Abstract
Background
MicroRNAs (miRNAs) are now recognized as critical regulators of diverse physiological and pathological processes; however, studies of miRNAs and arrhythmogenesis remain sparse. Connexin43 (Cx43), a major cardiac gap junction protein, has elicited great interest in its role in arrhythmias. Additionally, Cx43 was a potential target for miR-130a as predicted by several computational algorithms. This study investigates the effect of miR-130a overexpression in the adult heart and its effect on cardiac rhythm.
Methods and Results
Using a cardiac-specific inducible system, transgenic mice demonstrated both atrial and ventricular arrhythmias. We performed ventricular-programmed electrical stimulation and found that the αMHC-miR130a mice developed sustained ventricular tachycardia beginning 6 weeks after overexpression. Western blot analysis demonstrated a steady decline in Cx43 after 2 weeks of overexpression with over a 90% reduction in Cx43 levels by 10 weeks. Immunofluorescent staining confirmed a near complete loss of Cx43 throughout the heart. To validate Cx43 as a direct target of miR-130a, we performed in vitro target assays in 3T3 fibroblasts and HL-1 cardiomyocytes, both known to endogenously express miR-130a. Using a luciferase reporter fused to the 3’UTR of Cx43, we found a 52.9% reduction in luciferase activity in 3T3 cells (p<0.0001) and a 47.6% reduction in HL-1 cells (p=0.0056) compared to controls. Addition of an antisense miR-130a inhibitor resulted in a loss of inhibitory activity of the Cx43 3’UTR reporter.
Conclusions
We have identified an unappreciated role for miR-130a as a direct regulator of Cx43. Overexpression of miR-130a may contribute importantly to gap junction remodeling and to the pathogenesis of atrial and ventricular arrhythmias.
Keywords: MicroRNA, Ventricular Arrhythmia, Atrial Arrhythmia, Connexin43
1. INTRODUCTION
Cardiac arrhythmias are common and result in significant morbidity and mortality. Despite this, many questions remain unanswered about the molecular mechanisms involved in arrhythmia generation, propagation, and maintenance [1]. Electrical activation of the heart requires transfer of current from one discrete cardiac myocyte to another, a process that occurs at gap junctions, which are composed of connexin proteins. Remodeling of gap junction organization via connexin expression has been demonstrated in animal models of heart failure and myocardial infarction [2, 3]. A variety of connexins are expressed in cardiac myocytes; however, connexin43 (Cx43) is the predominant connexin expressed in both the atrium and ventricle [4–6]. Loss of Cx43 gap junction channels in the heart results in a marked increase in the incidence of spontaneous and inducible ventricular tachyarrhythmias and may also play an important role in the pathogenesis of atrial fibrillation (AF) [7]. Despite the great interest in connexins and arrhythmias, only recently has miRNA regulation of connexin expression been recognized.
MicroRNAs are a class of small noncoding RNAs that are important regulators of gene expression in diverse biological processes. They are generally regarded as negative regulators of gene expression that inhibit translation and/or promote mRNA degradation by base pairing to complementary sequences within the 3′ untranslated region (3′UTR) of protein-coding mRNA transcripts [8, 9]. Therefore, miRNAs can provide an additional layer of spatial and temporal control over complex genetic pathways, including cardiac excitation and arrhythmia.
To date, little is known about the cardiovascular function of microRNA-130a (miR-130a). MicroRNA-130a has been implicated in the angiogenic process [10, 11]; however, its role in adult myocardium is not known. Nevertheless, it has been shown to be upregulated in myocardium of patients with heart failure [12, 13]. Because miR-130a is predicted by several computational algorithms to target Cx43, we hypothesized that miR-130a overexpression would result in abnormalities in cardiac rhythm. Using an inducible transgenic model to induce miR-130a over-expression in the adult cardiomyocyte, we found both atrial and ventricular tachyarrhythmias in transgenic mice as Cx43 protein was lost throughout the myocardium. In addition, our data suggests that Cx43 is a direct target of miR-130a, thus contributing to the arrhythmia phenotype.
2. MATERIALS AND METHODS
2.1. Generation of miR-130a overexpression mice
All procedures were approved by and performed in accordance with the University of Chicago Institutional Animal Care and Use Committee. A genomic fragment encoding the miR-130a precursor was amplified by PCR using mouse genomic DNA and cloned into a tetracycline responsive promoter (TetO) vector (see supplemental methods for details). The TetO-miR130a construct was injected into the pronuclei of CD-1 embryos and implanted into pseudo-pregnant recipient females by the University of Chicago Transgenics Core. TetO-miR130a founder mice were established and crossed with CD-1 mice and expanded for a minimum of seven generations on the CD-1 background. Non-transgenic littermates (referred to as “control mice”) were compared with double-transgenic littermates genotyped αMHC-tTA/TetO-miR130a (referred to as “αMHC-miR130a mice”). Single-transgenic animals genotyped αMHC-tTA or TetO-miR-130a were analyzed and compared with both control and αMHC-miR130a mice (Supplemental Data, Figure S1, S2).
2.2. Western blotting
Murine hearts were harvested and protein lysates were prepared as previously described.[14] Twenty micrograms of whole heart lysate were resolved by 12% SDS-PAGE or 3–8% Tris-Acetate gel and transferred to a nitrocellulose membrane (GE Healthcare; Whatman, NJ). Antibodies used: Cx43 (3512S, Cell Signaling), phospho-Cx43 (Ser368, 3511, Cell Signaling) Cx40 (AB1726, Millipore), ryanodine receptor (RyR) (34C, Developmental Studies Hybridoma Bank), phospholamban (PLB) (8495, Cell Signaling), phospho-PLB (07–052, Millipore), SERCA2a (2862, Abcam), Amphiphysin 2 (Bin1) (H-100, SC-30099, Santa Cruz), DHPR (MA3-920, Thermo), Junctophilin2 (SC-51313, Santa Cruz), HRP-coupled goat anti-rabbit IgG (Jackson ImmunoResearch), and rabbit anti-gamma-tubulin (T6557, Sigma).
2.3. Immunofluorescence
Hearts were harvested and directly frozen in Tissue-Tek OCT compound (Electron Microscopy Sciences) at the time points specified. Frozen hearts were sectioned at 10 µm thickness and fixed with 100% methanol. Sections were then washed with phosphate buffered saline (PBS), and blocked in blocking buffer (PBS + 5% BSA + 0.2% Triton X-100) for 1 hour at room temperature. Sections were then incubated with a rabbit anti-Cx43 antibody (3512S, Cell Signaling) or goat anti-Cx40 antibody (SC-20466, Santa Cruz), followed by an Alexa-488-conjugated goat anti-rabbit IgG antibody (A11008, Invitrogen) or Alexa-594-conjugated donkey anti-goat IgG antibody (A11058, Invitrogen). Sections were mounted with VECTASHIELD® Mounting Media with DAPI (Vector Laboratories, CA), and visualized with a Zeiss Axiophot fluorescence microscope. Images were merged using ImageJ software.
2.4. Cell culture and luciferase assay
NIH 3T3 cells were maintained in 1X DMEM (11995, Gibco) supplemented with 10% fetal bovine serum, 1% penicillin/streptomycin. HL-1 cells were provided by W.C. Claycomb and maintained as per instructions. HL-1 cells were grown in flasks precoated with 0.02% gelatin and 5 µl/mL fibronectin. Cells were maintained at 37°C with 5% CO2 and 95% humidity in Claycomb medium (Sigma) supplemented with 10% FBS (Batch 12J001, Sigma), 0.1 mM norepinephrine, 2 mM L-glutamine, 100 U/mL penicillin, and 100 µg/mL streptomycin. Once the cells were confluent and beating, 5 × 105 cells were plated in 6-well plates precoated in gelatin/fibronectin [15].
For transfection assays, 3–5 × 105 cells were plated per well and transfected using Superfect (Qiagen) with 1 µg pMT01 (control vector), or Cx43 3’UTR reporter (GeneCopoeia, Maryland); which contain both firefly and renilla luciferase reporters. Luciferase activities were measured (Promega Glomax®-20/20 Luminometer) using the Dual Luciferase Reporter Assay System (Genecopia) with firefly luciferase activities calculated as the mean ± SD after being normalized by renilla luciferase activities. For inhibitor assays: a miRCURY LNA™ miR-130a inhibitor (412568-00, Exiqon) or a scramble control (199004-00, Exiqon) was co-transfected with the luciferase constructs ranging from 100–300 pmol.
2.5. RNA analysis
Total RNA was isolated using TRIzol (Invitrogen) according to manufacturers instructions. RNA was reversed transcribed (ABI TaqMan® RT kit; Applied Biosystems, CA) and then used for quantitative real-time PCR using SSo Advanced™ SYBR® Green MasterMix (Bio-Rad, CA) and BioRad CFX96 Thermocycler (Bio-Rad). MicroRNA-130a levels were detected using TaqMan® miRNA Assay Kit (4427975, Applied Biosystems) according to manufacturer’s protocol. GAPDH was used for normalization. Primer sequences for quantitative real-time PCR are provided in the data supplement.
2.6. Fluorescent in situ hybridization
Adult mouse hearts were harvested and directly frozen in Tissue-Tek OCT compound (Electron Microscopy Sciences, cat. no.62550-01). Frozen hearts were sectioned at 8–10 µm thickness and used for in situ hybridization according to established protocols [16] using specific probes for miR-130a (Exiqon, 38029-04; 7.5 pmol/slide), U6 (Exiqon, 99002-04; 5 pmol/slide), and scramble-miR (Exiqon, 99004-04; 5 pmol/slide).
