Abstract
Background
Previous studies suggest that the relationship between genetic risk and depression may be moderated by stressful life events (SLEs). The goal of this study was to assess whether SLEs moderate the association between polygenic risk of Major Depressive Disorder (MDD) and depressive symptoms in older adults.
Methods
We used logistic and negative binomial regressions to assess the associations between polygenic risk, SLEs and depressive symptoms in a sample of 8,761 participants from the Health and Retirement Study (HRS). Polygenic scores were derived from the Psychiatric Genomics Consortium (PGC) genome-wide association study (GWAS) of MDD. SLEs were operationalized as a dichotomous variable indicating whether participants had experienced at least 1 stressful event during the previous two years. Depressive symptoms were measured using an 8-item CES-D subscale and operationalized as both a dichotomous and a count variable.
Results
The odds of reporting ≥ 4 depressive symptoms were over twice as high among individuals who experienced at least one SLE (OR = 2.19, 95% CI = [1.86, 2.58]). Polygenic scores were significantly associated with depressive symptoms (β = .21, p = <.0001), although the variance explained was modest (Pseudo r2 = .0095). None of the interaction terms for polygenic scores and SLEs were statistically significant.
Conclusions
Polygenic risk and SLEs are robust, independent predictors of depressive symptoms in older adults. Consistent with an additive model, we found no evidence that SLEs moderated the association between common variant polygenic risk and depressive symptoms.
Keywords: depression, stress, genetics, polygenic score analysis
Introduction
Depression is known to have a measurable genetic component, with heritability estimated at 30–40% (Sullivan et al. 2000). But specific genes or variants that confer risk for depression have proven difficult to identify and/or replicate, despite multiple attempts using genome-wide association studies (GWAS) with large sample sizes and different characterizations of the phenotype (Bosker et al. 2011; Hek et al. 2013; Muglia et al. 2010; Rietschel et al. 2010; Ripke et al. 2013; Shi et al. 2011; Shyn et al. 2011; Sullivan et al. 2009; Terracciano et al. 2010; Wray et al. 2012). There is growing consensus that complex psychiatric disorders, including major depressive disorder (MDD), have a significant polygenic risk component, in which genetic liability is conferred by the combined additive effects of large numbers of variants with small effect sizes (Sullivan et al. 2012; Lee et al. 2013). Polygenic risk can be characterized in a genome-wide association study (GWAS) by creating a summary score of weighted allelic dosage across all single nucleotide polymorphisms (SNPs) associated with the outcome at a pre-specified significance threshold (Purcell et al. 2009). Previous GWAS studies using polygenic scores to characterize genetic liability for depression found that they explain between 0.2–1% of the variance (Chang et al. 2014; Demirkan et al. 2011; Ripke et al. 2013), suggesting that polygenic risk (as captured by current GWAS) has only a modest effect on depression. However these studies did not take into account the potential contribution of gene-x-environment (GXE) interactions.
A number of previous studies have found evidence suggesting that genetic factors may interact with stressful life events (SLEs) to increase the risk of depression by a larger amount than one would expect by the combined effects of each factor separately. In an early twin study, Kendler et al. (1995) found that exposure to a severe SLE increased the risk of depression to a larger degree among individuals with high genetic predisposition, compared with individuals with low genetic predisposition. In a widely publicized candidate gene study, Caspi et al. (2003) found that the effect of a functional polymorphism on the promoter region of the serotonin transporter gene (5-HTTPR) on depression risk increased with the number of SLEs. The findings of this study generated a great deal of interest in the 5-HTTPR-x-stress interaction as a potential contributing factor in the etiology of depression. Numerous attempts to replicate this finding over the past ten years yielded inconsistent results, however (Karg et al. 2011; Munafò et al. 2009; Risch et al. 2009), which has generated a great deal of discourse and conflicting opinions surrounding the validity or exact nature of this association (Brown & Harris 2008; Caspi et al. 2010; Uher & McGuffin 2010). Additional studies have found evidence supporting possible interactions between SLEs and genetic polymorphisms on other candidate genes, including the brain-derived neurotrophic factor (BDNF) gene (Bukh et al. 2009; Chen et al. 2012; Hosang et al. 2014; Kim et al. 2007), the dopamine receptor D2 (DRD2) gene (Elovainio et al. 2007), the corticotropin-releasing-hormone receptor type 1 (CRHR1) gene (Liu et al. 2013), the catechol-O-methyltransferase (COMT) gene (Mandelli et al. 2007), galanin and galanin receptor (GAL, GALR2-3) genes (Juhasz et al. 2014) and the hsp90 co-chaperone binding protein (FKBP5) gene (Lavebratt et al. 2010; Zimmermann et al. 2011). This suggests that the stress-x-gene interaction may involve a broad array of genetic factors beyond the 5-HTTPR locus.
Only one previous study has examined the interaction between polygenic risk (likely a better index of the overall genetic liability to depression than candidate genes) and SLEs (Peyrot et al. 2014). Peyrot and colleagues examined whether a history of childhood trauma moderated the association between polygenic risk scores and major depressive disorder in a sample of adult individuals from the Netherlands Study of Depression and Anxiety (NESDA). They found evidence for an interaction in that the effect of polygenic scores on major depression risk was stronger among individuals who reported a history of childhood trauma. To the best of our knowledge, no previous study has evaluated the extent to which either recent SLEs or SLEs experienced during adulthood moderate the association between polygenic risk and depression. The goal of the current study was to test the hypothesis that recent SLEs (experienced within the previous 2 years) moderate the association between polygenic risk scores and depressive symptoms among older adults. To accomplish this, we used data from the Psychiatric Genomics Consortium (Sullivan 2010), to derive polygenic scores in the Health and Retirement Study (HRS) and tested the association between these scores, SLEs and depressive symptoms.
Methods
Study Participants
The HRS is a longitudinal panel study designed to assess the effects of ageing and retirement on the physical and mental health of community dwelling older U.S. adults (HRS 2004). The HRS is sponsored by the National Institute on Aging (NIA; grant # NIA U01AG009740) and is conducted by the University of Michigan (Juster & Suzman 1995). The study began in 1992 with a nationally representative sample of older adults ages 50–60. In 1998 it expanded to incorporate additional cohorts of older adults, and since then new cohorts have been added at regular intervals to maintain the overall age range of the sample. At present the HRS consists of approximately 26,000 individuals between the ages of 50 and 90 years old. Information on a wide variety of physical, psychological, cognitive, social and economic variables is collected every 2 years (NIA 2007). Saliva samples were collected in 2006 and 2008, and DNA was obtained for GWAS.
For the current study, phenotype information was drawn from the year 2000 interview and genotype data was obtained from the HRS GWAS. The year 2000 interview was chosen both to maximize overlap with the genotyped sample and, as no new cohorts were added during that wave, to make it easier to establish the temporal relationship between SLEs and depressive symptoms. Participants were included in the current study if they met the following: 1) were present for the year 2000 interview and provided data on depressive symptoms and SLEs, 2) provided a saliva sample and had GWAS data available, 3) were genetically unrelated to all other HRS participants, and 4) reported no history of stroke, transient ischemic attack or memory related disease. A total of 8,761 participants meeting these criteria were included in the study sample.
