Abstract
Cyanobacteria represent a promising platform for the production of plant secondary metabolites. Their capacity to express plant P450 proteins, which have essential functions in the biosynthesis of many plant secondary metabolites, makes cyanobacteria ideal for this purpose, and their photosynthetic capability allows cyanobacteria to grow with simple nutrient inputs. This review summarizes the advantages of using cyanobacteria to transgenically produce plant secondary metabolites. Some techniques to improve heterologous gene expression in cyanobacteria are discussed.
Keywords: plant secondary metabolites, phenylpropanoid, cyanobacteria, P450 proteins, photosynthetic growth
Benefits of Plant Secondary Metabolites to Human Health
Secondary metabolites produced by plants confer protection against stresses such as infections, wounding, UV irradiation, and ozone (Douglas, 1996), and allow plants to adapt to continuously changing environmental conditions (Korkina, 2007). Secondary metabolites are largely derived from primary metabolites, such as amino acids and carbohydrates, which are modified by methylation, hydroxylation, or glycosylation (Crozier et al., 2006).
Increasing evidence suggests that plant secondary metabolites, especially the largest group, phenylpropanoids, and their derivatives, are powerful antioxidants that directly scavenge reactive oxygen and nitrogen species (ROS/RNS) (Perron and Brumaghim, 2009). A balance between oxidant and antioxidant systems is critical for maintaining cellular functions. The excessive production of ROS/RNS inside cells results in oxidative stress, loss of cell function, and apoptosis or necrosis. Detoxification of ROS/RNS by enzymatic and non-enzymatic antioxidants minimizes cell damage (Ratnam et al., 2006; Reuter et al., 2010). The antioxidant activity of five phenylpropanoids (i.e., verbascoside, forsythoside, arenareoside, ballotetraside, and caffeoyl malic acid) extracted from the perennial herb Ballota nigra were investigated against superoxide, hydrogen peroxide, hypochlorite, and hydroxyl radicals generated in cell-free systems (Fraga et al., 2010). The ability of these phenylpropanoids to scavenge free radicals was comparable to that of N-acetyl cysteine, an established antioxidant drug (Nordberg and Arner, 2001).
Plant secondary metabolites have become the focus of intensive research, due to their beneficial effects on human health as anticancer, antioxidant, anti-virus, and anti-inflammatory agents. However, these compounds are mainly isolated from plant extracts or from cultivated plant cells at relatively high cost and low yield. It is expensive and technically challenging to chemically synthesize these molecules. Therefore, there is a strong need to develop novel, efficient, and economical methods to produce beneficial plant secondary metabolites.
Cyanobacteria are Suitable for Producing Plant Secondary Metabolites
Several inherent properties of cyanobacteria make them attractive candidates for the biosynthesis of plant secondary metabolites, such as their photosynthetic activity, their amenability to genetic engineering, and their ability to live in tough environments.
Large-scale cyanobacterial cultivation is frequently performed in phototrophic conditions because this approach is cheaper, results in less contamination, and consumes CO2 (Chen et al., 2011). Commercially used open ponds or closed photobioreactors have been developed for large-scale biomass production (Olaizola, 2000; López et al., 2006; Eriksen, 2008; Ugwu et al., 2008; Rodolfi et al., 2009; Singh and Gu, 2010). The nutrient inputs for cyanobacteria are simple: sunlight, CO2, H2O, N, P, and a few mineral nutrients, without carbohydrate feedstocks (Yu et al., 2013).
A wide variety of enzymes and pathways are involved in plant secondary metabolite production. Of these enzymes, cytochrome P450 monooxygenases participate in the pathways to produce compounds such as phenylpropanoids, alkaloids, terpenoids, cyanogenic glycosides, and glucosinolates (Mizutani and Ohta, 2010). They contribute various oxidative modifications of the carbon skeleton using NADPH or NADH as reducing equivalents (Sligar, 1999). P450 sequences have been found in the genome of most known cyanobacterial species (Ke et al., 2005); Anabaena sp. PCC 7120 has six P450 genes (Robert et al., 2010) and Synechocystis sp. PCC 6803 has one (cyp120A1/slr0574) (Ke et al., 2005). Because most eukaryotic P450 proteins are membrane-bound proteins, it is challenging to heterologously express these proteins in other prokaryotes, such as E. coli, which lack developed internal membrane systems. By contrast, cyanobacteria have an intracellular membrane system, i.e., the thylakoids, which function in electron transport. This makes cyanobacteria highly suitable hosts in which to express P450 enzymes (Melis, 1999).
