF-2 antibody (dilution 1:100, Cell Signaling, Boston, MA) was incubated with the nuclear extracts of Ang II treated cells at 4°C overnight, and then incubated with the labeled arginase 1 promoter DNA probes for 30 m at room temperature. The DNA–protein complexes were resolved by electrophoresis on 6% polyacrylamide gels as described above in EMSA section (Sarge et al., 1993).
2.8 Chromatin Immunoprecipitation (ChIP) Assay
ChIP assay were performed in BAECs treated with Ang II versus vehicle treated control. The ChIP assay was performed using a kit (Upstate biotechnology, Temecula, CA) according to the manufacturer’s instructions. Cells were cross-linked, lysed, and sonicated three times at 4°C.
Two % of the sample was kept to be used as input DNA control. Remaining cell lysates were incubated with either ATF-2, c-Jun (Santa Cruz Biotechnology, Santa Cruz, CA) or IgG antibody (negative control) overnight with rotation, followed by 2 h incubation with protein A-agarose for ChIP.
The precipitated chromatin-protein complexes were washed and eluted, and then crosslinking was reversed. After isolation of the precipitated DNA, samples were quantified by qPCR. The following primers were custom designed for bovine arginase 1 promoter (IDT, Coralville, IA), forward GCTGGTCTGCTTGAGAAACTTAAAG and reverse GCAAAGGACAGGTCCCCATA. Analysis was performed using delta-delta Ct method.
2.9 Nitric Oxide (NO) Measurement
To measure NO, nitrite (NO2) the stable breakdown product of NO in the cell conditioned medium was analyzed using NO-specific chemiluminescence (Archer, 1993). After cells were treated, medium was replaced with fresh M199 for 30 m, and medium aliquots then were collected for basal reading. Cells were then exposed to the calcium ionophore ionomycin (1 µM) (Sigma Aldrich, St. Louis, MO) for 30 m and medium samples were collected.
In brief, samples containing NO2 were injected into glacial acetic acid containing sodium iodide. NO2 is quantitatively reduced to NO under these conditions, which can be quantified by a chemiluminescence detector after reaction with ozone in a NO analyzer (Sievers, Boulder, CO). The amount of NO generated is calculated as the difference in basal and ionomycin-stimulated NO levels.
2.10 Statistical Analysis
Data are given as mean ± S.E.M.. For multiple comparisons, statistical analysis was performed by one-way analysis of variance (ANOVA) with the Tukey post-test. For single comparisons, statistical differences were determined by the Student T test. Experiments were performed 3–6 times. All statistical analyses were performed with GraphPad Prism version 4.03 (San Diego, CA). Results were considered significant when P<0.05.
3. RESULTS
3.1 Arginase 1 promoter luciferase constructs and arginase 1 gene transcriptional activity in Ang II treated BAECs
We used a promoter driven luciferase reporter gene to determine the transcriptional activity of the arginase gene after Ang II treatment. The −4.78 kb Luc construct represents the full length arginase 1 promoter. 5′-deletion constructs at −3.29 and −2.78 kb were also used. These constructs have been used to assess arginase 1 transcriptional activity in macrophages (Gray et al., 2005) and in rat aortic endothelial cells (Zhu et al., 2009).
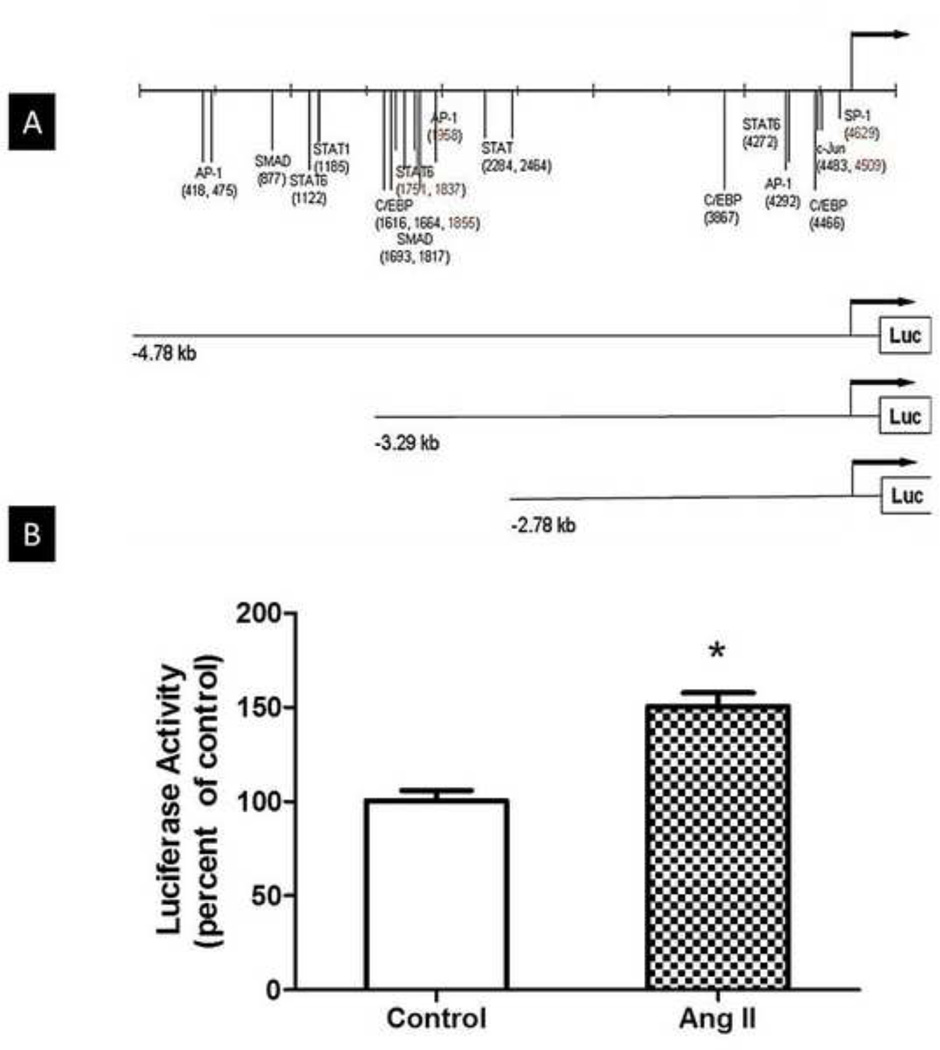
Fig. 1A shows a series of luciferase constructs driven by different lengths of the murine arginase 1 promoter and illustrates several putative transcription factor binding sequences in the 4.8 kb promoter region upstream of arginase 1 gene. This arginase promoter region is conserved among species and includes binding sites for AP-1, c-Jun, SMAD, C/EB, STAT1 and STAT6 (Gray et al., 2005; Serrat et al., 2012).
Fig 1. Arginase 1 promoter luciferase constructs and luciferase activity in Ang II treated BAECs.
(A) Scheme representing a series of luciferase constructs driven by different lengths of the murine arginase 1 promoter and illustrating several putative transcription factor binding sequences in the 4.8 kb promoter region upstream of arginase 1 gene. (B) BAECs were co-transfected with the full length luciferase construct- −4.78kb arginase 1 promoter-Luc for 48 h and Renilla luciferase gene plasmid to normalize for transfection efficiency. Cells were then treated with Ang II (0.1 µM, 24 h). Luciferase activity shows a 50% increase with Ang II treatment versus transfected untreated cells (control). n=4 times each performed in triplicates; *P<0.05 vs. control.
