Abstract
A label-free method for the measurement of the activity of HIV-1 protease is developed by real-time monitoring of the cleavage of a peptide substrate by HIV-1 protease in a nanopore. The method is rapid and sensitive: picomolar concentrations of HIV-1 protease could be detected in ~10 minutes. Simulated clinical samples are analyzed, and the activity of HIV-1 protease could be accurately detected. Our developed nanopore sensor design strategy should find useful application in the development of stochastic sensors for other proteases of medical, pharmaceutical, and biological importance.
Keywords: HIV-1 protease, proteolysis, peptide, biosensors, nanopore
1. Introduction
In spite of the significant advances in new diagnostic tools and therapeutic drugs, and better ways to prevent diseases, humans remain vulnerable to health threats posed by infectious diseases. One of such examples is the present pandemic of human immunodeficiency virus (HIV/AIDS), which originated in west-central Africa during the early twentieth century, and has caused nearly 30 million deaths since the first case of AIDS was reported on June 5, 1981 (De Cock et al., 2011). At present, the majority of the developed methods for HIV detection are based on the detection of the presence of antibodies that the patient’s body makes against HIV (Bhimji et al., 2013; Laird et al., 2013), direct molecular recognition of HIV and its components such as specific nucleic acid sequences or antigens (Zhang et al., 2013; Palmer et al., 2003; Yan et al., 2011), or measurement of the activity of HIV-1 protease (HIV-1 PR) (Esseghaier et al., 2013; Biswas et al., 2011; Davis et al., 2009), many of which are often laborious and time-consuming, and/or require the use of labels or sophisticated instruments.
Herein, we report a real-time nanopore sensing method for the sensitive detection of HIV-1 PR. Nanopore technology is an emerging label-free and amplification-free technique for measuring single molecules (Wang et al. 2013). By monitoring the ionic current modulations produced by the passage of the target analyte through a single nanopore at a fixed applied potential, the concentration of the analyte can be obtained by the frequency of occurrence (1/τon) of the blockage events, while its identity can be determined from the mean residence time (τoff) of the analyte coupled with the extent of current blockage (amplitude). Under experimental conditions of constant electrolyte pH, temperature, and applied potential bias, the event blockage amplitude is related to the size, structure, and/or conformation of the analyte molecule, while the event residence time depends on the length of the analyte and the strength of the interaction between the analyte and the nanopore. In addition to biosensing (Kang et al.. 2006; Liu et al., 2011; Siwy et al., 2005; Stefureac et al., 2010; Ying et al., 2012; Vercoutere et al., 2001; Schibel et al., 2010), nanopore analysis has been explored for a variety of other applications, including studying covalent and non-covalent bonding interactions (Zhao et al., 2008; Luchian et al., 2003), investigating biomolecular conformational changes (Talaga and Li, 2009; Ying et al., 2011), probing enzyme kinetics (Macrae et al., 2009; Majd et al., 2009; Zhao et al., 2009), DNA characterization and sequencing (Astier et al., 2006; Garaj et al., 2013; Squires et al., 2013; Stoloff et al., 2013), and so on.
2. Experimental
2.1. Materials and reagents
The HIV-1 protease substrate peptide with a sequence of FFSQNYPIVQ (> 98% pure) was purchased from Biomatik corporation (Wilmington, DE), while the HIV-1 protease was ordered from BioVendor Lab (Brno, Czech Republic). All the other chemicals were obtained from Sigma-Aldrich (St. Louis, MO). All of the peptide, protease, and chemicals were dissolved in HPLC-grade water (ChromAR, Mallinckrodt Baker). The stock solutions of the peptide and the protease were prepared at 10 mM, and 300 µg/mL, respectively, and were kept at −80 °C before and after use. The electrolyte used in this work contained 1.0 M NaCl buffered with 1 mM EDTA and 1 mM NaH2PO4, with the pH adjusted to 4.7 using H3PO4 solution. Lipid 1, 2-diphytanoylphosphatidylcholine was obtained from Avanti Polar Lipids (Alabaster, AL). Teflon film (25 µm thick) was purchased from Goodfellow (Malvern, PA).