Using Cyanobacteria to Produce Plant Secondary Metabolites
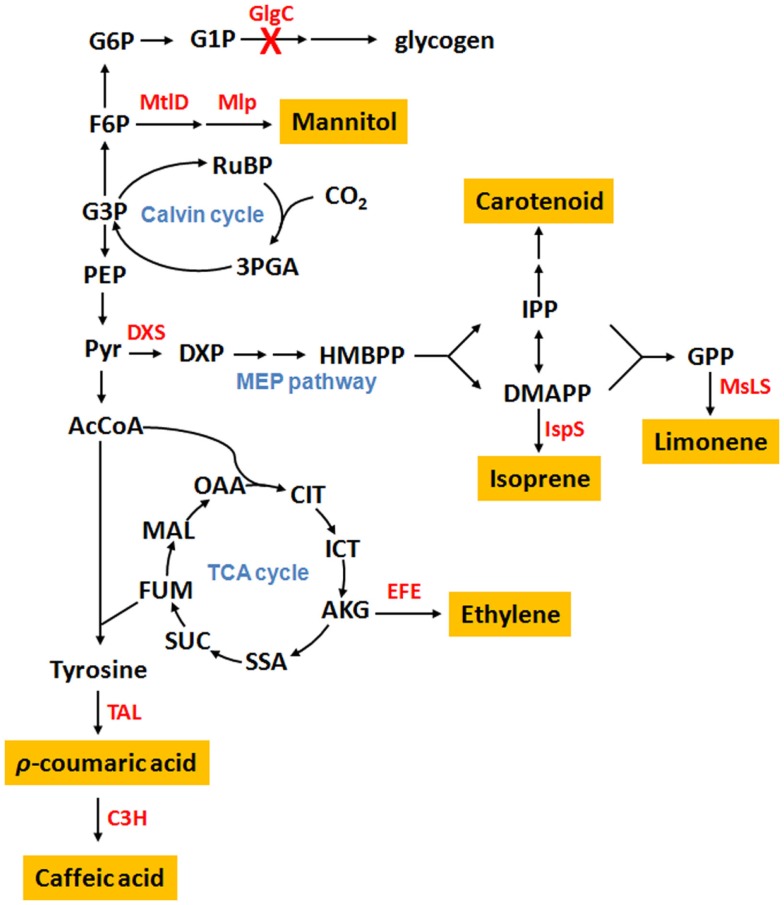
Several cyanobacteria have been engineered as cell factories for the production of plant secondary metabolites (Table 1). Metabolic distributions of produced plant secondary metabolites are summarized in Figure 1 based on their biosynthetic pathways.
Table 1.
Plant secondary metabolites produced by genetically engineered cyanobacteria.
| Products | Yield | Host | Reference |
|---|---|---|---|
| Ethylene | ~512 μg L−1 h−1 OD730−1b | S. elongatus PCC 7942 | Takahama et al. (2003) |
| ~3.9 μg L−1 h−1 OD730−1b | S. elongatus PCC 7942 | Jindou et al. (2014) | |
| Isoprene | 50 μg (g dry cell)−1 day−1 | Synechocystis sp. PCC 6803 | Lindberg et al. (2010) |
| ~125 μg (g dry cell)−1 day−1 | Synechocystis sp. PCC 6803 | Bentley et al. (2014) | |
| Caffeic acida | 7.2 mg L−1 | Synechocystis sp. PCC 6803 | Xue et al. (2014b) |
| ρ-coumaric acida | 82.6 mg L−1 | Synechocystis sp. PCC 6803 | Xue et al. (2014a) |
| Mannitol | 0.15 g L−1 day−1 | Synechococcus sp. PCC 7002 | Jacobsen and Frigaard (2014) |
| Limonene | 56 μg L−1 day−1 | Synechocystis sp. PCC 6803 | Kiyota et al. (2014) |
| 50 μg L−1 h−1 | Synechococcus sp. PCC 7002 | Davies et al. (2014) | |
| Carotenoida | 8.4 mg (g dry cell)−1 | Synechocystis sp. PCC 6803 | Kudoh et al. (2014) |
aTime over which the yield was achieved is not provided in the publication.
Figure 1.
