Abstract
Low rates of sequence evolution associated with purifying selection can be interrupted by episodic changes in selective regimes. Visual pigments are a unique system in which we can investigate the functional consequences of genetic changes, therefore connecting genotype to phenotype in the context of natural and sexual selection pressures. We study the RH2 and RH1 visual pigments (opsins) across 22 bird species belonging to two ecologically convergent clades, the New World warblers (Parulidae) and Old World warblers (Phylloscopidae), and evaluate rates of evolution in these clades along with data from 21 additional species. We demonstrate generally slow evolution of these opsins: both Rh1 and Rh2 are highly conserved across Old World and New World warblers. However, Rh2 underwent a burst of evolution within the New World genus Setophaga, where it accumulated substitutions at 6 amino acid sites across the species we studied. Evolutionary analyses revealed a significant increase in dN/dS in Setophaga, implying relatively strong selective pressures to overcome long-standing purifying selection. We studied the effects of each substitution on spectral tuning and found they do not cause large spectral shifts. Thus substitutions may reflect other aspects of opsin function, such as those affecting photosensitivity and/or dark-light adaptation. Although it is unclear what these alterations mean for color perception, we suggest that rapid evolution is linked to sexual selection, given the exceptional plumage colour diversification in Setophaga.
Keywords: birds, colour vision, Rh2 opsin, Parulidae, Phyllscopidae, spectral tuning, variable evolutionary rates
Introduction
The rate at which proteins evolve is often associated with particular selective regimes. Low rates of evolution are often the product of purifying selection interspersed with episodes of accelerated evolution that result from either positive selection or relaxation of purifying selection. It is generally believed that proteins are under strong constraint and evolve slowly (Dayhoff 1978), but some examples have been documented of proteins exhibiting a sudden burst of evolution, which results in clades accumulating many substitutions after otherwise slow rates of evolution (Wallis 1996; 2001). In most cases evolutionary rates are studied at the sequence level and no attempt is made to link sequence evolution to underlying functional changes. However, a thorough understanding of evolutionary processes and adaptation requires connecting DNA substitutions underlying protein function to an organism's phenotype and fitness (Dean & Thornton 2007; Barrett & Hoekstra 2011). Visual pigments are a system where such a synthetic approach can be employed, because after obtaining gene sequences the spectral sensitivity of visual pigments can be measured in vitro, thereby connecting genotype to phenotype. Tracing evolutionary trajectories allows us to dissect changes in evolutionary rates across species and investigate factors that may be responsible for their evolution.
Visual pigments mediate the first step of the visual transduction cascade in the eye (Baylor 1987; 1996). They consist of an opsin protein bound to a light-sensitive chromophore, which in vertebrates can be 11-cis-retinal or 3,4-dehydroretinal (see Bowmaker 2008 for a review). Vision starts when a photon hits and activates a visual pigment molecule, thereby isomerizing the associated chromophore. This results in a series of several conformational changes in the opsin protein (Imai et al. 2007), resulting in a metarhodopsin II form that activates the next protein in the phototransduction cascade, the G protein transducin, which ultimately leads to a signal that light has been perceived. In addition to rhodopsin (Rh1), responsible for scotopic “dim-light” vision, birds have a cone opsin from each of the four spectrally distinct vertebrate opsin classes involved in colour vision: a cone opsin maximally sensitive to long wavelengths (Lws), a medium-wavelength sensitive opsin (Rh2), and two types of short-wavelength sensitive opsins (Sws2 and Sws1). In birds, visual pigments are always associated with an 11-cis-retinal chromophore, and thus the only factor determining their wavelength of peak absorbance is the interaction between the chromophore and key amino acids residues of the opsin protein (Chang et al. 1995; Yokoyama 2000).
Many evolutionary studies of opsins have compared divergent taxa, coupled with statistical analysis to identify the residues responsible for the large spectral sensitivity differences among visual pigments (Yokoyama & Radlwimmer 2001; Shi et al. 2001; Yokoyama 2003; Hunt et al. 2009; Hauser et al. 2014). The functional and adaptive significance of different sites has sometimes been evaluated by using site-directed mutagenesis to measure the spectral shifts caused by each mutation (e.g. Yokoyama 2000; Yokoyama & Radlwimmer 2001; Bickelmann et al. 2012; van Hazel et al. 2013; Bloch et al. 2015). Such studies have contributed greatly to our understanding of visual pigment function and in some cases provided evidence for adaptability of the visual system to diverse environments (Yokoyama et al. 2008; Seehausen et al. 2008; Bickelmann et al. 2012). However, an important complementary approach lies in comparative analyses among closely related species, in which adaptive changes are more easily identified (Seehausen et al. 2008; Coyle et al. 2012). Here, we study sequence and functional evolution in the avian middle-wavelength sensitive visual pigments Rh2 and Rh1 within and between two clades of ecologically convergent bird families. These two opsins are the product of the most recent vertebrate opsin duplication (Lagman et al. 2013).
The New World warblers (Parulidae) and the Old World warblers (Phylloscopidae) occupy similar environments and are ecologically convergent (Price et al. 2000) having last shared a common ancestor ~30 Mya (± 1.8MY 95% confidence limits from a Bayesian analysis with fossil dates (Price et al. 2014); see Fig. 1)). The two clades differ strikingly in plumage patterns: the New World warblers are colourful and often sexually dimorphic, whereas the Old World warblers are dull and monomorphic (e.g., Fig. 1). We sequenced the Rh2 and Rh1 opsin genes in multiple species from these two clades, as well as additional species for use as outgroups. We then evaluated the adaptive significance of amino acid substitutions using evolutionary statistical tests (Yang & Bielawski 2000; Yang 2007). Finally, we assessed the functional consequences of the substitutions through in vitro expression and direct measurement of visual pigment spectral sensitivities (λmax). We place our results in context by considering the other species for which opsin sequence data has been previously obtained.
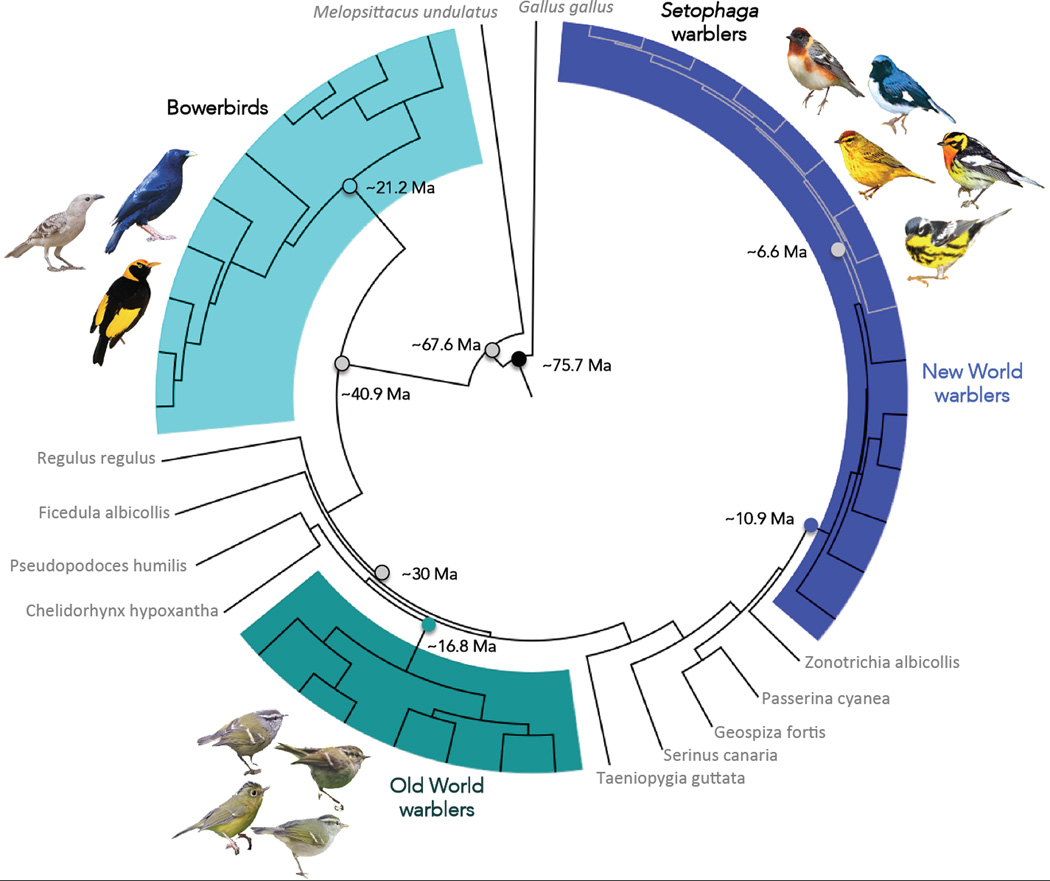
Figure 1.