BAECs were transfected with the −4.78 kb Luc construct and co-transfected a Renilla luciferase gene plasmid to normalize for transfection efficiency. We demonstrated that Ang II induces an increase in arginase gene expression using this Luciferase–based approach. Ang II (0.1 µM, 24 h) treatment resulted in a ~50% increase in luciferase activity compared to untreated transfected BAECs (Fig. 1B), indicating enhanced arginase 1 transcriptional activity in response to Ang II treatment. These results also indicate the presence of a responsive element within the arginase 1 promoter site.
3.2 Analysis of the arginase 1 promoter in response to Ang II
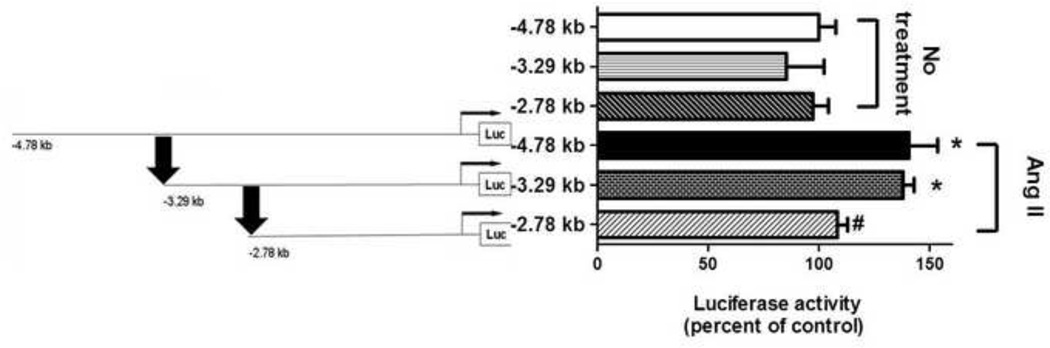
BAECs were next transfected with one of the three different arginase 1 promoter Luc constructs: the −4.78 kb Luc construct that represents the full length arginase 1 promoter, or 5′-deletion constructs at −3.29 and −2.78 kb. The cells were co-transfected with a Renilla luciferase gene plasmid to normalize for transfection efficiency. Cells then were treated with Ang II (0.1 µM, 24 h). Fig. 2 shows that Ang II caused an increase in luciferase activity of ~40% and ~55% versus the untreated controls in −4.78 and −3.29 arginase 1 promoter-Luc transfected cells, respectively (P<0.05). Ang II treatment of cells transfected with −2.78 kb arginase 1 promoter-Luc did not enhance luciferase activity. There was no difference in luciferase activity among cells transfected with any of the three different 5′-deletion arginase 1 promoter-Luc constructs under basal untreated conditions. These results showed that the Ang II-responsive element is located between −3.29 kb to −2.78 kb in the arginase 1 promoter region as indicated by the arrows (Fig. 2). This arginase promoter region includes binding sites for AP-1, SMAD, C/EBP,STAT1 and STAT6 (Gray et al., 2005; Serrat et al., 2012).
Fig 2. Analysis of the arginase 1 promoter in response to Ang II.
BAECs were co-transfected with one of three different deletion arginase 1 promoter-luciferase (Luc) constructs and Renilla luciferase gene plasmid to normalize for transfection efficiency. Luciferase activity induction showed that the Ang II-responsive element was located between −3.29 kb to −2.78 kb in the arginase 1 promoter as arrows indicate. n=3 each performed in triplicates; *P<0.05 vs. corresponding length of promoter (no treatment), #P<0.05 vs Ang II.
3.3 Role of AP-1 complex proteins: ATF-2 and c-Jun in Ang II-induced arginase 1 luciferase activity
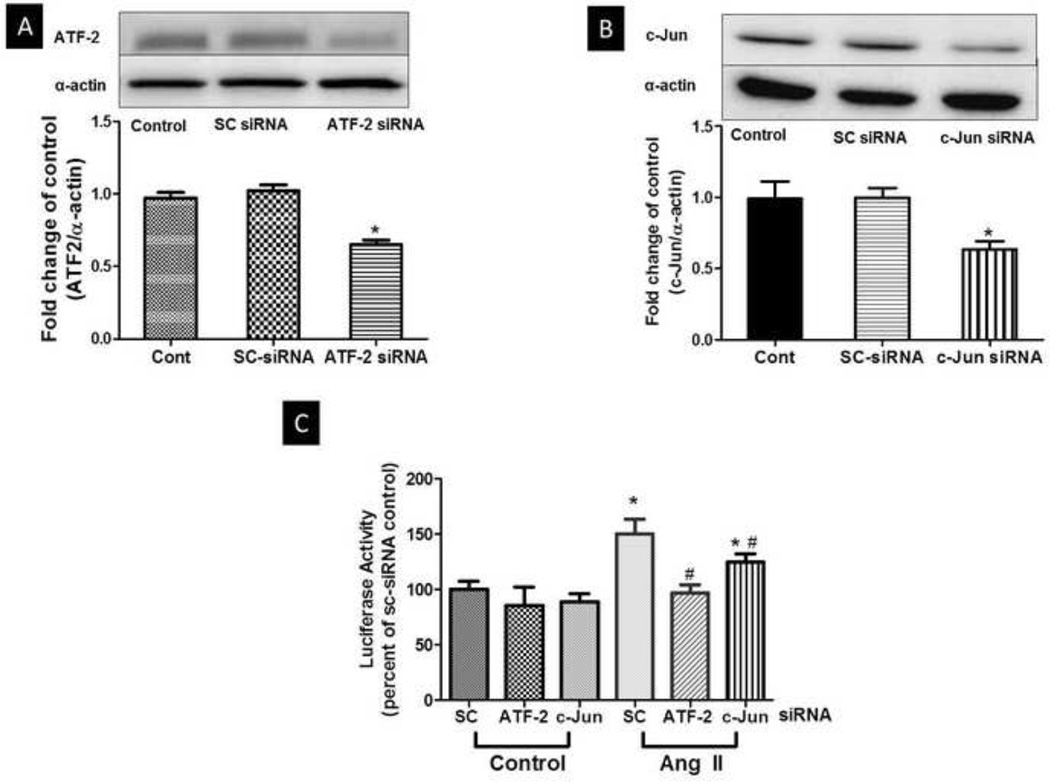
We next determined the role of the AP-1 protein complex in Ang II-induced arginase 1 luciferase activity with the full length −4.78 kb arginase 1 promoter-Luc construct. Both ATF-2 and c-Jun are members of activator protein 1 (AP-1) complex. To determine the role of ATF-2 and c-Jun in arginase 1 gene induction, we co-transfected BAECs with non-targeting scrambled siRNA (sc-siRNA) or siRNA targeting either ATF-2 or c-Jun in addition to transfection with the −4.78 kb arginase 1 promoter-Luciferase construct and the Renilla luciferase gene. Efficiency of the siRNA transfection for ATF-2 and c-Jun was determined by Western blot analysis. Analysis of the Western blots shows a 35% decrease in total ATF-2 protein expression, while c-Jun siRNA transfection produced 48% decrease in total c-Jun protein expression. SC siRNA transfection had no effect on ATF-2 or c-Jun protein levels. (Fig. 3A, 3B). After 48 h of transfection, cells were treated with Ang II (0.1 µM, 24 h) after which luciferase activity was measured.