2.2. Preparation and formation of protein pores
The mutant α-hemolysin (αHL) M113F gene was constructed by site-directed mutagenesis (Mutagenex, Piscataway, NJ) with a wild-type αHL gene in a T7 vector (pT7–αHL) (Song et al., 1996). The mutant αHL monomers were first synthesized by coupled in vitro transcription and translation (IVTT) using the E. Coli T7 S30 Extract System for Circular DNA from Promega (Madison, WI). Subsequently, they were assembled into homoheptamers by adding rabbit red cell membranes and incubating for 1~2 h. The heptamers were then purified by SDS-polyacrylamide gel electrophoresis and stored in aliquots at −80°C (Zhao et al., 2009).
2.3. Electrical recording
A bilayer of 1, 2-diphytanoylphosphatidylcholine was formed on an aperture (150 µm) in a Teflon septum that divided a planar bilayer chamber into cis and trans compartments. The formation of bilayer was achieved using Montal-Mueller method (Montal and Mueller, 1972). Unless otherwise noted, all the experiments were performed under a series of symmetrical buffer conditions with a 2.0 mL solution comprising 1 M NaCl, 1 mM EDTA, and 10 mM NaH2PO4 (pH 4.7) at 26 ± 1 °C. The αHL protein was added to the cis compartment, which was connected to “ground”, while the peptide substrate and HIV-1 protease were added to the trans compartment. The final concentration of the αHL proteins used for the single channel insertion was 0.2–2.0 ng·mL−1. Currents were recorded with a patch clamp amplifier (Axopatch 200B, Molecular Devices; Sunnyvale, CA, USA). They were low-pass filtered with a built-in four-pole Bessel filter at 5 KHz and sampled at 50 KHz by a computer equipped with a Digidata 1322A/D converter (Molecular Devices).
2.4. Data analysis
Data were analyzed with the following software: pClamp 10.3 (Molecular Devices), Origin 8.0 (Microcal, Northampton, MA), and SigmaPlot 12.0 (Systat Software Inc., San Jose, CA). Conductance values were obtained from the amplitude histograms after the peaks were fit to Gaussian functions. Values of τon and τoff for the peptide events were obtained from the open state (1) and close state (0) dwell time histograms, respectively by fitting the distributions to single exponential functions by the Levenberg-Marquardt procedure (Wang et al., 2013). The event frequency (f) was calculated by using the equation f = 1/τon.
3. Results and discussion
3.1. Principle for nanopore detection of HIV-1 PR
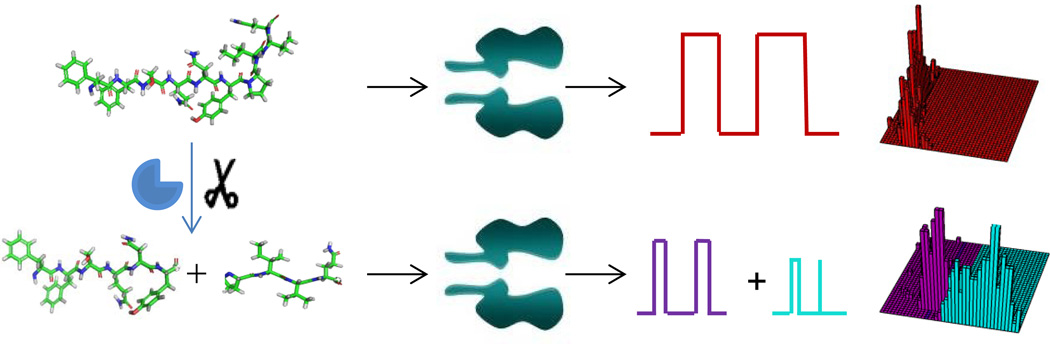
The activity of HIV-1 PR is measured by real-time monitoring of the ionic current modulations arising from the substrate-protease interactions. As shown in Figure 1, in the absence of HIV-1 PR, the peptide substrate produces only one major type of current modulation events during its translocation through the nanopore. However, in contrast, in the presence of the protease, the substrate is cleaved into two fragments. Since the substrate breakdown products have shorter lengths than the substrate, new blockage events having smaller residence times (τoff) and/or amplitudes from those of the substrate may be observed. Furthermore, with a constant substrate concentration and a fixed amount of recording (i.e., reaction) time, the concentration and hence the event frequency of the produced substrate cleavage products or the remaining substrate depends on the activity of the HIV-1 PR.
