Abstract
It is widely accepted that the process of retinal cell fate determination is under tight transcriptional control mediated by a combinatorial code of transcription factors. However, the exact repertoire of factors necessary for the genesis of each retinal cell type remains to be fully defined. Here we show that the HMG-box transcription factor, Sox9, is expressed in multipotent mouse retinal progenitor cells throughout retinogenesis. We also find that Sox9 is downregulated in differentiating neuronal populations, yet expression in Müller glial cells persists into adulthood. Furthermore, by employing a conditional knockout approach, we show that Sox9 is essential for the differentiation and/or survival of postnatal Müller glial cells.
Indexing terms: retina, transcription factors, differentiation, Sox2, Notch1
An essential aspect of the creation of mature, functional neural circuitry is the genesis of the correct proportions of neuronal cell types and localization of these cell types to the appropriate regions of the nervous system. During vertebrate retinogenesis all seven retinal cell types (six neuronal and one glial) are derived from a multipotent pool of retinal progenitor cells (RPCs) (Turner and Cepko, 1987). Neurogenesis proceeds in a stereotypical order that is conserved among most vertebrate species. Beginning around embryonic day 11 (E11) the ganglion cells (GCs) are the first retinal cell type to exit the cell cycle and begin the terminal differentiation program. Shortly thereafter, around E11.5, the cones, horizontal cells (HCs), and amacrine cells (ACs) exit the cell cycle and begin to differentiate. This is followed by a second wave of differentiation when the rods and bipolar (BP) cells are born, ending with the generation of the Müller glial (MG) cells (Carter-Dawson and LaVail, 1979; Young, 1985; Harman and Beazley, 1989; La Vail et al., 1991; Prada et al., 1991; Stiemke and Hollyfield, 1995; Rapaport et al., 2004).
It is now well established that the process of retinal cell fate determination is guided by transcription factor activity (intrinsic) in addition to signaling via extracellular (extrinsic) cues (Cepko et al., 1996; Livesey and Cepko, 2001; Hatakeyama and Kageyama, 2004). The time course of retinal cell fate determination led to the proposal of the “competence model of neuronal fate determination” (Cepko et al., 1996). This model describes RPCs as existing in various states of competency that change over time so that, during a particular period, only certain cell types can be generated. There is increasing evidence that competency is endowed upon a cell by expression of a combinatorial transcription factor network (Belliveau et al., 2000). These transcription factors likely regulate the expression of cell surface receptors or components of signal transduction cascades, thereby enabling the cells to respond to specific extrinsic cues (Cepko et al., 1996; Belliveau and Cepko, 1999; Hatakeyama and Kageyama, 2004). In order to obtain a thorough understanding of neuronal fate determination, deciphering this transcriptional network is of the utmost importance. Prime candidates for factors that control RPC competence states are the neuronal basic helix-loop-helix (bHLH) transcription factors (such as Mash1 and Math5), which are widely known to promote neurogenesis in both invertebrate and vertebrate species (Cepko et al., 1996; Belliveau and Cepko, 1999; Hatakeyama and Kageyama, 2004). However, it is becoming increasingly clear that bHLH factors alone cannot specify discrete neuronal subtypes. Rather, bHLH factors appear to function coordinately with other classes of transcription factors in order to specify particular neuronal subtypes (Hatakeyama et al., 2001; Hatakeyama and Kageyama, 2004).
Here we report the expression of HMG box transcription factor Sox9 in mitotic, multipotent RPCs as well as postmitotic and adult MG cells. Through co-immunofluorescence experiments we observed a down-regulation of Sox9 in differentiating, postnatal neuronal populations. These data potentially implicate Sox9 in the maintenance of the RPC pool and possibly in MG cell specification or terminal differentiation. In order to test this hypothesis we conditionally ablated Sox9 within the entire RPC population. Retinal neuronal specification appears to occur normally. However, postnatal MG cells are lost, thereby implicating Sox9 in MG cell genesis.
MATERIALS AND METHODS
Mouse strains
Sox9flox/flox (Akiyama et al., 2005) and Six3-Cretg/+ (Furuta et al., 2000) mice were maintained on a C57BL/6J, 129/SvEv mixed genetic background. Chx10-GFPCre+/tg (Rowan and Cepko, 2004) mice were maintained on an FVB, 129/SvEv, C57BL/6J, and SJL mixed genetic background. Analyses of wildtype mouse tissue was from a C57BL/6J, 129/SvEv mixed genetic background. For all embryonic time course studies, noon on the day when vaginal plugs were observed was designated E0.5. Furthermore, any analysis of mutant and control tissue samples was performed on littermates. All work reported was approved by the institutional animal care and use committee at the University of Texas, M. D. Anderson Cancer Center. Polymerase chain reaction (PCR) was used to genotype mice from all breeding experiments. Sox9flox/flox, Six3-Cretg/+, and Chx10-GFPCre+/tg mice were genotyped as described previously (Furuta et al., 2000; Rowan and Cepko, 2004; Akiyama et al., 2005).
Tissue processing
For embryonic retinae, whole heads were fixed in 4% paraformaldehyde (PFA) for 45 minutes at 4°C. For postnatal retinae, animals were transcardially perfused with 4% PFA and the eyes were carefully removed with rat tooth forceps and then submersion-fixed in 4% PFA for 30 minutes at 4°C. After fixation, all tissue was washed in 1× phosphate-buffered saline (PBS, pH 7.3) 3× 10 minutes at 4°C. Next the samples were cryoprotected by immersing in 15% and then 30% sucrose until the tissue sank to the bottom of the tubes. Subsequently the tissue was immersed in a 1:1 solution of 30% sucrose and OCT medium and left at 4°C for a couple of hours. Then the tissue was embedded in OCT on dry ice and stored at − 80°C prior to sectioning. Cryosections were cut at 20 µm on a cryostat and mounted on superfrost plus slides (VWR Brand, Westchester, PA). In order to make more direct comparisons between mutant and control retinal architecture, all sections were collected at the level of the optic nerve.
Immunofluorescence
Immunofluorescence on retinal cryosections was performed as described (Wang et al., 2001; Ohtoshi et al., 2004) with slight modifications. A table of all primary antibodies including dilutions and references can be found in Table 1. All secondary antibodies were Alexa Fluor-conjugated antibodies from Molecular Probes (Eugene, OR) and used at a dilution of 1:400. For immunohistochemistry, cryosections were allowed to dry completely. Slides were postfixed in 4% PFA for 10 minutes and then washed in 1× PBS-T (PBS + 0.1% Triton X-100) 3× 10 minutes at room temperature. Next the slides were blocked in 2% normal horse or goat serum diluted in 1× PBS for 1 hour at room temperature. Primary antibodies were diluted in the same blocking solution and incubated on the slides overnight at 4°C in a humid chamber. Next the slides were washed 4 times at room temperature in 1× PBS. Labeling with the secondary antibodies was performed using the same blocking solution (2% normal serum in 1× PBS) and slides were incubated for 1 hour at room temperature. All double-labeling experiments were performed by mixing primary antibodies followed by labeling with a mixture of the corresponding secondary antibodies. Slides were finally mounted with Vectashield Mounting Medium (Vector Laboratories, Burlingame, CA) containing DAPI nuclear stain. Negative controls, following the above protocol but without the primary antibodies, were run, consistently showing no cellular labeling.
TABLE 1.
Antibodies
| Antiserum | Dilution | Immunogen | Source |
|---|---|---|---|
| Mouse anti-Brn3 | 1:50 | aa. 397–410 of human BRN3B* | Santa Cruz sc-6026 |
| Mouse anti-calbindin | 1:500 | Purified bovine kidney calbindin-D-28 | Sigma C9848 |
| Mouse anti-calretinin | 1:100 | Recombinant rat calretinin | Chemicon MAB1568 |
| Sheep anti-Chx10 | 1:300 | Recombinant N-terminal aa. 1–131 of human CHX10 conjugated to GST | Exalpha Anti-Chx10 |
| Mouse anti-glutamine synthetase | 1:200 | aa. 1–373 of sheep glutamine synthetase | BD Transduction Labs 610517 |
| Mouse anti-p27Kip1 | 1:100 | Recombinant mouse p27Kip1 | BD Transduction Labs 610241 |
| Mouse anti-Pax6 | 1:20 | Recombinant chicken Pax6 | DSHB Pax6 |
| Mouse anti-rhodopsin | 1:400 | Rat retinal membranes | Sigma O4886 |
| Goat anti-Sox2 | 1:100 | Recombinant C-terminus aa. 277–293 of human SOX2* | Santa Cruz sc-17320 |
| Rabbit anti-Sox9 | 1:200 | Synthetic peptide VPSIPQTHSPQHWEQPVYTQLTRP from human SOX9 | Chemicon AB5535 |
| Mouse anti-Syntaxin-1 | 1:200 | Synaptosomal plasma fraction of rat hippocampus | Sigma S0664 |
Based on matrix-assisted laser desorption/ionization-time-of-flight (MALDI-TOF) and liquid chromatography (LC) mass spectrometry performed in this study.
Antibody characterization
Pax6 monoclonal antibody specificity was previously tested by Western blot of chicken E10 retinal extracts. In two separate experiments this antibody gave an identical staining pattern, detecting only a 48-kDa major band and an ≈34-kDa minor band (Kawakami et al., 1997; Bumsted-O’Brien et al., 2007). Furthermore, the Pax6 antibody stains with a cellular pattern and distribution that is consistent with previous studies in the mouse and chicken retina (Belecky-Adams et al., 1997; Marquardt et al., 2001).
