Abstract
The replication of herpes simplex virus type 1 (HSV-1) is associated with a high degree of homologous recombination, which is likely to be mediated, in part, by HSV-1-encoded proteins. We have previously shown that the HSV-1 encoded ICP8 protein and alkaline nuclease UL12 are capable of catalyzing an in vitro strand-exchange reaction. Here, we show, by electron microscopy, that the products of the strand exchange reaction between linear double-stranded DNA and circular single-stranded DNA consist of the expected joint molecule forms: sigma, alpha, and gapped circles. Other exonucleases, such as lambda Red α, which, like UL12, digests 5′-3′, as well as Escherichia coli exonuclease III (ExoIII), which digests 3′-5′, could substitute for UL12 in the strand exchange reaction by providing a resected DNA end. ICP8 generated the same intermediates and strand exchange products when the double-stranded DNA substrate was preresected by any of the nucleases. Using substrates with large regions of non-homology we found that pairing by ICP8 could be initiated from the middle of a DNA molecule and did not require a homologous end. In this reaction, the resection of a DNA end by the nuclease is required to reveal homologous sequences capable of being paired by ICP8. This study further illustrates the complexity of the multi-functional ICP8 protein.
Keywords: ICP8, UL12, herpes, recombinase, electron microscopy
Introduction
The herpes simplex virus type 1 (HSV-1) is a DNA virus that replicates in the nucleus of its host. The 152 kb double-stranded viral genome is packaged as a linear molecule containing both nicks and gaps that are randomly distributed along its length.1 The mechanism for the replication of HSV-1 DNA is not fully understood, but the complex nature of the replication intermediates implies that several steps are involved in this process. Until recently, it was thought that the replication of HSV-1 follows the pathway used by lambda phage in which the HSV-1 DNA circularizes upon entering the host nucleus, and replication begins in the theta mode.2 According to the model, at some point a switch occurs and head-to-tail concatemers of the genome are produced which are subsequently cleaved into unit-length linears. We have long held that homologous recombination is likely to be an integral part of the replication process,3 on the basis of several observations: (1) a high level of recombination is observed in cells infected with HSV-1, and it is specifically associated with viral DNA replication temporally and in its requirement for viral genes;4–8 (2) recombination rates between different strains co-infecting the same cells are high;5,9–12 (3) unique regions of the genome that are bounded by repeated elements invert relative to one another during replication and such inversions are detectable as soon as newly replicated DNA is detected;13–15 (4) replication intermediates in HSV-1-infected cells are present in a non-linear structure which cannot enter a pulsed-field gel, even after digestion with a restriction enzyme which has a single recognition site within the HSV genome;13,14,16,17 (5) the intermediates of HSV-1 replication are head-to-tail concatemers that are highly branched, with X and Y junctions;18–22 (6) we have shown previously that virus-encoded proteins are capable of participating in recombination events.23,24 It is possible that HSV-1 uses both viral and cellular recombinases to mediate the recombination associated with its replication.25 Here, we focus on the viral recombinase.
The HSV-1 protein that is clearly central to these recombinational events is ICP8, which is an extraordinary protein, in that it is the first example of a protein that shares properties with two different distinct classes of DNA-binding proteins: the single strand-binding proteins (SSBs) such as Escherichia coli SSB, T4 gene 32 protein, and RPA, and secondly, the recombinases such as RecA, UvsX and Rad 51. As an SSB, ICP8 stimulates HSV-1 replication in vitro and binds single-stranded DNA tightly, melting secondary structure.26,27 It also facilitates the reannealing of two complementary single-stranded DNAs, a hallmark of the SSBs.28 On the other hand, while the filaments formed by the classic SSBs on ssDNA are smooth or beaded in appearance, the ICP8-ssDNA filaments are helical, a structural feature of RecA, UvsX and Rad 51. Furthermore, ICP8 protein, like RecA, forms helical self-filaments in solution in the absence of DNA.29 In a study from one of our laboratories, ICP8 was found to catalyze strand transfer over homologous DNAs of ∼1 kb.24 While this activity may have been facilitated by an exonuclease activity, this study did reveal a recA-like property.
ICP8 interacts with many HSV-1 proteins, including the alkaline nuclease, encoded by the UL12 gene.30,31 UL12 is part of a family of exonucleases that share homology and that are now known to be encoded by many, if not all linear double-stranded viruses of plants, insects, bacteria and mammals.32,33 For instance, all herpes viruses sequenced to date, as well as the insect virus, baculovirus, encode a nuclease member of this family.32,34 This evolutionary conservation suggests that these nucleases play an important role in the life-cycle of dsDNA viruses with linear genomes. Several of these exonuclease family members have been shown to associate with strand-pairing proteins, forming a unit that has recombinase activity. The lambda Red alpha and beta proteins and E. coli RecE/T are examples of these protein pairs that have proven recombinase activity.35–37 Recently, UL12 and ICP8 have also been shown to work together to mediate a robust strand exchange activity in vitro.23 This activity, indicative of recombinases, suggests that UL12/ ICP8 constitute a recombinase and could contribute to the recombination associated with HSV-1 replication. While UL12 is not an essential gene, viruses lacking UL12 are severely defective, producing 100– 1000-fold fewer progeny virus.38 Furthermore, cells infected with the UL12 null virus contain replicating DNA that has an aberrant structure.17
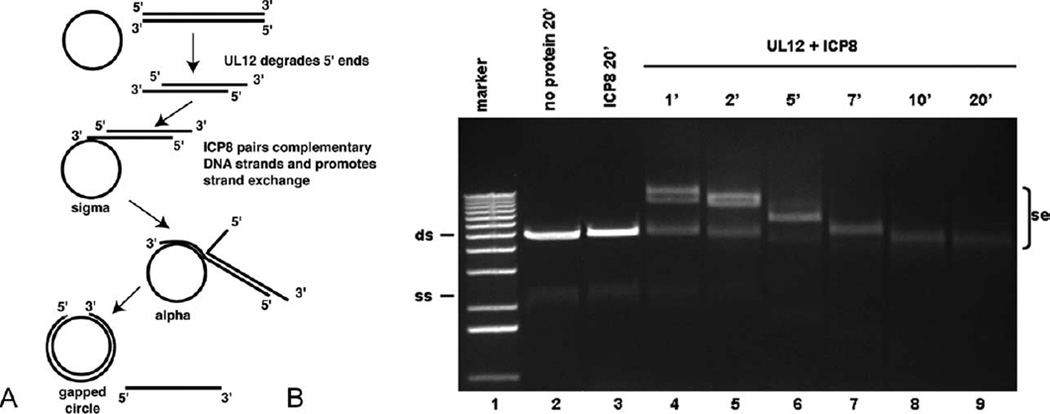
The strand exchange reaction catalyzed by UL12 and ICP8 involves the 5′′3′ resection by UL12 of the double-stranded DNA, followed by annealing of the revealed single-stranded tail to another homologous single-stranded region, which is mediated by ICP8. This reaction is depicted in Figure 1A. However, many details of its mechanism of action are still unknown. Here, we sought to examine the strand exchange reaction catalyzed by UL12 and ICP8 in more detail, including the examination of the reaction products by electron microscopy (EM). Our results verify that the strand exchange reaction mediated by UL12 and ICP8 produced intermediates that are typical of classical strand transfer reactions. We also show that the UL12/ICP8 strand exchange reaction could be separated into two parts, resection and annealing. Furthermore, other exonucleases could substitute for UL12 in the reaction, including those that digest in the 3′-5′ direction. ICP8 was also capable of pairing strands even when there was a region of non-homology blocking the end of the DNA to be transferred. Overcoming this block to pairing did, however, require the action of a nuclease, in order to reveal homologous single-stranded DNA beyond the non-homologous region.
Figure 1.
Strand exchange by UL12 and ICP8. A, A representation of the strand exchange reaction involving UL12, ICP8, and bacteriophage-derived ssDNA circles and linearized dsDNA. The products of the reaction, with structures referred to as sigma, alpha and gapped circle are shown. B, Strand exchange by UL12 and ICP8 using ϕX174 DNA as substrates. Assay conditions were as described in Materials and Methods, using 100 ng of each of the DNA substrates per 20 µl reaction. Incubations were at 37 °C for 1–20 minutes, as indicated. Lane 1, Invitrogen 1 kb ladder marker; lane 2, no protein control; lane 3, incubation of the DNA substrates with ICP8 only; lanes 4–9, incubation of the DNA substrates with ICP8 and UL12 for 1, 2, 5, 7, 10, and 20 minutes, respectively. A photograph of the ethidium bromide-stained gel is presented. Se, strand exchange products; ds, ϕX174 dsDNA linearized by XhoI; ss, ϕX174 ssDNA.
Results
EM visualization of the intermediates and products in a UL12/ICP8-catalyzed strand exchange reaction reveals structures typical of classic strand exchange reactions
We previously demonstrated that ICP8 and the UL12 alkaline nuclease together mediate a strand exchange reaction in vitro.23 The substrates used for the reaction were circular single-stranded M13 virion DNA, and linear double-stranded M13 DNA. The outline of this reaction and its products are shown in Figure 1A. Here, we have further analyzed this reaction by electron microscopy, and we have confirmed that the structures typical of classic strand exchange reactions are in fact produced by UL12/ICP8.