2.7. Transthoracic echocardiography
Mice were anesthetized with 1–2% isoflurane in 700 ml O2/min via a facemask. Temperature was monitored and maintained at 37°C using a heat pad and heat lamp. Heart rate was maintained at 400–450 bpm. Anesthetized mice were placed on a temperature-controlled platform and echocardiography was performed using a Vevo770 ultrasound system (VisualSonics). An echocardiographer blind to animal genotype captured M-mode and pulsed Doppler images.
2.8. Surface electrocardiograms and ambulatory ECG monitoring
Electrocardiograms (ECG) on anesthetized mice were obtained from needle electrodes inserted subcutaneously into each limb. Multiple leads were recorded for 3 minutes at 2 MHz. Electrocardiographic signals were amplified with an ADInstruments ECG Bioamplifier (Colorado Springs, CO), converted from analog to digital with an ACQ-16 Acquisition Interface and recorded with Ponemah Physiology Platform software (Gould; Valley View, OH). Recordings were analyzed using the ECG module of LabChart 5 software (AD Instruments).
For ambulatory ECG monitoring, 4- to 12-week-old mice were anesthetized with isofluorane, and telemetry transmitters (ETA-F10; DSI) were implanted in the back with leads tunneled to the right upper and left lower thorax. Heart rate, PR, and QRS intervals were calculated using Ponemah Physiology Platform (DSI) from 24-hour recordings.
2.9. Electrophysiological studies
After proper anesthetic induction using inhaled isoflurane, a jugular vein cutdown was performed and the vein isolated for direct endovascular access. We used both a 1.1 and 1.9 french octapolar catheter for study as used in reports of mouse electrophysiological studies [17, 18]. To minimize the effects of catheter placement on cardiac function or the effects of anesthesia on obtaining intracardiac electrograms, we used data obtained within the first 15 minutes of catheter placement. Electrograms were recorded with a BioAmp and Powerlab system with Chart5 software (ADInstruments). Programmed electrical stimulation (PES) studies were performed as described by Gutstein et al.[19] PES consisted of pacing with a train of twelve beats, followed by a double or triple extrastimulus at a cycle length of 20, 30, and 40 ms. We used the following criteria to categorize the ventricular arrhythmias: A) Non-sustained VT: 3–30 beats, B) Sustained VT: >30 beats, and C) Cycle Length: <100 ms [19, 20].
2.10. Statistics
Values are reported as means ± SEM unless indicated otherwise. The 2-tailed Mann-Whitney U test was used for comparing two means. Fisher’s exact test was used to compare inducibility of VT in control vs. transgenic mice (Prism; GraphPad). Values of P<0.05 were considered statistically significant.
3. RESULTS
3.1. MicroRNA-130a is expressed in adult cardiomyocytes
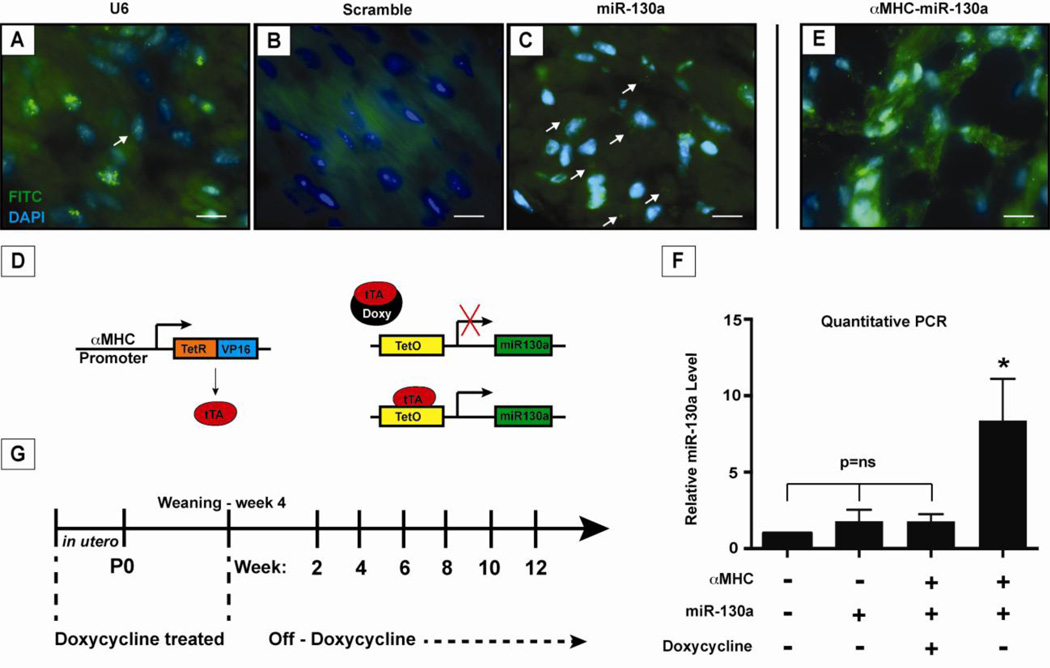
In a previous study, northern analysis on adult mouse tissue found the highest relative levels of miR-130a in the heart and lung, although levels in adult tissues were low [14]. To examine this more closely, we used fluorescent in situ hybridization to detect miR-130a expression in the adult heart. As expected, low levels of endogenous miR-130a were seen in normal adult cardiomyocytes (Figure 1, C). To investigate the role of miR-130a in cardiac remodeling in vivo, we generated an inducible transgenic mouse line carrying the mouse miR-130a gene specifically in the heart under the control of the α-myosin heavy chain (αMHC) promoter [21]. The overexpression strategy consisted of a transgene encoding miR-130a downstream of a tetracycline-responsive promoter (TetO–miR130a) and a second transgene encoding the tetracycline-controlled transactivator (tTA) protein driven by the αMHC promoter (αMHC-tTA) (Figure 1,D). Quantitative PCR results demonstrated that miR-130a was increased in the transgenic hearts by 8.38 ± 1.2 fold (Figure 1, F). This was also visualized using fluorescent in situ hybridization (Figure 1, E) on αMHC-miR130a hearts; which confirmed cardiomyocyte overexpression. Doxycycline administration was able to suppress miR-130a overexpression (1.0 vs 1.78 ± 0.22 fold, p=0.07) with no changes in cardiac function as assessed by echocardiography between controls and doxycycline treated αMHC-miR130a mice (Figure 2, A). Breeding pairs and offspring were maintained on doxycycline until weaning, at which time doxycycline was no longer added to the drinking water (Figure 1, G).
Figure 1. Expression of miR-130a in cardiomyocytes and induction of miR-130a in adult heart.
Localization of miR-130a using fluorescent in situ hybridization in adult cardiomyocytes. Panel (A) shows U6 stained nuclei as a positive control, panel (B) shows staining with a miR-scramble as a negative control, and panel (C) with miR-130a staining in cardiomyocytes. (D) Schematic of inducible transgenic system for miR-130a overexpression using the αMHC promoter to drive expression of the tTA protein. In panel (E), fluorescent in situ hybridization of αMHC-miR130a heart demonstrating increased miR-130a signal confirming overexpression of miR-130a. (F) Quantitative PCR of relative miR130a levels in transgenic heart 6–8 weeks after doxycycline is removed from water supply (n=5). (G) Schematic outline of doxycycline treatment and study time points. * indicates p<0.0001. Doxy: doxycycline; TetO: tetracycline responsive promoter; tTA: tetracycline-controlled transactivator.
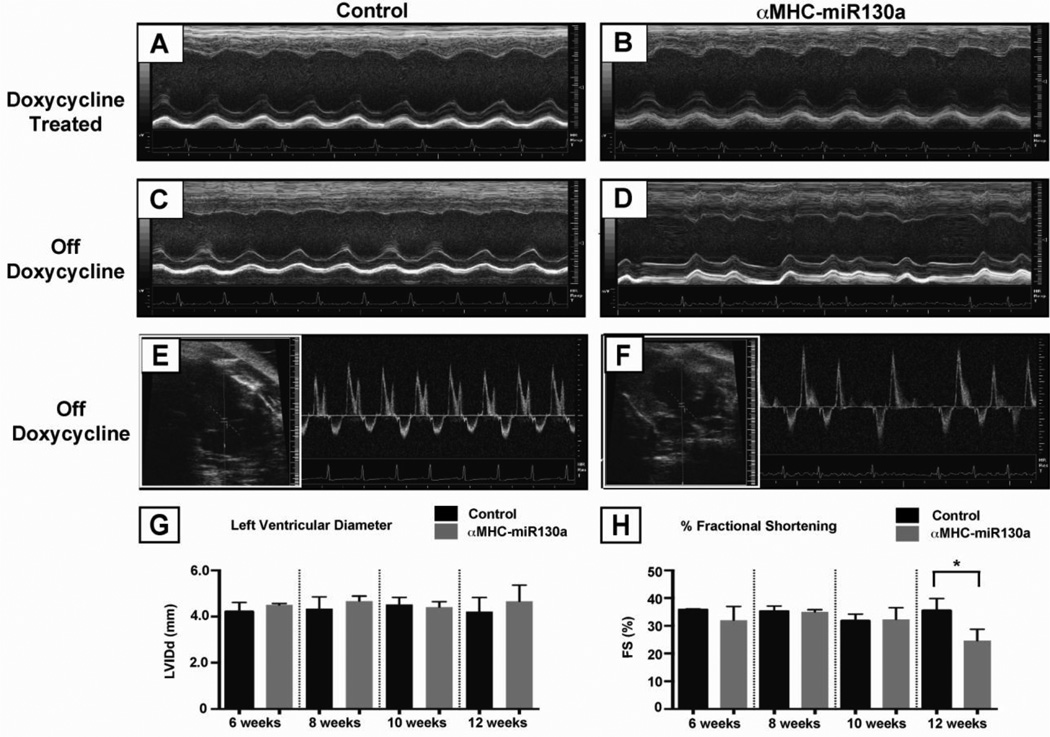
Figure 2. Echocardiography in αMHC-miR130a mice.
