Abstract
Advances in next generation sequencing (NGS) and mass spectrometry (MS) technologies have provided many new opportunities and angles for extending the scope of translational cancer research while creating tremendous challenges in data management and analysis. The resulting informatics challenge is invariably not amenable to the use of traditional computing models. Recent advances in scalable computing and associated infrastructure, particularly distributed computing for Big Data, can provide solutions for addressing these challenges. In this review, the next generation of distributed computing technologies that can address these informatics problems is described from the perspective of three key components of a computational platform, namely computing, data storage and management, and networking. A broad overview of scalable computing is provided to set the context for a detailed description of Hadoop, a technology that is being rapidly adopted for large-scale distributed computing. A proof-of-concept Hadoop cluster, set up for performance benchmarking of NGS read alignment, is described as an example of how to work with Hadoop. Finally, Hadoop is compared with a number of other current technologies for distributed computing.
Keywords: cancer, informatics, hadoop, high performance computing, gpu, cluster, cloud computing, big data, data storage, data management, scalable computing, NGS, genomics
Introduction
Recent advances in high-throughput technologies, including next generation sequencing (NGS), mass spectrometry (MS), and imaging assays and scans, are providing unprecedented capabilities for cancer researchers to interrogate biological systems of interest, while creating tremendous challenges with respect to data management, access, and analysis. The Cancer Genome Atlas (TCGA) project,1 for example, currently provides germline and tumor DNA-sequencing, RNA-sequencing, methylation, and imaging data from thousands of patients across multiple solid tumor and hematologic malignancies. Consequently, cancer researchers are faced with the formidable task of managing and integrating massive amounts of data, produced in structured as well as unstructured formats, to be positioned to use this treasure trove of data to push the scientific envelope. The requisite analyses are not confined to traditional assessment of differential expression but extend to integrative genomics including analysis of expression quantitative trait loci (eQTL2) linking DNA and RNA sequencing data.
In many cases, the data volume, velocity, and variety3 generated by these high-throughput platforms have collectively rendered the traditional single- and cluster-farm computing model, which was employed with great success in the microarray and genome-wide association studies (GWAS) era, technologically obsolete. Recent advances in computational technologies, especially distributed computing for “Big Data”, such as Hadoop, have shown great potential as technological solutions for addressing the challenges of the data deluge in next generation cancer research.
This paper provides an overview of scalable and distributed computing technologies with specific emphasis on the widely used open source Hadoop project. The presentation is organized as follows. In the next section, we provide an overview of the elements of scalable computing systems and provide a number of examples. Afterward, we provide an introduction to Hadoop as a full-featured distributed system for scalable computing and data storage and management. This section also includes an overview of the Hadoop ecosystem and specific examples of bioinformatics applications leveraging this technology. In the section that follows, we outline a proof-of-concept (POC) cluster to illustrate the design and implementation of a basic NGS data pre-processing system based on Hadoop. In the Discussion, we consider other available and widely used systems for distributed computing that could be used as an alternative to or in concert with Hadoop depending on the specific cancer informatics challenge at hand.
Scalable Computing Systems
Background
Computing models
Broadly speaking, computational systems can be grouped into two categories (see for example Refs.4,5):
Heterogeneous systems: These are typically single node workstations or servers for which computational power is scaled by upgrading or adding additional Central Processing Units (CPUs) or memory along with other components including Graphics Processing Units (GPUs) or Many-in-Core co-processors.
Homogeneous distributed systems: Another way to scale computation is by connecting several computers. If the computers are connected within the same administrative domain, the collective is referred to as a compute cluster. If connected across networks and administrative domains, it is referred to as a computer grid. The individual computers in the collective are called nodes. Scaling a cluster or grid is typically accomplished by adding nodes rather than adding components to the individual nodes.
Scaling of computation is accomplished through task or data parallelization.6,7 In task parallelization, a computational task is divided into several tasks to be run in parallel on the same dataset and the results are combined. For large datasets, this approach is often not feasible as the data may not fit into memory. In data parallelization, the data are divided into smaller sets and the same processing is applied to each subset after which the results are combined.
Traditionally only a single task or instruction could be carried out on one piece of data and the related CPU architecture is called Single Instruction Single Data (SISD8). In order to enable parallelization, other architectures have been developed. These include Single Instruction Multiple Data (SIMD8), which allows the same instruction or processing to be applied to different datasets, or Multiple Instruction Multiple Data (MIMD8) in which multiple instructions can be applied to different datasets.
In a distributed system, processing is done by the CPU in each of the nodes. Although the CPUs of the individual nodes are independent of each other the memory and storage could be shared among the nodes. The data stored on disk are made available to the CPU for processing through the memory, which requires a means of transferring the data through a communication channel such as memory bus and inter-node networking. If data are shared between the nodes, then coordination between the processes running on different nodes is required to maintain a consistent state of the data. The three common architectures for distributed systems are9:
Shared nothing, in which each node in the cluster works independent of other nodes with no inter-dependence for memory or storage. If required, coordination among processes running on different nodes is accomplished by passing messages among them or by underlying distributed system management software called middleware. An advantage of this configuration is that because of lack of dependence among different nodes, the cluster can scale indefinitely by simply adding more nodes to the cluster.
Shared memory, in which the nodes have access to a common memory that can be used for coordinating the processing tasks among the different nodes.
Shared disk, in which data processed by the different nodes are shared either through a central storage or by direct exchange with each other.
In distributed systems, the bandwidth of the communication channels used for sharing data and passing messages as well as the processing overhead of maintaining data in a consistent state can have a significant impact on the performance of the system.9 Each of the three distributed system architectures employs a different strategy for managing processing. The shared-nothing architecture has the advantage of minimizing network latency and process coordination for data consistency because of the independence of the nodes for memory and data.
Data storage and models
Efficient data storage, representation, and management is crucial for building a high performance scalable system. Data storage devices can be connected to a computer in one of three different ways:
Direct attached: Storage is attached directly to the computer through an interface including Serial AT Attachment (SATA) or Serial Attached SCSI (SAS).
Network attached: Storage is attached to a local network, such as a LAN, and other devices in the network can access the storage device through interfaces such as Fiber Channel.
Remote: Storage is physically located outside of the internal network and is accessed through the Internet as in cloud storage. Amazon Web Service, for example, provides cloud storage through their S3 service.10
Data required for active use are usually stored in “on-line” primary storage, which provides fast access and availability on demand. Data not requiring access for an extended period of time and that can be archived are usually stored on slower and relatively inexpensive “off-line” tertiary storage. For infrequent access, data can be stored in “near-line” secondary storage.11 Arrangement of data storage among these tiers optimizes the storage cost without compromising ready availability of data for processing. Hierarchical storage management (HSM) software, such as IBM Tivoli,12 automatically manages the storage and movement of data in a tiered hierarchy of devices. Data storage, access, and transfer are particularly challenging in systems that have distributed storage and clients that require data access.
A data model is a logical, rather than a physical, representation specifying the structure, types, and relationships among data elements. Applications can use data models to access data based on the logical representation without concern for the physical storage location or media. For example, in a Relational Database Management System (RDBMS), the Structured Query Language (SQL) is used for data access and manipulation based on the relational data model.13 Specific data models, along with tools and Application Programming Interfaces (APIs) to manipulate the data, have been created for scientific computing. Two data models that have been used extensively by the scientific community are the Network Common Data Form (NetCDF) and the Hierarchical Data Format (HDF15). Graph database models16,17 are used extensively for representing highly interconnected data. The Resource Description Framework (RDF18) provides a set of specifications for a graph data model for use on the web. The data elements are subject-predicate-object expressions called triples or triple stores.17 Each of the three components is a web resource represented by a Uniform Resource Identifier (URI). SPARQL19 is the standard querying language for an RDF triple store.