The syntaxin-1 monoclonal antibody was shown to detect a single band of 35 kDa on Western blots of rat retinal lysate (Barnstable et al., 1985). Also, this antibody is routinely employed to label mouse retinal amacrine cell processes and gives a staining pattern identical to results reported here (Dyer and Cepko, 2000b; Haverkamp and Wassle, 2000; Marquardt et al., 2001).
The calretinin monoclonal antibody has also been extensively used to label mouse amacrine cells and their stratifications as well as displaced amacrine cells and ganglion cells and gives a reproducible staining pattern (Haverkamp and Wassle, 2000; Heinze et al., 2007).
The calbindin monoclonal antibody detects a single 28 kDa band on Western blots (Gargini et al., 2007). This particular calbindin antibody is also routinely used as a retinal marker and gives consistent staining of horizontal cells, ganglion cells, and a subset of amacrine cells (Haverkamp and Wassle, 2000; Elshatory et al., 2007; Poche et al., 2007).
The Brn3 antibody specifically recognizes three bands on Western blot corresponding to the three Brn3 family members. Brn3a is 53 kDa, Brn3b is 51 kDa, and Brn3c is 42 kDa (manufacturer’s technical information). This antibody is also typically used as a Brn3b+ retinal ganglion cell-specific marker (Wagner et al., 2002; Elshatory et al., 2007). Based on matrix-assisted laser desorption/ ionization-time-of-flight (MALDI-TOF) and liquid chromatography (LC) mass spectrometry, we determined the Brn3 blocking peptide (Santa Cruz Biotechnology, Santa Cruz, CA) sequence to be from amino acid 397–410 (data not shown).
The Chx10 antibody recognizes a single 46-kDa band upon Western blot of mouse retinal lysate (manufacturer’s technical information; Elshatory et al., 2007). This particular antibody is also used as a standard marker for postnatal retinal bipolar cells and gives a staining pattern that matches the adult retinal RNA in situ pattern (Liu et al., 1994; Fu et al., 2006; Fujitani et al., 2006; Elshatory et al., 2007).
The glutamine synthetase (GS) antibody detects a single 45-kDa band upon Western blot of rat brain lysate (manufacturer’s technical information). Furthermore, this antibody is often used as a marker for postnatal Müller glial cells and gives consistent staining patterns in accordance with previous reports of mouse retinal GS expression (Linser et al., 1984; Haverkamp and Wassle, 2000; Greenberg et al., 2007).
The p27Kip1 antibody labels a single 27 kDa-band on Western blots of mouse retinal lysate (Dyer and Cepko, 2001). This antibody has been previously used for labeling of postnatal Müller glial cells and gives a regular pattern of expression (Dyer and Cepko, 2001; Elshatory et al., 2007).
The rhodopsin antibody specificity was determined via Western blot which shows detection of a closely spaced doublet of 39 kDa in addition to less intense bands of 78 kDa and 115 kDa representing monomers and aggregates (manufacturer’s technical information). This particular antibody has been used in numerous studies to label postnatal photoreceptors (Nishida et al., 2003; Fu et al., 2006; Fujitani et al., 2006; Koike et al., 2007).
The Sox2 antibody used our study detects a single, specific 34 kDa band in mouse embryonic stem cell lysate (manufacturer’s technical information). Sox2 protein was previously shown to be expressed in postnatal Müller glial cells and a subset of amacrine cells. In the same study, this expression pattern was also recapitulated in a Sox2+/GFP knockin mouse line (Taranova et al., 2006). The antibody used in our study results in a temporal and spatial staining pattern identical with these above-mentioned results. Based on MALDI-TOF and (LC) mass spectrometry, we determined the Sox2 blocking peptide (Santa Cruz) sequence to be from amino acid 277–293 (data not shown).
The Sox9 antibody detects the 60–65 kDa Sox9 protein and no other bands on Western blots of mouse brain lysate (manufacturer’s technical information). Furthermore, RNA in situ hybridization with a Sox9 probe that spans nucleotides 972–1508 (Wright et al., 1995) labels the E16.5 retina in an identical pattern to that of the Sox9 antibody (Fig. 1E of this study and data not shown). Additionally, conditional ablation of Sox9 in the developing retina abrogates Sox9 antibody staining, further supporting antibody specificity (Fig. 4 of this study).
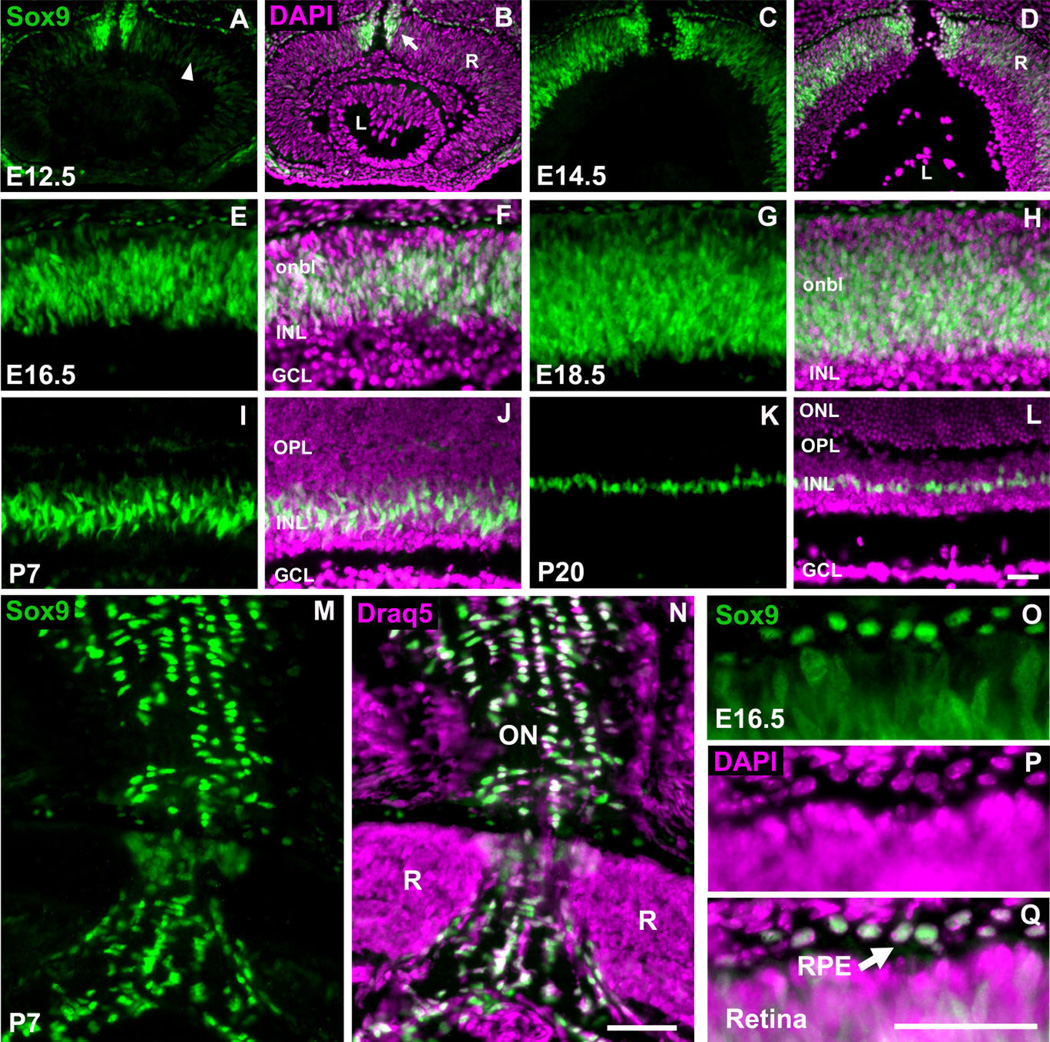
Fig. 1.
Sox9 expression throughout retinal development. Retinae from the indicated developmental stages were stained with an antibody against Sox9. All images represent the central retina at the level of the optic nerve. Sox9 is expressed throughout prenatal retinogenesis within the retinal neuroblastic layer (arrowhead in A–H). Sox9 is also expressed in cells that appear to be entering the optic nerve (arrow in B). As development proceeds into postnatal stages, Sox9 expression becomes restricted to discrete cells in the center of the inner nuclear layer (I–L). Sox9 expression is also observed in the optic nerve (M–N) and retinal pigment epithelium (O–Q, arrow). For a nuclear stain, 4′,6-diamidino-2-phenylindole (DAPI) (B,D,F,H,J,L,P) or Draq5 (N) was used. R, retina; L, lens; onbl, outer neuroblastic layer; INL, inner nuclear layer; GCL, ganglion cell layer; OPL, outer plexiform layer; ONL, outer nuclear layer; ON, optic nerve; RPE, retinal pigment epithelium. N = 3 mice per timepoint. Scale bars = 50 µm.