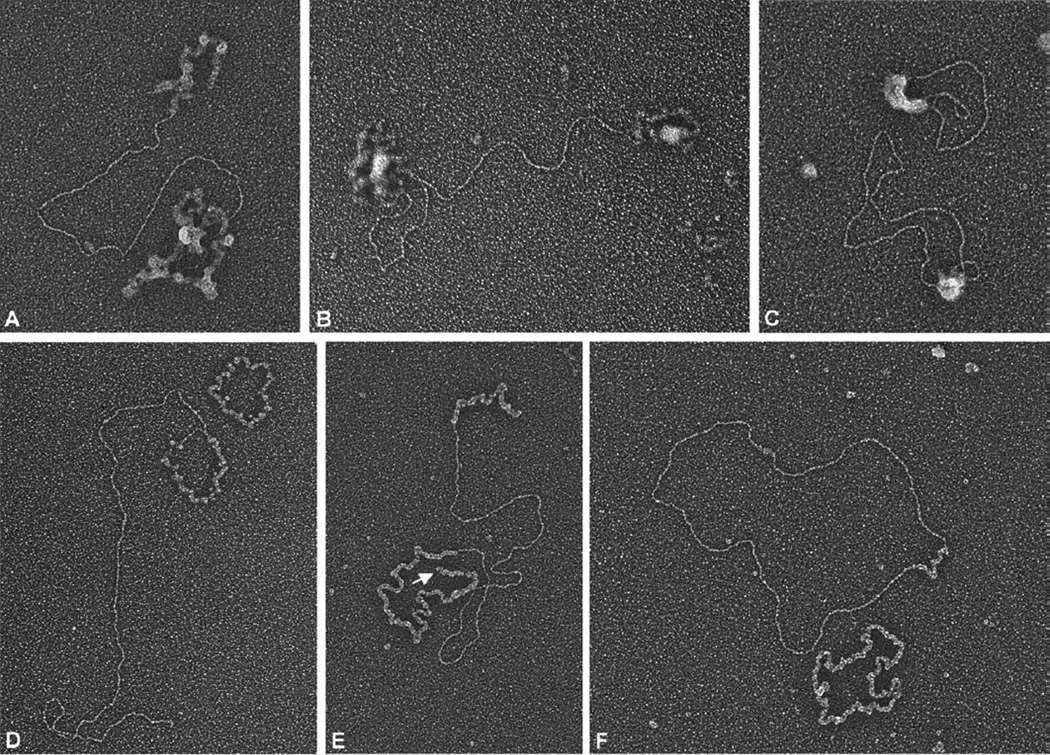
The DNA substrates used for this analysis consisted of the M13 substrates described above, as well as similar substrates prepared from bacteriophage ϕX174 DNA. Both sets of substrates gave rise to the same strand exchange products. The strand exchange reaction is carried out at a buffer pH that is less favorable for UL12 nuclease activity (pH 7.5 rather than pH 8–9), but is more physiological. In addition, the conditions supporting low nuclease activity prevent excessive degradation of the substrates and the products. The assay buffer includes 1 mM MgCl2 and 40 mM NaCl, conditions which support all of the processes necessary for strand exchange—resection, strand pairing, and strand melting. The molar ratio of ICP8 to UL12 is approximately 125:1. ICP8 is present at an amount sufficient to coat all of the ssDNA in the assay, at a ratio of ten bases per ICP8 molecule.39 This ratio of ICP8: ssDNA was found to be optimal for the strand exchange reaction.23 The ICP8 protein used in the assays was prepared by two different protocols.23,40 Both preparations of ICP8 yielded the same results. Figure 1B shows the products of the strand exchange assay using ϕX174 DNA as analyzed by agarose gel electrophoresis. ICP8 alone could not mediate strand exchange, even after an extended incubation (Figure 1B, lane 3). This demonstrates that ICP8 is unable to mediate pairing between long dsDNA substrates and ssDNA. When both UL12 and ICP8 were incubated with the DNA, strand exchange products were detectable as species that migrate more slowly than the original DNA substrates. The initial products (Figure 1B, lanes 4–6) migrate more slowly than the products found after a longer incubation (Figure 1B, lanes 7–9). This is consistent with their assignment as sigma and alpha forms which would be expected to migrate more slowly than the gapped circle form, the end-point of the strand exchange reaction. To confirm the identity of these various forms, we analyzed the products of the reaction by electron microscopy. Samples were prepared for visualization using two different protocols. In one, UL12 and ICP8 were not removed from the DNA prior to visualization (Figure 2A – C), and in the other, the proteins were removed by treatment with proteinase K. In the latter case, the DNA was subsequently coated with E. coli SSB, which extends and thickens the single-stranded DNA (SSB-bound ssDNA is extended to about one-third the length of an equivalent segment of dsDNA) (Figure 2D – F). ICP8-bound ssDNA tends to collapse and coil on itself, making it difficult to follow the contour of these structures. Nevertheless, the sigma (Figure 2A), alpha (Figure 2B), and gapped circle (Figure 2C) forms are readily seen. Deproteinization followed by complexing with SSB gave an even clearer picture of these structures. The sigma form (Figure 2D) represents the first step of pairing of the resected double-stranded molecule with the circular single-stranded DNA. As strand exchange progresses, the nonpairing strand of the dsDNA substrate is extruded, forming one tail of the alpha structure (indicated by an arrow in Figure 2E). This 5′ single-stranded tail is a substrate for the UL12 nuclease and, upon longer incubation, more of the alpha structures found had a shortened or eliminated 5′ tail (data not shown). The sigma and alpha forms were more predominant in reactions that were incubated for a short time, while the gapped circle form (Figure 2F) was the most common form found upon extended incubation (Table 1). The identification of these forms, which are characteristic of classic strand exchange reactions,41–43 demonstrates that UL12/ICP8 produces the same products as bona fide recombinases and supports our previous suggestion that this two-enzyme complex functions as such in infected cells.23
Figure 2.
EM analysis of strand exchange mediated by UL12 and ICP8. Strand exchange reactions were run as described for Figure 1 and further prepared for electron microscopy as described in Materials and Methods. A–C show examples of DNA/ICP8/UL12 complexes formed as a result of strand exchange. Images in D–F were obtained when strand exchange products were deproteinized and the ssDNA complexed with E. coli SSB (see Materials and Methods). The classic strand exchange structures, sigma (A and D), alpha (B and E), and gapped circle (C and F) are shown. The arrow in E points to the displaced strand of the alpha structure. ssDNA circles covered with SSB protein are seen in panels D and F. The scale bar represents the length of 1000 bp of dsDNA.
Table 1.
Electron microscopic analysis of the progression of strand exchange reactions as a function of incubation time
| Time (min) | Structures |
|||
|---|---|---|---|---|
| Linear dsDNA | Sigma | Alpha | Gapped circles | |
| 1 | 52 | 24 | 25 | 1 |
| 2 | 38 | 29 | 29 | 4 |
| 5 | 37 | 6 | 26 | 31 |
| 7 | 41 | 3 | 10 | 46 |
| 10 | 29 | 2 | 4 | 65 |
| 20 | 12 | 0 | 2 | 86 |
Strand exchange reactions using ϕX174 substrates as described in Figure 1 were analyzed by EM. At each time-point, 100 DNA molecules were counted and identified as either linear dsDNA (input dsDNA) or one of the strand exchange structures formed. ssDNA molecules were not included in the counts.
ICP8 alone catalyzes strand exchange when presented with preresected ends of dsDNA complementary to a ssDNA circle
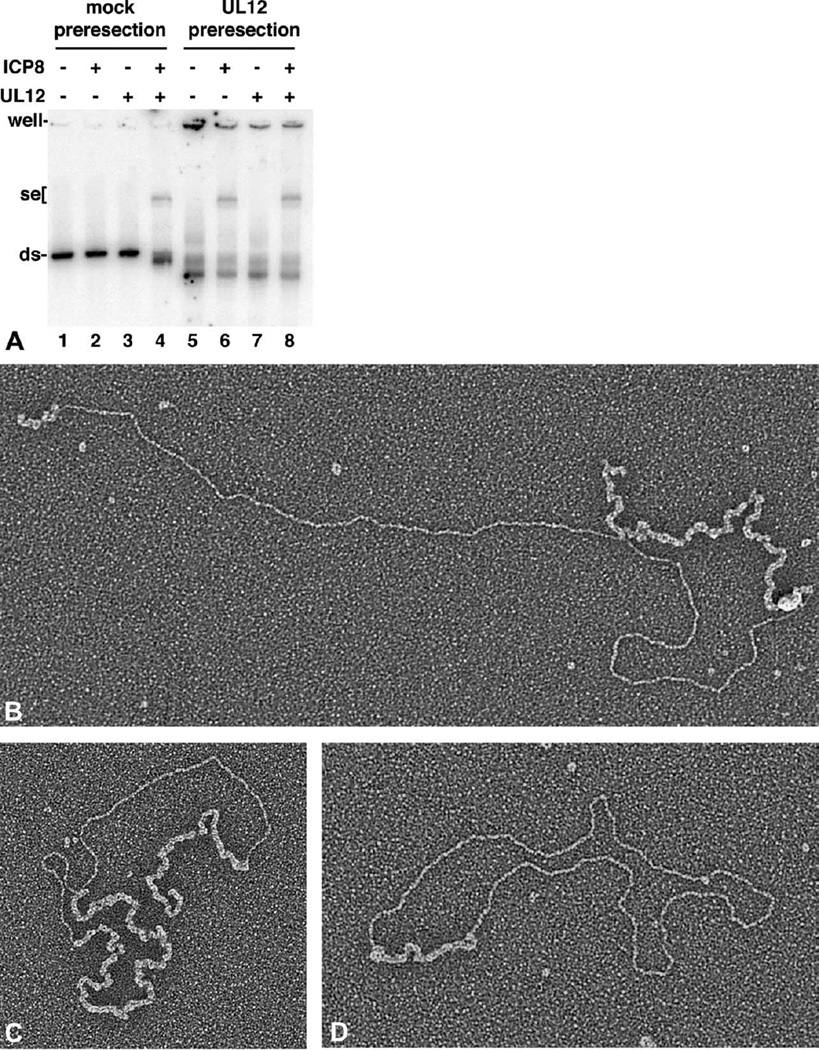
We have demonstrated that UL12 and ICP8 together mediate a robust strand exchange reaction, and ICP8 alone is unable to accomplish this over the full-length M13 and ϕX174 templates. However, the reaction consists of two elements, resection and annealing, and we wanted to test whether these two processes were separable. Such a separation has been demonstrated for RecE/T, where RecT is able to pair DNA that has been preresected by RecE.35 Figure 3A shows that ICP8 was also able to pair DNA that had been preresected by UL12. In this experiment, the dsDNA substrate was a 1.5 kb PCR-generated fragment that was labeled uniformly with [32P]dCTP during synthesis. Use of this substrate allowed the amount of digestion by UL12 to be monitored by measuring the acid-soluble radioactivity released during the course of the preresection. This nuclease assay showed that the DNA was resected an average of 330 bases per strand (data not shown). The resected DNA was deproteinized, purified and used for the strand exchange assay shown in Figure 3A. The ssDNA acceptor used was M13mp18 with the 1.5 kb fragment inserted into the BamHI and EcoRI sites, which is referred to as M13wins (for “M13 with insert”). UL12 and ICP8 produced a slowly migrating strand exchange product when the mock-preresected 1.5 kb fragment was used for strand exchange (Figure 3A, lane 4). The exchange product was not observed when the DNA substrates were incubated with UL12 (lane 3) or ICP8 (lane 2) alone. In contrast, when the DNA had been resected by nuclease, ICP8 alone was competent to produce strand exchange products (lane 6). These results were confirmed by electron microscopic analysis. In this experiment, full-length linear ϕX174 dsDNA was first resected by UL12, and the DNA was deproteinized and repurified. The DNA was then incubated with the ssDNA and ICP8 for strand exchange. For better visualization, following strand exchange, the DNA was deproteinized and coated with E. coli SSB. Figure 3B, C, and D, demonstrate that all of the joint molecule forms, sigma, alpha, and gapped circle, were produced by the incubation of ICP8 alone with the ssDNA and preresected dsDNA. In these experiments, the single-stranded tail of the alpha form was generally longer and was detectable upon extended incubation, in contrast to reactions conducted in the presence of UL12. The presence of the alpha form indicates that ICP8 was not merely pairing two complementary single-stranded regions, but was performing true strand exchange, with displacement of the non-pairing strand.