Cyanobacterial metabolic pathways for the production of plant secondary metabolites. Abbreviation: G6P, glucose-6-phosphate; G1P, glucose-1-phosphate; F6P, fructose-6-phosphate; G3P, glyceraldehyde-3-phosphate; RuBP, ribulose-1,5-diphosphate; 3PGA, 3-phosphoglycerate; PEP, phosphoenolpyruvate; Pyr, pyruvate; DXP, 1-deoxy-d-xylulose 5-phosphate; HMBPP, 4-hydroxy-3-methylbut-2-enyl diphosphate; IPP, isopentenyl pyrophosphate; DMAPP, dimethylallyl pyrophosphate; GPP, geranyl pyrophosphate; AcCoA, acetyl-CoA; OAA, oxoacetate; CIT, citrate; ICT, isocitrate; AKG, α-ketoglutarate; SSA, succinic semialdehyde; SUC, succinate; FUM, fumarate; MAL, malate; GlgC, ADP-glucose pyrophosphorylase; MtlD, mannitol-1-phosphate dehydrogenase; Mlp, mannitol-1-phosphatase; DXS, 1-deoxy-d-xylulose-5-phosphate synthase; IspS, isoprene synthase; MsLS, Mentha spicata limonene synthase; EFE, ethylene-forming enzyme; TAL, tyrosine ammonia-lyase; C3H, ρ-coumarate 3-hydroxylase; H.
Tricarboxylic Acid Cycle
To produce ethylene, an ethylene-forming enzyme gene (efe) from Pseudomonas syringae was inserted into the Synechococcus elongatus PCC 7942 chromosome at the psbAI locus, and the recombinant strain produced ~512 μg ethylene L−1 h−1 OD730−1 (Takahama et al., 2003). An artificial chimeric enzyme complex containing two ethylene-generating enzymes from Solanum lycopersicum (tomato) was introduced into S. elongatus PCC 7942, and the strain produced ethylene with a titer of ~3.9 μg ethylene L−1 h−1 OD730−1 (Jindou et al., 2014).
2-C-Methyl-d-Erythritol 4-Phosphate Pathway
Isoprene, which protects plants from abiotic stresses (Sharkey et al., 2008) and serves as a renewable biofuel, was produced in Synechocystis sp. PCC 6803 with a titer of 50 μg (g dry cell)−1 day−1 by expressing an isoprene synthase gene (ispS) from Pueraria Montana (Lindberg et al., 2010). By co-expressing seven genes of a heterologous mevalonic acid biosynthetic pathway from Enterococcus faecalis and Streptococcus pneumoniae in that ispS transformant, the yield of isoprene production increased 2.5-fold (Bentley et al., 2014). To produce limonene, a limonene synthase gene (LMS) from Schizonepeta tenuifolia was expressed in Synechocystis sp. PCC 6803, and the titer was 41 μg L−1 day−1. By overexpressing three genes (dxs, crtE, and ipi) in the 2-C-methyl-d-erythritol-4-phosphate (MEP) pathway to increase the supply of limonene substrate, geranyl pyrophosphate (GPP), limonene production was improved by 1.4-fold (Kiyota et al., 2014). In another study, 50 μg L−1 h−1 limonene was produced by a Synechococcus sp. PCC 7002 strain expressing codon optimized Mentha spicata limonene synthase gene (mslS) (Davies et al., 2014). Carotenoids are naturally produced terpenoid-type molecules by cyanobacteria through MEP pathway. Recently, Kai et al. overexpressed a key enzyme, 1-deoxy-d-xylulose-5-phosphate synthase (DXS), in Synechocystis sp. PCC 6803, and the carotenoid level in the strain was 1.5 times higher than that in the wild-type strain (Kudoh et al., 2014).
Other Metabolites
Our group recently constructed a Synechocystis sp. PCC 6803 strain genetically engineered to produce caffeic acid, in which an Arabidopsis ρ-coumarate 3-hydroxylase (encoded by ref8) was expressed. With the addition of substrate, ρ-coumaric acid, to the medium, the titer reached 7.2 mg L−1 (Xue et al., 2014b). In another report, the direct precursor of caffeic acid, ρ-coumaric acid, was produced at a concentration of 82.6 mg L−1 by a Synechocystis sp. PCC 6803 mutant, which harbored a tyrosine ammonia-lyase (TAL) gene (sam8) from Saccharothrix espanaensis and lacked a hypothetical laccase gene (Xue et al., 2014a). Mannitol was produced in Synechococcus sp. PCC 7002 with a titer of 0.15 g L−1 day−1 by heterologously expressing mannitol-1-phosphate dehydrogenase (mtlD) from E. coli and mannitol-1-phosphatase (mlp) from Eimeria tenella. By genetically inactivating glycogen synthesis, the yield increased 3.2-fold (Jacobsen and Frigaard, 2014).
