Abstract
The cuticle of the nematode Caenorhabditis elegans is a specialized extracellular matrix whose major component is collagen. Cuticle collagens are encoded by a large multi-gene family consisting of more than 150 members. Cuticle collagen genes are expressed in epidermis (hypodermis) and may be stage-specific or cyclically expressed. We identified cuticle collagen genes as transcriptional targets of the DBL-1 TGF-β-related signaling pathway. These studies prompted us to investigate the cis-regulatory sequences required for transcription of one of the target genes, col-41. We generated reporter constructs that reproduce stage- and tissue-specific expression of fluorescent markers. We identify four conserved sequence elements that are required for transcription of reporters. Finally, we provide evidence that col-41 expression is controlled by a sequence element containing two GATA sites and by the epidermal GATA transcription factors ELT-1 and ELT-3.
Keywords: TGFβ, GATA, hypodermis, epidermis
Introduction
In the nematode C. elegans, the cuticle serves as an exoskeleton required for proper growth and defense of the animal (Page and Johnstone, 2007). Collagen is the major component of the cuticle and is encoded by a large multi-gene family. Cuticle collagen genes are expressed in the epidermis (hypodermis), a large multinucleate tissue that surrounds the nematode body. Collagen is secreted from the apical surface of the epidermal cells to the exterior of the body. Cuticle collagen genes show highly specific temporal expression patterns that may be stage-specific or cycle in phase with the cuticle molts (Jackson et al., 2014; Johnstone and Barry, 1996; Park and Kramer, 1994). Additionally, levels and timing of cuticle collagen gene expression respond to cell signaling pathways, such as the TGFβ and Wnt pathways (Jackson et al., 2014; Liang et al., 2007; Roberts et al., 2010).
In our studies of the transcriptional response to the TGFβ family member DBL-1, we identified col-41 and other cuticle collagen genes as transcriptional targets whose expression levels depend on the activity of the signaling pathway (Liang et al., 2007). This finding prompted us to investigate the cis-regulatory requirements for accurate transcription of the col-41 gene. Although expression studies of cuticle collagen genes have been previously performed, little is known about the sequence elements that contribute to collagen gene regulation. In this study, we use transcriptional reporters to characterize col-41 regulatory sequences. We identify a 0.5kb promoter region that directs stage- and tissue-specific regulation of a fluorescent marker and additional upstream sequences that contribute to robust, reproducible expression. Within these regions, we identify four conserved sequence motifs that are required for optimal activation of the reporter, two of which are essential for any detectable expression. One of the essential elements contains two GATA motifs. We provide evidence that col-41 may be regulated by the epidermal-specific GATA factors ELT-1 and ELT-3.
Results and Discussion
Cuticle collagen genes were identified via microarray analysis as transcriptional targets of the DBL-1 signaling pathway (Liang et al., 2007). DBL-1 regulates multiple developmental and homeostatic functions in C. elegans, one of which is body size (Savage-Dunn et al., 2003; Wang et al., 2002). Since cuticle collagen genes are known to regulate body size and morphology, we were interested in these target genes as potential mediators of DBL-1 regulation of body size. Furthermore, recent evidence suggests that defects in the cuticle underlie multiple abnormalities seen in DBL-1 pathway mutants (Schultz et al., 2014). To investigate collagen gene expression in vivo, we generated transcriptional GFP reporters using upstream genomic sequences for the target genes col-37, col-41 and col-141. GFP expression was undetectable for col-37 and col-141 (data not shown), suggesting either that our constructs were missing critical regulatory sequences, or that expression levels were below our level of detection.
