Abstract
Although regulation of CXCR3 and CCR4 are related to Th1 and Th2 differentiation, respectively, many CXCR3+ and CCR4+ cells do not express IFN-γ and/or IL-4, suggesting that the chemokine receptor genes might be inducible by mechanisms that are lineage-independent. We investigated the regulation of CXCR3 versus IFNG, and CCR4 versus IL4 in human CD4+ T cells by analyzing modifications of histone H3. In naïve cord-blood cells, under non-polarizing conditions not inducing IL4, CCR4 was induced to high levels without many of the activation-associated changes in promoter histone H3 found for both IL4 and CCR4 in Th2 cells. Importantly, CCR4 expression was stable in Th2 cells, but fell in non-polarized cells after the cells were rested; this decline could be reversed by increasing histone acetylation using sodium butyrate. Patterns in histone H3 modifications in CXCR3+CCR4− and CXCR3−CCR4+ CD4+ T-cell subsets from adult blood matched those in cells cultured under polarizing conditions in vitro. Our data show that high-level lineage-independent induction of CCR4 can occur following T-cell activation without accessibility-associated changes in histone H3, but that without such changes expression is transient rather than persistent.
Keywords: human, T cells, chemokines, histones, epigenetics
Introduction
Trafficking of CD4+ helper T cells in vivo is coordinated in part by differential, subset-specific expression of chemokine receptors. Naïve CD4+ T cells have a narrow chemokine receptor repertoire limited to CXCR4 and CCR7, whereas effector/memory CD4+ T cell subsets have more complex and expanded receptor repertoires that allow efficient trafficking to peripheral tissues [1, 2]. The Th1, Th2 and Th17 subsets of effector/memory CD4+ T cells, defined by production of the signature cytokines IFN-γ, IL-4 and IL-17, respectively, are most strongly associated with expression of the receptors CXCR3, CCR4, and CCR6, respectively [1-5]. However, in each case, the corresponding cytokine-producing cells found in human blood are subsets of a larger pool of cells expressing these receptors [3-5]. These patterns suggest that regulation of the chemokine receptors and signature cytokines involves not only shared but also distinct determinants, and that conditions and mechanisms for induction of the chemokine receptor genes are less restricted than for the genes encoding the lineage-specific cytokines.
Among the shared factors, the master regulators of Th1 and Th2 differentiation, T-bet and GATA-3, respectively, are transcription factors that activate signature cytokine genes [6, 7], and have also been reported to drive expression of CXCR3 [8, 9] and CCR4 [8], respectively. For the cytokine genes, in addition to direct activation, T-bet and GATA-3 are also involved in the remodeling of the cytokines’ genetic loci through epigenetic processes, which are thought to be important for maintaining features of Th cell lineage commitment [6, 7].
Epigenetic elements linked to cell type-specific, inherited patterns of gene expression include DNA methylation and one or more covalent modifications of histones [10, 11]. Histone modifications are increasingly recognized as having broad roles in the control of transcription [10, 11]. The best-studied epigenetic modifications of histones include acetylation of lysines, methylation of lysines and arginines, phosphorylation of serines and threonines, and ubiquitinylation of lysines. In general, hyperacetylation of lysines at the 9th and 14th positions, and methylation of lysines at the 4th and 79th positions of histone H3 have been associated with “permissive” chromatin at active genes [12-14]. In particular, both di- and trimethyl histone H3K4, (H3K4me2 and H3K4me3, respectively) are highly enriched around the transcriptional start sites of active genes [13-15], and a number of proteins/protein complexes that support transcription bind to H3K4me3 directly [16]. Di- and/or trimethylated H3K79 (H3K79me2 and H3K79me3, respectively) are increased in a broad distribution at and downstream of promoters of active genes, associated particularly with transcriptional elongation [15, 17]. In contrast, methylation of H3K9 has been associated with “repressive” or “silenced” chromatin, particularly in the promoter regions through the recruitment of HP1 proteins [11, 13, 18]. Recently, gene-associated histone modifications have been correlated with transcriptional potentials and states based on genome-wide analyses of chromatin immunoprecipitations (ChIP) of various cell types, including mouse [19] and human [14, 18, 20] T cells. Mouse T cells were studied after activation and differentiation in vitro, with analysis of single permissive and repressive marks (see Discussion) [19]. Studies of human CD4+ T cells analyzed total cells, without separation into additional subsets [14, 20].
For cytokine genes, epigenetic regulation has been studied in detail principally in polarized mouse cells by analyzing DNAse I hypersensitive sites for CpG methylation and more recently the acetylation and methylation at histone H3 [6, 19, 21-24]. DNA sites with modified histones in the Il4 locus are essential for enabling cytokine expression after reactivation [25], and methylation of H3K4 has been directly implicated in maintaining Th2 “memory” [26]. Early studies of Il4 and Ifng made the important observation that permissive histone modifications were present at the cytokine loci in non-activated, differentiated cells that were not expressing cytokine genes, so that these modifications did not simply reflect transcriptional activity, but were instead indicators of stable states of locus accessibility and transcriptional competence [27]. Data on epigenetic modifications at the IL4 and IFNG loci in human cells are more limited [28, 29]. The goal of the current study was to analyze histone modifications at the promoters of CXCR3 and CCR4 in comparison with the promoters for IFNG and IL4 in order to understand the role of epigenetic regulation in the expression of the chemokine receptor genes, including mechanisms underlying their less restricted, lineage-independent expression. Our data provide new insights not only into the regulation of chemokine receptor and cytokine genes in human Th cells, but also more generally into pathways of Th cell differentiation and roles for histone modifications in patterns of gene expression following Th cell activation. One of our conclusions is that CCR4 can be induced on activated T cells in a lineage-independent fashion through a mechanism that is fundamentally different from that which occurs during Th2 differentiation, resulting in expression whose level is high, but which is unstable over time.
Results
Lack of a simple correlation between levels of promoter H3K9/14ac, H3K4me2, H3K9me2, H3K79me2 and expression of CCR4
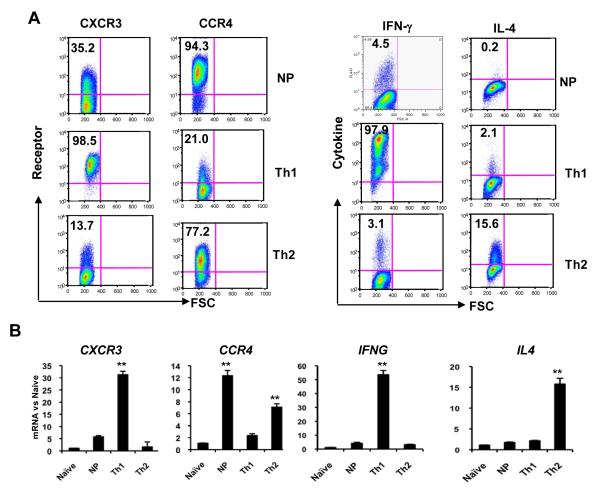
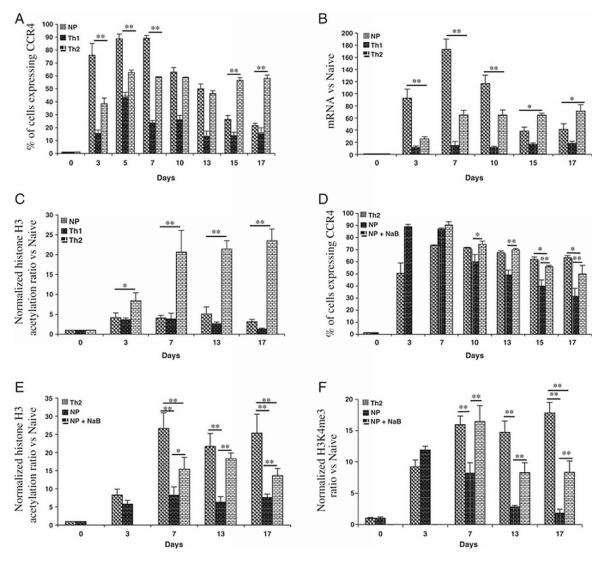
We began our investigation of histone modifications in the regulation of CXCR3 and CCR4 by using human umbilical cord blood as a source of naïve CD4+ T cells (Supplemental Fig. 1). After activating and culturing cells under non-polarizing conditions, very few cells were able to produced either IFN-γ or IL-4 (Fig. 1A). In contrast, Th1 and Th2 culture conditions yielded populations of cells able to produce preferentially IFN-γ or IL-4, respectively. Just as for the cytokines, freshly isolated naïve cells did not express CXCR3 or CCR4. In marked contrast, and unlike the cytokines, both receptors were expressed in cells cultured under non-polarizing conditions (~35% of the population for CXCR3 and ~95% for CCR4). When the cells were cultured under Th1 conditions, almost all expressed CXCR3 whereas only ~20% expressed CCR4. Conversely, under Th2 conditions, only ~15% expressed CXCR3 whereas ~75% expressed CCR4 (Fig. 1A and Supplemental Fig. 2). In all cultures, all the IFN-γ+ cells were CXCR3+CCR4− and all the IL-4+ cells were CXCR3−CCR4+ (data not shown). Our finding of a reciprocal pattern of expression for CXCR3 versus CCR4 cultured under Th1 versus Th2 conditions was as expected from previously published data [2], whereas expression under non-polarizing conditions of activation has not been well characterized. The relative levels of mRNA matched the pattern of protein expression for both signature cytokines and chemokine receptors (Fig. 1B), suggesting that the major determinant of reciprocal expression was at the level of transcription.