Some Considerations for the Genetic Manipulation of Cyanobacteria
Although many species of cyanobacteria have been successfully engineered to express heterologous genes and produce valuable compounds, as summarized above, this approach remains challenging due to epigenetic suppression, poor transcription, and protein post-translational modifications. Here, we highlight some factors that need to be considered when genetically engineering cyanobacteria to produce plant secondary metabolites.
Inactivation of Glycogen Synthesis Pathway
Glycogen is one of the dominant carbon sinks for cyanobacteria. Up to 60% of dry cell weight is converted to glycogen when cyanobacteria are growing in nitrogen-limited media (Allen and Smith, 1969). Therefore, inactivation of glycogen synthesis pathway should allow a greater proportion of carbon partitioning to non-native products. In a recent study, the glgC gene (coding for glucose-1-phosphate adenylyltransferase) was knocked out from S. elongatus PCC 7942 strain producing isobutanol. The amount of total fixed carbon flux toward isobutanol production was increased by 2.5-fold after deletion of glgC under constant high light condition (150 μE s−1 m−2) (Li et al., 2014). Another example is a Synechoccocuus sp. PCC 7002 strain expressing a mannitol biosynthetic pathway, which produces mannitol equivalent to 10% of cell dry weight under 250 μE s−1 m−2. By blocking glycogen biosynthesis, the yield increased to 32% (Jacobsen and Frigaard, 2014).
Laccase Knockout
Laccases are enzymes that oxidize many phenolic compounds and are present in a large variety of species. Oxidized substrates become free radicals, which are unstable and can be modified by other non-enzymatic reactions, such as hydration, polymerization, and disproportionation (Thurston, 1994). In a recent study, a putative laccase gene, slr1573, was found in Synechocystis sp. PCC 6803. After knocking out slr1573 from its genome, the production of p-coumaric acid increased by more than 25-fold in the strain expressing TAL enzyme (Xue et al., 2014a). Although reports of laccase functions in cyanobacteria are limited, this gene might be a barrier for phenylpropanoids production using cyanobacteria.
Codon Optimization
A codon usage bias exists for most protein-coding genes expressed in heterologous hosts. It is critical to optimize the codon usage in order to obtain high levels of overexpression of heterologous genes (Steen et al., 2010; Bond-Watts et al., 2011; Paddon et al., 2013). Several factors need to be considered when designing new gene sequences, including host codon usage frequency (Angov et al., 2011), AT/GC ratio (Gustafsson, 2009), mRNA secondary structure (Tang et al., 2011), repeat sequences (Li et al., 2011), and restriction sites for cloning (Raab et al., 2010). Our group recently constructed a transgenic Synechocystis sp. PCC 6803 that heterologously expressed ref8 from Arabidopsis thaliana, which encodes a P450 enzyme ρ-coumarate 3-hydroxylase, and was capable of producing caffeic acid (Xue et al., 2014b).
Transgene Stability
Several reports describe the instability of transgenes in genetically engineered cyanobacteria. During subculturing of an ethylene-producing S. elongatus PCC 7942 transformant, a duplicated sequence in the efe gene was found that resulted in a truncated and non-functional gene (Takahama et al., 2003). In another study, a Synechocystis sp. PCC 6803 strain was genetically modified to produce lactic acid by integrating a lactate dehydrogenase gene in the genome. Wild-type phenotypic colonies appeared during segregation, and further analysis identified a nonsense mutation in the transgene (Angermayr et al., 2012). Similarly, in an effort to produce isopropanol, four enzymes in the isopropanol biosynthetic pathway from Clostridium acetobutylicum, Clostridium beijerinckii, and E. coli were expressed in S. elongatus PCC 7942. The authors repeatedly found a missense mutation in one gene (atoD), which reduced the enzymatic activity (Kusakabe et al., 2013). The mechanism underlying the instability has not been determined. A common approach is to select transformants carrying single unrearranged transgenes and to keep these as backups.