Fig 3. Role of ATF-2 and c-Jun in Ang II-induced arginase 1 luciferase activity in BAECs.
BAECs were co-transfected with the full length luciferase construct- −4.78kb arginase 1 promoter-Luc for 48 h and Renilla luciferase gene plasmid to normalize for transfection efficiency. In addition co-transfection of BAECs with non-targeting siRNA (SC siRNA) or siRNA targeting either ATF-2 or c-Jun was performed. Cells were then treated with Ang II (0.1 µM, 24 h). (A) Representation of Western blot probed with ATF-2 antibody shows the efficiency of ATF-2 siRNA transfection. Band intensities from multiple experiments were quantified and normalized to α-actin. n=3 (B) Representation of Western blot probed with c-Jun antibody shows the efficiency of c-Jun siRNA transfection. Band intensities from multiple experiments were quantified and normalized to α-actin. n=3 (C) Luciferase activity was measured in cell lysate, normalized to Renilla luciferase and expressed as percent of scrambled (SC) siRNA transfected controls. n=3 each performed in triplicates; *P<0.05 vs. SC control, #P<0.05 vs. SC Ang II.
Luciferase activity was elevated in cells co-transfected with sc-siRNA and treated with Ang II as compared with sc-siRNA control cells (Fig. 3C) with levels being similar to those in cells not transfected with siRNA. On the other hand, knock-down of ATF-2 protein prevented the enhancement of luciferase activity induced by Ang II. c-Jun knockdown only partially inhibited (~25%) the Ang II-induced enhancement of luciferase activity (Fig. 3C). These results indicate a key role of ATF-2 in Ang II-induced arginase 1 gene transcription and also a potential cooperative role of c-Jun.
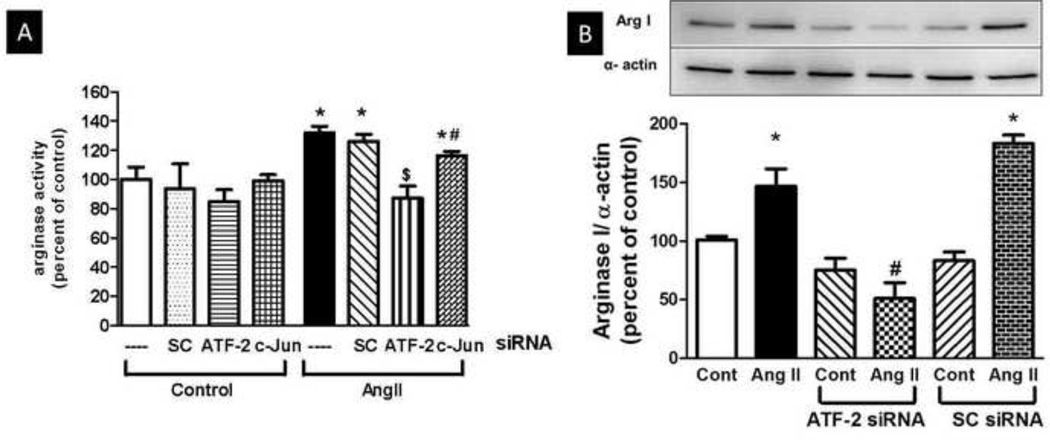
3.4 Role of ATF-2 in enhanced arginase activity and expression in endothelial cells
For this purpose, BAECs were transfected with either SC-siRNA or authentic siRNA targeting ATF-2 or c-Jun for 48 h and then treated with Ang II (0.1 µM, 24 h). Ang II treatment elevated arginase activity in non-transfected and sc-siRNA transfected cells. Reducing the levels of ATF-2 protein by siRNA prevented the Ang II-induced elevation of arginase activity (Fig. 4A). Transfection with c-Jun siRNA only partially reduced (~16%) the Ang II induced elevation of arginase activity (Fig. 4A). ATF-2 is also involved in upregulating arginase 1 protein expression under Ang II treatment. BAECs were transfected with either non targeting sc-siRNA or siRNA targeting ATF-2 for 48 h and then treated with Ang II (0.1 µM, 24 h). While Ang II caused elevation of arginase 1 expression in non-transfected and sc-siRNA transfected cells, it did not induce an increase in ATF-2 siRNA transfected cells (Fig. 4B). Moreover, knockdown of ATF-2 expression reduced arginase activity below non-siRNA control levels, both with and without Ang II treatment. These results indicate that ATF-2 controls arginase 1 protein levels (Fig. 4B) Efficiency of the transfection determined by Western blot of c-Jun and ATF-2 levels in cell lysates normalized to α-actin showed a marked reduction in expression levels of both proteins (see Fig. 3A and 3B).
Fig 4. Arginase Activity and Arginase 1 Expression in BAECs.
BAECs were transfected with either siRNA for ATF-2, c-Jun or scrambled (SC) siRNA. BAECs were then treated with Ang II (0.1 µM, 24h) (A) Elevation of arginase activity by exposure of BAECs to Ang II was prevented by transfection of cells with ATF-2 siRNA and partially prevented by c-Jun transfection. Transfection with sc-siRNA did not prevent response to Ang II. n= 3 *P<0.05 vs. control, #P<0.05 vs. SC siRNA for Ang II. (B) Elevation of arginase 1 expression by exposure of BAECs to Ang II (0.1 µM, 24 h) was prevented by transfection of cells with ATF-2 siRNA. Transfection with sc-siRNA did not prevent response to Ang II. Values are expressed as means of percent of control ± S.E.M..n= 3. *P<0.05 vs. control, #P<0.05 vs. SC siRNA for Ang II.
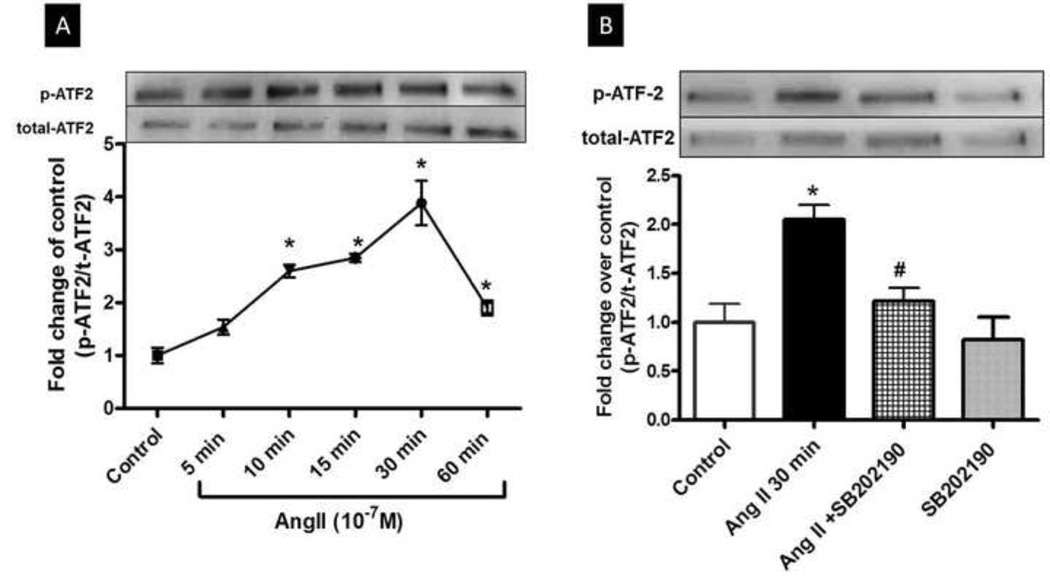
3.5 Ang II induces phosphorylation of activating transcription factor-2 (ATF-2)
BAECs were treated with Ang II for 5–60 m and effects on levels of phospho-ATF-2 and total ATF-2 were determined by Western blot. This analysis showed a time-dependent increase in phosphorylation which became evident at 10 m after exposure to Ang II and peaked at 30 m (Fig. 5A). Total ATF-2 appeared to increase in response to Ang II. Pretreatment with the p38 MAPK inhibitor SB-202190 (2 µM, 2 h) prevented these effect (Fig. 5B), indicating that ATF-2 is a downstream target of p38 MAPK. This finding agrees with reports indicating ATF-2 as a substrate for phosphorylation by p38 MAPK leading to its activation (Lopez-Bergami et al., 2010; Raingeaud et al., 1995).
