Abstract
Background
To determine the prevalence of RET rearrangement genes, RET copy number gains and expression in tumor samples from four Phase III non-small-cell lung cancer (NSCLC) trials of vandetanib, a selective inhibitor of VEGFR, RET and EGFR signaling, and to determine any association with outcome to vandetanib treatment.
Methods
Archival tumor samples from the ZODIAC (NCT00312377, vandetanib ± docetaxel), ZEAL (NCT00418886, vandetanib ± pemetrexed), ZEPHYR (NCT00404924, vandetanib vs placebo) and ZEST (NCT00364351, vandetanib vs erlotinib) studies were evaluated by fluorescence in situ hybridization (FISH) and immunohistochemistry (IHC) in 944 and 1102 patients.
Results
The prevalence of RET rearrangements by FISH was 0.7% (95% CI 0.3–1.5%) among patients with a known result. Seven tumor samples were positive for RET rearrangements (vandetanib, n = 3; comparator, n = 4). 2.8% (n = 26) of samples had RET amplification (innumerable RET clusters, or ≥7 copies in > 10% of tumor cells), 8.1% (n = 76) had low RET gene copy number gain (4–6 copies in ≥40% of tumor cells) and 8.3% (n = 92) were RET expression positive (signal intensity ++ or +++ in >10% of tumor cells). Of RET-rearrangement-positive patients, none had an objective response in the vandetanib arm and one patient responded in the comparator arm. Radiologic evidence of tumor shrinkage was observed in two patients treated with vandetanib and one treated with comparator drug. The objective response rate was similar in the vandetanib and comparator arms for patients positive for RET copy number gains or RET protein expression.
Conclusions
We have identified prevalence for three RET biomarkers in a population predominated by non-Asians and smokers. RET rearrangement prevalence was lower than previously reported. We found no evidence of a differential benefit for efficacy by IHC and RET gene copy number gains. The low prevalence of RET rearrangements (0.7%) prevents firm conclusions regarding association of vandetanib treatment with efficacy in the RET rearrangement NSCLC subpopulation.
Trial registration
Randomized Phase III clinical trials (NCT00312377, ZODIAC; NCT00418886, ZEAL; NCT00364351, ZEST; NCT00404924, ZEPHYR).
Electronic supplementary material
The online version of this article (doi:10.1186/s12885-015-1146-8) contains supplementary material, which is available to authorized users.
Keywords: RET rearrangement, Vandetanib, Non-small-cell lung cancer
Background
Cancer treatment paradigms are evolving to exploit the sensitivity of tumors to inhibitors that target the products of genes carrying driver mutations [1]. A number of genetic aberrations that drive and maintain tumorigenesis have recently been identified in non-small-cell lung cancer (NSCLC). These include fusion genes generated by chromosomal rearrangements between the rearranged during transfection (RET) gene and other genes, most commonly kinesin family 5B (KIF5B) and coiled coil domain containing-6 (CCDC6) [2-12]. These fusions lead to overexpression of truncated RET proteins containing the RET kinase domain, which can induce transformation and occur in tumors that rarely harbor mutations in other common drivers, ie epidermal growth factor receptor (EGFR), KRAS, human epidermal growth factor receptor, and anaplastic lymphoma receptor (ALK) genes. RET rearrangement was first shown to be associated with papillary thyroid carcinoma (PTC), leading to the fusion oncoprotein (RET/PTC) and constitutive activation of RET receptor tyrosine kinase in papillary cancer cells [13]. In addition, RET mutations are present in the germline of nearly all patients with hereditary forms of medullary thyroid cancer (MTC) [14-16] and approximately 50% of patients with sporadic MTC have somatic RET mutations that are associated with a worse outcome [17].
Vandetanib is a once-daily, oral anticancer agent that selectively inhibits vascular endothelial growth factor receptor (VEGFR), RET and EGFR signaling [18,19], and is licensed for the treatment of MTC in several geographical regions. Preclinical studies have demonstrated that vandetanib inhibits RET signaling arising from RET mutations in a MTC cell line and inhibits growth of human PTC cell lines that carry spontaneous RET/PTC rearrangements [19]. In addition, vandetanib has been shown to inhibit the proliferation of cells expressing RET-KIF5B [2,3] and a human lung adenocarcinoma cell line harboring an endogenous RET-CCDC6 [20].
Four randomized Phase III clinical trials have evaluated the efficacy of vandetanib in NSCLC in combination with docetaxel (NCT00312377; ZODIAC [21]), in combination with pemetrexed (NCT00418886; ZEAL [22]) or as a monotherapy (NCT00364351; ZEST [23] and NCT00404924; ZEPHYR [24]). These studies in unselected patient populations demonstrated antitumor activity of vandetanib, but there was no overall survival benefit when added to standard chemotherapy or as monotherapy versus erlotinib [21-24]. The aim of this study, a retrospective evaluation of tumor samples from the NSCLC Phase III program, was to determine the prevalence of RET rearrangements and other potential RET biomarkers within this population and to investigate any association with outcome to vandetanib treatment.