Higher level information about data is provided by metadata, which can be syntactic or semantic.20 Syntactic metadata provide information about the data structures, formats, and other physical characteristics of the data including file names and organizational hierarchy. Semantic metadata provide information about the meaning of the data elements particularly in the context of the knowledge domain. Both syntactic and semantic metadata can range from simple to complex. At a simple level, syntactic metadata are used by file systems to manage file storage and access. At a complex level, they are used by programs and databases to define data structures and schemas to be used by applications for data queries, manipulation, and analysis. Semantic metadata at a simple level provide definitions for data elements through naming classifications, data standards, and terminologies such as taxonomy, vocabulary, dictionary, and thesaurus.20 These can be used for effective data sharing, data integration, and interoperability among applications. More sophisticated semantic metadata, including ontologies, provide complex representations of the data to include relationships to be used for knowledge inference.21 The National Cancer Institute (NCI) has created a terminology called NCI Thesaurus (NCIt22) specifically for cancer research, which contains clinical and research terms related to cancer. Concepts for drugs, therapies, and genes, among many others, that are contained in the NCIt include terms, codes, synonyms, and relationships among the concepts. There are more than 200,000 relationships among more than 55,000 concepts, which can be used, among other things, for performing integrative analyses of data from different cancer research experiments.23,24 Several other thesauri are available for clinical research including the Unified Medical Language Systems (UMLS) meta-thesaurus and the Systemized Nomenclature of Medicine Clinical Terms (SNOMED CT).25 Gene Ontology is one of the most widely used semantic metadata in genomics research, which evolves dynamically with increase in knowledge about genes and proteins in eukaryotic cells.26
RDBMS13 has served as a predominant technology for data management. The hallmarks of an RDBMS are the atomicity, consistency, isolation, and durability (ACID27) properties. In these systems, the data are stored in a highly structured format subject to enforcement by a relational schema. An RDBMS is considered to be an Online Transaction Processing system (OLTP28). These systems are characterized by frequent reads and updates, referred to as transactions, of a relatively small number of data records. The performance of an RDBMS generally degrades as the number of data records or data fields increases. A key development to address the limitation of RDBMS with respect to scalability is the introduction of parallel databases using row based, also known as horizontal, partitioning.29 These databases distribute mutually exclusive sets of rows among the nodes of a cluster. The SQL query is applied to each partition on each of the nodes. The results of the partitioned queries are sent back to a single node to be merged to produce the final result. The RDBMS has been adopted for managing and querying GWAS data.30 An OLTP system is optimal for research projects that require a large number of small and simple queries, and frequent data updates. An On-line Analytical Processing (OLAP28) system on the other hand is optimal for projects requiring complex analytical queries but not frequent updates. The performance of an OLTP system is measured on the basis of its ability to maintain data consistency and integrity while maximizing the number of transactions per time unit. The performance of an OLAP system is measured on the basis of the throughput of the query and the corresponding response time.28 Data warehouses are OLAP systems developed to aggregate large amounts of data from multiple sources. Because of the heterogeneous sources of data, the incoming data are typically cleaned and transformed before they are loaded into the system. One of the key data warehouses in biomedical research is Informatics for Integrating Biology and the Bedside (i2B2)31,32 developed under the sponsorship of National Center for Biomedical Computing. The most common use of i2B2 is to repurpose data from Electronic Medical Records (EMRs) to be combined with clinical and genomics data.
Multi-core computing
Until the early 2000s, most motherboards housed a single CPU with a single core for processing. Later CPUs with multiple cores were developed to overcome the computational limitation of the single core design. The multi-core architecture has the advantage of providing higher clock rates because data do not have to travel across chip sets.33 Computational tasks are typically scaled through manual or programmatic forking of tasks to multiple cores, or by writing multi-threaded applications using the Open Multi-Processing (OpenMP34,35) API.
High performance computing
Two widely used approaches for distributed computing over a cluster or grid of server nodes or workstations are message passing and batch queuing. These fall under the category of High Performance Computing (HPC).36 Batch queuing systems simply distribute individual jobs as batches to the nodes. The job submission and management, and resource allocation are orchestrated by a batch submission engine such as the Simple Linux Utility for Resource Management (SLURM37). The most commonly used system for message passing is the Message Passing Interface (MPI38). This can be considered as a MIMD shared nothing distributed memory architecture as memory is not shared among the nodes and communication is done strictly through message passing. The batch queuing approach typically does not require specialized programming. MPI provides computational scaling at the cost of increased programming complexity. A number of bioinformatics applications developed for MPI-based distributed systems are listed in Table 1.
Table 1.
MPI-based applications for bioinformatics.
| CATEGORY | APPLICATION | DESCRIPTION |
|---|---|---|
| Alignment | mpiBLAST105 | Implementation of BLAST on MPI for parallel execution. |
| ClustalW-MPI106 | Implementation of Clustal-W, a multiple alignment tool, on MPI. | |
| mrNA107 | Short read alignment of NGS read. | |
| MrBayes 3108 | Bayesian phylogenetic analysis using MPI for parallelizing Markov chain Monte Carlo convergence. | |
| Proteomics | Parallel Tandem109 | Implementation of X!Tandem for MS/MS spectra search against a protein database. |
GPU computing
The GPU architecture is highly amenable to parallelized scientific computing.39,40 Development languages and APIs for General Purpose computing on GPU (GPGPU) include the Open Computing Language (OpenCL41,42), Compute Unified Device Architecture (CUDA43), and Brook for GPU (BrookGPU44). The CUDA language is intended for GPGPU on devices manufactured by NVIDIA. OpenCL is a standard adopted by several vendors including NVIDIA and AMD. A number of bioinformatics applications developed for leveraging GPUs are listed in Table 2. A typical approach for scientific computing on a GPU is to copy data from the host (CPU) memory to the memory on the device (GPU). The calculations are then carried out on the device after which the results are copied to the host. If the memory limitations of the device, relative to the size of the data, were to necessitate breaking up the data into smaller chunks, an overhead penalty is incurred because of repeated copies between the host and the device. Recent GPU card offerings provide larger amounts of memory, compared to early GPUs, rendering this technology now feasible for analysis of high-throughput data from NGS assays. For example, the NVIDIA K80 card45 consists of two GPUs collectively housing 24 GB of memory and 4,992 streaming cores providing peak single- and double-precision floating-point performance of 2.91 and 8.74 teraflops, respectively. The AMD FirePro W910046 card consists of a single GPU housing 16 GB of memory and 2,816 stream processors providing peak single- and double-precision floating-point performance of 5.24 and 2.62 teraflops, respectively. The GPU technology can be scaled further by installing multiple cards on the same motherboard.40
Table 2.
GPU applications for bioinformatics.
| CATEGORY | APPLICATION | DESCRIPTION |
|---|---|---|
| Alignment | CUSHAW110 | Short read alignment based on the Burrows-Wheeler transform (BWT111) and the Ferragina-Manzini index112 |
| SOAP3-dp113 | Short read alignment based on BWT with native BAM support | |
| Proteomics | FastPaSS114 | Spectra matching using spectral library searching implemented in CUDA |
| Tempest115 | Spectral matching using GPU-CPU | |
| Motif Discovery | GPUmotif116 | Motif scan and de novo motif finding using GPU |
| Epigenetics | GPU-BSM117 | GPU based tool for mapping whole genome bisulfite sequencing reads and estimating methylation levels |
| Systems Biology (see Ref.118 for a review) | ABC-SysBio119 | Simulate models written in the Systems Biology Markup Language (SBML120) format |
| PMCGPU121,122 | Parallel simulators for Membrane Computing on the GPU | |
| Genome-wide Inference | permGPU123 | Permutation resampling analysis for binary, quantitative, and censored time-to-event outcomes |
Cloud computing
The ability to conduct scalable computing requires the acquisition, installation, and ongoing management of a host of hardware and software resources. This may neither be a practically nor economically feasible proposition. Cloud computing has proven to be a powerful alternative for researchers to conduct scalable computing on a virtual computing infrastructure hosted and managed by a service provider. The infrastructure may consist of hardware resources including storage, CPU or GPU nodes, or software resources including middleware, applications and development frameworks provided to the researcher, who can provision the resources elastically depending on the research needs.47 There is no standard definition for cloud computing. The National Institute of Standards and Technology (NIST) defines cloud computing as a model48 with five essential characteristics. Resources can be provisioned on demand by the researcher without direct interaction with the service provider (On-demand self-service). The computing resources are accessible to the researcher through a variety of clients via a network (Broad network access). The resources are pooled by the service provider so as to be offered to multiple users with heterogeneous needs (Resource pooling). The resources can be provisioned on demand by the researcher depending on the demand (Rapid elasticity). The usage of the resources, in terms of storage or CPU cycles used, can be monitored and quantified in a transparent manner by both the researcher and service provider (Measured service). Cloud computing offers three service models.48 The Software as a Service (SaaS) model provides software applications hosted and managed by the service provider to be used by the researcher. The Platform as a Service (PaaS) model enables custom software application development and deployment by the researcher using programming languages, software libraries, and tools hosted and managed by the service provider. The Infrastructure as a Service (IaaS) model provides storage, computing and networking resources to the researcher for deployment of operating systems, applications, and development toolkits. The Amazon Elastic Compute cloud (EC2) was one of the early commercial offerings for cloud computing. Other current commercial offerings include Google Cloud, IBM Cloud, and Microsoft Azure.
Several bioinformatics applications including BLAST, genome assembly and alignment have been adapted for cloud computing. Some of these are listed in Table 3. Generally, these applications require the user to provision the computing resources on the cloud, and manage the application configuration and deployment. Several cloud manager tools have been developed enabling users to easily provision resources on the cloud and deploy one or more tools and data to work as a single unit. These tools in effect provide a “turnkey” solution to a complete bioinformatics data analysis platform. Cloudman49,50 is a manager initially developed to facilitate the deployment of the Galaxy platform51 for NGS data analysis on the cloud. It enables packaging of the data along with the analysis tools. Other cloud management software include StarCluster52 and elasticHPC.53
Table 3.
