Abstract
We demonstrate the ability to spatially pattern biochemical signals into nanofibrous materials using thiol-ene reactions of thiolated molecules to presented norbornene groups. This approach is used to pattern three molecules independently within one scaffold, pattern signals with depth through a scaffold, and to spatially control cell adhesion and morphology.
Keywords: nanofibers, patterning, electrospinning, tissue engineering, alignment
Biomaterials are being developed to investigate and control cellular interactions with their surroundings; however, the majority of engineered systems present signals (e.g. mechanics, topography, adhesion) in a spatially uniform manner, despite the role that spatially controlled signals – both biophysical and biochemical – play in a number of different processes in vivo.[1–3] For example, spatial organization of soluble signals occurs as early as gastrulation when morphogen gradients activate various signaling pathways (e.g. sonic hedgehog, WnT, activin)[4–7] to guide tissue development. This organization continues in adult tissues where spatial regulation of growth factors directly influences processes such as chondrogenesis[8], angiogenesis[9], and immune responses.[10] Beyond soluble factors, spatially controlled signaling of insoluble extracellular matrix (ECM) proteins is also important, as localized fibronectin deposition directs neural crest formation [11] and tumor angiogenesis,[12] vitronectin expression in the ventral neural tube promotes motor neuron differentiation,[13] and spatial alignment of collagen fibers contributes to soft tissue functions of the cornea,[14] articular cartilage,[15] and arterial wall.[16] Notably, these ECM proteins all contain amino acid sequences known to induce integrin-mediated cell adhesion,[17] suggesting a broader role for cell adhesion in spatially dictating cell behavior.
A variety of patterning techniques have been previously developed to engineer materials with precisely defined features, including the patterning of ECM proteins.[18] Studies using microcontact printing, a technique where proteins are “stamped” onto a substrate using a preformed master mold, have indicated that spatial patterning of cell adhesive proteins influences cellular processes including spreading, differentiation, proliferation, and death.[19, 20] Other patterning techniques such as soft lithography,[21] 3-D printing,[22] and microfluidic devices[23] have also been successful in forming patterns and gradients of ECM proteins to control cell-material interactions. In particular, photopatterning has emerged as a promising technique in which a desired reaction (e.g. crosslinking,[24] bond scission,[25] covalent attachment[26]) is spatially controlled to specific regions exposed to light,[27] without associated changes in surface topography that are typical of mechanical techniques like microcontact printing, 3-D printing, and soft lithography. Using photopatterning, hydrogel mechanics,[28, 29] biomolecule attachment,[30] and porosity,[25] have all been spatially patterned in 3-D.
While these material systems have allowed the spatial presentation of a range of biochemical and biophysical features, they are inherently non-fibrous, whereas natural ECM is composed of nanofibrous proteins that provide structural cues to guide cell behavior.[31] Studies have indicated that these structural features including fiber diameter, morphology, alignment, and stiffness affect cell adhesion, migration, differentiation, and proliferation[32–37]. Consequently, structural features have been patterned into nanofibrous materials – often through electrospinning – to control both the morphology of individual fibers (e.g. ribbon,[38] helical coils,[39] hollow core,[40] porous,[41]) and the topography of the entire nanofibrous scaffold. Several methods including selective deposition of fibers through specialized collectors,[42–44] microcontact printing,[45] direct melt writing,[46] photolithography,[47] and dissolution printing[48] have been used to control cell interactions with patterned electrospun fibers. Importantly, patterns formed by these techniques are characterized by differences in topography such that regions exist with and without fibers or with altered fiber orientation and fiber density. These biophysical patterns differ from biochemical patterns or gradients in which the pattern is formed from localization of biomolecules without associated changes in topography. Biochemical patterns may be more indicative of natural ECM signaling that occurs through gradients and spatial localization of biomolecules;[1–3, 48] however, current material systems are limited in their ability to incorporate nanofibrous architectures with spatially regulated biochemical features to investigate cell-material interactions.
In the present work, we utilize a norbornene-functionalized hyaluronic acid (HA, a linear polysaccharide composed of alternating d-glucuronic acid and N-acetyl-d-glucosamine) to generate electrospun nanofibrous hydrogels, spatially pattern biomolecule attachment via thiol-ene chemistry, and show how the unique structural and biochemical features of these scaffolds may be used to guide and investigate cell behavior. Notably, these electrospun scaffolds are formed from hydrophilic precursors, requiring crosslinking to stabilize the structure upon hydration. This results in a scaffold with water-swollen fibers, indicative of a hydrogel that mimics soft-tissue microenvironments. In contrast, commonly electrospun polymers (e.g. polycaprolactone, poly(lactic acid), poly(lactic-co-glycolic acid)) contain semi-crystalline/hydrophobic domains and remain rigid fibers with limited water absorption when placed in aqueous environments.