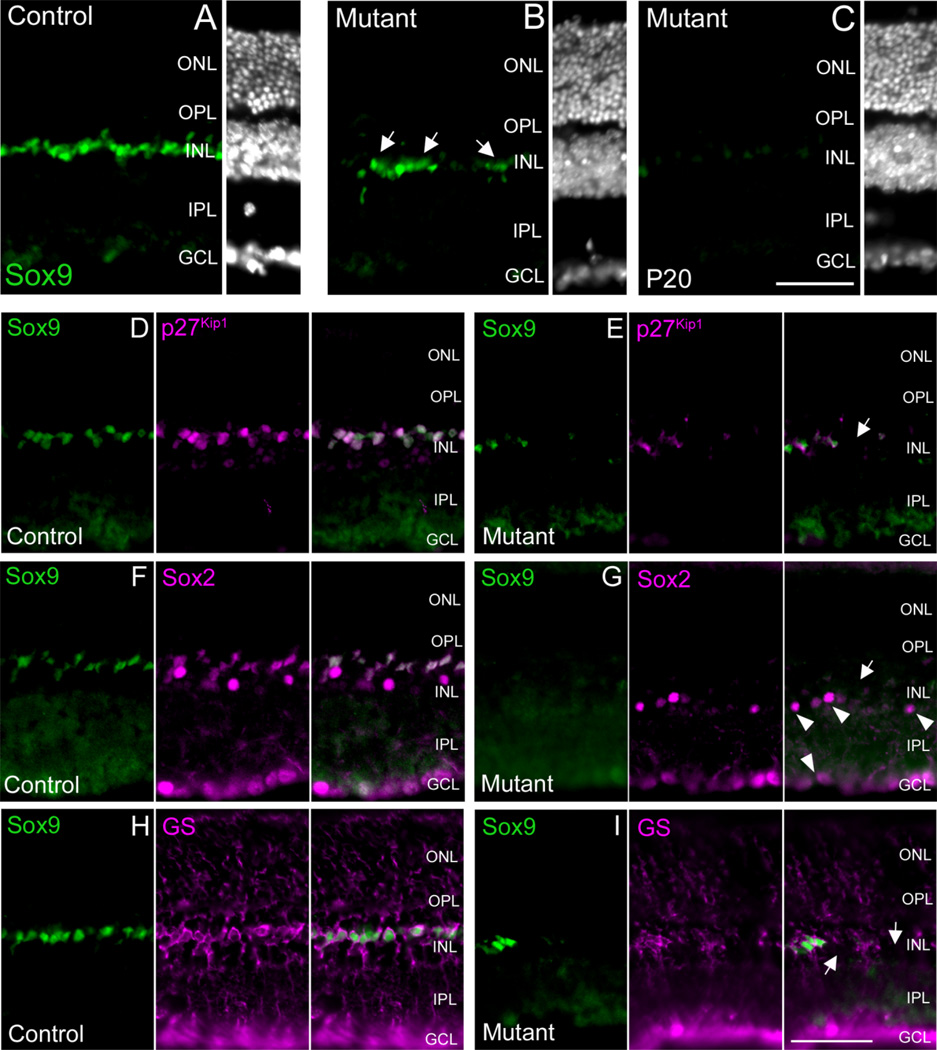
Fig. 4.
Mosaic deletion of Sox9 results in a loss of Müller glial cell markers. At P20, Chx10-Cre-mediated recombination of the Sox9-floxed allele resulted in retinae that exhibited residual Sox9 expression (compare A to B). However, within the same retinal sample, discrete patches showing profound reduction of Sox9 expression were evident (compare A to C). In the same retinal patches that lose Sox9 expression, a DAPI nuclear stain revealed that all retinal regions maintained a characteristic gross retinal architecture with a thick-ness comparable to controls (compare A–C). Colabeling for Sox9 with p27Kip1 (D,E, arrow), Sox2 (F,G, arrow), and GS (H,I, arrows) revealed that loss of Sox9 expression coincided with loss of p27Kip1, Sox2, and GS expression, indicating a loss of Müller glial cells. Sox2 + amacrine cells were not affected (arrowheads in G). ONL, outer nuclear layer, INL, inner nuclear layer; GCL, ganglion cell layer; OPL, outer plexiform layer; IPL, inner plexiform layer. N = 3 mice per genotype. Scale bars = 50 µm.
Image acquisition
All immunofluorescence staining was analyzed with a Leica DMR upright epifluorescence microscope equipped with a Photometrics CoolSnap ES, 12-bit, high resolution monochrome camera (Roper Scientific, Tucson, AZ). Blue, red, and far-red fluorescent channels were imaged separately as gray-scale images, pseudocolored, and merged using Metamorph image acquisition software (Molecular Devices, Palo Alto, CA). In Adobe Photoshop CS2 (San Jose, CA), all original digital images were cropped, scaled, and the resolution set to 300 pixels/inch. Depending on the intensity of the fluorescence for the particular antibody used, Adobe Photoshop CS2 was used to enhance the brightness and/or contrast. For comparisons of wildtype and mutant tissue, all image brightness/contrast enhancements were kept consistent for all genotypes.
RESULTS
Sox9 expression during ocular development
Immunostaining of wildtype mouse ocular cryosections with a Sox9 antibody revealed robust expression of Sox9 during eye development. At E12.5, low but detectable levels of Sox9 were observed within the neuroblastic layer of the developing retina in what appear to be mitotic RPCs (Fig. 1A,B). Intracellular localization of Sox9 did not appear to be exclusively nuclear, as some expression in the cytoplasm was observed. As embryonic development proceeds, Sox9 is upregulated in the neuroblastic layer (Fig. 1C–H). This expression pattern persists until early post-natal stages, when Sox9 expression becomes progressively restricted to the developing inner nuclear layer (INL) (Fig. 1I,J). Importantly, Sox9 in situ hybridization recapitulates the Sox9 antibody staining pattern thereby supporting antibody specificity (data not shown). By postnatal day 20 (P20), Sox9 expression is confined to a single layer of cells within the INL in a position indicative of MG cells (Fig. 1K,L). This overall temporal and spatial expression pattern is highly reminiscent of Notch1, Hes1, and Sox2.
In addition to its RPC and MG expression, nuclear Sox9 is also robustly expressed in the retinal pigment epithelium (RPE) throughout embryonic development and into postnatal stages (Fig. 1A–H,O–Q, and data not shown). Furthermore, at E12.5 and E14.5 intense Sox9 expression is observed in cells that appear to be entering the developing optic nerve (Fig. 1A–D). This expression persists throughout embryonic development (data not shown). At P7 and P20, Sox9 expression in astrocytes residing within the ganglion cell layer as well as in the optic nerve is readily apparent (Fig. 1M,N, and data not shown).
Sox9 is expressed in Sox2+ multipotent RPCs
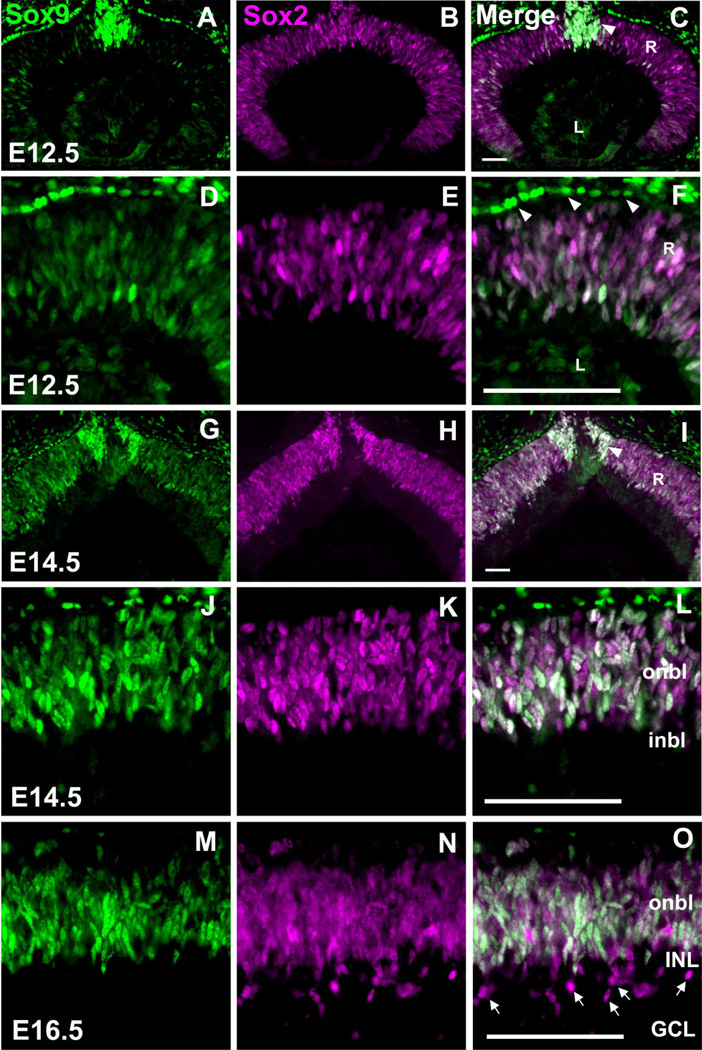
In order to determine whether Sox9 is indeed expressed in multipotent RPCs, we performed Sox9 co-immunofluorescence experiments with antibodies against related HMG-box transcription factor Sox2. Recently, Sox2 was shown to be expressed in proliferating, multipotent RPCs and its activity is essential for temporal and spatial regulation of RPC differentiation (Taranova et al., 2006). Sox9 colocalizes with Sox2 in RPCs. It should be noted that, at E12.5, Sox2 expression appears to be more intense and widespread than Sox9 (Fig. 2A–F). By E14.5, the staining for both antibodies appears comparable (Fig. 2G–L). At E16.5, Sox9 and Sox2 are still coexpressed in RPCs. However, at this time cells positive for Sox2 and negative for Sox9 are observed near the developing INL and ganglion cell layer (GCL) (Fig. 2M–O). These cells are likely the previously described Sox2+, postmitotic displaced amacrine cell subtypes (Taranova et al., 2006). In addition to Sox9/Sox2 coexpression in the retinal neuroblastic layer, these genes are also coexpressed in cells that appear to be entering the developing optic nerve (Fig. 2C,I). The Sox9 + RPE cells do not express Sox2 (Fig. 2F and data not shown).
Fig. 2.
