Abstract
Engineering sex-specific sterility is critical for developing transgene-based Sterile Insect Technology. Targeted genome engineering achieved by customized ZFN, TALENs or CRIPSPR/Cas9 systems has been exploited extensively in a variety of model organisms. However, screening mutated individuals without a detectable phenotype is still challenging. In addition, genetically recessive mutations only detectable in homozygotes make the experiments time-consuming. Here we model a novel genetic system in the silkworm, Bombyx mori, that results in female-specific sterility by combining transgenesis with transcription activator–like effector nucleases (TALENs) technologies. This system induces sex-specific sterility at a high efficiency by targeting the female-specific exon of the B. mori doublesex (Bmdsx) gene, which has sex-specific splicing isoforms regulating somatic sexual development. Transgenic animals co-expressing TALEN left and right arms targeting the female-specific Bmdsx exon resulted in somatic mutations and female mutants lost fecundity due to lack of egg storage and abnormal external genitalia. The wild-type sexual dimorphism of abdominal segment was not evident in mutant females. In contrast, there were no deleterious effects in mutant male moths. The current somatic TALEN technologies provide a promising approach in future insect functional genetics, thus providing the basis for the development of attractive genetic alternatives for insect population management.
Keywords: doublesex, gene targeting, genitalia, morphology
Introduction
The domesticated silkworm, Bombyx mori, is one of the most economically important insects and is the foundation of sericulture. A transgene-based genetic sexing system was established recently in this species, extending Sterile Insect Technology (SIT) into a non-pest insect in which sex separation is valuable (Tan et al., 2013). This female-specific lethality system is dependent on targeting the sex-specific alternative splicing modules of doublesex (dsx), a gene that controls somatic sex determination and differentiation in the final steps of the insect “sex-determination cascade” (Baker, 1989; Steinmann-Zwicky et al., 1990). The hierarchy of sex determination gene function in lepidopteran insects is poorly understood when compared with the model dipteran insect, Drosophila melanogaster (Harrison, 2007). Defining insect sex determination pathways, including functional analysis of key regulators such as dsx, is critical for developing SIT in lepidopteran insects.
Emerging genome engineering tools such as customized zinc-finger nucleases (ZFNs), TALENs or clustered, regularly interspaced, short palindromic repeats (CRISPR)/Cas9 endonuclease-mediated systems have been applied extensively in a wide range of model organisms (Urnov et al., 2005; Hockemeyer et al., 2011; Hwang et al., 2013). These tools provide the basis for developing new strategies for SIT. However, screening in vivo mutant alleles in genes with no a priori visible phenotypes is challenging for the targeted genome editing mediated by current customized nucleases systems. Furthermore, directing expression of these nucleases in tissue-, stage- and sex-specific manners is not trivial. Gene targeting technologies in B. mori have been established recently using ZFNs, TALENs and CRISPR/Cas9 systems (Takasu et al., 2010; Sajwan et al., 2013; Wang et al., 2013). Also, germline transformation technologies mediated by the piggybac transposon are well-established and numerous cis-regulatory elements have been identified (Tamura et al., 2000). Integrating genome engineering technologies with transgenic approaches may contribute to novel SIT approaches.
Here we report the establishment of a piggyBac-based binary transgenic system in B. mori in which custom designed TALEN left and right arm constructs are expressed separately. Sequence-specifically somatic mutagenesis is induced in the offspring following crossing of the two lines. We demonstrate that this method is highly efficient in inducing somatic mutagenesis when targeting the female-specific exon of Bmdsx. The results show this transgenic TALENs system has great potential in insect functional genetics.