Cloud-based applications for bioinformatics.
| CATEGORY | APPLICATION | DESCRIPTION |
|---|---|---|
| Alignment | CloudBrush124 | Distributed de novo genome assembler based on MapReduce which can be run on a cloud. |
| CloudBurst74 | Short read mapping software for Hadoop based on the RMAP125 mapping tool which can be run on a cloud. | |
| CloudBLAST126 | MapReduce based BLAST which can be run on a cloud. | |
| Proteomics | Integrated Proteomics Pipeline (IP2)127 | Proteomics data analysis pipeline also available on Amazon Web Service. |
| ProteoCloud128 | Proteomics computing pipeline system on the cloud for peptide and protein identifications available on Amazon Web Service. |
The Bio2RDF54 project facilitates integration across several databases by creating the datasets in a common RDF18 format to be pushed to the cloud for access. The data from the TCGA project are available as “Linked Data”55,56 and can be queried using SPARQL.19 The National Centers for Biomedical Ontology (NCBO) has created BioPortal57 for review and updates of various ontologies available to biomedical researchers. One of the key advantages of cloud computing is the provision of the infrastructure to make these federated biomedical data available to the research community for integrated data analysis.
Hadoop for Scalable Computing and Data Management
Background and core architecture
Figure 1 illustrates the two core components of the Hadoop architecture as two main layers.58 The MapReduce layer, shown above the dashed line, is responsible for computation and resource management while the Hadoop Distributed File System (HDFS) layer, shown below the dashed line, is responsible for storage and data management. Hadoop follows the Master–Slave architecture for managing computation and data in a distributed environment. The master node schedules and coordinates the computing tasks among the slave nodes. In a Hadoop system, the master node is referred to as the JobTracker while the slave nodes are referred to as TaskTrackers. The JobTracker also provides the software infrastructure for managing distributed computing such as resource scheduling and recovery from job failures. The master process for the HDFS layer is called the Name Node and it manages the metadata loaded and distributed to the slave nodes. The latter are called the Data nodes. One of the drawbacks in earlier versions of Hadoop was the tight coupling of the MapReduce engine and distributed computing services provided by the JobTracker.
Figure 1.
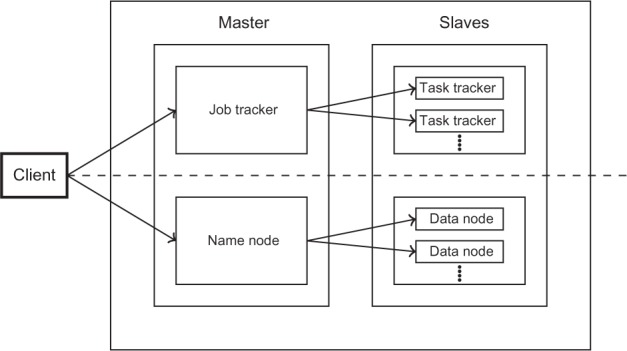
Core components of the Hadoop architecture. Adapted from Ref.58
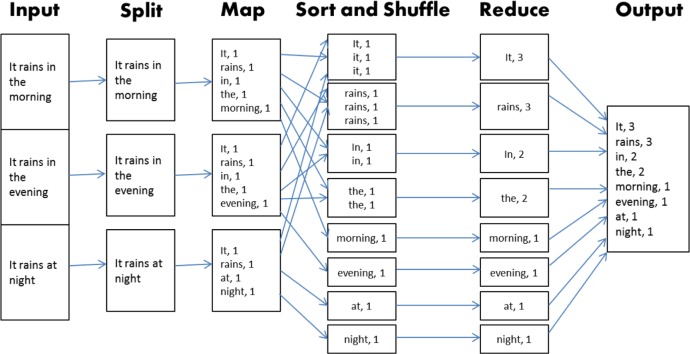
The computational layer of Hadoop is an implementation of the MapReduce algorithm.59 Any implementation of this algorithm requires the provision of two user-defined functions, Map and Reduce, as its name suggests. The primary task of the Map function is to generate a set of intermediate pairs of keys and values. Before these pairs are passed on to the Reduce function, they are binned according to the keys. The Reduce function then is applied to each bin. The algorithm is often illustrated using the example of counting the occurrence of each word in a text file from the MapReduce paper.59 Consider the example illustrated in Figure 2. The text file in this example consists of three sentences as illustrated under the Input. The file is split up along the three sentences each of which is passed on to the Map function. In this stage, the words are treated as the keys. The number of times each word appears in the sentence is the value corresponding to the key. Within each of the three bins in this stage, the keys are paired up with their corresponding value. In the next stage, Shuffle and Sort, the pairs from the previous step are grouped into bins by the keys. Each bin is passed on to the Reduce function, which adds the values from each pair therein. This effectively reduces the (key, value) pairs within each bin to a single (key, value) pair. Finally, these reduced pairs are passed on as output.
Figure 2.
Typical MapReduce algorithm workflow.
Hadoop manages the distribution of files on individual nodes through the HDFS. Since the data are spread over a set of nodes on a network, all the complications of network programming, such as node failure, have to be taken into account. The design of HDFS facilitates the key principles of Hadoop, namely storage and management of huge amounts of data on a cluster of commodity hardware with fast access. To enable these objectives, HDFS stores very large files as blocks rather than files and provides redundancy for the data by replicating each block. The metadata for the files is stored centrally in the Name Node, which uses this information to reconstruct the files from the blocks. HDFS also works on the master–worker pattern where the Name Node is the master server and Data Nodes the workers. The Name Node, through the metadata, manages the file system namespace and the information about which Data Node has the blocks for each of the files. A Map task receives blocks of data from the Data Node and works on one block at a time.60
Hadoop ecosystem
Decoupling of the MapReduce and the HDFS layers enables other tools to be built as higher level abstractions in other languages. Several tools have been developed to convert user applications written for these tools to MapReduce jobs for deployment and execution in a Hadoop cluster. Additionally, tools have been developed to facilitate pushing data in standard formats, including columnar data, into the HDFS layer whereby relieving the user of the burden of working with the native HDFS file format. Another major milestone in decoupling of these two layers was implemented by the introduction of the Yet Another Resource Negotiator (YARN61) in 2013.62 YARN decouples the distributed computing resource management from the MapReduce job execution engine and delegates many job flow control and scheduling functions to the individual application components. This refinement of the core architecture is expected to encourage wider adoption by allowing other computing paradigms to be implemented in Hadoop. The collection of tools built around the core infrastructure is referred to as the Hadoop ecosystem. A representative subset of tools in the ecosystem is shown in Figure 3. We provide additional details on some of the key tools next.
Figure 3.
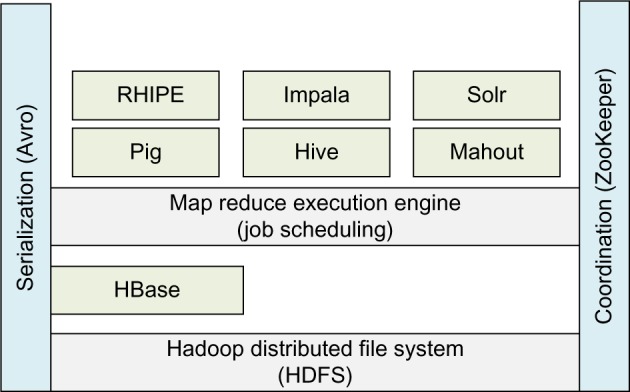
Representative subset of the Hadoop ecosystem.
HBase
The records in a traditional RDBMS are stored in a row format. This is generally optimal when the goal is to query a relatively small number of records (rows) consisting of large number of fields (columns).63 For certain applications, the goal is to query a small subset of columns from a large number of records. For these, it is preferable to store the data in column format so as to increase performance of the queries. HBase64 is a column-oriented63 database built on top of HDFS to provide high scalability and performance in a distributed computing environment. It is modeled after Google’s Big Table project.65 HBase, unlike HDFS, provides real time read and write capability with random access for large-scale data distributed in a cluster. Linear scaling is achieved in HBase by simply adding additional nodes to the cluster. Tables in HBase can be large and sparsely populated with billions of rows and millions of columns. Columns can be added dynamically to allow for changes in the data representation.60
Hive
Data in Hadoop are natively stored as files in HDFS. This structure does not enable convenient use of higher level languages for data queries. Apache Hive66 offers capabilities of a data warehouse by providing the ability to represent data as tables similar to those of a relational database.67 To this end, it provides the SQL-like language HiveQL67 to convert queries into a series of MapReduce jobs for execution in a Hadoop cluster. Within a traditional RDBMS, data schema constraints are enforced at load time. Hive, on the other hand, employs “schema-on-read,” which checks the constraints only when data are read by virtue of a query. This approach is optimal for loading large-scale data.60
Pig
Processing large datasets in Hadoop may require a series of data transformation steps that may be complicated to implement as Map and Reduce functions.60 Apache Pig68 provides higher level data structures and data transformation functions to facilitate the programming of these tasks. Pig includes the Pig Latin69 programming language and an execution environment for running the programs.60 The Pig execution engine converts the user written operations into MapReduce jobs at runtime. Additionally, Pig provides a set of built-in functions for a number of tasks including math and string processing. These can be modified or augmented with user-defined functions.68
Avro
In distributed computing systems, including Hadoop, data structured as objects are transferred on the network among different nodes as streams of bytes using the process of serialization. The process of converting the byte streams back to structured data is called deserialization.60,70 These two processes are also required for writing structured data to physical storage devices. Efficiency of these processes can have an impact on the performance of a distributed application.60 Apache Avro71 enables efficient implementation of serialization and deserialization for Hadoop-based distributed applications by providing an API for several programming languages. Serialization frameworks require a schema for representing data structures. Avro uses the widely used JavaScript Object Notation (JSON72) format71 to provide portability across a number of languages.