Panels (A) and (B), m-mode echocardiography of representative control vs. αMHC-miR130a mouse maintained on doxycycline at 10 weeks after weaning. Cardiac dimensions and function was preserved. In panels (C) and (D), representative M-mode echocardiography of control vs. αMHC-miR130a mouse at 10 weeks after doxycycline removal. Note the irregularity of ventricular contractions in the αMHC-miR130a mice. In panels (E) and (F), 2D–guided pulsed Doppler (see inset) of the mitral inflow in control and the αMHC-miR130a mice respectively. (G) and (H), serial assessment in LV diameter and % fractional shortening at 6, 8, 10, and 12 weeks. In each group, n = 5. * indicates p < 0.001.
3.2 Cardiac overexpression of microRNA-130a results in atrial arrhythmias
To evaluate the overall impact on cardiac function, we performed echocardiography in αMHC-miR130a and control littermate mice. No differences in left ventricular dimensions and fractional shortening (FS) were found from 4–10 weeks (Table 1). Beginning at 12 weeks, the fractional shortening showed a decline (35.5 ± 4.3% vs 24.7 ± 4.1%, p=0.004) (Figure 2, H). At that time, we also noted an irregularity in ventricular contraction often seen with arrhythmias (Figure 2, D). To look for evidence of atrial contractions, we performed pulsed-wave Doppler at the area of mitral inflow. As seen in control mice, a typical mitral inflow pattern in sinus rhythm comprises of an “E” wave and “A” wave indicating two distinct phases of blood flow (Figure 2, E). The E wave component, created by flow into the left ventricle during early diastolic filling, occurs prior to atrial contraction. The A wave component is created by flow into the left ventricle due to contraction of the atria. As seen in the αMHC-miR130a transgenic mice, pulse Doppler did not reveal the presence of an A wave generated by atrial contraction (Figure 2, F). As expected, evidence of early diastolic filling (independent of atrial contraction) was present, and the irregular timing was consistent with an atrial arrhythmia.
Table I.
Comparison of ECG and Echo Data in Control and αMHC-miR130a Mice
| 6 weeks | 10 weeks | 12 weeks | ||||
|---|---|---|---|---|---|---|
| Control | MHC-miR130a | Control | MHC-miR130a | Control | MHC-miR130a | |
| n | 5 | 5 | 5 | 5 | 5 | 3* |
| PR (ms) | 33.1±2.9 | 34.7±3.2 | 35.2±0.7 | 40.8±0.6† | 33.0±1.2 | 44.0±2.5† |
| QRS (ms) | 12.5±0.7 | 10.7±0.9 | 10.1±0.46 | 11.2±0.85 | 11.0±0.57 | 10.6±0.68 |
| QT (ms) | 19.3±0.7 | 19.0±1.2 | 18.7±0.5 | 20.0±0.9 | 19.0±0.6 | 20.4±0.7 |
| FS (%) | 35.8±0.2 | 32.0±2.8 | 31.8±1.1 | 32.3±1.7 | 35.5±2.2 | 24.7±1.7† |
| LVIDd (mm) | 4.3±0.2 | 4.6±0.1 | 4.5±0.2 | 4.4±0.1 | 4.2±0.3 | 4.6±0.3 |
| HR (bpm) | 534±98 | 515±105 | 512±113 | 519±96 | 514±89 | 459±113† |
2 of 5 mice in atrial arrhythmia, excluded from analysis of PR interval.
indicates p<0.01
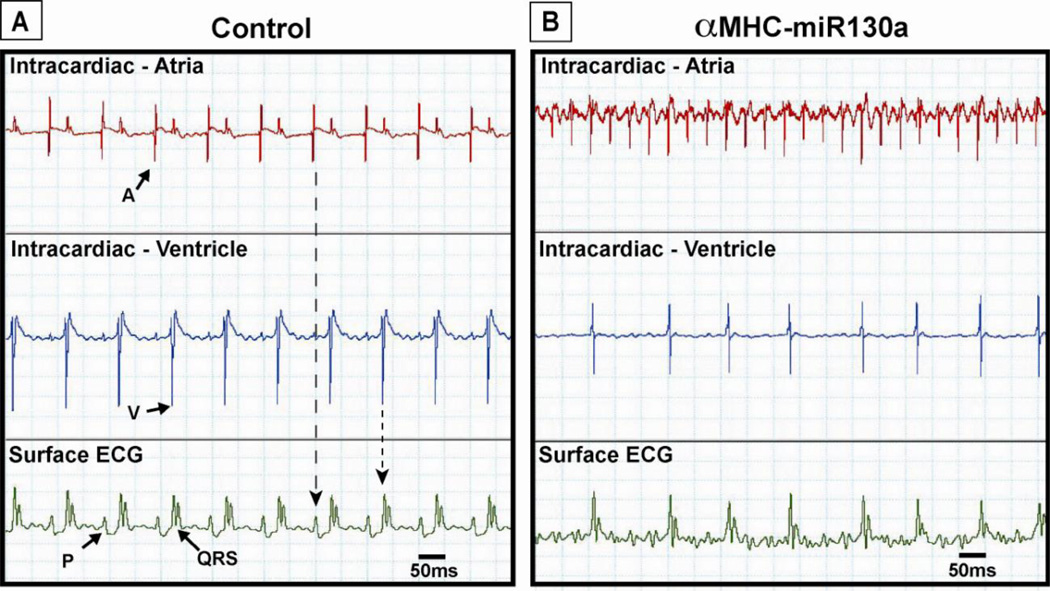
To investigate the possibility of an arrhythmia in more detail, we performed simultaneous atrial and ventricular intracardiac electrophysiologic recordings with simultaneous surface ECGs in control and αMHC-miR130a mice. After 10 weeks off doxycycline, the control mice demonstrated a regular P-QRS ECG pattern typical of sinus rhythm (Figure 3, A, bottom panel). The accompanying atrial electrogram displayed a regularly occurring high amplitude signal corresponding with the surface P wave. The ventricular electrogram displayed a high amplitude signal corresponding with the surface QRS complex. In contrast, surface ECG of αMHC-miR130a mice demonstrated an irregular ventricular rate with more rapid atrial activity, a hallmark for atrial tachyarrhythmias (Figure 3, B, bottom panel). The corresponding atrial electrogram demonstrated a rapid irregular atrial signal while the ventricular electrogram showed a slower irregular ventricular signal (Figure 3, B, top and middle panels). Intracardiac recordings were performed in eight mice in each group.
Figure 3. Simultaneous surface and intracardiac electrocardiograms in control and αMHC-miR130a mice.
In (A), simultaneous atrial (top panel), ventricular (middle panel) electrograms and surface ECG (bottom panel) of control mouse demonstrating normal sinus rhythm. Note the presence of regular P waves (P) representing atrial depolarization corresponding to high amplitude signal seen in the atrial recording. High amplitude ventricular signal corresponds to the QRS complex (QRS) on the surface ECG representing ventricular depolarization. In (B), simultaneous atrial, ventricular, and surface ECG in αMHC-miR130a mice. In contrast to the control mouse, the atrial electrogram (B, top panel) shows a rapid irregular atrial signal. The ventricular electrogram (B, middle panel) shows a high amplitude signal corresponding to QRS complexes on the surface ECG with a variable cycle length. Tracings obtained 10–12 weeks off doxycycline. Eight animals were studied in each group. A: atrial signal; V: ventricular signal.
Baseline electrocardiographic parameters were obtained in control and αMHC-miR130a transgenic mice after removal of doxycycline (Table 1). At 6 through 10 weeks, we did not detect any differences in heart rate between controls and transgenic mice. However, by 12 weeks, there was a significant reduction in heart rate in transgenic mice (514 ± 89 bpm vs. 459 ± 113 bpm, p<0.01). QRS duration and morphology did not change and was similar to controls (11.0 ± 0.58 ms vs. 10.6 ± 0.68 ms, p=ns) (Figure 4, A). QT interval also did not differ between groups. At 10 weeks off doxycycline, we noted an increase in the PR interval in the αMHC-miR130a transgenic mice (35.2 ± 0.7 ms vs 40.8 ± 0.6 ms, p=0.01) (Table 1).
