Abstract
Activating mutations in EGFR are present in a subset of lung cancers, and predict sensitivity to EGFR tyrosine kinase inhibitors (TKIs). Acquisition of EGFR T790M is the most common mechanism of resistance to EGFR TKIs and rarely is seen prior to treatment. Germline EGFR T790M mutations have been reported, although the penetrance and clinical significance of this mutation is unknown. We describe the identification of a patient with an EGFR T790M germline mutation and subsequent germline testing in her unaffected family members. Genetic testing revealed two additional EGFR T790M germline carriers, one of whom was subsequently diagnosed with metastatic lung adenocarcinoma.
Somatic, activating mutations in EGFR (Exon 19 deletions, as well as point mutations in L858R, G719, L861) promote oncogenesis in a specific subset of lung adenocarcinomas1. In patients with EGFR mutant lung cancer, EGFR tyrosine kinase inhibitors (TKI) are more effective than cytotoxic chemotherapy although patients develop resistance after a median of 12-16 months on therapy2. The most common mechanism of resistance to EGFR TKIs is the acquisition of the EGFR T790M point mutation which occurs in 60% of patients3,4. De novo EGFR T790M mutations are rarely seen by standard genotyping methods, and occur in <1% of all lung cancers and approximately 2% of all EGFR mutant lung cancers5.
Germline EGFR T790M mutations have been reported in association with familial non small cell lung cancer although the degree of risk, penetrance and the resultant clinical syndrome has not been fully elucidated6. Here we describe the results of comprehensive molecular testing on multiple synchronous lung tumors in a patient who had genetic testing revealing a germline EGFR T790M mutation. The identification of the germline mutation in the proband led to cohort testing of her unaffected relatives resulting in the discovery of two additional EGFR T790M germline carriers, one of whom was subsequently diagnosed with metastatic lung adenocarcinoma.
The index case is a 44 year old never smoker with no family history of lung cancer who initially presented with enlarged axillary lymph nodes. Imaging revealed multiple bilateral ground glass opacities within the lungs. She underwent right sided wedge biopsies, and biopsies of the right middle lobe and right lower lobe revealed well-differentiated adenocarcinoma. The right middle lobe nodule harbored both an EGFR T790M mutation and a 15bp EGFR exon 19 deletion. She underwent a left sided thoracotomy with multiple wedge biopsies of the left lower and left upper lobes to further define the extent of her disease. Four discrete left lower lobe (LLL) nodules and 1 left upper lobe (LUL) nodule were excised and were consistent with morphologically distinct adenocarcinomas indicating synchronous primary lung cancers rather than metastatic disease (Figure 1). The EGFR T790M mutation was identified in all samples upon routine diagnostic molecular testing. Four samples (3 from the LLL, 1 from the LUL) harbored an EGFR L858R point mutation. One sample from the LLL had a 3bp deletion in EGFR exon 19 found using fragment analysis and confirmed by Sanger sequencing (Table 1).
Table 1.
Synchronous tumors and resultant diagnostic molecular testing
| Tumor | Standard Molecular Path | NGS Mutation Profile |
|---|---|---|
| T-1 (RML) | EGFR T790M, 15bp exon 19 del | |
| T-2 (RUL) | EGFR T790M | EGFR T790M, ARID1A K1938N |
| T-3 (LLL) | EGFR T790M, L858R | EGFR T790M, L858R |
| T-4 (LLL) | EGFR T790M, L858R | EGFR T790M, L858R |
| T-5 (LLL) | EGFR T790M, L858R | |
| T-6 (LLL) | EGFR T790M, 3bp exon 19 del | |
| T-7 (LUL) | EGFR T790M, L858R |
Due to a strong family history of cancer (Table 2) and her new diagnosis of multiple primary lung cancers, she was referred to clinical genetics for evaluation for a germline susceptibility to cancer. She had testing performed on peripheral blood for both EGFR*2369C to T (T790M) and BRCA1/2 due to her family history of breast and ovarian cancer. She tested positive for the germline EGFR T790M mutation, and was also found to have a BRCA 2 variation of uncertain significance at L459S (1604 T to C). She is followed regularly with interval CT scans that indicate stable bilateral pulmonary nodules and ground glass opacities (Figure 2, Patient A). She continues to be monitored expectantly on no systemic therapy.
Table 2.