Results
Design the transgene-based TALENs system for B. mori
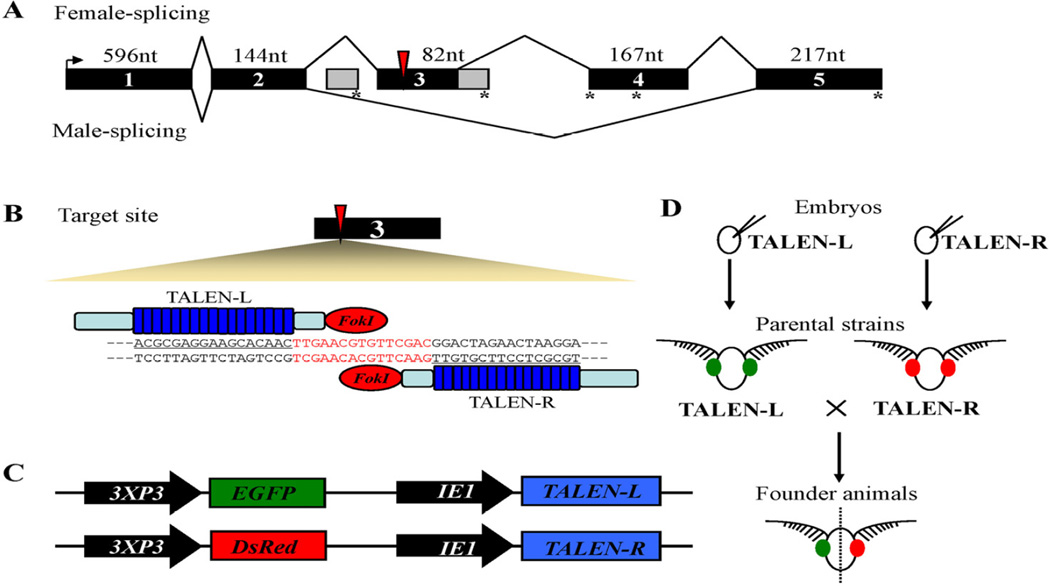
We established a piggybac-based, transgenic TALENs system in B. mori to exploit dsx gene function and its potential application in SIT. We designed TALENs targeting the sequence in Bmdsx exon 3 present only in the female-specific splicing isoforms (BmdsxF) (Fig. 1A). TALEN activity was validated in vitro using the luciferase SSA assay in 293T cells (Supplementary Table 1). Both TALEN left- (TALEN-L) and right-arm (TALEN-R) constructs had sequences encoding the FokI DNA endonuclease and a sequence-specific DNA recognition repeat domain, which targets the female-specific Bmdsx exon 3, and these were cloned into piggyBac-based transgenic vectors (Fig. 1B). An IE1 promoter was introduced to direct expression of TALENs in a ubiquitous manner and genes encoding fluorescent marker proteins also were integrated into the transgene vectors (enhanced green fluorescent protein [EGFP] for TALEN-L and the red fluorescent protein [DsRed2] for TALEN-R). Both EGFP and DsRed2 expression were under the control of the eye-specific promoter, 3×P3 (Fig. 1C). TALEN-L or TALEN-R plasmids were microinjected separately with helper plasmids into pre-blastoderm eggs and transgenic lines were established (Fig. 1d and Fig. S1 and S2). The resulting transgenic lines were viable and fertile, supporting the conclusion that the TALEN left- or right-arms alone were not functional.
Figure 1.
Sex-specific gene targeting using a transgenic TALENs system. (A) Alternative splicing of the Bombyx mori doublesex (Bmdsx) gene generates both female- and male-specific isoform. Black boxes, previously-reported canonical exons (15); numerals, length in nucleotides of canonical exons; grey boxes, newly-identified alternative exons in females; lines, introns; red arrows, TALENs target site; horizontal arrow, start codons; asterisks, stop codons. (B) TALENs and their DNA targets in exon 3 of the Bmdsx female-specific splice form. TALENs bind and cleave as dimers on a target DNA site. TALEN pairs were engineered to have 17 TALE repeats in left arm and 16 TALE repeats in right arm (dark blue boxes). The target sequences are underlined in black. Cleavage sequences are highlighted in red lettering. (C) The transgenic vectors TALEN-L and TALEN-R contain the full ORF of the TALENs protein driven by IE1 promoter, also with reporter genes EGFP or DsRed2, respectively, under the control of 3×P3 promoter. (D) Transgenic strains expressing TALEN left or TALEN right arms are established as parental strains. Somatic mutations are induced in F1 founder animals following crosses of the Left and Right Talen strains. Red and green fluorescence in the eyes confirm the presence of the appropriate transgenes construct.