Measures
Depression
The HRS includes an 8-item sub-scale of the Center for Epidemiological Studies Depression Scale (CES-D). This subscale has been used in previous studies to measure depression in older adults, and has demonstrated good internal consistency (Cronbach's α = .78) (Turvey et al.1999). We operationalized depression in two ways: as a count variable representing number of symptoms (range = 0 – 8) and as a dichotomous variable (1 = depressed, 0 = not depressed) with a cut-point of ≥ 4 symptoms. This cut-point was chosen because it corresponds to a score of 16 on the full CES-D scale, which indicates clinically relevant depressive symptoms (Steffick 2000).
Stressful life events
The HRS does not include any validated SLE scales or inventories, however it does include a number of questions related to health, job loss, bereavement and other significant life events, which correspond to items on well-validated stress measures such as the Social Readjustment Rating Scale (Holmes & Rahe 1967). We tested the associations between 13 of these events and clinically relevant depressive symptoms individually and used events associated with depressive symptoms at the p < .05 level to create a composite measure for evaluating moderation (See Table 1). As the number of individuals who reported experiencing more than 1 SLE within the previous two year period was small (n = 63) relative to the numbers of participants who reported 0 or 1 SLEs, we operationalized SLEs as a dichotomous variable: 0 = no SLEs and 1 = ≥ 1 SLE. We explored the possibility of weighting SLEs by their individual associations with depressive symptoms, however this approach did not improve the measure's ability to explain the variation in depression so for the sake of clarity and ease of interpretation we proceeded with the unweighted variable.
Table 1.
Associations between individual stressful life events and depression
| CES-D8 ≥ 4 (N=1,079) | CES-D8 < 4 (N = 7,682) | ||
|---|---|---|---|
|
|
|||
| Stressful life event | n (%) | n (%) | OR [95% CI] |
| Death of spouse | 43 (3.99) | 155 (2.02) | 2.00 [1.41, 2.84] * |
| Death of parent | 59 (5.47) | 277 (3.61) | 1.55 [1.17, 2.05] * |
| Death of parent-in-law | 31 (2.87) | 207 (2.70) | 1.13 [0.78, 1.63] |
| Heart attack | 24 (2.22) | 103 (1.34) | 1.67 [1.06, 2.65] * |
| Cancer | 24 (2.22) | 157 (2.04) | 1.06 [0.68, 1.65] |
| Nursing home stay | 8 (0.74) | 28 (0.36) | 1.71 [0.76,3.84] |
| Onset of disability | 66 (6.12) | 109 (1.42) | 4.42 [3.22, 6.06] * |
| Serious injury | 54 (5.00) | 211 (2.75) | 1.87 [1.37, 2.56] * |
| Retirement | 100 (9.27) | 764 (9.95) | 0.95 [0.76, 1.19] |
| Job loss | 10 (0.93) | 63 (0.82) | 1.13 [0.58, 2.23] |
| Marriage | 9 (0.83) | 45 (0.59) | 1.25 [0.60, 2.63] |
| Divorce | 19 (1.76) | 38 (0.49) | 3.71 [2.10, 6.57] * |
| Residential move | 96 (8.90) | 720 (9.37) | 0.88 [0.70, 1.11] |
Abbreviations: HRS = Heath and Retirement Study, OR = odds ratio, CI = confidence interval, CES-D8 = Centers for Epidemiologic Studies Depression Scale, 8 item version. Stressful life events occurred within the previous 2-year period. Events in bold were included in the composite measured used to test for moderation. Odds ratios compare the odds of depression among individuals who experienced the event to the odds of depression among individuals who did not experience the event.
p < .05
Polygenic Risk
Polygenic risk was measured using the polygenic score approach described by Purcell et al. (2009). This approach requires a discovery dataset and a target dataset. The discovery, or training, dataset is used to identify individual SNPs associated with the outcome at a chosen p value threshold: pT. Results from the discovery dataset are then used to derive polygenic scores in the target dataset. In this approach, the polygenic score is a weighted sum of all of the alleles that either confer risk for, or are protective against, the outcome of interest, significant at the pT threshold. For this study we used the results of the combined GWAS of MDD carried out by the Psychiatric Genomics Consortium (PGC-1) (Ripke et al. 2013) as our discovery dataset and the HRS as our target dataset.
The PGC (Sullivan 2010) is a consortium of research groups around the world carrying out combined analyses of existing GWAS to identify common variants associated with 5 different psychiatric disorders. The combined PGC MDD GWAS included 9 samples (Ising et al. 2009; Lewis et al. 2010; Muglia et al. 2010; Rietschel et al. 2010; Shi et al. 2011; Shyn et al. 2011; Sullivan et al. 2009; Wray et al. 2012) with a total of 9,240 MDD cases and 9,519 controls of European descent. All cases in the PGC MDD GWAS met DSM-IV criteria for lifetime history of MDD, measured either using structured diagnostic interviews administered by lay-interviewers or diagnostic checklists administered by clinicians. Samples were imputed against the European reference sample from HapMap3 (Altshuler et al. 2010). Polygenic scores were derived using the PGC's publically available, clean, linkage disequilibrium (LD) - pruned dataset (500 kb window, r2 = .25) consisting of 123,040 non-palindromic (i.e. AT or CG) SNPs in approximate linkage equilibrium with minor allele frequency (MAF) > 1% and INFO > 90%.
Genotyped data for HRS participants was downloaded from dbGAP. Genotyping of the HRS samples was performed by the Center for Inherited Disease Research (CIDR) using the Illumina Human Omni-2.5 Quad beadchip. HRS genotypes in dbGAP were available as genotypes and as 1000 genomes (McVean et al. 2012) imputed dosage files. Imputation was performed on 2,065,320 SNPs that both passed QC filters (described in detail in the HRS QC report (CIDR March 15 2012)) and overlapped with the 1000 genomes reference panel (CIDR August 14 2012). Of the 12,507 HRS participants with available GWAS data, imputed data was provided for 12,454 samples. The rest were excluded due to missing call rates > 2% (CIDR August 14 2012). For our primary analysis we used the imputed files, selected SNPs with INFO > .90 and converted them to PLINK files using GTOOL (Freeman & Marchini 2007). Although this method of analyzing imputed data does not account for uncertainly generated by the imputation process, simulation studies suggest that for SNPs with INFO > .90, this method does not result in a significant loss of power (Zheng et al. 2011).
Almost all the clumped MDD PGC SNPs were present in the imputed HRS GWAS (99%; 121,251 of 123,040 SNPs). We derived ten continuous polygenic score variables in PLINK using the following pT thresholds: pT < .10, pT < .20, pT < .30 and pT < .40, pT < .50, pT < .60, pT < .70, pT < .80, pT < .90, pT < 1.0. Each allele was weighted by the natural log (ln) of the odds ratio (OR) of its association with MDD in the discovery dataset.