Time calibrated phylogeny of the New World warblers, Old World warblers and Bowerbirds used in this study. Branch lengths are proportional to time. Males from a few species for each clade are illustrated. Species with names in grey are used as outgroups in our analysis. Grey branches within the New World warblers indicate species belonging to Setophaga. Mean node ages and corresponding 95% confidence intervals from the BEAST analysis are as follows: Setophaga warblers 6.62Ma [5.41–7.78]; New World warblers 10.89Ma [9.28–12.66]; Old World warblers 16.77Ma [14.30–19.51]; Bowerbirds 21.22Ma [17.69–24.58]; Last common ancestor to New and Old World warblers 30.02Ma [26.9–33.15]; Last common ancestor to bowerbirds and the rest of the passerines 40.9Ma [36.95–44.75]; Split between passerines and Psittaciformes 67.63Ma [64.44–72.42]; Root node (Black) 75.7Ma [66.44–86.71].
Materials and methods
Study system
Among the New World warblers we consider 15 species belonging to 6 genera (Cardellina, Geothlypis, Mniotilta, Oreothlypis, Seiurus, Setophaga). Among Old World warblers we consider 7 species that encompass two genera, the Phylloscopus and Seicercus. We sequenced complete opsin genes of several additional species to use as outgroups. These species are the White-Throated Sparrow (Zonotrichia albicolis) and the Indigo Bunting (Passerina cyanea), as outgroups to the New World warblers, the Yellow-bellied Fantail (Chelidorhynx hypoxantha) as outgroup to the Old World warblers and the Goldcrest (Regulus regulus), as outgroup to both warbler clades combined. We also used the available avian opsin gene sequences in GenBank as additional taxa for comparison (summarized in the Table S1). Bowerbirds (Ptilonorhynchidae) are the only clade of birds for which these visual pigments have been studied in a comparative framework, although most species have only partial sequences (Coyle et al. 2012; but see van Hazel et al. 2013; Table S1).
Tissue collection and opsin sequencing
Eye tissue was collected and preserved in RNAlater or liquid nitrogen in order to extract total RNA from the retinas of individual birds, as described in Bloch et al. (2015). Total RNA was extracted following TRIzol protocol (Life Technologies). In the 5 New World warbler species with the highest RNA integrity, we synthesized adaptor-ligated cDNA that was used in RACE-PCR (rapid amplification of cDNA ends; SMART RACE system – BD Clontech) to obtain full coding sequences for all opsins in these initial 5 species (Seiurus aurocapilla, Oreothlypis ruficapilla, Geothlypis philadelphia, Setophaga pensylvanica and S. palmarum). Based on an alignment of the resulting sequences we identified conserved regions and designed primers located in the 5’ and 3’ gene ends and/or UTRs of each opsin gene to amplify full coding sequences (primer sequences for every step are in Table S2). For all warblers we synthesized cDNA from total RNA extracted from retinas using oligo-dT primers (using Qiagen’s Omniscript RT kit), and used these primers to amplify full coding sequences in the species from this study. All sequences were deposited in GenBank (accession numbers: Rh2 KM977570-KM977595; Rh1 KM977596-KM977621) and on the Dryad digital repository at doi:10.5061/dryad.b2m1n
In vitro regeneration of visual pigments and spectral analyses
From the sequence data we identified opsin variants that are likely to differ in ëmax and subjected them to heterologous in vitro expression in mammalian cell culture, following detailed protocols in Bloch et al. (2015). Complete coding sequences for cone opsin genes were subcloned into the p1D4-hrGFP II expression vector (Morrow & Chang 2010), followed by transient transfection into HEK293T cells. Cells were harvested 48 h post-transfection and opsins were regenerated through incubation in 5 ìM 11-cis-retinal generously provided by Dr. Rosalie Crouch (Medical University of South Carolina). Visual pigments were solubilized in 1% N-dodecyl-D-maltoside (DM), and immunoaffinity purified using the 1D4 monoclonal antibody (Molday & MacKenzie 1983). The dark and light-bleached absorbance spectra of expressed visual pigments were recorded at 25°C using a Cary4000 double-beam absorbance spectrophotometer (Varian) and quartz absorption cuvettes (Helma). All λmax values were calculated after fitting data from multiple absorbance spectra of each visual pigment to a standard Govardovskii template for A1 visual pigments (Govardovskii et al. 2000, for further details on these methods, see Bloch et al. 2015). The λmax values we present correspond to the average of three separate measurements of the absorbance spectrum for each pigment expressed. Since substitution D332E in the C-terminus is unlikely to impact spectral tuning (Yokoyama et al. 2008), we did not express the Rh2 pigment of Setophaga caerulescens and Oreothlypis ruficapilla, which only differ by this substitution relative to the inferred ancestor and other warbler species.
Phylogeny
We estimated a time-calibrated phylogeny for species in this study in BEAST (Drummond & Rambaut 2007) following methods in Price et al. (2014). This phylogeny is based on up to 5 nuclear and 2 mitochondrial genes obtained from GenBank (see supplementary information, Table S3 for details and accession numbers), with several other taxa incorporated for the time calibration. The tree was dated using 7 fossil and 3 biogeographic calibrations (Table S4), which are the subset of those used in Price et al. (2014) relevant to our study. Bayesian inference of the time-calibrated phylogeny was performed by running BEAST for 30 million generations, sampling every 3,000 generations for a total of 10,000 trees of which 2,000 were discarded as burn-in. We used a separate analysis to estimate dates and relationships within the New World warblers, which have been generally difficult to resolve because of rapid divergence early in the clade’s history. For this clade, we used the tree published by Lovette et al. (2010), which was based on additional genes and has higher resolution. The date of insert of the New World clade into the larger phylogeny was estimated from Price et al. (2014), as in Bloch et al. (2015; Fig. 1), in order to incorporate the New World warbler phylogeny into the tree we constructed. For the New World warblers we estimated the age of the most recent common ancestor to the species in our study at ~11Ma (Fig. 1). The Old World warblers are older than the New World warblers, with a root ~16Ma (Price et al. 2014; Bloch et al. 2015).
Ancestral reconstructions
For the ancestral reconstructions and subsequent selection analyses, we considered two separate datasets. First, we built a dataset with New World and Old World warbler Rh2 sequences, including all the passerine outgroups we sequenced, for which we obtained complete open-reading frame sequences, plus an additional 7 complete Rh2 sequences available in GenBank. We refer to this dataset as the “main dataset”. We also performed ancestral reconstructions and selection analyses on a larger set of species that includes 10 Bowerbird species. Complete coding sequences are not available for most bowerbirds so we were only able to use 780 bases for these analyses, which means we had to exclude some of the warbler variable sites. We refer to this species set as the “limited sequence dataset”. In both cases, we used the budgerigar (Melopsittacus undulatus) and chicken (Gallus gallus) as outgroups to the passerines (see Fig. 1).
We performed ancestral sequence reconstructions with both parsimony, as implemented in Mesquite (Maddison & Maddison 2001), and likelihood/Bayesian approaches, as implemented in PAML, based on the phylogenetic relationships we estimated. Different reconstruction models are sensitive to different assumptions (Chang et al. 2002; Yang 2007; Chang et al. 2012), and thus we used nucleotide, codon-based and amino acid models relying on, where applicable, likelihood ratio tests to choose the best fitting models for each type of reconstruction. Parameters for each model choice are summarized in Table S5. We compared the ancestral reconstruction results to check for the robustness across methods and used posterior probabilities to determine the most likely opsin sequence at each ancestral node (Chang et al. 2002; Yang 2007; Chang et al. 2012).
Visual pigment molecular evolution
Selection tests
We relied on codon-based random sites models as implemented in PAML (Yang & Bielawski 2000; Yang 2007), using the topology shown in Fig. 1, to test for the presence of positively selected sites. Here estimates of ω = dN/dS are calculated in a likelihood framework using alternative models, which differentially constrain ω. The simplest M0 or “one-ratio” model assumes ω is constant for all sites in the alignment. Sets of nested models evaluate evidence for sites evolving under positive selection, M1a (neutral) against M2 (selection) and M7 (β-beta distribution for variable selection categories) against M8 (β+ω) (Shyue et al. 1998; Yang et al. 2000). While there have been past criticisms of PAML models (and other codon-based models) to detect selection (Friedman & Hughes 2007; Suzuki 2008; Nozawa et al. 2009), these criticisms have been largely refuted, and the models shown to have robust statistical properties (Yang et al. 2009; Yang & Reis 2011; Zhai et al. 2012; Gharib & Robinson-Rechavi 2013). We used likelihood ratio tests to evaluate whether the more parameter rich models (M2 and M8), fit the data better than the simpler models (M1 and M7 respectively), and used a Bayes Empirical Bayes (BEB) analysis to identify those sites inferred to have evolved under positive selection (Zhang 2005). To minimize the possibility of reaching local optima we ran all models with different starting ω values (ω = 0.01, 0.25, 0.5, 0.75, 1.0, 1.5, 2.0, 5.0).