Targeted mutagenesis in Bmdsx loci
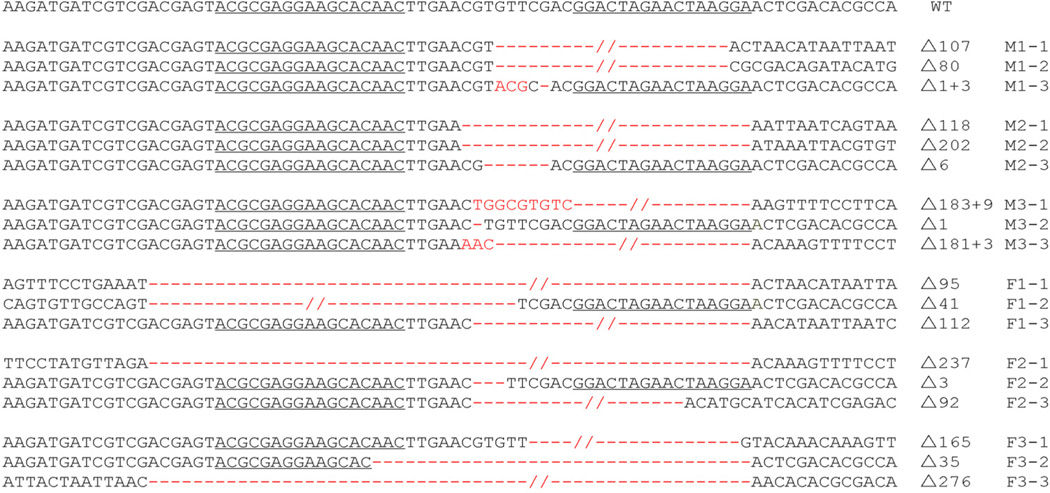
The TALEN-L and TALEN-R containing transgenic lines were crossed with each other and the F1 founder animals subjected to somatic mutagenesis analysis. Individual moths were sexed using B. mori W chromosome-specific primers, genomic DNA of F1 founder animals as templates and gene amplification analyses (Fig. S3). Remarkably, all (18/18) male and female animals analysed had mutations at the target site caused by non-homologous end joining (NHEJ)-induced indels (Fig. 2).
Figure 2.
Sequence of Bmdsx gene mutations in exon 3 induced by TALENs. TALEN-binding sequences are underlined in the wild type (WT) gene sequence. Deletions and insertions are indicated by red dashes and red letters, respectively. Sex and number of detection individuals marked with M1, 2, 3 or F1, 2, 3. The length of nucleotide deletions next to the delta character is listed for each individual mutant gene.