In contrast, transcriptional fusions for col-41 containing 1.6kb of upstream sequences fused to GFP or to nuclear localized 2xNLS-mCherry showed detectable expression. We chose 1.6kb since this was the distance to the nearest upstream transcript in the genome annotation at the time. (Current annotations show the presence of three small exons in this region from a gene transcribed in the opposite orientation). Reporter constructs were microinjected into the C. elegans gonad to create transgenic lines followed by transgene integration using γ-irradiation. Integrated col-41p::2xNLS::mCherry transgenes are expressed in a tissue-specific and stage-specific manner. mCherry expression is observed in epidermal nuclei in L3 and early L4 larval stage animals (Fig. 1a). mCherry is also expressed in the vulval precursor cells, a ventral epidermal lineage, during the L3 stage (Fig. 1b). Expression is absent from the lateral epidermal seam cells; this absence is more apparent in the GFP reporter, which is not nuclear localized and therefore shows diffuse fluorescence throughout the cytoplasm (Fig. 1c). The spatial expression pattern is very similar to that reported for the cuticle collagen gene dpy-7 (Gilleard et al., 1997). Temporally, however, COL-41 appears to be a larval-specific cuticle collagen, with expression absent from embryos and adults. This result is consistent with RNAseq analyses that demonstrate a peak of col-41 expression in L2 (Gerstein et al., 2010; Jackson et al., 2014). Expression of the fluorescent marker may be visible later than the peak in mRNA expression due to the time lag for protein synthesis and maturation, and/or perdurance of the mCherry protein. The observed expression timing is consistent with col-41 being identified as a target of the DBL-1 pathway at the L2 stage (Liang et al., 2007) but not at the L4 stage (Roberts et al., 2010).
Fig. 1.
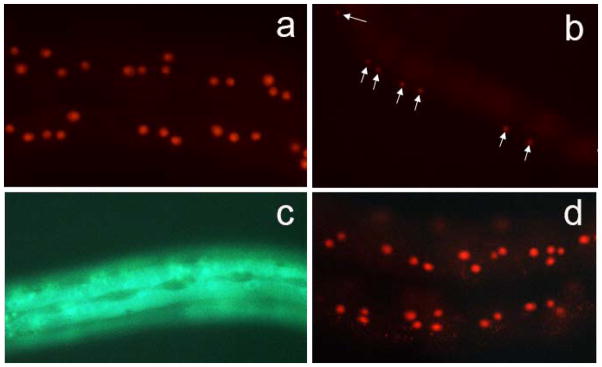
col-41 transcriptional reporters direct expression in the epidermis. (a) Expression of 1.6kb col-41p::2xNLS::mCherry reporter in epidermal nuclei during L3 stage. (b) Expression of 1.6kb col-41p::2xNLS::mCherry reporter in vulval precursor cells (marked by arrows) during L3 stage. (c) Expression of 1.6kb col-41p::GFP reporter in the epidermis is excluded from the lateral epidermal seam cells. (d) The 0.5kb col-41p::2xNLS::mCherry reporter directs expression in a similar stage- and tissue-specific manner.
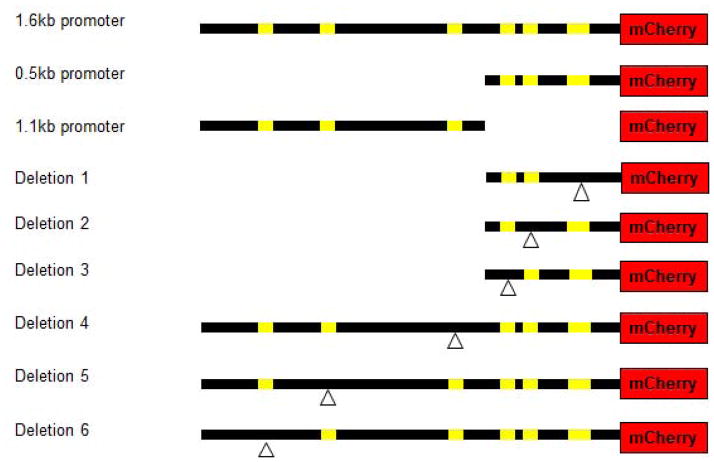
Next, we analyzed the putative regulatory sequences of col-41 in more detail. We aligned col-41 upstream sequences from three closely related nematodes, C. briggsae, C. remanei and C. elegans. Sequence conservation was most prominent in the proximal 0.5kb (Fig. 2). We therefore created separate mCherry reporter constructs containing the proximal 0.5kb and distal 1.1kb col-41 promoter fragments to generate transgenic animals as described above. In vivo, the reporter construct with the distal 1.1kb promoter fragment is not sufficient for expression (data not shown). In contrast, mCherry expression driven by the proximal 0.5kb promoter fragment results in tissue-specific (epidermal) and stage-specific (early larval) expression (Fig. 1d). However, expression appeared to be less robust from the 0.5kb construct, since in multiple integrated lines only ~60% of transgenic animals showed expression (four integrated lines were analyzed). We conclude that sequences in the proximal 1.1kb region contribute to reproducible expression of col-41. There is precedent for such mechanisms for producing robust and reproducible expression via redundant cis-acting elements and “shadow” enhancers (Lagha et al., 2012).