Markerless System
The standard method of genetically engineering cyanobacteria involves transformation of a plasmid carrying genes of interest and integration of these foreign genes into the cyanobacterial genome at specific sites through double-crossover homologous recombination (Vermaas, 1996). Antibiotic resistances are used as selectable markers for positive transformants. In some cases, when multiple gene integrations are required in one strain, the number of available antibiotic markers restricts the number of insertions, and thus markerless genomic mutations are desirable. Currently, the most widely used markerless technique for cyanobacteria was developed using a plasmid containing sacB (a levansucrase gene) and an antibiotic resistance cassette (Lagarde et al., 2000). In the first transformation, a target region in the genome is replaced by the sacB-antibiotic resistance cassette and antibiotic resistance is used for positive selection. The second transformation is performed by replacing the sacB-antibiotic resistance cassette with the gene of interest. Sucrose is added to the medium for negative selection. The levansucrase encoded by sacB converts sucrose to levans, a toxic polymer that kills the bacteria. Consequently, only markerless mutants can survive in the presence of sucrose. For example, a Synechocystis sp. PCC 6803 mutant was constructed for ethanol production by integrating pyruvate decarboxylase (pdc) and alcohol dehydrogenase II (adh) genes into the genome. An aphX/sacB selection cassette was used to generate a markerless transformant, which is able to produce ethanol at a titer of 5.2 mmol OD730 unit−1 L−1 day−1 (Dexter and Fu, 2009). Recently, an alternative strategy was developed using a one-step gene replacement approach (Viola et al., 2014). The plasmid designed for this strategy harbors an nptI (kanamycin resistance gene)-sacB selection cassette flanked by 5′and 3′ fragments of the gene of interest, which have overlapping segments. After transformation, the nptI-sacB cassette with the exogenous gene is integrated into the genome through double-crossover recombination, and complete segregations are selected based on kanamycin resistance. Then, a second single crossover event occurred between the overlapping fragments, leading to the excision of the nptI-sacB cassette. Mutants that had undergone the second recombination were screened on sucrose in the absence of kanamycin.
Another successful counter selection method based on acrylate toxicity was developed for Synechococcus sp. PCC 7002, in which sacB counter selection system did not work (Begemann et al., 2013). After one transformation step, a native acsA gene (encoding an acetyl-CoA ligase) is replaced by the gene of interest, and the loss of acetyl-CoA ligase function overcomes growth inhibition by acrylate. Thus, positive transformants can be screened on growth medium with addition of acrylate. After reinsertion of the acsA gene into a neutral site on genome, multiple gene integrations can be achieved.
Limitations of Cyanobacteria for Producing Plant Secondary Metabolites
In addition to the potential advantages of using cyanobacteria to produce plant secondary metabolites mentioned above, this technology is still in its infancy and numerous challenges need to be addressed. For instance, production titers from engineered cyanobacteria are much lower than that from heterotrophic fermentation, and efficient, large volume bioreactors need to be designed. Another consideration is the protein post-translational modifications. Because some plant enzymes in the secondary metabolite biosynthesis pathways require post-translational modifications (e.g., glycosylation), they could be non-functional when expressed in cyanobacteria that are not equipped with these machineries.
Conclusion
Cyanobacteria can be used as cell factories to convert solar energy into high value products, such as plant secondary metabolites, which are beneficial to human health. Their high photosynthetic efficiency and ease of genetic manipulation make cyanobacteria a better choice for this purpose than other organisms. Recently, researchers have put efforts into engineering cyanobacteria to produce plant secondary metabolites from sunlight and CO2. However, there are still challenges for engineering applications of cyanobacteria, such as improvement of product titers, bioprocess scale-up, and product recovery.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