Figure 1.
Patterns of receptor expression and cytokine production in cells activated in vitro. Naïve (CD45RA+CD62L+CXCR3− CCR4−) CD4+ T cells from cord blood were stimulated with plate-bound anti-CD3 and soluble anti-CD28 in non-polarizing, Th1, or Th2 conditions. (A) On day 12, cells were stained with FITC-anti-CD4 and PE-anti-CXCR3 or FITC-anti-CD4 and PE-anti-CCR4 antibodies or stimulated with the leukocyte activation cocktail for six hours, fixed, permeabilized and stained with FITC-anti-CD4 and APC-anti-IFN-γ or FITC-anti-CD4 and APC-anti-IL-4 antibodies before analysis. Quadrants were drawn based on the staining with isotype controls (see Supplemental Fig. 2) and the percentages of cells staining for chemokine receptor (left panel) or cytokine (right panel) are displayed. (B) Polarized cells were stimulated with PMA and ionomycin and the mRNA levels for chemokine receptor and cytokine genes were quantified using real-time RT-PCR. Values were normalized to GAPDH and then to the results for naïve cells. Data in (A) are from one donor, representative of five donors/experiments. Data in (B) are means + SEM of pooled results from three donors/experiments where RNA analysis was done together with receptor and cytokine staining, including the donor displayed in (A). **p<0.01 versus all other samples using Student’s t-test.
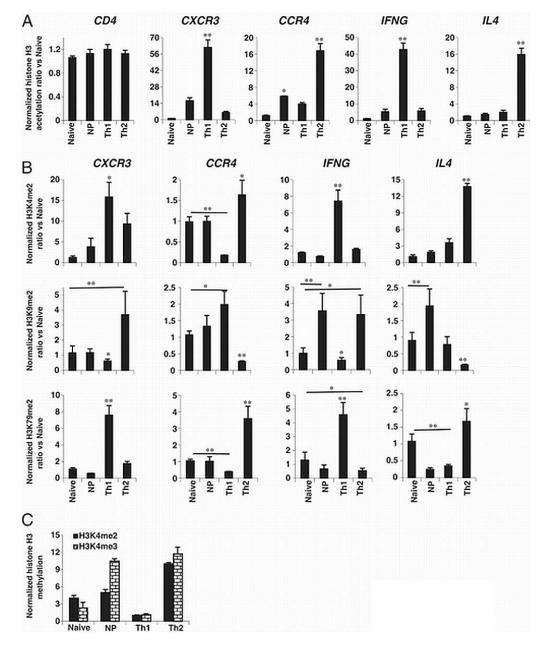
To test whether a similar pattern of epigenetic change was established for the cytokines and chemokine receptors, we analyzed acetylation by ChIP using antibodies that recognized actetylation of histone H3 at lysines 9 and/or 14 (denoted as H3K9/14ac). Lysine acetylation is the best characterized histone modification and H3 hyperacetylation is a well described determinant of transcriptionally competent chromatin [12, 13, 26, 29]. Quantification of amplicons after IP was done by capillary electrophoresis on an Agilent Bioanalyzer and each sample was normalized based on input (Fig. 2A). Negative controls using non-immune mouse IgG gave signals below those using antibodies against H3K9/14ac or other histone H3 modifications (data not shown). As a positive control, and as a test of consistency for the ChIP among samples/subsets, we confirmed that H3K9/14 was acetylated at the CD4 promoter at similar levels for all tested subsets of human CD4+ T cells.
Figure 2.
Lack of a simple correlation between levels of promoter acetylation, methylation and expression of CCR4 in cells activated in vitro. Naïve CD4+ T cells from cord blood were purified and cultured as in Fig. 1. On day 12, cells were analyzed for (A) H3K9/14ac at the CD4, CXCR3, CCR4, IFNG, and IL4 promoters; for (B) H3K4me2, H3K9me2 and H3K79me2 at the CXCR3, CCR4, IFNG, and IL4 promoters; and for (C) H3K4me3 at the CCR4 promoter. Shown are means + SEM of pooled results from three donors/experiments. Ratios of immunoprecipitated DNA over input DNA were normalized to the value for naive cells. *p<0.05, **p<0.01 versus all other samples using one-way ANOVA with Bonferroni correction. Additional, selected pair-wise comparisons are indicated by the lines above the bars (see text).
For IFNG and IL4, relative levels of H3K9/14ac correlated well with relative levels of gene induction. Importantly, in these and all other experiments, ChIP assays were done on cells before treatment with PMA and ionomycin, so that the results reflect the states of histones H3 prior to pharmacological activation. For CXCR3, relative levels of H3K9/14ac under the various culture conditions correlated with levels of CXCR3 expression. The results for CCR4 were, however, surprising. Although, levels of H3K9/14ac in the CCR4 promoter were higher under Th2 vs. Th1 conditions, the non-polarized cells showed levels of acetylation much lower than for the Th2-cultured cells, even though the non-polarized cells had the highest levels of CCR4 expression. We considered the possibility that these results might be confounded by the use of an alternate CCR4 promoter in the non-polarized cells, but analysis of the CCR4 transcriptional start site (TSS) by rapid amplification of 5′ cDNA ends showed that the site was identical in non-polarized and Th2-cultured cells (data not shown).
We next analyzed H3 methylation at each promoter in relation to gene expression. For permissive marks, we chose H3K4me2 and H3K79me2, and for a silencing mark we chose H3K9me2. H3K4me2 and H3K9me2 have been well studied in epigenetic regulation of cytokine genes during Th1/Th2 differentiation [26, 29, 30]. To our knowledge, no data are available for H3K79me2 in Th1/Th2 differentiation. However, given the recent information on the role of this modification in transcription elongation at active genes [15, 17], we thought it of interest to include this modification in our analysis. For the cytokine genes, in general and as expected, levels of H3K4me2 and H3K79me2 were increased when inducible expression was increased, whereas levels of H3K9me2 were increased when expression was decreased (Fig. 2B). In some cases the non-expressing naïve cells had patterns that were more permissive than in the polarized cells in which the given gene was silenced, suggesting a partially permissive state for the cytokine genes in the naïve cells. In addition, the patterns for the non-polarized cells, with high levels of H3K9me2, suggested that the cytokine genes in these cells had also been silenced and had not simply remained in their naïve configuration.
The pattern of histone H3 methylation at the CXCR3 and CCR4 promoters had a number of discordant features relative to expectations for permissive and silencing marks. For example, for CXCR3, based on levels of gene and protein expression, the expected rank order for H3K9me2 (silencing) was Naive>Th2>NP>Th1, whereas the observed order was Th2>>NaivêNP>Th1. For CCR4, based on levels of gene and protein expression, the expected rank order for H3K4me2 and H3K79me2 (permissive) was NP>Th2>Th1>Naive, whereas the observed orders were Th2>NP~Naive>>Th1 and Th2>>NP~Naive>Th1, respectively, and the expected rank order for H3K9me2 was Naive>Th1>Th2>NP, whereas the observed order was Th1>NP~Naive>Th2. As for the cytokine genes, these patterns showed a difference between the non-expressing naïve state and the silenced states, reflected in the high levels of H3K9me2 at CXCR3 in Th2-cultured cells, and the low levels of H3K4me2 and H3K79me2 and high levels of H3K9me2 at CCR4 in Th1-cultured cells.