To test for positive selection along particular branches (notably in this study along the terminal branch of the New World warbler phylogeny leading to S. fusca), we used branch models as implemented in PAML (Yang & Nielsen 1998; Yang 1998). Once more we used likelihood ratio tests to evaluate whether the more parameter rich model with a category of sites evolving under positive selection fits the data better than a simpler, null model (Nielsen & Yang 1998; Yang 1998; Morrow & Chang 2010). Here, our alternative model was one where S. fusca was assigned as the “foreground branch” and was therefore allowed to have a different ω value than the rest of the tree. We compared this model to the null model, where all branches share the same ω value to ask whether ω was significantly higher in the S. fusca lineage than the background, and whether it was significantly higher than 1.0.
In order to test for divergence in selective regimes in the Rh2 genes of New World warblers, Old World warblers and Bowerbirds we evaluated whether ω differs among clades using Clade model C (CmC) (Bielawski & Yang 2004; Yang et al. 2005; Weadick & Chang 2012). Clade model C (CmC) accommodates divergent selection by allowing ω to vary in a clade selected as “foreground clade” independent of the rest of the tree or “background”. To determine if this model is the best fit for the data we compared it against a recently improved null model proposed by Weadick and Chang (2012), M2a_rel, that does not allow among lineage variation, using a likelihood ratio test. If this likelihood ratio test is significant it implies the CmC model with different rates of evolution for each clade fits the data better than the null model in which ω remains constant across the tree. As for the models above, CmC models consider three classes of sites: a category of sites evolving under purifying selection with 0 < ω0 < 1, neutrality ω1 = 1 and a final category of sites that accounts for sites under divergent or positive selection ω2i, estimated separately for each predefined clade, i.
Clade model C can allows for more than two clades to be defined as separate partitions, estimating ω separately for each partition (Weadick & Chang 2012). This comparison of the signatures of selection in multiple clades allows for detection of more complex patterns of divergence across a phylogeny. We used this approach in a Clade model where both the Setophaga and the Old World warblers were set as partitions separate from the rest of the tree. Comparing this model against the simpler nested models and the null model, M2a_rel, provides a test of whether ω differs between Setophaga and Old World warblers, and between these clades and the rest of the tree. Finally, we evaluated the robustness of the parameter estimates and results of Clade model C to slight changes in model assumptions by performing a separate clade analysis using Clade model D (CmD) (Bielawski & Yang 2004; Weadick & Chang 2012). These clade models are very similar in that they model divergent selection between clades assuming three site classes, but in model CmD the ω values are not constrained to fall into one of three groups (0<ω <1, ω = 1, ω >1). Once again, we ran all models with different starting ω values to avoid local optima.
Results
Rh2 and Rh1 sequence variation in New and Old World warblers
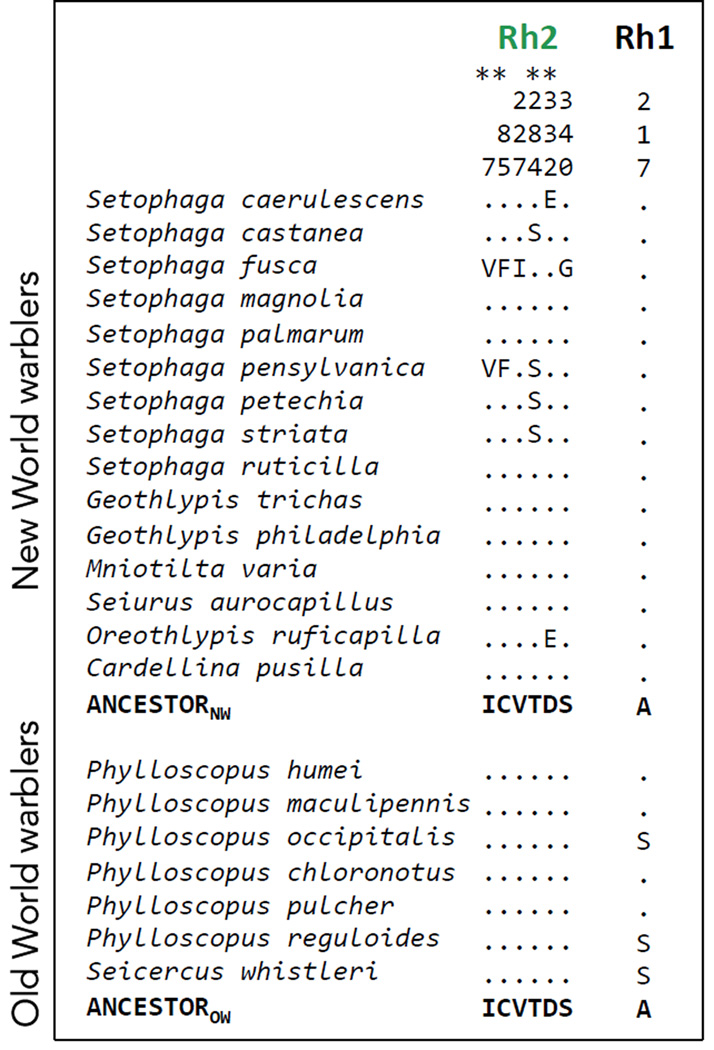
Rh1 was highly conserved across warblers with only one amino acid substitution in a subclade of the Old World warblers (Fig. 2). Rh2 is completely conserved across the 7 Old World warblers and that same sequence is shared by 8 of the 15 New World warblers we studied (Fig. 2, Fig. S1). Remarkably, in the other 7 New World warblers the Rh2 sequence differs, with amino acid substitutions occurring at 6 different sites (Fig. 2, Fig. S2 ). With the exception of one substitution along the lineage leading to Oreothlypis ruficapilla, all the substitutions are within the genus Setophaga.
Figure 2.
Variable amino acid residues in the coding sequences of Rh2 and Rh1 opsins in the New World warblers and the Old World warblers. Numbers correspond to amino acid positions standardized by the bovine rhodopsin (Genbank M21606). The first 15 species are New World warblers and the following 7 are Old World warblers. All variable sites are shown relative to the inferred ancestor, which is the same for both clades, as obtained by Empirical Bayes methods (Yang 2005). Dots indicate the identity of the amino acids with the ancestor at each site, thus species that only have dots match the ancestral amino acid sequence at all sites. Residues marked with an asterisk (*) are inferred to evolve under positive selection by maximum likelihood methods. See Fig. S2 for comparisons across all Passerines.
Ancestral reconstructions
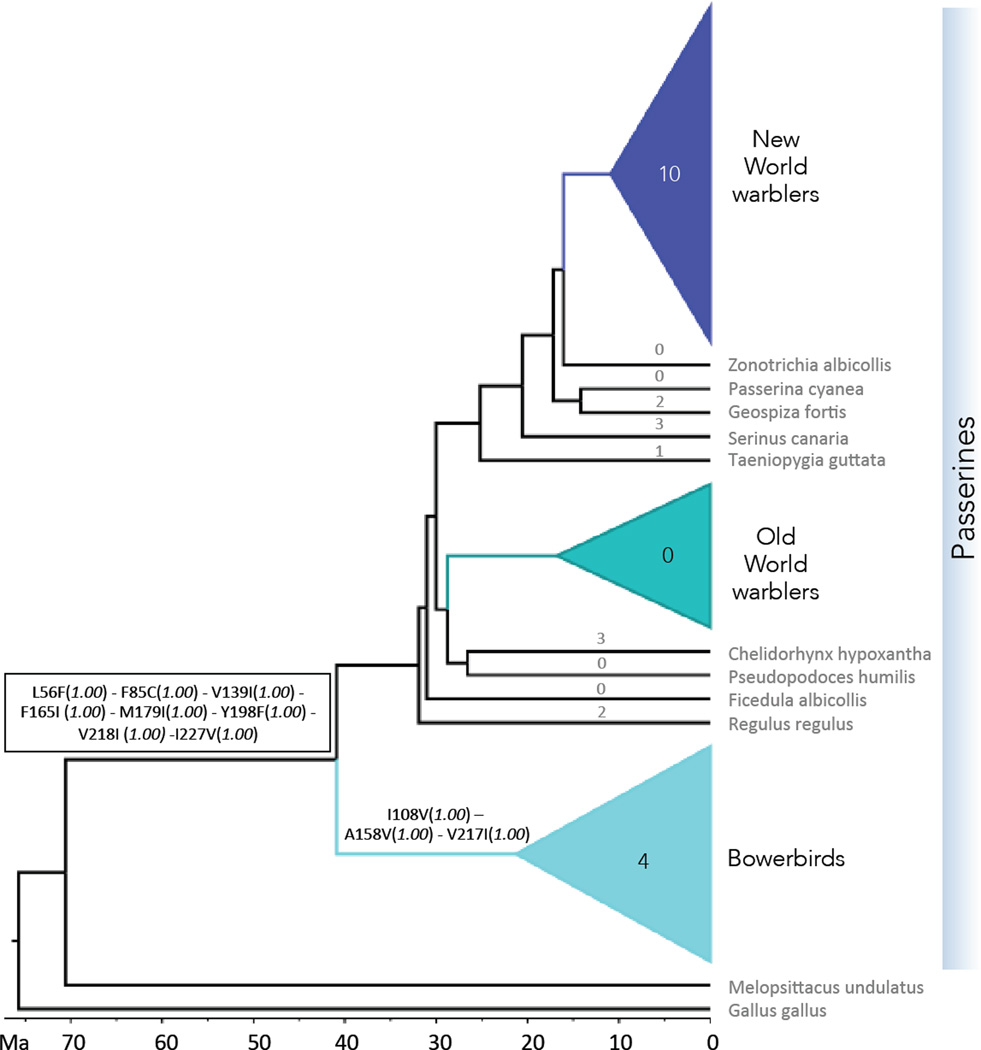
Nucleotide, codon-based, and amino acid models are in agreement for the amino acid identities at all ancestral nodes. Based on these reconstructions, the New World and Old World warbler ancestor share the same Rh2 amino acid sequence with each other and many present day species, implying conservation over >30 million years of evolution in branches leading to both warbler clades (last common ancestor of New and Old World warblers is inferred to be at about 30 Mya; Figs. 1, 3, S3). Moreover, this shared ancestral sequence is inferred to date back at least another 10 Mya (Fig. 3) as it was shared by the last common ancestor of bowerbirds and the other the passerine birds in the study (node labeled at 40.9 Mya in Fig. 1).