Figure 1.
Schematic representation of the principle for nanopore detection of the activity of HIV-1 PR. Without HIV-1 PR, the peptide substrate produces only one major type of current modulation events in the nanopore. With the protease, the substrate is cleaved into two fragments, which produce two new types of blockage events having smaller residence times and/or amplitudes than those of the substrate. By monitoring the frequency or counts of the produced new events, the activity of HIV-1 PR could be quantified.
3.2. Measurement of the activity of HIV-1 protease
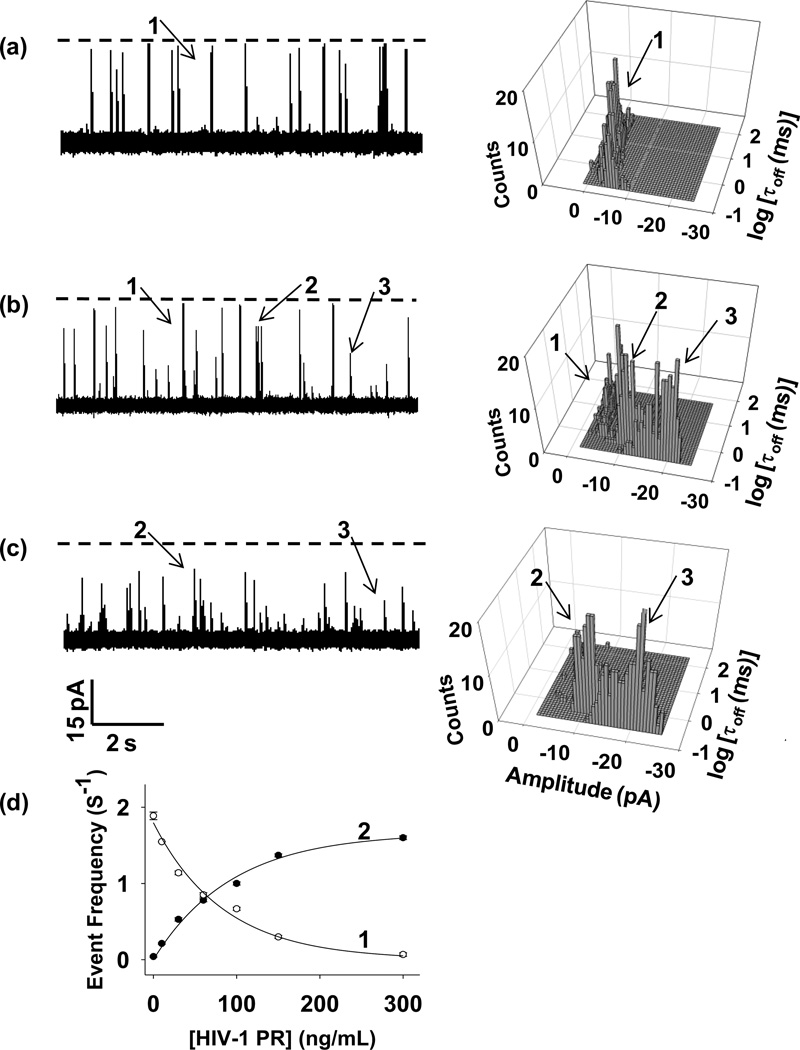
To demonstrate this concept, our initial experiment was carried out at an applied potential bias of −40 mV in an electrolyte solution comprising 1 M NaCl, 1 mM EDTA and 1 mM NaH2PO4 (pH 4.7). This voltage has been demonstrated appropriate for nanopore peptide analysis (Zhao et al., 2009), while pH 4.7 is the optimum solution pH for the detection of HIV-1 PR (Esseghaier et al., 2013). The nanopore sensing element used was a mutant αHL protein, (M113F)7, while a peptide having a sequence of FFSQNYPIVQ was employed as the substrate. It has been shown that the HL (M113F)7 protein could provide an enhanced resolution (e.g., prolonged residence time) for (bio-)molecule recognition compared with that observed with the wild-type αHL pore (Wang et al., 2014). Note that in our substrate design, two additional Phe amino acids were added to the sequence of a well-known HIV-1 PR substrate (Perez et al., 2010), SQNYPIVQ, for the purpose of creating two cleavage segments (i.e., FFSQNY and PIVQ) with different lengths. Unlike various conventional enzyme assays which detect the enzyme activity predominantly based on the signal decrease in the substrate, our nanopore sensor measures the HIV-1 PR activity based on the signal increase in the substrate degradation products. One significant advantage of such a sensor design strategy is that other interfering proteases (i.e., false positives) could be differentiated from the target HIV-1 PR if they cleave the peptide substrate at different positions, thus improving the sensor accuracy and selectivity. Since the molecular size (with dimensions of 45 × 23 × 25 Å) (Frutos et al., 2007) of the HIV-1 PR (a 99 amino acid aspartyl protease that functions as a homodimer with only one active site) is larger than that of the protein pore transmembrane domain (20 Å diameter) (Wang et al., 2013), it cannot enter the nanopore and hence cannot produce current blockage events that might interfere with