Methods
Overview of NSCLC studies
Study treatments and assessment of efficacy
All four studies are registered at www.clinicaltrials.gov and were Phase III, multicenter, randomized, double-blind studies conducted in 25 (ZODIAC), 21 (ZEAL), 22 (ZEPHYR) and 22 (ZEST) countries, respectively. Enrollment was conducted between 2006 and 2008. Full details of the study design and methodology have been reported previously [21-24]. Briefly, in ZODIAC (NCT00312377), patients received docetaxel in combination with oral vandetanib 100 mg/day (n = 694) or matched placebo (n = 697). Docetaxel 75 mg/m2 was administered as an intravenous infusion every 21 days, for a maximum of six cycles. In ZEAL (NCT00418886), patients received pemetrexed in combination with oral vandetanib, 100 mg/day (n = 256) or matched placebo (n = 278). Pemetrexed 500 mg/m2 was administered as an intravenous infusion every 21 days, for a maximum of six cycles. Vandetanib was evaluated as a monotherapy in two of the studies: in ZEPHYR (NCT00404924), patients received vandetanib 300 mg/day (n = 617) or placebo (n = 307); and in ZEST (NCT00364351), patients received vandetanib 300 mg/day (n = 623) or erlotinib 150 mg/day (n = 617). Objective tumor assessments were categorized using Response Evaluation Criteria in Solid Tumors (RECIST; version 1.0).
Patient eligibility
Data were evaluated from adult patients with histologically or cytologically confirmed locally advanced or metastatic (stage IIIB to IV) NSCLC after failure of: first-line anticancer therapy (ZODIAC and ZEAL); one or two chemotherapy regimens (ZEST); or an EGFR inhibitor following one or two chemotherapy regimens (ZEPHYR). For all studies, inclusion criteria included: World Health Organization performance status 0–2; life expectancy ≥12 weeks and no significant hematologic, hepatic, renal or cardiac abnormalities. Patients with squamous cell histology were also eligible. Prior treatment with VEGFR inhibitors (all studies), docetaxel (ZODIAC only), pemetrexed (ZEAL only) and EGFR inhibitors (ZEST only) was not permitted. Prior treatment with bevacizumab (all studies) and cetuximab (ZEST only) was allowed.
Ethics statements
All patients provided written informed consent, the trials were approved by all relevant institutional ethical committees or review bodies (The University of Texas MD Anderson Cancer Center Surveillance Committee, Houston, TX, USA; The Royal Melbourne Research Foundation, Melbourne, Australia; Institutional Review Board of National Cancer Center, Gyeonggi-do, Republic of Korea; Cedars-Sinai Medical Center Institutional Review Board, Beverly Hills, CA, USA) and was conducted in accordance with the Declaration of Helsinki, Good Clinical Practice, and the AstraZeneca policy on Bioethics [25]. Data were generated in accordance with the Medical and Healthcare Products Regulatory Agency Good Clinical Practice Guidelines for laboratories [26].
Samples
Archival tumor specimens were sampled prior to enrollment of patients onto study. Provision of these samples for genetic/immunohistochemical (IHC) assessment was not compulsory in any study, resulting in collection from approximately one third of patients. There was no observable bias to sampling consent. Tumor samples were supplied as formalin-fixed paraffin-embedded biopsies, resections or sections and could be obtained at any time during the respective study. Cell lines used as controls in IHC were supplied as cell pellets and stored at −80°C prior to use. All analyses were carried out in AstraZeneca laboratories in the UK and China.
Assay methods
Fluorescence in situ hybridization (FISH) analysis
RET-KIF5B, alternative RET rearrangements and RET gene amplifications were identified using a FISH probe set of four fluorescent-labeled bacterial artificial chromosome (BAC) clone-derived DNA probes designed in-house: RP11-124O11 (located upstream of RET) labeled with spectrum red; RP11-718J13 (located immediately downstream of RET) labeled with spectrum green (fluorescein isothiocyanate); RP11-983E11 (located upstream of KIF5B exon 2) labeled with spectrum gold (5[6]-carboxyrhodamine 6G deoxyuridine-5′-triphosphate [dUTP]); and centromere of chromosome 10 labeled with spectrum aqua (diethylaminocoumarin-5-dUTP; Figure 1). For tissue samples assessed at the UK site, these probes were sourced from Empire Genomics LLC, Buffalo, NY; for tissue samples assessed at the China site, the probes were generated in-house. BACs and labels used at both sites were identical. Sections were processed with reagents from the Histology FISH accessory kit (Dako, Dako Corporation, Carpinteria, CA, USA, cat #K5799) according to the manufacturer’s instructions. Briefly at the UK site, formalin-fixed, paraffin-embedded sections (4 μm) were mounted on glass slides. Sections were deparaffinized in xylene, hydrated through a graded ethanol series, and then incubated with the accessory kit pretreatment solution at 95–100°C for 10 minutes. The sections were then washed and pepsin digestion was carried out at 37°C in a Dako hybridizer for 5 minutes; the sections were dehydrated through ethanol and allowed to air dry. The fluorescent probe mix was applied and then the probe and section co-denatured in a Dako hybridizer at 83°C for 3 minutes followed by overnight incubation at 37°C. The sections were washed in 1x saline-sodium citrate (SSC)/0.3% Igepal at 72°C for 2 minutes, followed by 2x SSC at room temperature, before dehydration through ethanol. Sections were mounted in Dako antifade mounting media (Dako, #K5799). The procedure at the China site was similar, except that the sections were incubated with the Spotlight tissue pretreatment kit (Invitrogen, Carlsbad, CA, USA, cat #00-8401) and processed as previously described [27].
Figure 1.

Representative FISH images. (A) Unknown RET rearrangement, (B)RET-KIF5B fusion, (C)RET gene amplifications and (D) low RET gene copy number gain. (E) Loci for RET FISH probes.