Fig 5. Activation of ATF-2 by Ang II.
(A) BAECs exposed to Ang II (0.1 µM) for 5 to 60 mins show a time-dependent increase in phospho-ATF-2 expression first evident at 10 mins and peaking at 30 mins. (B) Pretreatment of BAEC with SB-202190 (2 µM, 2 h) blocked the phosphorylation of ATF-2 caused by Ang II (0.1 µM, 30 min). Representative autographs are shown. Values are expressed fold change over control ± S.E.M. of phospho-ATF-2/total-ATF-2 ratio. n=3 of independent experiments carried out in duplicate. *P<0.05 vs. control. #P<0.05 vs. Ang II.
3.6 Effect of Ang II on binding of nuclear proteins to the AP-1 site
We further performed the electrophoretic mobility shift assay (EMSA) to confirm that the AP-1 site in the arginase promoter was an Ang II-responsive regulatory element. Nuclear extracts were prepared from non-treated (control) or Ang II-treated BAECs. As stated in Materials and Methods, wild-type (WT) and mutant (MUT) double-stranded oligonucleotide probes were designed to encompass the AP-1 binding element and labeled with biotin. Nuclear extract treated with Ang II (0.1 µM, 30 m) showed enhanced binding complex with oligonucleotide containing the wild-type AP-1 site compared to untreated BAECs (Left panel, Fig. 6). The binding complex formation was determined by the presence of a band shift. No specific complex formation was observed for either nuclear extracts with the oligonucleotide containing the mutant AP-1 sequence (Right panel, Fig. 6).
Fig 6. Electrophoretic mobility shift assay in Ang II treated cells.
Electrophoretic mobility shift assay (EMSA) analysis was performed with nuclear extracts from control BAECs or Ang II treated BAECs (0.1 µM, 30 mins). Ang II treatment enhanced formation of binding complex between nuclear extracts with oligonucleotide containing the wild-type AP-1 site compared to untreated BAECs (Left panel). No specific complex formation was observed for either nuclear extracts with the oligonucleotide containing the mutant AP-1 sequence (Right panel). Result shown is representative of three independent experiments. n=3.
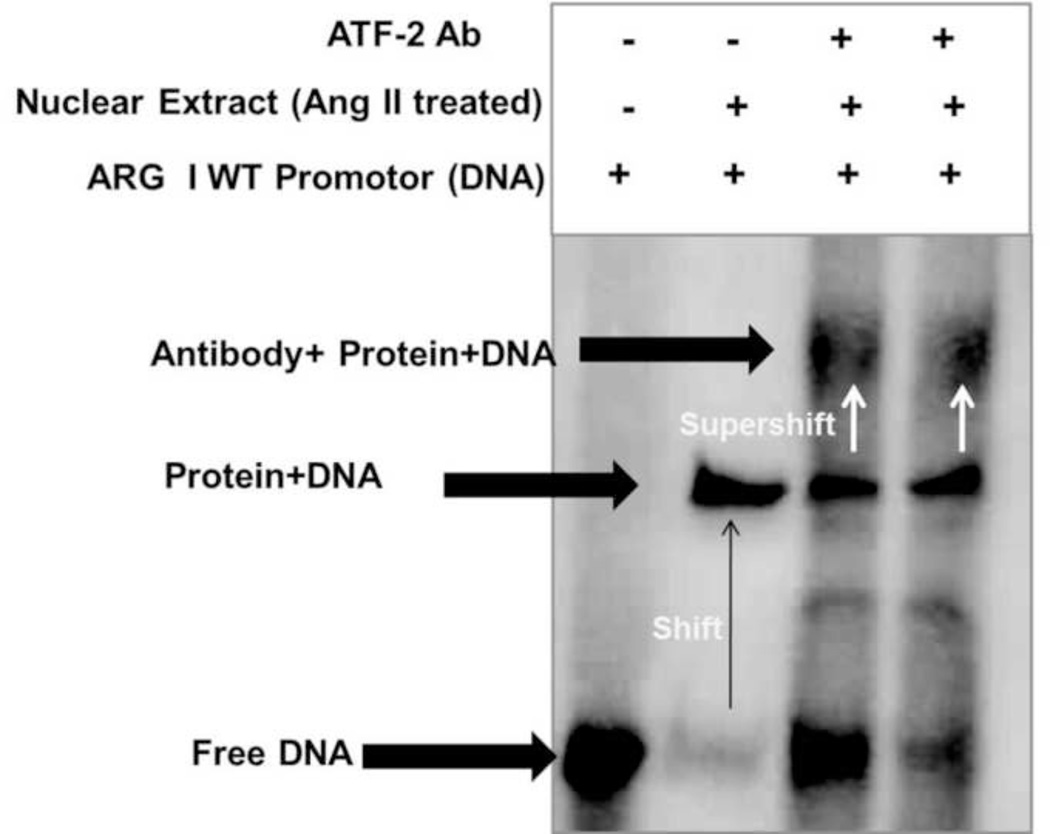
To determine whether ATF-2 binds to the arginase promoter in Ang II treated BAEC, supershift assays were performed with labeled wild type AP-1 probe. Ang II treated nuclear extracts were incubated with ATF-2 antibody before electrophoresis to determine if this treatment further decreases in electrophoretic mobility of the ATF-2 (supershift). Addition of ATF-2 antibody to the EMSA reaction mixture resulted in the formation of a larger complex of ATF-2 Ab/ATF-2 protein present in the nuclear extract and AP-1 probe. This new complex formation was detected by the appearance of another band that was shifted further (supershift) (Fig. 7). These results indicate that ATF-2 is binding to the AP-1 site in arginase promoter in BAEC.
Fig 7. Identification of activating transcription factor 2 (ATF-2) binding to AP-1 elements within the arginase 1 promoter using supershift assay.
Supershift assay was performed with nuclear extracts from BAECs treated with Ang II (0.1 µM, 30 mins) in the absence and presence ATF-2 specific antibody (1:100) before performing the EMSA with oligonucleotide containing the wild-type AP-1 site. Results shown are representative of three independent experiments. n=3.