Figure 3.
Evolution of Rh2 amino acid sequences inferred by ancestral reconstruction. Substitutions are shown above the branch where they occur. Numbers above branches correspond to the number of amino acid substitutions, for clades this number indicates the number of mutation events that caused the observed substitutions. For details on the identity of the substitutions refer to Fig. 2 and Fig. S2. Posterior probabilities for variable amino acid sites at every node from Empirical Bayes ancestral reconstructions are shown in parentheses. We show results for nucleotide Empirical Bayes models, which always had the most conservative posterior probabilities compared to codon and amino acid models. Melopsittacus undulatus (budgerigar) and Gallus gallus (chicken) were used as outgroups in ancestral reconstructions. Identity for variable residues at the root, and the corresponding posterior probabilities: L56 (0.998) - F85(0.999) - I108 (0.961) - V139(0.995) - A158 (0.998) - F165 (0.998) - M179 (1.00) - Y198(1.00) - V217(0.998) - V218 (0.995) - I227 (0.998)
Following prolonged stasis, Rh2 underwent a burst of evolution in the New World warblers (Figs. 2, 3, S2). Three substitutions, C85F, T284S and D332E show both reversals and parallel evolution within the New World warblers (Fig. S3) and two appear elsewhere: R. regulus, an outgroup to the clade containing both the New World and Old World warblers carries the T284S substitution (Figs. S2). D332E not only occurs twice within the New World warblers but 4 additional times in the branches leading to Z. albicollis, S. canaria, T. guttata and C. hypoxantha (Fig. S1, S2). Beyond the passerines, T284S is also found in the budgerigar and the chicken branches (Fig. 1, S2).
Selection tests
We identified four sites as having evolved under positive selection within New World warblers (sites 7, 85, 284 and 332; Table 1 and marked with an asterisk in Fig. 2). We confirmed Site 332 as being positively selected across all the Passerines included in the “limited sequence dataset” (Fig. S2, Table 1), i.e. in an analysis that included the Bowerbirds but was based on shorter Rh2 opsin sequences.
Table 1.
Results of PAML random sites models for Rh2 in New World warblers, Old World warblers and across Passerines, based on the main dataset, consisting of full opsin sequences. Likelihood ratio tests (LRT) were performed to test whether two more parameter rich models, M2a and M8, that allow for a category of positively selected sites, fit the data better than their nested, simpler counterparts, M1a and M7 respectively. The resulting test statistic was compared to a distribution both in the M2-M1a and M8-M7 comparison (Yang et al. 2000). Positively selected sites identified by Bayes Empirical Bayes factors (BEB) are shown. M0: One-ratio model; M1a neutral model, M2a: selection model; M7: Beta; M8:Beta+ω (Yang et al. 2000). ω= dN/dS
| Clade | Model | lnL | np | Parameter Estimates | Sites Identified by BEB |
p- value (LRT) |
|---|---|---|---|---|---|---|
| New World warblers |
M0 | −1891.15 | 31 | ω0=0.055 | - | |
| M1a | −1876.65 | 32 | p0= 0.9999, ωo=0.00001 | - | NS (0.14) |
|
| M2a | −1874.68 | 34 | p0= 0.978, ωo=0.000, p2= 0.022, ω2=2.731 | 7 85 284 332 | ||
| M7 | −1879.64 | 32 | p= 0.0097, q= 0.163 | - | 0.006 | |
| M8 | −1874.68 | 34 | p0= 0.978 (p1=0.022), ω=2.731 | 7 85 284 332 | ||
| Old World Warblers |
M0 | −1705.44 | 17 | ω0=0.0086 | - | |
| M1a | −1705.44 | 18 | p0=0.9999, ωo=0.0086 | - | NS (0.99) |
|
| M2a | −1705.44 | 20 | p0=1.00,ωo=0.0086, p2=0.00, ω2=1.00 | none | ||
| M7 | −1705.48 | 18 | p=0.90, q= 99.0 | - | NS (0.99) |
|
| M8 | −1705.49 | 20 | p0=0.999 (p1=0.001), ω=1.00 | none | ||
| Passerines (Main dataset) |
M0 | −3459.64 | 66 | ω0=0.017 | - | |
| M1a | −3428.71 | 67 | p0=0.979, ωo=0.01 | - | NS (0.99) |
|
| M2a | −3428.71 | 69 | p0=0.979,ωo=0.01, p2=0.021, ω2=1.00 | 332 | ||
| M7 | −3419.74 | 67 | p=0.06, q= 2.07 | - | 0.001 | |
| M8 | −3412.92 | 69 | p0=0.0994 (p1=0.006), ω=1.64 | 332 | ||
lnL the log value of the model’s likelihood score given the data and phylogeny.
NS = non-significant
np = number of parameters.
pi is the proportion of sites in each category.
p and q are shape parameters for the Beta distribution.
Using clade models and the main sequence dataset we compared Rh2 evolution across the New World warblers and the Old World warblers. We confirmed the presence of divergent selection within the Setophaga warblers. Estimates of ω were significantly higher in this clade than in the rest of passerines (when Setophaga warblers were set as the foreground clade ωSetophaga = 2.26 vs background ωbkgd = 0.141; 2Δl = 4.80, df =1, P = 0.028; Table 2). This is partly a consequence of multiple substitutions along the lineage leading to S. fusca (the ω = dN/dS ratio along this lineage (6.20) is significantly >1; LRT compared with the nested null model, 2Δl = 24.57, df = 2, P = 0.000). Conversely, divergence in Old World warblers was unusually low relative to other passerines (ωow = 0.000 vs ωbkgd = 0.25; 2Δl = 7.0, df = 1, P = 0.008; Table 2).
Table 2.
Parameter estimates for passerine Rh2 clade models (CmC). CmC: Clade model C, a model that allows for different ω between a clade set as foreground and the rest of the tree. In each case the clade associated with each test was set as foreground clade. ω3 is the ratio for the “foreground” clade. M2a_rel is the improved test for use as a null model in the LRT proposed by Weadick and Chang (2012). * Based on “limited sequence dataset”.
Tree length is defined as the sum of the branch lengths along the tree in substitutions per codon.
| RH2 | Purifying selection site class | Neutral site class | Divergent selection site class | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Model: Foreground clade | Tree length | κ | ω0 | p0 | ω1 | p1 | ω2 ω3 ω4 |
p2 | lnL | np |
p-value LRT vs M2a_rel (df) |
| CmC: Setophaga warblers | 1.77 | 6.02 | 0.006 | 0.976 | 1.0 | 0.008 | 0.141 2.26 |
0.016 | −2718.09 | 89 | 0.028 (1) |
| CmC: New World warblers | 1.71 | 5.99 | 0.005 | 0.956 | 1.0 | 0.000 | 0.207 0.401 |
0.044 | −2720.23 | 89 | 0.471 (1) |
| CmC: Old World warblers | 1.70 | 5.99 | 0.004 | 0.949 | 1.0 | 0.000 | 0.250 0.000 |
0.052 | −2716.97 | 89 | 0.008 (1) |
| CmC: Bowerbirds* | 1.69 | 5.73 | 0.004 | 0.949 | 1.0 | 0.000 | 0.187 0.323 |
0.051 | −2720.39 | 89 | 0.65 (1) |
| CmC: Setophaga + Old World warblers* |
1.71 | 5.99 | 0.004 | 0.954 | 1.0 | 0.000 | 0.233 0.604 0.0001 |
0.046 | −2715.81 | 90 | 0.009 (2) |
| M2a_rel |
1.74 | 5.96 | 0.0036 | 0.936 | 1.0 | 0.008 | 0.133 | 0.056 | −2720.49 | 88 | - |
κ (kappa) transition/transversion rate
pi refers to the proportion of sites inferred to be in each selection category.