The FISH gene fusion assay had previously been validated in a pilot study, which confirmed a FISH assay defined RET-KIF5B positive NSCLC tumor by sequencing (Additional file 1). Assessment of FISH signal was carried out by investigators blinded to clinical response. Preliminary assessment was performed at 60x magnification to identify any alterations in the distribution of the red and green signals. When these were observed, 50 tumor cells were analyzed at 100x magnification for the number of red, green, paired red/green, gold, paired green/gold and blue signals in 10–15 tumor cells from up to four regions of the tumor section. A further region was then analyzed by a second blinded observer. To describe tumor heterogeneity, estimation of the proportion and distribution of cells with RET events was determined independently by two observers.
Immunohistochemistry
Sections were deparaffinized and rehydrated as described above. Antigen retrieval was performed at 110°C for 5 minutes in Target Retrieval Solution (pH 9.0; Dako, #S2367) in a RHS-2 microwave processor (Milestone, Sorisole, Italy) within a pressurized reaction vessel. Endogenous peroxidase activity was quenched by incubating the sections in 3% hydrogen peroxide for 20 minutes at room temperature and non-specific binding was blocked by incubating in serum-free protein block (Dako, #X0909) for 20 minutes at room temperature. Sections were labeled with a rabbit anti-RET monoclonal antibody (1:1000 dilution, clone EPR2871, Epitomics, Burlingame, CA, USA; see Additional file 1 for antibody specificity and immunohistochemistry validation; Additional file 1: Table S1) and RET expression was visualized with EnVision™ FLEX+ (Dako, #K8012). Sections were counterstained with hematoxylin. Cell lines (TT, MiaPaCa, SKNMC and Panc1) and tissue samples (human non-inflamed appendix and in-house NSCLC tissue microarray) were used as controls for RET immunostaining.
Statistical analysis
For FISH analysis, tumors were categorized as positive for RET rearrangement if >10% of the tumor cells presented with broken-apart red and green RET signals; this could be further classified as positive for RET-KIF5B if the broken-apart red/green signal was accompanied by a paired green/gold signal. Tumors were categorized as low RET gene copy number gain if ≥40% of tumor cells had 4–6 copies of red/green RET signals. Tumors were categorized as amplified if >10% of tumor cells had ≥7 red/green signals or signal clusters. In the IHC analysis, tumor samples with >100 intact tumor cells were assessed by a system similar to that described previously [8]. Tumors were categorized as RET expression positive where the staining signal intensity was ++ or +++ (0 to +++ scale) in >10% of tumor cells. The objective response rate (ORR) was presented by RET biomarker status and treatment arm with corresponding 95% confidence intervals (CIs). Prevalence rates were estimated across all patients with a known result (including those not randomized to treatment) and were presented as a percentage with corresponding 95% CIs.
Results
Patients
From 4089 patients across the four NSCLC studies, 1291 and 1234 screened patients had tumor samples available for FISH and IHC analysis, respectively. Evaluable data were obtained for 944 (FISH; Additional file 1: Table S2) and 1102 (IHC; Additional file 1: Table S3) patients, with seven and eight patients donating FISH and IHC samples, respectively, not randomized to treatment. Failure rates in the IHC analysis (10.7%) were largely due to an inadequate number of tumor cells in the samples, whereas in the FISH analysis, failure rates (26.9%) were largely due to an inadequate number of tumor cells or sample quality. The median age of patients was 61 years; approximately two-thirds were white, with the remainder predominantly of Asian origin. Most patients (61%) presented with adenocarcinoma. Patient demographics and baseline characteristics for patients with tumor samples evaluable for FISH or IHC analysis and clinicopathologic characteristics of patients and their RET biomarker status are outlined in Additional file 1: Tables S2–S4, respectively.
Prevalence of RET biomarkers
In this NSCLC patient population, the overall prevalence of RET rearrangements was 0.7% (95% CI 0.3–1.5) among patients with a known result. Seven tumor samples were classified as positive for RET rearrangements (vandetanib treatment, n = 3; comparator treatment, n = 4; Table 1; Figure 1a and b). Five of the seven RET rearrangements were RET-KIF5B and the other two had unknown fusion partners with RET. Single red or green signals without a corresponding 3′ or 5′ RET signal were occasionally seen in samples but were not scored as rearrangements.
Table 1.
Frequency of RET biomarkers in vandetanib Phase III NSCLC trial program
| Clinical study | RET biomarker, n (%) [95% CI] | |||||||
|---|---|---|---|---|---|---|---|---|
| RETrearrangement | RETamplification | LowRETgene copy number gain | RET expression | |||||
| Van | Comp | Van | Comp | Van | Comp | Van | Comp | |
| ZODIAC + ZEAL | 1 | 2** | 6 | 7 | 14 | 14 | 16 | 24 |
| ZEPHYR | 2 | 1 | 2 | 3 | 19 | 5 | 18 | 10 |
| ZEST | 0 | 1** | 4 | 2 | 8 | 14 | 12 | 10 |
| Untreated* | – | 2 | 2 | 2 | ||||
| Non-Asian | 5/632 (0.8) | 19/632 (3.0) | 49/632 (7.7) | 52/756 (6.9) | ||||
| [0.3–1.8] | [1.8–4.6] | [5.7–10.0] | [5.2–8.9] | |||||
| Asian | 2/305 (0.7) | 7/305 (2.3) | 27/305 (8.8) | 40/346 (11.6) | ||||
| [0.1–2.3] | [0.9–4.6] | [5.9–12.5] | [8.4–15.4] | |||||
| Overall | 7/937 (0.7) | 26/937 (2.8) | 76/937 (8.1) | 92/1102 (8.3) | ||||
| [0.3–1.5] | [1.8–4.0] | [6.4–10.0] | [6.8–10.1] | |||||
Comp, comparator; Van, vandetanib. *One patient randomized to ZODIAC and one randomized to ZEAL did not receive treatment. **One RET rearrangement with an unknown, non-KIF5B fusion partner was identified in the ZEAL comparator and one was identified in the ZEST comparator arm.