- Allen M. M., Smith A. J. (1969). Nitrogen chlorosis in blue-green algae. Arch. Mikrobiol. 69, 114–120 10.1007/BF00409755 [DOI] [PubMed] [Google Scholar]
- Angermayr S. A., Paszota M., Hellingwerf K. J. (2012). Engineering a cyanobacterial cell factory for production of lactic acid. Appl. Environ. Microbiol. 78, 7098–7106. 10.1128/AEM.01587-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angov E., Legler P. M., Mease R. M. (2011). “Adjustment of codon usage frequencies by codon harmonization improves protein expression and folding,” in Heterologous Gene Expression in E. coli, eds Evans T. C., Jr, Xu M. Q. (Berlin: Springer; ), 1–13. [DOI] [PubMed] [Google Scholar]
- Begemann M. B., Zess E. K., Walters E. M., Schmitt E. F., Markley A. L., Pfleger B. F. (2013). An organic acid based counter selection system for cyanobacteria. PLoS One 8:e76594. 10.1371/journal.pone.0076594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentley F. K., Zurbriggen A., Melis A. (2014). Heterologous expression of the mevalonic acid pathway in cyanobacteria enhances endogenous carbon partitioning to isoprene. Mol. Plant. 7, 71–86. 10.1093/mp/sst134 [DOI] [PubMed] [Google Scholar]
- Bond-Watts B. B., Bellerose R. J., Chang M. C. (2011). Enzyme mechanism as a kinetic control element for designing synthetic biofuel pathways. Nat. Chem. Biol. 7, 222–227. 10.1038/nchembio.537 [DOI] [PubMed] [Google Scholar]
- Chen C.-Y., Yeh K.-L., Aisyah R., Lee D.-J., Chang J.-S. (2011). Cultivation, photobioreactor design and harvesting of microalgae for biodiesel production: a critical review. Bioresour. Technol. 102, 71–81. 10.1016/j.biortech.2010.06.159 [DOI] [PubMed] [Google Scholar]
- Crozier A., Jaganath I. B., Clifford M. N. (2006). “Phenols, polyphenols and tannins: an overview,” in Plant Secondary Metabolites: Occurrence, Structure and Role in the Human Diet, eds Crozier A., Clifford M. N., Ashihara H. (Oxford, UK: Blackwell Publishing Ltd; ), 1–24. [Google Scholar]
- Davies F. K., Work V. H., Beliaev A. S., Posewitz M. C. (2014). Engineering limonene and bisabolene production in wild type and a glycogen-deficient mutant of Synechococcus sp. PCC 7002. Front. Bioeng. Biotechnol. 2:21. 10.3389/fbioe.2014.00021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dexter J., Fu P. (2009). Metabolic engineering of cyanobacteria for ethanol production. Energy Environ. Sci. 2, 857–864 10.1039/b811937f [DOI] [Google Scholar]
- Douglas C. J. (1996). Phenylpropanoid metabolism and lignin biosynthesis: from weeds to trees. Trends Plant Sci. 1, 171–178 10.1016/1360-1385(96)10019-4 [DOI] [Google Scholar]
- Eriksen N. T. (2008). The technology of microalgal culturing. Biotechnol. Lett. 30, 1525–1536. 10.1007/s10529-008-9740-3 [DOI] [PubMed] [Google Scholar]
- Fraga C. G., Galleano M., Verstraeten S. V., Oteiza P. I. (2010). Basic biochemical mechanisms behind the health benefits of polyphenols. Mol. Aspects Med. 31, 435–445. 10.1016/j.mam.2010.09.006 [DOI] [PubMed] [Google Scholar]
- Gustafsson C. (2009). “Tools designed to regulate translational efficiency,” in The Metabolic Pathway Engineering Handbook, ed. Smolke C. D. (Boca Raton, FL: CRC Press; ), 9–14. [Google Scholar]
- Jacobsen J. H., Frigaard N.-U. (2014). Engineering of photosynthetic mannitol biosynthesis from CO2 in a cyanobacterium. Metab. Eng. 21, 60–70. 10.1016/j.ymben.2013.11.004 [DOI] [PubMed] [Google Scholar]
- Jindou S., Ito Y., Mito N., Uematsu K., Hosoda A., Tamura H. (2014). Engineered platform for bioethylene production by a cyanobacterium expressing a chimeric complex of plant enzymes. ACS Synth. Biol. 3, 487–496. 10.1021/sb400197f [DOI] [PubMed] [Google Scholar]
- Ke N., Baudry J., Makris T. M., Schuler M. A., Sligar S. G. (2005). A retinoic acid binding cytochrome P450: CYP120A1 from Synechocystis sp. PCC 6803. Arch. Biochem. Biophys. 436, 110–120. 10.1016/j.abb.2005.01.011 [DOI] [PubMed] [Google Scholar]
- Kiyota H., Okuda Y., Ito M., Hirai M. Y., Ikeuchi M. (2014). Engineering of cyanobacteria for the photosynthetic production of limonene from CO2. J. Biotechnol. 185, 1–7. 10.1016/j.jbiotec.2014.05.025 [DOI] [PubMed] [Google Scholar]