The multiple foreground clade model, in which we defined both Setophaga and the Old World warblers as foreground clades, confirms the rate heterogeneity in Rh2 evolution, indicating Rh2 has evolved faster within the Setophaga and slower within Old World warblers (ωSetophaga = 0.604, ωow = 0.0001 vs ωbkgd =0.226; LRT compared to corresponding M2a_rel model 2Δl = 9.36, df =2, P <0.009; Table 2).
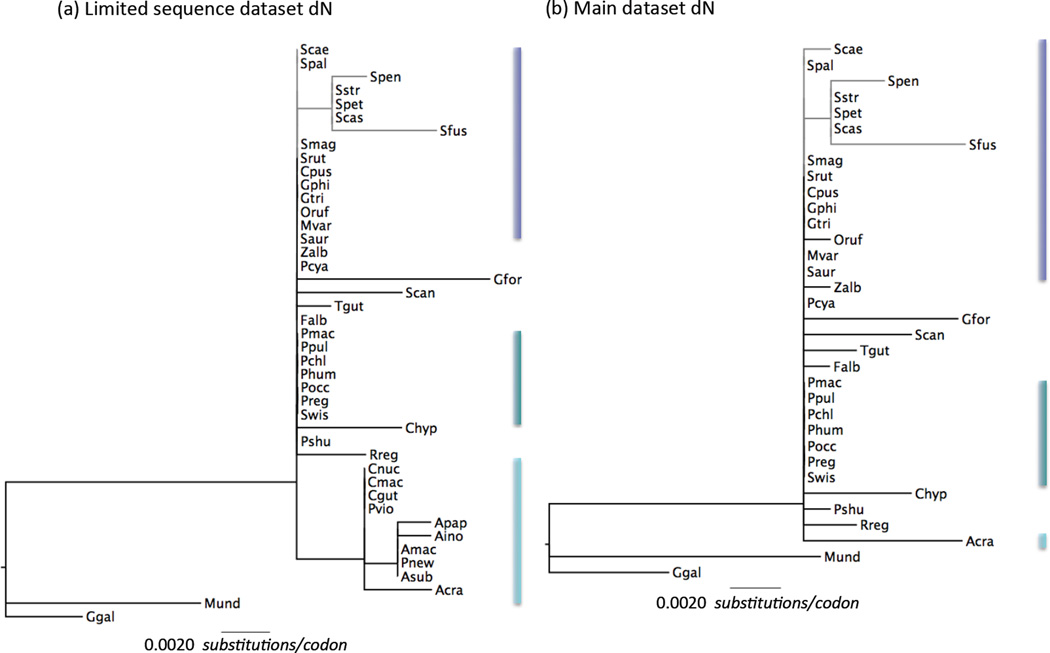
Incorporating the bowerbirds using the limited sequence data set, indicates higher ω in this clade than background branches (ωBW = 0.32 vs ωbkgd = 0.19), but this difference is not statistically significant (Table 2). Bowerbirds have similar numbers of non-synonymous substitutions as the Setophaga warblers (Fig. 4), but more synonymous substitutions, presumably reflecting their greater age (crown group ages ~21.2 Mya against ~6.6 Mya, Fig. 1).
Figure 4.
Variation in Rh2non-synonymous rates of evolution (dN) across Passerines. Gene tree for Rh2 where branch lengths are scaled by dN rate (substitutions per codon) estimated along that branch by a free-ratio model as implemented in PAML (parameters specified in Table S8 for the best fitting codon model; Yang 2007). (a) the limited sequence dataset including bowerbirds for which incomplete coding sequences were used and (b) the full sequence dataset. Bars next to clades are colored following the scheme in Figs. 1 and 3 to highlight the position of each clade on the tree. Scale bar indicates value of dN and is the same in both trees. Species names are abbreviated as the first letter of the genus and the three first letters of the species (i.e. Setophaga castanea is Scas, see Figs. 1 and 2 for full species names).
Parameter estimates for all clade models reanalyzed under the CmD framework were broadly similar to those obtained under CmC models (Table S6). The only exception is the ω2 estimates for the CmD model with New World warblers as foreground, which were higher than in the equivalent CmC model and also higher than one (ω2 = 1.27).
The acceleration in the rates of evolution we observe is particular to Rh2 and not observed for Rh1 (Table S7). For Rh1, very low rates of evolution are observed throughout passerines and there are no episodes of accelerated evolution (Table S7).
In vitro expression: Rh2 spectral tuning
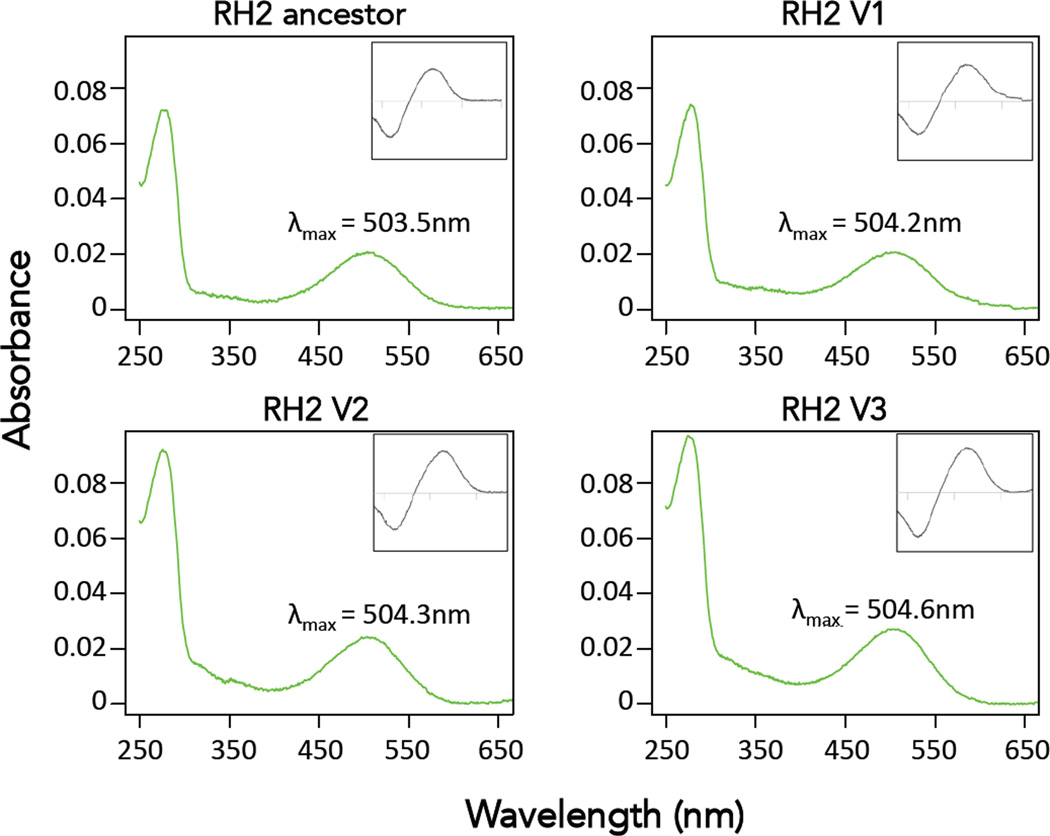
The effects of the Rh2 substitutions we documented in warblers on spectral tuning are unknown. Substitutions observed at sites 227 and 332 have been previously studied in other groups and not thought to impact spectral tuning (Yokoyama et al. 2008). The effects of the other substitutions on spectral tuning have never been studied. To study the functional effects of the observed Rh2 substitutions we expressed reconstituted Rh2 visual pigments in vitro. The absorbance curves we obtained were of high quality and allowed us to obtain accurate values of λmax for each visual pigment (Fig. 5, Table 3). Despite having accumulated multiple substitutions, spectral sensitivities of RH2 pigments are largely similar (point estimates of peak absorbance vary by about 1nm) and not significantly different from each other (Table 3). Therefore, the observed substitutions in the RH2 pigments of Setophaga warblers do not appear to strongly impact spectral tuning, an unexpected finding given 4 of these sites are inferred to have evolved under positive selection.
Figure 5.
Absorbance spectra of the regenerated New World warbler and Old World warbler Rh2 visual pigments. Main figure shows dark spectra and insets correspond to dark-light difference spectra for Rh2. The x-axis for insets has the same range as the main graph in all cases. Wavelengths of maximum absorbance (λmax) shown are the average of 3 different absorbance measurements. In addition to the ancestral Rh2 pigment present in most species, we expressed all the Rh2 visual pigments that had substitutions, except for D332E, which is not thought to affect spectral tuning (Yokoyama et al. 2008). Rh2 variant V1 corresponds to the one present in Setophaga pensylvanica, Rh2 V2 to that in S. fusca and Rh2 V3 is in S. castanea S. petechia and S. striata (See Fig. 2 and Table 3)
Table 3.