the identification of the target peptide(s) (Supporting Information, Figure S1). Our experimental results showed that, in the absence of the HIV-1 PR, the buffer solution containing the substrate peptide produced only one major type of current blockage with a mean residual current around −3.5 pA (Figure 2a). However, upon addition of HIV-1 PR to the solution, two new types of blockage events having mean residual currents of −9.5 pA, and −18.0 pA were observed (they were labeled as “type 2” and “type 3” events in Figure 2b and 2c). Note that in addition to their amplitude difference, these two new types of events also showed significantly different residence times. Furthermore, with an increase in the concentration of the added protease, the event frequency of the new events increased, while that of the substrate decreased, clearly suggesting that the new events are attributed to the proteolytic cleavage process, specifically from the produced two fragments FFSQNY and PIVQ. It should be noted that since the smaller amplitude blockage events of the substrate degradation products partially overlapped with the background current spikes (~ one event per ten seconds) of the (M113F)7 pore, only the larger amplitude new type of (i.e., type 2) events were included in the data analysis of the event frequency. The detection limit (defined as the concentration corresponding to three times the standard deviation of a blank signal) for HIV-1 PR in a 1-h enzymatic reaction was 0.47 ng/mL (equivalent to 47 pM).
Figure 2.
Measurement of the activity of HIV-1 PR in a nano-cavity. (a) 0 ng/mL HIV-1 PR; (b) 60 ng/mL HIV-1 PR; and (c) 300 ng/mL HIV-1 PR. (Left) Typical single-channel current recording trace segments after 55 min proteolytic reactions. Dashed lines represent the levels of zero current; (Right) the corresponding 3D plots of event count vs. residence time vs. blockage amplitude. Labels 1, 2, and 3 in Figs. 2a–c represent events attributed to the substrate, and the two degradation products, respectively. d) Plot of event frequency versus the concentration of HIV-1 PR. Labels 1 and 2 represent the substrate and the cleavage product, respectively. The experiments were performed by real-time monitoring of the substrate-protease interaction continuously for 1 h at −40 mV in 1 M NaCl solution buffered with 1 mM EDTA and 1 mM NaH2PO4 (pH 4.7). The concentration of the substrate peptide was 5 µM. Event counts and event frequency in Figs. 2a–d were calculated based on the last 5 min trace segment of a 60 min single channel recording.