Analyses
We evaluated the associations between SLEs, polygenic scores and depressive symptoms using logistic and negative binomial regression models. Because the range of polygenic scores across individuals was narrow, we multiplied the scores by 10,000 to make differences in beta coefficients between models observable. Next we estimated the main effects of both SLEs and polygenic scores together in the same models before including interaction terms to test for moderation. All regression models were adjusted for age (centered), sex, education and ancestry (using self-reported race (black, white, Hispanic) in models without polygenic score variables and the first 3 principal components (PCs) in models that included polygenic score variables (Price et al. 2006)). We adjusted for the first 3 PCs based on an examination of eigenvalues and a scree plot from a PC analysis conducted as part of the HRS GWAS quality control. We also ran the models with up to 8 PCs and observed no meaningful difference in the results. Interaction models were additionally adjusted for all covariate-x-environment and covariate-x-polygenic score interactions (Keller 2014).
Analyses were conducted in SAS 9.3. An alpha level of p < .05 was used to judge statistical significance. Because the results were similar for all ten polygenic score variables, we report only the results for the pT < 0.40 – the threshold after which the proportion of variance explained by polygenic scores stopped increasing (See Supplementary Figure 1). Complete results for all ten polygenic scores thresholds are shown in Supplementary Tables 1–2.
We conducted a number of additional analyses to test the robustness of the findings, including calculating polygenic scores using only directly assayed SNPs from the HRS GWAS sample, testing for interaction on the additive scale using linear models and testing the interactions between SLEs and various categorical measures of polygenic score. We also ran the regression models in the sample of HRS participants who self-reported their race as `White/Caucasian' (see Supplementary Tables 3 and 4). None of these additional analyses produced results that differed significantly from the main analyses, although the proportion of variance explained by the polygenic score variables was slightly smaller in the White/Caucasian sample compared with the full sample.
Results
Participant Characteristics
The study sample was primarily white (86%) and female (62%), with an average age of 64 years. Of the 8,761 participants, 1,079 (12.3%) met criteria for clinically relevant depressive symptoms (CES-D8 score ≥ 4). Participants with clinically relevant depressive symptoms were significantly younger and more likely to be female, black or Hispanic, and to have no education degree (See Table 2). Approximately 12.6% (n = 1,101) of participants reported experiencing at least one SLE within the previous 2 years. The most common SLE reported in this sample was the death of a parent (3.83%, n = 336) followed by a serious injury (3.02%, n = 265). The least common SLE was a nursing home stay, reported by only 0.41% (n = 36) of the sample.
Table 2.
Sample characteristics for participants with and without clinically relevant depressive symptoms
| Characteristic | CES-D8 ≥ 4 (N=1,079) | CES-D8 < 4 (N = 7,682) | Total (N =8,761) |
|---|---|---|---|
| Age (M, SD)* | 62.9 (9.6) | 64.1 (8.9) | 63.9 (9.0) |
| Women (N, %)* | 812 (75.3) | 4,619 (60.1) | 5,431 (62.0) |
| Race (N, %)* | |||
| White | 836 (77.5) | 6,692 (87.1) | 7,528 (85.9) |
| Black | 203 (18.8) | 852 (11.1) | 1,055 (12.0) |
| Hispanic | 163 (15.1) | 495 (6.4) | 658 (7.5) |
| Other | 40 (3.7) | 138 (1.8) | 178 (2.0) |
| Education (N, %)* | |||
| No degree | 387 (35.9) | 1,349 (17.6) | 1,736 (19.8) |
| High School/GED | 526 (48.8) | 4,237 (55.2) | 4,763 (54.4) |
| College Degree | 120 (11.1) | 1,340 (17.4) | 1,460 (16.7) |
| Master's Degree | 38 (3.5) | 568 (7.4) | 606 (6.9) |
| Professional Degree | 8 (0.74) | 188 (2.5) | 196 (2.2) |
Abbreviations: M = mean, SD = standard deviation, CES-D8 = Centers for Epidemiologic Studies Depression Scale, 8 item version.
t or chi squared test comparing depressed vs. non-depressed participants significant at the p < .05 level.
SLEs and depression
The odds of experiencing ≥ 4 depressive symptoms were over twice as high among individuals who reported experiencing at least one SLE (OR = 2.19, 95% CI = [1.86, 2.58]; p < .0001). SLEs explained ~1% of the variance in depressive symptoms (Pseudo r2 = .0108). The pattern of results was similar when depressive symptoms were modeled as a count variable (See Table 3).
Table 3.
Results of regression models examining the associations between polygenic scores, stressful life events, and depressive symptoms
| Clinically Relevant Depressive Symptomsa | # of Depressive Symptomsb | ||||
|---|---|---|---|---|---|
|
|
|||||
| Model | Variable/Statistic | β (SE) | p value | β (SE) | p value |
| Polygenic Score Only | Polygenic Score | .21 (.05) | <.0001 | .09 (.02) | <.0001 |
| Pseudo r2 | .0095 | - | |||
| Life Stress Only | SLEs | .78 (.08) | <.0001 | .46 (.04) | <.0001 |
| Pseudo r2 | .0108 | - | |||
| Polygenic score and life stress | Polygenic Score | .21 (.05) | <.0001 | .09 (.02) | <.0001 |
| (Main effects model) | SLEs | .79 (.08) | <.0001 | .46 (.04) | <.0001 |
| Pseudo r2 | .0201 | - | |||
| Polygenic score and life stress | Polygenic Score | −.02 (.18) | .91 | −.007 (.07) | .93 |
| (Interaction model) | SLEs | 1.11 (.41) | .009 | .74 (.19) | .0002 |
| PG score*SLEs | −.06 (.13) | .65 | −.0003 (.06) | .996 | |
| Pseudo r2 | .0201 | - | |||
Abbreviations: SLE = Stressful life event, PG = polygenic score. All regression analyses adjusted for age (centered), sex, education and ancestry (self-reported race (white, black, Hispanic, other) in the ‘Life Stress Only’ model, first 3 principal components in all other models). Interaction models adjusted for all PG x covariate and SLE x covariate interaction terms. Polygenic scores presented in the table were generated using a pT threshold of p < .40. The polygenic score variable generated in PLINK was multiplied by 10,000 for the regression analyses in order to make changes in beta coefficients between models observable. Pseudo r2 values were ascertained by fitting models without demographic covariates.
Depression = 1 if CES-D8 score ≥ 4; model type = logistic.
Depressive symptoms = CES-D8 score, range = 1–8; model type = negative binomial.
Polygenic scores and depressive symptoms
Polygenic scores were significantly associated depressive symptoms: as polygenic score increased, so did the odds of experiencing ≥ 4 depressive symptoms (See Table 3). Polygenic scores explained ~1% of the variance in depressive symptoms (Pseudo r2 = .0095). Again, the pattern of results was similar when depressive symptoms were modeled as a count variable.