Spectral sensitivities for RH2 visual pigments expressed in vitro. Substitutions and spectral shifts are relative to the ancestral sequence, which is common to many species in both the New World warblers and the Old World warblers. Positive shifts correspond to changes in λmax toward longer wavelengths. All the observed shifts are small and spectral sensitivities of Rh2 pigments are not significantly different from each other. Residues in bold are inferred to have evolved under positive selection.
| Substitutions | Estimated† λmax (nm) |
Shift (nm) |
Species‡ |
|---|---|---|---|
| 503.5 ± 0.4 | - | Ancestor | |
| I7V; C85F; T284S | 504.2 ± 1.0 | + 0.7 | Spen |
| I7V; C85F; V227I; S340G | 504.3 ± 0.1 | + 0.8 | Sfus |
| T284S | 504.6 ± 0.4 | + 1.1 | Scas, Spet, Sstr |
Standard errors are calculated from curve fits of 3 different absorbance measurements.
Species that carry the sequence variant (see Fig. 2 for abbreviations).
Discussion
We traced sequence and functional evolution of the Rh2 visual pigments in two clades of warblers and found strong conservatism over much of the ~60 million years of evolution that separate species in these two clades. This conservatism was mirrored in the rod pigment, Rh1, used in dim light vision, during diversification within the clades. While Rh2 was generally conserved in our dataset and subject to purifying selection (ω ≈ 0.2), it experienced an increase in its rate of evolution in Setophaga, exhibiting evidence for positive selection (ω = 2.26 in clade models, Table 2). Ten substitutions occurred at the 6 variable sites among the New World warbler species we studied. Some substitutions in distantly related clades parallel those in the New World warblers. Parallel evolution and evolutionary reversal are not uncommon in the opsins (Yokoyama & Yokoyama 1990; Yokoyama 2008; Hofmann et al. 2012; Bloch et al. 2015).
The generally slow evolution of the Rh2 visual pigment extends beyond the passerines. Sequences of 44 avian genomes across the bird tree, representing all major clades, indicate slow rates of Rh2 evolution, with mean ω values below 0.25 (Zhang et al. 2014). The only exception was in the penguins (Sphenisciformes), in which two species show fast rates of evolution, apparently due to relaxed selection after loss of function of the Rh2 gene (Zhang et al. 2014). Slow opsin evolution may be explained in two complementary ways. The first is that many sites are subject to purifying selection for functional reasons (Jordan et al. 2002; Wall et al. 2005; Zhang et al. 2006). Evolutionary constraints associated with the function and complexity of the visual system could result in purifying selection across the entire molecule. Opsin evolution may be constrained by multiple selective pressures imposed by the environment and an organism’s ecology (diet, predators, etc.), in addition to discrimination of conspecific colours during social interactions. Further, spectral tuning is only one aspect of opsin function (see below). The spectral sensitivity of visual pigments therefore may not be able to shift in response to one selection pressure without becoming maladaptive for another aspect of vision.
The second factor that may contribute to slow opsin evolution is high expression levels in the retina. High expression is a generally strong correlate of slow protein evolution (Pal et al. 2001; Drummond et al. 2005). The most widely accepted explanation is that mutations affect protein folding, thereby causing cell toxicity and toxicity increases in proportion to quantity of misfolded protein. For highly expressed proteins, selection may be sufficiently strong that adjustments in the cell machinery come to make one or a few related sequences evolve to fold particularly efficiently (Drummond et al. 2005; Serohijos et al. 2012). In consequence, any mutations in highly expressed proteins are strongly selected against. All opsins are very highly expressed in the bird retina (Bloch 2015). Hence slow evolutionary rates in the opsins may be partly a result of this basic constraint.
Accelerated evolution of Rh2 in the New World warbler genus Setophaga indicates constraints on opsin gene evolution, associated with either visual pigment function or protein folding, have to some extent been overcome in this clade. The Setophaga warblers are particularly colourful and sexually dimorphic species (Fig. 1), suggesting they are subject to relatively strong sexual selection pressures. Bowerbirds are known to be strongly sexually selected, using colour in their displays in plumage and/or bower decorations (Uy & Borgia 2000; Endler et al. 2005). The Bowerbirds have also accumulated multiple non-synonymous substitutions in the Rh2 gene, albeit over a longer time period than the New World warblers. By comparison, the dull and monomorphic Old World warblers, which are of intermediate age, show no substitutions in the Rh2 opsin at all, at least among those species we have been able to study. Considering the multiple hypotheses linking the visual sensory system to colour pattern evolution that have been developed (Lande 1981; Boughman 2002; Bradbury & Vehrencamp 2011), we suggest accelerated rates of Rh2 opsin evolution are related to sexual selection pressures.
The most obvious way in which color perception should vary as a result of opsin sequence changes is by altered spectral tuning. However, our results imply tuning differences among the Rh2 opsins of warblers are small. Rh2 spectral tuning is relatively invariant across the passerine birds as a whole: λmax measurements, mostly based in microspectrophotometry of whole retinas, range between 499 and 504nm in other species (Hart & Hunt 2007), except for bowerbirds where it is estimated at around 510nm (Coyle et al. 2012) the long-wavelength shift in the bowerbirds could be a result of the unique A158V substitution that occurred in the branch leading to this clade (Fig. 3), but this needs to be tested.
In Setophaga warblers evidence for positive selection, and differences from the pattern of stasis across long timescales, suggest that some aspects of opsin function are affected, aside from spectral sensitivity. To date Characterization of visual pigments for functions other than spectral tuning has been limited to a few model species (Sakmar et al. 2002; Sugawara et al. 2005; Bickelmann et al. 2012; Morrow & Chang in review). Molecular genetic studies have shown that substitutions at certain amino acid residues are responsible for changes in the efficiency of transducin activation, and/or rate of chromophore release when photoactivated (Yan et al. 2002; Sugawara et al. 2005; Bickelmann et al. 2012). Recent studies examining the evolution of cichlid fish rhodopsins identified a number of positively selected sites that were attributed to aspects of function other than spectral tuning (Schott et al. 2014). These changes could contribute to visual adaptation to different environments, but how they feedback on to colour perception and other aspects of sexual selection, such as distinguishing subtle difference in male displays, is completely unknown. Bloch (2015) found that opsin expression levels in the retina vary greatly among Setophaga species. One possible impact of high opsin expression is altered optical density, which changes the shape of the cone’s spectral sensitivity function and thus, is likely to affect color perception (Thomas et al. 2011). Similar principles may apply to opsin structure, and opsins may also evolve directly in response to new selection pressures resulting from altered expression levels.
Whatever the causes of the sudden burst of Rh2 evolution in the New World warblers, it is not a phenomenon common to all opsins in this clade. Rh1 evolution remains very slow throughout passerines (Table S7; Fig. S4), and shows no evidence of accelerated evolution in Setophaga. On the other hand, the Sws2 opsin diverged significantly between the New World and Old World warblers, accumulating several substitutions that lead to small spectral shifts (Bloch et al. 2015). The Sws2 opsin has also evolved within both the New World and Old World warblers (Bloch et al. 2015).
In summary, examining RH2 sequence and spectral tuning evolution in New World and Old World warblers shows this visual pigment experienced an episode of accelerated evolution in the especially colourful, sexually dimorphic, Setophaga warblers. The period of long stasis followed by a burst of evolution, together with evidence for positive selection implies that even highly expressed proteins subject to strong selective constraints can rapidly evolve when selective regimes are changed. This change in selective constraints may be associated with changes in the intensity of sexual selection on colour patterns. Increased understanding of the way in which alterations in the opsin protein translates into colour perception should lead to much better understanding of the co-evolution of female preferences for male colour and the colours themselves, the essential components of many models of sexual selection.
Supplementary Material
Acknowledgements
We especially thank J. Morrow and I. van Hazel for their invaluable help with visual pigment in vitro expression and Daniel Hooper for help with constructing the BEAST tree. We thank K. Marchetti for advice and for providing us with Old World Warbler retinas, J. Endler for comments and discussion during the initial stages of this project, as well as the Associate Editor and three anonymous reviewers for a constructive review of this manuscript. We gratefully acknowledge the Field Museum of Natural History and the Chicago Bird Collision Monitors for all their help collecting New World warbler specimens. This work was supported by the National Institute of Health NRSA 1F31EY020105 (to NIB) and National Science Foundation 1209876 (to NIB), and the Natural Sciences and Engineering Research Council (to BSWC).
Footnotes
DATA AVAILABILITY
All sequences have been deposited on GenBank (accession numbers: Rh2 KM977570-KM977595; Rh1 KM977596-KM977621). Tree files for phylogenies as well as nucleotide and amino acid alignments are available on Dryad (doi:10.5061/dryad.b2m1n). Additional data are presented as supporting information.