BmDSXF mutation induced female genital abnormality and egg-free females
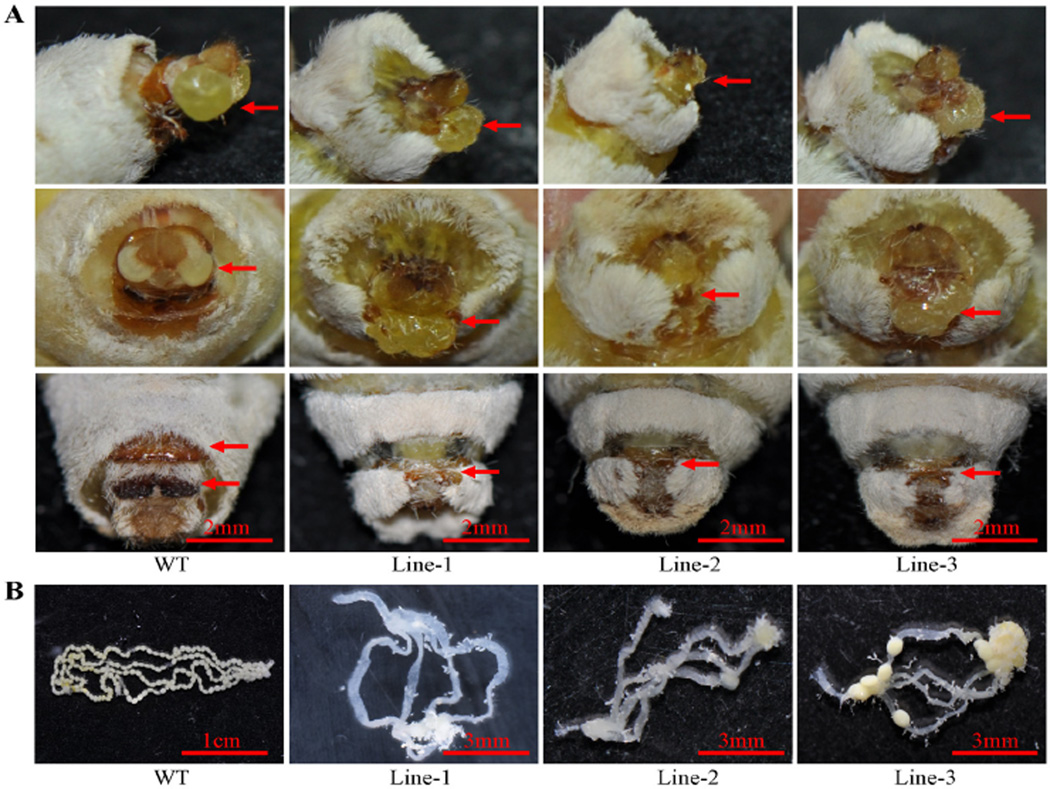
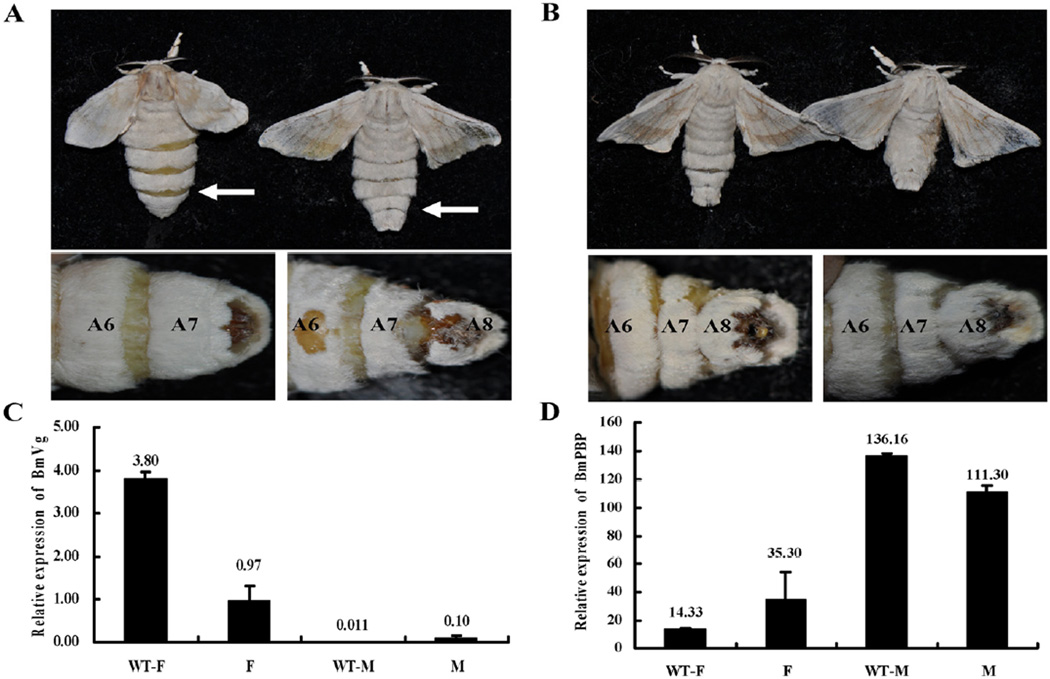
All F1 founder animals of either sex developed to adults without any apparent distortion of the sex-ratio despite the high mutagenesis efficiency. However, one of the conserved biological functions of insect dsx genes is the regulation of genital disc development (Vincent et al., 2001), and we therefore investigated the genital morphology of the transgenic females. Distinct morphological defects were seen in the external genitalia of transgenic females. The dorsal chitin plate was absent and the genital papilla exhibited severe morphological abnormalities (Fig. 3A and Fig. S4). Furthermore, the gross overall morphology of transgenic females resembled that of wild-type (WT) males. Specifically, their abdomens were not enlarged and appeared to be free of developed ovaries. Dissection of these females confirmed that no or only few eggs were present in the ovaries (Fig. 3B). Importantly, transgenic F1 females were sterile and did not lay eggs (Fig. 4A). We also observed that the wild-type sexually-dimorphic abdominal segment number, seven, was absent in mutant females, and they had eight segments (Fig. 4A and Fig. S5). However, there was no difference between mutant and WT males, which all had eight abdominal segments (Fig. 4B and Fig. S4).
Figure 3.
Photographs of external genitalia and ovipositors of mutant females. (A) External genitalia of mutant females: lateral view (upper panel) and front view (middle panel) show the genital morphology of wild-type and mutant line 1–3 individuals. Red arrows indicate morphology of genital papilla. The lower panel shows that the female chitin plate is absent in the mutant individuals. Red arrows indicate morphology of chitin plate. (B) Mutant females have no (lines -1 and -2) or few (line-3) eggs in their ovarioles.
Figure 4.