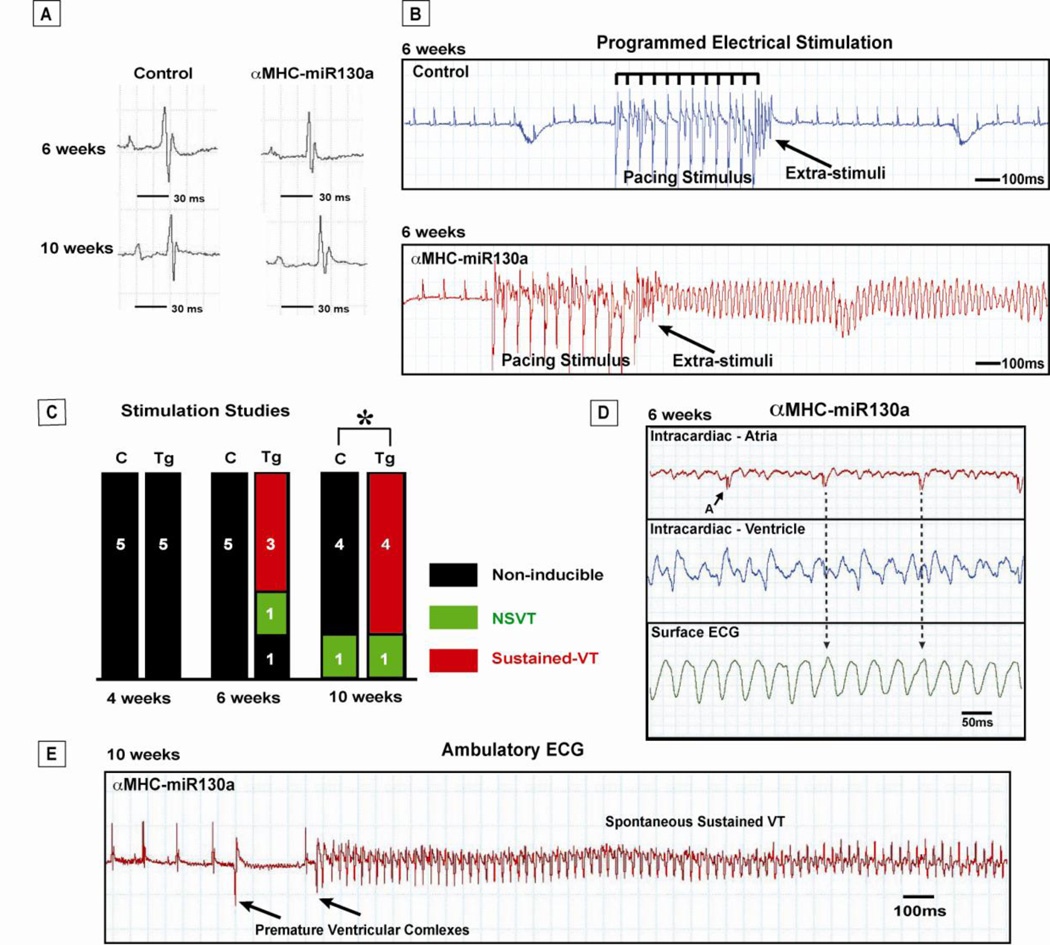
Figure 4. Propensity for ventricular arrhythmias in αMHC-miR130a mice.
In panel (A), representative P-QRS complexes in control and αMHC-miR130a mice at 6 and 10 weeks after doxycycline removal. In panel (B), representative programmed electrical ventricular stimulation studies in control mice (top panel) and αMHC-miR130a transgenic mice (bottom panel). In the control mouse, pacing stimulus could not induce VT. In contrast, sustained VT is seen in the αMHC-miR130a mouse following pacing stimulus. In (C), summary of programmed electrical stimulation studies with the number of inducible VT episodes in control (C) and miR-130a transgenic (Tg) animal cohorts at 4, 6, and 10 weeks. Sustained ventricular arrhythmias could not be induced in any of the controls. Beginning at 6 weeks after doxycycline removal, αMHC-miR130a mice could be induced into monomorphic VT. In panel (D), simultaneous intracardiac and surface ECG in αMHC-miR130a mice during VT episode. Atrial electrogram (top panel) demonstrates a slower atrial signal with a regular cycle length. Ventricular electrogram (middle panel) demonstrates a rapid high amplitude signal corresponding with the rapid wide QRS complexs on surface ECG. Arrows indicate potential atrial signal and corresponding surface ECG location within the QRS complex consistent with atrioventricular dissociation. These findings are supportive of VT. Shown in panel (E), an example of spontaneous VT initiated after premature ventricular complexes in αMHC-miR130a mice during ambulatory ECG monitoring 10 weeks after removal of doxycycline. Five animals in each group was studied for both stimulation studies and ambulatory ECG monitoring. * indicates p=0.026. NSVT: nonsustained ventricular tachycardia; A: atrial signal; VT: ventricular tachycardia; Tg: transgenic; C: control littermate.
3.3. MicroRNA-130a overexpression induces ventricular tachycardia
While performing surface ECGs, we detected periodic spontaneous nonsustained ventricular tachycardia (NSVT). To explore the arrhythmic susceptibility of the ventricle, we performed programmed electrical stimulation (PES) studies 4 through 10 weeks after removal of doxycycline. Hearts were paced using a subdiaphragmatic approach previously described [19]. A train of 12 stimuli was delivered at pacing cycle length of 120, 100, and 80 ms followed by double extrastimuli at cycle lengths of 20, 30, and 40 ms to assess the inducibility of ventricular tachyarrhythmias. For comparison of the inducibility in each mouse, programmed extrastimulation techniques and stimulation duration of ventricular burst pacing were the same in all mice. Mice were categorized as having no ventricular tachycardia (VT), NSVT, or sustained VT (Figure 4, B). We used the following criteria to categorize the ventricular arrhythmias: A) Non-sustained VT: 3–30 beats, B) Sustained VT: >30 beats, and C) Cycle Length: <100ms [19, 20].
At all time points between 4 and 10 weeks, sustained VT could not be induced in any of the control mice (Figure 4, B). At 4 weeks after doxycyline removal, αMHC-miR130a mice could not be induced into VT by any pacing maneuver. However, beginning at 6 weeks after doxycycline removal, sustained VT was induced in 3 of 5 of the αMHC-miR130a mice (Figure 4, B,C). To confirm the presence of VT, we performed simultaneous atrial and ventricular intracardiac recordings with surface ECG recordings after VT induction. As expected, we identified a rapid (approximately 40 ms cycle length), regular signal in the ventricular electrogram and a slower, regular high amplitude signal in the atrial electrogram (Figure 4, D). The atrial signal did not correspond with any ventricular activity corroborating an independent ventricular arrhythmia. Induction of either sustained VT with PES was accomplished in 4 of 5 animals studied and was found to be statistically significant by Fisher’s exact test (p=0.026). Interestingly, 10 weeks after removal of doxycycline, we observed frequent premature ventricular contractions and the development of spontaneous sustained ventricular tachycardia (3 of 5 animals) on ambulatory ECG monitoring (Figure 4, E).
3.4. Reduced Cx43 protein in αMHC-miR130a transgenic hearts
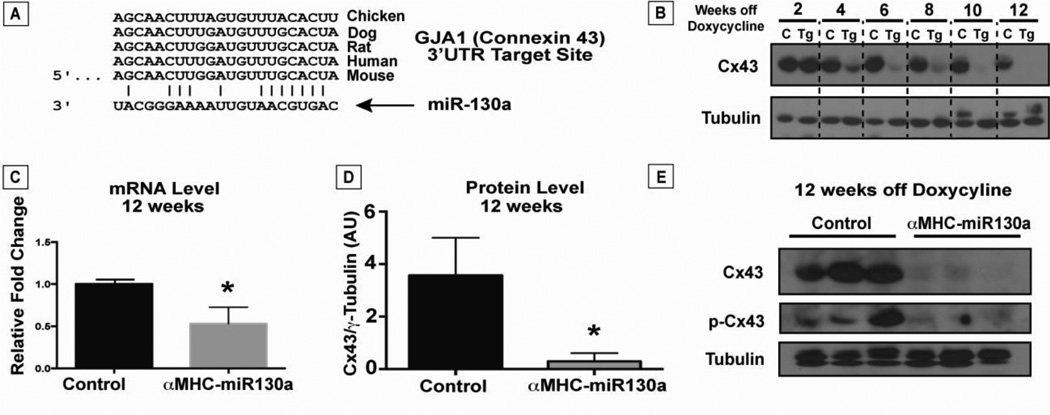
To explore the potential mechanism of the observed cardiac arrhythmias, we screened candidate miR-130a targets using known microRNA target databases. PicTar, TargetScan, PITA, and DIANA-microT 3.0 databases all predicted a conserved region in the 3’UTR of GJA1, encoding Cx43, to be a target of miR-130a (Figure 5, A) [22–25]. To examine the effect of miR-130a overexpression on connexin43 protein expression over time, we studied Cx43 levels on ventricular lysates from 2 weeks after doxycycline removal through 12 weeks. We noted a stepwise reduction in Cx43 protein levels starting from 4 weeks to a near complete loss of Cx43 by 10 weeks (Figure 5, B). Western blot analysis of ventricular lysates at 12 weeks off doxycycline revealed an approximate 90% reduction in Cx43 levels in the αMHC-miR130a hearts (n=10, each group) with a 52% reduction in mRNA levels compared with control littermates (n=5, each group) (Figure 5, C and D). Phosphorylation of Cx43 has been correlated with changes in assembly and degradation of Cx43 gap junction channels. Therefore, we also examined for changes in phosphorylated connexin43 (p-Cx43) levels in αMHC-miR130a hearts [26]. As shown in Figure 5E, there was a similar reduction in p-Cx43 levels in αMHC-miR130a mice compared with control littermate mice.