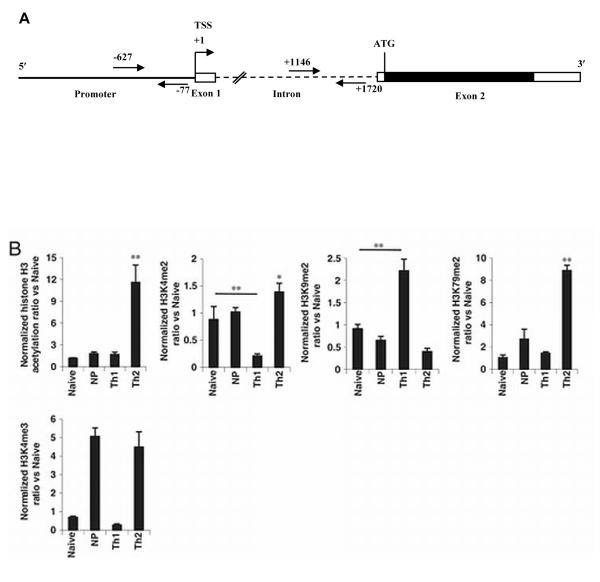
Most surprisingly, but similar to the results for H3K9/14ac, the CCR4 promoter in naïve and non-polarized cells had similar H3 methylation patterns, with much lower levels of H3K4me2 and H3K79me2 and a higher level of H3K9me2 as compared with Th2-cultured cells - as if despite the high level of CCR4 expression in the non-polarized cells, little remodeling had occurred at the CCR4 locus. In genome-wide studies, H3K4me3, which binds to TFIID [16] among other factors, is the histone H3 methylation that shows the most consistently elevated level localized at the start sites of actively transcribed genes [20, 31]. Alone among the modifications that we analyzed, and in contrast to H3K4me2, the level of H3K4me3 was similar in the non-polarized and Th2-cultured cells (Fig. 2C). It is notable, however, that even for this H3K4me3 modification, the CCR4 promoter was “under-modified” in the non-polarized cells, since expression of CCR4 in the non-polarized cells was significantly higher as compared with the Th2-cultured cells (Fig.1 and see Fig. 5 below). In order to rule out that the discordance between histone H3 modification and expression of CCR4 was not confined to the gene’s promoter region, we also analyzed the patterns of histone H3 acetylation and methylation from the body of CCR4 at approximately 1500 bp downstream of the transcription start site (Fig. 3A). Here too we found relatively little modification at CCR4 in non-polarized cells. There were no significant differences between the results for H3K9/14ac, H3K4me2, H3K9me2, and H3K79me2 in the naïve versus non-polarized cells, and H3K4me3 was increased in the non-polarized and Th2-cultured cells (Fig. 3B). Overall, the levels of H3K79me2 were higher, and the levels of H3K4me3 were lower than at the promoter, consistent with the existing data on relative levels of these modifications across active genes [11].
Figure 3.
For CCR4, the pattern of histone H3 modifications in the body of the gene match the pattern at the promoter in cells activated in vitro. (A) Schematic diagram showing genomic organization of the CCR4 locus and the positions of the primers used in ChIP assays. The solid line represents the promoter region, boxes represent exons, and the dashed line represents an intron. Open boxes show non-translated regions and the filled box shows the open reading frame, beginning with the ATG codon. Positions are numbered based on +1 for the transcription start site, which we identified using 5′ RACE, and corresponds to position 32993130 in NCBI Reference Sequence: NC_000003.11. The positions of the 21-nucleotide primers used to amplify promoter and down-stream sequences are indicated by arrows. (B) Naïve CD4+ T cells from cord blood were cultured and analyzed as in Fig. 2, except that the primers used for amplification began at positions +1146 (5′) and +1720 (3′) as shown in A. Shown are means + SEM of pooled results from three donors/experiments. Data were analyzed and displayed just as in Fig. 2.
Hyperacetylation can drive expression of cytokine and chemokine receptor genes
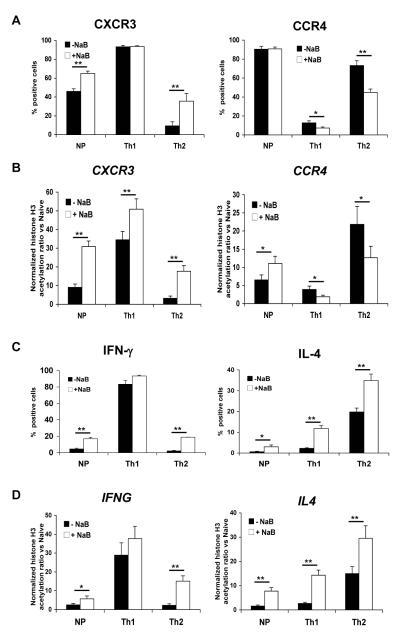
Histone acetylation can be augmented through the use of agents such as sodium butyrate, an inhibitor of class I and IIa histone deacetlylases [32]. We used sodium butyrate, therefore, to test whether acetylation of H3 is functionally important for expression of CXCR3, CCR4, IFNG and IL4 during CD4+ T cell differentiation. As shown in Fig. 4A and C and Supplemental Fig. 3, treatment with sodium butyrate either increased the percentage of cells expressing CXCR3, IFN-γ or IL-4 when the control percentage was less than 100%, or caused no change when the control percentage was already maximal. Consistent with these effects, we found that sodium butyrate increased levels of H3K9/14ac in the corresponding genes (Fig. 4B and D). We were surprised, however, to find that for CCR4, under Th1 and Th2, but not non-polarizing conditions, sodium butyrate treatment led to a decrease in the percentage of positive cells. These changes were associated with decreased levels of H3K9/14ac at the CCR4 promoter (Fig. 4B). By contrast, the level of H3K9/14ac at the CCR4 promoter was increased in the non-polarized cells. Although we have no explanation for this paradoxical action of sodium butyrate at CCR4 in the Th1- and Th2-cultured cells, we presume that the effects were indirect, for example by increasing expression of a butyrate-resistant histone deacetylase active at the CCR4 promoter. Together with the increases in expression of the other genes, the effect of sodium butyrate on both expression and histone H3 acetylation of CCR4 is an “exception that proves the rule”, strengthening the evidence for a causal connection between levels of H3K9/14ac at these promoters and gene expression. In addition, the contrasting effect of sodium butyrate on CCR4 in the Th1- and Th2-cultured vs. non-polarized cells is consistent with the data in Fig. 1 showing a basic difference in how CCR4 expression was regulated in the Th1/Th2 cultured vs. non-polarized cells.
Figure 4.
Hyperacetylation can drive expression of chemokine receptor and cytokine genes. Naïve CD4+ T cells from cord blood were purified and cultured as in Fig. 1. and sodium butyrate (NaB) was added on day 9. On day 12, cells were harvested, treated and analyzed as in Fig. 1A. For determining percentages of cells staining positive, quadrants were drawn based on isotype controls (see Supplemental Fig 3). The percentages of cells expressing (A) chemokine receptors, or (C) producing cytokines are shown for each culture condition in sodium butyrate-treated cells and untreated cells. (B, D) Cells were analyzed for H3K9/14ac at the CXCR3, CCR4, IFNG, and IL4 promoters as in Fig. 2. Shown are means + SEM of pooled results from three donors/experiments. Comparisons between sodium butyrate-treated cells and untreated cells that yielded significant differences are indicated by the lines above the bars. *p<0.05, **p<0.01, using Student’s t-test.
Patterns of histone H3 modifications at the CCR4 promoter correlate with the kinetics of CCR4 expression
Although the histone H3 modifications at the CCR4 promoter in the Th2-cultured cells showed the pattern expected for an active gene, results with the cells cultured under non-polarizing conditions showed that these changes were not required for high induction of CCR4. Given the expression of CCR4 on resting Th2 cells in peripheral blood [4] and the apparent stability in patterns of chemokine receptor expression reported for individual donors over time [33], we hypothesized that differences in CCR4 histone modifications in the Th2-cultured versus non-polarized cells might affect stability of gene expression. We tested this by activating naïve cells for three days under non-polarizing, Th1 or Th2 conditions, and then left cells in IL-2 alone while following CCR4 expression.