Gross morphology of segment of wild-type and transgenic F1 founder moths and expression analysis of downstream target genes. (A) A wild-type (WT) female moth with a large abdomen filled with mature eggs (left); transgenic F1 female moth with a small abdomen without eggs (right). Numbers in the lower panel show the abdominal segments. The abdomens are indicated by white arrows. (B) WT moth and transgenic F1 male moths. (C and D). Relative mRNA expression of vitellogenin (BmVg) and pheromone binding protein (BmPBP) encoding genes in WT animals and TALEN-induced mutants. The mean is an average value of mRNA measurements from three individuals.
BmDSXF mutation blocked female mating behavior but not courtship
We investigated transcription levels of genes with sex-specific expression that are expected to be regulated by dsx. The expression level in mutant females of the vitellogenin (Vg) gene, which encodes a protein essential for oogenesis, was only 25% of that seen in WT females (Fig. 4C). In contrast, Vg expression levels in mutant males were 10-fold higher than that seen in WT males. This finding supports the conclusion that female-specific isoform, BmdsxF, contributes to Vg suppression in males. The expression level of a male-specific pheromone-binding protein (BmPBP) was high in transgenic females but not males, supporting the conclusion that BmdsxF suppresses PBP in females (Fig. 4D).
We crossed egg-free F1 females with WT or transgenic males to investigate whether courtship behaviour also is affected. Mutant females could attract mutant or WT males and the time taken by the male to recognize the female and begin courtship was comparable with WT animals (Movie. S1–4). The results support the conclusion that female infertility most likely results from copulation failure caused by the abnormal external genitalia.
Discussion
This transgene-based, somatic mutagenesis technology provides a fast and robust gene analysis tool. Distinct from other transgenic TALENs or CRISPR/CAS systems in which germ-cell specific promoters were used (Kondo & Ueda, 2013; Treen et al., 2014), our system allows somatic mutagenesis analyses to be performed in F1 animals. Importantly, the observed 100% efficiency in generating mutations is useful particularly for analysis of those genes without visible phenotypes. This system is anticipated to contribute to functional gene analyses and provide the basis for generating novel applications in SIT for agricultural and medical insect pest management.