Figure 3.
ICP8 mediates strand exchange of preresected dsDNA. A, The 1.5 kb 32P-labeled dsDNA fragment was incubated in strand exchange buffer for 20 minutes in the presence (for lanes 5–8) or absence (for mock, lanes 1–4) of UL12. DNA was deproteinized with proteinase K, extracted with phenol/chloroform, and ethanol-precipitated. This material was resuspended in low TE (10 mM Tris–HCl (pH 7.5), 0.1 mM EDTA) and used in the strand exchange assay. The strand exchange reaction was performed as described in Materials and Methods with 1.6 nM ssM13wins DNA (100 ng) and 1 nM (approximately 20 ng) 1.5 kb 32P-labeled dsDNA as substrates. Incubation was for 20 minutes at 37 °C. The phosphorimage of the dried gel is presented. B–D, Linear double-stranded ϕX174 DNAwas preresected with UL12 and then incubated with circular ϕX174 ssDNA in the presenceofICP8in strand exchange buffer for 10–20 minutes at 37 °C. The samples were deproteinized and complexed with E. coli SSB to extend the single-stranded segments and further prepared for EM as described in Materials and Methods. The expected strand exchange products are seen: alpha (B), sigma (C), and gapped circle (D). The scale bar represents the length of 1000 bp of dsDNA.
Other exonucleases can substitute for UL12 in the strand exchange reaction
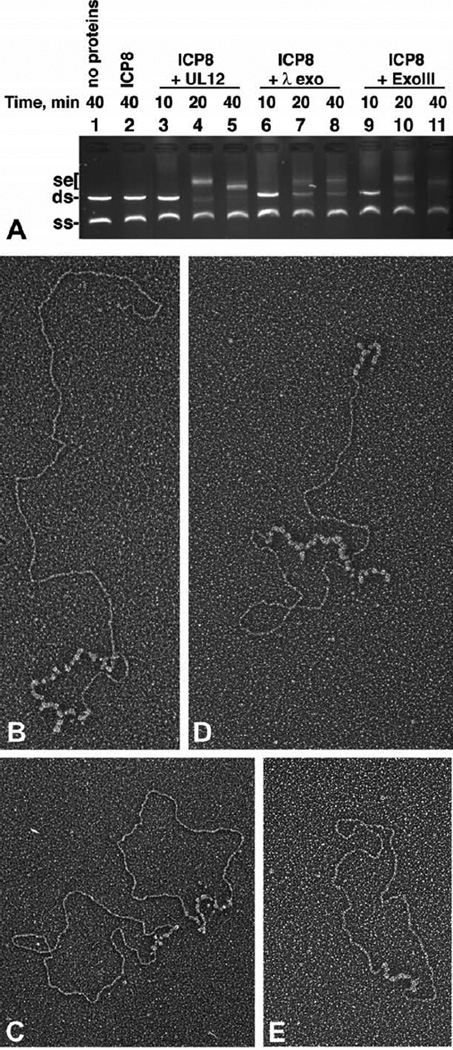
We have demonstrated that the strand exchange reaction could be separated into two steps, resection and annealing. We therefore asked whether other exonucleases could substitute for UL12. We used both phage lambda Red α (lambda exo), which digests 5′-3′, as well as E. coli exonuclease III (ExoIII), which digests 3′-5′. Figure 4A demonstrates that all of the exonucleases could enable ICP8 to catalyze strand exchange, seen clearly at the 20 and 40 minute time-points (Figure 4A, lanes 4–5, 7–8, and 10–11). For electron microscopic analysis, ϕX174 DNA that had been preresected by lambda exo and by ExoIII was incubated with ICP8. Figure 4 (B–E) presents examples of the resulting products. These products appear identical with those seen with UL12 and include all of the joint molecule forms. This shows that other exonucleases are capable of providing ICP8 with the single-stranded DNA necessary to initiate strand exchange. It also demonstrates that ICP8 can mediate strand exchange irrespective of the polarity of the single-stranded end; pairing can begin with either a 3′ or a 5′ end.
Figure 4.
Other exonucleases can perform strand exchange with ICP8. A, Strand exchange with full-length M13mp18 substrates was performed as described in Materials and Methods. Incubations were at 37 °C for 10–40 minutes, as indicated. All of the lanes included 100 ng of ssM13mp18 DNA and 100 ng of dsM13mp18 DNA linearized by EcoRI. Lane 1, no protein control; lane 2, 40 minutes incubation with ICP8 only; lanes 3–5, incubation with ICP8 and 13.9 nM UL12 for 10, 20, and 40 minutes, respectively; lanes 6–8, incubation with ICP8 and five units of lambda exonuclease for 10, 20, and 40 minutes, respectively; lanes 9–11, incubation with ICP8 and 100 units of ExoIII for 10, 20, and 40 minutes, respectively. A photograph of the ethidium bromide-stained gel is presented. Se, strand exchange products; ds, M13mp18 dsDNA linearized by EcoRI; ss, M13mp18 ssDNA. B–E, Visualization of ICP8 catalyzed strand exchange reactions using dsDNA preresected with lambda exonuclease and ExoIII. Linear double-stranded ϕX174 DNA was subjected to digestion by lambda exonuclease (B and C) or ExoIII (D and E) as described in Materials and Methods. The nuclease-treated DNA was then used in strand exchange reactions. The classic strand exchange products are seen: sigma (B), alpha (D), and gapped circles (C and E). The scale bar represents the length of 1000 bp of dsDNA.