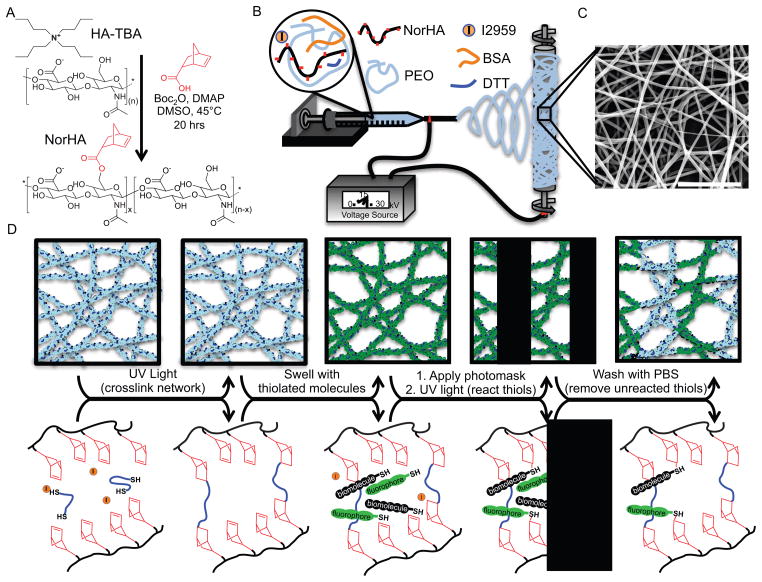
Norbornene-functionalized HA (NorHA, Figure 1A) was synthesized by an esterification of the primary hydroxyl group of HA with 5-norbornene-2-carboxylic acid (~20% of HA repeat units modified with norbornene groups calculated from 1H NMR spectroscopy). HA was chosen as the starting material due to the ease of chemical modification of the repeat structure and naturally low cell adherence in the absence of adsorbed or covalently attached cell-adhesion molecules. To generate nanofibrous hydrogels, solutions of NorHA (3.25 wt%), polyethylene oxide (PEO, 2.75 wt%), bovine serum albumin (BSA, 1 wt%), Irgacure 2959 (I2959 – UV initiator, 0.05 wt%), and dithiothreitol (DTT) in phosphate buffered saline (PBS) were electrospun (Figure 1B) as thin films onto thiolated coverslips (to facilitate scaffold attachment). PEO was included as a carrier polymer to improve fiber formation during electrospinning, and BSA was included to limit non-specific adsorption of thiolated peptides during patterning. SEM imaging of fibers (Figure 1C) confirmed smooth fiber morphology, random fiber orientation, and submicron fiber diameters (220 ± 50 nm). The electrospun scaffolds were crosslinked and patterned with biomolecules via a process outlined in Figure 1D. Compared to the specificity of photoreactive groups like benzophenones, arylazides, and diazirines,[49] norbornene functional groups are specific in their selectivity to thio-radicals as compared to norbornene radicals or non-radical thiols, which permits selective crosslinking (through the number of di-thiols) and patterning (through the number of mono-thiols) of biomolecules in a step-wise process.[50, 51] First, crosslinking was initiated in dry scaffolds by exposure to UV light (320–390 nm) under inert atmosphere. Scaffolds that were not exposed to UV light dissolved upon hydration. By controlling the thiol:norbornene ratio of the initial solution (0.4) via the concentrations of DTT and NorHA, only a portion of the norbornene groups within the fibers were consumed during di-thiol mediated crosslinking, which retained norbornene groups for subsequent patterning. Next, the scaffolds were hydrated in a solution containing the photoinitiator I2959 and a mono-thiolated biomolecule and exposed to UV light through a photomask, permitting thiol-ene reactions at locations exposed to light. Subsequent washes with PBS removed any thiolated biomolecules, yielding a patterned nanofibrous hydrogel.
Figure 1.
Electrospun nanofibrous hydrogel formation, crosslinking, and biochemical ligand patterning. (A) Synthesis of norbornene-hyaluronic acid (NorHA) with norbornene group shown in red. (B) Electrospinning process and (C) morphology of nanofibers post electrospinning (Scale bar: 5μm). (D) Schematic illustrating gross fiber appearance (top row) and the corresponding molecular reactions (bottom row) associated with the steps to crosslink and pattern biochemical ligands in nanofibrous hydrogels. Crosslinking occurs in the dry state via UV light initiated thiol-ene reactions of a di-thiol and norbornene groups on NorHA (to stabilize the nanofibrous structure upon hydration). Subsequent patterning is achieved by exposing scaffolds to UV light through a photomask in the presence of a UV initiator and thiolated biomolecules to react with remaining norbornene groups.