AUTHORS' CONTRIBUTIONS
NIB carried out the sequencing, in vitro expression of visual pigments, all the analyses and drafted the manuscript. NIB and TDP conceived of the study. TDP participated in study design and oversaw all aspects of the study. BSWC participated in the design of the study, oversaw the in vitro expression of visual pigments and molecular analyses. All authors contributed to drafting the manuscript and have read and approved the final manuscript.
References
- Barrett RDH, Hoekstra HE. Molecular spandrels: tests of adaptation at the genetic level. Nature Reviews Genetics. 2011;12:767–780. doi: 10.1038/nrg3015. [DOI] [PubMed] [Google Scholar]
- Baylor D. Photoreceptor Signals and Vision. Investigative Ophthalmology & Visual Science. 1987;28:34–49. [PubMed] [Google Scholar]
- Baylor D. How photons start vision. Proceedings of the National Academy of Sciences USA. 1996;93:560–565. doi: 10.1073/pnas.93.2.560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bickelmann C, Morrow JM, Müller J, Chang BS. Functional characterization of the rod visual pigment of the echidna (Tachyglossus aculeatus), a basal mammal. Visual Neuroscience. 2012;29:211–217. doi: 10.1017/S0952523812000223. [DOI] [PubMed] [Google Scholar]
- Bielawski JP, Yang Z. A maximum likelihood method for detecting functional divergence at individual codon sites, with application to gene family evolution. Journal of Molecular Evolution. 2004;59:121–132. doi: 10.1007/s00239-004-2597-8. [DOI] [PubMed] [Google Scholar]
- Bloch NI. Evolution of opsin expression in birds driven by sexual selection and habitat. Proceedings of The Royal Society of London Series B. 2015;282:20142321–20142321. doi: 10.1098/rspb.2014.2321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloch NI, Morrow JM, Chang BS, Price TD. SWS2 visual pigment evolution as a test of historically contingent patterns of plumage color evolution in warblers. Evolution. 2015 doi: 10.1111/evo.12572. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boughman JW. How sensory drive can promote speciation. Trends in Ecology & Evolution. 2002;17:571–577. [Google Scholar]
- Bowmaker JK. Evolution of vertebrate visual pigments. Vision Research. 2008;48:2022–2041. doi: 10.1016/j.visres.2008.03.025. [DOI] [PubMed] [Google Scholar]
- Bradbury JW, Vehrencamp SL. Principles of Animal Communication. Sunderland, MA: Sinauer Associates; 2011. [Google Scholar]
- Chang BS, Crandall KA, Carulli JP, Hartl DL. Opsin phylogeny and evolution: a model for blue shifts in wavelength regulation. Molecular Phylogenetics and Evolution. 1995;4:31–43. doi: 10.1006/mpev.1995.1004. [DOI] [PubMed] [Google Scholar]
- Chang BS, Du J, Weadick C, et al. The future of codon models in studies of molecular evolution: Ancestral reconstruction, and clade models of functional divergence. In: Cannarozzi GM, Schneider A, editors. Codon Evolution: Mechanisms and Models. Oxford University Press; 2012. pp. 145–163. [Google Scholar]
- Chang BS, Jonsson K, Kazmi M, Donoghue M, Sakmar T. Recreating a functional ancestral archosaur visual pigment. Molecular Biology and Evolution. 2002;19:1483–1489. doi: 10.1093/oxfordjournals.molbev.a004211. [DOI] [PubMed] [Google Scholar]
- Coyle BJ, Hart NS, Carleton KL, Borgia G. Limited variation in visual sensitivity among bowerbird species suggests that there is no link between spectral tuning and variation in display colouration. Journal of Experimental Biology. 2012;215:1090–1105. doi: 10.1242/jeb.062224. [DOI] [PubMed] [Google Scholar]
- Dayhoff MO. Atlas of protein sequence and structure: observed frequencies of amino acid replacements between closely related proteins. Washington DC: National Biomedical Research Foundation; 1978. [Google Scholar]
- Dean AM, Thornton JW. Mechanistic approaches to the study of evolution: the functional synthesis. Nature Reviews Genetics. 2007;8:675–688. doi: 10.1038/nrg2160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond AJ, Rambaut A. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evolutionary Biology. 2007;7:214. doi: 10.1186/1471-2148-7-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond DA, Bloom JD, Adami C, Wilke CO, Arnold FH. Why highly expressed proteins evolve slowly. Proceedings of the National Academy of Sciences USA. 2005;102:14338–14343. doi: 10.1073/pnas.0504070102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endler JA, Westcott DA, Madden JR, Robson T. Animal visual systems and the evolution of color patterns: sensory processing illuminates signal evolution. Evolution. 2005;59:1795–1818. doi: 10.1554/04-669.1. [DOI] [PubMed] [Google Scholar]
- Friedman R, Hughes AL. Likelihood-ratio tests for positive selection of human and mouse duplicate genes reveal nonconservative and anomalous properties of widely used methods. Molecular Phylogenetics and Evolution. 2007;42:388–393. doi: 10.1016/j.ympev.2006.07.015. [DOI] [PubMed] [Google Scholar]
- Gharib WH, Robinson-Rechavi M. The branch-site test of positive selection is surprisingly robust but lacks power under synonymous substitution saturation and variation in GC. Molecular Biology and Evolution. 2013;30:1675–1686. doi: 10.1093/molbev/mst062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hart NS, Hunt DM. Avian visual pigments: Characteristics, spectral tuning, and evolution. The American Naturalist. 2007;169:S7–S26. doi: 10.1086/510141. [DOI] [PubMed] [Google Scholar]
- Hauser FE, van Hazel I, Chang BS. Spectral tuning in vertebrate short wavelength-sensitive 1 (SWS1) visual pigments: can wavelength sensitivity be inferred from sequence data? Journal of Experimental Zoology. Part B, Molecular and developmental evolution. 2014 doi: 10.1002/jez.b.22576. [DOI] [PubMed] [Google Scholar]
- Hofmann CM, Marshall NJ, Abdilleh K, et al. Opsin evolution in damselfish: convergence, reversal, and parallel evolution across tuning sites. Journal of Molecular Evolution. 2012;75:79–91. doi: 10.1007/s00239-012-9525-0. [DOI] [PubMed] [Google Scholar]
- Hunt DM, Carvalho LS, Cowing JA, Davies WL. Evolution and spectral tuning of visual pigments in birds and mammals. Philosophical Transactions of the Royal Society B. 2009;364:2941–2955. doi: 10.1098/rstb.2009.0044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai H, Kefalov V, Sakurai K, et al. Molecular properties of rhodopsin and rod function. The Journal of Biological Chemistry. 2007;282:6677–6684. doi: 10.1074/jbc.M610086200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan IK, Rogozin IB, Wolf YI, Koonin EV. Essential genes are more evolutionarily conserved than are nonessential genes in bacteria. Genome Research. 2002;12:962–968. doi: 10.1101/gr.87702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagman D, Ocampo Daza D, Widmark J, et al. The vertebrate ancestral repertoire of visual opsins, transducin alpha subunits and oxytocin/vasopressin receptors was established by duplication of their shared genomic region in the two rounds of early vertebrate genome duplications. BMC Evolutionary Biology. 2013;13:238. doi: 10.1186/1471-2148-13-238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lande R. Models of speciation by sexual selection on polygenic traits. Proceedings of the National Academy of Sciences USA. 1981;78:3721–3725. doi: 10.1073/pnas.78.6.3721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddison WP, Maddison DR. Mesquite: a modular system for evolutionary analysis. 2001 [Google Scholar]
- Molday RS, MacKenzie D. Monoclonal antibodies to rhodopsin: characterization, cross-reactivity, and application as structural probes. Biochemistry. 1983;22:653–660. doi: 10.1021/bi00272a020. [DOI] [PubMed] [Google Scholar]
- Morrow JM, Chang BS. The p1D4-hrGFP II expression vector: a tool for expressing and purifying visual pigments and other G protein-coupled receptors. Plasmid. 2010;64:162–169. doi: 10.1016/j.plasmid.2010.07.002. [DOI] [PubMed] [Google Scholar]
- Morrow JM, Chang BS. Molecular mechanisms mediating the natural variation of retinal release in rhodopsin. in review. [Google Scholar]
- Nielsen R, Yang Z. Likelihood models for detecting positively selected amino acid sites and applications to the HIV-1 envelope gene. Genetics. 1998;148:929–936. doi: 10.1093/genetics/148.3.929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nozawa M, Suzuki Y, Nei M. Reliabilities of identifying positive selection by the branch-site and the site-prediction methods. Proceedings of the National Academy of Sciences USA. 2009;106:6700–6705. doi: 10.1073/pnas.0901855106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pal C, Papp B, Hurst LD. Highly expressed genes in yeast evolve slowly. Genetics. 2001;158:927–931. doi: 10.1093/genetics/158.2.927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price TD, Hooper DM, Buchanan CD, et al. Niche filling slows the diversification of Himalayan songbirds. Nature. 2014;509:222–225. doi: 10.1038/nature13272. [DOI] [PubMed] [Google Scholar]