Strand exchange by ICP8 past regions of non-homology
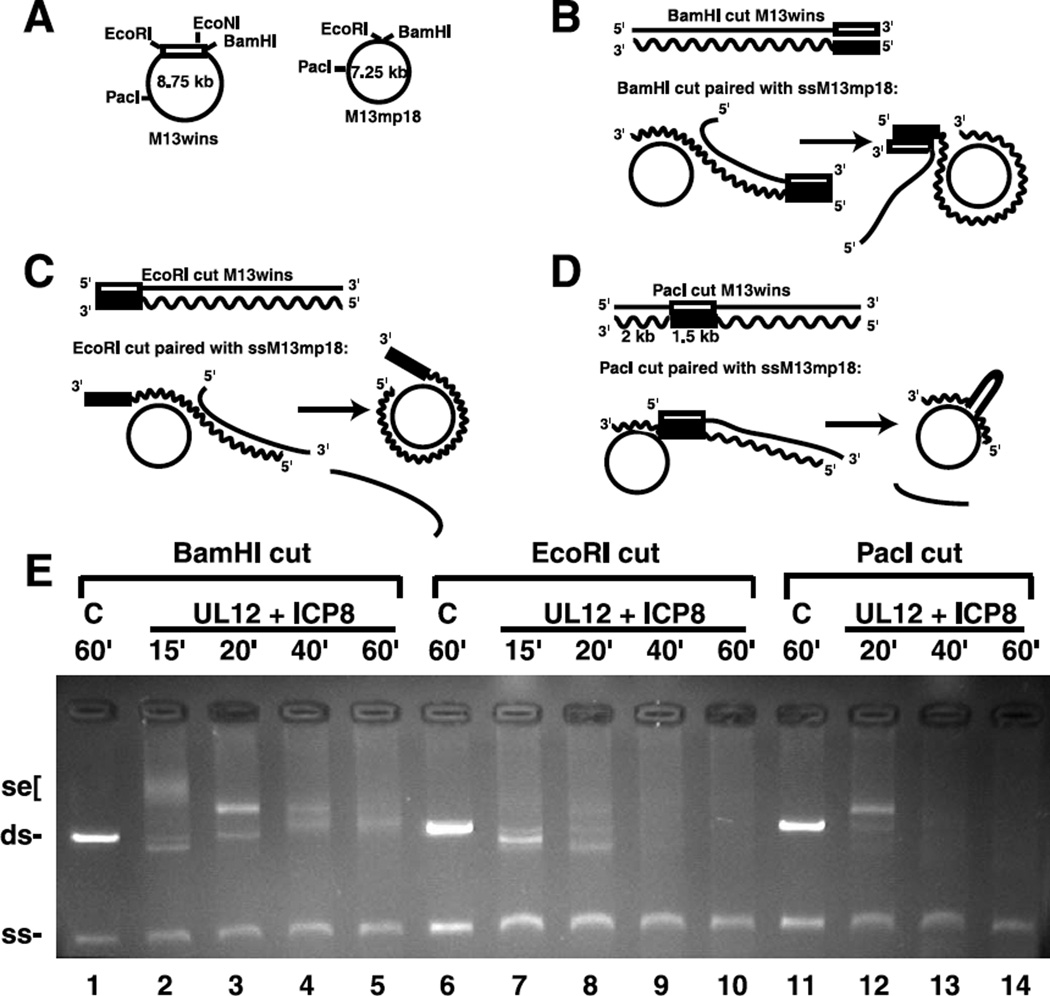
We have shown that ICP8 is capable of mediating strand exchange when it is presented with either a 3′ or 5′ single-stranded end. We next asked whether ICP8 required a homologous end in order to initiate the pairing reaction. Previous studies have shown that joint molecule formation by RecA is blocked by non-homology at the 3′ end of the pairing strand.44–46 To examine this, we used substrates with regions of terminal non-homology to determine whether these regions would block strand transfer by ICP8. Figure 5A – D outlines the substrates, based on M13wins, used for these experiments. The rectangle represents the 1.5 kb insert region. When the RF DNA is cut with EcoRI, the 1.5 kb insert is at the 3′ end of the pairing strand, which would represent a block of non-homology when UL12/ICP8 are used to pair this DNA with ssM13mp18. Cutting the dsDNA with BamHI results in the positioning of the 1.5 kb region at the 5′ end of the pairing strand, and digestion with PacI leaves 2 kb of homology at the 3′ end before the homology is interrupted by the 1.5 kb insert. These substrates were used for strand exchange mediated by UL12 and ICP8. When the dsM13wins substrates were paired with the homologous ssM13wins DNA, the normal strand exchange products were seen by agarose gel analysis, all displaying the same kinetics (data not shown). However, when the dsM13wins substrates were paired with ssM13mp18 DNA, different patterns of strand exchange products were produced, depending upon the position of the region of homology within the dsDNA. Figure 5E shows that the DNA digested with BamHI (lanes 1–5) displays normal kinetics, with early strand exchange products migrating more slowly than later products. With this substrate, pairing begins with DNA that shares homology with M13mp18, so there is no delay in producing a strand exchange product (Figure 5B). The final strand exchange products would be expected to have a 1.5 kb unpaired region at the 5′ end of the pairing strand. The products of this reaction, as analyzed by EM, were practically indistinguishable from reactions with perfectly homologous DNA pairs. The expected 1.5 kb tail was most likely removed by the action of the UL12 nuclease, since it was not usually found on the gapped circles produced (Figure 6A, left, and B). However, a rare example of a late alpha form shown on the right in Figure 6A provides a view of the strand exchange process with this substrate. It presents an interesting example in which the strand exchange reaction has almost gone to completion judging from the very small section of ssDNA left in the circle. At the same time, a dsDNA tail of aproximately 1.6 kb is still attached to the strand exchange junction, as is the SSB-coated ssDNA displaced strand. It appears that in this case UL12 digestion at the 5′ non-homologous end was very limited. These results taken together confirm that strand exchange by UL12/ICP8 progresses without interference when substrates possessing non-homology at the 5′ end of the pairing strand are used.
Figure 5.
Strand exchange using substrates with interrupted homology. A–D, A representation of the DNA substrates used for the strand exchange assay. The rectangle represents the 1.5 kb region derived from HSV-1 DNA. The wavy line and filled rectangle represent the strand complementary to the ssDNA substrates. B–D, Schematic representations of the pairing of ssM13mp18 DNA with dsM13wins DNA linearized by BamHI, EcoRI, and PacI, respectively. E, Strand exchange was performed by UL12 and ICP8 as described in Materials and Methods with 100 ng of ssM13mp18 DNA (2 nM) and 100 ng dsM13wins DNA (0.8 nM) linearized by the following restriction endonucleases: lanes 1–5, BamHI; lanes 6–10, EcoRI; lanes 11–14, PacI. Reactions were incubated for 15–60 minutes, as indicated. C, No protein control.
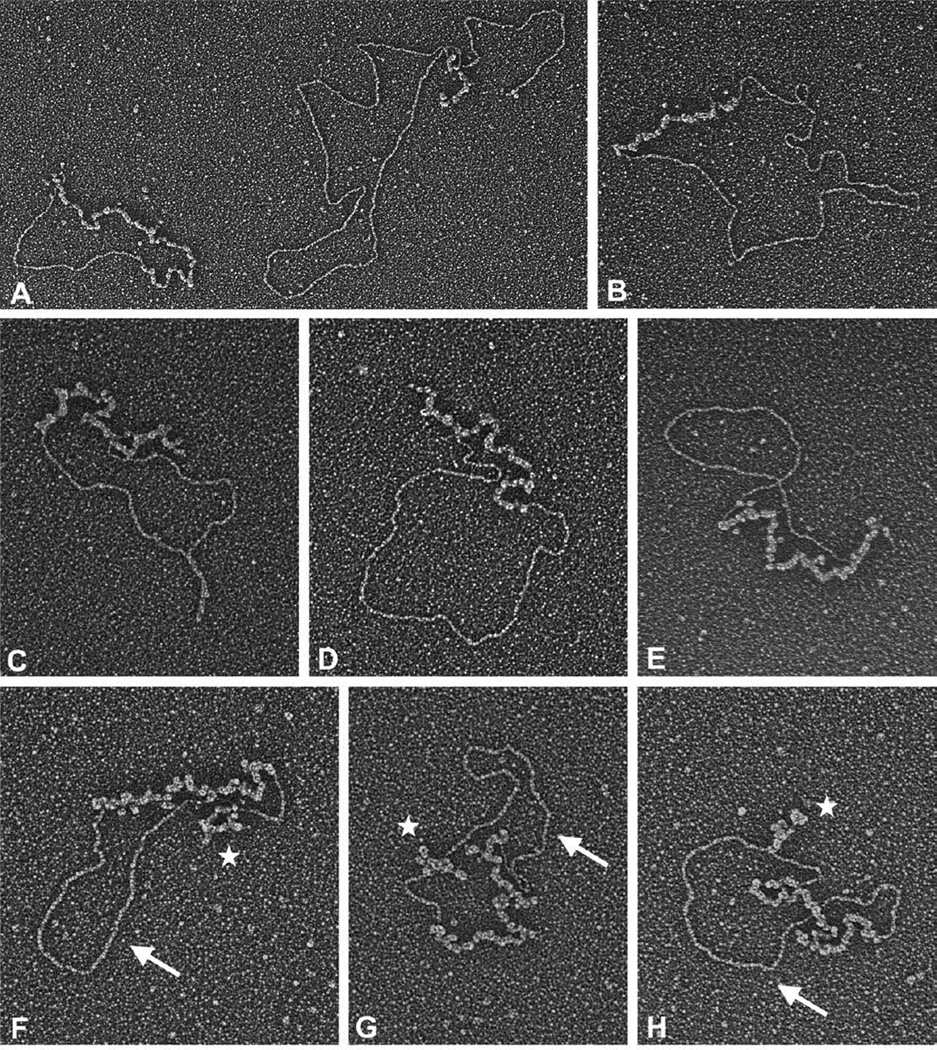
Figure 6.
Visualization of strand exchange products using substrates with interrupted homology. M13wins dsDNA was linearized with BamHI (A and B), EcoRI (C–E), or PacI (F–H). Strand exchange reactions using these DNAs, ssM13mp18 DNA, UL12 and ICP8, as described in Materials and Methods, were examined. Examples of an alpha structure (A) and gapped circles (A–H) are shown to illustrate how the position of the non-homologous 1.5 kb fragment within the dsDNA substrate affected the resulting strand exchange products. Arrows in F–H point to a 2 kb region of dsDNA followed by ssDNA loops (identified by stars) which are formed by the non-homologous DNA within the gapped circle product. The scale bar represents 700 bp (A and B) and 1000 bp (C–H) of dsDNA.
Strand exchange with EcoRI-cut M13wins resulted in a different distribution of products on the agarose gel. The early time-points indicate that the DNA underwent more degradation than strand exchange, and diffuse strand exchange products were produced only at later time-points (Figure 5E, lanes 6–10). Thus, the 1.5 kb region of non-homology at the 3′ end of the pairing strand appeared to present a block to strand transfer, but only a temporary one. Ultimately, strand exchange products were produced that could be visualized by EM, although they were difficult to discern on the agarose gel (Figure 5E, lanes 8–10). EM analysis of these products clearly showed the unpaired 1.5 kb region as a tail on the gapped circle molecules (Figure 6C – E). This demonstrates that ICP8 does not require a homologous end in order to initiate strand pairing and transfer.