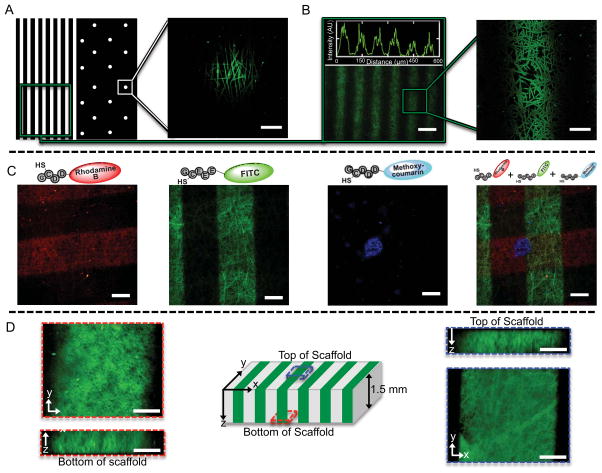
To demonstrate patterning, thiolated peptide fluorophores (Red: GCDD-Rho, Green: GCEEE-FITC, Blue: GCDDD-Methoxycoumarin) were synthesized via solid-phase peptide synthesis and used in all subsequent studies to confirm the photopatterning reaction. When patterned using a photomask with either 50 μm diameter circles or line widths, the measured width of the scaffold pattern was 57 ± 5 μm for circles and 55 ± 6 μm for lines, which is consistent for high fidelity between the photomask and visualized pattern (Figure 2A–B). A horizontal line intensity profile within a patterned region showed intensity peaks and valleys that were consistent with the nanofibrous heterogeneity of the scaffold (Figure 2B). Altering experimental photopatterning conditions (e.g. UV light intensity, biomolecule concentration, exposure time to UV light) allowed for precise control over the surface density of biomolecules (Figure S1, Supporting Information). Hydrated fiber diameters were measured (740 ± 140 nm) by confocal microscopy and confirmed the swollen nature of the nanofibers when compared to the initial dry fiber diameters (220 ± 50 nm). While previous approaches to pattern electrospun scaffolds have focused on altering topography by forming grooves or pores,[42–47, 52] the presented technique spatially patterns biomolecule attachment without changes in surface topography. To ensure that nanofibrous architecture was unchanged between regions with patterned biomolecules (areas in green in Figure 2A–B) and regions without biomolecules (areas in black in Figure 2A–B), an electrospun scaffold was formed with a thiolated peptide-fluorophore in the initial solution so that the hydrated fiber diameters could be measured without any secondary patterning. These scaffolds were swollen in buffer and fiber diameters (730 ± 110 nm) were similar to fiber diameters observed in patterned scaffolds, indicating similar fiber dimensions regardless of the patterned biomolecule attachment.
Figure 2.
Spatial resolution of biomolecules within a nanofibrous scaffold. (A–B) Photomask and corresponding patterns after UV light mediated covalent attachment of a thiolated peptide fluorophore (GCEEE-FITC) to an electrospun NorHA hydrogel. (A) and (B-right) are each one focal plane in a confocal image. (B-left) Intensity profile of a horizontal line across the image. Scale bars: (A,B-right) 25 μm, (B-left) 100 μm. (C) Demonstration of patterning multiple ligands onto the same electrospun NorHA hydrogel. Patterning included 200 μm lines of thiolated peptide fluorophore (GCDD-Rho), 200 μm lines of a second thiolated peptide fluorophore (GCEEE-FITC), and 100 μm circles of a third thiolated peptide fluorophore (GCEE-Methoxycoumarin). The overlaid image demonstrates patterning of three different biomolecules onto the same nanofibrous scaffold. Scale bars: 100 μm. (D) Patterns with depth in a thick scaffold. 200 μm lines were patterned through the top of the scaffold and x-y and z-projections are shown for the top (right) and bottom (left) of the scaffold indicating patterning to a depth of 1.5 mm. Scale bars: 50 μm.
Cellular processes are often influenced by more than one biomolecule,[53, 54] which motivates the localization of several biomolecules into scaffolds for more complex cell-material studies. Therefore, we evaluated the patterning of NorHA scaffolds with multiple biomolecules by photopatterning three thiolated peptide fluorophores. Specifically, GCDD-Rho was patterned onto a scaffold, the scaffold was washed with PBS to remove unreacted thiolated peptides, and the patterning/washing process was repeated for both GCEEE-FITC and GCDDD-Methoxycoumarin. Wide field fluorescent microscopy (Figure 2C) illustrates the patterning of multiple biomolecules, as all three peptide fluorophores retained their distinct pattern within the same scaffold. Notably, regions where the patterns overlap indicate that unreacted norbornene groups were available after initial thiol-ene patterning for subsequent patterning of additional biomolecules.
While the majority of electrospun scaffolds are opaque upon hydration (due to hydrophobic and/or semi-crystalline materials), electrospun NorHA scaffolds transform from opaque films to swollen, translucent hydrogels after crosslinking and hydration (Figure S2, Supporting Information). This property suggests that patterns can be transmitted throughout the bulk of the scaffold. To determine pattern fidelity with depth, thick scaffolds (~1.5 mm thick after hydration) were incubated in a thiolated fluorophore solution for 30 minutes and then photopatterned with 200 μm lines through the entire scaffold. Increased light from out of plane fluorescence and a limited depth of focus prohibited imaging within the interior of the bulk scaffold; however, the scaffold was imaged from the top and bottom surfaces to show the depth of the pattern. Visible heterogeneity from the nanofibrous architecture is visible throughout the scaffold (Figure 2D) with confocal images showing that the pattern was maintained through the thickness of the scaffold, albeit with more diffuse pattern boundaries at the greatest depths from the original light exposure.