Figure 5. Reduced connexin43 in αMHC-miR130a mice.
In panel (A), computational alignment of a potential conserved target site in the Cx43 3’UTR and miR-130a. In (B), representative timecourse of Cx43 protein loss after doxycycline removal in αMHC-miR130a hearts compared to controls (n=3, each group). After 2 weeks, we found a steady reduction of Cx43 with near complete loss by 10 weeks after doxycycline removal. In (C), quantitative PCR demonstrated a 52% reduction in Cx43 mRNA at 12 weeks after doxycycline removal (n=5, each group). Western analysis on ventricular lysates (panel D) at the same time point shows >90% reduction in Cx43 protein in αMHC-miR130a hearts compared to controls (n=10 in each group). In panel (E), western blot for both Cx43 and phosphorylated-Cx43 at 12 weeks after doxycycline removal demonstrates significant reduction in both forms of Cx43. * indicates p < 0.001.
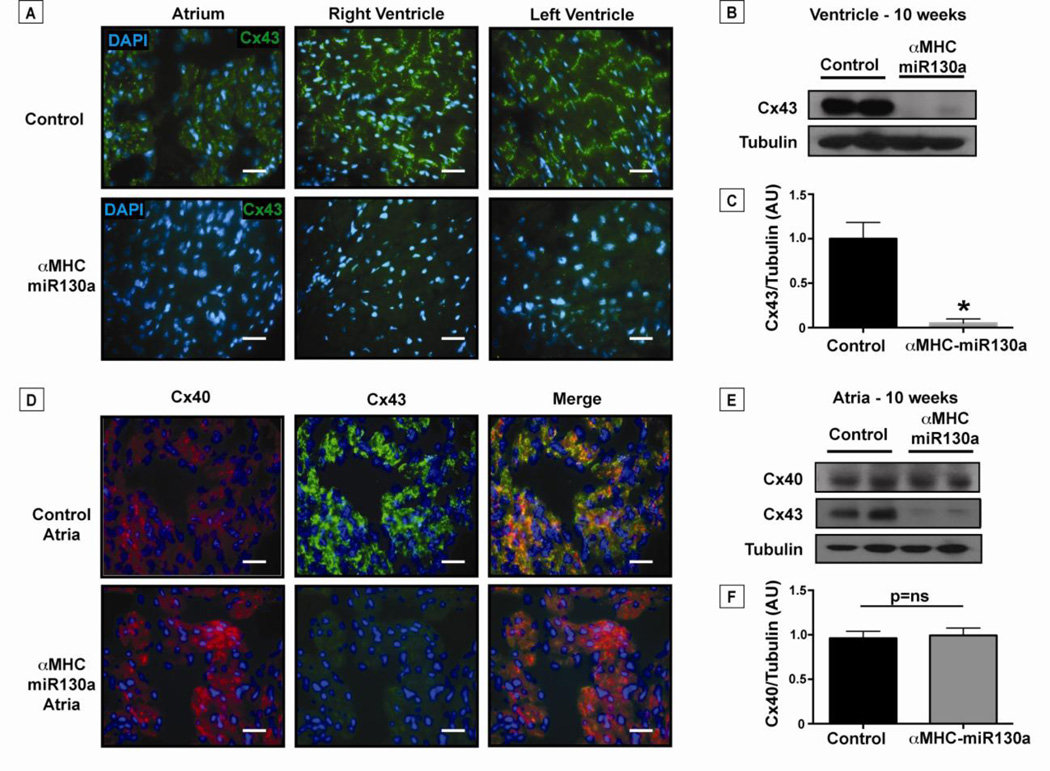
Given the significant reduction in Cx43 protein in heart lysates, we next visualized the distribution of Cx43 loss using immunofluorescence at 10 weeks after doxycycline removal. Not surprisingly, Cx43 was decreased throughout the atria and the ventricles in 16/16 transgenic animals studied (Figure 6, A). Unlike ventricular tissue, the atria express both Cx43 and connexin40 (Cx40), but unlike Cx43, Cx40 is not predicted to be a target for miR-130a. Given the atrial arrhythmias noted in the αMHC-miR130a mice, we performed immunofluorescence to assess the presence of Cx40 in the atria. We found that the atria of the αMHC-miR130a mice continued to express Cx40 (Figure 6, D). Atrial lysates were used for western blotting which confirmed preserved Cx40 levels in αMHC-miR130a atria compared to controls (Figure 6, E and F). As expected, there was near complete loss of Cx43 in the atria, both by immunofluorescence and western analysis (Figure 6, D and E), supporting the notion that the atrial arrhythmias observed were due to changes in Cx43 expression. Finally, we examined morphology of the intercalated disc using transmission electron microscopy (EM). The αMHC-miR130a hearts were notable for the near-complete absence of gap junctions (n=3, control and transgenic groups). Structures that appeared to be adherens-type junctions and desmosomes in the αMHC-miR130a heart sections were unchanged in their localization, organization, and general appearance, when compared with the controls (Supplemental Data, Figure S3).
Figure 6. Loss of connexin43 in the atria and ventricles of αMHC-miR130a mice.
In panel (A), representative immunofluorescent staining of Cx43 (green) in adult control atria and ventricles compared to αMHC-miR130a atria and ventricles (n=16 hearts studied). Cx43 is reduced throughout the myocardium of αMHC-miR130a mice. (B) Western blot of ventricular lysate at 10 weeks off doxycycline demonstrating >90% reduction in Cx43 levels as quantified in (C) (n=6 per group). In panel (D), representative immunofluorescent staining of atrial tissue for Cx40 (red) and Cx43 (green) at 10 weeks off doxycycline. Note the loss of Cx43 in αMHC-miR130a atria with preserved Cx40 expression. In panel (E), atrial lysates were used for western blotting confirming preserved Cx40 levels in αMHC-miR130a atria compared to controls and quantified in (F) (n=4 per group). Consistent with the ventricular lysates, Cx43 levels were significantly reduced in the atrial lysates. Nuclei are counterstained with DAPI (blue). * indicates p < 0.001.
3.5. MicroRNA-130a directly targets connexin43
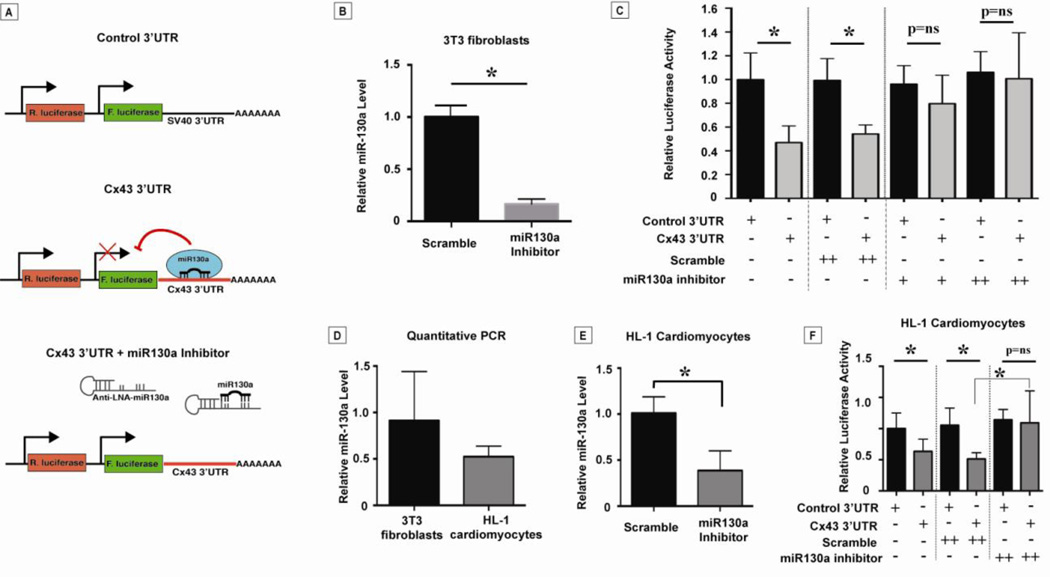
Because multiple targets may be affected by miR-130a overexpression in vivo, we sought to determine the relevance of the 3′UTR of Cx43 for translational regulation by miR-130a using in vitro transfection assays. We utilized a reporter construct in the vector pMT01 which contains both firefly and renilla luciferase reporters. The firefly luciferase coding region was fused to a control 3’UTR or the 3’UTR of Cx43 (Figure 7, A). Renilla luciferase provided a measure of transfection efficiency and was used to normalize firefly luciferase measurements. We transfected these constructs into NIH 3T3 fibroblasts, which were previously shown to endogenously express miR-130a [14, 27]. We predicted that binding of miR-130a would inhibit translation of the firefly luciferase (Figure 7, A, middle panel) and that addition of a specific miR-130a inhibitor would permit translation of the firefly luciferase (Figure 7, A, bottom panel). The 3T3 cells transfected with the Cx43 3′UTR construct showed 52.9% less luciferase activity compared with controls (0.998 ± 0.05 vs 0.469 ± 0.03, p<0.0001) (Figure 7, C, columns 1–2), indicating that the 3′ UTR of Cx43 was able to mediate translational repression.
Figure 7. MicroRNA-130a mediates translational repression via the Cx43 3’UTR.