The substrate cut with PacI is useful for testing whether ICP8, once it has initiated strand transfer, is capable of driving the transfer past large regions of non-homology. The agarose gel shows that strand exchange products are produced early (Figure 5E, lane 12). This is expected, since the 3′ end of the pairing strand begins with 2 kb of sequence homologous to M13mp18 (Figure 5D). Later products are more diffuse, and it appears that much of the DNA has been degraded (Figure 5E, lanes 13– 14). EM analysis of this reaction revealed gapped circle molecules with a looped-out region of single-stranded DNA (Figure 6F – H). Measurement of the various segments of these molecules showed that the initial 2 kb were paired in each of the molecules (marked by an arrow), followed by the 1.5 kb single-stranded region (marked by a star). The region that is paired beyond the 1.5 kb loop was found to be short, due to extensive digestion of the 5′ end of this DNA by UL12. The extensive digestion seen at the 5′ end of the pairing strand suggests that more UL12 digestion was also needed at the 5′ end of the displaced strand to obtain strand exchange final products with the PacI cut DNA than was required when the other substrates were used. This implies that UL12 digestion beyond the 1.5 kb region of non-homology was required in order for ICP8 to continue pairing the homologous DNA. It appears that ICP8 was able to mediate strand transfer past a region of non-homology, but only if the complementary DNA to be paired was revealed as single-stranded DNA by further nuclease action.
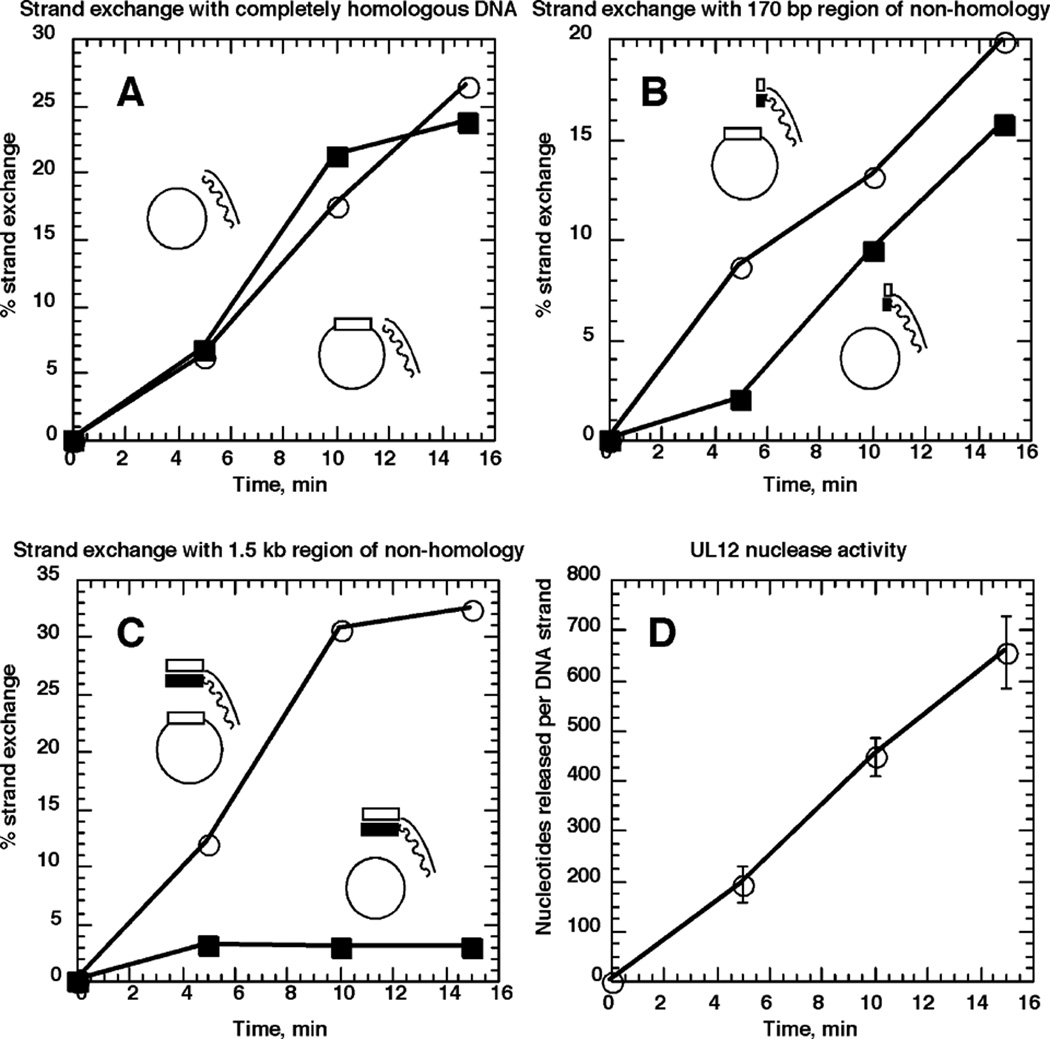
In order to test this hypothesis, we used a 3.5 kb substrate which was generated and 32P-labeled by PCR using the M13wins DNA as template. The substrate has the 1.5 kb non-M13 region at the 3′ end of the pairing strand, and the remaining 2 kb is M13 sequence. Two more substrates were also used, one with the entire 1.5 kb fragment removed, leaving only the 2 kb region, and another truncated such that 170 bp of the insert sequence remains at the 3′ end of the pairing strand. The 3.5 kb substrate does produce a stable “strand exchange” product when boiled and allowed to anneal with M13mp18 (data not shown). These substrates were paired with either M13mp18 or M13wins ssDNA in a UL12/ICP8-mediated strand exchange reaction. Samples were removed for assay of nuclease digestion at each time-point. The results of these experiments are presented in Figure 7. These data show that when the 2 kb substrate is used, which is completely homologous to both M13mp18 and M13wins, the strand exchange products with both acceptor ssDNAs are produced with the same kinetics (Figure 7A). When there is a 170 bp region of non-homology at the 3′ end of the pairing strand, there is a delay in pairing this DNA with M13mp18 (Figure 7B). The delay appears to correlate with the amount of time required to remove the 170 bp of non-homology and reveal homologous DNA (Figure 7D). It should be noted that the rate of nuclease digestion in these experiments is slower than in the other experiments presented, due to the lower concentration of the dsDNA substrates. When the 3.5 kb fragment is used, which has the 1.5 kb region at the 3′ end of the pairing strand, no significant strand exchange is detected when this substrate is paired with M13mp18 (Figure 7C). During the time-course of this experiment, nuclease digestion did not progress past the 1.5 kb region of non-homology. In addition, since the 3.5 substrate is shorter than the full-length M13wins substrates used earlier, digestion of the substrate to remove the 1.5 kb region of non-homology would leave the DNA with little left to pair, assuming that UL12 digestion is occurring on both ends of the molecule. These results imply that in order to initiate strand exchange, ICP8 requires that two complementary regions be single-stranded. Once strand exchange has begun, ICP8 does use strand melting to displace the non-pairing strand. However, it appears that ICP8 does not use strand melting to find regions of homology when it is bound at a stretch of non-homologous DNA.
Figure 7.
Non-homology at the 3′ end of the pairing strand delays strand exchange. Strand exchange was performed by UL12 and ICP8 as described in Materials and Methods with either ssM13mp18 (filled squares) or ssM13wins (open circles). The dsDNA substrates used were prepared by PCR as 32P-labeled 3.5 kb fragments using M13wins as template. Both [α-32P]dCTP and [α-32P]dATP were used for labeling of this substrate. The 1.5 kb block of HSV-1 sequence, represented by rectangles, is at the 3′ end of the pairing strand, and the remaining 2 kb are M13mp18 sequence. The wavy line/filled rectangle represents the strand complementary to the ssDNA substrates. The 3.5 kb fragment was cut with BamHI to completely remove the 1.5 kb region of non-homology, and the 2 kb fragment was purified. This substrate was used in A. The 3.5 kb fragment was also cut with EcoNI, to create a substrate possessing 170 bases of DNA non-homologous to M13mp18. The 2.17 kb fragment was purified and used for strand exchange illustrated in B. In C, the full-length 3.5 kb fragment was used, which has 1.5 kb of DNA non-homologous to M13mp18. Percentage strand exchange was calculated as the percentage of radioactivity in slowly migrating species representing strand exchange products, out of the total radioactivity in the lane. Results are the averages of two independent experiments. D, The results of the nuclease digestion of the dsDNA substrates during the course of the strand exchange assay. Results are the averages of at least three DNA samples per time-point.
Discussion
The strand transfer reaction mediated by the HSV-1 UL12 and ICP8 proteins generates products typical of classical strand exchange reactions, thus confirming the identification of UL12/ICP8 as a recombinase. We showed that this reaction can be separated into two steps in vitro, and that other exonucleases can replace UL12. Although the in vitro results do not demonstrate any preference toward the UL12 nuclease in this assay, it is possible and even likely that such a preference does occur in vivo. This was shown to be true for the analogous proteins lambda Red α/β and E. coli RecE/T. Although these protein pairs have similar activity and Red mutants can be compensated for by RecE/T, in vivo recombination requires that each synaptase be paired with its cognate exonuclease. In other words, RecT does not mediate exchange in vivo with Red a.37 Other observations suggest that UL12 and ICP8 work in concert in vivo. UL12 and ICP8 bind each other in solution,30,31,47 and we have found that ICP8 specifically modulates UL12 activity whereas, other SSBs do not (N.B.R. and S.K.W., unpublished results).