3.3. Effect of proteolytic reaction time on the nanopore HIV-1 sensor
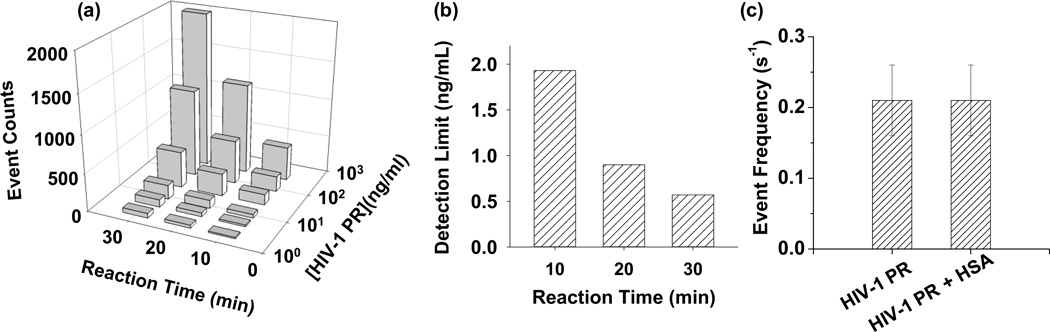
To examine whether sensitive detection of HIV-1 PR can be achieved rapidly, the effect of the monitoring time (i.e., the reaction time) on the detection limit for protease detection was systematically studied. It should be noted that, in a proteolytic reaction, the instantaneous concentrations and hence the event frequencies of the substrate and the degradation products vary with the reaction time until all the substrate is degraded (Supporting Information, Figure S2). Hence, unlike the conventional nanopore sensing, where the event frequency is used as a parameter in the dose-response curve, the number of events (i.e., event counts) is used instead in our reaction time effect study. One advantage of replacing the event frequency with the number of events in the data analysis is that this approach can remedy the small event frequency issue, which is due to the low concentration of the degradation products at the early stage of an enzymatic reaction (especially for a reaction with a low concentration of protease), so that a long recording time is needed to collect enough events for statistical analysis. Our experimental results (Figure 3) showed that, although decreasing the recording time resulted in a worse sensor sensitivity, sensitive detection of HIV-1 PR was still able to be accomplished even in a 10 minute recording with a detection limit as low as 1.93 ng/mL (equivalent to 193 pM).
Figure 3.
Characteristics of the HIV-1 PR nanopore sensor. (a) Dose-response curve for HIV-1 PR detection at various reaction times; (b) The effect of proteolytic reaction time on the detection limit; and (c) Interference study for the HIV-1 PR nanopore detection system. The experiments were performed at −40 mV in 1 M NaCl solution buffered with 1 mM EDTA and 1 mM NaH2PO4 (pH 4.7) in the presence of 5 µM peptide FFSQNYPIVQ. The event counts in Fig. 3a were obtained after deducting the background signals. The concentrations of HIV-1 PR and HSA used in Fig. 3c were 30 ng/mL, and 20 µM, respectively. The event frequency in Fig. 3c was calculated based on the total number of peptide events having mean residual currents of −9.5 pA collected in a 10 min single-channel recording.
3.4. Simulated clinical sample analysis
To demonstrate the application of our nanopore sensor in the analysis of samples resembling those relevant for clinical analysis, three samples were examined. One sample contained only HIV-1 PR, while the other two contained a mixture of human serum albumin (HSA, a dominant protein in human blood) and HIV-1 PR, and a mixture of human serum and HIV-1 PR, respectively. Our experimental results (Figure 3c) showed that the event frequencies of the HIV-1 PR and the HIV-1 PR/HSA mixture samples were not significantly different, suggesting HSA would not affect HIV-1 PR detection. However, in the case of the HIV-1 PR spiked serum sample, we found that serum disturbed lipid bilayer, making long-time real-time monitoring of the protease-substrate interaction extremely difficult. In order to evaluate the effect of human serum on HIV-1 PR detection, the HIV-1 PR spiked serum sample and the substrate were incubated for 20 minutes before addition to the nanopore sensing chamber for single channel recording. As a control, a HIV-1 PR standard solution was analysed similarly. Our results showed that these two samples produced similar frequencies for the target peptide events (Supporting Information, Fig. S4), suggesting our developed nanopore sensor can effectively detect HIV-1 PR in the presence of human serum.