Starfish
The core components of Hadoop, namely the MapReduce execution engine and the HDFS distributed storage, are extensible through their pluggable architecture. In addition, there are various procedural and declarative interfaces for interaction with Hadoop. Such an architecture provides great flexibility for extending and building distributed applications in Hadoop. At the same time, it creates an enormous challenge for performance tuning, which is required for optimal use of resources. In order for the system to adapt to changing user needs and system workloads, several system parameters require tuning. There are over 190 configuration parameters that control the running of MapReduce jobs,73 the default values of which may not be optimal in all cases. Manually tuning the parameters for performance optimization requires expertise and can be a daunting task of trial and error. Starfish is a self-tuning system built within the architecture of Hadoop that can be used for automatic or “self” tuning.73 It enables applications and users of Hadoop to get good performance throughout the life cycle of the data processing jobs without the need for manual performance tuning. The performance tuning is not limited to a single MapReduce job, and can be extended to workflows and workloads consisting of multiple MapReduce jobs.
Bioinformatics applications based on Hadoop
Cloudburst, an application for mapping short reads to a reference genome and released in 2009, was one of the first bioinformatics applications developed using Hadoop.74 Since then several other applications have been developed.75 Many of these were developed for the purpose of conducting short read alignments, while some are for further downstream processing such as RNA-seq and variant calling. In addition, some general purpose applications have been developed based on the Hadoop ecosystem that can be used for developing custom bioinformatics applications. Table 4 summarizes a number of applications useful for cancer research. Detailed information regarding five other applications is also provided.
Table 4.
Bioinformatics applications for Hadoop.
| CATEGORY | APPLICATION | DESCRIPTION |
|---|---|---|
| Alignment and Assembly Genome Assembly | Jnomics88 | Command line driven alignment on Hadoop cluster for a number of alignment software |
| Contrail | de novo assembly without reference genome | |
| DistMap129 | Perl tool, works with 9 different aligners, very easy client-only installation | |
| Seal130 | Alignment tool based on Pydoop (a python based API for developing Hadoop applications131) | |
| RNA-seq | Eoulsan132 | In additional to alignment, complete RNA-seq pipeline |
| Myrna133 | Complete RNA-seq pipeline, from alignment to differential expression, written in Perl, integrates R79 and Bioconductor81 for downstream statistical analysis | |
| Variant Calling | Crossbow134 | Performs alignment and SNP genotyping |
Hadoop-BAM
Hadoop-BAM is an application enabling access to the contents of a BAM file76 within HDFS by virtue of providing an API as a Java library. BAM files are treated as Hadoop input and output formats. Picard77 is a full-featured set of tools for processing NGS data. It provides extensive support for processing reads stored in the Sequence Alignment/Mapping (SAM) or BAM formats. Hadoop-BAM is built on the top of the Picard for accessing and manipulating BAM files. This feature facilitates the Picard API to be used directly within Hadoop. By distributing the processing of BAM files across a Hadoop cluster, Hadoop-BAM enables significant gains in processing time as well as linear scalability with the number of nodes in a cluster.76
SeqPig
SeqPig78 is an extension of the Apache Pig scripting language for development of analysis pipelines for manipulation and analysis of NGS data on Hadoop. SeqPig scripts are converted to MapReduce tasks to operate on sequencing data stored in HDFS. It leverages Hadoop-BAM76 to provide import and export functions for common NGS data formats including FASTQ, FASTA, SAM, and BAM. In addition to input and output functionality, it offers facilities for computing base-level statistics and pile-up among other things.
RHIPE
R79 is an open source environment for statistical computing and graphics widely used by the research community. Its powerful analytical and graphical facilities are extended by a large library of user-contributed extension packages hosted by the Comprehensive R Archive Network (CRAN80). The Bioconductor project81 provides a large collection of extension packages for genomic research. R Hadoop Integrated Programming Environment (RHIPE) is an R extension package implementing the Divide and Recombine (D&R) method on a Hadoop cluster for parallel statistical computation.82 The D&R algorithm divides the data into subsets on which the statistical or computational procedure of interest is applied. The results from the subsets are then recombined. Hadoop is used to orchestrate these two steps while providing resource and fault tolerance management. BlueSNP83 is an R extension package developed for GWAS on a Hadoop cluster utilizing the D&R framework provided by RHIPE. For the statistical analysis, in addition to the tests provided, users can define and implement their own tests as R functions. BlueSNP is especially useful for performing computationally intensive statistical tests for GWAS including large-scale inference using permutation resampling and eQTL analyses. Performance benchmarking of BlueSNP suggested linear scaling in proportion to the number of nodes.83
Hydra
Mass spectroscopy based assays used in proteomics and metabolomics research generate raw data rivaling in size of those generated by NGS assays. The identification of peptide sequences from the spectra84 is one of the major computational challenges in this field. This is accomplished by matching the spectra against known sequences in a large database. Most matching algorithms developed for this problem are amenable to parallelization. Hydra,85 an open source Hadoop application, performs scalable peptide database search. The Each query is based on matching the mass to charge ratio score of the source spectra against those of the target peptides in the database. The latter is generated once up front and distributed to HDFS so as to be reused for all matching tasks.
Hadoop Cluster Setup
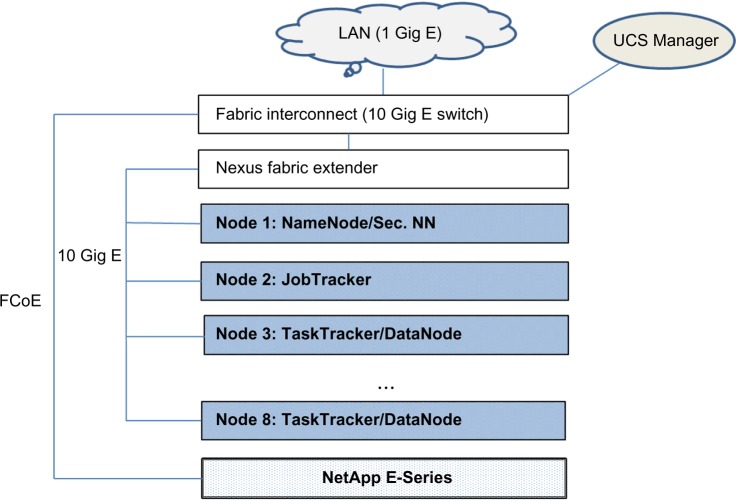
In this section, we outline a POC Hadoop cluster to illustrate the design and implementation of a basic NGS data pre-processing pipeline for RNA Sequencing data. A Hadoop cluster requires a minimum of two nodes for a distributed architecture. It can be set up on a single node in a “pseudo-distributed” mode to be primarily used for learning and testing purposes. The POC was designed using the FlexPod product line, which is an integrated commercial distributed computing system developed by Cisco and Netapp.86 Each of the eight nodes in the cluster was a dual eight-core CPU server, providing 16 cores per node, with 128 GB RAM. One of the eight nodes was dedicated as the Name Node and another as the JobTracker. The other six nodes served as worker nodes, each functioning as a TaskTracker and a Data Node. The combined internal storage across the eight nodes was 40 TB and used for HDFS storage. A Netapp E-series storage server with 100 TB capacity was included in the POC. Since Hadoop works most efficiently with local storage rather than network storage, to avoid the latency of moving data during a MapReduce job, the latter was primarily used as a staging area for large datasets. The process of transferring data from the staging area to HDFS for processing is called ingestion. The nodes were rack mounted and connected with a Cisco Fabric Interconnect 10 Gig switch. The latter was connected to an internal 1 Gig LAN. It should be noted that since most of the communication during the execution of Hadoop jobs is among the nodes, the network throughput during job processing is not limited by the bandwidth of LAN and will take advantage of the higher bandwidth of the switch. The external storage server communicates with the cluster using the Fiber Channel over Ethernet (FCoE) protocol. Cluster management for the Flexpod line is carried out using the UCS Manager, which is cluster administrative software providing features such as addition or removal of nodes. The design of the POC is illustrated in Figure 4.