Sox9 and Sox2 are coexpressed in multipotent retinal progenitor cells. Retinae from the indicated developmental stages were colabeled with antibodies against Sox9 and Sox2. At E12.5, Sox9 and Sox2 are expressed throughout the RPCs (A–F) and the developing optic nerve (C, arrowhead) while Sox9 is expressed distinctively in the retinal pigment epithelium (arrowheads in F). By E14.5, Sox9, and Sox2 are coexpressed at comparable levels in the RPCs (J–L). At E16.5, discrete Sox2 +, but Sox9-negative cells appear within the vitreal side of the retina (M–O, arrows). R, retina; L, lens; onbl, outer neuroblastic layer; inbl, inner neuroblastic layer; INL, inner nuclear layer; GCL, ganglion cell layer. N = 3 mice per timepoint. Scale bars = 50 µm.
Sox9 is downregulated in developing retinal neurons, whereas expression in postnatal Müller glia persists
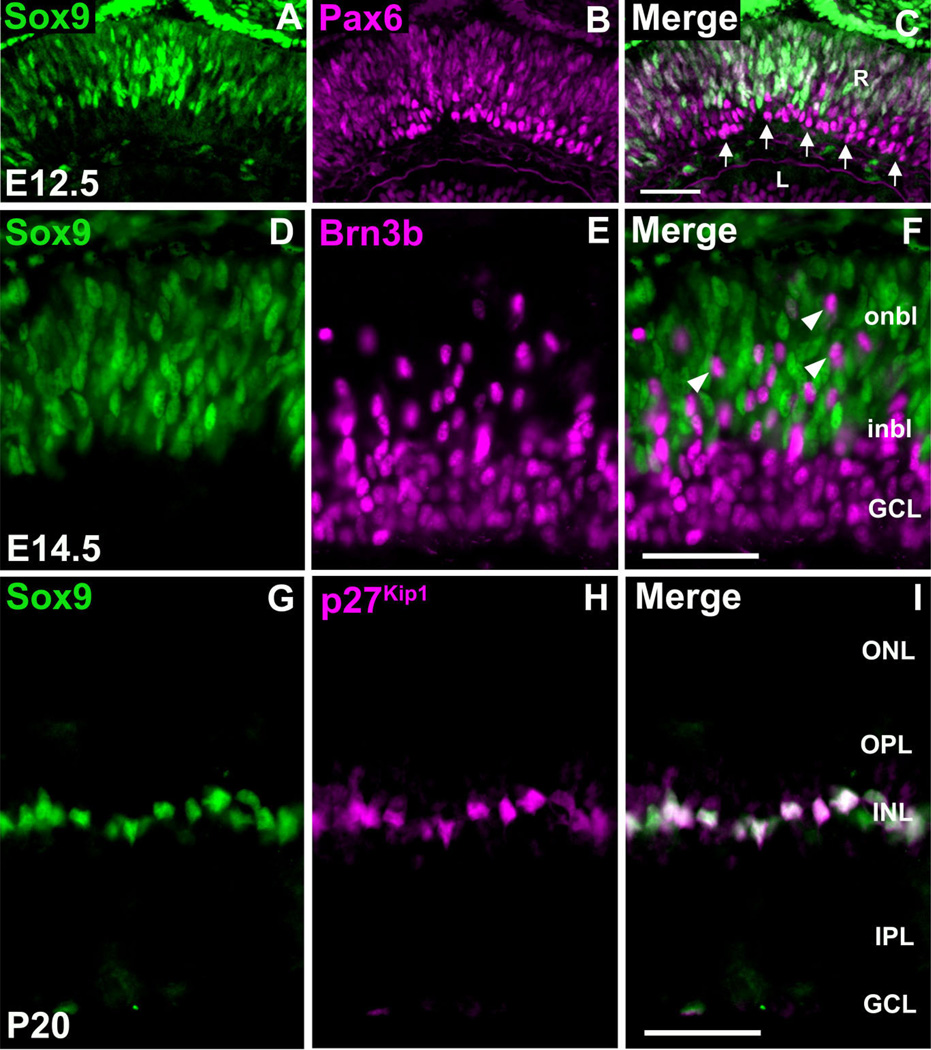
In our time course analyses of Sox9 expression it was noticed that, as development proceeds toward postnatal stages, Sox9 expression becomes progressively restricted to the developing INL (Fig. 1I,J). In order to investigate this observation further we performed Sox9/Pax6 co-immunofluorescence. Pax6 is expressed in mitotic RPCs, but is also highly expressed in the postmitotic retinal ganglion cells (RGCs) settling into the prospective GCL (Marquardt et al., 2001; de Melo et al., 2003). At E12.5, Sox9 and Pax6 are coexpressed in the neuroblastic layer in a similar fashion to Sox2, thereby confirming that Sox9 is expressed in RPCs (Fig. 3A–C). However, cells that are negative for Sox9 and positive for Pax6 were observed in the vitreal side of the retina within the developing GCL (Fig. 3C). These data indicate that Sox9 is turned off once RPCs have exited the cell cycle and are undergoing a differentiation program. As a confirmation of these results, we colabeled retinae for Sox9 and Brn3b. Brn3b is specifically expressed in recently postmitotic, differentiating RGCs (Gan et al., 1996). At E14.5 there is no overlap between Sox9 and Brn3b staining (Fig. 3D–F). We ob-served cells positive for only Brn3b residing within the Sox9+ neuroblastic layer. These cells are likely recently postmitotic RGCs in transit to the developing GCL (Fig. 3F).
Fig. 3.
Sox9 is not expressed in retinal ganglion cells, while expression in postnatal Müller glial cells persists. E12.5 retinae were colabeled with antibodies against Sox9 and Pax6 (A–C). Clear colo-calization is observed within the retinal ventricular zone. Within the developing ganglion cell layer, which consists of Pax6 + differentiating retinal neurons, Sox9 is not expressed (arrow in C). Colabeling for Sox9 and Brn3b (D–F) confirms the above-mentioned results as Brn3b + ganglion cells do not express Sox9. These cells appear to be traversing the Sox9+ ventricular zone toward the developing GCL (arrowheads in F). At P20, Sox9 protein expression is confined to the inner nuclear layer (G). Colabeling with Sox9 and p27Kip1 revealed that postnatal retinal expression of Sox9 is specific to Müller glial cells (G–I). R, retina; L, lens; onbl, outer neuroblastic layer; inbl, inner neuroblastic layer; ONL, outer nuclear layer, INL, inner nuclear layer; GCL, ganglion cell layer; OPL, outer plexiform layer; IPL, inner plexiform layer. N = 3 mice per timepoint. Scale bars = 50 µm.
At P20, Sox9 expression is restricted to a discrete layer within the central INL in a position indicative of MG cells (Fig. 1K,L). In order to confirm the identity of these cells we performed Sox9 co-immunofluorescence with the Müller glia markers p27Kip1 and GS (Linser et al., 1984; Dyer and Cepko, 2000a). Sox9 and p27Kip1 are coexpressed in all MG cell nuclei (Fig. 3G–I). Additionally, Sox9+ nuclei precisely localize to the cell bodies of GS + Müller glia (Fig. 4H). Thus, adult retinal expression of Sox9 is confined specifically to the Müller glial cells.
Sox9 conditional mutant retinae
Our Sox9 expression data indicate a potential role for Sox9 in regulating RPC proliferation and/or differentiation as well as possibly postmitotic MG cell terminal differentiation or survival. However, the Sox9-null allele results in perinatal haploinsufficient lethality and is lethal by E11.5 in the homozygous state (Bi et al., 2001; Chaboissier et al., 2004). Thus, in order to test these hypotheses, we employed a Sox9 floxed allele and a Chx10-CreGFP transgenic line, thereby producing retinal-specific Sox9 mutant mice (Rowan and Cepko, 2004; Akiyama et al., 2005). Chx10-Cre becomes active at E10.5 and exhibits widespread Cre-mediated recombination shortly thereafter, leading to fate mapping of all retinal cell types (Rowan and Cepko, 2004).
We were successful in generating Chx10-Cre; Sox9flox/flox mice that were viable with no obvious gross ocular or behavioral abnormalities. It should be noted that we also attempted to create Sox9 mutant mice using the RPC-expressed Six3-Cre transgenic line (Furuta et al., 2000). However, this line resulted in high levels of residual Sox9 expression, likely due to mosaic activity of the Cre recombinase (data not shown). Furthermore, Chx10-Cre; Sox9flox/flox and Six3-Cre; Sox9flox/flox mice were obtained at frequencies of ≈9% and 10%, respectively, which is below the expected Mendelian ratio of 25%. We interpret these results to indicate that early embryonic (possibly ectopic) Chx10-Cre and Six3-Cre activity resulted in early recombination of the Sox9-floxed allele (Furuta and Behringer, 2005). We believe that this apparent early Cre activity, combined with the haploinsufficient nature of the Sox9-null allele, caused the majority of Chx10-Cre; Sox9flox/flox and Six3-Cre; Sox9flox/flox mice to die in utero or at perinatal stages.
In order to assess the efficiency of Chx10-Cre in recombining the Sox9 locus, we performed Sox9 immunofluorescence on P20 retinal cryosections. In all samples analyzed a mosaic pattern of Sox9 deletion was evident. Throughout the entire retina, large patches of Sox9 expression persisted with a level of intensity comparable to that of the control animals (Fig. 4A,B). These data are in accordance with the previously reported mosaic activity of the Chx10-Cre transgene (Rowan and Cepko, 2004; Johnson et al., 2006). Nevertheless, we were able to identify discrete patches of the retina that exhibited significant loss of Sox9 expression (Fig. 4A,C). For all subsequent analyses we will focus on these regions of unambiguous loss of Sox9 expression compared to control samples.