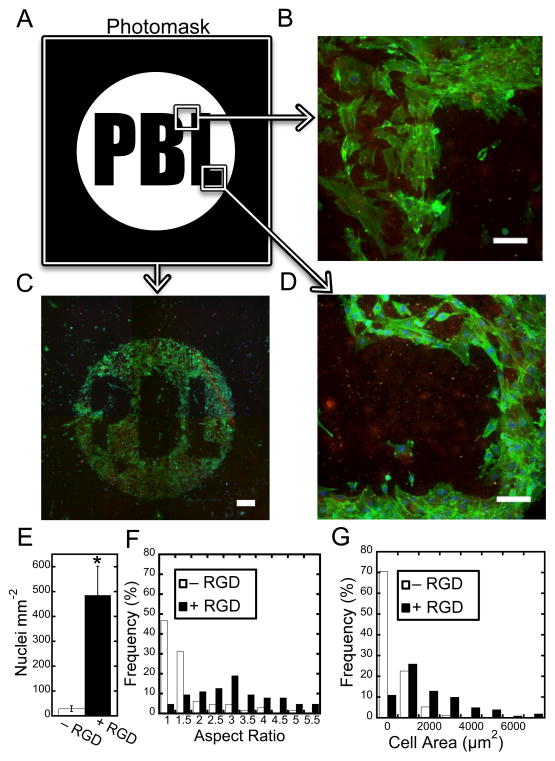
One of the most ubiquitously studied biomolecules in cell-material interactions is RGD, an amino acid sequence found in ECM proteins such as fibronectin, vitronectin, laminin, and collagen that is known to induce integrin-mediated adhesion of cells to surfaces. Given its importance in various cellular processes,[17] we photopatterned regions of RGD in a nanofibrous scaffold to alter cellular adhesion and morphology. A thiolated RGD peptide was included with GCDD-Rho to visually indicate pattern formation (Figure 3A, PBL-Polymeric Biomaterials Lab). 3T3 fibroblasts were then seeded onto scaffolds, cultured for three days, and stained for F-actin (FITC-phalloidin) and nuclei (DAPI). Fluorescent images illustrate cells adhered preferentially to regions containing RGD, replicating the shape of the pattern with high fidelity (Figure 3B–D). Quantification of cell density further confirmed the observed differences in cell adhesion (Figure 3E, S3, Supporting Information). For the limited number of cells that were adherent on regions without RGD, the cell aspect ratio (measure of cell elongation, Figure 3F) and cell area (Figure 3G) was drastically different than regions containing RGD. Cells were considerably more elongated with larger cell areas in regions containing RGD compared to regions without RGD, agreeing with previous studies of cell adhesion on isotropic, non-fibrous hydrogels containing RGD.[55, 56] Images of scaffolds prior to and after incubation in 3T3 growth media for three days (Figure S4, Supporting Information) indicated that fiber morphology was unaltered for the duration of the study. This general cell behavior was also observed in scaffolds seeded with human umbilical vein endothelial cells (HUVECs –Figure S5, Supporting Information), offering evidence for the broad applicability of the material system to investigate other cell types.
Figure 3.
Patterning of biochemical ligands alters cell attachment and morphology. (A) Photomask (white areas indicate areas permitting transmittance of UV light) used to covalently attach thiolated RGD to the nanofibrous scaffold (PBL – Polymeric Biomaterials Lab). (B–D) 3T3 fibroblasts adhere to the RGD pattern with high fidelity. Scale bars: (B,D) 100 μm, (C) 500 μm. Quantification of the number of cells per area (E), aspect ratio (F), and cell area (G) shows differences in cell adhesion, elongation, and morphology between nanofiber regions patterned with RGD and nanofiber regions without RGD. (p<0.001)
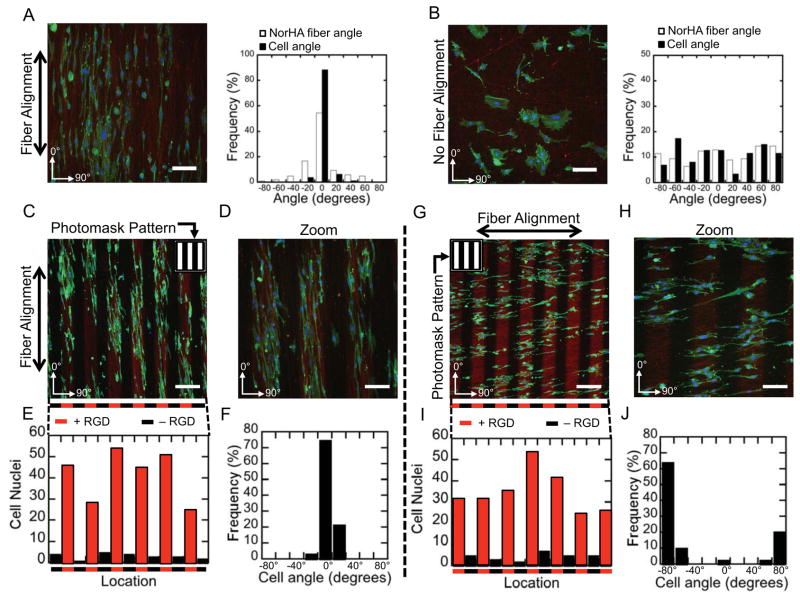
In addition to spatially regulated biomolecule attachment, the nanofibrous architecture of electrospun NorHA scaffolds facilitates control of cell morphology through structural cues. To evaluate the influence of these structural features, scaffolds were generated with unaligned nanofibers (random fiber orientation as in previous examples) or aligned nanofibers (formed through increased mandrel speed during fiber collection), and thiolated RGD was uniformly presented throughout the scaffold. The angles of the longitudinal axis of fibers were measured for scaffolds and confirmed alignment or random orientation depending on the speed of the mandrel during fiber collection (Figure 4A, B). Cells were preferentially elongated in the direction of fiber alignment (Figure 4A), whereas cells were randomly oriented in the absence of fiber alignment (Figure 4B). This ability to modulate cell alignment via structural features was then combined with the ability to spatially