Consistent with our hypothesis, after levels of CCR4 mRNA and protein reached their maxima at days 5-7 for both non-polarized and Th2-cultured cells, levels in the non-polarized cells fell when cultured in IL-2 alone, whereas levels in the Th2-cultured cells remained stable (Fig. 5A and B and Supplemental Fig. 4). The kinetics in the Th2-cultured cells were associated with significant and sustained hyperacetylation of histone H3 at the CCR4 promoter, while, as we found previously, acetylation in the non-polarized cells was low, similar to that seen under Th1 conditions (Fig. 5C). If H3 hyperacetylation at the CCR4 promoter in Th2-cultured cells were important for the sustained expression of CCR4, then treating the non-polarized cells with sodium butyrate would be expected to mitigate the decline in CCR4 expression, and this is what was observed (Fig. 5D). While treatment of non-polarized cells with sodium butyrate increased the level of H3K9/14ac at the CCR4 promoter, levels of acetylation as well as gene expression were not brought up to those seen in the Th2-cultured cells (Fig. 5E). Consistent with the inability of sodium butyrate to stabilize CCR4 expression fully, the compound produced only modest or no changes in H3K4me2, H3K9me2, and H3K79me2 at the CCR4 promoter (data not shown). For H3K4me3, as we showed in Fig. 2 and 3, levels rose significantly along with induction of CCR4 under both non-polarizing and Th2 conditions, and the overall trends in H3K4me3 matched expression CCR4 gene and protein patterns (Fig. 5F). However, it is evident from all the data at multiple time points that even for H3K4me3 the non-polarized cells were relatively “under-modified”. Together, these data suggest that modification of histone H3 at the CCR4 promoter has a role in determining the kinetics, as opposed to the levels per se, of CCR4 expression in the non-polarized vs. Th2-cultured cells–transient (although high) in the former, and sustained in the latter.
Figure 5.
H3K9/14ac at the CCR4 promoter correlates with rates of loss of CCR4 expression in cells activated in vitro. Naïve CD4+ T cells from cord blood were purified and cultured initially as in Fig. 1. On day 3, cells were transferred to IL-2-containing medium without polarizing cytokines or antibodies. On day 6, cells were either treated with sodium butyrate (NaB) or left untreated. Samples were analyzed at various days as shown. (A, D) Cells were harvested and stained with FITC-anti-CD4 and PE-anti-CCR4 antibody. For determining percentages of cells staining positive, quadrants were drawn based on isotype controls (see Supplemental Fig. 4). (B) mRNA levels for CCR4 were quantified as in Fig. 1. (C, E) Cells were analyzed for H3K9/14ac and (F) for H3K4me3 at the CCR4 promoter as in Fig. 2. Shown are the means + SEM of pooled results from three donors/experiments in (A-C), from three additional donors/experiments in (D and E), and from three additional donors/experiments in (F). Selected comparisons are indicated by the lines above the bars. *p<0.05, **p<0.01, using Student’s t-test.
Differences in chemokine receptor expression and cytokine production in CXCR3+CCR4-and CXCR3−CCR4+ versus CXCR3+CCR4+ subsets
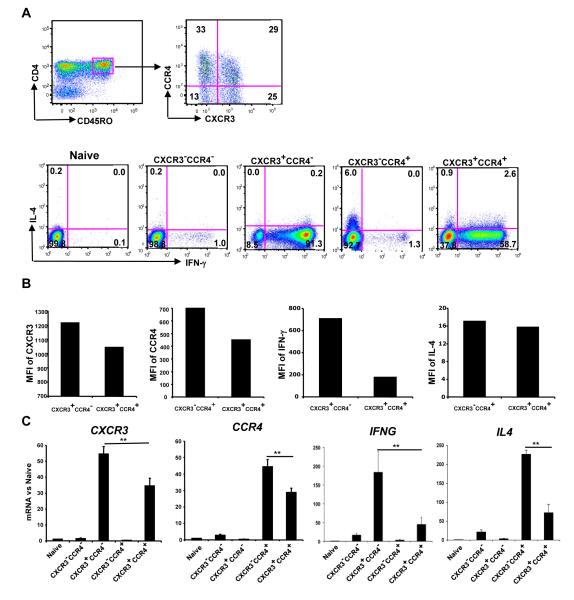
We extended our analyses from cord blood-derived, in vitro-polarized effector/memory CD4+ T cells to subsets of bona fide memory cells purified from peripheral blood. As shown in Fig. 6A and Supplemental Fig. 5, we identified naïve cells as CD4+CD45RO−CD62L+CXCR3−CCR4− and effector/memory cells as CD4+CD45RO+. The CD4+CD45RO+ (effector/memory) cells were further separated into the four subsets as defined by CXCR3 and CCR4 expression. Unlike cord blood-derived cells, because the subsets from peripheral blood were sorted based on receptor expression, each subset was homogeneous with regard to the presence or absence of CXCR3 and/or CCR4. Almost all CXCR3- and CCR4-expressing CD4+ T cells are in the CD+CD45RO+ population, and we have shown previously that the CXCR3+ and CCR4+ cells in the CD4+CD45RO− population are, in fact, effector/memory cells [34]. Therefore, for purposes of cell sorting, we routinely removed any CXCR3+ or CCR4+ CD4+CD45RO− cells from our naïve samples. As described previously [3, 4, 34, 35] and as shown in Fig. 6A, and Supplemental Fig. 5B, IFN-γ-producing cells were found wholly within the CXCR3+ populations, and IL-4-producing cells were found wholly within the CCR4+ populations.
Figure 6.
Using CXCR3 and CCR4 to purify subsets of CD4+CD45RO+ memory T cells reveals differences in chemokine receptor expression and/or cytokine production in single-positive vs. CXCR3+CCR4+ cells. (A) Top panels: Cells enriched for CD4+ T cells from PBMC of healthy donors were stained for CD4, CD45RO, CD62L, CXCR3 and CCR4. Staining for CXCR3 and CCR4 is shown for the effector/memory (CD45RO+) T cells. Bottom panels, CD4+ T cells were sorted into naïve (CD4+CD45RO−CD62L+CXCR3−CCR4−) and the four effector/memory (CD4+CD45RO+) subsets based on expression of CXCR3 and CCR4. Sorted cells were stimulated immediately as in Fig. 1 and stained with FITC-anti-IFN-γ and APC-anti-IL-4 antibodies before analysis. In both sets of panels, quadrants were drawn based on the staining with isotype controls (see Supplemental Fig. 5) and the percentage of cells in each quadrant is displayed. (B) Mean fluorescence intensities (MFI) are shown in arbitrary units for CXCR3 and IFN-γ in CXCR3+CCR4− and CXCR3+CCR4+ cells and for CCR4 and IL-4 in CXCR3−CCR4+ and CXCR3+CCR4+ cells. (C) mRNA levels for chemokine receptor and cytokine genes were quantified using real-time RT-PCR as in Fig. 1. Results in (A and B) are shown for one donor/experiment representative of three and the results in (C), which shows means + SEM for the levels of mRNA, are pooled from four (for CXCR3, IFNG and IL4) and five (for CCR4) donors/experiments, including the donor used for (A and B). Selected comparisons are indicated by the lines above the bars, and **p<0.01 using Student’s t-test.
Two observations from these expression data are of interest. As judged by fluorescent intensities, levels of surface CXCR3 and CCR4 were lower in the CXCR3+CCR4+ cells than in their respective single-positive (CXCR3+CCR4−, CXCR3−CCR4+) counterparts and, similarly, levels of intracellular IFN-γ (as well as numbers of IFN-γ+ cells) were lower in the CXCR3+CCR4+ versus the CXCR3+CCR4− subset. Although there were fewer IL-4+ cells in the CXCR3+CCR4+ versus the CXCR3−CCR4+ subset, levels of IL-4 per cell were not obviously different (Fig. 6B). Relative levels of cytokine and chemokine receptor mRNA among the subsets, as shown in Fig. 6C, generally matched the protein expression data.