Using a DAPI stain, we first analyzed P20 Sox9 mutant retinae for gross defects in retinal architecture. In order to make direct comparisons with control retinae, all images were taken at the level of the optic nerve. Sox9 mutant retinae exhibited normal lamination as well as nuclear layer thickness comparable to control samples (Fig. 4A–C). These data indicate that loss of Sox9 alone likely does not affect global retinal fate determination, as disruptions in this process would likely result in a thin, poorly defined retina as seen for the Sox2 conditional mutant (Taranova et al., 2006). Additionally, Sox9 likely does not affect retinal lamination, as this would be expected to lead to a broad disorganization of the retina, possibly with formation of rosette structures (Fu et al., 2006).
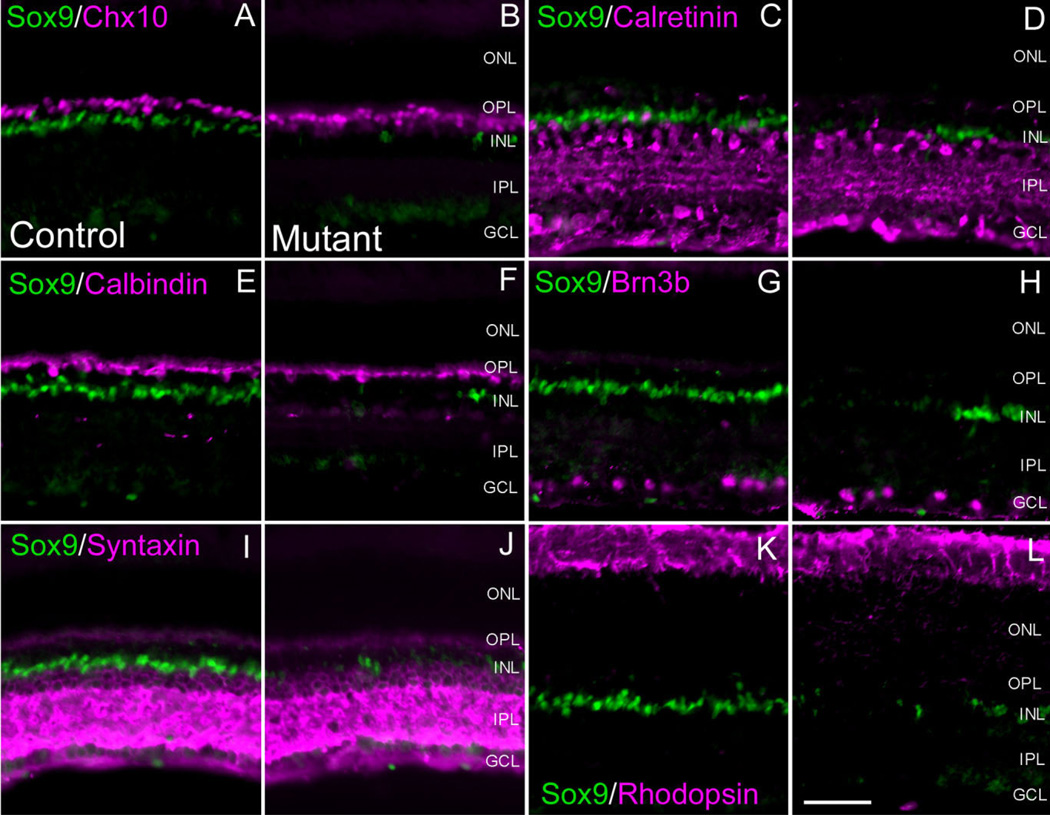
In order to determine whether a more discrete disruption of retinal fate determination or lamination had occurred, we analyzed P20 Sox9 mutant retinae for various retinal markers (Table 1). We began our analyses by checking for defects in Müller gliogenesis by performing Sox9/p27Kip1, Sox9/Sox2, and Sox9/GS co-immunofluorescence. In retinal patches that display loss of Sox9 expression, we also observed corresponding loss of Sox2, p27Kip1, and GS expression (Fig. 4D–I). These data suggest that MG cells are not present in retinal areas that lose Sox9 expression, thereby implying that Sox9 is necessary for MG cell differentiation and/or survival. Importantly, in Sox2 labeled mutant retinae, Sox2 expression in the lower INL and GCL persists, further indicating that loss of Sox2 is due to an absence of MG cells (Fig. 4G). Subsequently, we performed co-labeling experiments for Sox9 and markers corresponding to all other retinal cell types. We did not observe any alterations in the expression of these markers coincident with loss of Sox9 (Fig. 5A–L). Therefore, we conclude that Sox9 loss-of-function has no obvious consequence on retinal neurogenesis, but specifically affects Müller gliogenesis.
Fig. 5.
Retinal neuron populations are not affected by loss of Sox9. Co-immunofluorescence for Sox9 and retinal neuron markers was performed in adult conditional mutant retinae and compared to controls. In the conditional mutant retinae, expression of all markers appeared comparable to control littermates. Chx10 is a pan-bipolar cell marker (A,B). Calretinin (C,D) and Syntaxin (I,J) were used to label amacrine cell populations. Calbindin was used to assess horizontal cells (E,F). Brn3b marks retinal ganglion cells (G,H) and rhodopsin marks photoreceptor segments (K,L). ONL, outer nuclear layer, INL, inner nuclear layer; GCL, ganglion cell layer; OPL, outer plexiform layer; IPL, inner plexiform layer. N = 3 mice per genotype. Scale bar = 50 µm.
DISCUSSION
Sox9 expression is consistent with a possible role in retinal fate determination
All seven retinal cell types are derived from a common progenitor pool (Turner and Cepko, 1987). Throughout the entire period of retinogenesis, RPCs proliferate while simultaneously giving rise to specific subtypes over time. Therefore, it is absolutely essential to maintain multipotent RPCs until late in development so that there is a large enough pool of RPCs still available for “late” cell type specification in the appropriate proportions. The bHLH repressors Hes1 and Hes5 are necessary for maintaining RPC potency over time (Tomita et al., 1996; Hatakeyama and Kageyama, 2004). The recognition of Hes1 and Hes5 as factors essential for RPC maintenance immediately implicated Notch signaling as Hes1 and Hes5 are direct downstream targets of the Notch pathway (Ohtsuka et al., 1999; Hatakeyama and Kageyama, 2004). Once activated by the Notch pathway, Hes1 and Hes5 negatively regulate the expression of bHLH proneural genes, thereby inhibiting neuronal differentiation. Indeed, an early retinal-specific knockout of Notch1 recapitulates some aspects of the Hes1-null phenotype (Jadhav et al., 2006b; Yaron et al., 2006). In one of these studies, as expected, Notch1 inactivation resulted in a downregulation of Hes1 and Hes5 expression and the consequential upregulation of proneural transcription factors leading to precocious differentiation and RPC depletion (Yaron et al., 2006). Surprisingly, both studies also found that the Notch1 conditional mutant retinae exhibit a dramatic increase in photoreceptors at the expense of other retinal cell types. These data suggest that, in addition to its role as a negative regulator of neural induction, Notch1 signaling contributes to retinal cell diversification by inhibiting the acquirement of the photoreceptor fate. It was initially reported, through data generated in the chick, that Notch1 knockdown results in an excess of the earliest retinal cell types, the ganglion cells (Austin et al., 1995). These data conceptually make sense. If Notch1 functions to repress neurogenesis, upon early alleviation of that repression one would expect the retinal developmental program to proceed in a normal histogenic order with the genesis of the ganglion cells (the first specified retinal neurons) followed by the other cell types. In the Notch1 conditional mutants, however, there was actually a reduction in ganglion cell numbers (Jadhav et al., 2006b; Yaron et al., 2006). The reason for the discrepancy between the mouse and chicken data is not currently clear. It is also unclear whether Hes genes are functioning downstream of Notch1 in the mouse to repress photoreceptor production.
In this article we have shown that Sox9 is expressed in proliferating RPCs in a very similar temporal and spatial manner to that of Hes1, Hes3, Notch1, and Sox2. Of particular note is our finding that Sox9 expression is down-regulated in differentiating retinal neurons (summarized in Fig. 6). These data suggest that Sox9 might be important in maintaining progenitor cell status within the retina, possibly functioning within the Notch pathway, rather than having a direct role in neurogenesis. Recently, it was reported that Sox9 regulates pancreatic expression of Hes1 and is required for the maintenance of the mouse pancreatic progenitor pool (Seymour et al., 2007). Thus, Sox9 might be a Notch1 transcriptional cofactor in activating Hes1 expression or Sox9 might directly promote Notch1 expression. In terms of retinal fate determination, Sox2 has recently been identified as a direct upstream component of the Notch1 pathway (Taranova et al., 2006). These data, in light of our finding that Sox9 is expressed in the Sox2+ RPC population, suggest that Sox9 might function in a similar capacity as Sox2 in regulating the competence status of RPCs by promoting Notch1 expression.
Fig. 6.
Summary of Sox9 expression and hypothetical placement of Sox9 within the Müller glial cell developmental genetic pathway. In early stages of retinal development, Sox9 is expressed in all retinal progenitor cells (RPCs) (A). As progenitor cells exit the cell cycle and differentiate into neurons (clear cells), Sox9 is downregulated (B). Once retinal neurogenesis is complete, Sox9 is specifically expressed in postmitotic MG cells (C). Data presented in this article raise the possibility that Sox9 functions in RPC maintenance and/or MG cell terminal differentiation or survival. In the bottom panel a developmental genetic pathway of MG cell fate determination and differentiation is presented. This figure is not meant to suggest that MG cells are derived from a separate RPC lineage, but rather to outline the emerging genetic pathway leading to the MG cell fate to now include Sox9 (D). In order for MG cells to be specified (or to differentiate), some proportion of the RPC population must maintain Sox9 expression, whereas RPCs giving rise to neurons lose Sox9 expression. Furthermore, Sox9 might function in a similar capacity as Sox2 in regulating Notch1 expression in retinal development (see Discussion for details).