regulate RGD attachment. Specifically, 100 μm lines of RGD were patterned parallel to fiber alignment or perpendicular to fiber alignment. When RGD was patterned parallel to fiber alignment, cells elongated in the direction of fiber alignment and localized into regions containing RGD (Figure 4C–F). Interestingly, when RGD was patterned perpendicular to fiber alignment, cells maintained their preferential elongation in the direction of fiber alignment and localized into patterned regions containing RGD (Figure 4G–J). Thus, cell morphology and elongation may be tuned independent of the spatial localization of cells. The observation of cells maintaining elongated morphology in the direction of nanofiber alignment when presented with a perpendicular pattern of cell adhesion further points to the importance of including structural features with spatially localized biochemical features in studies investigating cell behavior.
Figure 4.
Altered cell behavior in the presence of topographical cues and spatial patterning of RGD. (A) Cells orient and elongate with aligned nanofibrous topography on NorHA scaffolds with uniform RGD, (B) whereas cell orientation and elongation is abrogated in the absence of aligned nanofibers. Scale bars: 100 μm. (C–D) Cells orient and elongate with aligned nanofibrous topography on 100 μm wide cell adhesive lines of RGD parallel to nanofiber orientation. Scale bars: (C) 200 μm, (D) 100 μm. (E) Counts of nuclei (DAPI) as a function of horizontal position, and (F) angle of cells from image (C). (G–H) Cells elongate and orient with aligned nanofibrous topography (horizontal) but are spatially restricted (vertically) when RGD is patterned in 100 μm lines perpendicular to nanofiber orientation. Scale bars: (G) 200 μm, (H) 100 μm. (I) Counts of nuclei (DAPI) as a function of horizontal position, and (J) angle of cells from image (G). Staining in all images: Green: F-Actin (FITC-Phalloidin), Blue: Nuclei (DAPI), Red: RGD (GCDD-Rho).
In this communication we present a novel technique to generate, crosslink, and spatially pattern electrospun hydrogels with biomolecules to guide cell behavior. Patterns with spatial resolution as low as 50 μm were achieved and the technique was amenable to patterning multiple biomolecules into a single scaffold. Smaller patterns may be possible with improved photomasks or other patterning techniques (e.g., multi-photon patterning). Patterns were not limited to the surface of scaffolds as patterns were able to form through the depth of thicker scaffolds (~1.5 mm). Cell adhesion and morphology were modulated by spatially patterned RGD such that cell density, elongation, and area increased in regions with RGD, while cells retained their orientation with nanofiber alignment within patterned regions. Importantly, this platform allows for independent control of biophysical cues (nanofibrous structure, alignment) and spatially altered biochemical cues (RGD, thiolated peptide) such that more complex cell-material interactions may be investigated in a synthetically produced material system. The technology is amenable to a range of other signals, such as fiber mechanics, the incorporation of electrostatic interactions, and the binding of full proteins (after diffusion into the scaffold), in a spatially patterned manner.
Experimental Section
NorHA synthesis and electrospinning parameters
Experimental procedures for NorHA synthesis, thiolated peptide fluorophore synthesis, and electrospun scaffold formation are provided in the Supporting Information.