In panel (A), schematic of luciferase reporter constructs used for in vitro transfection assays. Firefly luciferase (F. luciferase) coding region is fused to the control 3’UTR vs. Cx43 3’UTR. Binding of miR-130a is predicted to inhibit translation of F. luciferase. Addition of a specific miR-130a inhibitor is predicted to allow translation of F. luciferase. As shown in (B), administration of a miR-130a inhibitor (300 picomoles) reduced miR-130a levels in 3T3 fibroblasts approximately 83%. Reporter constructs were transfected into NIH 3T3 fibroblasts (known to endogenously express miR-130a). In (C), luciferase assay with luciferase reporter constructs and miR-130a inhibitor vs. scramble. Inhibition of miR-130a had no effect on the translational efficiency of the control reporter. In contrast, translational repression of the reporter construct was relieved in a dose-dependent fashion. Co-transfection with a scramble inhibitor was performed as a control. Luciferase experiments were also carried out in HL-1 cardiomyocytes (also known to endogenously express miR-130a). Using quantitative PCR in (D), HL-1 cells contained 52.4% of the miR-130a compared to 3T3 cells. Administration of the miR-130a inhibitor resulted in a 73.2% reduction in miR-130a (E). Similar to 3T3 cells, luciferase assay demonstrated reduced luciferase activity in the presence of the Cx43 3’UTR reporter construct. Translational repression of the reporter construct was relieved with administration of a miR-130a inhibitor. Transfection with a scramble inhibitor was performed as a control. Experiments performed in triplicate × three independent experiments. * indicates p<0.01. 3’UTR: 3’ untranslated region; LNA: locked nucleic acid.
To demonstrate that miR-130a directly targeted the Cx43 3′UTR, we utilized a LNA-antisense oligonucleotide to specifically block miR-130a. As a preliminary step, we demonstrated that the miR-130a inhibitor was able to reduce endogenous miR-130a by 83% compared with the scramble control (Figure 7, B). Next, we transfected oligonucleotides along with our reporter constructs into 3T3 fibroblasts. As expected, miR-130a inhibition had no effect on the translational efficiency of the control reporter. In contrast, translational repression was relieved in a dose-dependent fashion with the addition of increasing amounts of anti-miR-130a (100–300 pmol) (Figure 7, C, columns 5–8). Co-transfection with a scramble oligonucleotide (300 pmol) was performed as a control and demonstrated no effect on luciferase activity. These observations strongly suggest that miR-130a is acting directly on the Cx43 3′UTR to mediate translational repression.
To validate these luciferase reporter assay findings in cardiomyocytes, we used HL-1 cardiomyocytes, which have also been shown to endogenously express miR-130a [28]. First, we performed quantitative PCR to measure miR-130a levels in HL-1 cells (Figure 7,D). HL-1 cells had approximately 47.6% less miR-130a than 3T3 fibroblasts. Addition of a miR-130a inhibitor reduced miR-130a levels by 73% (Figure 7,E). Co-transfection experiments confirmed our findings in the 3T3 cells with a reduction in luciferase activity compared with the control vector (0.997 ± 0.03 (control) vs. 0.576 ± 0.05 (Cx43 3’UTR), p<0.0001). Addition of anti-miR-130a oligonucleotide (300 pmol) produced an increase in luciferase activity (Figure 7,F) similar to our findings in 3T3 cells. Taken together, endogenously expressed miR-130a in both 3T3 cells and HL-1 cardiomyocytes was sufficient to target the 3’UTR of Cx43 and effect a reduction in luciferase activity. The repression of luciferase activity was attenuated by a specific miR-130a inhibitor implicating a direct effect of miR-130a on the 3’UTR of Cx43.
4. DISCUSSION
Using a conditional transgenic system, we have shown that overexpression of miR-130a in adult cardiomyocytes results in atrial and ventricular arrhythmias. Our data indicate that loss of a miR-130a target, Cx43, may contribute to this phenotype. MicroRNA-130a has been identified in a wide array of cell types and more is being uncovered about its function in disease states [29–31]. In endothelial cells, miR-130a was found to down-regulate the transcription factors GAX and HOXA5, thereby attenuating the anti-angiogenic activities of these genes [32]. We previously studied the role of miR-130a during cardiac development and found that miR-130a targeted Friend of GATA-2 (FOG-2 or ZFPM2), a gene critical for normal heart development [14]. As demonstrated by Thum et al., a microRNA network regulates the development and function of the heart from the early stages of cardiac development to adulthood and also appears to play a role in adult cardiovascular disease [12]. In human heart failure, levels of miR-130a were shown to be elevated compared with normal hearts [12, 13]. Therefore, we hypothesized that overexpression of miR-130a in adult cardiomyocytes would result in abnormal cardiac function.
Although we did detect LV dysfunction 12 weeks after miR-130a induction, the early appearance of cardiac arrhythmias before structural changes highlighted one predicted target -Cx43. Gap junction channels are responsible for cell-to-cell communication and intercellular propagation of electrical signals throughout the heart. A variety of connexins are expressed in cardiac myocytes; however, Cx43 is the predominant connexin expressed in both the atria and ventricle [4–6]. In advanced stages of heart disease, connexin expression and intercellular coupling are diminished and gap junction channels become redistributed. These changes have been strongly implicated in the pathogenesis of lethal arrhythmias through the creation of a proarrhythmic substrate [33, 34].
With the dramatic loss of Cx43 seen in our miR-130a transgenic mice, it is not surprising to find sustained arrhythmias. With increasing duration of miR-130a induction, there was a steady reduction in connexin43. Inducibility of ventricular arrhythmias began between 4 and 6 weeks after miR-130a overexpression and spontaneous ventricular arrhythmias between by 10 weeks of induction. Although LV dysfunction alone may alter Cx43 expression and thereby provoke arrhythmias, we consistently found that the onset of ventricular arrhythmias occurred before changes in cardiac function. As reported by Gutstein et al., cardiac deletion of Cx43 resulted in spontaneous ventricular arrhythmias without LV dysfunction, similar to what was seen in the miR-130a mice in our study [7].
Interestingly, miR-130a transgenic mice also developed spontaneous atrial arrhythmias. One aspect of atrial fibrillation that has elicited great interest has been changes in connexin protein expression. Several studies have linked decreased Cx43 levels and atrial fibrillation [35–37]. In our model, spontaneous atrial arrhythmias occurred at timepoints ranging from 8–12 weeks after miR-130a induction. The atrial arrhythmias appeared later than the development of ventricular arrhythmias. This may be partially because of preserved expression of Cx40 in the atria of the miR-130a transgenic mice. It is conceivable that the presence of Cx40 was able to compensate for the loss of Cx43 in the early stages of miR-130a induction and thus account for the later appearance of the atrial arrhythmia [38].
Aside from the generation of atrial and ventricular arrhythmias, we also noted prolongation of the PR interval in the miR-130a transgenic mice. Within the AV node, Cx43 expression is highly suppressed, with levels increasing after the compact portion of the AV node [39]. PR prolongation may be attributable to the suppression of Cx43 within this portion of the conduction system; however, a secondary mechanism cannot be excluded.
It has been shown in our model and other studies that gap junction remodeling can reduce intercellular coupling in ways that contribute to the pathogenesis of re-entrant arrhythmias. Atrial and ventricular arrhythmias are complex phenomena, and are often thought of as arising from multiple input determinants. The electrophysiological mechanisms by which miR-130a initiates arrhythmias are uncertain. Cellular uncoupling resulting from loss of Cx43 gap junction channels may unmask ectopic foci or enhance the generation of early afterdepolarizations [40]. In a chimeric mouse model of focal Cx43 loss in the myocardium, areas of focal uncoupling were associated with a significant increase in spontaneous ectopic events, possibly by unmasking intrinsic automaticity or by disrupting the conduction wave front leading to wave breaks [41]. Additional effects of miR-130a on targets not yet identified may also contribute to triggered activity perpetuated by gap junction remodeling.
Several recent studies have highlighted the role of miRNAs in the regulation of gap junction proteins and point towards a network of miRNAs critical for cardiac rhythm. In murine models of myocardial infarction and myocarditis, miR-1 was found to regulate Cx43 [42, 43]. Recently, the microRNA cluster miR-17–92 has been implicated in cardiomyopathy and arrhythmia. The authors demonstrated that Cx43 is a direct target of miR-19a/b and that the expression of Cx43 was suppressed in transgenic hearts [44]. In this study, we provided data demonstrating that miR-130a also targets Cx43 resulting in atrial and ventricular arrhythmias attributable to the loss of this important gap junction protein. These findings recapitulate the arrhythmogenic myocardial substrate seen in the Cx43 knockout studies and highlight the complex molecular mechanisms regulating Cx43 expression in the heart.
4.1. LIMITATIONS
We cannot definitively exclude the possibility that the arrhythmias seen are due to secondary effects of Cx43 reduction, independent of gap junction remodeling. Previous work has demonstrated a highly arrhythmogenic substrate when Cx43 alone is lost in the myocardium. Recapitulation of these findings, therefore, highlights a novel role for miR-130a in arrhythmia. In addition, it is likely that effects on other targets of miR-130a may contribute to the eventual reduction in left ventricular function and further provoke arrhythmias [1].