Fig. 2.
Alignment of upstream sequences of col-41 orthologs from C. elegans, C. briggsae, and C. remanei. Alignment was performed using Clustalw. * indicates identical nucleotides. Red boxes indicate conserved regions that were deleted in reporter constructs.
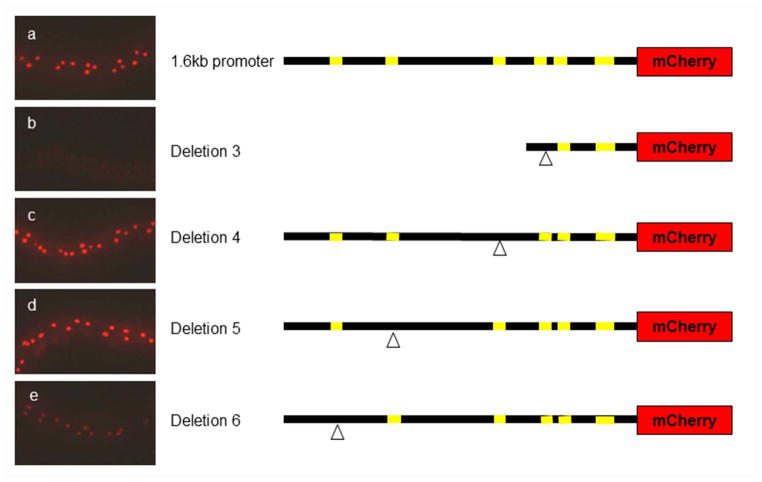
From the sequence alignment, we identified 6 conserved sequence elements (Fig. 2). We deleted the three proximal elements in the context of the 0.5kb promoter fragment and the three distal elements in the context of the 1.6kb promoter (Fig. 3). Deletion constructs were microinjected into nematodes, transgenes were integrated, and expression was analyzed by fluorescence microscopy. Images in Fig. 4 were taken on the same day with the same exposure times. Deletions 1 and 2 show no detectable mCherry expression (data not shown). Deletions 3 and 6 have reduced levels of mCherry expression (Fig. 4b,e). Deletions 4 and 5 show levels of expression similar to the wild-type 1.6kb construct (Fig. 4a,c,d). None of these deletions produced any ectopic expression, either temporally or spatially. Taken together, we conclude that elements 1, 2, 3, and 6, but not elements 4 and 5, are required for col-41 expression. Additionally, since deletion 6 in the context of the full 1.6kb promoter has a lower level of expression than the 0.5kb promoter (that lacks element 6), we conclude that the 1.1kb region also contains an undefined repressor element.
Fig. 3.
col-41 promoter constructs. Yellow bars indicate conserved regions. Deletions are indicated by the delta (inverted triangle) symbol.
Fig. 4.
Expression of col-41 deletion series reporters. (a) 1.6kb promoter control; (b) Deletion 3 has no detectable expression; (c) Deletion 4 has normal levels of expression; (d) Deletion 5 has normal levels of expression; (e) Deletion 6 has reduced levels of expression.
We examined sequence elements 1, 2, 3, and 6 for potential transcription factor binding sites (Fig. 2). Since we identified col-41 as a transcriptional target of DBL-1, a TGFβ-related ligand (Suzuki et al., 1999), we first searched for sequences related to TGFβ signaling. TGFβ responses are mediated by Smad transcription factors, which bind two types of sequences: GTCT Smad Binding Elements (SBEs) or GRCGNC GC-rich sequences (Gao et al., 2005; Shi et al., 1998; Zawel et al., 1998). Element 6 contains one potential GTCT Smad Binding Element (SBE). Most Smad target genes, however, contain more than one SBE, and the GTCT upstream of col-41 is not conserved in C. remanei (Fig. 2). Thus, at this time, we do not have evidence to distinguish between col-41 as a direct or indirect target of Smads.