Crosslinking and patterning of NorHA nanofibrous hydrogels
To crosslink and attach fibers to a glass substrate for imaging and cell culture experiments, NorHA was electrospun (~50 μm thick after swelling) onto thiolated glass coverslips (Supporting Information) and exposed (in the dry state) to UV light (Omnicure s1000 – 10 mW cm−2, 320–390 nm) for 15 minutes under inert atmosphere (N2). For photopatterning of thick scaffolds, fibers were collected as free-swelling films and exposed to equivalent crosslinking and patterning conditions as fibers collected on thiolated glass. Scaffolds were sterilized via germicidal UV light for 45 minutes and maintained under sterile conditions thereafter. Scaffolds were then inverted and hydrated face down into a custom-built shallow well (1.5 ml) with a solution containing the thiolated molecule to be patterned. For non-cell culture experiments, solutions contained thiolated peptide fluorophore (0.25 mM GCDD-Rho, 0.25 mM GCEEE-FITC, or 2.5 mM GCDDD-Methoxycoumarin), 1 wt% bovine serum albumin (BSA – to limit non-specific attachment of thiolated peptides), and 0.05% (v/v) I2959 in PBS. For cell-culture studies, 0.5 mM thiolated RGD (sequence: GCGYGRGDSPG, GenScript) was included as a cell adhesive biomolecule and 0.25 mM GCDD-Rho was included to indicate RGD location. Photomask transparencies (CAD/Art Services, Inc.) were placed on the backside of the inverted coverslip and the scaffold was irradiated through the photomask and glass coverslip for 90 seconds with UV light (10 mW cm−2). Patterned scaffolds were moved into individual wells of a 6 well plate, incubated at 37°C in PBS, and washed three times daily for two days to remove any unreacted thiolated molecules. The ability to control surface density of biomolecules was demonstrated by altering UV intensity, fluorophore concentration, or exposure time to UV light while keeping all other conditions equivalent and analyzing fluorescence intensity of 10 patterned sections (Figure S1, Supporting Information). To demonstrate the patterning of multiple ligands, previously patterned scaffolds (after several washes) were again inverted face down into a well of a different thiolated fluorophore and the photopatterning process (UV irradiation and washing) was repeated to sequentially introduce thiolated peptide fluorophores.
Scaffold characterization
To image electrospun NorHA hydrogels, fibers were collected onto aluminum foil and imaged (dry) with a FEI Quanta 600 environmental scanning electron microscope (SEM), or collected onto thiolated coverslips and imaged (hydrated) using confocal laser fluorescent microscopy (Zeiss LSM 510 Meta Confocal Microscope) or wide field fluorescent microscopy (Olympus BX51). To determine fiber diameters, scaffolds were imaged in 6 distinct scaffold areas and fiber diameters were quantified using Image J (>25 fibers per image, 63× magnification – confocal, 9500× magnification – SEM). Fluorescence intensity profiles were generated by drawing a horizontal line across images and analyzing pixel intensity using ImageJ. Fiber angle was quantified by measuring the direction of individual fibers (40× wide field, >20 fibers per image, >150 fibers) in relation to a standard vertical line arbitrarily set at 0 degrees.
Cell culture
NIH 3T3 fibroblasts or HUVECs (Lonza) were seeded at a density of 5,000–10,000 cells cm−2 in growth media (3T3 – DMEM 1× +Glutamax supplemented with 10% fetal bovine serum and 1% penicillin/streptomycin: HUVECs – EGM™-2 Bulletkit™, Lonza), rinsed with PBS after 30 minutes incubation at 37°C, and cultured for 3 days in growth media (refreshed on day 2). Cells were fixed in 4% formalin and stained for F-actin (FITC-phalloidin, Life Technologies) and nuclei (DAPI, Life Technologies) and imaged by wide field fluorescent microscopy. Cell counts per area (nuclei mm−2) were measured by thresholding images of nuclei (20× magnification, DAPI) in ImageJ and analyzing the number of nuclei via the built in function in ImageJ (6 distinct images per region of interest: – RGD, + RGD). Cell aspect ratio, defined as the maximum orthogonal length of a cell to the width of each cell, was manually measured in ImageJ (20× magnification, >15 cells per image, >60 cells per region of interest) and reported as a histogram. Cell area was measured by thresholding images (F-actin, nuclei) in ImageJ and analyzing the area of individual cells via the built in function in ImageJ (20× magnification, >15 cells per image, >60 cells per region of interest).
Statistical analysis
Statistical differences between compared groups were determined using single factor ANOVA with a p value less than 0.05 indicating significance.
Supplementary Material
Acknowledgments
The authors acknowledge funding from a National Science Foundation graduate research fellowship (RJW) and a MRSEC grant at the University of Pennsylvania, as well as National Institutes of Health grants R01 HL107938 and R01 AR056624.