Figure 4.
Cisco FlexPod cluster architecture.
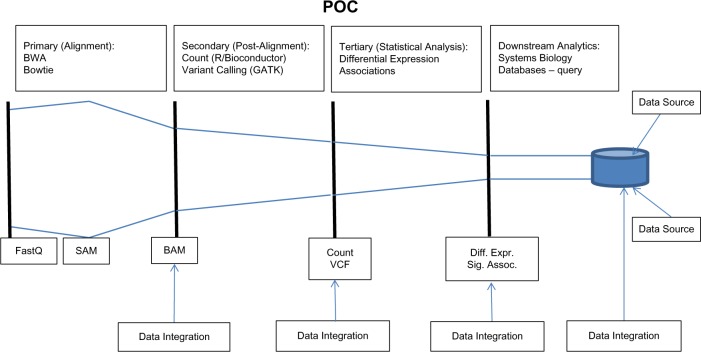
The POC was designed to provide a scalable computing solution for a representative basic three-stage data analysis pipeline for cancer research using high-throughput sequencing assays. The primary stage is devoted to initial processing of the raw reads. These, typically stored using the FASTQ format, are aligned against a reference genome. If a reference genome is not available or a custom reference is needed, de novo genome assembly may be performed at this stage. The resulting data are captured in the SAM file87 format. For efficient downstream processing, the file size is reduced by converting to a BAM file, a binary compressed version of the SAM file. In the second stage, the aligned reads are further processed to obtain summary information including variant calls for DNA sequencing, or gene or isoform level counts for RNA sequencing data. The third stage is devoted to downstream statistical analyses including differential expression, eQTL, and feature discovery analysis. The pipeline is conceptualized in Figure 5.
Figure 5.
Genomics data analysis pipeline.
A preliminary performance benchmarking for the primary data analysis stage was carried out for comparing the FASTQ alignment performance on a single node with that on the POC Hadoop cluster. The single node was a Linux server with similar Alignment of FASTQ files on the Hadoop cluster done using Jnomics88 and bowtie289 on the single node. FASTQ files of size 4 GB, 9 GB, and 65 GB were considered.
Discussion
To address the computational needs of a given research problem requiring a scalable computational framework, Hadoop is neither the exclusive nor necessarily always the optimal solution. Some of these are geared toward specific application areas while others are designed to provide a more generic framework. They can be used as an alternative to or in concert with Hadoop. The Genome Analysis Toolkit (GATK90,91) is a powerful and feature-rich framework for NGS variant discovery. It employs a custom MapReduce framework along with CPU threading features to facilitate variant discovery and calling. GATK uses aligned and pre-processed reads using the BAM87 format as input and returns the results, including annotation, quality score, number of variant base calls, depth, and genotype call in the Variant Calling Format (VCF92). The input BAM files can be produced using existing Hadoop applications for processing raw reads and then processed by GATK.
Genomics research in cancer often involves repeated application of computationally intensive statistical inference methods or learning algorithms on the same dataset. A performance penalty is incurred if data are written to disk after each job and reloaded from the disk into memory for the next. Spark93 is a system for scalable computing that replaces the MapReduce layer of Hadoop while leveraging its HDFS layer. Spark enables memory resident cyclic data flow to optimize performance.94,95 The compute tasks are carried out on Resilient Distributed Datasets (RDD96). An RDD is a read-only collection of objects, which can be partitioned across nodes in a cluster. The RDDs are cached in memory across the cluster nodes and can be re-used when needed in parallel without the need for writing the data to disk between individual jobs. It also provides comprehensive fault tolerance.
In case a partition is lost, it can be rebuilt from the information in the handle to the RDD about how the partition was built in the first place. The RDDs are cached in memory across the cluster nodes and can be re-used in MR jobs executed in parallel without the need for materializing the data in the RDDs to disk between individual jobs. Spark natively supports three programming languages, Scala, Python, and Java, for development and provides a library of machine learning algorithms (MLib). This library includes function for basic statistics methods, including summary statistics and random number generation, class discovery methods (eg, k-means clustering and principal components analysis97), and supervised learning methods (eg, support vector machines, decision tress, and random forests98). It also provides numerical algorithms for matrices (eg, singular value decomposition99) and optimization (eg, gradient descent and BGFS100). This library can be used as a tool kit for development of a scalable pipeline for downstream analyses. It should be noted that the HAMA project101 provides matrix operation capabilities for Hadoop. Currently, the most widely used format for storing aligned reads from NGS assays is the SAM or its binary counterpart BAM. While there are Hadoop applications supporting this format, as previously discussed, this format may not be optimally designed for distributed computing. ADAM102 provides a set of data formats for specifying large-scale NGS data along with a set of APIs for accessing and processing the data efficiently:
A data format and access API, built on top of Apache Avro, for transforming the general purpose genomic data formats into the ADAM format and Apache Parquet for accessing the data.
A data transformation API, built using the Scala programming language and implemented on Apache Spark, for transforming and working with the data specified in the ADAM format.
Hadoop is not a turnkey technology. To gauge the performance of the POC, we considered short read sequence files of size 4, 9, and 65 GB, respectively. The maximum gain in performance was observed for the medium sized files (21 versus 50 minutes with a relative gain of 58%). The gain for the large size files was modest (227 versus 241; a relative gain of 6%), whereas for the small sized files, a degradation in performance was observed (25 versus 16 minutes; a relative loss of 56%). This degradation may be attributed to the startup overhead for operating a Hadoop cluster. It also suggests that tuning of operational parameters may be required to optimize performance for this particular size dataset. For the large size data files, the startup overhead is likely to be irrelevant. To increase performance, tuning or addition of more nodes will be required.
The computational facilities of Hadoop can be extended using other hardware and software technologies. For example, GPUs can be used in the compute nodes of a Hadoop cluster to provide a second level of parallelization. JCuda,103 a java binding for CUDA, can be used to communicate with programs written in CUDA.104
Conclusions
Considering the unrelenting onslaught of massive amounts of genomics, clinical, and EHR data, Hadoop provides a powerful computational framework for integrative data management and analysis in cancer research. While a number of bioinformatics tools with applications for cancer research have been developed on the basis of this technology, further refinement and development of de novo tools leveraging the collective spectrum of the Hadoop ecosystem is needed to reap its full potential. As with any other system for distributed computing, Hadoop is not a turnkey system. Developing a highly optimized solution using this technology requires understanding of computer algorithms, specifically for writing custom Map and Reduce functions, and understanding of the interaction among hardware and software components. Other technologies for scalable and distributed computing, including GPU, MPI, and Spark, are available. These can be used in concert with or as an alternative to Hadoop depending on the research problem at hand. The choice of a distributed system must be carefully evaluated during the early design stage and be periodically reevaluated throughout the course of the research.
Acknowledgments
The authors thank the reviewers for insightful and helpful comments.
Footnotes
ACADEMIC EDITOR: J.T. Efird, Editor in Chief
FUNDING: Authors disclose no funding sources.
COMPETING INTERESTS: Authors disclose no potential conflicts of interest.
Paper subject to independent expert blind peer review by minimum of two reviewers. All editorial decisions made by independent academic editor. Upon submission manuscript was subject to anti-plagiarism scanning. Prior to publication all authors have given signed confirmation of agreement to article publication and compliance with all applicable ethical and legal requirements, including the accuracy of author and contributor information, disclosure of competing interests and funding sources, compliance with ethical requirements relating to human and animal study participants, and compliance with any copyright requirements of third parties. This journal is a member of the Committee on Publication Ethics (COPE). Published by Libertas Academica. Learn more about this journal
Author Contributions
Designed the POC and carried out the pre-processing: PA. Wrote the manuscript: PA. Made critical revisions: KO. Both authors reviewed and approved the final manuscript.