3.7 Chromatin Immunoprecipitation (ChIP) analysis of ATF-2 association with the arginase 1 promoter
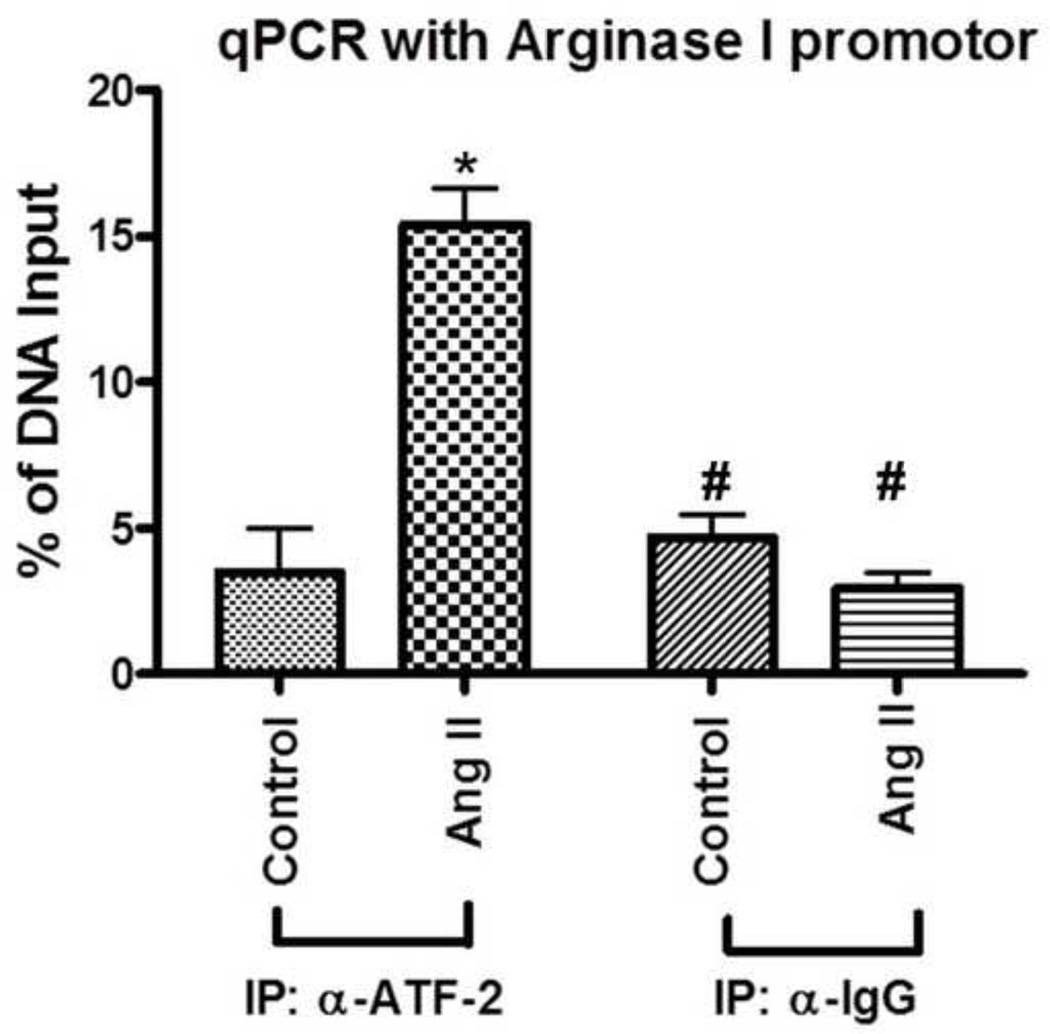
ChIP assays were performed with ATF-2 antibody and IgG (as a control) by using chromatin from untreated and Ang II-treated BAECs. We used bovine arginase 1 DNA primers and amplified DNA fragments from anti-ATF-2 chromatin precipitates in Ang II-treated cells. qPCR was performed with primers for the 18S promoter and for arginase 1 gene. The qPCR analysis was performed using delta-delta cycle times. Our results showed that anti-ATF-2 precipitates from untreated control BAECs resulted in little or no DNA amplification. However in cells treated with Ang II, anti-ATF-2 chromatin precipitates showed an amplification of DNA of about 5 folds (Fig. 8). These results demonstrate that Ang II increased specific binding of ATF-2 to the arginase 1 promoter in BAECs.
Fig 8. ChIP assay determines ATF-2 association with the arginase 1 promoter.
ChIP assays were performed in BAECs treated with Ang II versus untreated control. anti-ATF-2 antibody and arginase 1 PCR primer were used. IgG (5 µg/IP) was used as negative IP control. qPCR was performed with primers for the 18S promoter and for bovine arginase 1 gene. qPCR analysis was performed using delta-delta circle time (Ct). Fig. shows percentage of DNA input (relative amount of immunoprecipitated DNA compared to input DNA after qPCR analysis normalized to the internal control 18S). IP; Immunoprecipation, α-ATF-2; anti-ATF-2. n=3 of independent experiments. *P<0.05 vs. control, #P<0.05 vs. Ang II IP: α-ATF-2.
3.8 Role of ATF-2 in limiting NO production in to Ang II
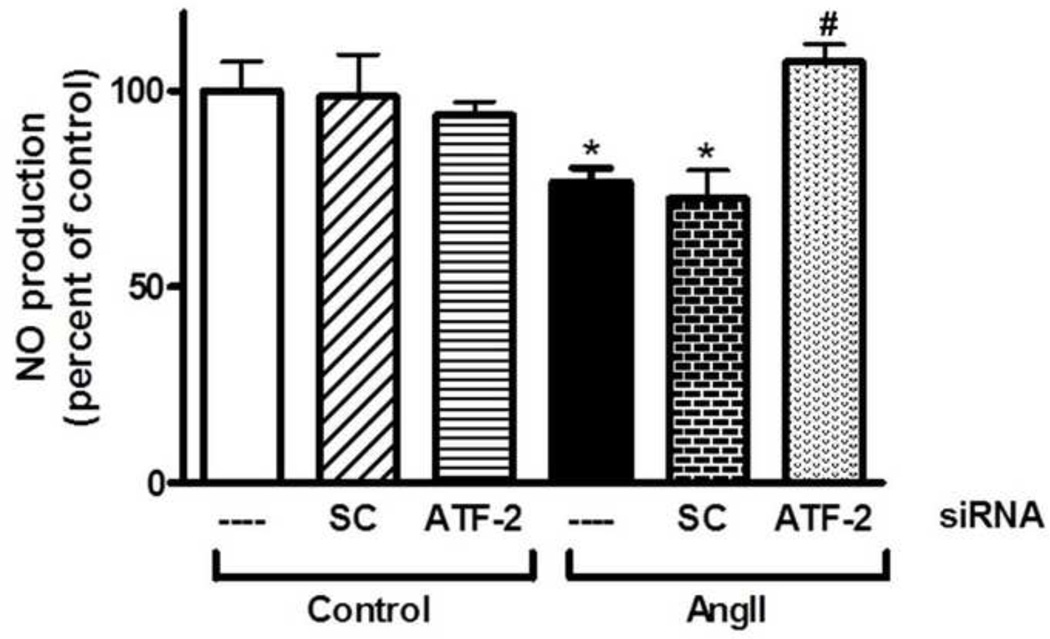
Since arginase and NOS share the same substrate, we determined the effect of Ang II on NO production from endothelial cells and the role of ATF-2 in this effect. BAECs were subjected to the same transfection treatments as in Fig. 4. Stimulated NO production measured in the cell media confirmed the role of ATF-2-induced arginase expression in reducing NO levels, showing a decrease in NO production with Ang II treatment that was also prevented with ATF-2 siRNA transfection (Fig. 9).