The in vitro experiments demonstrated the ability of miR-130a to target the Cx43 3’UTR; however, it is difficult to exclude the possibility of secondary effects of miR-130a overexpression. There may be effects of miR-130a on important regulators of calcium homeostasis not yet described and thus contribute to arrhythmia generation. In an effort to examine this important aspect of arrhythmia, we pursued analysis at the protein level of several calcium homeostasis proteins. We chose to perform western analysis at the 8-week time point to reflect a time in our model when the ventricular arrhythmias are evident without changes in LV function. We performed western analysis on several proteins known to regulate calcium homeostasis including: Serca2a, phospholamban and phosphorylated-phospholamban, ryanodine receptor, the calcium channel protein Cav1.1, BIN1–a protein that assists in L-type calcium channels targeting to T-tubules in cardiac myocytes, and junctophilin 2, a membrane-binding protein believed to keep the plasma membrane and sarcoplasmic reticulum within a fixed proximity to one another [45–47]. At 8 weeks after doxycycline removal, we did not find any significant protein reductions greater than 50% (Supplemental Data, Figure S4). When investigating the predicted miR-130a targets using the databases that predicted Cx43, we did not identify a viable target influencing calcium homeostasis. We acknowledge that discerning microRNA targets based solely on target prediction databases has inherent limitations, thus identification of additional targets important for arrhythmia remains a focus of future studies.
5. CONCLUSIONS
Electrical remodeling and structural remodeling are well-established mechanisms of arrhythmia generation, particularly in the setting of heart failure. However, published studies linking miRNAs and cardiac conduction are sparse. Recent studies have highlighted the role of miRNAs in cardiac rhythm through the regulation of key ion channels, transporters, and cellular proteins in arrhythmogenic conditions [42, 48–52]. Studies have shown that misregulation of these critical “rhythmirs” are sufficient to induce a variety of conduction disturbances and arrhythmias. Further study of these miRNAs will hopefully reveal a regulatory network for cardiac rhythm by linking previously disparate signaling pathways and identifying novel targets.
As the study of miRNAs begins to move into the arena of therapeutics, a thorough understanding of the broad functions of any particular miRNA will be necessary to avoid harmful “off target” effects. As mentioned previously, miR-130a has been described in its role as a potential regulator of angiogenesis. In a recent study, treatment with a miR-130a mimic showed a trend toward improved capacity for CD34+ cells to stimulate cardiac repair and neovascularization [11]. While a miR-130a mimic may hold promise in angiogenesis, a better understanding of elevating miR-130a levels and its effects on organ function is critical. A potential therapeutic application of miR-130a mimics for heart failure patients would warrant caution because of possible pro-arrhythmic effects, based on possible effects on Cx43.
Supplementary Material
Highlights.
We used an inducible cardiac-specific model to study microRNA-130a in adult heart.
Overexpression of miR-130a results in atrial and ventricular tachyarrhythmias.
Connexin43 is reduced throughout the heart after miR-130a overexpression.
MicoRNA-130a directly targets the 3’UTR of connexin43.
We report a novel role for miR-130a in gap junction remodeling and arrhythmias.
Acknowledgements
We are grateful for the assistance of John Fahrenbach in programming of electrical stimulation studies.
Sources of Funding:
Work is supported by NIH-K08-HL098565 (GHK) and the Institute for Cardiovascular Research, University of Chicago.
GLOSSARY
- αMHC
alpha myosin heavy chain
- 3’UTR
3’ untranscribed region
- AF
atrial fibrillation
- AV
atrioventricular
- Cx
connexin
- ECG
electrocardiogram
- LV
left ventricle
- miR
microRNA
- PES
programmed electrical stimulation
- VT
ventricular tachycardia
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Disclosures:
No competing potential conflicts exist.
References
- 1.Nattel S, Maguy A, Le Bouter S, Yeh YH. Arrhythmogenic ion-channel remodeling in the heart: heart failure, myocardial infarction, and atrial fibrillation. Physiol Rev. 2007;87:425–456. doi: 10.1152/physrev.00014.2006. [DOI] [PubMed] [Google Scholar]
- 2.Peters NS, Coromilas J, Severs NJ, Wit AL. Disturbed connexin43 gap junction distribution correlates with the location of reentrant circuits in the epicardial border zone of healing canine infarcts that cause ventricular tachycardia. Circulation. 1997;95:988–996. doi: 10.1161/01.cir.95.4.988. [DOI] [PubMed] [Google Scholar]
- 3.Yao JA, Hussain W, Patel P, Peters NS, Boyden PA, Wit AL. Remodeling of gap junctional channel function in epicardial border zone of healing canine infarcts. Circ Res. 2003;92:437–443. doi: 10.1161/01.RES.0000059301.81035.06. [DOI] [PubMed] [Google Scholar]
- 4.Kar R, Batra N, Riquelme MA, Jiang JX. Biological role of connexin intercellular channels and hemichannels. Arch Biochem Biophys. 2012;524:2–15. doi: 10.1016/j.abb.2012.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Willecke K, Eiberger J, Degen J, Eckardt D, Romualdi A, Guldenagel M, et al. Structural and functional diversity of connexin genes in the mouse and human genome. Biol Chem. 2002;383:725–737. doi: 10.1515/BC.2002.076. [DOI] [PubMed] [Google Scholar]
- 6.Davis LM, Rodefeld ME, Green K, Beyer EC, Saffitz JE. Gap junction protein phenotypes of the human heart and conduction system. J Cardiovasc Electrophysiol. 1995;6:813–822. doi: 10.1111/j.1540-8167.1995.tb00357.x. [DOI] [PubMed] [Google Scholar]
- 7.Gutstein DE, Morley GE, Tamaddon H, Vaidya D, Schneider MD, Chen J, et al. Conduction slowing and sudden arrhythmic death in mice with cardiac-restricted inactivation of connexin43. Circ Res. 2001;88:333–339. doi: 10.1161/01.res.88.3.333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kim GH. MicroRNA regulation of cardiac conduction and arrhythmias. Transl Res. 2012 doi: 10.1016/j.trsl.2012.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 10.Chen Y, Gorski DH. Regulation of angiogenesis through a microRNA (miR-130a) that down-regulates antiangiogenic homeobox genes GAX and HOXA5. Blood. 2008;111:1217–1226. doi: 10.1182/blood-2007-07-104133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jakob P, Doerries C, Briand S, Mocharla P, Krankel N, Besler C, et al. Loss of angiomiR-126 and 130a in angiogenic early outgrowth cells from patients with chronic heart failure: role for impaired in vivo neovascularization and cardiac repair capacity. Circulation. 2012;126:2962–2975. doi: 10.1161/CIRCULATIONAHA.112.093906. [DOI] [PubMed] [Google Scholar]
- 12.Thum T, Galuppo P, Wolf C, Fiedler J, Kneitz S, van Laake LW, et al. MicroRNAs in the human heart: a clue to fetal gene reprogramming in heart failure. Circulation. 2007;116:258–267. doi: 10.1161/CIRCULATIONAHA.107.687947. [DOI] [PubMed] [Google Scholar]
- 13.Matkovich SJ, Van Booven DJ, Youker KA, Torre-Amione G, Diwan A, Eschenbacher WH, et al. Reciprocal regulation of myocardial microRNAs and messenger RNA in human cardiomyopathy and reversal of the microRNA signature by biomechanical support. Circulation. 2009;119:1263–1271. doi: 10.1161/CIRCULATIONAHA.108.813576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim GH, Samant SA, Earley JU, Svensson EC. Translational control of FOG-2 expression in cardiomyocytes by microRNA-130a. PLoS One. 2009;4:e6161. doi: 10.1371/journal.pone.0006161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Claycomb WC, Lanson NA, Jr, Stallworth BS, Egeland DB, Delcarpio JB, Bahinski A, et al. HL-1 cells: a cardiac muscle cell line that contracts and retains phenotypic characteristics of the adult cardiomyocyte. Proc Natl Acad Sci U S A. 1998;95:2979–2984. doi: 10.1073/pnas.95.6.2979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Silahtaroglu AN, Nolting D, Dyrskjot L, Berezikov E, Moller M, Tommerup N, et al. Detection of microRNAs in frozen tissue sections by fluorescence in situ hybridization using locked nucleic acid probes and tyramide signal amplification. Nat Protoc. 2007;2:2520–2528. doi: 10.1038/nprot.2007.313. [DOI] [PubMed] [Google Scholar]