REFERENCES
- 1.The Cancer Genome Atlas. 2014. [Accessed December 10, 2014]. Available at: http://cancergenome.nih.gov/
- 2.Sun W. A statistical framework for eqtl mapping using RNA-seq data. Biometrics. 2012;68(1):1–11. doi: 10.1111/j.1541-0420.2011.01654.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Demchenko Y, Grosso P, de Laat C, Membrey P. Addressing big data issues in scientific data infrastructure; Collaboration Technologies and Systems (CTS), 2013 International Conference; 2013. pp. 48–55. [Google Scholar]
- 4.Kunzman DM, Kale LV. Programming heterogeneous systems; Parallel and Distributed Processing Workshops and Phd Forum (IPDPSW), 2011 IEEE International Symposium; 2011. pp. 2061–4. [Google Scholar]
- 5.Suri PK, Mitall S. A comparative study of various computing processing environments: a review. Int J Comp Sci Inf Technol. 2012;3(5):5215–8. [Google Scholar]
- 6.Daniel Hillis W, Steele GL., Jr Data parallel algorithms. Commun ACM. 1986;29(12):1170–83. [Google Scholar]
- 7.Andrade D, Fraguela BB, Brodman J, Padua D. Task-parallel versus data-parallel library-based programming in multicore systems; Parallel, Distributed and Network-based Processing, 2009 17th Euromicro International Conference; 2009. pp. 101–10. [Google Scholar]
- 8.Eijkhout V. Introduction to High Performance Scientific Computing. Creative Commons Attribution 3.0 Unported license. 2014 [Google Scholar]
- 9.Kale V. Guide to Cloud Computing for Business and Technology Managers. Chapman and Hall/CRC; Boca Raton, FL: 2014. pp. 33487–2742. (CRC Press, Taylor & Francis Group, 6000 Broken Sound Parkway NW, Suite 300). [Google Scholar]
- 10.Amazon S3. 2014. [Accessed November 19, 2014]. Available at: http://aws.amazon.com/s3/
- 11.Erbacci G, Sbrighi M, Crosbie A, et al. Data management in HPC. A joint scientific and technological study undertaken by CINECA and TCD for the ENACTS network. Sectoral Report. 2003 [Google Scholar]
- 12.Tivoli Storage Manager for Space Management. 2014. [Accessed November 18, 2014]. Available at: http://www-03.ibm.com/software/products/en/tivostormanaforspacmana.
- 13.Codd E. A relational model of data for large shared data banks. Commun ACM. 1970;13(6):377–87. [PubMed] [Google Scholar]
- 14.Rew R, Davis G. NetCDF: an interface for scientific data access. Comput Graph Appl IEEE. 1990;10(4):76–82. [Google Scholar]
- 15.Folk R, McGrath RE, Yeager N. HDF: an update and future directions. Geosci Remote Sens Symp IEEE. 1999;1:273–5. [Google Scholar]
- 16.Angles R, Gutierrez C. Survey of graph database models. ACM Comput Surv. 2008;40(1):1:1–1:39. [Google Scholar]
- 17.Angles R. A comparison of current graph database models; Data Engineering Workshops (ICDEW), 2012 IEEE 28th International Conference; 2012. pp. 171–7. [Google Scholar]
- 18.Resource Description Framework (RDF) Model and Syntax Specification. 2014. [Accessed December 10, 2014]. Available at: http://www.w3.org/TR/PR-rdf-syntax/
- 19.SPARQL Query Language for RDF. 2014. [Accessed December 10, 2014]. Available at: http://www.w3.org/TR/rdf-sparql-query/
- 20.Duval E, Hodgins W, Sutton S, et al. Metadata principles and practicalities. D-Lib Magazine. 2002;8 [Google Scholar]
- 21.Floridi L, editor. The Blackwell Guide to the Philosophy of Computing and Information, chapter Ontology. Blackwell Publishing Limited; Malden, MA, USA: 2008. [Google Scholar]
- 22.NCI Thesaurus (NCIt) 2014. [Accessed December 21, 2014]. Available at: https://wiki.nci.nih.gov/display/EVS/NCI+Thesaurus+%28NCIt%29.
- 23.Sioutos N, de Coronado S, Haber MW, Hartel FW, Shaiu WL, Wright LW. NCI thesaurus: a semantic model integrating cancer-related clinical and molecular information. J Biomed Inform. 2007;40:30–43. doi: 10.1016/j.jbi.2006.02.013. [DOI] [PubMed] [Google Scholar]
- 24.Golbeck J, Fragoso G, Hartel F, et al. The National Cancer Institute’s Thésaurus and Ontology. Web Semantics: Science, Services and Agents on the World Wide Web 1. 2003:75–80. [Google Scholar]
- 25.Richesson RL, Nadkarni P. Data standards for clinical research data collection forms: current status and challenges. J Am Med Inform Assoc. 2011;18(3):341–6. doi: 10.1136/amiajnl-2011-000107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ashburner M, Ball CA, Blake JA, et al. Gene ontology: tool for the unification of biology. Nat Genet. 2000;25:25–9. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Haerder T, Reuter A. Principles of transaction-oriented database recovery. ACM Comput Surv. 1983;15(4):287–17. [Google Scholar]
- 28.Chaudhuri S, Dayal U. An overview of data warehousing and OLAP technology. ACM Sigmod Record. 1997 Mar;26(1):65–74. [Google Scholar]
- 29.Stonebraker M, Abadi D, DeWitt DJ, et al. Mapreduce and parallel DBMSs: friends of foes? Commun ACM. 2010;53(1):64–71. [Google Scholar]
- 30.Mitha F, Herodotou H, Borisov N, Jiang C, Yoder J, Owzar K. SNPpy – database management for SNP data from genome wide association studies. PLoS ONE. 2011;6(10):e24982. doi: 10.1371/journal.pone.0024982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Murphy S, Churchill S, Bry L, et al. Instrumenting the health care enterprise for discovery research in the genomic era. Genome Res. 2009;19(9):1675–81. doi: 10.1101/gr.094615.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Murphy SN, Weber G, Mendis M, et al. Serving the enterprise and beyond with informatics for integrating biology and the bedside (i2b2) J Am Med Inform Assoc. 2010 Mar-Apr;17(2):124–30. doi: 10.1136/jamia.2009.000893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vajda A. Programming Many-Core Chips. Springer Science+Business Media, LLC 2011, 233 Spring Street; New York, NY 10013, USA: [Google Scholar]
- 34.Dagum L, Menon R. OpenMP: an industry standard api for shared-memory programming. Comput Sci Eng IEEE. 1998;5(1):46–55. [Google Scholar]
- 35.OpenMP Architecture Review Board OpenMP application program interface version 3.0. 2008.
- 36.Severance C, Dowd K. High Performance Computing. O’Reilly Media; 1998. Inc. [Google Scholar]
- 37.Yoo AB, Jette MA, Grondona M. Slurm: simple linux utility for resource management. In: Feitelson D, Rudolph L, Schwiegelshohn U, editors. Job Scheduling Strategies for Parallel Processing, Volume 2862 of Lecture Notes in Computer Science, pages 44–60. Berlin, HD: Springer; 2003. [Google Scholar]
- 38.Message P Forum . MPI: a message-passing interface standard. Technical report. Knoxville, TN: 1994. [Google Scholar]
- 39.Fung J, Tang F, Mann S. Mediated Reality Using Computer Graphics Hardware for Computer Vision; Wearable Computers, 2002. Proceedings. Sixth International Symposium; IEEE; 2002. pp. 83–9. [Google Scholar]
- 40.Fung J, Mann S. Using multiple graphics cards as a general purpose parallel computer: applications to computer vision; Pattern Recognition, 2004. Proceedings of the 17th International Conference; ICPR; Publisher: IEEE; 2004. pp. 805–8. [Google Scholar]
- 41.Jääskeläinen PO, de la Lama CS, Huerta P, Takala JH. Opencl-based design methodology for application-specific processors; Embedded Computer Systems (SAMOS), 2010 International Conference; 2010. pp. 223–30. [Google Scholar]
- 42.Khronos OpenCL Registry. 2014. [Accessed December 10, 2014]. Available at: https://www.khronos.org/registry/cl/
- 43.Halfhill TR. Parallel processing with CUDA. 2008. (Microprocessor Report). Available at: http://www.nvidia.com/docs/I0/55972/220401_Reprint.pdf.
- 44.Buck I, Foley T, Horn D, et al. Brook for GPUs: stream computing on graphics hardware. ACM Trans Graph. 2004;23(3):777–86. [Google Scholar]
- 45.NVIDIA 2014. [Accessed December 10, 2014]. Available at: http://international.download.nvidia.com/pdf/kepler/BD-07317-001_v04.pdf. TESLA K80 GPU ACCELERATOR, BD-07317-001_v04 edition, 2014.
- 46.AMD FireProTM W9100 Professional Graphics. 2014. [Accessed December 10, 2014]. Available at: http://www.amd.com/en-us/products/graphics/workstation/firepro-3d/9100#.