Fig 9. Role of ATF-2 in limiting NO production in endothelial cells in response to Ang II.
lonomycin stimulated NO production was determined in non-transfected, scrambled (SC) or ATF-2 siRNA transfected cell lysate with or without exposure to Ang II (0.1 µM, 24 h) as nitrites released in cell media of the treated cells. Basal control NO production was 60.5 pmol/ml/hr. Ang II decreased NO production. This effect was prevented with ATF-2 siRNA transfection. Data represent mean ± S.E.M. n=4 independent experiments carried out in triplicates. *P<0.05 vs. control. #P<0.05 vs. Ang II.
4. DISCUSSION
Elevated arginase activity/expression is involved in vascular endothelial dysfunction in many disease states, such as hypertension, diabetes, atherosclerosis, ischemic reperfusion injury and inflammation (Demougeot et al., 2005; Gao et al., 2007; Yang and Ming, 2006). In these conditions, arginase competes with NOS for their common substrate, L-arginine. Reduction in L-arginine availability to NOS can lead to decreased production of NO, NOS uncoupling and increased superoxide formation. Competition between arginase and NOS is involved in disease states such as hypertension and vascular complications of diabetes (Demougeot et al., 2005; Romero et al., 2008) and in aging (Berkowitz et al., 2003).
We previously reported a role for angiotensin II (Ang II), a peptide heavily linked to endothelial and vascular dysfunction, in increased arginase activity/expression in BAEC (Shatanawi et al., 2011). This elevation was mediated through a signaling pathway that involved activation of RhoA/Rho kinase and subsequent activation of p38 MAPK. In a model of hypertension and vascular dysfunction in mice infused with Ang II, we also evaluated Ang II effect on arginase levels. Our findings showed that Ang II elevates arginase activity and expression in mice aortas, and this effect is mediated through p38 MAPK. This increase in arginase activity was associated with endothelial dysfunction and was blocked by prior treatment with p38 MAPK inhibitor. Ang II-induced vascular dysfunction and hypertension was also prevented in transgenic mice that lacked one copy of the arginase 1 gene and both copies of the arginase 2 gene (Shatanawi et al., 2011).
Blockade of essential physiological functions of arginase could limit the clinical usefulness of arginase inhibitors to control endothelial arginase activity. However, identifying signaling pathways that lead to enhanced arginase activity in endothelial cells can provide more specific signaling targets to control/limit arginase activity in pathologic conditions, without disrupting upstream pathways essential for normal organ functions.
We sought to determine downstream signaling events from p38 MAPK that are involved in increased arginase 1 expression and activity. As our results show, arginase activity appears to be proportional to the amount of arginase protein, which, in turn, is determined primarily by transcription of the arginase genes. Elevation of arginase protein levels result from increased transcriptional activity of the arginase genes. We demonstrated that treatment with siRNA for ATF-2, which markedly reduced ATF-2 expression, completely blocked the Ang II-induced increase in arginase 1-linked luciferase activity, prevented the rise in arginase expression and activity and reduced arginase 1 protein levels even below those in the untreated cells. Our results provide very strong evidence for the critical involvement of ATF-2 in arginase 1 gene transcription.
Recent examination of the transcriptional regulation of arginase 1 gene in endothelial cells in response to thrombin has indicated involvement of AP-1 transcription factors proteins, ATF-2 and c-Jun (Zhu et al., 2009). Our results indicate the involvement of AP-1 proteins in Ang II-induced upregulation of arginase 1. We have shown that arginase 1 gene transcriptional activity is elevated in response to Ang II as measured by luciferase activity. Elevation of luciferase activity in response to Ang II was lost when cells were transfected with a luciferase arginase 1 promoter construct missing the AP-1 consensus sequence. These results show that an Ang II responsive element in BAECs is likely present in the arginase promoter region between −3.29 and −2.78 kb that encompasses the AP-1 binding site. These data indicate the importance of AP-1 proteins in enhancing arginase 1 transcription in response to Ang II. Two AP-1 proteins appear involved, ATF-2 and c-Jun. Knockdown of ATF-2 completely prevented the Ang II-induced activation of arginase 1 transcription, while c-Jun knockdown was only partially effective.
The actions of AP-1 proteins are known to occur through dimerization of two of the AP-1 family proteins. This could involve homodimerization of one transcription factor or heterodimerization of two different factors of family members which include Jun, Fos, ATF and Maf (Lopez-Bergami et al., 2010). Dimeric combinations and transcriptional activity observed in vivo are largely influenced by tissue-specific expression patterns of the individual proteins, and by specific activating mechanisms. Thus, while both ATF-2 and c-Jun seem to be equally important in activation of arginase 1 in isolated endothelial cells by thrombin (Zhu et al., 2009), effects of Ang II on arginase 1 transcription appear to mainly require actions of ATF-2 with only a partial contribution of c-Jun protein.
It is important to note here that analysis of the arginase 1 promoter region reveals several putative transcription factor binding sequences in the promoter region between −3.29 and −2.78 kb upstream of the transcription start site that are conserved among species (Gray et al., 2005). Studies of the structure and promoter of human liver-type arginase (arginase 1) indicate that there is a 84% homology with the murine arginase 1 gene, and that the promoter region used in our study is present in the promoter of human arginase 1 (Serrat et al., 2012; Takiguchi et al., 1988). These include binding sites for AP-1, c-Jun, SMAD, C/EBP, STAT1 and STAT6. Involvement of these transcription factors in arginase 1 upregulation in endothelial cells should be studied further. We focused on ATF-2, a known target of p38 MAPK (Lopez-Bergami et al., 2010), and determined its involvement in Ang II-induced arginase expression/activity.
Our results indicate that Ang II causes activation of ATF-2 as shown by increased levels of phospho-ATF-2. Phosphorylation of ATF-2 either at Thr69, Thr71 or Thr73 is known to stimulate its transcriptional capacity (Livingstone et al., 1995). ATF-2 phosphorylation was maximal within 30 m of treatment which runs in line with the increase of p38 MAPK activation that peaked by 15 m of Ang II treatment, but was still elevated at 30 m. Activation of ATF-2 was prevented by pretreatment with a p38 MAPK inhibitor, indicating that ATF-2 activation is a downstream event of p38 MAPK that regulates its phosphorylation. Our prior studies using the p38 MAPK inhibitor SB-203580 have demonstrated its ability to inhibited Ang II-induced phosphorylation of p38 (Shatanawi et al., 2011; Toque et al., 2010). As explained earlier, the MAPK family regulates activity of several AP-1 proteins. ERK, JNK and p38 MAPK are predominantly responsible for phosphorylation and activation of Fos, Jun and ATF-2, respectively, in response to stress, mitogens, hormones or oncogene activation (Lopez-Bergami et al., 2010; Niwano et al., 2006). Our results support these findings in relation to p38 MAPK and ATF-2. We did note an apparent increase in ATF-2 expression in response to Ang II for 60 m. While we did not expect to see this change in expression early on as early as 60 m, others have shown that Ang II can induce expression of transcription factors of the AP-1 family in a period of 45 m and lasting at least 2 h (Sharma et al., 1994). So our results show that not only increasing the phosphorylation of ATF-2 can play a role in arginase expression but also increasing levels of the ATF-2 protein as well.