In order to test our hypothesis that Sox9 functions to maintain RPC competence, we genetically ablated Sox9 within multipotent RPCs. We were able to discern discrete regions of postnatal mutant retinae that showed significant loss of Sox9 expression and these tissue samples were further evaluated for defects in retinogenesis. In regions of unambiguous Sox9 loss, mature retinal architecture was intact and there was no dramatic decrease in retinal thickness. These data indicate that Sox9 alone does not play a role in maintaining RPC competence. Nevertheless, one caveat in the interpretation of these data is that we were not able to achieve high levels of Sox9 loss-of-function. Furthermore, since the Sox9 conditional mutants did not exhibit defects in retinal neurogenesis, it is formally possible that Sox2 may be able to compensate for the loss of Sox9 resulting in appropriate maintenance of the RPC pool. Whatever the case, our data do not entirely fit within a model whereby Sox9 regulates the Notch pathway, as we obviously did not recapitulate all aspects of the mouse Sox2 or Notch1 loss-of-function phenotypes. However, we believe that definitive confirmation of a role for Sox9 in RPC maintenance awaits a more robust knockout of Sox9.
Sox9 in Müller glial cell development
In this study we showed that postnatal expression of Sox9 is confined to the MG cells. Thus, it is possible that Sox9 directly promotes MG cell specification or regulates MG cell differentiation. In order to test this hypothesis we analyzed Sox9 conditional mutant retinae for expression of MG cell-specific markers. Interestingly, we observed a loss of MG marker p27Kip1 coincident with loss of Sox9 expression. We also observed a loss of Sox2 and GS within the MG cell layer. Our data suggest that MG cells are not present in regions of the mutant retinae that exhibit a profound loss of Sox9. Thus, it is possible that Sox9 directly promotes Müller gliogenesis or is responsible for MG cell differentiation and survival. Consistent with this idea is our finding that loss of Sox9 expression does not affect postnatal retinal neuron populations. An alternative interpretation of our data is that, in the absence of Sox9, MG cells are still present but remain in an undifferentiated state whereby they fail to express markers of differentiated MG cells (such as GS and p27Kip1). While certainly possible, we believe this conclusion to be less likely since we also fail to detect Sox2 expression in retinal patches that lost Sox9. If Sox9-mutant cells remained as undifferentiated progenitor cells, which would have otherwise formed mature MG cells, we would expect these cells to maintain expression of Sox2 (Taranova et al., 2006).
In this article we speculate that Sox2 might be able to compensate for Sox9 function in RPC maintenance. However, it is curious that Sox9 does not appear to compensate for the loss of Sox2 (Taranova et al., 2006). Furthermore, it does not appear that Sox2 can compensate for Sox9 in MG differentiation and/or survival. Further experimental confirmation of this relationship would suggest the role of Sox9 in Müller glia formation or survival is distinct from any role it has in RPCs. Future studies aimed at precisely defining the genetic interaction (if any) between Sox9 and Sox2 will be most informative.
It is also important to note that, in the absence of MG cells, deficiencies in retinal homeostasis would likely result (Bringmann et al., 2006). Nevertheless, we observed no gross retinal degeneration or structural defects. At present, we believe this discrepancy to be due to the mosaic nature of the Chx10-Cre transgene. Perhaps, in the Sox9 mutants, there is a large enough population of MG cells still present that functionally compensate for a partial loss of the MG population. A more thorough knockout of Sox9 would address this possibility.
If Sox9 is indeed functioning in direct MG cell specification, it is possible that some interplay with Notch signaling is also important in this context, as Hes5 has already been implicated in MG fate specification (Furukawa et al., 2000; Hojo et al., 2000). In addition to the Hes-family members, similar results were observed by forced expression of homeodomain transcription factor Rx as well as Notch1 in the rat retina (Furukawa et al., 2000). In these experiments, Rx and Notch1 lead to the induction of MG cell markers. However, it was not clear whether these cells completed the full MG cell differentiation program. The overall interpretation of these results was that Rx lies at the top of a genetic hierarchy which leads to activation of the Notch pathway, thereby promoting MG specification. However, these data can also be interpreted to suggest that MG cells and RPCs are closely related to one another. It is possible that cells that do not downregulate Notch1 in the postnatal retina eventually adopt the MG cell (last cell) fate. It is also possible that, for an RPC to become an MG cell, it must initially downregulate Notch1 whereby it exits the cell cycle and subsequently reactivates Notch1 (once all neuronal populations are specified) in order to directly promote MG fate. In a very recent study this model was tested by acute reactivation of Notch1 in what were described as newly postmitotic retinal cells (Jadhav et al., 2006a). Interestingly, it was shown that a significant subset of these Notch-reactivated cells expressed glial markers and exhibited morphology reminiscent of MG cells. If this model of Notch pathway reactivation in instructing MG cell specification is correct, at least conceptually, Sox9 could fit nicely within such a mechanism. In the developing gonad, before the onset of sex determination, Sox9 protein appears cytoplasmic in both sexes. However, once Sry-induced male sex determination begins, Sox9 is imported into the nucleus in a male-specific manner, thereby regulating genes necessary for testis development (Gasca et al., 2002; Argentaro et al., 2003; Malki et al., 2005). This apparent capacity of the Sox9 protein for nuclear shuttling could provide a rapid regulatory mechanism to control Notch activity. Once all neuronal populations are specified, Sox9 might briefly leave the nucleus of the remaining RPCs which could possibly result in a downregulation of Notch activity. Then Sox9 might reenter the nucleus shortly thereafter leading to a reactivation of Notch activity and specification of the remaining RPCs to an MG cell fate. Consistent with this idea, in the developing retinae, Sox9 expression does not appear to be exclusively nuclear.
Future efforts should be focused on a more complete assessment of Sox9 function by generating conditional mutants with a greater level of recombination. Furthermore, in order to determine whether Sox9 directly promotes Müller gliogenesis, experiments which force ectopic expression of Sox9 in RPCs should be performed in vivo.
Sox9 expression in other ocular cell types
In addition to its expression in retinal cell types, we also observed Sox9 expression in glial cells of the optic nerve. Significantly, conditional ablation of Sox9 in the developing spinal cord revealed defects in the specification of gray matter astrocytes and oligodendrocytes as indicated by severe loss of these glial cell types (Stolt et al., 2003). The authors interpreted these results to mean that Sox9 determines the switch from spinal cord neurogenesis to gliogenesis. Furthermore, the authors also transfected Sox9 into Neuro2a cells and observed coincident induction of glial cell markers, thereby suggesting that Sox9 is instructive for the glial cell fate. This study did not investigate glial cells of the optic nerve, but it is likely that Sox9 is playing a similar role within this context. Furthermore, these findings allude to a possible role for Sox9 in directly specifying MG cell fate in a similar manner to spinal cord glial fate. One other point of interest is that Sox9 expression in the spinal cord ventricular zone precedes the onset of gliogenesis (Stolt et al., 2003). This expression profile is similar to the developing retina where Sox9 is expressed at early stages of retinogenesis well before the generation of MG cells. The significance of this expression pattern as it relates to gliogenesis remains to be further defined.
Another interesting location of Sox9 expression described in this study is within the embryonic and mature RPE. In human melanocytes it was recently reported that SOX9 can directly bind to and regulate the microphthalmia-associated transcription factor (MITF) promoter, thereby increasing the expression of melano-genic proteins leading to the stimulation of pigmentation (Passeron et al., 2007). In addition to its role in the development of neural-crest-derived melanocytes, Mitf is essential for the specification of the RPE versus neuroretina, as both tissues are derived from an initially bipotential optic neuroepithelium. Evidence for this mechanism comes from studies of Mitf mutant mouse embryos that exhibit transdifferentiation of the prospective RPE into a second neuroretina (Hodgkinson et al., 1993; Steingrimsson et al., 1994; Nguyen and Arnheiter, 2000; Bumsted-O’Brien et al., 2007). Our observation that Sox9 is expressed in the developing and mature RPE and the evidence that Sox9 is a direct regulator of Mitf makes it formally possible that Sox9 is playing a role in either RPE specification or pigmentation. Consistent with this idea, a transgene insertion within a 0.98 Mb region upstream of Sox9 results in microphthalmia and pigmentation defects (Bishop et al., 2000; Qin et al., 2004). Future studies aimed at conditional ablation of Sox9 within the RPE should be performed.
ACKNOWLEDGMENTS
We thank Sheldon Rowan and Connie Cepko for the Chx10-GFPCretg/+ mice, and Xiuqian Mu, Steven F. Wang, and William Klein for antibodies. We also thank Paul Overbeek and Naomi Iwai for critical reading of the article and Henry Adams for advice on image acquisition. Finally, we would like to give a very special thanks to David Hawke and the U.T.M.D. Anderson Proteomics Facility.
Grant sponsor: National Eye Institute; Grant numbers: T32 EY07102 (to R.A.P.), RO1 EY013128 (to Y.F.); Grant sponsor: Ben F. Love Endowment (to R.R.B.) and HD30284.