Family history of proband
| Family member | Breast | Ovarian Ca | Pancreatic Cancer | Prostate Cancer | Leukemia |
|---|---|---|---|---|---|
| Father | X | ||||
| Maternal aunt #1 | X (pre-menopausal) | ||||
| Maternal aunt #2 | X (pre-menopausal) | X | |||
| Maternal aunt #3 | X (post-menopausal) | ||||
| Maternal aunt #4 | X | ||||
| Maternal cousin | X (pre-menopausal) | ||||
| Maternal grandfather | X |
Further molecular analysis of this patient's synchronous tumors was performed in order to gain insight on the molecular progression from a germline susceptibility mutation, EGFR T790M, to clinically evident malignancy. Massively parallel sequencing 7 was performed of all exons of 230 cancer genes (see supplementary data for gene list). The technique used to sequence exons of genes of interest7 and the technical implementation of an oncogene screening profile8 have been described previously. Exonic DNA was captured via solution-based hybrid selection9 and sequenced on the Illumina HiSeq platform. The sequencing data was analyzed for base mutations, insertions, deletions, copy number alterations and genomic rearrangements in all target genes.
A peripheral blood sample and three tumor samples (T-2, T-3, T-4) yielded adequate DNA for analysis on this platform. The EGFR T790M variant was detected in all three tumor samples (T-2, T-3, T-4) and the normal blood at allele frequencies of 48%, 51%, 48% and 52%, respectively, as expected for a germline variant. The BRCA2 L459S germline variant was identified in the peripheral blood normal tissue in 413 of 868 reads. T-2 had an additional ARID1A K1938N somatic point mutation at an allele frequency of 19%, T-3 and T-4 had EGFR L858R somatic point mutations at allele frequencies of 11% and 16%, respectively. No additional insertions, deletions, point mutations or rearrangements were identified in the 230 cancer genes analyzed (Figure 3).
After our proband was identified as a carrier of a germline EGFR T790M mutation, her mother was referred to clinical genetics for germline EGFR T790M testing to ascertain whether the mutation in the proband was a de novo event or an inherited EGFR 2369C>T (T790M) germline mutation. After the initial clinical genetics evaluation, the proband's mother was diagnosed with metastatic lung adenocarcinoma presenting with bilateral ground glass opacities and pulmonary nodules similar to the proband. She returned to clinical genetics and was informed of her positive EGFR 2369C>T (T790M) germline mutation. BRCA 2 testing for the variation of uncertain significance at L459S was not performed due to lack of insurance coverage for the requested test.
Due to the T790M germline mutation found in two family members, we offered all interested family members genetic counseling and germline testing. Eleven family members (Figure 4) came to an informational session where voluntary germline testing was offered. Testing was performed on seven family members. Of the seven people tested, the proband's daughter was the only one found to carry the EGFR 2369C>T (T790M) germline mutation (Table 3).
Table 3.
Germline testing results
| Location on Pedigree | Relationship to Proband | Tested (result) |
|---|---|---|
| 1 | Mother | Yes (positive) |
| 2 | Daughter | Yes (positive) |
| 3 | Sister | Yes (negative) |
| 4 | Brother | Yes (negative) |
| 5 | Niece | Yes (negative) |
| 6 | Maternal uncle | Yes (negative) |
| 7 | 1st cousin | Yes (negative) |
| 8 | 1st cousin | Yes (negative) |
| 9 | 1st cousin | No |
| 10 | 1st cousin | No |
| Not pictured | 2nd cousin | No |
| Not pictured | 3rd cousin | No |
Germline EGFR T790M mutation carriers need to be prospectively studied in order to better understand the clinical implications of this germline mutation. In mouse models, the presence of the EGFR T790M mutation induces tumor formation, suggesting that the mutation is sufficient for oncogenesis10. Clinically, the presence of germline EGFR T790M is associated with the development of lung adenocarcinoma, although the penetrance and clinical significance of this mutation is not fully elucidated. Screening for germline T790M mutations in unselected populations has not been fruitful11,12. However, germline EGFR T790M mutations are present in approximately 50% of all patients with baseline EGFR T790M identified in their pre-treatment tumor specimens13. We have recommended that all patients with baseline EGFR T790M identified in their lung tumor tissue be referred to clinical genetics to discuss EGFR T790M germline testing13.
In our practice, patients with germline EGFR T790M mutations can present radiographically with bilateral ground glass opacities and pulmonary nodules (Figure 2), and often have an indolent disease course. Two of our four patients with confirmed germline EGFR T790M are being monitored expectantly and have gone as long as six years off treatment with stable disease. This is consistent with cell line data that EGFR mutant cell lines that acquire EGFR T790M mutation display indolent growth14. However, this is not uniformly so, as the other two patients had more aggressive, widely metastatic disease treated with chemotherapy and both eventually died from their disease. All four patients with germline EGFR T790M were never-smokers, and information regarding response to erlotinib in germline EGFR T790M carriers is not available as none were treated with erlotinib monotherapy.