4. Conclusions
In summary, by direct monitoring of the events produced by the translocation of the substrate degradation products through a nanopore, a real-time, label-free method for the measurement of the activity of HIV-1 PR is developed. Compared with various protease detection techniques developed thus far, our nanopore sensor has several advantages. For example, the traditional antibody-based protease assay such as ELISA is laborious and time-consuming, while the standard methods to measure protease activity require the use of fluorescence- or radio-labelled substrates. Given the high sensitivity of the method and its potential to discriminate the target protease from false positives, our nanopore sensor design strategy should find useful application in the development of stochastic sensors for other proteases, especially those have potential clinical values for disease diagnosis or prognosis, and have become priority targets for new pharmaceuticals.
Supplementary Material
Highlights.
We developed a label-free method for measuring the activity of HIV-1 protease.
The method could detect picomolar concentrations of HIV-1 protease in ~10▒minutes.
The method has the potential to discriminate target proteases from false positives.
Simulated clinical samples were successfully analyzed.
Acknowledgements
This work was financially supported by the National Institutes of Health (1R15GM110632), and Department of Homeland Security (HSHQDC-09-C-00091).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Astier Y, Braha O, Bayley H. J. Am. Chem. Soc. 2006;128:1705–1710. doi: 10.1021/ja057123+. [DOI] [PubMed] [Google Scholar]
- Bhimji A, Zaragoza A, Live L, Kelley S. Anal. Chem. 2013;14:6813–6819. doi: 10.1021/ac4009429. [DOI] [PubMed] [Google Scholar]
- Biswas P, Cella L, Kang S, Mulchandani A, Yates M, Chen W. Chem. Commun. 2011;47:5259–5261. doi: 10.1039/c1cc10648a. [DOI] [PubMed] [Google Scholar]
- De Cock KM, Curran JW, Jaffe HW. Emerg. Infect. Dis. 2011;17:1044–1048. doi: 10.3201/eid1706.100184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis D, Tebbs I, Daniels S, Stahl S, Kaufman J, Wingfield P, Bowman M, Chmielewski J, Yarchoan R. Biochem. J. 2009;419:497–506. doi: 10.1042/BJ20082068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esseghaier C, Ng A, Zourob M. Biosens. Bioelectron. 2013;41:335–341. doi: 10.1016/j.bios.2012.08.049. [DOI] [PubMed] [Google Scholar]
- Frutos S, Rodriguez-Mias RA, Madurga S, Collinet B, Reboud-Ravaux M, Ludevid D, Giralt E. Biopolymers. 2007;88:164–173. doi: 10.1002/bip.20685. [DOI] [PubMed] [Google Scholar]
- Garaj S, Liu S, Golovchenko J, Branton D. Proc. Natl. Acad. Sci. USA. 2013;110:12192–12196. doi: 10.1073/pnas.1220012110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang X, Cheley S, Guan X, Bayley H. J. Am. Chem. Soc. 2006;128:10684–10685. doi: 10.1021/ja063485l. [DOI] [PubMed] [Google Scholar]
- Laird G, Eisele E, Rabi S, Lai J, Chioma S, Blankson J, Siliciano J, Siliciano R. PLoS Pathog. 2013;9:e1003398. doi: 10.1371/journal.ppat.1003398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu A, Zhao Q, Krishantha DM, Guan X. J. Phys. Chem. Lett. 2011;2:1372–1376. doi: 10.1021/jz200525v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luchian T, Shin SH, Bayley H. Angew. Chem. Int. Ed. 2003;42:3766–3771. doi: 10.1002/anie.200351313. [DOI] [PubMed] [Google Scholar]
