Abstract
SUMMARY
We present a pipeline named BIR (Blast, Identify and Realign) developed for phylogenomic analyses. BIR is intended for the identification of gene sequences applicable for phylogenomic inference. The pipeline allows users to apply their own manually curated sequence alignments (seed) in search for homologous genes in sequence databases and available genomes. BIR automatically adds the identified sequences from these databases to the seed alignments and reconstruct a phylogenetic tree from each. The BIR pipeline is an efficient tool for the identification of orthologous gene copies because it expands user-defined sequence alignments and conducts massive parallel phylogenetic reconstruction. The application is also particularly useful for large-scale sequencing projects that require management of a large number of single-gene alignments for gene comparison, functional annotation, and evolutionary analyses.
AVAILABILITY
The BIR user manual is available at http://www.bioportal.no/ and can be accessed through Lifeportal at https://lifeportal.uio.no. Access is free but requires a user account registration using the link “Register for BIR access” from the Lifeportal homepage.
Keywords: phylogenetics, phylogenomics, genomics, transcriptomics, ortholog prediction, alignment construction
Introduction
Over the last decade phylogenomics has been used to infer the global phylogenetic tree and reconstruct the evolutionary relationship among the major assemblages of eukaryote lineages.1–10 The success of such analyses is based on the construction of a large number of sequence alignments and the concatenation of multiple single-gene alignments into one supermatrix. This laborious and time-consuming enterprise involves sequence annotations at the genomic or transcriptomal level, construction of single-gene alignments, identification of ortholog sequences, and the final inference of multigene phylogenetic trees. The workload further increases as new generations of efficient sequencing techniques are being developed and the amount of available sequence data is growing rapidly.11,12 Construction of sequence alignments can be laborious because it requires, for instance, collection of relevant data, manual adjustment of indels, and removal of ambiguously aligned sites. Hence, curated alignments (hereafter referred to as seed alignments) are natural starting points for phylogenomic inferences and for inclusion of newly sequenced genomes or transcriptomes.
In phylogenomic inferences, a critical step is the identification of ortholog gene copies, ie, genes that have been vertically inherited from a single origin. In contrast, paralogous gene copies generated by gene duplications within a genome may have diverged substantially by acquiring new functions. Inclusion of paralogous gene copies in sequence alignments can therefore seriously mislead the phylogenetic inferences of species.13,14 There is no simple solution to distinguish orthologs from paralogs, and often several different approaches are used.15–17 Most usually however, a phylogenetic tree is used to detect paralogs. If homologous sequences for selected species are scattered into different clades in a phylogenetic tree with robust statistical value (eg, .70% bootstrap support3,18), it can indicate the presence of paralog copies that should be removed before the concatenation of genes. Phylogenetic methods are preferred over the simple pairwise clustering strategy or similarity searches such as BLAST, because such methods include models of sequence evolution that better accommodate the evolutionary history of the sequences.
Phylogenomic analyses are often done on data that are based on hundreds of different single-gene alignments.2,5,6,8,9,19 Hence, there is an obvious need for an efficient process to construct alignments and perform single-gene phylogenetic inferences for paralog detection and definition of ortholog sequences.
In this paper, we present a bioinformatics pipeline called BIR (BLAST, Identify, Re-align) for the preparation of phylogenomic data. BIR produces two sets of data that provide a basis for the determination of orthologous gene sequences: 1) single-gene trees and statistical estimation of the robustness of the branch pattern that are inferred from single-gene alignments and 2) clustering of sequences into orthologous groups. Together, these enable the user to select the correct ortholog of interest. To ensure the usability and flexibility of the pipeline, we have designed a web page where the user can set all the parameters according to his or her own analyses. The pipeline is installed on Lifeportal (https://lifeportal.uio.no/root), where several other relevant programs are available for the users of BIR, such as BLAST,20 modeltest,21 mrbayes,22 phylobayes,23 RAxML,24 and the AIR package.25 All programs are implemented on a high-performance computing cluster to ensure high speed of the analysis and easy access to other relevant bioinformatic software. Altogether, the BIR pipeline is therefore an efficient and user-friendly tool for the massive parallel construction of alignments and identification of orthologs, in particular useful for the annotation of genes and the initial steps of phylogenomic inference.
Method
BIR is implemented on the Lifeportal web portal
BIR is written in Perl v5.8 and implemented on the web-based Lifeportal bioinformatics service at the University of Oslo. Initially, the user provides two files (both in FASTA format): 1) a ZIP-compressed file containing the query files. These typically consist of non-annotated sequences generated in genome and transcriptome sequencing projects, and 2) a ZIP file containing all the seed alignments.