Histone H3 modifications at promoters for CXCR3, CCR4, IFNG, and IL4 in CD4+ T cells from blood
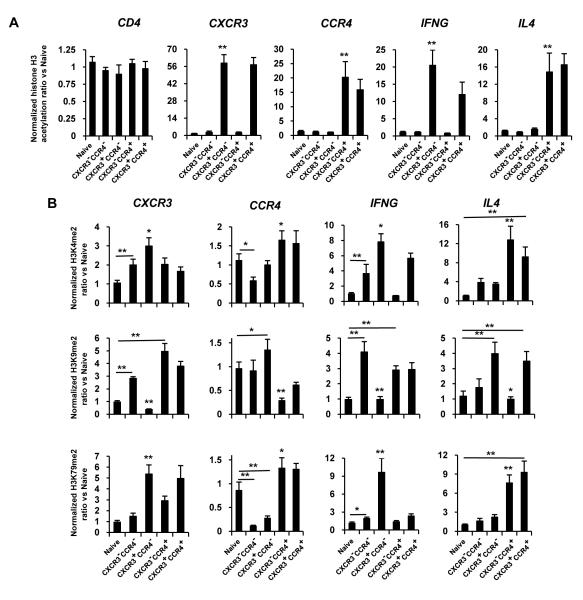
Fig. 7 shows the results of ChIP assays for the cells from peripheral blood, which were analyzed without activation ex vivo. The pattern for acetylation was strongly polarized. Naïve cells and CXCR3−CCR4− memory cells lacked significant H3K9/14ac at the promoter of either of the receptor genes. In marked contrast, CXCR3+CCR4− and CXCR3−CCR4+ memory cells had high levels of H3K9/14ac on the promoters of CXCR3 and CCR4, respectively, and CXCR3+CCR4+ memory cells had high levels of H3K9/14ac at both receptor genes (Fig. 7A). Further, the CXCR3+CCR4− subset, which had high inducible expression of IFNG, but not IL4, showed high H3K9/14ac on the promoter for IFNG but not IL4. The CXCR3−CCR4+ subset showed a strongly polarized reciprocal pattern. Of additional interest, the CXCR3+CCR4+ subset, which had much lower levels of cytokine gene expression and frequencies of cytokine-producing cells and lower levels of inducible IFN-γ per IFN-γ+ cell as compared with the single receptor positive subsets, showed levels of H3K9/14ac at the cytokine gene promoters that were not significantly different from what was found in the single-receptor positive cells. These latter data suggested subset-specific differences in the relationship between histone H3 acetylation and permissiveness for gene activation.
Figure 7.
Combinatorial patterns of promoter histone H3 modifications distinguish subsets of primary CD4+ T cells with varying levels of chemokine receptor and cytokine gene expression. Naïve and effector/memory CD4+ T cells were sorted from peripheral blood as in Fig. 6. Cells were analyzed for (A) H3K9/14ac at the CD4, CXCR3, CCR4, IFNG, and IL4 promoters and for (B) H3K4me2, H3K9me2, and H3K79me2 at the CXCR3, CCR4, IFNG, and IL4 promoters and the results from three donors/experiments were analyzed and displayed as in Fig. 2. For (A) **p<0.01, significant differences between CXCR3+CCR4− or CXCR3−CCR4+ and all other subsets, with the exception of the CXCR3+CCR4+ cells, for which differences were not significant. For (B) *p<0.05, **p<0.01, significant differences versus all other samples, except in the cases of H3K79me2 at CXCR3 in CXCR3+CCR4− versus CXCR3+CCR4+ cells, H3K4me2 and H3K79me2 at CCR4 in CXCR3−CCR4+ versus CXCR3+CCR4+ cells, and H3K4me2 and H3K79me2 at IL4 in CXCR3−CCR4+ versus CXCR3+CCR4+ cells. Additional, selected pair-wise comparisons are indicated by the lines above the bars (see text) using one-way ANOVA with Bonferroni correction.
For the histone H3 methylations, significant differences were found among the cell subsets, although as compared with the acetylation data, the magnitudes of the differences were less pronounced (Fig. 7B). At the CXCR3 promoter, the CXCR3+CCR4− cells had the highest levels of H3K4me2 and H3K79me2 (permissive) and the lowest level of H3K9me2 (silencing). The “oppositely” polarized CXCR3−CCR4+ cells were most notable for the high level of H3K9me2 at the CXCR3 promoter, suggesting silencing and not merely non-expression. Naïve (CXCR3−) cells were generally low in both permissive and silencing modifications, including levels of H3K9me2 much lower than in the other CXCR3− subsets, whereas the CXCR3−CCR4-memory cells showed a “balanced” increase in both classes of methylations at the CXCR3 promoter. The pattern for CXCR3 in CXCR3+CCR4+ cells was of particular interest. Although the level of H3K79me2 in these cells was equal to the level in the CXCR3+CCR4− subset, the level of H3K4me2 was lower and the level of H3K9me2 was much higher, similar to the CXCR3− subsets, creating a unique, “mixed” pattern that correlated with an intermediate level of CXCR3 expression (Fig. 6).
For CCR4, the CXCR3−CCR4+ cells had a polarized pattern analogous to that of CXCR3 in the CXCR3+CCR4− subset – high H3K4me2 and H3K79me2 and low H3K9me2 – and the CCR4− memory subsets showed patterns consistent with a silenced CCR4. Naïve (CCR4−) cells had levels of H3K4me2 and H3K79me2 higher than for one or both of the other CCR4− subsets, along with a high level of the silencing modification, H3K9me2. For CCR4, analogous to but less dramatic than for CXCR3, the CXCR3+CCR4+ cells differed from the CXCR3−CCR4+ principally in having higher levels of H3K9me2, correlating with an intermediate level of CCR4 expression (Fig. 6B).
For the IFNG and IL4 promoters, the CXCR3+CCR4− and CXCR3−CCR4+ cells showed highly polarized patterns, consistent with the CXCR3+CCR4− and CXCR3−CCR4+ subsets containing IFN-γ+IL-4− and IFN-γ−IL-4+ cells, respectively. These findings are particularly notable for the IL4 gene in the CXCR3−CCR4+ cells, since only a small percentage of these cells are IL-4 producers after activation ex vivo (Fig. 6A), suggesting that the IL4 gene has been at least partially enabled in many more of these cells than can be shown to be IL-4 producers. Changes at the IFNG promoter in the CXCR3−CCR4− cells, which are not able to express either IFN-γ or IL-4, resembled those at the CXCR3 promoter in these same cells, with increases in both permissive and silencing modifications versus naïve cells. Of particular interest were the patterns for cytokine genes in the CXCR3+CCR4+ cells. The promoters at both cytokine genes showed a “mixed” permissive/silent pattern: at the IFNG promoter an intermediate level of H3K4me2, a high level of H3K9me2, and a low level of H3K79me2, and at the IL4 promoter high levels of H3K4me2, H3K9me2, and H3K79me2. Inspection of the data in Fig. 7 suggests that the patterns at the cytokine gene promoters in the CXCR3+CCR4+ subset could not have resulted simply from mixtures of cells that displayed the other, polarized patterns, and that these data revealed combinations of modifications unique to the CXCR3+CCR4+ cells.
Discussion
For CXCR3 and CCR4, very little is known about how expression is regulated, and in particular how epigenetic mechanisms control expression in relation to changes at the cytokine genes during Th differentiation. CXCR3 and CCR4 are clearly associated with Th1 and Th2 pathways of differentiation, respectively. The histone H3 modifications at the CCR4 promoter in the CXCR3+CCR4− and Th1-cultured cells, as well as the pattern at the CXCR3 promoter in the CXCR3−CCR4+ and Th2-cultured cells, strengthens the connections between these chemokine receptors and polarizing pathways of Th cell lineages - like the cytokine genes, CCR4 and CXCR3 are not merely non-expressed in cells of the “opposite” lineage, but show evidence of silencing.
For the CCR4 and IL4 promoters, we found similarities in the modifications of histone H3, particularly in the highly polarized populations–Th2- and Th1-polarized cultured cells, and in the CXCR3−CCR4+ and CXCR3+CCR4− subsets from blood. In addition, in naïve cells promoters for both CCR4 and IL4 showed higher levels of permissive modifications and/or lower levels of silencing modifications as compared with Th1-polarized cells, suggesting that the promoter histones in naive cells are in a relatively permissive state – and not simply unmodified –with the genes poised to be rapidly induced after T cell activation [36]. In spite of these points of convergence, it is equally clear that regulation of CCR4 and IL4 differ. Our clearest example was the high expression of CCR4, but not IL4, under non-polarizing culture conditions - and we found, in fact, that CCR4 was regulated by mechanisms that were different in the non-polarized versus Th2-cultured cells. Of the five modifications of histone H3 that we analyzed, only H3K4me3, which is found reliably at the transcriptional start sites of active genes [20, 31], was elevated at the CCR4 promoter in the non-polarized cells at levels similar to what we found in Th2 cells. This latter result is consistent with our finding of the same transcriptional start site for CCR4 in the non-polarized and Th2-cultured cells. Nonetheless, even for this modification CCR4 was “under-modified” in the non-polarized as compared with Th2-cultured cells.