Polygenic scores, SLEs and depressive symptoms
Both polygenic scores and SLEs were significant predictors of depressive symptoms in the combined main effects regression models (See Table 3). The magnitudes of the coefficients for polygenic scores and SLEs did not change when both predictors were included in the same models, suggesting that SLEs did not mediate or confound the association between polygenic scores and depressive symptoms. We also failed to find evidence for gene-environment correlation: polygenic scores did not predict SLEs in a separate logistic regression model (β = .02, SE = .02, p = .33). There was no significant interaction between the effects of SLE's and polygenic scores for either clinically relevant depressive symptoms (β = −.06, SE = .13, p = .65) or number of depressive symptoms (β = −.0003, SE = .06, p = .996).
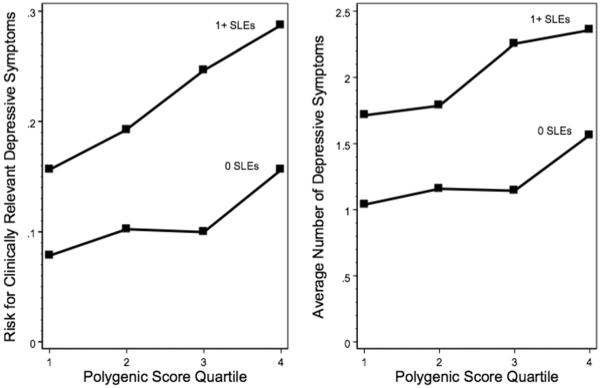
Figure 1 shows the unadjusted associations between polygenic score quartiles and risk (i.e. probability) for depressive symptoms separately for individuals who experienced 0 and 1+ SLEs. Risk for experiencing ≥ 4 depressive symptoms ranged from 8% among individuals in the lowest polygenic score quartile who did not experience an SLE to 29% among individuals in the top polygenic score quartile who experienced 1 or more SLEs. Average symptom count ranged from 1.04 among individuals in the lowest polygenic score quartile who did not experience an SLE to 2.36 among individuals in the top polygenic score quartile who experienced 1 or more SLEs. Consistent with an additive model, there was an overall increase in depression risk associated with experiencing 1 or more SLEs, however the associations between polygenic scores and depressive symptoms were similar regardless of whether or not individuals had experienced an SLE.
Figure 1. Polygenic risk and depressive symptoms in the Health and Retirement Study, stratified by stressful life even.
Abbreviations: SLE = Stressful life event. Polygenic scores were derived in study participants using SNPs associated with major depressive disorder at the p < .40 level in the PGC MDD combined GWAS. Individuals were considered to have clinically relevant depressive symptoms if they scored ≥ 4 on the CES-D8.
Discussion
In this study, we examined the associations between polygenic risk scores, recent adult onset stressful life events and depressive symptoms in a large U.S. sample of older adults. We found that both SLEs and polygenic scores were significantly and independently associated with depressive symptoms, but found no evidence that SLEs moderated the association between polygenic risk and depressive symptoms. Instead, the combined effect of SLEs and polygenic scores on depressive symptoms was equal to the sum of the individual effects, as predicted by an additive model.
The overall effect of polygenic scores on depressive symptoms in this study was modest; polygenic risk explained less than 1% of the variance in depressive symptoms, which is consistent with results from other samples (Chang et al. 2014; Demirkan et al. 2011; Ripke et al. 2013). However while polygenic scores may not be especially predictive of depression risk at the population level, our results suggest that the combined effects of SLEs and polygenic risk may have a relevant, measurable impact on depression risk that could potentially be harnessed for preventative purposes.
The results of this study are consistent with a version of the diathesis-stress model in which environmental stress and biological diatheses are viewed as additive components in the etiology of psychopathology (Ingram & Luxton, 2005). According to this model, the same degree of depression could theoretically be precipitated in anyone regardless of genetic susceptibility, but the degree of stress required to precipitate a given level of depression will vary depending on an individual's level on the underlying diathesis (in this case, the polygenetic risk). Based on these findings, it appears that older adults with lower genetic risk profiles may have about the same probability of experiencing clinically relevant depressive symptomatology as individuals with higher genetic risk profiles if they are exposed to more stress. It is worth noting, however, that the polygenic scores used in this study have only measured the additive effects of common genetic variants. Other sources of genetic variation that may be relevant for assessing gene-x-stress interactions, such as rare variation or gene-x-gene interactions (Conway et al. 2010; Kaufman et al. 2006; Kim et al. 2007; Priess-Groben & Hyde 2013) were not considered. Consequently, our findings do not rule out the possibility that other sources of genetic variation interact with stressful life events to increase depression risk. This may explain the discrepancy between our findings and the findings of Kendler et al. (1995), who used an indirect measure of genetic predisposition that would have incorporated all possible sources of genetic variation.
Our results appear inconsistent with those of Caspi et al. (2003), as well as many other published candidate gene GXE studies (although we did not specifically test any associations with the candidate gene polymorphisms). However, the candidate gene approach for studying GXE interactions may lack statistical power, which raises that possibility that previous positive findings may have been false positives (Duncan & Keller 2011). Contrary to our initial hypothesis, our results are also inconsistent with the findings of Peyrot et al. (2014), who reported a significant interaction between polygenic risk and childhood trauma. This suggests that polygenic risk-x-stress effects could be specific to certain developmental periods - a hypothesis which has also been suggested as a possible explanation for the inconsistent replication of Caspi et al.'s (2003) 5-HTTPR-x-stress interaction (Brown & Harris 2008; Karg et al. 2011).
We found no evidence to suggest that SLEs mediate the association between genetic risk and depression, which has been suggested elsewhere (Kendler 2001; Kendler & Baker 2007; Kendler & Karkowski-Shuman 1997). It should be noted, however, that our measure of SLEs assessed only whether an SLE had occurred, without inquiring about how the participant experienced the event. Previous research suggests that it is the subjective appraisal of SLEs that may mediate the association between genetic risk and depression (Conway et al. 2012), which we would not have been able to detect in this study.
The results of this study should be interpreted in light of several potential limitations. First, depression was assessed differently in our discovery and target datasets: The PGC samples used structured interviews to identify depression cases, while depression symptoms in HRS participants were measured using a shortened version of the CES-D. These two methods for case definition yield results that are correlated, but not equivalent (Radloff 1977; Turvey et al. 1999). Another limitation of the CES-D is that it only assesses depressive symptoms within the past week. As such, it may have missed depressive symptoms that occurred in the wake of a stressful event but had already dissipated by the time the HRS interview was given.
Second, our measure of SLEs included all events experienced within the past 2 years. Previous research suggests that the depression-inducing effects of SLEs may be more limited to the month in which the event occurred (Kendler et al. 1995). Hence, it is possible that the effect of SLEs, and any interaction with polygenic risk, may have been attenuated by the length of time between our assessment of depression and the experience of the stressful event(s). In addition, although it is clear that the SLEs preceded our measure of depression, we cannot be certain that the onset of the depression did not precede the SLE. This limits our ability make inferences regarding whether or not the association between SLEs and depression is causal. Previous research has suggested that depression may in fact increase the probability of experiencing a SLE, particularly an SLE that is influenced by the actions or behavior of the individual (such as divorce or job loss) (Hammen 1991, 2006; Liu & Alloy 2010; Middeldorp et al. 2008).