- 47.Dustdar S, Guo Y, Satzger B, Truong HL. Principles of elastic processes. IEEE Internet Comput. 2011;15(5):66–71. [Google Scholar]
- 48.Mell P, Grance T. The NIST Definition of Cloud Computing. Gaithersburg, MD: National Institute of Standards and Technology; 2011. 20899 8930, special publication 800 145 edition. [Google Scholar]
- 49.Afgan E, Baker D, Coraor N, Chapman B, Nekrutenko A, Taylor J. Galaxy CloudMan: delivering cloud compute clusters. BMC Bioinformatics. 2010;11(suppl 12):S4. doi: 10.1186/1471-2105-11-S12-S4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Afgan E, Chapman B, Taylor J. Cloudman as a platform for tool, data, and analysis distribution. BMC Bioinformatics. 2012;(13):315. doi: 10.1186/1471-2105-13-315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Giardine B, Riemer C, Hardison RC, et al. Galaxy: a platform for interactive large-scale genome analysis. Genome Res. 2005;15:1451–5. doi: 10.1101/gr.4086505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Starcluster. 2014. [Accessed November 9, 2014]. Available at: http://star.mit.edu/cluster/
- 53.El-Kalioby M, Abouelhoda M, Krüger J, et al. Personalized cloud-based bioinformatics services for research and education: use cases and the elasticHPC package. BMC Bioinformatics. 2012;13(suppl 17):S22. doi: 10.1186/1471-2105-13-S17-S22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Belleau F, Nolin MA, Tourigny N, Rigault P, Morissette J. Bio2rdf: towards a mashup to build bioinformatics knowledge systems. J Biomed Inform. 2008;41(5):706–16. doi: 10.1016/j.jbi.2008.03.004. [DOI] [PubMed] [Google Scholar]
- 55.Deus HF, Veiga DF, Freire PR, Weinstein JN, Mills GB, Almeida JS. Exposing the cancer genome atlas as a SPARQL endpoint. J Biomed Inform. 2010;43(6):998–1008. doi: 10.1016/j.jbi.2010.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Saleem M, Padmanabhuni S, Ngomo AN, et al. TopFed: TCGA tailored federated query processing and linking to LOD. J Biomed Semantics. 2014;5(1):47. doi: 10.1186/2041-1480-5-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Whetzel PL, Noy NF, Shah NH, et al. BioPortal: enhanced functionality via new Web services from the National Center for Biomedical Ontology to access and use ontologies in software applications. Nucleic Acids Res. 2011;39:W541–5. doi: 10.1093/nar/gkr469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Prajapati V. Big Data Analytics with R and Hadoop. Birmingham: Packt Publishing; 2013. [Google Scholar]
- 59.Dean J, Ghemawat S. Mapreduce: simplified data processing on large clusters. Commun ACM. 2008;51(1):13. [Google Scholar]
- 60.White T. Hadoop: The Definitive Guide. Second ed. Sebastopol, CA: O’Reilly Media; 2010. [Google Scholar]
- 61.Vavilapalli VK, Murthy AC, Douglas C, et al. Apache Hadoop yarn: yet another resource negotiator; Proceedings of the 4th Annual Symposium on Cloud Computing, SOCC ’13; New York, NY: ACM. 2013. pp. 1–5.pp. 16 [Google Scholar]
- 62.Hadoop Releases. 2014. [Accessed December 10, 2014]. Available at: http://hadoop.apache.org/releases.html#15+October%2C+2013%3A+Release+2.2.0+available.
- 63.Abadi DJ, Madden SR, Hachem N. Column-stores vs. row-stores: how different are they really?; Proceedings of the 2008 ACM SIGMOD International Conference on Management of Data, SIGMOD ’08; New York, NY: ACM. 2008. pp. 967–80. [Google Scholar]
- 64.Apache HBase. 2014. [Accessed November 15, 2014]. Available at: http://hbase.apache.org/
- 65.Chang F, Dean J, Ghemawat S, et al. Bigtable: A distributed storage system for structured data. ACM Transactions on Computer Systems. 26(2) Article 4, Pub. date: June 2008. [Google Scholar]
- 66.Apache Hive. 2014. [Accessed November 15, 2014]. Available at: https://hive.apache.org/
- 67.Thusoo A, Sarma JS, Jain N, et al. Hive: a warehousing solution over a map-reduce framework. Proc VLDB Endow. 2009;2(2):1626–9. [Google Scholar]
- 68.Apache Pig. 2014. [Accessed November 15, 2014]. Available at: http://pig.apache.org/
- 69.Olston C, Reed B, Srivastava U, Kumar R, Tomkins A. Pig Latin: a not-so-foreign language for data processing; Proceedings of the 2008 ACM SIGMOD International Conference on Management of Data, SIGMOD ’08; New York, NY: ACM. 2008. pp. 1099–110. [Google Scholar]
- 70.Hericko M, Juric MB, Rozman I, Beloglavec S, Zivkovic A. Object serialization analysis and comparison in java and .net. Sig plan Not. 2003;38(8):44–54. [Google Scholar]
- 71.Apache Avro. 2014. [Accessed November 15, 2014]. Available at: http://avro.apache.org/
- 72.JSON. 2014. [Accessed November 15, 2014]. Available at: http://www.json.org//
- 73.Herodotou H, Lim H, Luo G, et al. Starfish: a self-tuning system for big data analytics; Proceedings of the 5th Conference on Innovative Data Systems Research; 2011. [Google Scholar]
- 74.Schatz MC. Cloudburst: highly sensitive read mapping with MapReduce. Bioinformatics. 2009;25(11):1363–9. doi: 10.1093/bioinformatics/btp236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Taylor RC. An overview of the hadoop/mapreduce/hbase framework and its current applications in bioinformatics. BMC Bioinformatics. 2010;11(suppl 12):S1. doi: 10.1186/1471-2105-11-S12-S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Niemenmaa M, Kallio A, Schumacher A, Klemelä P, Korpelainen E, Heljanko K. Hadoop-BAM: directly manipulating next generation sequencing data in the cloud. Bioinformatics. 2012;28(6):876–7. doi: 10.1093/bioinformatics/bts054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Picard. 2014. [Accessed November 15, 2014]. Available at: http://broadinstitute.github.io/picard/
- 78.Schumacher A, Pireddu L, Niemenmaa M, et al. Seqpig: simple and scalable scripting for large sequencing data sets in hadoop. Bioinformatics. 2014;30(1):119–20. doi: 10.1093/bioinformatics/btt601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.R Core Team . R: A Language and Environment for Statistical Computing. Vienna, Austria: R Foundation for Statistical Computing; 2014. [Google Scholar]
- 80.The Comprehensive R Archive Network. 2014. [Accessed December 10, 2014]. Available at: http://cran.r-project.org/
- 81.Gentleman RC, Carey VJ, Bates DM, et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Guha S, Hafen R, Rounds J, et al. Large complex data: divide and recombine D&R with RHIPE. Statistics. 2012;1:53–67. [Google Scholar]
- 83.Huang H, Tata S, Prill RJ. BlueSNP: R package for highly scalable genome-wide association studies using hadoop clusters. Bioinformatics. 2013;29(1):135–6. doi: 10.1093/bioinformatics/bts647. [DOI] [PubMed] [Google Scholar]
- 84.Mann M, Wilm M. Error-tolerant identification of peptides in sequence databases by peptide sequence tags. Anal Chem. 1994;66(24):4390–9. doi: 10.1021/ac00096a002. [DOI] [PubMed] [Google Scholar]
- 85.Lewis S, Csordas A, Killcoyne S, et al. Hydra: a scalable proteomic search engine which utilizes the Hadoop distributed computing framework. BMC Bioinformatics. 2012;(13):324. doi: 10.1186/1471-2105-13-324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.The Cisco FlexPod Product Line. 2014. [Accessed December 10, 2014]. Available at: http://www.cisco.com/c/en/us/solutions/data-center-virtualization/flexpod/index.html.
- 87.Li H, Handsaker B, Wysoker A, et al. The sequence alignment/map format and SAMtools. Bioinformatics. 2009;25(16):2078–9. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Jnomics. 0000. Available at: https://github.com/jgurtowski/jnomics.
- 89.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Met. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.McKenna A, Hanna M, Banks E, et al. The genome analysis toolkit: a Map-Reduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20(9):1297–303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.DePristo MA, Banks E, Poplin R, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 2011;43(5):491–8. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Danecek P, Auton A, Abecasis G, et al. 1000 Genomes Project Analysis Group. The variant call format and VCFtools. Bioinformatics. 2011;27(15):2156–8. doi: 10.1093/bioinformatics/btr330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Spark. 2014. [Accessed November 15, 2014]. Available at: https://spark.apache.org/
- 94.Engle C, Lupher A, Xin R, et al. Shark: fast data analysis using coarse-grained distributed memory; Proceedings of the 2012 ACM SIGMOD International Conference on Management of Data; 2012. pp. 689–92. [Google Scholar]
- 95.Zaharia M, Chowdhury M, Das T, et al. Resilient Distributed Datasets: A Fault-Tolerant Abstraction for In-Memory Cluster Computing. USENIX NSDI; San Jose, CA: 2012. [Google Scholar]
- 96.Zaharia M, Chowdhury M, Das T, et al. Resilient distributed datasets: a fault-tolerant abstraction for in-memory cluster computing; Proceedings of the 9th USENIX Conference on Networked Systems Design and Implementation, NSDI’12; Berkeley, CA: USENIX Association; 2012. p. 2. [Google Scholar]
- 97.Mardia KV, Kent JT, Bibby JM. Multivariate Analysis. Academic Press, A HarcourtScience and Technology Company; San Diego, CA, USA: 1979. [Google Scholar]
- 98.Hastie T, Tibshirani R, Friedman J. The Elements of Statistical Learning. 2 ed. Springer; Springer Science+Business Media, LLC 2011 233 Spring Street, New York, NY 10013, USA: 2009. [Google Scholar]
- 99.Golub GH, Van Loan CF. Matrix Computations. 3rd Ed. ed. Baltimore, MD: Johns Hopkins University Press; 1996. [Google Scholar]
- 100.Lange K. Optimization. Springer Texts in Statistics. Springer; Springer-Verlag, NY, LLC Part of Springer Science+Business Media, LLC 2011 233 Spring Street, New York, NY 10013, USA: 2004. [Google Scholar]
- 101.Seo S, Yoon EJ, Kim J, et al. HAMA: an efficient matrix computation with the MapReduce framework; 2010 IEEE Second International Conference on Cloud Computing Technology and Science. CloudCom; IEEE Computer Society; 2010. pp. 721–6. [Google Scholar]
- 102.Massie M, Nothaft F, Hartl C, et al. ADAM: Genomics Formats and Processing Patterns for Cloud Scale Computing. Technical Report UCB/EECS-2013–207. Berkeley: EECS Department, University of California; 2013. [Google Scholar]
- 103.Yan Y, Grossman M, Sarkar V. Jcuda: A Programmer-Friendly Interface for Accelerating Java Programs with Cuda; Proceedings of the 15th International Euro-Par Conference on Parallel Processing, Euro-Par ’09; Berlin, HD: Springer-Verlag; 2009. pp. 887–99. [Google Scholar]
- 104.CUDA on Hadoop. 2014. [Accessed November 11, 2014]. Available at: http://wiki.apache.org/hadoop/CUDA%200n%20Hadoop.