Extending seed alignments in five steps
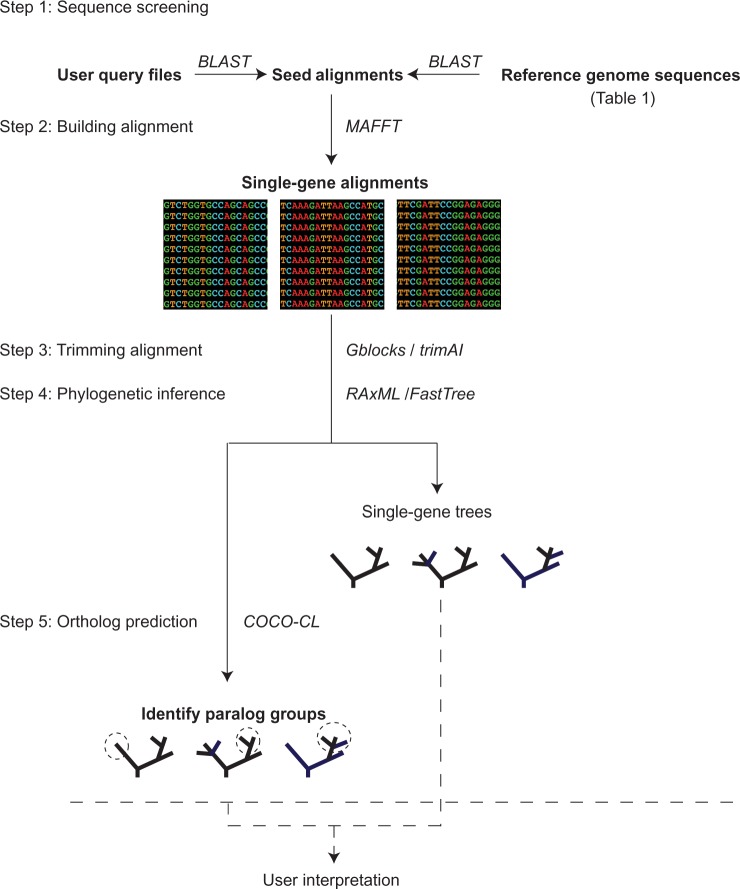
The procedure to generate extended single-gene alignments is divided into five steps (fig. 1). First, the query files are matched against the seed alignment using the BLAST algorithm.20 Based on the BLAST search and the quality criteria set by the user (ie, query coverage percentage, subject coverage percentage, identity percentage, score, and e-value), sequences from the query files are added to the seed alignments with the best match. Using the same approach, the user can increase the probability of detecting hidden paralogs in the seed alignments and query sequences by incorporating sequences from other available genomes representing all the major groups of eukaryotes (see Table 1 for detailed information about these genomes). The user can define the maximum number of sequences to be added from each of the selected genomes. BLAST result files often contain multiple high-scoring pair (HSP) sequences that describe regions of similarity between query and hit sequences. In contrast, BIR uses a combination of alignment length, identity percentage, score, and e-value statistics to calculate sequence similarity.
Figure 1.
Overview of steps in BIR pipeline. 1) The user provides a zipped file with the query sequences and another zipped file with the seed alignments. The sequence and alignments should be in FASTA format. Additionally, protein sequences from completely sequenced genomes (Table 1) can be added. Sequences from query files and selected reference genomes are added to the seed alignments with highest match using BLAST. 2) The modified seed alignments can be realigned using MAFFT. 3) Gblocks or trimAl can be used for removal of unambiguously aligned regions. 4) Phylogenetic trees can be inferred with FastTree or RaxML. 5) Paralog prediction is done by the COCO-CL program. Putative paralogs are marked in circles with a dashed line. The resulting phylogenetic trees can then further be assessed and interpreted using any tree-viewing software.
Table 1.