- Korkina L. (2007). Phenylpropanoids as naturally occurring antioxidants: from plant defense to human health. Cell. Mol. Biol. 53, 15–25. 10.1170/T772 [DOI] [PubMed] [Google Scholar]
- Kudoh K., Kawano Y., Hotta S., Sekine M., Watanabe T., Ihara M. (2014). Prerequisite for highly efficient isoprenoid production by cyanobacteria discovered through the over-expression of 1-deoxy-d-xylulose 5-phosphate synthase and carbon allocation analysis. J. Biosci. Bioeng. 118, 20–28. 10.1016/j.jbiosc.2013.12.018 [DOI] [PubMed] [Google Scholar]
- Kusakabe T., Tatsuke T., Tsuruno K., Hirokawa Y., Atsumi S., Liao J. C., et al. (2013). Engineering a synthetic pathway in cyanobacteria for isopropanol production directly from carbon dioxide and light. Metab. Eng. 20, 101–108. 10.1016/j.ymben.2013.09.007 [DOI] [PubMed] [Google Scholar]
- Lagarde D., Beuf L., Vermaas W. (2000). Increased production of zeaxanthin and other pigments by application of genetic engineering techniques to Synechocystis sp. strain PCC 6803. Appl. Environ. Microbiol. 66, 64–72. 10.1128/AEM.66.1.64-72.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan E. I., Liao J. C. (2011). Metabolic engineering of cyanobacteria for 1-butanol production from carbon dioxide. Metab. Eng. 13, 353–363. 10.1016/j.ymben.2011.04.004 [DOI] [PubMed] [Google Scholar]
- Li W., Ng I.-S., Fang B., Yu J., Zhang G. (2011). Codon optimization of 1, 3-propanediol oxidoreductase expression in Escherichia coli and enzymatic properties. Electron. J. Biotechnol. 14, 7–7 10.2225/vol14-issue4-fulltext-9 [DOI] [Google Scholar]
- Li X., Shen C. R., Liao J. C. (2014). Isobutanol production as an alternative metabolic sink to rescue the growth deficiency of the glycogen mutant of Synechococcus elongatus PCC 7942. Photosyn. Res. 120, 301–310. 10.1007/s11120-014-9987-6 [DOI] [PubMed] [Google Scholar]
- Lindberg P., Park S., Melis A. (2010). Engineering a platform for photosynthetic isoprene production in cyanobacteria, using Synechocystis as the model organism. Metab. Eng. 12, 70–79. 10.1016/j.ymben.2009.10.001 [DOI] [PubMed] [Google Scholar]
- López M., Sánchez E., Lopez J., Fernández F., Sevilla J., Rivas J., et al. (2006). Comparative analysis of the outdoor culture of Haematococcus pluvialis in tubular and bubble column photobioreactors. J. Biotechnol. 123, 329–342. 10.1016/j.jbiotec.2005.11.010 [DOI] [PubMed] [Google Scholar]
- Melis A. (1999). Photosystem-II damage and repair cycle in chloroplasts: what modulates the rate of photodamage in vivo? Trends Plant Sci. 4, 130–135. 10.1016/S1360-1385(99)01387-4 [DOI] [PubMed] [Google Scholar]
- Mizutani M., Ohta D. (2010). Diversification of P450 genes during land plant evolution. Annu. Rev. Plant Biol. 61, 291–315. 10.1146/annurev-arplant-042809-112305 [DOI] [PubMed] [Google Scholar]
- Nordberg J., Arner E. S. (2001). Reactive oxygen species, antioxidants, and the mammalian thioredoxin system. Free Radic. Biol. Med. 31, 1287–1312. 10.1016/S0891-5849(01)00724-9 [DOI] [PubMed] [Google Scholar]
- Olaizola M. (2000). Commercial production of astaxanthin from Haematococcus pluvialis using 25,000-liter outdoor photobioreactors. J. Appl. Phycol. 12, 499–506 10.1023/A:1008159127672 [DOI] [Google Scholar]
- Paddon C., Westfall P., Pitera D., Benjamin K., Fisher K., Mcphee D., et al. (2013). High-level semi-synthetic production of the potent antimalarial artemisinin. Nature 496, 528–532. 10.1038/nature12051 [DOI] [PubMed] [Google Scholar]
- Perron N. R., Brumaghim J. L. (2009). A review of the antioxidant mechanisms of polyphenol compounds related to iron binding. Cell Biochem. Biophys. 53, 75–100. 10.1007/s12013-009-9043-x [DOI] [PubMed] [Google Scholar]
- Raab D., Graf M., Notka F., Schödl T., Wagner R. (2010). The GeneOptimizer Algorithm: using a sliding window approach to cope with the vast sequence space in multiparameter DNA sequence optimization. Syst. Synth. Biol. 4, 215–225. 10.1007/s11693-010-9062-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratnam D. V., Ankola D., Bhardwaj V., Sahana D. K., Kumar M. (2006). Role of antioxidants in prophylaxis and therapy: a pharmaceutical perspective. J. Control. Release 113, 189–207. 10.1016/j.jconrel.2006.04.015 [DOI] [PubMed] [Google Scholar]