Our results show that DNA preresected by several different nucleases can be paired by ICP8; however, the efficiency of this process is dependent upon the particular substrate used. When full-length M13mp18 cut at the polylinker region was preresected by UL12 and then incubated with ICP8, significant strand exchange products were not detected (data not shown). However, as we have shown, ICP8 was capable of pairing other preresected DNA substrates. The likely explanation is that the M13 origin region contains a cluster of inverted repeats which form highly stable hairpins in the single-stranded viral DNA. Indeed, in early studies of strand transfer by the Rad 51 protein, which had traditionally utilized M13 substrates, it was the use of ϕX174 templates that made it possible to show significant recombinase activity for Rad 51.48 It was for this reason that the ϕX174 templates were used for the EM studies presented here. In contrast to the preresected M13 DNA, when M13 DNA is incubated with UL12 and ICP8 together, a robust strand exchange reaction is achieved.23 In this situation, it is likely that UL12 digestion reveals the single-stranded 3′ end of the DNA to ICP8, which binds it before it has the opportunity to form a hairpin structure. It is possible that such a coordination of activities in vivo also prevents secondary structure of ssDNA from forming in the viral DNA, thus facilitating efficient strand pairing.
We have demonstrated that ICP8 can initiate pairing with either a 3′ or a 5′ single-stranded tail, and can pair internal stretches of complementary DNA when the end is blocked by non-homology. Nimonkar et al. have also found that ICP8 is capable of using either a 3′ or 5′ end to initiate D-loop formation, although non-homology at the ends blocks this process.49 The structures of the joint molecules produced by either UL12 or lambda exo, both 5′-3′ exonucleases, or by ExoIII, a 3′-5′ exonuclease, in conjunction with ICP8 suggest that pairing, once initiated, continues in the same direction in which it was begun. The mechanism appears to be one of continuous pairing, as alpha structures were formed in all the reactions tested. The alpha structures are proof of the strand displacement aspect of the reaction, showing that as one strand is paired another is displaced. Since alpha structures were formed regardless of the direction of the strand pairing, we conclude that pairing is continuous in both directions. However, despite the ability of ICP8 to pair DNA when provided with a 5′ single-stranded end, it is unlikely that this reaction would be utilized productively in vivo. The annealing of a 3′ end to a single-stranded region allows this end to be used as a primer for DNA synthesis. Such recombination-directed replication has been proposed to be an important part of HSV-1 replication,3 and has been observed in an in vitro assay model.50 The annealing of a 5′ end has no such opportunity. Replicating DNA in cells infected with a UL12 null virus is in a fragile, aberrant form, and few viral progeny are produced.17,38 It is possible that in the absence of UL12, cellular nucleases could resect the HSV-1 DNA. If viral DNA is resected by a 3′ to 5′ cellular nuclease and ICP8 anneals the resulting 5′ tails, a dead-end product would be generated, likely to be oddly branched and ineligible for further replication and packaging. This scenario is consistent with the defective phenotype of the UL12 null mutants.
Strand exchange using the substrates with regions of non-homology demonstrated that pairing by ICP8 could be initiated at an internal site, but also shed light on other requirements for pairing by ICP8. The data suggest that the pairing mechanism requires two complementary single-stranded regions for ICP8 to begin the strand exchange process. Since ICP8 is capable of strand melting, it might have been assumed to be sufficient to provide ICP8 with a region of single-stranded DNA to bind. We therefore expected that an internal complementary region within the double stranded portion of the molecule would be revealed by ICP8 strand melting and paired. Results with the partially non-homologous substrates showed, however, that this is not the case. It appears rather that nuclease digestion is required to reveal complementary single-stranded DNA for pairing. This is likely to hold true even when ICP8 has successfully initiated strand exchange, as in the case of M13wins cut with PacI. Instead of ICP8 melting of DNA beyond the non-homologous region, it appears that nuclease digestion is required to again reveal homologous DNA beyond the 1.5 kb block. This mechanism is similar to that of RecA, where strand transfer is blocked by regions of non-homology. For instance, joint molecule formation by RecA is blocked by non-homology at the 3′ end of the pairing strand.44–46 RecJ, a 5′-3′ DNA exonuclease, greatly enhances the efficiency with which RecA is able to traverse regions of non-homology, allowing it to pair substrates with a 187-base region of non-homology which otherwise blocked strand pairing.51 It was suggested that exonucleases might enhance the strand pairing by RecA in vivo by helping it to continue pairing past regions of non-homology, as well as by preventing the renaturation of the original substrate.51 The blocks of non-homology used in our studies were rather large, from 170 bp to 1.5 kb. It will be interesting to see how smaller interruptions of homology affect strand pairing by ICP8, and how tolerant ICP8 is in pairing semi-complementary DNA.
This study has demonstrated the potent capability of ICP8 to pair and promote strand exchange when presented with two complementary single-stranded regions. This could give the impression that the source of the single-stranded DNA is unimportant, yet several observations point to the importance of a nuclease in this process. Nimonkar et al. demonstrated that ICP8 could mediate strand exchange of short double-stranded substrates with circular ssDNA in conjunction with the HSV-1 helicase/primase.52 However, this reaction had to be performed in two steps, with a change of reaction conditions midway. In addition, the yield of strand exchange products was much lower than that we have reported with UL12 and ICP8. In our hands, the helicase/primase along with ICP8 did not promote strand exchange when the full-length M13 substrate was used; furthermore, the addition of helicase/primase to the ICP8/UL12 reaction did not increase the yield or rate of appearance of strand exchange products (N.B.R. and S.K.W., unpublished results). Therefore, the presence of a nuclease is critical for ICP8 to promote strand exchange using long and potentially complex double-stranded substrates such as M13. These data suggest that although ICP8 can participate in limited strand exchange either in the presence of the helicase-primase or when sufficient single-stranded DNA is present, a more robust reaction will likely require the presence of nuclease activity. On the basis of previously demonstrated interaction between UL12 and ICP8,30,31,47 it is reasonable to assume that UL12 is likely to be the nuclease responsible for allowing ICP8 to display its potent recombinase activity; however, we cannot rule out the participation of host nucleases. As discussed above, nuclease action has also been shown to enhance recombination by RecA. In addition, in the case of the lambda Red and RecE/Trecombinases, it is clear that the nuclease partners play an important role in the recombination mediated by the pairing proteins. In summary, on the basis of data presented here, combined with work previously reported by us and others, we propose that during HSV infection, UL12 and ICP8 function as a potent two-component recombinase and that these activities are essential for efficient and normal DNA replication leading to the production of longer than unit-length viral DNA suitable for processing by the viral encapsidation machinery. This model is supported by the observation that aberrant DNA is produced in cells infected with a UL12 null mutant virus.17,38
Materials and Methods
Materials
α[32P]ATP and α[32P]dCTP were from Dupont-NEN. All other materials were reagent grade.
DNA
M13mp18 replicative form (RF) was purified from infected E. coli UT481 [Δ(lac-pro)hsdS(r−m−)lacIqlacZ] cells using the Qiagen maxi plasmid kit. M13mp18 ssDNA was purified from M13 phage-infected UT481 cells according to standard protocols.53 Phage ϕX174 RF and virion DNA were from New England Biolabs (NEB). DNA fragments were purified from agarose gels using the GeneClean® Spin kit (Bio-101) or the Qiagen Qiaquick gel extraction kit.
Enzymes and proteins
Restriction endonucleases and ExoIII were from New England Biolabs. Lambda exonuclease was generously provided by Richard S. Myers. Some experiments were also performed with lambda exonuclease purchased from Novagen. Proteinase K was from Roche. The UL12 protein was prepared by Joshua Goldstein from insect cells infected with recombinant baculoviruses as described.54 ICP8 was also purified from recombinant baculovirus-infected insect cells in each of our laboratories by two different protocols, as described.23,40 In both preparations, the protein was greater than 95% pure as determined by SDS-PAGE and Coomassie staining and did not contain detectable levels of nuclease activity.
Substrates for strand exchange assays
M13 with a 1.5 kb insert was constructed by cloning the BamHI-EcoRI fragment from pFastBacUL12.547 (coresponding to nucleotides 25007–26510 of the HSV-1 genome with flanking sequences added encoding the restriction sites) into the same sites in M13mp18, to create M13wins. Full-length unlabeled dsDNA substrates were prepared by cleaving M13mp18, M13wins, and ϕX174 RF DNA with restriction endonucleases (as indicated in the Figure legends) and the DNA fragments were purified from agarose gels. To obtain preresected ϕX174 dsDNA, the following enzymes and protocols were used: for UL12 preresection, the enzyme was incubated with XhoI-cut linear dsDNA in strand exchange buffer for 10–20 minutes at 37 °C at a ratio of 100 ng of DNA to 18.8 ng of UL12. Preresection with lambda exonuclease and ExoIII was done for two minutes at 37 °C at a ratio of five units of lambda exo or 100 units of Exo III per 1.5 mg of dsDNA. Digested DNA was deproteinized with 100 ng/µl of proteinase K, 0.5% (w/v) SDS and 50 mM EDTA for one hour at 558C and cleaned through a QIAquick spin column. Radiolabeled double-stranded fragments were prepared by PCR using the M13wins RF DNA as template. The primers used for amplification were phosphorylated at the 5′ end. The dNTP mixes used for PCR included α[32P]dCTP or both α[32P]dCTP and α[32P]dATP. For the 1.5 kb fragment, the primers used for amplification were: primer A, 5′ ATGTGGTCGGCGTCGGTGATC; primer B, 5′TCAGC GAGACGACCTCCCCGTC. The 3.5 kb fragment was amplified using the same template, and the following primers: primer C, 5′TGACCTTCATCAAGAGTAATC; primer D, 5′AGCGGATAACAATTTCACACAG. In order to prepare substrates with varying amounts of non-homology, the 3.5 kb labeled substrate was prepared. Portions of this substrate were cleaved with either BamHI or EcoNI, and the 2.0 kb and 2.17 kb fragments were gel-purified.