Our results from EMSA and supershift indicate that Ang II enhances binding of nuclear proteins to the arginase promoter region encompassing an AP-1 binding site. Supershift assay identified ATF-2 as the nuclear protein in Ang II treated cells. Our results reveal that after exposure of endothelial cell to Ang II, ATF-2 is recruited to the AP-1 consensus sequence in the arginase 1 promoter leading to transcriptional upregulation of arginase 1. Results from our ChIP assay confirm the EMSA results. We observed that Ang II enhances binding between ATF-2 and the arginase 1 promoter.
We also examined the functional role of ATF-2 on arginase activity and NO production. Decreasing levels of ATF-2 protein by siRNA prevented the Ang II-induced increase in arginase activity. This effect was associated with restoration of NO production levels that were blunted in response to Ang II. Since arginase and NOS share the same substrate L-arginine, Ang II-induced decreases in NO production can be attributed to increases in the activity of arginase and thus limitation of L-arginine availability. Our results confirm a functional role of ATF-2 in limiting NO production. Our NO production results represent an important contribution to vascular function in intact tissue. We have previously reported the role of arginase 1 in Ang II-induced vascular endothelial dysfunction (Shatanawi et al., 2011).
ATF-2 also seems to be involved in repressing eNOS expression in IL-1β treated endothelial cells. Interestingly, this effect of ATF-2 was shown to be also mediated through p38 MAPK activation (Niwano et al., 2006). Repression of eNOS gene transcription by ATF-2 also could limit or decrease NO production. This mechanism represents another means for maintaining NOS function and NO production by limiting activity of ATF-2.
5. CONCLUSIONS
Collectively, our results indicate that ATF-2 is a central factor in regulating arginase transcription and activity in response to Ang II in endothelial cells. Involvement of ATF-2 also has implications for NO production. As discussed and presented, Ang II can impair vascular endothelial function by limiting NO availability through elevated arginase activity. Thus, identifying mediators of this pathway may not only shed light on the signal pathway of arginase upregulation, but may also provide additional targets for novel therapies that limit vascular dysfunction and damage resulting from excessive arginase activity in conditions of elevated Ang II, such as hypertension and diabetes.
Continued research is needed to identify inhibitors for transcription factors such as ATF-2 involved in pathological conditions. Piperine, the phenolic component of black pepper, is a reported potent inhibitor of several transcription factors in melanoma cells, among which is ATF-2 (Pradeep and Kuttan, 2004). Several inhibitors are being developed. Interferon regulatory factor-2-binding protein-1, reported to be a transcriptional co-repressor of interferon regulatory factor-2, also has been shown to repress ATF-2 -mediated transcriptional activation (Kimura, 2008). Selectively targeting transcription factors involved in arginase upregulation in endothelial cells could be a novel therapeutic strategy to limit vascular dysfunction in conditions of elevated Ang II. Since many factors upregulate endothelial arginase 1 expression causing vascular dysfunction, the signal transduction pathway for arginase being defined for Ang II may also apply to other stimuli.
ACKNOWLEDGEMENTS
This work was supported by National Institutes of Health Grants HL70215 (to RWC), EY11766 (to RBC and RWC), The University of Jordan research deanship grant (to AS), the 2012 L’Oréal-UNESCO For Women in Science Pan-Arab Regional Fellowship and the 2014 L’Oréal-UNESCO For Women in Science International Fellowship (to AS). The authors thank Dr. Sydney M. Morris, Jr. for kindly providing luciferase constructs of differing lengths of the murine arginase 1 promoter.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Archer S. Measurement of nitric oxide in biological models. Faseb J. 1993;7:349–360. doi: 10.1096/fasebj.7.2.8440411. [DOI] [PubMed] [Google Scholar]
- Bachetti T, Comini L, Francolini G, Bastianon D, Valetti B, Cadei M, Grigolato P, Suzuki H, Finazzi D, Albertini A, Curello S, Ferrari R. Arginase pathway in human endothelial cells in pathophysiological conditions. J Mol Cell Cardiol. 2004;37:515–523. doi: 10.1016/j.yjmcc.2004.05.004. [DOI] [PubMed] [Google Scholar]
- Bagnost T, Berthelot A, Bouhaddi M, Laurant P, Andre C, Guillaume Y, Demougeot C. Treatment with the arginase inhibitor N(omega)-hydroxy-nor-L-arginine improves vascular function and lowers blood pressure in adult spontaneously hypertensive rat. Journal of hypertension. 2008;26:1110–1118. doi: 10.1097/HJH.0b013e3282fcc357. [DOI] [PubMed] [Google Scholar]
- Berkowitz DE, White R, Li D, Minhas KM, Cernetich A, Kim S, Burke S, Shoukas AA, Nyhan D, Champion HC, Hare JM. Arginase reciprocally regulates nitric oxide synthase activity and contributes to endothelial dysfunction in aging blood vessels. Circulation. 2003;108:2000–2006. doi: 10.1161/01.CIR.0000092948.04444.C7. [DOI] [PubMed] [Google Scholar]
- Bivalacqua TJ, Burnett AL, Hellstrom WJ, Champion HC. Overexpression of arginase in the aged mouse penis impairs erectile function and decreases eNOS activity: influence of in vivo gene therapy of anti-arginase. Am J Physiol Heart Circ Physiol. 2007;292:H1340–1351. doi: 10.1152/ajpheart.00121.2005. [DOI] [PubMed] [Google Scholar]
- Chandra S, Romero MJ, Shatanawi A, Alkilany AM, Caldwell RB, Caldwell RW. Oxidative species increase arginase activity in endothelial cells through the RhoA/Rho kinase pathway. Br J Pharmacol. 2012;165:506–519. doi: 10.1111/j.1476-5381.2011.01584.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corraliza IM, Campo ML, Soler G, Modolell M. Determination of arginase activity in macrophages: a micromethod. Journal of immunological methods. 1994;174:231–235. doi: 10.1016/0022-1759(94)90027-2. [DOI] [PubMed] [Google Scholar]
- Demougeot C, Prigent-Tessier A, Marie C, Berthelot A. Arginase inhibition reduces endothelial dysfunction and blood pressure rising in spontaneously hypertensive rats. Journal of hypertension. 2005;23:971–978. doi: 10.1097/01.hjh.0000166837.78559.93. [DOI] [PubMed] [Google Scholar]
- Gao X, Xu X, Belmadani S, Park Y, Tang Z, Feldman AM, Chilian WM, Zhang C. TNF-alpha contributes to endothelial dysfunction by upregulating arginase in ischemia/reperfusion injury. Arterioscler Thromb Vasc Biol. 2007;27:1269–1275. doi: 10.1161/ATVBAHA.107.142521. [DOI] [PubMed] [Google Scholar]