LITERATURE CITED
- Akiyama H, Kim JE, Nakashima K, Balmes G, Iwai N, Deng JM, Zhang Z, Martin JF, Behringer RR, Nakamura T, de Crombrugghe B. Osteo-chondroprogenitor cells are derived from Sox9 expressing precursors. Proc Natl Acad Sci U S A. 2005;102:14665–14670. doi: 10.1073/pnas.0504750102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Argentaro A, Sim H, Kelly S, Preiss S, Clayton A, Jans DA, Harley VR. A SOX9 defect of calmodulin-dependent nuclear import in cam-pomelic dysplasia/autosomal sex reversal. J Biol Chem. 2003;278:33839–33847. doi: 10.1074/jbc.M302078200. [DOI] [PubMed] [Google Scholar]
- Austin CP, Feldman DE, Ida JA, Jr, Cepko CL. Vertebrate retinal ganglion cells are selected from competent progenitors by the action of Notch. Development. 1995;121:3637–3650. doi: 10.1242/dev.121.11.3637. [DOI] [PubMed] [Google Scholar]
- Barnstable CJ, Hofstein R, Akagawa K. A marker of early amacrine cell development in rat retina. Brain Res. 1985;352:286–290. doi: 10.1016/0165-3806(85)90116-6. [DOI] [PubMed] [Google Scholar]
- Belecky-Adams T, Tomarev S, Li HS, Ploder L, McInnes RR, Sundin O, Adler R. Pax-6, Prox 1, and Chx10 homeobox gene expression correlates with phenotypic fate of retinal precursor cells. Invest Ophthalmol Vis Sci. 1997;38:1293–1303. [PubMed] [Google Scholar]
- Belliveau MJ, Cepko CL. Extrinsic and intrinsic factors control the genesis of amacrine and cone cells in the rat retina. Development. 1999;126:555–566. doi: 10.1242/dev.126.3.555. [DOI] [PubMed] [Google Scholar]
- Belliveau MJ, Young TL, Cepko CL. Late retinal progenitor cells show intrinsic limitations in the production of cell types and the kinetics of opsin synthesis. J Neurosci. 2000;20:2247–2254. doi: 10.1523/JNEUROSCI.20-06-02247.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi W, Huang W, Whitworth DJ, Deng JM, Zhang Z, Behringer RR, de Crombrugghe B. Haploinsufficiency of Sox9 results in defective cartilage primordia and premature skeletal mineralization. Proc Natl Acad Sci U S A. 2001;98:6698–6703. doi: 10.1073/pnas.111092198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop CE, Whitworth DJ, Qin Y, Agoulnik AI, Agoulnik IU, Harrison WR, Behringer RR, Overbeek PA. A transgenic insertion upstream of sox9 is associated with dominant XX sex reversal in the mouse. Nat Genet. 2000;26:490–494. doi: 10.1038/82652. [DOI] [PubMed] [Google Scholar]
- Bringmann A, Pannicke T, Grosche J, Francke M, Wiedemann P, Skatch-kov SN, Osborne NN, Reichenbach A. Muller cells in the healthy and diseased retina. Prog Retin Eye Res. 2006;25:397–424. doi: 10.1016/j.preteyeres.2006.05.003. [DOI] [PubMed] [Google Scholar]
- Bumsted-O’Brien KM, Hendrickson A, Haverkamp S, Ashery-Padan R, Schulte D. Expression of the homeodomain transcription factor Meis2 in the embryonic and postnatal retina. J Comp Neurol. 2007;505:58–72. doi: 10.1002/cne.21458. [DOI] [PubMed] [Google Scholar]
- Carter-Dawson LD, LaVail MM. Rods and cones in the mouse retina. II. Autoradiographic analysis of cell generation using tritiated thymi-dine. J Comp Neurol. 1979;188:263–272. doi: 10.1002/cne.901880205. [DOI] [PubMed] [Google Scholar]
- Cepko CL, Austin CP, Yang X, Alexiades M, Ezzeddine D. Cell fate determination in the vertebrate retina. Proc Natl Acad Sci U S A. 1996;93:589–595. doi: 10.1073/pnas.93.2.589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaboissier MC, Kobayashi A, Vidal VI, Lutzkendorf S, van de Kant HJ, Wegner M, de Rooij DG, Behringer RR, Schedl A. Functional analysis of Sox8 and Sox9 during sex determination in the mouse. Development. 2004;131:1891–1901. doi: 10.1242/dev.01087. [DOI] [PubMed] [Google Scholar]
- de Melo J, Qiu X, Du G, Cristante L, Eisenstat DD. Dlx1, Dlx2, Pax6, Brn3b, and Chx10 homeobox gene expression defines the retinal ganglion and inner nuclear layers of the developing and adult mouse retina. J Comp Neurol. 2003;461:187–204. doi: 10.1002/cne.10674. [DOI] [PubMed] [Google Scholar]
- Dyer MA, Cepko CL. Control of Muller glial cell proliferation and activation following retinal injury. Nat Neurosci. 2000a;3:873–880. doi: 10.1038/78774. [DOI] [PubMed] [Google Scholar]
- Dyer MA, Cepko CL. p57(Kip2) regulates progenitor cell proliferation and amacrine interneuron development in the mouse retina. Development. 2000b;127:3593–3605. doi: 10.1242/dev.127.16.3593. [DOI] [PubMed] [Google Scholar]
- Dyer MA, Cepko CL. p27Kip1 and p57Kip2 regulate proliferation in distinct retinal progenitor cell populations. J Neurosci. 2001;21:4259–4271. doi: 10.1523/JNEUROSCI.21-12-04259.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elshatory Y, Deng M, Xie X, Gan L. Expression of the LIM-homeodomain protein Isl1 in the developing and mature mouse retina. J Comp Neurol. 2007;503:182–197. doi: 10.1002/cne.21390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu X, Sun H, Klein WH, Mu X. Beta-catenin is essential for lamination but not neurogenesis in mouse retinal development. Dev Biol. 2006;299:424–437. doi: 10.1016/j.ydbio.2006.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujitani Y, Fujitani S, Luo H, Qiu F, Burlison J, Long Q, Kawaguchi Y, Edlund H, Macdonald RJ, Furukawa T, Fujikado T, Magnuson MA, Xiang M, Wright CV. Ptf1a determines horizontal and amacrine cell fates during mouse retinal development. Development. 2006;133:4439–4450. doi: 10.1242/dev.02598. [DOI] [PubMed] [Google Scholar]
- Furukawa T, Mukherjee S, Bao ZZ, Morrow EM, Cepko CL. rax, Hes1, and notch1 promote the formation of Muller glia by postnatal retinal progenitor cells. Neuron. 2000;26:383–394. doi: 10.1016/s0896-6273(00)81171-x. [DOI] [PubMed] [Google Scholar]
- Furuta Y, Behringer RR. Recent innovations in tissue-specific gene modifications in the mouse. Birth Defects Res C Embryo Today. 2005;75:43–57. doi: 10.1002/bdrc.20036. [DOI] [PubMed] [Google Scholar]
- Furuta Y, Lagutin O, Hogan BL, Oliver GC. Retina- and ventral forebrain-specific Cre recombinase activity in transgenic mice. genesis. 2000;26:130–132. [PubMed] [Google Scholar]
- Gan L, Xiang M, Zhou L, Wagner DS, Klein WH, Nathans J. POU domain factor Brn-3b is required for the development of a large set of retinal ganglion cells. Proc Natl Acad Sci U S A. 1996;93:3920–3925. doi: 10.1073/pnas.93.9.3920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gargini C, Terzibasi E, Mazzoni F, Strettoi E. Retinal organization in the retinal degeneration 10 (rd10) mutant mouse: a morphological and ERG study. J Comp Neurol. 2007;500:222–238. doi: 10.1002/cne.21144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasca S, Canizares J, De Santa Barbara P, Mejean C, Poulat F, Berta P, Boizet-Bonhoure B. A nuclear export signal within the high mobility group domain regulates the nucleocytoplasmic translocation of SOX9 during sexual determination. Proc Natl Acad Sci U S A. 2002;99:11199–11204. doi: 10.1073/pnas.172383099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenberg KP, Geller SF, Schaffer DV, Flannery JG. Targeted trans-gene expression in Muller glia of normal and diseased retinas using lentiviral vectors. Invest Ophthalmol Vis Sci. 2007;48:1844–1852. doi: 10.1167/iovs.05-1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harman AM, Beazley LD. Generation of retinal cells in the wallaby, Setonix brachyurus (quokka) Neuroscience. 1989;28:219–232. doi: 10.1016/0306-4522(89)90246-7. [DOI] [PubMed] [Google Scholar]
- Hatakeyama J, Kageyama R. Retinal cell fate determination and bHLH factors. Semin Cell Dev Biol. 2004;15:83–89. doi: 10.1016/j.semcdb.2003.09.005. [DOI] [PubMed] [Google Scholar]
- Hatakeyama J, Tomita K, Inoue T, Kageyama R. Roles of homeobox and bHLH genes in specification of a retinal cell type. Development. 2001;128:1313–1322. doi: 10.1242/dev.128.8.1313. [DOI] [PubMed] [Google Scholar]
- Haverkamp S, Wassle H. Immunocytochemical analysis of the mouse retina. J Comp Neurol. 2000;424:1–23. [PubMed] [Google Scholar]
- Heinze L, Harvey RJ, Haverkamp S, Wassle H. Diversity of glycine receptors in the mouse retina: localization of the alpha4 subunit. J Comp Neurol. 2007;500:693–707. doi: 10.1002/cne.21201. [DOI] [PubMed] [Google Scholar]
- Hodgkinson CA, Moore KJ, Nakayama A, Steingrimsson E, Copeland NG, Jenkins NA, Arnheiter H. Mutations at the mouse microphthalmia locus are associated with defects in a gene encoding a novel basic-helix-loop-helix-zipper protein. Cell. 1993;74:395–404. doi: 10.1016/0092-8674(93)90429-t. [DOI] [PubMed] [Google Scholar]