Strand exchange assay
The reaction was carried out in a final volume of 20 µl and consisted of: 100 ng of circular single-stranded M13mp18 DNA, M13wins, or ϕX174 DNA, 7–200 ng of linear double-stranded DNA (as indicated in the Figure legends), 18.8 ng of UL12 (13.9 nM), 4.5 µg of ICP8 (1.75 µM), 20 mM Tris–HCl (pH 7.5), 40 mM NaCl, 1 mM MgCl2, and 1 mM DTT. The reaction mixture was incubated at 37 °C for the times indicated in the Figure legends and stopped, for visualization on agarose gels, by adding 5 µl of 5× stop buffer (50% (v/v) glycerol, 50 mM EDTA, 1% SDS, 0.2% (w/v) bromphenol blue). Samples were electrophoresed on a 1% (w/v) agarose gel with 0.7 µg/ml of ethidium bromide, using TAE buffer (0.04 M Tris–acetate, 0.001 M EDTA). Gels were photographed, and those with radiolabeled DNA were dried and exposed to phosphorimager screens (National Diagnostics). The ImageQuant version 5.0 software package was used for quantification of the results.
Nuclease assay
DNA and proteins were incubated in 20 µl reaction volumes as in the strand exchange assay. The assay was terminated by the addition of 5 µl of 3.5 mg/ml of salmon sperm DNA and 25 µl of 20% TCA. After ten minutes on ice, samples were centrifuged for ten minutes at 14,000g, and radioactivity in 25 µl of the supernatant fraction was determined by scintillation counting. The results presented are averages of duplicate determinations.
Preparation of strand exchange complexes for electron microscopy
The reactions were carried out as described above, with substrates and incubation times as indicated in the Figure legends. The reactions were stopped with 50 mM EDTA when observation of DNA-ICP8 complexes was desired. The EDTA incubation was done for 30 minutes on ice to remove magnesium ions and open the ICP8 complexes for better visualization by EM. When complete removal of UL12 and ICP8 proteins was desired, the reactions were deproteinized with 50 mM EDTA, 1% SDS and 100 ng/µl of proteinase K, at 55 °C for one hour. The deproteinized samples were then cleaned through a 2 ml Biogel A50-m column and eluted in TE buffer (10 mM Tris–HCl pH 7.5), 0.5 mM EDTA. Fractions containing DNA were further prepared for EM analysis by incubation with 40 ng/µl of E. coli SSB for 15 minutes at room temperature, for visualization of single-stranded DNA within the expected structures.
Electron microscopy
To eliminate unwanted buffers and/or free protein from samples, the reaction mixtures were loaded onto 2 ml Biogel A50-m columns previously equilibrated in TE buffer. The same buffer was then used to elute the samples from the columns and 250 µl fractions were collected. Aliquots of the protein–DNA containing fractions were mixed with a buffer containing spermidine55 for three seconds and quickly applied to a mesh copper grid coated with a thin carbon film, glow-charged shortly before sample application. Following adsorption of the samples to the EM support for two to three minutes, the grids were dehydrated through a graded water–ethanol series to 100% ethanol and then air-dried. The samples were then rotary shadowcast with tungsten at 10−7 Torr and examined in a Tecnai TEM instrument at 40 kV. Micrographs, taken at 46,000×, were scanned using a Nikon LS-4500AF film scanner and panels were arranged using Adobe Photoshop.
Acknowledgements
This work was supported, in part, by grants to S.K.W. (A121747, A137549) and to J.D. G (GM31819, CA19014). N.B.R. was supported by a grant from the Damon Runyon Cancer Research Fund (DRG-1625). We thank Richard S. Myers and members of our laboratories for their helpful discussions.
Abbreviations used
- HSV-1
herpes simplex virus type 1
- SSBs
single strand-binding proteins
- ExoIII
exonuclease III
- RF
replicative form
- EM
electron microscope
References
- 1.Fields BN, Knipe DM, Howley PM, Griffin DE. tComputer laser optical disc. Fields Virology. 4th. Philadelphia, PA: Lippincott Williams & Wilkins; 2002. 2002. p. 1. [Google Scholar]
- 2.Lehman IR, Boehmer PE. Replication of herpes simplex virus DNA. J. Biol. Chem. 1999;274:28059–28062. doi: 10.1074/jbc.274.40.28059. [DOI] [PubMed] [Google Scholar]
- 3.Weller SK. Herpes simplex virus DNA replication and genome maturation. In: Cooper GM, Temin RG, Sugden B, editors. The DNA Provirus: Howard Temin’s Scientific Legacy. Washington, DC: ASM Press; 1995. pp. 189–213. [Google Scholar]
- 4.Dutch RE, Bianchi V, Lehman IR. Herpes simplex virus type 1 DNA replication is specifically required for high-frequency homologous recombination between repeated sequences. J. Virol. 1995;69:3084–3089. doi: 10.1128/jvi.69.5.3084-3089.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Honess RW, Buchan A, Halliburton IW, Watson DH. Recombination and linkage between structural and regulatory genes of herpes simplex virus type 1: study of the functional organization of the genome. J. Virol. 1980;34:716–742. doi: 10.1128/jvi.34.3.716-742.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Smiley JR, Duncan J, Howes M. Sequence requirements for DNA rearrangements induced by the terminal repeat of herpes simplex virus type 1 KOS DNA. J. Virol. 1990;64:5036–5050. doi: 10.1128/jvi.64.10.5036-5050.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dutch RE, Bruckner RC, Mocarski ES, Lehman IR. Herpes simplex virus type 1 recombination: role of DNA replication and viral a sequences. J. Virol. 1992;66:277–285. doi: 10.1128/jvi.66.1.277-285.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Weber PC, Challberg MD, Nelson NJ, Levine J, Glorioso JC. Inversion events in the HSV-1 genome are directly mediated by the viral DNA replication machinery and lack sequence specificity. Cell. 1988;54:369–381. doi: 10.1016/0092-8674(88)90200-0. [DOI] [PubMed] [Google Scholar]
- 9.Schaffer PA, Tevethia MJ, Benyesh-Melnick M. Recombination between temperature-sensitive mutants of herpes simplex virus type 1. Virology. 1974;58:219–228. doi: 10.1016/0042-6822(74)90156-1. [DOI] [PubMed] [Google Scholar]
- 10.Wildy P. Recombination with herpes simplex virus. J. Gen. Microbiol. 1955;13:346–360. doi: 10.1099/00221287-13-2-346. [DOI] [PubMed] [Google Scholar]
- 11.Umene K. Intermolecular recombination of the herpes simplex virus type 1 genome analysed using two strains differing in restriction enzyme cleavage sites. J. Gen. Virol. 1985;66:2659–2670. doi: 10.1099/0022-1317-66-12-2659. [DOI] [PubMed] [Google Scholar]
- 12.Brown SM, Subak-Sharpe JH, Harland J, MacLean AR. Analysis of intrastrain recombination in herpes simplex virus type 1 strain 17 and herpes simplex virus type 2 strain HG52 using restriction endonuclease sites as unselected markers and temperature-sensitive lesions as selected markers. J. Gen. Virol. 1992;73:293–301. doi: 10.1099/0022-1317-73-2-293. [DOI] [PubMed] [Google Scholar]
- 13.Severini A, Morgan AR, Tovell DR, Tyrrell DL. Study of the structure of replicative intermediates of HSV-1 DNA by pulsed-field gel electrophoresis. Virology. 1994;200:428–435. doi: 10.1006/viro.1994.1206. [DOI] [PubMed] [Google Scholar]
- 14.Zhang X, Efstathiou S, Simmons A. Identification of novel herpes simplex virus replicative intermediates by field inversion gel electrophoresis: Implications for viral DNA amplification strategies. Virology. 1994;202:530–539. doi: 10.1006/viro.1994.1375. [DOI] [PubMed] [Google Scholar]
- 15.Lamberti C, Weller SK. The herpes simplex virus type 1 UL6 protein is essential for cleavage and packaging but not for genomic inversion. Virology. 1996;226:403–407. doi: 10.1006/viro.1996.0668. [DOI] [PubMed] [Google Scholar]
- 16.Bataille D, Epstein A. Herpes simplex virus replicative concatemers contain L components in inverted orientation. Virology. 1994;203:384–388. doi: 10.1006/viro.1994.1498. [DOI] [PubMed] [Google Scholar]
- 17.Martinez R, Sarisky RT, Weber PC, Weller SK. Herpes simplex virus type 1 alkaline nuclease is required for efficient processing of viral DNA replication intermediates. J. Virol. 1996;70:2075–2085. doi: 10.1128/jvi.70.4.2075-2085.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Severini A, Scraba DG, Tyrrell DLJ. Branched structures in the intracellular DNA of herpes simplex virus type 1. J. Virol. 1996;70:3169–3175. doi: 10.1128/jvi.70.5.3169-3175.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jacob RJ, Roizman B. Anatomy of herpes simplex virus DNA. VIII. Properties of the replicating DNA. J. Virol. 1977;23:394–411. doi: 10.1128/jvi.23.2.394-411.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shlomai J, Friedmann A, Becker Y. Replication intermediates of herpes simplex virus DNA. Virology. 1976;69:647–659. doi: 10.1016/0042-6822(76)90493-1. [DOI] [PubMed] [Google Scholar]