The low levels of the other enabling histone H3 modifications at both at the promoter and in the body of the CCR4 gene in the non-polarized cells, suggested that, in these cells, CCR4 remained in a relatively inactive configuration. This supposition was supported by our finding that without continued activation, levels of CCR4 expression in the non-polarized cells fell over time. By contrast, levels of CCR4 were maintained on the Th2-cultured cells, which showed a highly active pattern of CCR4 histone H3 modifications. Of particular importance, inhibiting deacetylases with sodium butyrate had a mitigating effect on the fall in CCR4 expression in the non-polarized cells, consistent with a causal role for the modified histones in maintaining gene activity.
The high level of expression of CCR4 in the cells cultured under non-polarizing conditions may seem at odds with the results for the CXCR3−CCR4− subset in adult blood, which had a non-polarized phenotype. One potential explanation is that the vast majority of cells in blood are resting, and we showed that as non-polarized cells come to rest they lose CCR4. It is possible, therefore, that the CXCR3−CCR4− cells in blood represent the memory equivalents of cells that had been activated under non-polarizing conditions. In fact, we did not identify a CCR4+ subset from blood that corresponded to the cells that were activated in vitro under non-polarizing conditions. Although in this report we have described and characterized epigenetic regulation of a pathway of CCR4 induction separate from Th2 differentiation, which we presume is relevant to a subset of acutely activated cells in vivo, our data do not in themselves explain the broader pattern of expression of CCR4 vis a vis IL4 in cells in peripheral blood. CCR4+ cells in the CXCR3−CCR4+ subset from adult blood, only few of which are demonstrably Th2 cells, showed the same pattern of histone H3 modifications that we found on cells cultured under Th2 conditions in vitro. Within the limits of our analysis then, the critical role for epigenetic regulation of CCR4 appears to be in determining that activation-induced, lineage-independent expression is transient and that lineage-associated expression, which our data suggest includes expression in the CXCR3−CCR4+ subset from blood, is persistent.
It is of interest that despite the small number of IL-4+ cells in the CXCR3−CCR4+ subset, the IL4 promoter in these cells showed the same permissive pattern of histone modifications as for CCR4. These findings are consistent with a model where promoters for both IL4 and CCR4 have been modified to favor expression in the CXCR3−CCR4+ cells, but that for IL4 these changes alone are not sufficient for transcriptional competence. The changes at the IL4 gene suggest that the promoter has been pre-configured in these cells as an early component of a stepwise process, and the data help in understanding the previous observation that CXCR3-CCR4+ central-memory T cells consist of “pre-Th2” cells that differentiate into fully competent Th2 cells after additional activation [37].
Patterns of expression for CXCR3 and IFNG in both the in vitro activated and peripheral blood cells were more uniformly concordant than for CCR4 and IL4. One mechanism, surely, is the genes’ shared use of T-bet, which can induce H3K4me2 and histone H3 hyperacetylation at the cxcr3 promoter in mouse cells [38, 39]. Nonetheless, CXCR3 and IFNG were not regulated identically, and expression of CXCR3 was more permissive than for IFNG. Of potential relevance to this observation, even in “oppositely” polarized cells that expressed neither CXCR3 nor IFN-γ (in vitro Th2 cultured and CXCR3−CCR4+ cells from blood), we found more mixed patterns of histone H3 modification at the CXCR3 promoter as compared with IFNG, suggesting that CXCR3 was not fully silenced.
Where mixed permissive/non-permissive patterns of histone H3 modifications were most striking was in the CXCR3+CCR4+ subset from blood. These cells showed lower levels of surface CXCR3 and mRNA for CXCR3 and IFNG as compared with the CXCR3+CCR4− cells, and lower levels of surface CCR4 and mRNA for CCR4 and IL4 as compared with CXCR3-CCR4+ cells. For IFN-γ-producing cells, intracellular staining showed not only fewer positive cells, but lower MFI’s for IFN-γ in the CXCR3+CCR4+ versus CXCR3+CCR4− cells. Although, in an analogous fashion, the IL-4-producing cells in the CXCR3+CCR4+ subset may have made less IL-4 per cell vs. the CXCR3−CCR4+ cells, this could not be demonstrated from the staining, perhaps due to the low-intensity signal and poor dynamic range for measuring intracellular IL-4. For all four genes, the promoter histones H3 showed mixed permissive/silenced patterns of modifications whose levels could not have resulted from simple mixtures of cells from polarized subsets, but rather suggested unique patterns. The most consistent repressive modification at each of the genes in the CXCR3+CCR4+ cells was H3K9me2. Within the Th1 memory population, the IFN-γ-producing CXCR3+CCR4+ cells therefore represent a distinct subset, which presumably arises under activating conditions that differ from those found in highly polarizing type 1 environments. The recognition of this subset reinforces the observations that pathways of Th differentiation are highly branched, with under-appreciated levels of heterogeneity [34], and that schemes describing mutually exclusive early lineage commitments are oversimplifications of events in vivo.
For CCR4, an important conclusion from our analysis is that the roles for histone H3 modifications in gene expression can be highly dependent on the overall transcriptional environment, producing significant discordance between modifications at promoter histones H3 and expression – even for a single gene. In fact, we are not aware of another example of a single gene that can be highly induced with very different patterns of promoter histone modifications, nor where such patterns can be implicated in determining transient versus persistent expression. One interpretation of these findings is that levels and/or the identities of factors present in cells activated under non-polarizing conditions are extremely potent in driving CCR4 expression and do not require the permissive modifications of histone H3 to enhance the formation of transcriptional complexes. The lack of these histone modifications in the non-polarized cells assured that when the environment became less favorable, i.e. when the cells were allowed to rest, CCR4 expression was lost. By contrast, the permissive modifications in the Th2-cultured cells (and CCR4+ memory cells in peripheral blood) guaranteed continued expression. Together, our data suggest, as have others [40], that the histone code hypothesis, which proposes that specific combinations of histone modifications specify unique downstream functions, may not best conceptualize the role of histones in transcriptional regulation – at least in the dynamic environment of activated, differentiating T cells.
Materials and methods
Reagents
Anti-CD3, clone OKT3, was obtained from Ortho Biotech, and anti-CD28, clone CK248, was a gift from Calman Prussin, NIH, Bethesda, MD. Anti-IL-4. anti-IL-12, anti-IFN-γ, and all antibodies used for flow cytometry were from BD Biosciences. IL-2 was from Hoffmann-La Roche, and IL-4, IL-12 and TGF-β1.2 were from R&D Systems. Monensin was from Calbiochem. Chromatin immunoprecipitation (ChIP) assay kits, sodium butyrate, and all the ChIP-grade antibodies were purchased from Upstate Biotechnology. A 5′ Rapid Amplification of cDNA Ends (5′ RACE) kit was from Invitrogen.
In vitro activation of naïve cord blood CD4+ T lymphocytes
Human cord blood was obtained at Shady Grove Adventist Hospital (Gaithersburg, MD) as approved by the hospital’s Institutional Review Board. Naïve CD4+ T lymphocytes were isolated by negative selection using RosetteSep CD4+ T cell enrichment cocktail (StemCell Technologies). Naïve CD4+ T cells were further purified by positive selection using CD45RA microbeads (Miltenyi Biotec) and negative selection using PE-anti-CXCR3 and PE-anti-CCR4 plus anti-PE microbeads (Miltenyi Biotec). Stimulation of the CD4+ T cells was performed as describe previously [35] using plate-bound anti-CD3 (10 μg/ml), soluble anti-CD28 (1 μg/ml) in non-polarizing conditions (200 IU/ml rIL-2, 0.4 μg/ml anti-IL-4, 2 μg/ml anti-IL-12, 8 μg/ml anti-IFN-γ, and 10 ng/ml TGF-β1.2), Th1 conditions (200 IU/ml rIL-2, 2 ng/ml rIL-12 and 0.4 μg/ml anti-IL-4), or Th2 conditions (200 IU/ml rIL-2, 4 ng/ml rIL-4, 2 μg/ml anti-IL-12, and 8 μg/ml anti-IFN-γ). After 3 days, cells were washed and expanded under the same conditions in the absence of anti-CD3 and anti-CD28, or only in IL-2 as indicated in the figure legends.
Purification and sorting of lymphocytes subsets
Elutriated lymphocytes were obtained from healthy donors by the Department of Transfusion Medicine, National Institutes of Health (Bethesda, MD) under a protocol approved by the Institutional Review Board. CD4+ T cells were purified by negative selection using RosetteSep as above. Naïve and effector/memory subsets of CD4+ T cells were isolated by cell sorting using a FACS Aria flow cytometer (BD Biosciences) as described previously [34] and shown in Supplemental Fig. 5. The purity of sorted populations was 95-99%.