- 105.mpiBLAST Open-Source Parallel BLAST. 2014. [Accessed November 23, 2014]. Available at: http://www.mpiblast.org/
- 106.Li KB. ClustalW-MPI: ClustalW analysis using distributed and parallel computing. Bioinformatics. 2003;19(12):1585–6. doi: 10.1093/bioinformatics/btg192. [DOI] [PubMed] [Google Scholar]
- 107.Del Fabbro C, Vezzi F, Policriti A. mrNA: The MPI randomized Numerical Aligner. Pennsylvania: IEEE International Conference on Bioinformatics and Biomedicine (BIBM); 2011. pp. 139–42. [Google Scholar]
- 108.Ronquist F, Huelsenbeck JP. MrBayes 3: bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19(12):1572–4. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- 109.Duncan DT, Craig R, Link AJ. Parallel Tandem: a program for parallel processing of tandem mass spectra using PVM or MPI and X!tandem. J Proteome Res. 2005;4(5):1842–7. doi: 10.1021/pr050058i. [DOI] [PubMed] [Google Scholar]
- 110.Liu Y, Schmidt B, Maskell DL. CUSHAW: a CUDA compatible short read aligner to large genomes based on the burrows-wheeler transform. Bioinformatics. 2012;28(14):1830–7. doi: 10.1093/bioinformatics/bts276. [DOI] [PubMed] [Google Scholar]
- 111.Burrows M, Wheeler DJ. A Block Sorting Lossless Data Compression Algorithm. Technical Report, Digital Equipment Corporation, 1994. 1994. Available at: http://www.hpl.hp.com/techreports/Compaq-DEC/SRC-RR-124.pdf.
- 112.Ferragina P, Manzini G. Opportunistic data structures with applications In foundations of computer science; Proceedings of 41st Annual Symposium IEEE; 2000. pp. 390–8. [Google Scholar]
- 113.Luo R, Wong T, Zhu J, et al. SOAP3-dp: fast, accurate and sensitive GPU-based short read aligner. PLoS One. 2013;8(5):e65632. doi: 10.1371/journal.pone.0065632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Baumgardner LA, Shanmugam AK, Lam H, Eng JK, Martin DB. Fast parallel tandem mass spectral library searching using GPU hardware acceleration. J Proteome Res. 2011;10(6):2882–8. doi: 10.1021/pr200074h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Milloy JA, Faherty BK, Gerber SA. Tempest: GPU-CPU computing for high-throughput database spectral matching. J Proteome Res. 2012;11(7):3581–91. doi: 10.1021/pr300338p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Zandevakili P, Hu M, Qin Z. GPUmotif: an ultra-fast and energy-efficient motif analysis program using graphics processing units. PLoS One. 2012;7(5):e36865. doi: 10.1371/journal.pone.0036865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Manconi A, Orro A, Manca E, Armano G, Milanesi L. GPU-BSM: a GPU-based tool to map bisulfite-treated reads. PLoS One. 2014;9(5):e97277. doi: 10.1371/journal.pone.0097277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Dematté L Prandi D. GPU computing for systems biology. Brief Bioinform. 2010;11(3):323–33. doi: 10.1093/bib/bbq006. [DOI] [PubMed] [Google Scholar]
- 119.Liepe J, Barnes C, Cule E, et al. SysBio – approximate Bayesian computation in python with GPU support. Bioinformatics. 2010;26(14):1797–9. doi: 10.1093/bioinformatics/btq278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Hucka M, Finney A, Sauro HM, et al. SBML Forum The systems biology markup language (SBML): a medium for representation and exchange of biochemical network models. Bioinformatics. 2003;19(4):524–31. doi: 10.1093/bioinformatics/btg015. [DOI] [PubMed] [Google Scholar]
- 121.Cecilia JM, García JM, Guerrero GD, Martínez-del-Amor MA, Pérez-Hurtado I, Pérez-Jiménez MJ. Simulation of P systems with active membranes on Cuda. Brief Bioinform. 2009;11:313–22. doi: 10.1093/bib/bbp064. [DOI] [PubMed] [Google Scholar]
- 122.Cecilia JM, García JM, Guerrero GD, Martínez-del-Amor MA, Pérez-Hurtado I, Pérez-Jiménez MJ. Implementing P systems parallelism by means of GPUs. In: Pâun G, Pérez-Jiménez MJ, Riscos-Núñez A, Rozenberg G, Salomaa A, editors. Membrane Computing, Lecture Notes in Computer Science. Vol. 5957. Berlin HD: Springer; 2010. pp. 227–41. [Google Scholar]
- 123.Shterev ID, Jung SH, George SL, Owzar K. permGPU: using graphics processing units in RNA microarray association studies. BMC Bioinformatics. 2010;11(1):329. doi: 10.1186/1471-2105-11-329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Chang YJ, Chen CC, Chen CL, Ho JM. A de novo next generation genomic sequence assembler based on string graph and MapReduce cloud computing framework. BMC Genomics. 2012;13(S28):Sul7. doi: 10.1186/1471-2164-13-S7-S28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Smith AD, Xuan Z, Zhang MQ. Using quality scores and longer reads improves accuracy of Solexa read mapping. BMC Bioinformatics. 2008;(9):128. doi: 10.1186/1471-2105-9-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Matsunaga A, Tsugawa M, Fortes J. Cloudblast: Combining Mapreduce and Virtualization on Distributed Resources for Bioinformatics Applications; Proceedings of Fourth IEEE CS International Conference. eScience (ESCIENCE ’,08); IEEE Computer Society; 2008. pp. 222–9. [Google Scholar]
- 127.Integrated Proteomics Pipeline. 2014. [Accessed November 11, 2014]. Available at: http://integratedproteomics.com/
- 128.Muth T, Peters J, Blackburn J, Rapp E, Martens L. Proteocloud: a full-featured open source proteomics cloud computing pipeline. J Proteomics. 2013;88:104–8. doi: 10.1016/j.jprot.2012.12.026. [DOI] [PubMed] [Google Scholar]
- 129.Pandey RV, Schlötterer C. DistMap: a toolkit for distributed short read mapping on a hadoop cluster. PLoS One. 2013;8(8):e72614. doi: 10.1371/journal.pone.0072614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Pireddu L, Leo S, Zanetti G. SEAL: a distributed short read mapping and duplicate removal tool. Bioinformatics. 2011;27:2159–60. doi: 10.1093/bioinformatics/btr325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Leo S, Zanetti G. Pydoop: a Python MapReduce and HDFS API for Hadoop; Proceedings of the 19th ACM International Symposium on High Performance Distributed Computing, HPDC ’10; New York, NY, USA: ACM. 2010. pp. 819–25. [Google Scholar]
- 132.Jourdren L, Bernard M, Dillies MA, Le Crom S. Eoulsan: a cloud computing-based framework facilitating high throughput sequencing analyses. Bioinformatics. 2012;28(11):1542–3. doi: 10.1093/bioinformatics/bts165. [DOI] [PubMed] [Google Scholar]
- 133.Langmead B, Hansen KD, Leek JT. Cloud-scale RNA-sequencing differential expression analysis with Myrna. Genome Biol. 2010;11(R83):1–11. doi: 10.1186/gb-2010-11-8-r83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Gurtowski J, Schatz MC, Langmead B. Genotyping in the cloud with Crossbow. Curr Protoc Bioinformatics. 2012 doi: 10.1002/0471250953.bi1503s39. Chapter 15:Unit15.3. [DOI] [PMC free article] [PubMed] [Google Scholar]