Molecular testing of our patient revealed few concurrent somatic mutations. Besides the EGFR exon 19 deletion identified in one lesion, concurrent EGFR L858R and EGFR T790M mutations were identified in 2 of the samples which has been previously described6. Another sample had a concurrent ARID1A mutation which was recently reported in lung adenocarcinoma and is known to be oncogenic in other malignancies15. Newer tyrosine kinase inhibitors, such as AP26113 and CO-1686, have selectivity for EGFR T790M over wild-type EGFR and are currently in early phase studies (NCT01449461 and NCT01526928, respectively). Chemoprevention using an EGFR T790M selective agent could be studied in unaffected T790M germline carriers, similar to the use of tamoxifen in the breast cancer high risk population. In addition, screening CT scans for patients with germline EGFR T790M mutations may be warranted as is recommended for other high-risk populations.
The highest incidence of germline EGFR T790M mutations have been identified in patients with pre-treatment somatic EGFR T790M mutations, and continued referral of these patients to clinical genetics is important to discuss germline testing. Carriers of EGFR T790M need to be followed prospectively in order to learn the clinical significance of this germline mutation. Other somatic mutations (EGFR L858R, EGFR exon 19 deletions, ARID1A K1938N) occur concurrently with EGFR T790M, and analysis of these germline carriers may provide insight into the oncogenic process from germline mutation carrier to clinically evident malignancy. The ultimate goal of this line of research is to understand the cancer risk associated with this germline mutation, identify carriers and provide them with interventions that will reduce their risk of cancer-related mortality.
Supplementary Material
Figure 1.
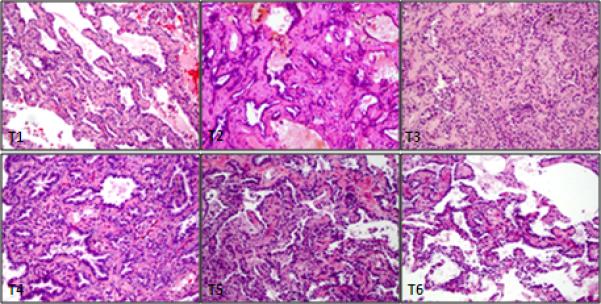
H&E stained sections of 6 tumors at intermediate magnification show adenocarcinomas of mixed phenotype with varying proportions of lepidic, bronchoalveolar, acinar and papillary patterns. All tumors appeared morphologically distinct, favoring separate primaries.
Figure 2.
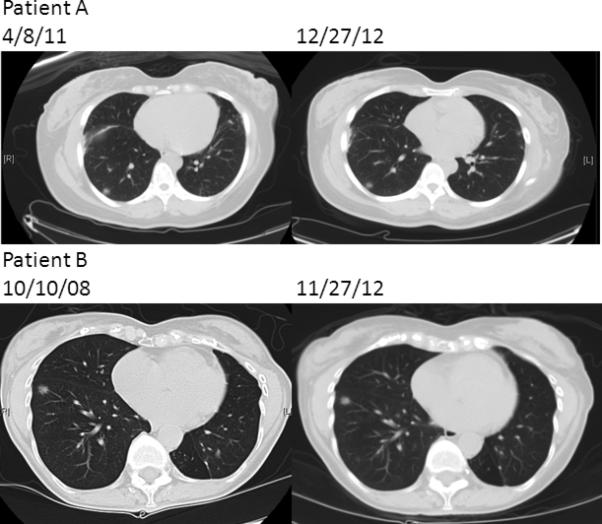
serial interval CT scans of patients with germline EGFR T790M
Figure 3.
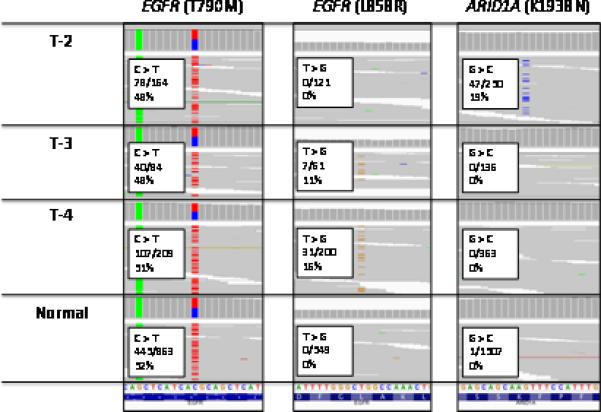
The results from next generation sequencing on Tumor 2 (T2), Tumor 3 (T3), Tumor 4 (T4) and normal blood. The EGFR T790M point mutation was identified in all samples. In addition to EGFR T790M, T2 had evidence of an ARID1A K1938N mutation, and T3 and T4 had evidence of an EGFR L858R point mutation.
Figure 4.