Flow cytometry and analysis for intracellular cytokines
For staining surface antigens, cells were incubated for 30 min at room temperature. For analyzing cytokine production, cells were stimulated with 20 ng/ml PMA, and 1 mM ionomycin in the presence of 2 mM monensin or Leukocyte Activation Cocktail, with GolgiPlus™ (BD Pharmingen) for 6 h at 37 °C before being stained with APC-anti-IFN-γ and APC-anti-IL-4 using Cytofix/CytoPerm Plus kit (BD Pharmingen), Cells were analyzed on a FACSCalibur cytometer with CellQuest (BD Biosciences), and data were analyzed using FlowJO (Tree Star).
Real-time RT-PCR
RNA was isolated using Trizol (Invitrogen). Real-time RT-PCR was performed with 50 ng of RNA using SuperScript One Step RT-PCR kit (Invitrogen). Primer and probe sets (FAM/MGB-labeled) were purchased from Applied Biosystems. Real-time PCR analysis was performed in duplicate using an ABI 7700 Sequencer System (Applied Biosystems). Concentrations of input RNA and primers were adjusted to assure that threshold cycles were within the exponential phase of amplification. Results were normalized based on the values for GAPDH mRNA, detected using TaqMan GAPDH Control reagents (Applied Biosystems), and expressed as noted in the figure legends.
Chromatin immunoprecipitation
ChIP experiments were performed using EZ ChIP™ kit per the manufacturer’s instructions (Upstate Biotechnology). T cells were treated with 1% formaldehyde for 10 min at room temperature followed by addition of glycine. Cells were resuspended in SDS lysis buffer and incubated on ice for 10 min before sonication to generate DNA fragments between 200–1000 base pairs. Samples in ChIP dilution buffer were pre-cleared by incubating with protein G agarose/salmon sperm DNA for 1 h at 4 °C. After removing 1% of the sample for analyzing input DNA, the pre-cleared chromatin was incubated at 4 °C overnight with anti-histone H3 antibodies or normal mouse IgG (for the negative control). Complexes were immunoprecipitated using protein G agarose. After washing with low salt, high salt, and LiCl immune complex wash buffers and TE, the precipitates were eluted twice with elution buffer. Cross-linking was reversed by incubation at 65 °C overnight with 0.2 M NaCl followed by digestions with RNase A and proteinase K. DNA was purified either by phenol-chloroform extraction followed by ethanol precipitation or using spin columns provided with the EZ ChIP™ kit.
PCR assay and quantification of immunprecipitated DNA
For CXCR3 and CCR4, PCR primers were 21-mers located either immediately upstream of the transcriptional start sites or in the body of the gene (see Fig. 3). Primers for CD4, IFNG, and IL4 promoters were as described [28]. PCR conditions and serial tenfold dilutions of input DNA were used to insure non-saturation kinetics and similar amplification efficiencies for all amplicons within the reactions. PCR products were analyzed in two ways: 1) agarose gel electrophoresis and staining with ethidium bromide, and 2) quantification by capillary electrophoresis using DNA 1000 LabChip Kit and Agilent 2100 Bioanalyzer (Agilent Technologies, Palo Alto, CA). As indicated in the figure legends, data were normalized to the input DNA and, in some cases, normalized additionally to values from naïve cells.
5′ rapid amplification of cDNA ends (5′ RACE)
5′ RACE was performed using 5′ RACE system kit (Invitogen) according to the manufacturer’s protocol. Sequences of gene-specific primers are available on request. Products were purified by agarose gel electrophoresis and sequenced.
Statistical analysis
Statistical analysis was done using either the Student’s t-test or one-way ANOVA with Bonferroni correction as noted in the figure legends.
Supplementary Material
Acknowledgments
We are grateful to Lori Wilkinson and Brian De for assistance in ChIP assays, Calvin Eigsti and other members of the Research Technology Branch, NIAID, for their help with cell sorting, Keji Zhao for helpful discussions, and Philip Murphy and John O’Shea for critical review of the manuscript. The Intramural Research Program of NIAID, NIH, supported this research and authors S.P.S., H.H.Z., J.F.F., M.N.H., and J.M.F. M.M.C. is supported by the “Young Investigators Program” from FAPESP (01/02584-2).
Footnotes
Conflict of interest The authors declare no financial or commercial conflict of interest.
References
- 1.Bonecchi R, Bianchi G, Bordignon PP, D’Ambrosio D, Lang R, Borsatti A, Sozzani S, Allavena P, Gray PA, Mantovani A, Sinigaglia F. Differential expression of chemokine receptors and chemotactic responsiveness of type 1 T helper cells (Th1s) and Th2s. J Exp Med. 1998;187:129–134. doi: 10.1084/jem.187.1.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sallusto F, Lenig D, Mackay CR, Lanzavecchia A. Flexible programs of chemokine receptor expression on human polarized T helper 1 and 2 lymphocytes. J Exp Med. 1998;187:875–883. doi: 10.1084/jem.187.6.875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kim CH, Rott L, Kunkel EJ, Genovese MC, Andrew DP, Wu L, Butcher EC. Rules of chemokine receptor association with T cell polarization in vivo. J Clin Invest. 2001;108:1331–1339. doi: 10.1172/JCI13543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yamamoto J, Adachi Y, Onoue Y, Adachi YS, Okabe Y, Itazawa T, Toyoda M, Seki T, Morohashi M, Matsushima K, Miyawaki T. Differential expression of the chemokine receptors by the Th1- and Th2-type effector populations within circulating CD4+ T cells. J Leukoc Biol. 2000;68:568–574. [PubMed] [Google Scholar]
- 5.Singh SP, Zhang HH, Foley JF, Hedrick MN, Farber JM. Human T cells that are able to produce IL-17 express the chemokine receptor CCR6. J Immunol. 2008;180:214–221. doi: 10.4049/jimmunol.180.1.214. [DOI] [PubMed] [Google Scholar]
- 6.Ansel KM, Djuretic I, Tanasa B, Rao A. Regulation of Th2 differentiation and Il4 locus accessibility. Annu. Rev. Immunol. 2006;24:607–656. doi: 10.1146/annurev.immunol.23.021704.115821. [DOI] [PubMed] [Google Scholar]
- 7.Wilson CB, Merkenschlager M. Chromatin structure and gene regulation in T cell development and function. Curr Opin Immunol. 2006;18:143–151. doi: 10.1016/j.coi.2006.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sundrud MS, Grill SM, Ni D, Nagata K, Alkan SS, Subramaniam A, Unutmaz D. Genetic reprogramming of primary human T cells reveals functional plasticity in Th cell differentiation. J Immunol. 2003;171:3542–3549. doi: 10.4049/jimmunol.171.7.3542. [DOI] [PubMed] [Google Scholar]
- 9.Lord GM, Rao RM, Choe H, Sullivan BM, Lichtman AH, Luscinskas FW, Glimcher LH. T-bet is required for optimal proinflammatory CD4+ T-cell trafficking. Blood. 2005;106:3432–3439. doi: 10.1182/blood-2005-04-1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Berger SL. The complex language of chromatin regulation during transcription. Nature. 2007;447:407–412. doi: 10.1038/nature05915. [DOI] [PubMed] [Google Scholar]
- 11.Li B, Carey M, Workman JL. The role of chromatin during transcription. Cell. 2007;128:707–719. doi: 10.1016/j.cell.2007.01.015. [DOI] [PubMed] [Google Scholar]
- 12.Agalioti T, Chen G, Thanos D. Deciphering the transcriptional histone acetylation code for a human gene. Cell. 2002;111:381–392. doi: 10.1016/s0092-8674(02)01077-2. [DOI] [PubMed] [Google Scholar]
- 13.Bernstein BE, Kamal, Lindblad-Toh K, Bekiranov S, Bailey DK, Huebert DJ, McMahon S, Karlsson EK, Kulbokas EJ, III, Gingeras TR, Schreiber SL, Lander ES. Genomic maps and comparative analysis of histone modifications in human and mouse. Cell. 2005;120:169–181. doi: 10.1016/j.cell.2005.01.001. [DOI] [PubMed] [Google Scholar]