- 17.Westphal C, Spallek B, Konkel A, Marko L, Qadri F, DeGraff LM, et al. CYP2J2 overexpression protects against arrhythmia susceptibility in cardiac hypertrophy. PLoS One. 2013;8:e73490. doi: 10.1371/journal.pone.0073490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ye L, Zhu W, Backx PH, Cortez MA, Wu J, Chow YH, et al. Arrhythmia and sudden death associated with elevated cardiac chloride channel activity. Journal of cellular and molecular medicine. 2011;15:2307–2316. doi: 10.1111/j.1582-4934.2010.01243.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gutstein DE, Danik SB, Sereysky JB, Morley GE, Fishman GI. Subdiaphragmatic murine electrophysiological studies: sequential determination of ventricular refractoriness and arrhythmia induction. Am J Physiol Heart Circ Physiol. 2003;285:H1091–H1096. doi: 10.1152/ajpheart.00100.2003. [DOI] [PubMed] [Google Scholar]
- 20.Vaidya D, Morley GE, Samie FH, Jalife J. Reentry and fibrillation in the mouse heart. A challenge to the critical mass hypothesis. Circ Res. 1999;85:174–181. doi: 10.1161/01.res.85.2.174. [DOI] [PubMed] [Google Scholar]
- 21.Sanbe AGJ, Hanks MC, Liang Q, Osinska H, Robbins J. Reengineering Inducible Cardiac-Specific Transgenesis With an Attenuated Myosin Heavy Chain Promoter Circ Res. 2003;92:609–616. doi: 10.1161/01.RES.0000065442.64694.9F. [DOI] [PubMed] [Google Scholar]
- 22.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 23.Krek A, Grun D, Poy MN, Wolf R, Rosenberg L, Epstein EJ, et al. Combinatorial microRNA target predictions. Nat Genet. 2005;37:495–500. doi: 10.1038/ng1536. [DOI] [PubMed] [Google Scholar]
- 24.Kertesz M, Iovino N, Unnerstall U, Gaul U, Segal E. The role of site accessibility in microRNA target recognition. Nat Genet. 2007;39:1278–1284. doi: 10.1038/ng2135. [DOI] [PubMed] [Google Scholar]
- 25.Maragkakis M, Alexiou P, Papadopoulos GL, Reczko M, Dalamagas T, Giannopoulos G, et al. Accurate microRNA target prediction correlates with protein repression levels. BMC Bioinformatics. 2009;10:295. doi: 10.1186/1471-2105-10-295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Remo BF, Qu J, Volpicelli FM, Giovannone S, Shin D, Lader J, et al. Phosphatase-resistant gap junctions inhibit pathological remodeling and prevent arrhythmias. Circ Res. 2011;108:1459–1466. doi: 10.1161/CIRCRESAHA.111.244046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Houbaviy HB, Murray MF, Sharp PA. Embryonic stem cell-specific MicroRNAs. Dev Cell. 2003;5:351–358. doi: 10.1016/s1534-5807(03)00227-2. [DOI] [PubMed] [Google Scholar]
- 28.Humphreys DT, Hynes CJ, Patel HR, Wei GH, Cannon L, Fatkin D, et al. Complexity of murine cardiomyocyte miRNA biogenesis, sequence variant expression and function. PLoS One. 2012;7:e30933. doi: 10.1371/journal.pone.0030933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wu WH, Hu CP, Chen XP, Zhang WF, Li XW, Xiong XM, et al. MicroRNA-130a mediates proliferation of vascular smooth muscle cells in hypertension. Am J Hypertens. 2011;24:1087–1093. doi: 10.1038/ajh.2011.116. [DOI] [PubMed] [Google Scholar]
- 30.Sepramaniam S, Ying LK, Armugam A, Wintour EM, Jeyaseelan K. MicroRNA-130a represses transcriptional activity of aquaporin 4 M1 promoter. J Biol Chem. 2012;287:12006–12015. doi: 10.1074/jbc.M111.280701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tang L, Pu Y, Wong DK, Liu T, Tang H, Xiang T, et al. The hepatitis B virus-associated estrogen receptor alpha (ERalpha) was regulated by microRNA-130a in HepG2.2.15 human hepatocellular carcinoma cells. Acta Biochim Biophys Sin (Shanghai) 2011;43:640–646. doi: 10.1093/abbs/gmr051. [DOI] [PubMed] [Google Scholar]
- 32.Chen YGD. Regulation of angiogenesis through a microRNA (miR-130a) that down-regulates antiangiogenic homeobox genes GAX and HOXA5. Blood. 2008;111:1217–1226. doi: 10.1182/blood-2007-07-104133. PMCID: PMC2214763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Remo BF, Giovannone S, Fishman GI. Connexin43 cardiac gap junction remodeling: lessons from genetically engineered murine models. J Membr Biol. 2012;245:275–281. doi: 10.1007/s00232-012-9448-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Severs NJ, Bruce AF, Dupont E, Rothery S. Remodelling of gap junctions and connexin expression in diseased myocardium. Cardiovasc Res. 2008;80:9–19. doi: 10.1093/cvr/cvn133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Igarashi T, Finet JE, Takeuchi A, Fujino Y, Strom M, Greener ID, et al. Connexin gene transfer preserves conduction velocity and prevents atrial fibrillation. Circulation. 2012;125:216–225. doi: 10.1161/CIRCULATIONAHA.111.053272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Luo MH, Li YS, Yang KP. Fibrosis of collagen I and remodeling of connexin 43 in atrial myocardium of patients with atrial fibrillation. Cardiology. 2007;107:248–253. doi: 10.1159/000095501. [DOI] [PubMed] [Google Scholar]
- 37.Thibodeau IL, Xu J, Li Q, Liu G, Lam K, Veinot JP, et al. Paradigm of genetic mosaicism and lone atrial fibrillation: physiological characterization of a connexin 43-deletion mutant identified from atrial tissue. Circulation. 2010;122:236–244. doi: 10.1161/CIRCULATIONAHA.110.961227. [DOI] [PubMed] [Google Scholar]
- 38.Thomas SA, Schuessler RB, Berul CI, Beardslee MA, Beyer EC, Mendelsohn ME, et al. Disparate effects of deficient expression of connexin43 on atrial and ventricular conduction: evidence for chamber-specific molecular determinants of conduction. Circulation. 1998;97:686–691. doi: 10.1161/01.cir.97.7.686. [DOI] [PubMed] [Google Scholar]
- 39.Temple IP, Inada S, Dobrzynski H, Boyett MR. Connexins and the atrioventricular node. Heart Rhythm. 2013;10:297–304. doi: 10.1016/j.hrthm.2012.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Saiz J, Ferrero JM, Jr, Monserrat M, Ferrero JM, Thakor NV. Influence of electrical coupling on early afterdepolarizations in ventricular myocytes. IEEE transactions on bio-medical engineering. 1999;46:138–147. doi: 10.1109/10.740876. [DOI] [PubMed] [Google Scholar]
- 41.Gutstein DE, Danik SB, Lewitton S, France D, Liu F, Chen FL, et al. Focal gap junction uncoupling and spontaneous ventricular ectopy. Am J Physiol Heart Circ Physiol. 2005;289:H1091–H1098. doi: 10.1152/ajpheart.00095.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yang B, Lin H, Xiao J, Lu Y, Luo X, Li B, et al. The muscle-specific microRNA miR-1 regulates cardiac arrhythmogenic potential by targeting GJA1 and KCNJ2. Nat Med. 2007;13:486–491. doi: 10.1038/nm1569. [DOI] [PubMed] [Google Scholar]
- 43.Xu HF, Ding YJ, Shen YW, Xue AM, Xu HM, Luo CL, et al. MicroRNA- 1 represses Cx43 expression in viral myocarditis. Mol Cell Biochem. 2012;362:141–148. doi: 10.1007/s11010-011-1136-3. [DOI] [PubMed] [Google Scholar]
- 44.Danielson LS, Park DS, Rotllan N, Chamorro-Jorganes A, Guijarro MV, Fernandez-Hernando C, et al. Cardiovascular dysregulation of miR-17–92 causes a lethal hypertrophic cardiomyopathy and arrhythmogenesis. FASEB J. 2013;27:1460–1467. doi: 10.1096/fj.12-221994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Beavers DL, Wang W, Ather S, Voigt N, Garbino A, Dixit SS, et al. Mutation E169K in junctophilin-2 causes atrial fibrillation due to impaired RyR2 stabilization. J Am Coll Cardiol. 2013;62:2010–2019. doi: 10.1016/j.jacc.2013.06.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Luo M, Anderson ME. Mechanisms of altered Ca(2)(+) handling in heart failure. Circ Res. 2013;113:690–708. doi: 10.1161/CIRCRESAHA.113.301651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hong TT, Smyth JW, Gao D, Chu KY, Vogan JM, Fong TS, et al. BIN1 localizes the L-type calcium channel to cardiac T-tubules. PLoS biology. 2010;8:e1000312. doi: 10.1371/journal.pbio.1000312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Adam O, Lohfelm B, Thum T, Gupta SK, Puhl SL, Schafers HJ, et al. Role of miR-21 in the pathogenesis of atrial fibrosis. Basic Res Cardiol. 2012;107:278. doi: 10.1007/s00395-012-0278-0. [DOI] [PubMed] [Google Scholar]
- 49.Belevych AE, Sansom SE, Terentyeva R, Ho HT, Nishijima Y, Martin MM, et al. MicroRNA-1 and −133 increase arrhythmogenesis in heart failure by dissociating phosphatase activity from RyR2 complex. PLoS One. 2011;6:e28324. doi: 10.1371/journal.pone.0028324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Care A, Catalucci D, Felicetti F, Bonci D, Addario A, Gallo P, et al. MicroRNA-133 controls cardiac hypertrophy. Nat Med. 2007;13:613–618. doi: 10.1038/nm1582. [DOI] [PubMed] [Google Scholar]
- 51.Lu Y, Zhang Y, Wang N, Pan Z, Gao X, Zhang F, et al. MicroRNA-328 contributes to adverse electrical remodeling in atrial fibrillation. Circulation. 2010;122:2378–2387. doi: 10.1161/CIRCULATIONAHA.110.958967. [DOI] [PubMed] [Google Scholar]
- 52.Rau F, Freyermuth F, Fugier C, Villemin JP, Fischer MC, Jost B, et al. Misregulation of miR-1 processing is associated with heart defects in myotonic dystrophy. Nat Struct Mol Biol. 2011;18:840–845. doi: 10.1038/nsmb.2067. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.