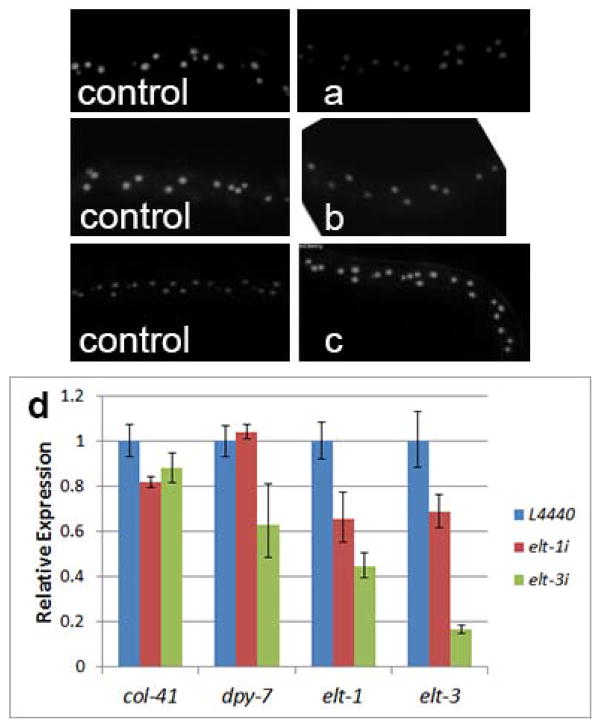
Element 2 contains two potential GATA sites. The GATA motif is a common binding site for transcription factors, including GATA factors. In C. elegans, specific GATA factor genes act as master regulators of intestinal and epidermal fates: elt-2 and elt-7 in the intestine (Sommermann et al., 2010) and elt-1 and elt-3 in the epidermis (Gilleard and McGhee, 2001). We therefore tested whether GATA factors play a role in the transcriptional regulation of col-41. We used RNAi by feeding to inactivate the GATA factor genes elt-1, elt-2, and elt-3 in animals carrying the 1.6k col-41 reporter construct. We found that inhibition of the intestinal GATA factor gene elt-2 had no effect on expression, as expected (Fig. 5b). On the other hand, inhibition of the epidermal GATA factor gene elt-1 caused a reduction in mCherry expression (Fig. 5a). Somewhat surprisingly, inhibition of elt-3, another epidermal GATA factor gene, resulted in increased mCherry expression (Fig. 5c). We therefore conclude that ELT-1 plays a role in transcriptional activation of col-41 in the epidermis. ELT-3, on the other hand, may be a transcriptional repressor of col-41 or have a more complex regulatory interaction with col-41. Interestingly, the GATA factor GATA-4 has been identified as a transcriptional repressor for human COL1A2 (Antoniv et al., 2005), which could indicate a conserved regulatory mechanism for invertebrate and vertebrate collagens.
Fig. 5.
Depletion of epidermal, but not intestinal, GATA factors results in altered expression of cuticle collagen genes. (a–c) Expression of col-41 transcriptional reporter. The respective empty vector control for each experiment is shown to the left. (a) elt-1 RNAi (epidermal); (b) elt-2 RNAi (intestinal); (c) elt-3 RNAi (epidermal). (d) Quantitative RT-PCR analysis of col-41, dpy-7, elt-1 and elt-3 upon elt-1 and elt-3 RNAi. col-41 expression is dependent on ELT-1 and, to a lesser extent, ELT-3. dpy-7 expression is dependent on ELT-3. Data are averages of trials from three biologically independent replicates; error bars show standard deviation. Transcript levels are determined in L2 stage; note that elt-1 transcript levels and functional activity are higher during embryogenesis.
To clarify these regulatory interactions, we used qRT-PCR to determine transcript levels of the endogenous col-41 gene upon RNAi of elt-1 and elt-3. elt-1 is required in embryos to specify epidermal fates and is a transcriptional activator of elt-3 (Gilleard et al., 1999; Page et al., 1997). Null mutations in elt-1 are lethal, but partial loss of function by RNAi is viable. As expected, elt-1(RNAi) caused reduced transcript levels for both elt-1 and elt-3 (Fig. 5d). Additionally, consistent with the transgenic reporter results, we found that col-41 transcript levels are reduced in elt-1(RNAi) animals (Fig. 5d). elt-3 is required for maintenance of epidermal fates and null alleles are viable (Gilleard et al., 1999). Due to the lack of lethality, elt-3(RNAi) efficiently reduced elt-3 transcript levels (Fig. 5d). It also reduced elt-1 transcript levels, suggesting that ELT-3 is required for maintenance of elt-1 expression, which has not previously been reported. Unexpectedly, we saw a slight reduction in col-41 transcript levels in elt-3(RNAi) animals (Fig. 5d), as opposed to the increase seen in the transgenic reporter. This result suggests that our transgenic reporter may be missing sequences required for the correct direction of the regulatory interaction. Consistent with the hypothesis of direct regulation of col-41 by ELT-3 is the identification of ELT-3 binding upstream of col-41 by ChIP-seq analysis (Niu et al., 2011).