RET gene amplifications and low RET gene copy number gains were reported in 26 (2.8%; 95% CI 1.8–4.0) and 76 (8.1%; 95% CI 6.4–10.0) patients, respectively (Table 1; Figure 1c and d). RET expression was positive in samples from 92 (8.3%; 95% CI 6.8–10.1) patients (Table 1). Tumor cell immunostaining was generally cytoplasmic and diffuse (Figure 2). The prevalence of RET rearrangement, RET protein expression, RET amplification or low RET gene copy number gain was similar for Asian and non-Asian populations (Table 1).
Figure 2.
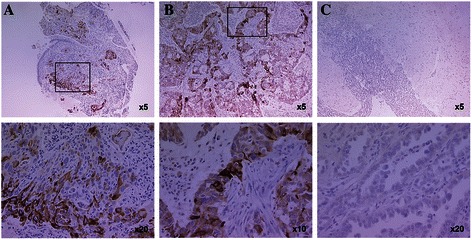
Representative IHC images positive for RET expression. (A) Tumor biopsies and (B) resections. (C) Negative (weak) staining.
In our study, four out of seven tumors that were positive for RET rearrangement expressed RET when evaluated with IHC. Of the remaining three samples, one RET-KIF5B and two RET-other rearrangements did not express any detectable RET protein when evaluated with IHC (Additional file 1: Table S5). In all IHC-positive cases, RET was predominantly cytoplasmic and typically of moderate to weak intensity. In two cases, RET was only detected in the focal areas of tumor cells. Weak staining for RET was also observed in the stroma of three RET-rearrangement-positive tumors. Of the seven RET-rearrangement-positive tumors, only one showed amplification which was also positive when evaluated with IHC; this case also showed weak RET staining across all of the stroma present in the sample.
Clinical outcome by positive RET biomarker status
None of the three vandetanib-treated RET-rearrangement-positive patients had an objective response (Table 2). Radiologic evidence of tumor shrinkage was observed in two of these patients (ZEPHYR, 23% and 33% shrinkage of target lesions). However, a response could not be confirmed at the next visit. One patient in ZODIAC who received docetaxel alone had a confirmed objective response and 32% shrinkage of target lesions at day 85 (Table 3). The ORRs were similar for the vandetanib and comparator arms for patients positive for RET amplification (8.3% vs 8.3%), those with low RET gene copy number gain (9.8% vs 9.1%) or those positive for RET protein expression (15.2% vs 13.6%, Table 2). In conclusion, we considered there were too few RET-rearrangement-positive patients to draw definitive conclusions regarding efficacy in this patient population. However, the lack of additional benefit observed in the higher prevalence biomarker-positive groups of RET amplification and low RET gene copy number gain is consistent with these biomarkers having little predictive utility to identify those patients who will benefit from vandetanib therapy.
Table 2.
ORRs (RECIST) in patients positive for RET biomarkers
| Clinical study | Objective responses, n/N (%) | |||||||
|---|---|---|---|---|---|---|---|---|
| RETrearrangement | RETamplification | LowRETgene copy number gain | RET expression | |||||
| Van | Comp | Van | Comp | Van | Comp | Van | Comp | |
| ZODIAC + ZEAL | 0/1 | 1/2 | 1/6 | 1/7 | 2/14 | 2/14 | 5/16 | 3/24 |
| ZEPHYR | 0/2 | 0/1 | 0/2 | 0/3 | 0/19 | 0/5 | 0/18 | 0/10 |
| ZEST | 0/0 | 0/1 | 0/4 | 0/2 | 2/8 | 1/14 | 2/12 | 3/10 |
| Overall | 0/3 (0.0) | 1/4 (25.0) | 1/12 (8.3) | 1/12 (8.3) | 4/41 (9.8) | 3/33 (9.1) | 7/46 (15.2) | 6/44 (13.6) |
Table 3.
Clinicopathologic characteristics of seven patients positive for RET rearrangements
| Study | Age (years) | Sex | Race | Smoking status* | Histology | EGFRstatus | KRASstatus | Dose/day | Exposure | RECIST response | Tumor shrinkage | Reason for discontinuation | RET partner | % cells with rearrangements detected |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Vandetanib | ||||||||||||||
| ZODIAC | 68 | F | Asian | Non-smoker | Adenocarcinoma | Mutation negative | Negative | 100 mg | 21 days | Progressive disease | – | Progressive disease | KIF5B | 50% |
| ZEPHYR | 69 | F | White | Non-smoker | Adenocarcinoma | Mutation negative; amplification positive | Negative | 300 mg | 180 days | Stable disease | 23% shrinkage of target lesions | Adverse event | KIF5B | 75–100% |
| ZEPHYR | 59 | F | Asian | Non-smoker | Adenocarcinoma | Mutation negative; amplification positive | Negative | 300 mg | 57 days | Progressive disease | 33% shrinkage of target lesions (progressive disease in non-target lesions) | Progressive disease | KIF5B | 50–75% |
| Comparator | ||||||||||||||
| ZODIAC | 59 | F | White | Ex-smoker | Adenocarcinoma | Mutation negative | Unknown | – | Six cycles docetaxel | Partial response (day 85); progressive disease (day 210) | 32% shrinkage of target lesions at day 85 | Completed six cycles | KIF5B | 75–100% |
| ZEAL** | 58 | M | White | Ex-smoker | Large cell carcinoma | Mutation negative | Negative | – | Five cycles pemetrexed | Progressive disease (day 245) | None | Completed five cycles | Not known | 75–100% |
| ZEPHYR | 57 | M | White | Ex-smoker | Adenocarcinoma | Mutation negative | Negative | – | 26 days placebo | Progressive disease (day 25) | None | Progressive disease | KIF5B | 75–100% |
| ZEST** | 70 | M | White | Ex-smoker | Adenocarcinoma | Mutation negative; amplification positive | Negative | – | 315 days erlotinib | Progressive disease (day 166) | None | Progressive disease | Not known | 25–50% |
F, female; M, male. *Non-smoker = never smoked >20 g tobacco in lifetime; ex-smoker = stopped smoking ≥1 year ago; occasional smoker = <1 tobacco product per day; habitual smoker = ≥1 tobacco product per day. **RET rearrangements with unknown, non-KIF5B fusion partners.