- Price TD, Lovette I, Bermingham E, Gibbs H, Richman A. The imprint of history on communities of North American and Asian warblers. The American Naturalist. 2000;156:354–367. doi: 10.1086/303397. [DOI] [PubMed] [Google Scholar]
- Sakmar TP, Menon ST, Marin EP, Awad ES. Rhodopsin: insights from recent structural studies. Annual Review of Biophysics and Biomolecular Structure. 2002;31:443–484. doi: 10.1146/annurev.biophys.31.082901.134348. [DOI] [PubMed] [Google Scholar]
- Schott RK, Refvik SP, Hauser FE, López-Fernández H, Chang BS. Divergent positive selection in rhodopsin from lake and riverine cichlid fishes. Molecular Biology and Evolution. 2014;31:1149–1165. doi: 10.1093/molbev/msu064. [DOI] [PubMed] [Google Scholar]
- Seehausen O, Terai Y, Carleton KL, et al. Speciation through sensory drive in cichlid fish. Nature. 2008;455:620–626. doi: 10.1038/nature07285. [DOI] [PubMed] [Google Scholar]
- Serohijos AWR, Rimas Z, Shakhnovich EI. Protein biophysics explains why highly abundant proteins evolve slowly. Cell Reports. 2012;2:249–256. doi: 10.1016/j.celrep.2012.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y, Radlwimmer FB, Yokoyama S. Molecular genetics and the evolution of ultraviolet vision in vertebrates. Proceedings of the National Academy of Sciences USA. 2001;98:11731–11736. doi: 10.1073/pnas.201257398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shyue SK, Boissinot S, Schneider H, et al. Molecular genetics of spectral tuning in New World monkey color vision. Journal of Molecular Evolution. 1998;46:697–702. doi: 10.1007/pl00006350. [DOI] [PubMed] [Google Scholar]
- Sugawara T, Terai Y, Imai H, et al. Parallelism of amino acid changes at the RH1 affecting spectral sensitivity among deep-water cichlids from Lakes Tanganyika and Malawi. Proceedings of the National Academy of Sciences USA. 2005;102:5448–5453. doi: 10.1073/pnas.0405302102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki Y. False-positive results obtained from the branch-site test of positive selection. Genes & genetic systems. 2008;83:331–338. doi: 10.1266/ggs.83.331. [DOI] [PubMed] [Google Scholar]
- Thomas PBM, Formankiewicz MA, Mollon JD. The effect of photopigment optical density on the color vision of the anomalous trichromat. Vision Research. 2011;51:2224–2233. doi: 10.1016/j.visres.2011.08.016. [DOI] [PubMed] [Google Scholar]
- Uy JA, Borgia G. Sexual selection drives rapid divergence in bowerbird display traits. Evolution. 2000;54:273–278. doi: 10.1111/j.0014-3820.2000.tb00027.x. [DOI] [PubMed] [Google Scholar]
- van Hazel I, Sabouhanian A, Day L, Endler JA, Chang BS. Functional characterization of spectral tuning mechanisms in the great bowerbird short-wavelength sensitive visual pigment (SWS1), and the origins of UV/violet vision in passerines and parrots. BMC Evolutionary Biology. 2013;13:250. doi: 10.1186/1471-2148-13-250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wall DP, Hirsh AE, Fraser HB, et al. Functional genomic analysis of the rates of protein evolution. Proceedings of the National Academy of Sciences USA. 2005;102:5483–5488. doi: 10.1073/pnas.0501761102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallis M. The molecular evolution of vertebrate growth hormones: a pattern of near-stasis interrupted by sustained bursts of rapid change. Journal of Molecular Evolution. 1996;43:93–100. doi: 10.1007/BF02337353. [DOI] [PubMed] [Google Scholar]
- Wallis M. Episodic evolution of protein hormones in mammals. Journal of Molecular Evolution. 2001;53:10–18. doi: 10.1007/s002390010187. [DOI] [PubMed] [Google Scholar]
- Weadick CJ, Chang BS. An Improved Likelihood Ratio Test for Detecting Site-Specific Functional Divergence among Clades of Protein-Coding Genes. Molecular Biology and Evolution. 2012;29:1297–1300. doi: 10.1093/molbev/msr311. [DOI] [PubMed] [Google Scholar]
- Yan ECY, Kazmi MA, De S, et al. Function of extracellular loop 2 in rhodopsin: glutamic acid 181 modulates stability and absorption wavelength of metarhodopsin II. Biochemistry. 2002;41:3620–3627. doi: 10.1021/bi0160011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z. Likelihood ratio tests for detecting positive selection and application to primate lysozyme evolution. Molecular Biology and Evolution. 1998;15:568–573. doi: 10.1093/oxfordjournals.molbev.a025957. [DOI] [PubMed] [Google Scholar]
- Yang Z. PAML 4: Phylogenetic analysis by maximum likelihood. Molecular Biology and Evolution. 2007;24:1586–1591. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
- Yang Z, Bielawski J. Statistical methods for detecting molecular adaptation. Trends in Ecology & Evolution. 2000;15:496–503. doi: 10.1016/S0169-5347(00)01994-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Nielsen R. Synonymous and nonsynonymous rate variation in nuclear genes of mammals. Journal of Molecular Evolution. 1998;46:409–418. doi: 10.1007/pl00006320. [DOI] [PubMed] [Google Scholar]
- Yang Z, Reis dos M. Statistical properties of the branch-site test of positive selection. Molecular Biology and Evolution. 2011;28:1217–1228. doi: 10.1093/molbev/msq303. [DOI] [PubMed] [Google Scholar]
- Yang Z, Nielsen R, Goldman N. In defense of statistical methods for detecting positive selection. Proceedings of the National Academy of Sciences USA. 2009;106 doi: 10.1073/pnas.0904550106. E95–author reply E96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Nielsen R, Goldman N, Pedersen AM. Codon-substitution models for heterogeneous selection pressure at amino acid sites. Genetics. 2000;155:431–449. doi: 10.1093/genetics/155.1.431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Wong WSW, Nielsen R. Bayes empirical bayes inference of amino acid sites under positive selection. Molecular Biology and Evolution. 2005;22:1107–1118. doi: 10.1093/molbev/msi097. [DOI] [PubMed] [Google Scholar]
- Yokoyama R, Yokoyama S. Convergent evolution of the red- and green-like visual pigment genes in fish, Astyanax fasciatus, and human. Proceedings of the National Academy of Sciences USA. 1990;87:9315–9318. doi: 10.1073/pnas.87.23.9315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoyama S. Molecular evolution of vertebrate visual pigments. Progress in Retinal and Eye Research. 2000;19:385–419. doi: 10.1016/s1350-9462(00)00002-1. [DOI] [PubMed] [Google Scholar]
- Yokoyama S. The spectral tuning in the short wavelength-sensitive type 2 pigments. Gene. 2003;306:91–98. doi: 10.1016/s0378-1119(03)00424-4. [DOI] [PubMed] [Google Scholar]
- Yokoyama S. Evolution of dim-light and color vision pigments. Annual review of genomics and human genetics. 2008;9:259–282. doi: 10.1146/annurev.genom.9.081307.164228. [DOI] [PubMed] [Google Scholar]
- Yokoyama S, Radlwimmer FB. The molecular genetics and evolution of red and green color vision in vertebrates. Genetics. 2001;158:1697–1710. doi: 10.1093/genetics/158.4.1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoyama S, Tada T, Zhang H, Britt L. Elucidation of phenotypic adaptations: Molecular analyses of dim-light vision proteins in vertebrates. Proceedings of the National Academy of Sciences USA. 2008;105:13480–13485. doi: 10.1073/pnas.0802426105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai W, Nielsen R, Goldman N, Yang Z. Looking for Darwin in genomic sequences--validity and success of statistical methods. Molecular Biology and Evolution. 2012;29:2889–2893. doi: 10.1093/molbev/mss104. [DOI] [PubMed] [Google Scholar]
- Zhang G, Li C, Li Q, et al. Comparative genomics reveals insights into avian genome evolution and adaptation. 2014;346:1311–1320. doi: 10.1126/science.1251385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J. Evaluation of an improved branch-site likelihood method for detecting positive selection at the molecular level. Molecular Biology and Evolution. 2005;22:2472–2479. doi: 10.1093/molbev/msi237. [DOI] [PubMed] [Google Scholar]
- Zhang J, Liao B-Y, Scott NM. Impacts of gene essentiality, expression pattern, and gene compactness on the evolutionary rate of mammalian proteins. Molecular Biology and Evolution. 2006;23:2072–2080. doi: 10.1093/molbev/msl076. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.