Despite the large sample sizes of both our discovery and target datasets, it is still possible that we were underpowered to detect a significant interaction effect. Simulation studies indicate that detecting statistically significant GXE interactions may require even larger sample sizes than those required to detect significant main effects, particularly when the environmental factor is measured with more than a small amount of error (Luan et al. 2001). Moreover, the predictive ability of our polygenic scores may have been limited by the sample size of the discovery dataset (Chatterjee et al. 2013; Dudbridge 2013) and the inherent heterogeneity of the MDD phenotype (Levinson et al. 2014), which likely contributed to the modest polygenic effects.
Finally, our sample consisted almost entirely of older adults, with an average age of 64 years. Considering that the median age-of-onset for depression is approximately 32 years (Kessler et al. 2005), few if any of our cases are likely to be first-onset. This could potentially impact the generalizability of the findings, as there is evidence to suggest that the association between life stress and depression is different for first onset vs. recurrent episodes (Post 1992; Stroud et al. 2008). Several previous studies have detected significant GXE interactions among older adult populations, however (Kim et al. 2007; Nakatani et al. 2005). It is also possible that among older adults, depressive symptoms may be more commonly attributable to organic causes (such as cerebrovascular or neurodegenerative disease) than to genetic factors relative to younger individuals. We attempted to minimize this difference by excluding HRS participants who reported a history of stroke, TIA or memory related disease.
In conclusion, the results of this study suggest that polygenic risk and recent, adult onset SLEs are each significant, independent predictors of depressive symptoms, with no evidence to suggest that the association between polygenic risk and depressive symptoms is moderated by SLEs. Future research should determine the extent to which this finding generalizes to samples of other ages, and also investigate further the extent to which life stress moderates the association between other sources of genetic variance (e.g. rare variants, copy number variation, gene-x-gene interactions) and depression.
Supplementary Material
Acknowledgements
The authors would like to thank Dr. Tamar Mendelson, Pia Mauro, Sarah Murray, Angela Lee Winn, Stacey Lloyd and Emma Stapp for their help editing the manuscript. This study used publically available data from the psychiatric GWAS consortium, which was supported by grant funding from the National Institute of Mental Health (NIMH MH085520 and NIMH MH080403), and the Health and Retirement Study, which was supported by grant funding from the National Institute of Aging (NIA U01AG009740). While conducting the analyses described in the manuscript, the first author received funding for doctoral training from the National Institute of Mental Health Psychiatric Epidemiology Institutional Training Grant (NIMH 2T32MH014592-36).
Footnotes
Conflicts of interest: The authors have no conflicts of interest, financial or otherwise, to disclose.
References
- Altshuler DM, Gibbs RA, Peltonen L, Dermitzakis E, Schaffner SF, Yu F, Bonnen PE, de Bakker PI, Deloukas P, Gabriel SB, Gwilliam R, Hunt S, Inouye M, Jia X, Palotie A, Parkin M, Whittaker P, Chang K, Hawes A, Lewis LR. Integrating common and rare genetic variation in diverse human populations. Nature. 2010;467:52–58. doi: 10.1038/nature09298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosker FJ, Hartman CA, Nolte IM, Prins BP, Terpstra P, Posthuma D, van Veen T, Willemsen G, DeRijk RH, de Geus EJ, Hoogendijk WJ, Sullivan PF, Penninx BW, Boomsma DI, Snieder H, Nolen Poor replication of candidate genes for major depressive disorder using genome-wide association data. Molecular Psychiatry. 2011;16:516–532. doi: 10.1038/mp.2010.38. [DOI] [PubMed] [Google Scholar]
- Brown GW, Harris TO. Depression and the serotonin transporter 5-HTTLPR polymorphism: A review and a hypothesis concerning gene-environment interaction. Journal of Affective Disorders. 2008;111:1–12. doi: 10.1016/j.jad.2008.04.009. [DOI] [PubMed] [Google Scholar]
- Bukh JD, Bock C, Vinberg M, Werge T, Gether U, Kessing LV. Interaction between genetic polymorphisms and stressful life events in first episode depression. Journal of Affective Disorders. 2009;119:107–115. doi: 10.1016/j.jad.2009.02.023. [DOI] [PubMed] [Google Scholar]
- Caspi A, Hariri AR, Holmes A, Uher R, Moffitt TE. Genetic sensitivity to the environment: The case of the serotonin transporter gene and its implications for studying complex diseases and traits. The American Journal of Psychiatry. 2010;167:509–527. doi: 10.1176/appi.ajp.2010.09101452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caspi A, Sugden K, Moffitt TE, Taylor A, Craig IW, Harrington H, McClay J, Mill J, Martin J, Braithwaite A, Poulton R. Influence of life stress on depression: Moderation by a polymorphism in the 5-HTT gene. Science. 2003;301:386–389. doi: 10.1126/science.1083968. [DOI] [PubMed] [Google Scholar]
- Chang SC, Glymour MM, Walter S, Liang L, Joenen KC, Tchetgen EJ, Cornelis MC, Kawachi I, Rimm E, Kubzansky LD. Genome-wide polygenic scoring for a 14-year long-term average depression phenotype. Brain and Behavior. 2014;4:298–311. doi: 10.1002/brb3.205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterjee N, Wheeler B, Sampson J, Hartge P, Chanock SJ, Park J. Projecting the performance of risk prediction based on polygenic analyses of genome-wide association studies. Nature Genetics. 2013;45:400–405. doi: 10.1038/ng.2579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Li X, McGue M. Interacting effect of BDNF Val66Met polymorphism and stressful life events on adolescent depression. Genes Brain and Behavior. 2012;11:958–965. doi: 10.1111/j.1601-183X.2012.00843.x. [DOI] [PubMed] [Google Scholar]
- CIDR . Health and Retirement Study. Ann Arbor, MI: Mar 15, 2012. Quality control report for genotypic data. [Google Scholar]
- CIDR . Health and Retirement Study. Ann Arbor, MI: Aug 14, 2012. Health and retirement study imputation report – 1000 genomes project reference panel. [Google Scholar]
- Conway CC, Hammen C, Brennan PA, Lind PA, Najman JM. Interaction of chronic stress with serotonin transporter and catechol-O-methyltransferase polymorphisms in predicting youth depression. Depression and Anxiety. 2010;27:737–745. doi: 10.1002/da.20715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conway CC, Hammen C, Espejo EP, Wray NR, Najman JM, Brennan PA. Appraisals of stressful life events as a genetically-linked mechanism in the stress–depression relationship. Cognitive Therapy and Research. 2012;36:338–347. [Google Scholar]
- Demirkan A, Penninx BW, Hek K, Wray NR, Amin N, Aulchenko YS, van Dyck R, de Geus EJ, Hofman A, Uitterlinden AG, Hottenga J, Nolen WA, Oostra BA, Sullivan PF, Willemsen G, Zitman FG, Tiemeier H, Janssens AC, Boomsma DI, van Duijn CM, Middeldorp CM. Genetic risk profiles for depression and anxiety in adult and elderly cohorts. Molecular Psychiatry. 2011;16:773–783. doi: 10.1038/mp.2010.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudbridge F. Power and predictive accuracy of polygenic risk scores. PLOS Genetics. 2013;9:e1003348. doi: 10.1371/journal.pgen.1003348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan LE, Keller MC. A critical review of the first 10 years of candidate gene-by-environment interaction research in psychiatry. The American Journal of Psychiatry. 2011;168:1041–1049. doi: 10.1176/appi.ajp.2011.11020191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elovainio M, Kokela M, Kivimaki M, Pulkki-Raback L, Lehtimaki T, Airla N, Keltikangas-Jarvinen L. Genetic variants in the DRD2 gene moderate the relationship between stressful life events and dperessive symptoms in adults: Cardiovascular risk in young Finns study. Psychosomatic Medicine. 2007;69:391–395. doi: 10.1097/psy.0b013e31806bf365. [DOI] [PubMed] [Google Scholar]
- Freeman C, Marchini J. [Accessed 20 September 2014];GTOOL. 2007 http://www.well.ox.ac.uk/~cfreeman/software/gwas/gtool.html#Contact.