Discussion
In this retrospective study, the overall prevalence of RET rearrangements within the Phase III vandetanib NSCLC clinical program was determined as 0.7% among patients with a known RET rearrangement status. We found consistent frequencies of RET rearrangement in Asian (0.7%) and non-Asian patients (0.8%). In general, RET rearrangement prevalence rates may be considered as low and may vary according to the proportions of smokers, racial origin and histological subtype in the study population. Prevalence rates of RET rearrangement in Asian populations have been reported at 1–2% for NSCLC [5,11] and lung adenocarcinoma [2,3,7,12], and were estimated as high as approximately 6% in lung adenocarcinoma [4]. Our own study contains a high proportion of non-Asian patients (67%, Additional file 1: Table S2) and smokers/ex-smokers (77%, Additional file 1: Table S2), in contrast to previous reports on RET rearrangement prevalence rates [3,5,7,11,12], in which study populations were either entirely or largely Asian and non-smokers.
RET rearrangements have previously been reported in squamous cell carcinoma [5], lung neuroendocrine tumor [5] and adenosquamous tumor [11]; however, the majority occur in adenocarcinomas. This is consistent with our findings, which show a higher prevalence of RET rearrangements in patients with adenocarcinoma (1.2%, 6/510) compared with those in other histology types (0.2%, 1/427). Our data are not in agreement with a number of studies that report a higher frequency of RET rearrangements in non-smokers compared with smokers/ex-smokers; in our study, we have observed three and four RET rearrangements, respectively (Table 3) [2,5,11]. As in previous studies, we did not observe co-occurrence of RET rearrangements with EGFR and KRAS mutations. Interestingly, one of the three RET-KIF5B tumors reported by Go et al. [28] in lung adenocarcinomas negative for KRAS and EGFR mutations and ALK rearrangements was from a smoker. However, all of these observations should be interpreted with caution given the small numbers.
The techniques used to identify RET rearrangement genes in previously reported studies were sequencing [2-4] or reverse transcription-polymerase chain reaction followed by verification with FISH [11], sequencing [5,12] or differential expression of 3′ and 5′ RET exons [7,9,11]. Some of these techniques may underestimate the frequency of RET rearrangement genes by failing to detect fusions to partner genes other than KIF5B. We used a four-probe FISH assay to detect RET rearrangements. This technique is highly sensitive in detecting chromosomal rearrangements and has the advantage of detecting other partners or isoforms, though it is not known whether all these rearrangements are functional. For example, in a study using a split FISH assay to evaluate 1528 lung cancers, 22 tumors were detected with split RET signals, of which 12 were confirmed as fusions with KIF5B and one with CCDC6 [10] and the remaining nine tumors showed little or no expression of the RET kinase domain.
Although the prevalence of RET rearrangements in NSCLC patients is low, RET inhibition may be efficacious within a subset of patients who carry these genetic aberrations. In this study, there were too few vandetanib-treated patients with RET rearrangements to form conclusions regarding association with efficacy. A number of studies have reported increased expression of RET protein in NSCLC tumor cells, not necessarily associated with RET rearrangements [2,3,8,11]. This led us to investigate both IHC and RET copy number gain as possible predictive biomarkers for vandetanib response. No difference was observed in the ORRs between vandetanib and comparator arms for IHC and copy number analyses.
In our study, we observed RET-KIF5B and other RET rearrangements in samples that were negative for RET protein expression. This observation is in line with previous studies of NSCLC samples which have used a range of anti-RET antibodies (including the Epitomics EPR2871 antibody we have used here) and differing IHC techniques [2,3,8,28]. Sasaki et al. [8] reported three cases of RET translocation (from 371 NSCLC samples), all of which had weak positive cytoplasmic staining when evaluated with a 3F8 anti-RET mouse monoclonal antibody (Vector Laboratories, Peterborough, UK). In contrast, when using the EPR2871 antibody used in our study, weak, moderate and strong staining were reported for the three RET translocation positive samples; this suggests that apparent RET expression is dependent on both the antibody and the local IHC protocol used. Another study has reported weak to strong RET expression with IHC when using the 3F8 anti-RET antibody; however, only one of the RET IHC positive cases was also RET-KIF5B positive [3]. Using the EPR2871 antibody, Kohno et al. reported 48/222 NSCLC cases to express RET in the absence of a RET fusion; all cases of RET-KIF5B were also RET positive with IHC [2]. Go et al. [28] used three different anti-RET antibodies to screen 53 NSCLC cases for RET protein expression. RET IHC positive cases were defined as those with >30% of cells expressing cytoplasmic RET. Three samples that were RET-fusion positive with whole transcript sequencing were negative for RET with IHC, whereas RET protein was identified in four samples, none of which harbored a RET-KIF5B rearrangement. Taken together, these NSCLC studies, along with our results, suggest that not all cases of RET-KIF5B or other RET rearrangements express RET protein when evaluated with IHC. RET protein appears to be largely cytoplasmic; however, considerable inter-patient variation and heterogeneity among tumor cells within individual tumors is observed.