Contributor Information
Ryan J. Wade, Department of Materials Science and Engineering, Department of Bioengineering, University of Pennsylvania, Philadelphia, PA 19103, USA
Ethan J. Bassin, Department of Bioengineering, University of Pennsylvania, Philadelphia, PA 19103, USA
Prof. William M. Gramlich, Department of Chemistry, University of Maine, Orono, ME 04469, USA
Prof. Jason A. Burdick, Department of Bioengineering, Department of Materials Science and Engineering, University of Pennsylvania, Philadelphia, PA 19103, USA
References
- 1.Lutolf M, Hubbell J. Nat Biotechnol. 2005;23:47. doi: 10.1038/nbt1055. [DOI] [PubMed] [Google Scholar]
- 2.Stevens MM, George JH. Science. 2005;310:1135. doi: 10.1126/science.1106587. [DOI] [PubMed] [Google Scholar]
- 3.Burdick JA, Murphy WL. Nat Commun. 2012;3:1269. doi: 10.1038/ncomms2271. [DOI] [PubMed] [Google Scholar]
- 4.Harfe BD, Scherz PJ, Nissim S, Tian H, McMahon AP, Tabin CJ. Cell. 2004;118:517. doi: 10.1016/j.cell.2004.07.024. [DOI] [PubMed] [Google Scholar]
- 5.Gao B, Song H, Bishop K, Elliot G, Garrett L, English MA, Andre P, Robinson J, Sood R, Minami Y. Dev Cell. 2011;20:163. doi: 10.1016/j.devcel.2011.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gurdon J, Harger P, Mitchell A, Lemaire P. 1994 doi: 10.1038/371487a0. [DOI] [PubMed] [Google Scholar]
- 7.Gurdon J, Bourillot P-Y. Nature. 2001;413:797. doi: 10.1038/35101500. [DOI] [PubMed] [Google Scholar]
- 8.Heldin C-H, Miyazono K, Ten Dijke P. Nature. 1997;390:465. doi: 10.1038/37284. [DOI] [PubMed] [Google Scholar]
- 9.Ruhrberg C, Gerhardt H, Golding M, Watson R, Ioannidou S, Fujisawa H, Betsholtz C, Shima DT. Genes Dev. 2002;16:2684. doi: 10.1101/gad.242002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rot A, von Andrian UH. Annu Rev Immunol. 2004;22:891. doi: 10.1146/annurev.immunol.22.012703.104543. [DOI] [PubMed] [Google Scholar]
- 11.Bronner-Fraser M. Dev Biol. 1986;117:528. doi: 10.1016/0012-1606(86)90320-9. [DOI] [PubMed] [Google Scholar]
- 12.Kim S, Bell K, Mousa SA, Varner JA. The American journal of pathology. 2000;156:1345. doi: 10.1016/s0002-9440(10)65005-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Martínez-Morales JR, Barbas JA, Martí E, Bovolenta P, Edgar D, Rodríguez-Tébar A. Development. 1997;124:5139. doi: 10.1242/dev.124.24.5139. [DOI] [PubMed] [Google Scholar]
- 14.Maurice DM. The Journal of physiology. 1957;136:263. doi: 10.1113/jphysiol.1957.sp005758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim IL, Mauck RL, Burdick JA. Biomaterials. 2011;32:8771. doi: 10.1016/j.biomaterials.2011.08.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gasser TC, Ogden RW, Holzapfel GA. J Royal Soc Interface. 2006;3:15. doi: 10.1098/rsif.2005.0073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ruoslahti E, Pierschbacher MD. Science. 1987;238:491. doi: 10.1126/science.2821619. [DOI] [PubMed] [Google Scholar]
- 18.Khademhosseini A, Langer R, Borenstein J, Vacanti JP. Proc Natl Acad Sci USA. 2006;103:2480. doi: 10.1073/pnas.0507681102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen CS, Mrksich M, Huang S, Whitesides GM, Ingber DE. Science. 1997;276:1425. doi: 10.1126/science.276.5317.1425. [DOI] [PubMed] [Google Scholar]
- 20.McBeath R, Pirone DM, Nelson CM, Bhadriraju K, Chen CS. Dev Cell. 2004;6:483. doi: 10.1016/s1534-5807(04)00075-9. [DOI] [PubMed] [Google Scholar]
- 21.Kane RS, Takayama S, Ostuni E, Ingber DE, Whitesides GM. Biomaterials. 1999;20:2363. doi: 10.1016/s0142-9612(99)00165-9. [DOI] [PubMed] [Google Scholar]
- 22.Pati F, Jang J, Ha D-H, Kim SW, Rhie J-W, Shim J-H, Kim D-H, Cho D-W. Nat Commun. 2014:5. doi: 10.1038/ncomms4935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jeon NL, Dertinger SK, Chiu DT, Choi IS, Stroock AD, Whitesides GM. Langmuir. 2000;16:8311. [Google Scholar]
- 24.Khetan S, Burdick JA. Biomaterials. 2010;31:8228. doi: 10.1016/j.biomaterials.2010.07.035. [DOI] [PubMed] [Google Scholar]