- Hammen C. Generation of stress in the course of unipolar depression”. Journal of Abnormal Psychology. 1991;100:555–561. doi: 10.1037//0021-843x.100.4.555. [DOI] [PubMed] [Google Scholar]
- Hammen C. Stress generation in depression: Reflections on origins, research, and future directions. Journal of Clinical Psychology. 2006;62:1065–1082. doi: 10.1002/jclp.20293. [DOI] [PubMed] [Google Scholar]
- Health and Retirement Study . HRS 2000 Core, Final V1.0, public use dataset. Produced and distributed by the University of Michigan with funding from The National Institute on Aging; Ann Arbor, MI: 2004. grant number NIA U01AG009740. [Google Scholar]
- Hek K, Demirkan A, Lahti J, Terracciano A, Teumer A, Cornelis MC, Amin N, Bakshis E, Baumert J, Ding J, Liu Y, Marciante K, Meirelles O, Nalls MA, Sun YV, Vogelzangs N, Yu L, Bandinelli S, Benjamin EJ, Bennett DA, Boomsma D, Cannas A, Coker LH, de Geus E, De Jager PL, Diez-Roux A, Purcell S, Hu FB, Rimm EB, Hunter DJ, Jensen MK, Curhan G, Rice K, Penman AD, Rotter JI, Sotoodehnia N, Emeny R, Eriksson JG, Evans DA, Ferrucci L, Fornage M, Gudnason V, Hofman A, Illig T, Kardia S, Kelly-Hayes M, Koenen K, Kraft P, Kuningas M, Massaro JM, Melzer D, Mulas A, Mulder CL, Murray A, Oostra BA, Palotie A, Penninx B, Petersmann A, Pilling LC, Psaty B, Rawal R, Reiman EM, Schulz A, Shulman JM, Singleton AB, Smith AV, Sutin AR, Uitterlinden AG, Völzke H, Widen E, Yaffe K, Zonderman AB, Cucca F, Harris T, Ladwig K, Llewellyn DJ, Räikkönen K, Tanaka T, van Duijn CM, Grabe HJ, Launer LJ, Lunetta KL, Mosley TH, Jr, Newman AB, Tiemeier H, Murabito J. A genome-wide association study of depressive symptoms. Biological Psychiatry. 2013;73:667–678. doi: 10.1016/j.biopsych.2012.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes TH, Rahe RH. The social readjustment rating scale. Journal of Psychosomatic Research. 1967;11:213–221. doi: 10.1016/0022-3999(67)90010-4. [DOI] [PubMed] [Google Scholar]
- Hosang GM, Shiles C, Tansey KE, KcGuffin P, Uher R. Interaction between stress and the BDNF Val66Met polymorphism in depression: A systematic review and meta analysis. BMC Medicine. 2014;12 doi: 10.1186/1741-7015-12-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingram RE, Luxton DD. Vulnerability-stress models. In: Hankin BL, Abela JR, editors. Development of Psychopathology: A Vulnerability-Stress Perspective. Sage Publications, Inc, Thousand Oaks; CA US: 2005. pp. 32–46. [Google Scholar]
- Ising M, Lucrae S, Binder EB, Bettecken T, Uhr M, Ripke S, Kohli MA, Hennings JM, Horstmann S, Kloiber S, Menke A, Bondy B, Rupprecht R, Domschke K, Baune BT, Arolt V, Rush JA, Holsboer F, Muller-Myhsok B. A genomewide association study points to multiple loci that predict antidepressant drug treatment outcome in depression. Archives of General Psychiatry. 2009;66:966–975. doi: 10.1001/archgenpsychiatry.2009.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juhasz G, Hullam G, Eszlari N, Gonda X, Antal P, Anderson IM, Hokfelt TG, Deakin JF, Bagdy G. Brain galanin system genes interact with life stresses in depression-related phenotypes. Proceedings of the National Academy of Sciences. 2014;111:E1666–E1673. doi: 10.1073/pnas.1403649111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juster FT, Suzman R. An overview of the Health and Retirement Study. Journal of Human Resources. 1995;30:S7–S56. [Google Scholar]
- Karg K, Burmeister M, Shedden K, Sen S. The serotonin transporter promoter variant (5-HTTLPR), stress, and depression meta-analysis revisited: evidence of genetic moderation. Archives of General Psychiatry. 2011;68:444–454. doi: 10.1001/archgenpsychiatry.2010.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufman J, Yang BZ, Douglas-Palumberi H, Grasso D, Lipschitz D, Houshyar S, Krystal JH, Gelernter J. Brain-derived neurotrophic factor-5-HTTLPR gene interactions and environmental modifiers of depression in children. Biological Psychiatry. 2006;59:673–680. doi: 10.1016/j.biopsych.2005.10.026. [DOI] [PubMed] [Google Scholar]
- Keller MC. Gene x environment interaction studies have not properly controlled for potential confounders: The problem and the (simple) solution. Biological Psychiatry. 2014;75:18–24. doi: 10.1016/j.biopsych.2013.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kendler KS. Twin studies of psychiatric illness: An update. Archives of General Psychiatry. 2001;58:1005–1014. doi: 10.1001/archpsyc.58.11.1005. [DOI] [PubMed] [Google Scholar]
- Kendler KS, Baker JH. Genetic influences on measures of the environment: a systematic review. Psychological medicine. 2007;37:615–626. doi: 10.1017/S0033291706009524. [DOI] [PubMed] [Google Scholar]
- Kendler KS, Karkowski-Shuman L. Stressful life events and genetic liability to major depression: genetic control of exposure to the environment? Psychological Medicine. 1997;27:539–547. doi: 10.1017/s0033291797004716. [DOI] [PubMed] [Google Scholar]
- Kendler KS, Kessler RC, Walters EE, MacLean C, Neale MC, Heath AC, Eaves LJ. Stressful life events, genetic liability, and onset of an episode of major depression in women. The American Journal of Psychiatry. 1995;152:833–842. doi: 10.1176/ajp.152.6.833. [DOI] [PubMed] [Google Scholar]
- Kessler RC, Berglund P, Demler O, Jin R, Merikangas KR, Walters EE. Lifetime prevalence and age-of-onset distributions of DSM-IV disorders in the National Comorbidity Survey Replication. Archives of General Psychiatry. 2005;62:593–602. doi: 10.1001/archpsyc.62.6.593. [DOI] [PubMed] [Google Scholar]
- Kim JM, Stewart R, Kim SW, Yang SJ, Shin IS, Kim YH, Yoon JS. Interactions between life stressors and susceptibility genes (5-HTTLPR and BDNF) on depression in korean elders. Biological Psychiatry. 2007;62:423–428. doi: 10.1016/j.biopsych.2006.11.020. [DOI] [PubMed] [Google Scholar]