We were interested to determine whether regulation of cuticle collagen genes by GATA factors is a general phenomenon. Expression analyses of dpy-7 collagen have identified a GATA sequence in the minimal promoter region (Gilleard et al., 1997). We therefore determined dpy-7 transcript levels in elt-1 and elt-3 knockdowns. We find that ELT-3, but not ELT-1, is required for normal expression of dpy-7 (Fig. 5d). Furthermore, like col-41, the dpy-7 locus is associated with direct binding of ELT-3 by ChIP-seq (Niu et al., 2011).
In conclusion, we have generated transcriptional reporters for col-41 that can be used to mark larval epidermal and vulval precursor nuclei in C. elegans. We have identified four conserved elements that are required for expression of col-41. One of these elements contains two GATA sites. In vertebrates, GATA factors play critical roles in specification of hematopoietic and endoderm lineages (Burch, 2005; Wolff and Humeniuk, 2013). Similarly, in C. elegans GATA factors can specify intestinal and epidermal fates (Gilleard and McGhee, 2001; Sommermann et al., 2010). In our experiments, depletion of the epidermal-specific GATA factor ELT-1 reduced expression of col-41, suggesting that the cuticle collagen gene is a direct target of a tissue-specific master regulator. Furthermore, expression of the cuticle collagen gene dpy-7 is dependent on the GATA factor ELT-3, suggesting that this may be a widespread mechanism for the tissue-specific regulation of this multi-gene family.
Methods
Molecular cloning and sequencing
The col-41 transcriptional GFP reporter was generated by fusion PCR (Hobert, 2002) using 1.6kb upstream promoter region and GFP coding region from pPD117.01 as the template. The col-41 2xNLS::mCherry transcriptional reporter was generated by PCR cloning of col-41 upstream sequences into 2xNLS-mCherry (optimized) vector (converted from pPD95.75). The col-41 deletion series constructs were generated by using Phusion Site-Directed Mutagenesis Kit (NEB) and confirmed by sequencing.
Transgenic animals
Transgenic nematodes were generated by microinjection of plasmid DNA (10 ng/μl) with myo-2::GFP (5 ng/μl) as a marker (Mello et al., 1991). Arrays were integrated into chromosomes using γ-irradiation. 2–3 independent lines were generated for each construct, and all showed comparable results.
qRT–PCR
Total RNA was isolated by Trizol (GibcoBRL) and using the Qiagen RNeasy Mini kit from N2 (wild-type) worms treated with L4440 (empty vector control), elt-1(RNAi) and elt-3(RNAi). Reverse transcription was done using enzyme from SUPERSCRIPT™ III Two-Step qRT–PCR (Invitrogen). Real-time PCR was performed using PWR SYBR Green PCR Master Mix on Applied Biosystem StepOne 48 well machine (Lifetechnologies). Data Analysis software used was StepOne Software version2.2.2. Gene act-1 was used as endogenous control. The intron primer of act-1 gene was used to test for the presence of genomic DNA. N2 animals were synchronized to late L2 stage with a 4 hour synchronization. Three biologically independent samples were collected. Each sample was analyzed in two technical replicates. A total of 4 genes (col-41, dpy-7, elt-1, and elt-3) was examined in L4440, elt-1, and elt-3 RNAi backgrounds. Results are shown as relative quantitation compared with L4440 control.
RNAi
RNAi feeding was performed as described in (Kamath et al., 2003; Liang et al., 2013). For transgene reporter experiments, 12 L4 animals were transferred to feeding plates, incubated overnight, transferred to fresh RNAi feeding plates to lay eggs for 4 hours and the stage-synchronized progeny were scored. For qRT-PCR, 50 L4 animals were transferred to RNAi plates and allowed to lay eggs overnight. Adults were washed off the plates, leaving unhatched embryos. After 4 hours, newly hatched L1 animals were transferred to new RNAi plates and allowed to develop until late L2 stage.