- Macrae MX, Blake S, Jiang X, Capone R, Estes DJ, Mayer M, Yang J. ACS Nano. 2009;11:3567–3580. doi: 10.1021/nn901231h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majd S, Yusko EC, MacBriar AD, Yang J, Mayer M. J. Am. Chem. Soc. 2009;131:16119–16126. doi: 10.1021/ja904072s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montal M, Mueller P. Proc. Natl Acad. Sci. U.S. A. 1972;69:3561. doi: 10.1073/pnas.69.12.3561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer S, Wiegand A, Maldarelli F, Bazmi H, Mican J, Polis M, Dewar R, Planta A, Liu S, Metcalf J, Mellors J, Coffin J. J. Clin. Microbiol. 2003;41:4531–4536. doi: 10.1128/JCM.41.10.4531-4536.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez MA, Fernandes PA, Ramos MJ. J. Phys. Chem. B. 2010;114:2525–2532. doi: 10.1021/jp910958u. [DOI] [PubMed] [Google Scholar]
- Siwy Z, Trofin L, Kohli P, Baker LA, Trautmann C, Martin CR. J. Am. Chem. Soc. 2005;127:5000–5001. doi: 10.1021/ja043910f. [DOI] [PubMed] [Google Scholar]
- Stefureac RI, Madampage CA, Andrievskaia O, Lee J. Biochem. Cell. Biol. 2010;88:347–358. doi: 10.1139/o09-176. [DOI] [PubMed] [Google Scholar]
- Schibel AE, An N, Jin Q, Fleming AM, Burrows CJ, White HS. J. Am. Chem. Soc. 2010;132:17992–17995. doi: 10.1021/ja109501x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song LZ, Hobaugh MR, Shustak C, Cheley S, Bayley H, Gouaux JE. Science. 1996;274:1859. doi: 10.1126/science.274.5294.1859. [DOI] [PubMed] [Google Scholar]
- Squires A, Meller A. Biophys. J. 2013;105:543–544. doi: 10.1016/j.bpj.2013.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoloff DH, Wanunu M. Curr. Opin. Biotechnol. 2013;24:699–704. doi: 10.1016/j.copbio.2012.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talaga DS, Li J. J. Am. Chem. Soc. 2009;131:9287–9297. doi: 10.1021/ja901088b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vercoutere W, Winters-Hilt S, Olsen H, Deamer D, Haussler D, Akeson M. Nat Biotechnol. 2001;19:248–252. doi: 10.1038/85696. [DOI] [PubMed] [Google Scholar]
- Wang G, Wang L, Han Y, Zhou S, Guan X. Acc. Chem. Res. 2013;46:2867–2877. doi: 10.1021/ar400031x. [DOI] [PubMed] [Google Scholar]
- Wang G, Wang L, Han Y, Zhou S, Guan X. Biosens. Bioelectron. 2014;53:453–458. doi: 10.1016/j.bios.2013.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G, Zhao Q, Kang X, Guan X. J. Phys. Chem. B. 2013;117:4763. doi: 10.1021/jp309541h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan N, O`Day E, Wheeler L, Engelman A, Lieberman J. Proc. Natl. Acad. Sci. USA. 2011;108:9244–9249. doi: 10.1073/pnas.1102943108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ying Y, Li D, Li Y, Dey SK, Kraatz HB, Long Y. Chem. Commun. 2012;48:8784–8786. doi: 10.1039/c2cc32636a. [DOI] [PubMed] [Google Scholar]
- Ying Y, Wang H, Sutherland TC, Long Y. Small. 2011;7:87–94. doi: 10.1002/smll.201001428. [DOI] [PubMed] [Google Scholar]
- Zhang D, Zhao M, He H, Guo S. Anal. Biochem. 2013;440:120–122. doi: 10.1016/j.ab.2013.05.032. [DOI] [PubMed] [Google Scholar]
- Zhao Q, Jayawardhana DA, Guan X. Biophys. J. 2008;94:1267–1275. doi: 10.1529/biophysj.107.117598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Q, Wang D, Jayawardhana DA, de Zoysa RS, Guan X. J. Am. Chem. Soc. 2009;131:6324–6325. doi: 10.1021/ja9004893. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.