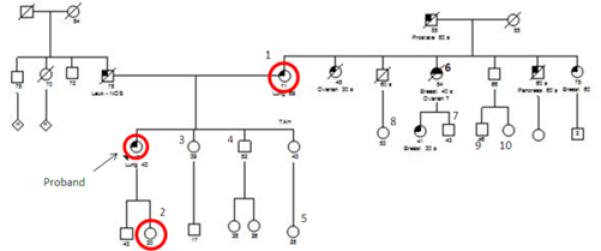
Pedigree with labeled family members and germline testing results
Footnotes
Gregory Riely has consulted for Ariad, Abbott Molecular, Foundation Medicine and Celgene, and has received research support from Infinity Pharmaceuticals, Bristol-Myers Squibb, Novartis, Chugai, Pfizer, Millennium and GlaxoSmithKline. Marc Ladanyi has consulted for Novartis and NanoString. The other others report no conflicts of interest.
References
- 1.Lynch TJ, Bell DW, Sordella R, et al. Activating Mutations in the Epidermal Growth Factor Receptor Underlying Responsiveness of Non-Small-Cell Lung Cancer to Gefitinib. N Engl J Med. 2004;350:2129–39. doi: 10.1056/NEJMoa040938. [DOI] [PubMed] [Google Scholar]
- 2.Rosell R, Carcereny E, Gervais R, et al. Erlotinib versus standard chemotherapy as first-line treatment for European patients with advanced EGFR mutation-positive non-small-cell lung cancer (EURTAC): a multicentre, open-label, randomised phase 3 trial. The lancet oncology. 2012;13:239–46. doi: 10.1016/S1470-2045(11)70393-X. [DOI] [PubMed] [Google Scholar]
- 3.Pao W MV, Politi KA, Riely GJ, Somwar R, Zakowski MF, Kris MG, Varmus H. Acquired resistance of lung adenocarcinomas to gefitinib or erlotinib is associated with a second mutation in the EGFR kinase domain. PLoS Med. 2005;2:225–35. doi: 10.1371/journal.pmed.0020073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yu H, Arcila ME, Rekhtman N, et al. Analysis of Mechanisms of Acquired Resistance to EGFR TKI therapy in 155 patients with EGFR-mutant Lung Cancers. Clinical cancer research. 2013;19(8):2240–7. doi: 10.1158/1078-0432.CCR-12-2246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Riely G, Yu H, Arcila M, et al. Response to Erlotinib and Prognosis in Patients with De Novo Epidermal Growth Factor Receptor T790M Mutations. J Clin Oncol. 2013:31. (suppl; abstr 8018) [Google Scholar]
- 6.Bell DW, Gore I, Okimoto RA, et al. Inherited susceptibility to lung cancer may be associated with the T790M drug resistance mutation in EGFR. Nature genetics. 2005;37:1315–6. doi: 10.1038/ng1671. [DOI] [PubMed] [Google Scholar]
- 7.Meyerson M, Gabriel S, Getz G. Advances in understanding cancer genomes through second-generation sequencing. Nature reviews Genetics. 2010;11:685–96. doi: 10.1038/nrg2841. [DOI] [PubMed] [Google Scholar]
- 8.Wagle N, Emery C, Berger MF, et al. Dissecting therapeutic resistance to RAF inhibition in melanoma by tumor genomic profiling. J Clin Oncol. 2011;29:3085–96. doi: 10.1200/JCO.2010.33.2312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gnirke A, Melnikov A, Maguire J, et al. Solution hybrid selection with ultra-long oligonucleotides for massively parallel targeted sequencing. Nat Biotechnol. 2009;27:182–9. doi: 10.1038/nbt.1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Regales L, Balak MN, Gong Y, et al. Development of new mouse lung tumor models expressing EGFR T790M mutants associated with clinical resistance to kinase inhibitors. PLoS ONE. 2007;2:e810. doi: 10.1371/journal.pone.0000810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vikis H, Sato M, James M, et al. EGFR-T790M is a rare lung cancer susceptibility allele with enhanced kinase activity. Cancer Res. 2007;67:4665–70. doi: 10.1158/0008-5472.CAN-07-0217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Girard N, Lou E, Azzoli CG, et al. Analysis of genetic variants in never-smokers with lung cancer facilitated by an Internet-based blood collection protocol: a preliminary report. Clin Cancer Res. 2010;16:755–63. doi: 10.1158/1078-0432.CCR-09-2437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Oxnard GR, Miller VA, Robson ME, et al. Screening for germline EGFR T790M mutations through lung cancer genotyping. Journal of thoracic oncology. 2012;7:1049–52. doi: 10.1097/JTO.0b013e318250ed9d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chmielecki J, Foo J, Oxnard GR, et al. Optimization of dosing for EGFR-mutant non-small cell lung cancer with evolutionary cancer modeling. Science translational medicine. 2011;3:90ra59. doi: 10.1126/scitranslmed.3002356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Imielinski M, Berger AH, Hammerman PS, et al. Mapping the hallmarks of lung adenocarcinoma with massively parallel sequencing. Cell. 2012;150:1107–20. doi: 10.1016/j.cell.2012.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