Investigation of RET inhibitors in NSCLC patients with a documented confirmation of a RET rearrangement is an active area of research with three clinical studies currently ongoing (NCT01639508, NCT01823068 and NCT01813734). Results from a study on the use of vandetanib in RET-rearrangement-positive NSCLC patients (NCT01823068) should provide further insight into the role of vandetanib in this patient population. Preliminary data from NCT01639508, a prospective Phase II trial investigating the use of cabozantinib, a small-molecule inhibitor of MET, VEGFR2 and RET, has been published [6]. For the first three patients treated with cabozantinib, two patients showed confirmed partial clinical responses and the third patient had prolonged stable disease approaching 8 months [6]. A case study in a patient with poorly differentiated lung adenocarcinoma, positive for a RET-KIF5B and refractory for previous chemotherapy, is also noteworthy. In this patient, 4 weeks of treatment with vandetanib 300 mg once daily produced a fluorodeoxyglucose-positron emission tomography/computed tomography response [29]. In addition, in a preliminary study in which two heavily pretreated patients with confirmed RET rearrangements were treated with vandetanib, stable disease was observed following treatment [30].
Conclusions
This study has demonstrated an overall prevalence of 0.7% for RET rearrangements in a large Phase III NSCLC patient population among patients with a known determination of RET rearrangement status composed predominantly of non-Asian patients and smokers. RET rearrangements were found most frequently, but not exclusively, in adenocarcinomas and occurred in tumors negative for other driver mutations, in agreement with previous reports in predominantly Asian populations. The prevalence of RET rearrangement is too low in this unselected population to determine whether RET-rearrangement-positive patients can be effectively treated with RET inhibitors, such as vandetanib. Changes in RET gene copy number and level of RET protein expression are more frequent aberrations than RET rearrangement in this NSCLC population, but also do not provide predictive markers for response to vandetanib. Results from additional studies, specifically in RET-rearrangement-positive NSCLC patients, are needed to determine whether this patient population can be effectively treated with RET inhibitors, such as vandetanib. If these studies support RET rearrangements as a clinically relevant target, then screening of NSCLC patients for RET rearrangements may become part of standard care.
Acknowledgments
We dedicate this article to the memory of our colleague Neil Gray, FIBMS.
The authors would like to acknowledge John Williams for facilitating the delivery of the contracts with external parties to time, quality and cost, Darren Hodgson for clinical data management, transmission, reporting and critical review of data analysis files, and Jennifer Bradford for assistance with analysis and interpretation.
This study was sponsored by AstraZeneca. Medical writing assistance was provided by Claire Routley from Mudskipper Business Ltd and funded by AstraZeneca.
Financial support
This study was sponsored by AstraZeneca.
Previously presented at American Society for Clinical Oncology (ASCO) 2013; J Clin Oncol 31:(suppl; abst 8045).
Additional file
Supporting information for FISH assay validation, Western blotting/IHC antibody specificity, localization of RET in tumor tissue sections, patient demographics and baseline characteristics for patients with tumor samples evaluable for FISH and/or IHC analysis.
Footnotes
Please see related article: http://dx.doi.org/10.1186/s12885-015-1147-7
Neil Gray deceased.
Competing interests
G Bigley, A Dale, S Fan, H Fu, Q Ji, A Platt, J Read, X Su, V Williams, Q Ye, L Zheng and T Zhang are employed (other than primary affiliation; e.g., consulting) by AstraZeneca. L Blockley, E Donald, P Elvin, G Harrod, J Stevens, J Morten, C Cresswell, A Davies, A Gladwin and C Womack are employed by and own shares in AstraZeneca. J-S Lee has received research funding from AstraZeneca. R de Boer has received research funding and honoraria as a consultant from AstraZeneca. J Vasselli is employed by MedImmune. R Herbst has no potential conflicts of interest.
Authors’ contributions
AP, PE, JM, ED, AG, CW and QJ were involved in the conception and design of the study. XS, ED, VW, GB and CW developed the methodology for the study. All authors obtained data for the study. The analysis and interpretation of the data was performed by AP, JM, GB, LB, CC, AD, AD, NG, SF, HF, GH, JR, JS, VW, QY, TZ, QJ, XS, LZ, ED, CW and PE. All authors contributed to the writing and critical review of the manuscript. All authors read and approved the final manuscript.
Authors’ information
Dr Womack was employed by AstraZeneca at the time of the study.
Contributor Information
Adam Platt, Email: adam.platt@astrazeneca.com.
John Morten, Email: john.morten@astrazeneca.com.
Qunsheng Ji, Email: qunsheng.ji@astrazeneca.com.
Paul Elvin, Email: paul.elvin@astrazeneca.com.
Chris Womack, Email: chris.womack@astrazeneca.com.
Xinying Su, Email: xinying.su@astrazeneca.com.
Emma Donald, Email: emma.donald@astrazeneca.com.