Completely sequenced genomes from the eukaryotic super groups available in the BIR pipeline.
| ORGANISM | SUPERGROUP | SIZE (MB) | GC% | #AA | BIOPROJECT |
|---|---|---|---|---|---|
| Arabidopsis thaliana | Plantae | 119.67 | 36.1 | 35375 | PRJNA116, PRJNA10719 |
| Bigelowiella natans | SAR* (Rhizaria) | 0.17 | 29.7 | 136 | PRJNA27939, PRJNA27935 |
| Dictyostelium discoideum | Amoebozoa | 34.2 | 22.5 | 13315 | PRJNA13925, PRJNA201 |
| Guillardia theta | Hacrobia | 0.3 | 29.2 | 309 | PRJNA210, PRJNA20389, PRJNA27847 |
| Homo sapiens | Opisthokonta | 3224.46 | 41.7 | 34931 | PRJNA168, PRJNA31257 |
| Monosiga brevicollis | Opisthokonta | 38.73 | 54.8 | 9203 | PRJNA28133, PRJNA19045 |
| Naegleria gruberi | Excavata | 36.3 | 33.1 | 15759 | PRJNA43691, PRJNA14010 |
| Paramecium tetraurelia | SAR* (Alveolata) | 72.07 | 28.1 | 40043 | PRJNA19409, PRJNA18363 |
| Saccharomyces cerevisiae | Opisthokonta | 12.16 | 38.2 | 5909 | PRJNA128, PRJNA13838, PRJNA43747 |
| Thalassiosira pseudonana | SAR* (Stramenopila) | 32.44 | 46.9 | 11849 | PRJNA34119, PRJNA191 |
Note:
SAR = Stramenopila, Alveolata, Rhizaria. #AA = Number of protein sequences in each genome.
In the second step, the user can decide to align the sequences in the seed alignments by either realigning all sequences (implemented by choosing progressive or iterative methods in MAFFT26), or alternatively to preserve the original seed alignment and only add the newly identified sequences. In the third step, the main task is to remove ambiguously aligned characters. This can be done by using either the Gblocks27 or the trimAI programs.28 Gblocks and trimAl parameters can be modified by the user; parameters for the removal of columns can be set to conservative (strict) or liberal (relaxed). In the forth step, phylogenetic trees for each single-gene alignment are inferred by RAxML24 or FastTree.29 For RAxML, the user can select the evolutionary model and define the number of pseudo-replicates for bootstrapping analyses. Only the “-f a” option is implemented in BIR pipeline, but other options can be used in a separate installation of RAxML on Lifeportal. In the last step of the pipeline, orthologous groups of sequences are predicted by hierarchical clustering with COCO-CL.30 This algorithm requires a similarity distance matrix that is calculated separately using ClustalW.31 The pipeline provides alignments for users who want to use other bioinformatic tools or tree inferring programs, such as PhyloBayes,23 RAxML,24 and MrBayes22 (also available on Lifeportal under the Bioportal Phylogeny25 Tools section).
Results and Discussions
Fast and easy addition of sequences to seed alignments
BIR allows the fast and easy screening of high numbers of sequences against custom-defined seed alignments using BLAST. 20 Sequences with similarities higher than the user-defined cutoffs are automatically added to the seed alignments. Additionally, homologous amino acid sequences from representative species of all eukaryotic supergroups (for details, see Table 1) can optionally be added to the seed alignments, so as to better recover hidden paralog sequences in the input data. New sequences are aligned to the seed alignment using MAFFT. 26 The Gblocks27 or trimAl28 programs are included for the visualization and removal of ambiguously aligned sites. In the final steps of the pipeline, phylogenies from each singlegene alignments are generated by FastTree29 or RAxML.24 These phylogenetic trees, together with the prediction of orthologs by COCO-CL,30 provide the user with ample information so as to select true ortholog sequences. Upon completion of data processing, the download section of Lifeportal provides several output files. These include log files, result files, alignment files, and tree files. The generated files can be downloaded and interpreted manually, and tree files can be visualized separately by using one of the many available graphical programs such as FigTree32 and TreeView.33,34 For ease of visualization, sequences added to seed alignments from either the queries or the genomes, as well as the predicted paralogs from COCO-CL, are marked with *_Q, *_G, and *_C, respectively.
Unique aspects of BIR
Some of the steps in the BIR pipeline are similar to those of the other bioinformatics applications such as PyPhy,35 PhyloGena,36 Hal,37 and bioinformatics services such as phylogeny.fr38 and PALM.39 However, BIR is unique in providing a sequence screening using seed alignments to generate gene alignments. Also, it is the only program that adds amino acid sequences from representative eukaryote genomes that belong to all eukaryotic supergroups. Other programs and pipelines such as PyPhy, PhyloGena, and Hal are stand-alone tools that require the installation of third-party software and databases prior to use. Hal is currently only available as a command line program without graphical user interface, while online programs such as phylogeny.fr and PALM are specified for phylogenetic inference and selection detection, and they have strict limitations on number and length of the sequences. In contrast, BIR is a web-based bioinformatics service installed on a high-performance computing cluster, thus avoiding installations on local computational resources. Since the query files and the alignment files can be in nucleotide or protein sequences, it gives the user the added flexibility to use either type of data. However, the quality of the generated alignments is, in general, dependent on how conserved the input seed alignments are. BIR is linked to several other bioinformatics applications on Lifeportal for upstream and downstream data processing such as contig assembly, annotation, statistical analyses, and phylogenetic inferences. Hence, BIR can easily be combined with many other relevant applications in many fields of genomics and evolutionary biology.