- 14.Wang Z, Zang C, Rosenfeld JA, Schones DE, Barski A, Cuddapah S, Cui K, Roh TY, Peng W, Zhang MQ, Zhao K. Combinatorial patterns of histone acetylations and methylations in the human genome. Nat Genet. 2008;40:897–903. doi: 10.1038/ng.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guenther MG, Levine SS, Boyer LA, Jaenisch R, Young RA. A chromatin landmark and transcription initiation at most promoters in human cells. Cell. 2007;130:77–88. doi: 10.1016/j.cell.2007.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vermeulen M, Mulder KW, Denissov S, Pijnappel WW, van Schaik FM, Varier RA, Baltissen MP, Stunnenberg HG, Mann M, Timmers HT. Selective anchoring of TFIID to nucleosomes by trimethylation of histone H3 lysine 4. Cell. 2007;131:58–69. doi: 10.1016/j.cell.2007.08.016. [DOI] [PubMed] [Google Scholar]
- 17.Steger DJ, Lefterova MI, Ying L, Stonestrom AJ, Schupp M, Zhuo D, Vakoc AL, Kim JE, Chen J, Lazar MA, Blobel GA, Vakoc CR. DOT1L/KMT4 recruitment and H3K79 methylation are ubiquitously coupled with gene transcription in mammalian cells. Mol Cell Biol. 2008;28:2825–2839. doi: 10.1128/MCB.02076-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Roh TY, Cuddapah S, Cui K, Zhao K. The genomic landscape of histone modifications in human T cells. Proc Natl Acad Sci U S A. 2006;103:15782–15787. doi: 10.1073/pnas.0607617103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wei G, Wei L, Zhu J, Zang C, Hu-Li J, Yao Z, Cui K, Kanno Y, Roh TY, Watford WT, Schones DE, Peng W, Sun HW, Paul WE, O’Shea JJ, Zhao K. Global mapping of H3K4me3 and H3K27me3 reveals specificity and plasticity in lineage fate determination of differentiating CD4+ T cells. Immunity. 2009;30:155–167. doi: 10.1016/j.immuni.2008.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 21.Wilson CB, Makar KW, Shnyreva M, Fitzpatrick DR. DNA methylation and the expanding epigenetics of T cell lineage commitment. Semin Immunol. 2005;17:105–119. doi: 10.1016/j.smim.2005.01.005. [DOI] [PubMed] [Google Scholar]
- 22.Lee GR, Kim ST, Spilianakis CG, Fields PE, Flavell RA. T helper cell differentiation: regulation by cis elements and epigenetics. Immunity. 2006;24:369–379. doi: 10.1016/j.immuni.2006.03.007. [DOI] [PubMed] [Google Scholar]
- 23.Bowen H, Kelly A, Lee T, Lavender P. Control of cytokine gene transcription in Th1 and Th2 cells. Clin Exp Allergy. 2008;38:1422–1431. doi: 10.1111/j.1365-2222.2008.03067.x. [DOI] [PubMed] [Google Scholar]
- 24.Wilson CB, Rowell E, Sekimata M. Epigenetic control of T-helper-cell differentiation. Nat Rev Immunol. 2009;9:91–105. doi: 10.1038/nri2487. [DOI] [PubMed] [Google Scholar]
- 25.Tykocinski LO, Hajkova P, Chang HD, Stamm T, Sozeri O, Lohning M, Hu-Li J, Niesner U, Kreher S, Friedrich B, Pannetier C, Grutz G, Walter J, Paul WE, Radbruch A. A critical control element for interleukin-4 memory expression in T helper lymphocytes. J Biol Chem. 2005;280:28177–28185. doi: 10.1074/jbc.M502038200. [DOI] [PubMed] [Google Scholar]
- 26.Yamashita M, Hirahara K, Shinnakasu R, Hosokawa H, Norikane S, Kimura MY, Hasegawa A, Nakayama T. Crucial role of MLL for the maintenance of memory T helper type 2 cell responses. Immunity. 2006;24:611–622. doi: 10.1016/j.immuni.2006.03.017. [DOI] [PubMed] [Google Scholar]
- 27.Avni O, Lee D, Macian F, Szabo SJ, Glimcher LH, Rao A. T(H) cell differentiation is accompanied by dynamic changes in histone acetylation of cytokine genes. Nat Immunol. 2002;3:643–651. doi: 10.1038/ni808. [DOI] [PubMed] [Google Scholar]
- 28.Messi M, Giacchetto I, Nagata K, Lanzavecchia A, Natoli G, Sallusto F. Memory and flexibility of cytokine gene expression as separable properties of human T(H)1 and T(H)2 lymphocytes. Nat Immunol. 2003;4:78–86. doi: 10.1038/ni872. [DOI] [PubMed] [Google Scholar]
- 29.Kaneko T, Hosokawa H, Yamashita M, Wang CR, Hasegawa A, Kimura MY, Kitajiama M, Kimura F, Miyazaki M, Nakayama T. Chromatin remodeling at the Th2 cytokine gene loci in human type 2 helper T cells. Mol Immunol. 2007;44:2249–2256. doi: 10.1016/j.molimm.2006.11.004. [DOI] [PubMed] [Google Scholar]
- 30.Schoenborn JR, Dorschner MO, Sekimata M, Santer DM, Shnyreva M, Fitzpatrick DR, Stamatoyannopoulos JA, Wilson CB. Comprehensive epigenetic profiling identifies multiple distal regulatory elements directing transcription of the gene encoding interferon-gamma. Nat Immunol. 2007;8:732–742. doi: 10.1038/ni1474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schneider R, Bannister AJ, Myers FA, Thorne AW, Crane-Robinson C, Kouzarides T. Histone H3 lysine 4 methylation patterns in higher eukaryotic genes. Nat Cell Biol. 2004;6:73–77. doi: 10.1038/ncb1076. [DOI] [PubMed] [Google Scholar]
- 32.Walkinshaw DR, Yang XJ. Histone deacetylase inhibitors as novel anticancer therapeutics. Curr Oncol. 2008;15:237–243. doi: 10.3747/co.v15i5.371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kivisakk P, Trebst C, Lee JC, Tucky BH, Rudick RA, Campbell JJ, Ransohoff RM. Expression of CCR2, CCR5, and CXCR3 by CD4+ T cells is stable during a 2-year longitudinal study but varies widely between individuals. Journal of Neurovirology. 2003;9:291–299. doi: 10.1080/13550280390201001. [DOI] [PubMed] [Google Scholar]
- 34.Song K, Rabin RL, Hill BJ, De Rosa SC, Perfetto SP, Zhang HH, Foley JF, Reiner JS, Liu J, Mattapallil JJ, Douek DC, Roederer M, Farber JM. Characterization of subsets of CD4+ memory T cells reveals early branched pathways of T cell differentiation in humans. Proc Natl Acad Sci U S A. 2005;102:7916–7921. doi: 10.1073/pnas.0409720102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rabin RL, Alston MA, Sircus JC, Knollmann-Ritschel B, Moratz C, Ngo D, Farber JM. CXCR3 is induced early on the pathway of CD4+ T cell differentiation and bridges central and peripheral functions. J Immunol. 2003;171:2812–2824. doi: 10.4049/jimmunol.171.6.2812. [DOI] [PubMed] [Google Scholar]
- 36.Grogan JL, Mohrs M, Harmon B, Lacy DA, Sedat JW, Locksley RM. Early transcription and silencing of cytokine genes underlie polarization of T helper cell subsets. Immunity. 2001;14:205–215. doi: 10.1016/s1074-7613(01)00103-0. [DOI] [PubMed] [Google Scholar]
- 37.Rivino L, Messi M, Jarrossay D, Lanzavecchia A, Sallusto F, Geginat J. Chemokine receptor expression identifies Pre-T helper (Th)1, Pre-Th2, and nonpolarized cells among human CD4+ central memory T cells. J Exp Med. 2004;200:725–735. doi: 10.1084/jem.20040774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Beima KM, Miazgowicz MM, Lewis MD, Yan PS, Huang TH, Weinmann AS. T-bet binding to newly identified target gene promoters is cell type-independent but results in variable context-dependent functional effects. J Biol Chem. 2006;281:11992–12000. doi: 10.1074/jbc.M513613200. [DOI] [PubMed] [Google Scholar]
- 39.Lewis MD, Miller SA, Miazgowicz MM, Beima KM, Weinmann AS. T-bet’s ability to regulate individual target genes requires the conserved T-box domain to recruit histone methyltransferase activity and a separate family member-specific transactivation domain. Mol Cell Biol. 2007;27:8510–8521. doi: 10.1128/MCB.01615-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schreiber SL, Bernstein BE. Signaling network model of chromatin. Cell. 2002;111:771–778. doi: 10.1016/s0092-8674(02)01196-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.