Neil Gray, Email: adam.platt@astrazeneca.com.
Jessica Read, Email: jessica.read@astrazeneca.com.
Graham Bigley, Email: graham.bigley@astrazeneca.com.
Laura Blockley, Email: laura.blockley@astrazeneca.com.
Carl Cresswell, Email: carl.cresswell@astrazeneca.com.
Angela Dale, Email: angela.dale@astrazeneca.com.
Amanda Davies, Email: amanda.davies@astrazeneca.com.
Tianwei Zhang, Email: tianwei.zhang@astrazeneca.com.
Shuqiong Fan, Email: shuqiong.fan@astrazeneca.com.
Haihua Fu, Email: haihua.fu@astrazeneca.com.
Amanda Gladwin, Email: amanda.gladwin@astrazeneca.com.
Grace Harrod, Email: grace.harrod@astrazeneca.com.
James Stevens, Email: james.stevens@astrazeneca.com.
Victoria Williams, Email: victoria.williams@astrazeneca.com.
Qingqing Ye, Email: qingqing.ye@astrazeneca.com.
Li Zheng, Email: li.zheng@astrazeneca.com.
Richard de Boer, Email: Richard.DeBoer@wh.org.au.
Roy S Herbst, Email: roy.herbst@yale.edu.
Jin-Soo Lee, Email: jslee@ncc.re.kr.
James Vasselli, Email: vassellija@MedImmune.com.
References
- 1.Oxnard GR, Binder A, Janne PA. New targetable oncogenes in non-small-cell lung cancer. J Clin Oncol. 2013;31:1097–104. doi: 10.1200/JCO.2012.42.9829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kohno T, Ichikawa H, Totoki Y, Yasuda K, Hiramoto M, Nammo T, et al. KIF5B-RET fusions in lung adenocarcinoma. Nat Med. 2012;18:375–7. doi: 10.1038/nm.2644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lipson D, Capelletti M, Yelensky R, Otto G, Parker A, Jarosz M, et al. Identification of new ALK and RET gene fusions from colorectal and lung cancer biopsies. Nat Med. 2012;18:382–4. doi: 10.1038/nm.2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ju YS, Lee WC, Shin JY, Lee S, Bleazard T, Won JK, et al. A transforming KIF5B and RET gene fusion in lung adenocarcinoma revealed from whole-genome and transcriptome sequencing. Genome Res. 2012;22:436–45. doi: 10.1101/gr.133645.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cai W, Su C, Li X, Fan L, Zheng L, Fei K, et al. KIF5B-RET fusions in Chinese patients with non-small cell lung cancer. Cancer. 2013;119:1486–94. doi: 10.1002/cncr.27940. [DOI] [PubMed] [Google Scholar]
- 6.Drilon A, Wang L, Hasanovic A, Suehara Y, Lipson D, Stephens P, et al. Response to cabozantinib in patients with RET fusion-positive lung adenocarcinomas. Cancer Discov. 2013;3:630–5. doi: 10.1158/2159-8290.CD-13-0035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li F, Feng Y, Fang R, Fang Z, Xia J, Han X, et al. Identification of RET gene fusion by exon array analyses in “pan-negative” lung cancer from never smokers. Cell Res. 2012;22:928–31. doi: 10.1038/cr.2012.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sasaki H, Shimizu S, Tani Y, Maekawa M, Okuda K, Yokota K, et al. RET expression and detection of KIF5B/RET gene rearrangements in Japanese lung cancer. Cancer Med. 2012;1:68–75. doi: 10.1002/cam4.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Suehara Y, Arcila M, Wang L, Hasanovic A, Ang D, Ito T, et al. Identification of KIF5B-RET and GOPC-ROS1 fusions in lung adenocarcinomas through a comprehensive mRNA-based screen for tyrosine kinase fusions. Clin Cancer Res. 2012;18:6599–608. doi: 10.1158/1078-0432.CCR-12-0838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Takeuchi K, Soda M, Togashi Y, Suzuki R, Sakata S, Hatano S, et al. RET, ROS1 and ALK fusions in lung cancer. Nat Med. 2012;18:378–81. doi: 10.1038/nm.2658. [DOI] [PubMed] [Google Scholar]
- 11.Wang R, Hu H, Pan Y, Li Y, Ye T, Li C, et al. RET fusions define a unique molecular and clinicopathologic subtype of non-small-cell lung cancer. J Clin Oncol. 2012;30:4352–9. doi: 10.1200/JCO.2012.44.1477. [DOI] [PubMed] [Google Scholar]
- 12.Yokota K, Sasaki H, Okuda K, Shimizu S, Shitara M, Hikosaka Y, et al. KIF5B/RET fusion gene in surgically-treated adenocarcinoma of the lung. Oncol Rep. 2012;28:1187–92. doi: 10.3892/or.2012.1908. [DOI] [PubMed] [Google Scholar]
- 13.Lanzi C, Borrello MG, Bongarzone I, Migliazza A, Fusco A, Grieco M, et al. Identification of the product of two oncogenic rearranged forms of the RET proto-oncogene in papillary thyroid carcinomas. Oncogene. 1992;7:2189–94. [PubMed] [Google Scholar]