Performance of the BIR pipeline – creating alignments for phylogenomic analyses
We developed a test case to demonstrate the usefulness and speed of analysis performed by the BIR program. As a starting point, we used the 124 single-gene alignments for phylogenomic analysis of the genus named Collodictyon. This genus was recently suggested to constitute one of the earliest branching eukaryote lineages based on phylogenomic analyses of 124 genes (published by Zhao et al.3). These single-gene alignments contained between 36 and 77 taxa, and varied in length from 53 to 975 AA (Table S1). From each of the 124 alignments, we randomly extracted 2–10 taxa with a total of 429 sequences; the lengths were in the range 36–975 amino acids (Table S2). These sequences were placed in one Fasta file, together with 1000 randomly generated protein sequences. The lengths of the randomly generated sequences varied from 96 to 1000 amino acids. The resulting file with 1429 sequences was then used as the BIR query file. The 124 single-gene alignments were zipped together and used as seed alignments. We subsequently ran BIR with the default settings. In less than 10 minutes, all 429 of the extracted sequences where added to the corresponding seed alignments, they were realigned using “add to existing alignments” option, and a phylogenetic tree for each alignment was produced using FastTree. All of the identified sequences from the query files were placed in the same alignment from where they had originated, and none of the randomly generated sequences were picked out. Several sequences were marked as possible paralogs by COCO-CL (Table S3). However, most of these were found to be either from species known to be hard to place phylogenetically because of long branches (eg, the parasitic taxa Entamoeba, Lesihmania, and Trypanosoma; see Zhao et al.3), or sequences with a high proportion of missing data. Since COCO-CL uses a phylogenetic framework to mark dubious sequences, it is natural that taxa with long branches should be marked as possible paralogs. The effect of removing these sequences from the analysis is discussed in Zhao et al.3
Conclusion
BIR provides a simple, fast, and user-friendly Web-based pipeline installed on a high-performance computing resource. The pipeline can create a massive number of alignments highly useful for sequence annotation and the identification of paralogs. Hence it can be used in many different bioinformatics disciplines including key steps in phylogenomic analyses and other comparative and functional studies.
Supplementary File
Table S1. Information about the single-gene alignments used in the test case.
Table S2. Sequences randomly extracted from the singlegene alignments.
Table S3. Results from COCO-CL.
Acknowledgments
We thank Åsmund Skjæveland, Jon Bråte, Russell J. S. Orr, Thomas Haverkamp, Othilde Elise Håvelsrud, and Roberto Sierra for useful discussions and valuable comments on manuscript.
Footnotes
ACADEMIC EDITOR: Jike Cui, Associate Editor
FUNDING: This work is supported by University of Oslo fellowships and project funding for SK, AKK, SZ, RSN, and XZ. KS-T thanks University of Oslo and the Molecular Life Science Board for starting grant and grants for the development of Bioportal and Lifeportal bioinformatics services. The authors confirm that the funder had no influence over the study design, content of the article, or selection of this journal.
COMPETING INTERESTS: Authors disclose no potential conflicts of interest.
Paper subject to independent expert blind peer review by minimum of two reviewers. All editorial decisions made by independent academic editor. Upon submission manuscript was subject to anti-plagiarism scanning. Prior to publication all authors have given signed confirmation of agreement to article publication and compliance with all applicable ethical and legal requirements, including the accuracy of author and contributor information, disclosure of competing interests and funding sources, compliance with ethical requirements relating to human and animal study participants, and compliance with any copyright requirements of third parties. This journal is a member of the Committee on Publication Ethics (COPE).
Author Contributions
All programming, analysis, and implementation on Lifeportal, drafting, and writing the manuscript: SK, AKK, RSN. Contributed in preparing and analyzing test case: AKK, RSN. Contributed in testing the programs: XZ, SZ, RSN. Contributed in implementation on Lifeportal: SK, KM. Conceived, designed, and wrote the manuscript: KST. All authors reviewed and approved of the final manuscript.