Acknowledgments
We thank Iva Greenwald for the 2xNLS-mCherry vector and for use of a Cs source. We thank Nick Palmisano and Kristina Ames for critical reading of the manuscript. This work was supported by NIH grant 1R15GM073678-01 and PSC-CUNY award TRADB-42-267 to C.S.D. Some of the experiments were done with equipment from the Core Facilities for Imaging, Cellular and Molecular Biology at Queens College. This work was carried out in partial fulfillment of the requirements for the Ph.D. degree from the Graduate Center of City University of New York (J.Y. and U.M.).
NIH Grant Support: 1R15GM073678-01, 1R15GM097692-01
Other Grant Support: PSC-CUNY TRADB-42-267
References
- Antoniv TT, Tanaka S, Sudan B, De Val S, Liu K, Wang L, Wells DJ, Bou-Gharios G, Ramirez F. Identification of a repressor in the first intron of the human alpha2(I) collagen gene (COL1A2) The Journal of biological chemistry. 2005;280:35417–35423. doi: 10.1074/jbc.M502681200. [DOI] [PubMed] [Google Scholar]
- Burch JB. Regulation of GATA gene expression during vertebrate development. Seminars in cell & developmental biology. 2005;16:71–81. doi: 10.1016/j.semcdb.2004.10.002. [DOI] [PubMed] [Google Scholar]
- Gao S, Steffen J, Laughon A. Dpp-responsive silencers are bound by a trimeric Mad-Medea complex. The Journal of biological chemistry. 2005;280:36158–36164. doi: 10.1074/jbc.M506882200. [DOI] [PubMed] [Google Scholar]
- Gerstein MB, Lu ZJ, Van Nostrand EL, Cheng C, Arshinoff BI, Liu T, Yip KY, Robilotto R, Rechtsteiner A, Ikegami K, et al. Integrative analysis of the Caenorhabditis elegans genome by the modENCODE project. Science. 2010;330:1775–1787. doi: 10.1126/science.1196914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilleard JS, Barry JD, Johnstone IL. cis regulatory requirements for hypodermal cell-specific expression of the Caenorhabditis elegans cuticle collagen gene dpy-7. Molecular and cellular biology. 1997;17:2301–2311. doi: 10.1128/mcb.17.4.2301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilleard JS, McGhee JD. Activation of hypodermal differentiation in the Caenorhabditis elegans embryo by GATA transcription factors ELT-1 and ELT-3. Molecular and cellular biology. 2001;21:2533–2544. doi: 10.1128/MCB.21.7.2533-2544.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilleard JS, Shafi Y, Barry JD, McGhee JD. ELT-3: A Caenorhabditis elegans GATA factor expressed in the embryonic epidermis during morphogenesis. Developmental biology. 1999;208:265–280. doi: 10.1006/dbio.1999.9202. [DOI] [PubMed] [Google Scholar]
- Hobert O. PCR fusion-based approach to create reporter gene constructs for expression analysis in transgenic C. elegans. BioTechniques. 2002;32:728–730. doi: 10.2144/02324bm01. [DOI] [PubMed] [Google Scholar]
- Jackson BM, Abete-Luzi P, Krause MW, Eisenmann DM. Use of an activated beta-catenin to identify Wnt pathway target genes in caenorhabditis elegans, including a subset of collagen genes expressed in late larval development. G3. 2014;4:733–747. doi: 10.1534/g3.113.009522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnstone IL, Barry JD. Temporal reiteration of a precise gene expression pattern during nematode development. The EMBO journal. 1996;15:3633–3639. [PMC free article] [PubMed] [Google Scholar]
- Kamath RS, Fraser AG, Dong Y, Poulin G, Durbin R, Gotta M, Kanapin A, Le Bot N, Moreno S, Sohrmann M, et al. Systematic functional analysis of the Caenorhabditis elegans genome using RNAi. Nature. 2003;421:231–237. doi: 10.1038/nature01278. [DOI] [PubMed] [Google Scholar]
- Lagha M, Bothma JP, Levine M. Mechanisms of transcriptional precision in animal development. Trends in genetics : TIG. 2012;28:409–416. doi: 10.1016/j.tig.2012.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang J, Xiong S, Savage-Dunn C. Using RNA-mediated interference feeding strategy to screen for genes involved in body size regulation in the nematode C. elegans. Journal of visualized experiments : JoVE. 2013 doi: 10.3791/4373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang J, Yu L, Yin J, Savage-Dunn C. Transcriptional repressor and activator activities of SMA-9 contribute differentially to BMP-related signaling outputs. Developmental biology. 2007;305:714–725. doi: 10.1016/j.ydbio.2007.02.038. [DOI] [PubMed] [Google Scholar]