- 14.Carlson KM, Dou S, Chi D, Scavarda N, Toshima K, Jackson CE, et al. Single missense mutation in the tyrosine kinase catalytic domain of the RET protooncogene is associated with multiple endocrine neoplasia type 2B. Proc Natl Acad Sci U S A. 1994;91:1579–83. doi: 10.1073/pnas.91.4.1579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Donis-Keller H, Dou S, Chi D, Carlson KM, Toshima K, Lairmore TC, et al. Mutations in the RET proto-oncogene are associated with MEN 2A and FMTC. Hum Mol Genet. 1993;2:851–6. doi: 10.1093/hmg/2.7.851. [DOI] [PubMed] [Google Scholar]
- 16.Mulligan LM, Kwok JB, Healey CS, Elsdon MJ, Eng C, Gardner E, et al. Germ-line mutations of the RET proto-oncogene in multiple endocrine neoplasia type 2A. Nature. 1993;363:458–60. doi: 10.1038/363458a0. [DOI] [PubMed] [Google Scholar]
- 17.Elisei R, Cosci B, Romei C, Bottici V, Renzini G, Molinaro E, et al. Prognostic significance of somatic RET oncogene mutations in sporadic medullary thyroid cancer: a 10-year follow-up study. J Clin Endocrinol Metab. 2008;93:682–7. doi: 10.1210/jc.2007-1714. [DOI] [PubMed] [Google Scholar]
- 18.Wedge SR, Ogilvie DJ, Dukes M, Kendrew J, Chester R, Jackson JA, et al. ZD6474 inhibits vascular endothelial growth factor signaling, angiogenesis, and tumor growth following oral administration. Cancer Res. 2002;62:4645–55. [PubMed] [Google Scholar]
- 19.Carlomagno F, Vitagliano D, Guida T, Ciardiello F, Tortora G, Vecchio G, et al. ZD6474, an orally available inhibitor of KDR tyrosine kinase activity, efficiently blocks oncogenic RET kinases. Cancer Res. 2002;62:7284–90. [PubMed] [Google Scholar]
- 20.Matsubara D, Kanai Y, Ishikawa S, Ohara S, Yoshimoto T, Sakatani T, et al. Identification of CCDC6-RET fusion in the human lung adenocarcinoma cell line, LC-2/ad. J Thorac Oncol. 2012;7:1872–6. doi: 10.1097/JTO.0b013e3182721ed1. [DOI] [PubMed] [Google Scholar]
- 21.Herbst RS, Sun Y, Eberhardt WE, Germonpre P, Saijo N, Zhou C, et al. Vandetanib plus docetaxel versus docetaxel as second-line treatment for patients with advanced non-small-cell lung cancer (ZODIAC): a double-blind, randomised, phase 3 trial. Lancet Oncol. 2010;11:619–26. doi: 10.1016/S1470-2045(10)70132-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.de Boer RH, Arrieta Ó, Yang C-H, Gottfried M, Chan V, Raats J, et al. Vandetanib plus pemetrexed for the second-line treatment of advanced non-small-cell lung cancer: a randomized, double-blind Phase III trial. J Clin Oncol. 2011;29:1067–74. doi: 10.1200/JCO.2010.29.5717. [DOI] [PubMed] [Google Scholar]
- 23.Natale RB, Thongprasert S, Greco FA, Thomas M, Tsai C-M, Sunpaweravong P, et al. Phase III trial of vandetanib compared with erlotinib in patients with previously treated advanced non-small-cell lung cancer. J Clin Oncol. 2011;29:1059–66. doi: 10.1200/JCO.2010.28.5981. [DOI] [PubMed] [Google Scholar]
- 24.Lee JS, Hirsh V, Park K, Qin S, Blajman CR, Perng R-P, et al. Vandetanib versus placebo in patients with advanced non-small-cell lung cancer after prior therapy with an epidermal growth factor receptor tyrosine kinase inhibitor: a randomized, double-blind Phase III trial (ZEPHYR) J Clin Oncol. 2012;30:1114–21. doi: 10.1200/JCO.2011.36.1709. [DOI] [PubMed] [Google Scholar]
- 25.AstraZeneca. Global Policy: Bioethics. 2011. Available at: http://www.astrazeneca.com/Responsibility/Code-policies-standards/Our-global-policies.
- 26.MHRA. Good Clinical Practice for Clinical Laboratories. 2009. Available at: http://webarchive.nationalarchives.gov.uk/20141205150130/http://www.mhra.gov.uk/home/groups/is-insp/documents/websiteresources/con051910.pdf.
- 27.Xie L, Su X, Zhang L, Yin X, Tang L, Zhang X, et al. FGFR2 gene amplification in gastric cancer predicts sensitivity to the selective FGFR inhibitor AZD4547. Clin Cancer Res. 2013;19:2572–83. doi: 10.1158/1078-0432.CCR-12-3898. [DOI] [PubMed] [Google Scholar]
- 28.Go H, Jung YJ, Kang HW, Park IK, Kang CH, Lee JW, et al. Diagnostic method for the detection of KIF5B-RET transformation in lung adenocarcinoma. Lung Cancer. 2013;82:44–50. doi: 10.1016/j.lungcan.2013.07.009. [DOI] [PubMed] [Google Scholar]
- 29.Gautschi O, Zander T, Keller FA, Strobel K, Hirschmann A, Aebi S, et al. A patient with lung adenocarcinoma and RET fusion treated with vandetanib. J Thorac Oncol. 2013;8:e43–4. doi: 10.1097/JTO.0b013e31828a4d07. [DOI] [PubMed] [Google Scholar]
- 30.Varella-Garcia M, Xue LG, Mahale S, Berge EM, Bennati C, Le AT, et al. RET rearrangements detected by FISH in “pan-negative” lung adenocarcinoma. J Clin Oncol. 2013;31:abstr 8024. [Google Scholar]