REFERENCES
- 1.Shalchian-Tabrizi K, Minge MA, Espelund M, et al. Multigene phylogeny of choanozoa and the origin of animals. PLoS One. 2008;3(5):e2098. doi: 10.1371/journal.pone.0002098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Burki F, Inagaki Y, Bråte J, et al. Large-scale phylogenomic analyses reveal that two enigmatic protist lineages, telonemia and centroheliozoa, are related to photosynthetic chromalveolates. Genome Biol Evol. 2009;1:231–8. doi: 10.1093/gbe/evp022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhao S, Burki F, Brate J, Keeling PJ, Klaveness D, Shalchian-Tabrizi K. Collodictyon – an ancient lineage in the tree of eukaryotes. Mol Biol Evol. 2012;29(6):1557–68. doi: 10.1093/molbev/mss001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Burki F. The eukaryotic tree of life from a global phylogenomic perspective. Cold Spring Harb Perspect Biol. 2014;6(5):a016147. doi: 10.1101/cshperspect.a016147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Burki F, Okamoto N, Pombert JF, Keeling PJ. The evolutionary history of haptophytes and cryptophytes: phylogenomic evidence for separate origins. Proc Biol Sci. 2012;279(1736):2246–54. doi: 10.1098/rspb.2011.2301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dunn CW, Hejnol A, Matus DQ, et al. Broad phylogenomic sampling improves resolution of the animal tree of life. Nature. 2008;452(7188):745–9. doi: 10.1038/nature06614. [DOI] [PubMed] [Google Scholar]
- 7.Burki F, Shalchian-Tabrizi K, Pawlowski J. Phylogenomics reveals a new ‘mega-group’ including most photosynthetic eukaryotes. Biol Lett. 2008;4(4):366–9. doi: 10.1098/rsbl.2008.0224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rodríguez-Ezpeleta N, Brinkmann H, Burger G, et al. Toward resolving the eukaryotic tree: the phylogenetic positions of jakobids and cercozoans. Curr Biol. 2007;17(16):1420–5. doi: 10.1016/j.cub.2007.07.036. [DOI] [PubMed] [Google Scholar]
- 9.Philippe H, Telford MJ. Large-scale sequencing and the new animal phylogeny. Trends Ecol Evol. 2006;21(11):614–20. doi: 10.1016/j.tree.2006.08.004. [DOI] [PubMed] [Google Scholar]
- 10.Bapteste E, Brinkmann H, Lee JA, et al. The analysis of 100 genes supports the grouping of three highly divergent amoebae: Dictyostelium, Entamoeba, and Mastigamoeba. Proc Natl Acad Sci U S A. 2002;99(3):1414–9. doi: 10.1073/pnas.032662799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Philippe H, Roure B. Difficult phylogenetic questions: more data, maybe; better methods, certainly. BMC Biol. 2011;9:91. doi: 10.1186/1741-7007-9-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Parfrey LW, Grant J, Tekle YI, et al. Broadly sampled multigene analyses yield a well-resolved eukaryotic tree of life. Syst Biol. 2010;59(5):518–33. doi: 10.1093/sysbio/syq037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Philippe H, Brinkmann H, Lavrov DV, et al. Resolving difficult phylogenetic questions: why more sequences are not enough. PLoS Biol. 2011;9(3):e1000602. doi: 10.1371/journal.pbio.1000602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fitch WM. Homology a personal view on some of the problems. Trends Genet. 2000;16(5):227–31. doi: 10.1016/s0168-9525(00)02005-9. [DOI] [PubMed] [Google Scholar]
- 15.Kuzniar A, van Ham RC, Pongor S, Leunissen JA. The quest for orthologs: finding the corresponding gene across genomes. Trends Genet. 2008;24(11):539–51. doi: 10.1016/j.tig.2008.08.009. [DOI] [PubMed] [Google Scholar]
- 16.Chen F, Mackey AJ, Vermunt JK, Roos DS. Assessing performance of orthology detection strategies applied to eukaryotic genomes. PLoS One. 2007;2(4):e383. doi: 10.1371/journal.pone.0000383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li L, Stoeckert CJ, Jr, Roos DS. OrthoMCL: identification of ortholog groups for eukaryotic genomes. Genome Res. 2003;13(9):2178–89. doi: 10.1101/gr.1224503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Philippe H, Delsuc F, Brinkmann H, Lartillot N. Phylogenomics. Annu Rev Ecol Evol Syst. 2005;36:541–62. [Google Scholar]