- 21.Friedmann A, Shlomai J, Becker Y. Electron microscopy of herpes simplex virus DNA molecules isolated from infected cells by centrifugation in CsCl density gradients. J. Gen. Virol. 1977;34:507–522. doi: 10.1099/0022-1317-34-3-507. [DOI] [PubMed] [Google Scholar]
- 22.Hirsch I, Cabral G, Patterson M, Biswal N. Studies on the intracellular replicating DNA of herpes simplex virus type 1. Virology. 1977;81:48–61. doi: 10.1016/0042-6822(77)90057-5. [DOI] [PubMed] [Google Scholar]
- 23.Reuven NB, Staire AE, Myers RS, Weller SK. The herpes simplex virus type 1 alkaline nuclease and single-stranded DNA binding protein mediate strand exchange in vitro. J. Virol. 2003;77:7425–7433. doi: 10.1128/JVI.77.13.7425-7433.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bortner C, Hernandez TR, Lehman IR, Griffith J. Herpes simplex virus 1 single-strand DNA-binding protein (ICP8) will promote homologous pairing and strand transfer. J. Mol. Biol. 1993;231:241–250. doi: 10.1006/jmbi.1993.1279. [DOI] [PubMed] [Google Scholar]
- 25.Wilkinson DE, Weller SK. The role of DNA recombination in herpes simplex virus DNA replication. IUBMB Life. 2003;55:451–458. doi: 10.1080/15216540310001612237. [DOI] [PubMed] [Google Scholar]
- 26.Ruyechan WT, Weir AC. Interaction with nucleic acids and stimulation of the viral DNA polymerase by the herpes simplex virus type 1 major DNA-binding protein. J. Virol. 1984;52:727–733. doi: 10.1128/jvi.52.3.727-733.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.O’Donnell ME, Elias P, Lehman IR. Processive replication of single-stranded DNA templates by the herpes simplex virus-induced DNA polymerase. J. Biol. Chem. 1987;262:4252–4259. [PubMed] [Google Scholar]
- 28.Dutch RE, Lehman IR. Renaturation of complementary DNA strands by herpes simplex virus type 1 ICP8. J. Virol. 1993;67:6945–6949. doi: 10.1128/jvi.67.12.6945-6949.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.O’Donnell ME, Elias P, Funnell BE, Lehman IR. Interaction between the DNA poly-merase and single-stranded DNA-binding protein (infected cell protein 8) of herpes simplex virus 1. J. Biol. Chem. 1987;262:4260–4266. [PubMed] [Google Scholar]
- 30.Thomas MS, Gao M, Knipe DM, Powell KL. Association between the herpes simplex virus major DNA-binding protein and alkaline nuclease. J. Virol. 1992;66:1152–1161. doi: 10.1128/jvi.66.2.1152-1161.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vaughan PJ, Banks LM, Purifoy DJ, Powell KL. Interactions between herpes simplex virus DNA-binding proteins. J. Gen. Virol. 1984;65:2033–2041. doi: 10.1099/0022-1317-65-11-2033. [DOI] [PubMed] [Google Scholar]
- 32.Myers RS, Rudd KE. Miami, Florida: Proceedings of the Miami Nature Biotechnology Series; 1998. [Google Scholar]
- 33.Aravind L, Makarova KS, Koonin EV. Holliday junction resolvases and related nucleases: identification of new families, phyletic distribution and evolutionary trajectories. Nucl. Acids Res. 2000;28:3417–3432. doi: 10.1093/nar/28.18.3417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mikhailov VS, Okano K, Rohrmann GF. Baculovirus alkaline nuclease possesses a 5′/3′ exonuclease activity and associates with the DNA-binding protein LEF-3. J. Virol. 2003;77:2436–2444. doi: 10.1128/JVI.77.4.2436-2444.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hall SD, Kolodner RD. Homologous pairing and strand exchange promoted by the Escherichia coli RecT protein. Proc. Natl Acad. Sci. USA. 1994;91:3205–3209. doi: 10.1073/pnas.91.8.3205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stahl MM, Thomason L, Poteete AR, Tarkowski T, Kuzminov A, Stahl FW. Annealing versus invasion in phage lambda recombination. Genetics. 1997;147:961–977. doi: 10.1093/genetics/147.3.961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Muyrers JP, Zhang Y, Buchholz F, Stewart AF. RecE/RecT and Redalpha/Redbeta initiate double-stranded break repair by specifically interacting with their respective partners. Genes Dev. 2000;14:1971–1982. [PMC free article] [PubMed] [Google Scholar]
- 38.Weller SK, Seghatoleslami MR, Shao L, Rowse D, Carmichael EP. The herpes simplex virus type 1 alkaline nuclease is not essential for viral DNA synthesis: isolation and characterization of a lacZ insertion mutant. J. Gen. Virol. 1990;71:2941–2952. doi: 10.1099/0022-1317-71-12-2941. [DOI] [PubMed] [Google Scholar]
- 39.Gourves AS, Tanguy Le Gac N, Villani G, Boehmer PE, Johnson NP. Equilibrium binding of single-stranded DNA with herpes simplex virus type I-coded single-stranded DNA-binding protein, ICP8. J. Biol. Chem. 2000;275:10864–10869. doi: 10.1074/jbc.275.15.10864. [DOI] [PubMed] [Google Scholar]
- 40.Boehmer PE, Lehman IR. Physical interaction between the herpes simplex virus 1 origin-binding protein and single-stranded DNA-binding protein ICP8. Proc. Natl Acad. Sci. USA. 1993;90:8444–8448. doi: 10.1073/pnas.90.18.8444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Register JC, III, Christiansen G, Griffith J. Electron microscopic visualization of the RecA protein-mediated pairing and branch migration phases of DNA strand exchange. J. Biol. Chem. 1987;262:12812–12820. [PubMed] [Google Scholar]
- 42.Radding CM, Flory J, Wu A, Kahn R, DasGupta C, Gonda D, Bianchi M, Tsang SS. Three phases in homologous pairing: polymerization of recA protein on single-stranded DNA, synapsis, and polar strand exchange. Cold Spring Harbor Symp. Quant. Biol. 1983;47:821–828. doi: 10.1101/sqb.1983.047.01.094. [DOI] [PubMed] [Google Scholar]
- 43.Stasiak A, Stasiak AZ, Koller T. Visualization of RecA–DNA complexes involved in consecutive stages of an in vitro strand exchange reaction. Cold Spring Harbor Symp. Quant. Biol. 1984;49:561–570. doi: 10.1101/sqb.1984.049.01.063. [DOI] [PubMed] [Google Scholar]
- 44.Cox MM, Lehman IR. Directionality and polarity in recA protein-promoted branch migration. Proc. Natl Acad. Sci. USA. 1981;78:6018–6022. doi: 10.1073/pnas.78.10.6018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kahn R, Cunningham RP, DasGupta C, Radding CM. Polarity of heteroduplex formation promoted by Escherichia coli recA protein. Proc. Natl Acad. Sci. USA. 1981;78:4786–4790. doi: 10.1073/pnas.78.8.4786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.West SC, Cassuto E, Howard-Flanders P. Heteroduplex formation by recA protein: polarity of strand exchanges. Proc. Natl Acad. Sci. USA. 1981;78:6149–6153. doi: 10.1073/pnas.78.10.6149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Reuven NB, Antoku S, Weller SK. The UL12.5 gene product of herpes simplex virus type 1 exhibits nuclease and strand exchange activities but does not localize to the nucleus. J. Virol. 2004;78:4599–4608. doi: 10.1128/JVI.78.9.4599-4608.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sung P. Catalysis of ATP-dependent homologous DNA pairing and strand exchange by yeast RAD51 protein. Science. 1994;265:1241–1243. doi: 10.1126/science.8066464. [DOI] [PubMed] [Google Scholar]
- 49.Nimonkar AV, Boehmer PE. The herpes simplex virus type-1 single-strand DNA-binding protein (ICP8) promotes strand invasion. J. Biol. Chem. 2003;278:9678–9682. doi: 10.1074/jbc.m212555200. [DOI] [PubMed] [Google Scholar]
- 50.Nimonkar AV, Boehmer PE. Reconstitution of recombination-dependent DNA synthesis in herpes simplex virus 1. Proc. Natl Acad. Sci. USA. 2003;100:10201–10206. doi: 10.1073/pnas.1534569100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Corrette-Bennett SE, Lovett ST. Enhancement of RecA strand-transfer activity by the RecJ exonuclease of Escherichia coli. J. Biol. Chem. 1995;270:6881–6885. doi: 10.1074/jbc.270.12.6881. [DOI] [PubMed] [Google Scholar]
- 52.Nimonkar AV, Boehmer PE. In vitro strand exchange promoted by the herpes simplex virus type-1 single strand DNA-binding protein (ICP8) and DNA helicase-primase. J. Biol. Chem. 2002;277:15182–15189. doi: 10.1074/jbc.M109988200. [DOI] [PubMed] [Google Scholar]
- 53.Maniatis T, Fritsch EF, Sambrook J. Cold Spring Harbor, NY: Molecular Cloning: A Laboratory Manual, Cold Spring Harbor Laboratory Press; 1982. [Google Scholar]
- 54.Goldstein JN, Weller SK. The exonuclease activity of HSV-1 UL12 is required for in vivo function. Virology. 1998;244:442–457. doi: 10.1006/viro.1998.9129. [DOI] [PubMed] [Google Scholar]
- 55.Griffith JD, Christiansen G. Electron microscope visualization of chromatin and other DNA–protein complexes. Annu. Rev. Biophys. Bioeng. 1978;7:19–35. doi: 10.1146/annurev.bb.07.060178.000315. [DOI] [PubMed] [Google Scholar]