- Laurie CC, Doheny KF, Mirel DB, Pugh EW, Bierut LJ, Bhangale T, Boehm F, Caporaso NE, Cornelis MC, Edenberg HJ, Gabriel SB, Harris EL, Hu FB, Jacobs KB, Kraft P, Landi MT, Lumley T, Manolio TA, McHugh C, Painter I, Paschall J, Rice JP, Rice KM, Zheng X, Weir BS. Quality control and quality assurance in genotypic data for genome-wide association studies. Genetic Epidemiology. 2010;34:591–602. doi: 10.1002/gepi.20516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavebratt C, Aberg E, Sjoholm LK, Forsell Y. Variations in FKBP5 and BDNF genes are suggestively associated with depression in a swedish population-based cohort. Journal of Affective Disorders. 2010;125:249–255. doi: 10.1016/j.jad.2010.02.113. [DOI] [PubMed] [Google Scholar]
- Lee SH, et al. Cross-Disorder Group of the Psychiatric Genomics Consortium & International Inflammatory Bowel Disease Genetics Consortium (IIBDGCT) (2013). Genetic relationship between five psychiatric disorders estimated from genome-wide SNPs. Nature Genetics. 45:984–995. doi: 10.1038/ng.2711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levinson DF, Mostafavi S, Milaneschi Y, Rivera M, Ripke S, Wray NR, Sullivan PF. Genetic studies of major depressive disorder: Why are there no genome-wide association study findings and what can we do about it? Biological Psychiatry. 2014;76:510–512. doi: 10.1016/j.biopsych.2014.07.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis CM, Ng MY, Butler AW, Cohen-Woods S, Uher R, Pirlo K, Weale ME, Schosser A, Paredes UM, Rivera M, Craddock N, Owen MJ, Jones L, Jones I, Korszun A, Aitchison KJ, Shi J, Quinn JP, Mackenzie A, Vollenweider P, Waeber G, Heath S, Lathrop M, Muglia P, Barnes MR, Whittaker JC, Tozzi F, Holsboer F, Preisig M, Farmer AE, Breen G, Craig IW, McGuffin P. Genome-wide association study of major recurrent depression in the U.K. population. The American Journal of Psychiatry. 2010;167:949–957. doi: 10.1176/appi.ajp.2010.09091380. [DOI] [PubMed] [Google Scholar]
- Liu Z, Liu W, Yao L, Yang C, Xiao L, Wan Q, Gao K, Wang H, Zhu F, Wang G, Xiao Z. Negative life events and corticotropin-releasing-hormone receptor1 gene in recurrent major depressive disorder. Scientific Reports. 2013;3 doi: 10.1038/srep01548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu RT, Alloy LB. Stress generation in depression: A systematic review of the empirical literature and recommendations for future study. Clinical Psychology Review. 2010;30:582–593. doi: 10.1016/j.cpr.2010.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luan JA, Wong MY, Day NE, Wareham NJ. Sample size determination for studies of gene-environment interaction. International Journal of Epidemiology. 2001;30:1035–1040. doi: 10.1093/ije/30.5.1035. [DOI] [PubMed] [Google Scholar]
- Mandelli L, Serretti A, Marino E, Pirovano A, Calati R, Colombo C. Interaction between serotonin transporter gene, catechol-O-methyltransferase gene and stressful life events in mood disorders. International Journal of Neuropsychopharmacology. 2007;10:437–447. doi: 10.1017/S1461145706006882. [DOI] [PubMed] [Google Scholar]
- Middeldorp CM, Cath DC, Beem AL, Willemsen G, Boomsma DI. Life events, anxious depression and personality: a prospective and genetic study. Psychological Medicine. 2008;38:1557–1565. doi: 10.1017/S0033291708002985. [DOI] [PubMed] [Google Scholar]
- Muglia P, Tozzi F, Galwey NW, Francks C, Upmanyu R, Kong XQ, Antoniades A, Domenici E, Perry J, Rothen S, Vandeleur CL, Mooser V, Waeber G, Vollenweider P, Preisig M, Lucae S, Müller-Myhsok B, Holsboer F, Middleton LT, Roses AD. Genome-wide association study of recurrent major depressive disorder in two European case-control cohorts. Molecular Psychiatry. 2010;15:589–601. doi: 10.1038/mp.2008.131. [DOI] [PubMed] [Google Scholar]
- Munafò MR, Durrant C, Lewis G, Flint J. Gene X environment interactions at the serotonin transporter locus. Biological Psychiatry. 2009;65:211–219. doi: 10.1016/j.biopsych.2008.06.009. [DOI] [PubMed] [Google Scholar]
- Nakatani D, Sato H, Sakata Y, Shiotani U, Kinjo K, Mizuno H, Shimizu M, Ito H, Koretsune Y, Hirayama A, Hori M. Influence of serotonin transporter gene polymorphism on depressive symptoms and new cardiac events after acute myocardial infarction. American Heart Journal. 2005;150:652–658. doi: 10.1016/j.ahj.2005.03.062. [DOI] [PubMed] [Google Scholar]
- National Institute of Aging . Growing Old in America: The Health and Retirement Study (HRS Databook) 2007. [Google Scholar]
- Peyrot WJ, Milaneschi Y, Abdellaoui A, Sullivan PF, Hottenga JJ, Boomsma DI, Penninx BW. Effect of polygenic risk scores on depression in childhood trauma. The British Journal of Psychiatry. 2014;205:113–119. doi: 10.1192/bjp.bp.113.143081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Post RM. Transduction of psychosocial stress into the neurobiology of recurrent affective disorder. The American Journal of Psychiatry. 1992;149:999–1010. doi: 10.1176/ajp.149.8.999. [DOI] [PubMed] [Google Scholar]
- Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nature Genetics. 2006;38:904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- Priess-Groben H, Hyde JS. 5-HTTLPR X stress in adolescent depression: moderation by MAOA and gender. Journal of Abnormal Child Psychology. 2013;41:281–294. doi: 10.1007/s10802-012-9672-1. [DOI] [PubMed] [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ, Sham PC. PLINK: A Tool Set for Whole-Genome Association and Population-Based Linkage Analyses. American Journal of Human Genetics. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell S, et al. The International Schizophrenia Consortium Common polygenic variation contributes to risk of schizophrenia and bipolar disorder. Nature. 2009;460:748–752. doi: 10.1038/nature08185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radloff LS. The CES-D Scale: A self-report depression scale for research in the general population. Applied Psychological Measurement. 1977;1:385–401. [Google Scholar]