- Hojo M, Ohtsuka T, Hashimoto N, Gradwohl G, Guillemot F, Kageyama R. Glial cell fate specification modulated by the bHLH gene Hes5 in mouse retina. Development. 2000;127:2515–2522. doi: 10.1242/dev.127.12.2515. [DOI] [PubMed] [Google Scholar]
- Jadhav AP, Cho SH, Cepko CL. Notch activity permits retinal cells to progress through multiple progenitor states and acquire a stem cell property. Proc Natl Acad Sci U S A. 2006a;103:18998–19003. doi: 10.1073/pnas.0608155103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jadhav AP, Mason HA, Cepko CL. Notch 1 inhibits photoreceptor production in the developing mammalian retina. Development. 2006b;133:913–923. doi: 10.1242/dev.02245. [DOI] [PubMed] [Google Scholar]
- Johnson DA, Donovan SL, Dyer MA. Mosaic deletion of Rb arrests rod differentiation and stimulates ectopic synaptogenesis in the mouse retina. J Comp Neurol. 2006;498:112–128. doi: 10.1002/cne.21059. [DOI] [PubMed] [Google Scholar]
- Kawakami A, Kimura-Kawakami M, Nomura T, Fujisawa H. Distributions of PAX6 and PAX7 proteins suggest their involvement in both early and late phases of chick brain development. Mech Dev. 1997;66:119–130. doi: 10.1016/s0925-4773(97)00097-x. [DOI] [PubMed] [Google Scholar]
- Koike C, Nishida A, Ueno S, Saito H, Sanuki R, Sato S, Furukawa A, Aizawa S, Matsuo I, Suzuki N, Kondo M, Furukawa T. Functional roles of Otx2 transcription factor in postnatal mouse retinal development. Mol Cell Biol. 2007;27:8318–8329. doi: 10.1128/MCB.01209-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Vail MM, Rapaport DH, Rakic P. Cytogenesis in the monkey retina. J Comp Neurol. 1991;309:86–114. doi: 10.1002/cne.903090107. [DOI] [PubMed] [Google Scholar]
- Linser PJ, Sorrentino M, Moscona AA. Cellular compartmentalization of carbonic anhydrase-C and glutamine synthetase in developing and mature mouse neural retina. Brain Res. 1984;315:65–71. doi: 10.1016/0165-3806(84)90077-4. [DOI] [PubMed] [Google Scholar]
- Liu IS, Chen JD, Ploder L, Vidgen D, van der Kooy D, Kalnins VI, McInnes RR. Developmental expression of a novel murine homeobox gene (Chx10): evidence for roles in determination of the neuroretina and inner nuclear layer. Neuron. 1994;13:377–393. doi: 10.1016/0896-6273(94)90354-9. [DOI] [PubMed] [Google Scholar]
- Livesey FJ, Cepko CL. Vertebrate neural cell-fate determination: lessons from the retina. Nat Rev Neurosci. 2001;2:109–118. doi: 10.1038/35053522. [DOI] [PubMed] [Google Scholar]
- Malki S, Nef S, Notarnicola C, Thevenet L, Gasca S, Mejean C, Berta P, Poulat F, Boizet-Bonhoure B. Prostaglandin D2 induces nuclear import of the sex-determining factor SOX9 via its cAMP-PKA phos-phorylation. EMBO J. 2005;24:1798–1809. doi: 10.1038/sj.emboj.7600660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marquardt T, Ashery-Padan R, Andrejewski N, Scardigli R, Guillemot F, Gruss P. Pax6 is required for the multipotent state of retinal progenitor cells. Cell. 2001;105:43–55. doi: 10.1016/s0092-8674(01)00295-1. [DOI] [PubMed] [Google Scholar]
- Nguyen M, Arnheiter H. Signaling and transcriptional regulation in early mammalian eye development: a link between FGF and MITF. Development. 2000;127:3581–3591. doi: 10.1242/dev.127.16.3581. [DOI] [PubMed] [Google Scholar]
- Nishida A, Furukawa A, Koike C, Tano Y, Aizawa S, Matsuo I, Furukawa T. Otx2 homeobox gene controls retinal photoreceptor cell fate and pineal gland development. Nat Neurosci. 2003;6:1255–1263. doi: 10.1038/nn1155. [DOI] [PubMed] [Google Scholar]
- Ohtoshi A, Wang SW, Maeda H, Saszik SM, Frishman LJ, Klein WH, Behringer RR. Regulation of retinal cone bipolar cell differentiation and photopic vision by the CVC homeobox gene Vsx1. Curr Biol. 2004;14:530–536. doi: 10.1016/j.cub.2004.02.027. [DOI] [PubMed] [Google Scholar]
- Ohtsuka T, Ishibashi M, Gradwohl G, Nakanishi S, Guillemot F, Kageyama R. Hes1 and Hes5 as notch effectors in mammalian neuronal differentiation. EMBO J. 1999;18:2196–2207. doi: 10.1093/emboj/18.8.2196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Passeron T, Valencia JC, Bertolotto C, Hoashi T, Le Pape E, Takahashi K, Ballotti R, Hearing VJ. SOX9 is a key player in ultraviolet B-induced melanocyte differentiation and pigmentation. Proc Natl Acad Sci U S A. 2007;104:13984–13989. doi: 10.1073/pnas.0705117104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poche RA, Kwan KM, Raven MA, Furuta Y, Reese BE, Behringer RR. Lim1 is essential for the correct laminar positioning of retinal horizontal cells. J Neurosci. 2007;27:14099–14107. doi: 10.1523/JNEUROSCI.4046-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prada C, Puga J, Perez-Mendez L, Lopez And R, Ramirez G. Spatial and temporal patterns of neurogenesis in the chick retina. Eur J Neurosci. 1991;3:1187. doi: 10.1111/j.1460-9568.1991.tb00053.x. [DOI] [PubMed] [Google Scholar]
- Qin Y, Kong LK, Poirier C, Truong C, Overbeek PA, Bishop CE. Long-range activation of Sox9 in Odd Sex (Ods) mice. Hum Mol Genet. 2004;13:1213–1218. doi: 10.1093/hmg/ddh141. [DOI] [PubMed] [Google Scholar]
- Rapaport DH, Wong LL, Wood ED, Yasumura D, LaVail MM. Timing and topography of cell genesis in the rat retina. J Comp Neurol. 2004;474:304–324. doi: 10.1002/cne.20134. [DOI] [PubMed] [Google Scholar]
- Rowan S, Cepko CL. Genetic analysis of the homeodomain transcription factor Chx10 in the retina using a novel multifunctional BAC transgenic mouse reporter. Dev Biol. 2004;271:388–402. doi: 10.1016/j.ydbio.2004.03.039. [DOI] [PubMed] [Google Scholar]
- Seymour PA, Freude KK, Tran MN, Mayes EE, Jensen J, Kist R, Scherer G, Sander M. SOX9 is required for maintenance of the pancreatic progenitor cell pool. Proc Natl Acad Sci U S A. 2007;104:1865–1870. doi: 10.1073/pnas.0609217104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steingrimsson E, Moore KJ, Lamoreux ML, Ferre-D’Amare AR, Burley SK, Zimring DC, Skow LC, Hodgkinson CA, Arnheiter H, Copeland NG, et al. Molecular basis of mouse microphthalmia (mi) mutations helps explain their developmental and phenotypic consequences. Nat Genet. 1994;8:256–263. doi: 10.1038/ng1194-256. [DOI] [PubMed] [Google Scholar]
- Stiemke MM, Hollyfield JG. Cell birthdays in Xenopus laevis retina. Differentiation. 1995;58:189–193. doi: 10.1046/j.1432-0436.1995.5830189.x. [DOI] [PubMed] [Google Scholar]
- Stolt CC, Lommes P, Sock E, Chaboissier MC, Schedl A, Wegner M. The Sox9 transcription factor determines glial fate choice in the developing spinal cord. Genes Dev. 2003;17:1677–1689. doi: 10.1101/gad.259003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taranova OV, Magness ST, Fagan BM, Wu Y, Surzenko N, Hutton SR, Pevny LH. SOX2 is a dose-dependent regulator of retinal neural progenitor competence. Genes Dev. 2006;20:1187–1202. doi: 10.1101/gad.1407906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomita K, Ishibashi M, Nakahara K, Ang SL, Nakanishi S, Guillemot F, Kageyama R. Mammalian hairy and Enhancer of split homolog 1 regulates differentiation of retinal neurons and is essential for eye morphogenesis. Neuron. 1996;16:723–734. doi: 10.1016/s0896-6273(00)80093-8. [DOI] [PubMed] [Google Scholar]
- Turner DL, Cepko CL. A common progenitor for neurons and glia persists in rat retina late in development. Nature. 1987;328:131–136. doi: 10.1038/328131a0. [DOI] [PubMed] [Google Scholar]
- Wagner KD, Wagner N, Vidal VP, Schley G, Wilhelm D, Schedl A, Englert C, Scholz H. The Wilms’ tumor gene Wt1 is required for normal development of the retina. EMBO J. 2002;21:1398–1405. doi: 10.1093/emboj/21.6.1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang SW, Kim BS, Ding K, Wang H, Sun D, Johnson RL, Klein WH, Gan L. Requirement for math5 in the development of retinal ganglion cells. Genes Dev. 2001;15:24–29. doi: 10.1101/gad.855301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright E, Hargrave MR, Christiansen J, Cooper L, Kun J, Evans T, Gangadharan U, Greenfield A, Koopman P. The Sry-related gene Sox9 is expressed during chondrogenesis in mouse embryos. Nat Genet. 1995;9:15–20. doi: 10.1038/ng0195-15. [DOI] [PubMed] [Google Scholar]
- Yaron O, Farhy C, Marquardt T, Applebury M, Ashery-Padan R. Notch1 functions to suppress cone-photoreceptor fate specification in the developing mouse retina. Development. 2006;133:1367–1378. doi: 10.1242/dev.02311. [DOI] [PubMed] [Google Scholar]
- Young RW. Cell differentiation in the retina of the mouse. Anat Rec. 1985;212:199–205. doi: 10.1002/ar.1092120215. [DOI] [PubMed] [Google Scholar]