- Gray MJ, Poljakovic M, Kepka-Lenhart D, Morris SM., Jr Induction of arginase I transcription by IL-4 requires a composite DNA response element for STAT6 and C/EBPbeta. Gene. 2005;353:98–106. doi: 10.1016/j.gene.2005.04.004. [DOI] [PubMed] [Google Scholar]
- Hultstrom M, Helle F, Iversen BM. AT(1) receptor activation regulates the mRNA expression of CAT1, CAT2, arginase-1, and DDAH2 in preglomerular vessels from angiotensin II hypertensive rats. Am J Physiol Renal Physiol. 2009;297:F163–168. doi: 10.1152/ajprenal.00087.2009. [DOI] [PubMed] [Google Scholar]
- Iyer RK, Yoo PK, Kern RM, Rozengurt N, Tsoa R, O'Brien WE, Yu H, Grody WW, Cederbaum SD. Mouse model for human arginase deficiency. Mol Cell Biol. 2002;22:4491–4498. doi: 10.1128/MCB.22.13.4491-4498.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeyabalan G, Klune JR, Nakao A, Martik N, Wu G, Tsung A, Geller DA. Arginase blockade protects against hepatic damage in warm ischemia-reperfusion. Nitric Oxide. 2008;19:29–35. doi: 10.1016/j.niox.2008.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura M. IRF2-binding protein-1 is a JDP2 ubiquitin ligase and an inhibitor of ATF2-dependent transcription. FEBS Lett. 2008;582:2833–2837. doi: 10.1016/j.febslet.2008.07.033. [DOI] [PubMed] [Google Scholar]
- Livingstone C, Patel G, Jones N. ATF-2 contains a phosphorylation-dependent transcriptional activation domain. EMBO J. 1995;14:1785–1797. doi: 10.1002/j.1460-2075.1995.tb07167.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Bergami P, Lau E, Ronai Z. Emerging roles of ATF2 and the dynamic AP1 network in cancer. Nat Rev Cancer. 2010;10:65–76. doi: 10.1038/nrc2681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris CR, Kato GJ, Poljakovic M, Wang X, Blackwelder WC, Sachdev V, Hazen SL, Vichinsky EP, Morris SM, Jr, Gladwin MT. Dysregulated arginine metabolism, hemolysis-associated pulmonary hypertension, and mortality in sickle cell disease. Jama. 2005;294:81–90. doi: 10.1001/jama.294.1.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niwano K, Arai M, Koitabashi N, Hara S, Watanabe A, Sekiguchi K, Tanaka T, Iso T, Kurabayashi M. Competitive binding of CREB and ATF2 to cAMP/ATF responsive element regulates eNOS gene expression in endothelial cells. Arterioscler Thromb Vasc Biol. 2006;26:1036–1042. doi: 10.1161/01.ATV.0000215179.76144.39. [DOI] [PubMed] [Google Scholar]
- Pradeep CR, Kuttan G. Piperine is a potent inhibitor of nuclear factor-kappaB (NF-kappaB), c-Fos, CREB, ATF-2 and proinflammatory cytokine gene expression in B16F-10 melanoma cells. Int Immunopharmacol. 2004;4:1795–1803. doi: 10.1016/j.intimp.2004.08.005. [DOI] [PubMed] [Google Scholar]
- Raingeaud J, Gupta S, Rogers JS, Dickens M, Han J, Ulevitch RJ, Davis RJ. Pro-inflammatory cytokines and environmental stress cause p38 mitogen-activated protein kinase activation by dual phosphorylation on tyrosine and threonine. J Biol Chem. 1995;270:7420–7426. doi: 10.1074/jbc.270.13.7420. [DOI] [PubMed] [Google Scholar]
- Romero MJ, Platt DH, Caldwell RB, Caldwell RW. Therapeutic use of citrulline in cardiovascular disease. Cardiovascular drug reviews. 2006;24:275–290. doi: 10.1111/j.1527-3466.2006.00275.x. [DOI] [PubMed] [Google Scholar]
- Romero MJ, Platt DH, Tawfik HE, Labazi M, El-Remessy AB, Bartoli M, Caldwell RB, Caldwell RW. Diabetes-induced coronary vascular dysfunction involves increased arginase activity. Circulation research. 2008;102:95–102. doi: 10.1161/CIRCRESAHA.107.155028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarge KD, Murphy SP, Morimoto RI. Activation of Heat-Shock Gene-Transcription by Heat-Shock Factor-I Involves Oligomerization, Acquisition of DNA-Binding Activity, and Nuclear-Localization and Can Occur in the Absence of Stress (Vol 13, Pg 1394, 1993) Molecular and Cellular Biology. 1993;13:3838–3839. doi: 10.1128/mcb.13.3.1392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satoh M, Fujimoto S, Arakawa S, Yada T, Namikoshi T, Haruna Y, Horike H, Sasaki T, Kashihara N. Angiotensin II type 1 receptor blocker ameliorates uncoupled endothelial nitric oxide synthase in rats with experimental diabetic nephropathy. Nephrol Dial Transplant. 2008;23:3806–3813. doi: 10.1093/ndt/gfn357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serrat N, Pereira-Lopes S, Comalada M, Lloberas J, Celada A. Deacetylation of C/EBPbeta is required for IL-4-induced arginase-1 expression in murine macrophages. European journal of immunology. 2012;42:3028–3037. doi: 10.1002/eji.201242413. [DOI] [PubMed] [Google Scholar]
- Sharma HS, Vanheugten HAA, Goedbloed MA, Verdouw PD, Lamers JMJ. Angiotensin-li-lnduced Expression of Transcription Factors Precedes Increase in Transforming Growth-Factor-Beta-1 Messenger-Rna in Neonatal Cardiac Fibroblasts. Biochem Bioph Res Co. 1994;205:105–112. doi: 10.1006/bbrc.1994.2636. [DOI] [PubMed] [Google Scholar]
- Shatanawi A, Romero MJ, Iddings JA, Chandra S, Umapathy NS, Verin AD, Caldwell RB, Caldwell RW. Angiotensin II-induced vascular endothelial dysfunction through RhoA/Rho kinase/p38 mitogen-activated protein kinase/arginase pathway. Am J Physiol Cell Physiol. 2011;300:C1181–1192. doi: 10.1152/ajpcell.00328.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takiguchi M, Haraguchi Y, Mori M. Human liver-type arginase gene: structure of the gene and analysis of the promoter region. Nucleic Acids Res. 1988;16:8789–8802. doi: 10.1093/nar/16.18.8789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toque HA, Romero MJ, Tostes RC, Shatanawi A, Chandra S, Carneiro ZN, Inscho EW, Webb RC, Caldwell RB, Caldwell RW. p38 Mitogen-activated protein kinase (MAPK) increases arginase activity and contributes to endothelial dysfunction in corpora cavernosa from angiotensin-II-treated mice. The journal of sexual medicine. 2010;7:3857–3867. doi: 10.1111/j.1743-6109.2010.01996.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Lewis CM, Chandrasekharan UM, Kinney CM, Dicorleto PE, Kashyap VS. Arginase activity is increased by thrombin: a mechanism for endothelial dysfunction in arterial thrombosis. Journal of the American College of Surgeons. 2006;203:817–826. doi: 10.1016/j.jamcollsurg.2006.08.023. [DOI] [PubMed] [Google Scholar]
- Yang Z, Ming XF. Endothelial arginase: a new target in atherosclerosis. Current hypertension reports. 2006;8:54–59. doi: 10.1007/s11906-006-0041-8. [DOI] [PubMed] [Google Scholar]
- Zhu W, Chandrasekharan UM, Bandyopadhyay S, Morris SM, Jr, DiCorleto PE, Kashyap VS. Thrombin induces endothelial arginase through AP-1 activation. Am J Physiol Cell Physiol. 2009;298:C952–960. doi: 10.1152/ajpcell.00466.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]