- Rietschel M, Mattheisen M, Frank J, Treutlein J, Degenhardt F, Breuer R, Steffens M, Mier D, Esslinger C, Walter H, Kirsch P, Erk S, Schnell K, Herms S, Wichmann H, Schreiber S, Jöckel K, Strohmaier J, Roeske D, Haenisch B, Gross M, Hoefels S, Lucae S, Binder EB, Wienker TF, Schulze TG, Schmäl C, Zimmer A, Juraeva D, Brors B, Bettecken T, Meyer-Lindenberg A, Müller-Myhsok B, Maier W, Nöthen M, Cichon S. Genome-wide association-, replication-, and neuroimaging study implicates HOMER1 in the etiology of major depression. Biological Psychiatry. 2010;68:578–585. doi: 10.1016/j.biopsych.2010.05.038. [DOI] [PubMed] [Google Scholar]
- Ripke S, et al. The Major Depressive Disorder Work Group of the Psychiatric GWAS Consortium A mega-analysis of genome-wide association studies for major depressive disorder. Molecular Psychiatry. 2013;18:497–511. doi: 10.1038/mp.2012.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Risch N, Herrell R, Lehner T, Liang K, Eaves L, Hoh J, Griem A, Kovacs M, Ott J, Merikangas KR. Interaction between the serotonin transporter gene (5-HTTLPR), stressful life events, and risk of depression: a meta-analysis. The Journal Of The American Medical Association. 2009;301:2462–2471. doi: 10.1001/jama.2009.878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi J, Potash JB, Knowles JA, Weissman MM, Coryell W, Scheftner WA, Lawson WB, DePaulo J, Gejman PV, Sanders AR, Johnson JK, Adams P, Chaudhury S, Jancic D, Evgrafov O, Zvinyatskovskiy A, Ertman N, Gladis M, Neimanas K, Goodell M, Hale N, Ney N, Verma R, Mirel D, Holmans P, Levinson DF. Genome-wide association study of recurrent early-onset major depressive disorder. Molecular Psychiatry. 2011;16:193–201. doi: 10.1038/mp.2009.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shyn SI, Shi J, Kraft JB, Potash JB, Knowles JA, Weissman MM, Garriock HA, Yokoyama JS, McGrath PJ, Peters EJ, Scheftner WA, Coryell W, Lawson WB, Jancic D, Gejman PV, Sanders AR, Holmans P, Slager SL, Levinson DF, Hamilton SP. Novel loci for major depression identified by genome-wide association study of Sequenced Treatment Alternatives to Relieve Depression and meta-analysis of three studies. Molecular Psychiatry. 2011;16:202–215. doi: 10.1038/mp.2009.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steffick DE. Documentation of Affective Functioning Measures in the Health and Retirement Study. 2000 Available: http://hrsonline.isr.umich.edu/sitedocs/userg/dr-005.pdf [2013, March 10]
- Stroud CB, Davila J, Moyer A. The relationship between stress and depression in first onsets versus recurrences: A meta-analytic review. Journal of Abnormal Psychology. 2008;117:206–213. doi: 10.1037/0021-843X.117.1.206. [DOI] [PubMed] [Google Scholar]
- Sullivan PF. The psychiatric GWAS consortium: big science comes to psychiatry. Neuron. 2010;68:182–186. doi: 10.1016/j.neuron.2010.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan PF, Daly MJ, O'Donovan M. Genetic architectures of psychiatric disorders: the emerging picture and its implications. Nature Reviews Genetics. 2012;13:537–551. doi: 10.1038/nrg3240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan PF, de Geus EJ, Willemsen G, James MR, Smit JH, Zandbelt T, Arolt V, Baune BT, Blackwood D, Cichon S, Coventry WL, Domschke K, Farmer A, Fava M, Gordon SD, He Q, Heath AC, Heutink P, Holsboer F, Hoogendijk WJ, Hottenga JJ, Hu Y, Kohli M, Lin D, Lucae S, Macintyre DJ, Maier W, McGhee KA, McGuffin P, Montgomery GW, Muir WJ, Nolen WA, Nöthen MM, Perlis RH, Pirlo K, Posthuma D, Rietschel M, Rizzu P, Schosser A, Smit AB, Smoller JW, Tzeng J, van Dyck R, Verhage M, Zitman FG, Martin NG, Wray NR, Boomsma DI, Penninx BW. Genome-wide association for major depressive disorder: a possible role for the presynaptic protein piccolo. Molecular Psychiatry. 2009;14:359–375. doi: 10.1038/mp.2008.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan PF, Neale MC, Kendler KS. Genetic epidemiology of major depression: review and meta-analysis. The American Journal of Psychiatry. 2000;157:1552–1562. doi: 10.1176/appi.ajp.157.10.1552. [DOI] [PubMed] [Google Scholar]
- Terracciano A, Tanaka T, Sutin AR, Sanna S, Deiana B, Lai S, Uda M, Schlessinger D, Abecasis GR, Ferrucci L, Costa PT. Genome-wide association scan of trait depression. Biological Psychiatry. 2010;68:811–817. doi: 10.1016/j.biopsych.2010.06.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turvey CL, Wallace RB, Herzog R. A revised CES-D measure of depressive symptoms and a DSM-based measure of major depressive episodes in the elderly. International Psychogeriatrics. 1999;11:139–148. doi: 10.1017/s1041610299005694. [DOI] [PubMed] [Google Scholar]
- Uher R, McGuffin P. The moderation by the serotonin transporter gene of environmental adversity in the etiology of depression: 2009 update. Molecular Psychiatry. 2010;15:18–22. doi: 10.1038/mp.2009.123. [DOI] [PubMed] [Google Scholar]
- Wray NR, Pergadia ML, Blackwood DH, Penninx BW, Gordon SD, Nyholt DR, Ripke S, MacIntyre DJ, McGhee KA, Maclean AW, Smit JH, Hottenga JJ, Willemsen G, Middeldorp CM, de Geus EJ, Lewis CM, McGuffin P, Hickie IB, van den Oord EJ, Liu JZ, Macgregor S, McEvoy BP, Byrne EM, Medland SE, Statham DJ, Henders AK, Heath AC, Montgomery GW, Martin NG, Boomsma DI, Madden PA, Sullivan PF. Genome-wide association study of major depressive disorder: new results, meta-analysis, and lessons learned. Molecular Psychiatry. 2012;17:36–48. doi: 10.1038/mp.2010.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng J, Li Y, Abecasis GR, Scheet P. Comparison of approaches to account for uncertainty in analysis if imputed genotypes. Genetic Epidemiology. 2011;35:102–110. doi: 10.1002/gepi.20552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann P, Bruckl T, Nocon A, Pfister H, Binder EB, Uhr M, Lieb R, Moffitt TE, Caspi A, Holsboer F, Ising M. Interaction of FKBP5 gene variants and adverse life events in predicting depression onset: results from a 10-year prospective community study. American Journal of Psychiatry. 2011;168:1107–1116. doi: 10.1176/appi.ajp.2011.10111577. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.