- Mello CC, Kramer JM, Stinchcomb D, Ambros V. Efficient gene transfer in C.elegans: extrachromosomal maintenance and integration of transforming sequences. The EMBO journal. 1991;10:3959–3970. doi: 10.1002/j.1460-2075.1991.tb04966.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu W, Lu ZJ, Zhong M, Sarov M, Murray JI, Brdlik CM, Janette J, Chen C, Alves P, Preston E, et al. Diverse transcription factor binding features revealed by genome-wide ChIP-seq in C. elegans. Genome research. 2011;21:245–254. doi: 10.1101/gr.114587.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page AP, Johnstone IL. The cuticle. WormBook : the online review of C elegans biology. 2007:1–15. doi: 10.1895/wormbook.1.138.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page BD, Zhang W, Steward K, Blumenthal T, Priess JR. ELT-1, a GATA-like transcription factor, is required for epidermal cell fates in Caenorhabditis elegans embryos. Genes & development. 1997;11:1651–1661. doi: 10.1101/gad.11.13.1651. [DOI] [PubMed] [Google Scholar]
- Park YS, Kramer JM. The C. elegans sqt-1 and rol-6 collagen genes are coordinately expressed during development, but not at all stages that display mutant phenotypes. Developmental biology. 1994;163:112–124. doi: 10.1006/dbio.1994.1127. [DOI] [PubMed] [Google Scholar]
- Roberts AF, Gumienny TL, Gleason RJ, Wang H, Padgett RW. Regulation of genes affecting body size and innate immunity by the DBL-1/BMP-like pathway in Caenorhabditis elegans. BMC developmental biology. 2010;10:61. doi: 10.1186/1471-213X-10-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savage-Dunn C, Maduzia LL, Zimmerman CM, Roberts AF, Cohen S, Tokarz R, Padgett RW. Genetic screen for small body size mutants in C. elegans reveals many TGFbeta pathway components. Genesis. 2003;35:239–247. doi: 10.1002/gene.10184. [DOI] [PubMed] [Google Scholar]
- Schultz RD, Bennett EE, Ellis EA, Gumienny TL. Regulation of Extracellular Matrix Organization by BMP Signaling in Caenorhabditis elegans. PloS one. 2014;9:e101929. doi: 10.1371/journal.pone.0101929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y, Wang YF, Jayaraman L, Yang H, Massague J, Pavletich NP. Crystal structure of a Smad MH1 domain bound to DNA: insights on DNA binding in TGF-beta signaling. Cell. 1998;94:585–594. doi: 10.1016/s0092-8674(00)81600-1. [DOI] [PubMed] [Google Scholar]
- Sommermann EM, Strohmaier KR, Maduro MF, Rothman JH. Endoderm development in Caenorhabditis elegans: the synergistic action of ELT-2 and -7 mediates the specification-->differentiation transition. Developmental biology. 2010;347:154–166. doi: 10.1016/j.ydbio.2010.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki Y, Yandell MD, Roy PJ, Krishna S, Savage-Dunn C, Ross RM, Padgett RW, Wood WB. A BMP homolog acts as a dose-dependent regulator of body size and male tail patterning in Caenorhabditis elegans. Development. 1999;126:241–250. doi: 10.1242/dev.126.2.241. [DOI] [PubMed] [Google Scholar]
- Wang J, Tokarz R, Savage-Dunn C. The expression of TGFbeta signal transducers in the hypodermis regulates body size in C. elegans. Development. 2002;129:4989–4998. doi: 10.1242/dev.129.21.4989. [DOI] [PubMed] [Google Scholar]
- Wolff L, Humeniuk R. Concise review: erythroid versus myeloid lineage commitment: regulating the master regulators. Stem cells. 2013;31:1237–1244. doi: 10.1002/stem.1379. [DOI] [PubMed] [Google Scholar]
- Zawel L, Dai JL, Buckhaults P, Zhou S, Kinzler KW, Vogelstein B, Kern SE. Human Smad3 and Smad4 are sequence-specific transcription activators. Molecular cell. 1998;1:611–617. doi: 10.1016/s1097-2765(00)80061-1. [DOI] [PubMed] [Google Scholar]