- Reuter S., Gupta S. C., Chaturvedi M. M., Aggarwal B. B. (2010). Oxidative stress, inflammation, and cancer: how are they linked? Free Radic. Biol. Med. 49, 1603–1616. 10.1016/j.freeradbiomed.2010.09.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert F. O., Pandhal J., Wright P. C. (2010). Exploiting cyanobacterial P450 pathways. Curr. Opin. Microbiol. 13, 301–306. 10.1016/j.mib.2010.02.007 [DOI] [PubMed] [Google Scholar]
- Rodolfi L., Chini Zittelli G., Bassi N., Padovani G., Biondi N., Bonini G., et al. (2009). Microalgae for oil: strain selection, induction of lipid synthesis and outdoor mass cultivation in a low-cost photobioreactor. Biotechnol. Bioeng. 102, 100–112. 10.1002/bit.22033 [DOI] [PubMed] [Google Scholar]
- Sharkey T. D., Wiberley A. E., Donohue A. R. (2008). Isoprene emission from plants: why and how. Ann. Bot. 101, 5–18 10.1093/aob/mcm240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen C. R., Liao J. C. (2012). Photosynthetic production of 2-methyl-1-butanol from CO2 in cyanobacterium Synechococcus elongatus PCC7942 and characterization of the native acetohydroxyacid synthase. Energy Environ. Sci. 5, 9574–9583 10.1039/c2ee23148d [DOI] [Google Scholar]
- Singh J., Gu S. (2010). Commercialization potential of microalgae for biofuels production. Renew. Sustain. Energ. Rev. 14, 2596–2610 10.1016/j.rser.2010.06.014 [DOI] [Google Scholar]
- Sligar S. (1999). Nature s universal oxygenases: the cytochromes P450. Essays Biochem. 34, 71–83. [DOI] [PubMed] [Google Scholar]
- Steen E. J., Kang Y., Bokinsky G., Hu Z., Schirmer A., Mcclure A., et al. (2010). Microbial production of fatty-acid-derived fuels and chemicals from plant biomass. Nature 463, 559–562. 10.1038/nature08721 [DOI] [PubMed] [Google Scholar]
- Takahama K., Matsuoka M., Nagahama K., Ogawa T. (2003). Construction and analysis of a recombinant cyanobacterium expressing a chromosomally inserted gene for an ethylene-forming enzyme at the psbAI locus. J. Biosci. Bioeng. 95, 302–305. 10.1016/S1389-1723(03)80034-8 [DOI] [PubMed] [Google Scholar]
- Tang L., Jiang R., Zheng K., Zhu X. (2011). Enhancing the recombinant protein expression of halohydrin dehalogenase HheA in Escherichia coli by applying a codon optimization strategy. Enzyme Microb. Technol. 49, 395–401. 10.1016/j.enzmictec.2011.06.021 [DOI] [PubMed] [Google Scholar]
- Thurston C. F. (1994). The structure and function of fungal laccases. Microbiology 140, 19–26 10.1099/13500872-140-1-19 [DOI] [Google Scholar]
- Ugwu C., Aoyagi H., Uchiyama H. (2008). Photobioreactors for mass cultivation of algae. Bioresour. Technol. 99, 4021–4028. 10.1016/j.biortech.2007.01.046 [DOI] [PubMed] [Google Scholar]
- Vermaas W. (1996). Molecular genetics of the cyanobacterium Synechocystis sp. PCC 6803: principles and possible biotechnology applications. J. Appl. Phycol. 8, 263–273 10.1007/BF02178569 [DOI] [Google Scholar]
- Viola S., Rühle T., Leister D. (2014). A single vector-based strategy for markerless gene replacement in Synechocystis sp. PCC 6803. Microb. Cell Fact. 13, 1–12. 10.1186/1475-2859-13-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B., Wang J., Zhang W., Meldrum D. R. (2012). Application of synthetic biology in cyanobacteria and algae. Front. Microbiol. 3:344. 10.3389/fmicb.2012.00344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue Y., Zhang Y., Cheng D., Daddy S., He Q. (2014a). Genetically engineering Synechocystis sp. Pasteur Culture Collection 6803 for the sustainable production of the plant secondary metabolite p-coumaric acid. Proc. Natl. Acad. Sci. U S A 111, 9449–9454. 10.1073/pnas.1323725111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue Y., Zhang Y., Grace S., He Q. (2014b). Functional expression of an Arabidopsis p450 enzyme, p-coumarate-3-hydroxylase, in the cyanobacterium Synechocystis PCC 6803 for the biosynthesis of caffeic acid. J. Appl. Phycol. 26, 219–226 10.1007/s10811-013-0113-5 [DOI] [Google Scholar]
- Yu Y., You L., Liu D., Hollinshead W., Tang Y. J., Zhang F. (2013). Development of Synechocystis sp. PCC 6803 as a phototrophic cell factory. Mar. Drugs 11, 2894–2916. 10.3390/md11082894 [DOI] [PMC free article] [PubMed] [Google Scholar]