- 19.Hampl V, Hug L, Leigh JW, et al. Phylogenetic analyses support the monophyly of Excavata and resolve relationships among eukaryotic “supergroups”. Proc Natl Acad Sci U S A. 2009;106(10):3859–64. doi: 10.1073/pnas.0807880106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Altschul SF, Madden TL, Schäfer AA, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25(17):3389–402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Posada D. Using MODELTEST and PAUP* to select a model of nucleotide substitution. Curr Protoc Bioinformatics. 2003 doi: 10.1002/0471250953.bi0605s00. Chapter 6: Unit 6.5. [DOI] [PubMed] [Google Scholar]
- 22.Huelsenbeck JP, Ronquist F. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics. 2001;17(8):754–5. doi: 10.1093/bioinformatics/17.8.754. [DOI] [PubMed] [Google Scholar]
- 23.Lartillot N, Lepage T, Blanquart S. PhyloBayes 3: a Bayesian software package for phylogenetic reconstruction and molecular dating. Bioinformatics. 2009;25(17):2286–8. doi: 10.1093/bioinformatics/btp368. [DOI] [PubMed] [Google Scholar]
- 24.Stamatakis A. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006;22(21):2688–90. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- 25.Kumar S, Skjaeveland A, Orr RJ, et al. AIR: a batch-oriented web program package for construction of supermatrices ready for phylogenomic analyses. BMC Bioinformatics. 2009;10:357. doi: 10.1186/1471-2105-10-357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Katoh K, Kuma K, Toh H, Miyata T. MAFFT version 5: improvement in accuracy of multiple sequence alignment. Nucleic Acids Res. 2005;33(2):511–8. doi: 10.1093/nar/gki198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Castresana J. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol. 2000;17(4):540–52. doi: 10.1093/oxfordjournals.molbev.a026334. [DOI] [PubMed] [Google Scholar]
- 28.Capella-Gutierrez S, Silla-Martinez JM, Gabaldon T. trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics. 2009;25(15):1972–3. doi: 10.1093/bioinformatics/btp348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Price MN, Dehal PS, Arkin AP. FastTree 2 – approximately maximum-likelihood trees for large alignments. PLoS One. 2010;5(3):e9490. doi: 10.1371/journal.pone.0009490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jothi R, Zotenko E, Tasneem A, Przytycka TM. COCO-CL: hierarchical clustering of homology relations based on evolutionary correlations. Bioinformatics. 2006;22(7):779–88. doi: 10.1093/bioinformatics/btl009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Larkin MA, Blackshields G, Brown NP, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23(21):2947–8. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 32.2009. http://tree.bio.ed.ac.uk/software/figtree/
- 33.Page RD. TreeView: an application to display phylogenetic trees on personal computers. Comput Appl Biosci. 1996;12(4):357–8. doi: 10.1093/bioinformatics/12.4.357. [DOI] [PubMed] [Google Scholar]
- 34.Page RD. Space, time, form: viewing the tree of life. Trends Ecol Evol. 2012;27(2):113–20. doi: 10.1016/j.tree.2011.12.002. [DOI] [PubMed] [Google Scholar]
- 35.Sicheritz-Ponten T, Andersson SG. A phylogenomic approach to microbial evolution. Nucleic Acids Res. 2001;29(2):545–52. doi: 10.1093/nar/29.2.545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hanekamp K, Bohnebeck U, Beszteri B, Valentin K. PhyloGena – a user-friendly system for automated phylogenetic annotation of unknown sequences. Bioinformatics. 2007;23(7):793–801. doi: 10.1093/bioinformatics/btm016. [DOI] [PubMed] [Google Scholar]
- 37.Robbertse B, Yoder RJ, Boyd A, Reeves J, Spatafora JW. Hal: an automated pipeline for phylogenetic analyses of genomic data. PLoS Curr. 2011;3:RRN1213. doi: 10.1371/currents.RRN1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dereeper A, Guignon V, Blanc G, et al. Phylogeny.fr: robust phylogenetic analysis for the non-specialist. Nucleic Acids Res. 2008;36(Web Server issue):W465–9. doi: 10.1093/nar/gkn180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen SH, Su SY, Lo CZ, et al. PALM: a paralleled and integrated framework for phylogenetic inference with automatic likelihood model selectors. PLoS One. 2009;4(12):e8116. doi: 10.1371/journal.pone.0008116. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Information about the single-gene alignments used in the test case.
Table S2. Sequences randomly extracted from the singlegene alignments.
Table S3. Results from COCO-CL.