- 25.DeForest CA, Anseth KS. Nat Chem. 2011;3:925. doi: 10.1038/nchem.1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.DeForest CA, Polizzotti BD, Anseth KS. Nat Mater. 2009;8:659. doi: 10.1038/nmat2473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Khetan S, Burdick JA. Soft Matter. 2011;7:830. [Google Scholar]
- 28.Guvendiren M, Perepelyuk M, Wells RG, Burdick JA. Journal of the mechanical behavior of biomedical materials. 2013 doi: 10.1016/j.jmbbm.2013.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gramlich WM, Kim IL, Burdick JA. Biomaterials. 2013;34:9803. doi: 10.1016/j.biomaterials.2013.08.089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hahn MS, Miller JS, West JL. Adv Mater. 2006;18:2679. [Google Scholar]
- 31.Wade RJ, Burdick JA. Mater Today. 2012;15:454. [Google Scholar]
- 32.Doyle AD, Wang FW, Matsumoto K, Yamada KM. J Cell Biol. 2009;184:481. doi: 10.1083/jcb.200810041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Oh S, Brammer KS, Li YJ, Teng D, Engler AJ, Chien S, Jin S. Proc Natl Acad Sci. 2009;106:2130. doi: 10.1073/pnas.0813200106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Christopherson GT, Song H, Mao H-Q. Biomaterials. 2009;30:556. doi: 10.1016/j.biomaterials.2008.10.004. [DOI] [PubMed] [Google Scholar]
- 35.Kim IL, Khetan S, Baker BM, Chen CS, Burdick JA. Biomaterials. 2013;34:5571. doi: 10.1016/j.biomaterials.2013.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dang JM, Leong KW. Adv Mater. 2007;19:2775. doi: 10.1002/adma.200602159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sundararaghavan HG, Saunders RL, Hammer DA, Burdick JA. Biotechnol Bioeng. 2013;110:1249. doi: 10.1002/bit.24788. [DOI] [PubMed] [Google Scholar]
- 38.Rockwood DN, Chase DB, Akins RE, Jr, Rabolt JF. Polymer. 2008;49:4025. [Google Scholar]
- 39.Kessick R, Tepper G. Applied physics letters. 2004;84:4807. [Google Scholar]
- 40.Li D, Xia Y. Nano Lett. 2004;4:933. [Google Scholar]
- 41.Lin J, Ding B, Yu J. ACS Appl Mater. 2010;2:521. doi: 10.1021/am900736h. [DOI] [PubMed] [Google Scholar]
- 42.Zhang D, Chang J. Nano Lett. 2008;8:3283. doi: 10.1021/nl801667s. [DOI] [PubMed] [Google Scholar]
- 43.Ding Z, Salim A, Ziaie B. Langmuir. 2009;25:9648. doi: 10.1021/la901924z. [DOI] [PubMed] [Google Scholar]
- 44.Liu Y, Zhang L, Li H, Yan S, Yu J, Weng J, Li X. Langmuir. 2012;28:17134. doi: 10.1021/la303490x. [DOI] [PubMed] [Google Scholar]
- 45.Shi J, Wang L, Chen Y. Langmuir. 2009;25:6015. doi: 10.1021/la900811k. [DOI] [PubMed] [Google Scholar]
- 46.Brown TD, Dalton PD, Hutmacher DW. Adv Mater. 2011;23:5651. doi: 10.1002/adma.201103482. [DOI] [PubMed] [Google Scholar]
- 47.Lee HJ, Kim H-S, Kim HO, Koh W-G. Lab Chip. 2011;11:2849. doi: 10.1039/c1lc20186g. [DOI] [PubMed] [Google Scholar]
- 48.Jia C, Yu D, Lamarre M, Leopold PL, Teng YD, Wang H. Adv Mater. 2014:n/a. doi: 10.1002/adma.201403509. [DOI] [PubMed] [Google Scholar]
- 49.Dubinsky L, Krom BP, Meijler MM. Bioorg Med Chem. 2012;20:554. doi: 10.1016/j.bmc.2011.06.066. [DOI] [PubMed] [Google Scholar]
- 50.Reddy SK, Cramer NB, Bowman CN. Macromolecules. 2006;39:3681. [Google Scholar]
- 51.Hoyle CE, Bowman CN. Angewandte Chemie International Edition. 2010;49:1540. doi: 10.1002/anie.200903924. [DOI] [PubMed] [Google Scholar]
- 52.Liu Y, Zhang L, Wei J, Yan S, Yu J, Li X. J Mater Chem B. 2014;2:3029. doi: 10.1039/c3tb21779e. [DOI] [PubMed] [Google Scholar]
- 53.Avigdor A, Goichberg P, Shivtiel S, Dar A, Peled A, Samira S, Kollet O, Hershkoviz R, Alon R, Hardan I. Blood. 2004;103:2981. doi: 10.1182/blood-2003-10-3611. [DOI] [PubMed] [Google Scholar]
- 54.Massagué J, Wotton D. The EMBO journal. 2000;19:1745. doi: 10.1093/emboj/19.8.1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shu XZ, Ghosh K, Liu Y, Palumbo FS, Luo Y, Clark RA, Prestwich GD. J Biomed Mater Res A. 2004;68:365. doi: 10.1002/jbm.a.20002. [DOI] [PubMed] [Google Scholar]
- 56.Hersel U, Dahmen C, Kessler H. Biomaterials. 2003;24:4385. doi: 10.1016/s0142-9612(03)00343-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.