The current study provides the first evidence that transgenic TALENs efficiently induced female-specific sterility in insect through targeting Bmdsx. Insect dsx is a well characterized double-switch gene that produces sex-specific transcription factors via alternative splicing of its transcripts, which function at the final step in the sex-determination cascade. Genetic null mutants of dsx in Drosophila affect sex differentiation and induce intersexual phenotypes in both males and females (Waterbury et al., 1999). Transgenic analysis showed that female D. menalogaster Dsx protein (DsxF) functions as a positive regulator of female differentiation and a negative regulator of male differentiation (Waterbury et al., 1999). Transgenic expression of BmdsxF in B. mori males induces Vg that is expressed specifically in WT females. It also represses the pheromone-binding protein (BmPBP) gene that is expressed dominantly in males, although transgenic animals display normal morphological characteristics (Suzuki et al., 2003).
Ectopic expression of BmdsxM in females resulted in abnormal differentiation of female-specific genital organs and caused partial male differentiation in female genitalia. This latter phenotype is in contrast to what is seen with ectopic expression of DsxF (Suzuki et al., 2003; Duan et al., 2014). These reports support the conclusion that the products of Bmdsx play an important sex-specific role in sex determination and differentiation. Furthermore, our somatic knock-out analysis showed that the chitin plate structure completely disappeared in the DsxF mutant females, not an intermediate phenotype reported before (Duan et al., 2014). This result indicated that loss-of-function analysis is needed to fully exploit the mechanism of Bmdsx mode of action and the transgenic TALEN technologies will greatly contribute to this achievement.
Mutation of BmdsxF by somatic TALEN technologies caused severely deleterious defect in oogenesis development. Transgenic females have normal oviducts with no or a few eggs and this is correlated with significantly decreased Vg expression. In contrast, although transgenic females with BmdsxF deletion had abnormal external genitalia and copulation failure, they still showed normal sexual behavior, indicating that Bmdsx was not involved in sexual behavior regulation. Sexual orientation and courtship behavior in D. melanogaster are controlled by fruitless (fru), the first gene in a branch of the sex-determination hierarchy functioning specifically in the central nervous system (Kimura et al., 2008; Kohatsu et al., 2011). The phenotypes of loss-of-function fru mutants encompassed nearly all aspects of male sexual behavior (Ito et al., 1996; Anand et al., 2001). Other genes such as intersex (ix) and hermaphrodite (her) also act independently or dependently to regulate some aspects of sexual differentiation in D. melanogaster (Waterbury et al., 1999; Ito et al., 2012; Garrett-Engele et al., 2002). Whether these homologous genes regulate B. mori sexual behavior need further investigation.
The regulation of customized nucleases expression in sex-, tissue- or stage-specific manners is still challenging despite of its wide application in genome editing. Screening gene knock-out mutants is dependent largely on visible phenotypes, or large-scale sequencing of many putative mutant insects, which is costly in labor and resources (Li et al., 2012; Katsuyama et al., 2013; Sajwan et al., 2013). Limited success was reported for gene knock-in strategies based on homologous recombination of donor templates (Auer et al., 2014). Genetic transformation technologies mediated by piggybac have been established in many insect species including B. mori. Thus, it will be a fast and efficient strategy to apply somatic nucleases technologies, including ZFN, CRISPR/Cas (Ren et al., 2013; Kondo et al., 2013) system and current TALENs technologies in insect genome editing. These technologies will be useful particularly for dissecting genes without visible phenotypes when mutated. In addition, by using different promoters to regulate TALEN expression, it will be easy to dissect gene function with sex, stage and tissue specificity. The somatic TALEN technologies established here will not only provide a powerful tool for dissecting the sex-specific regulatory mechanism of Bmdsx, but also will greatly facilitate future insect functional gene analysis.
Experimental procedures
Silkworm strains
The B. mori strain used in this study is a multivoltine, nondiapausing silkworm strain, Nistari. Larvae were reared on fresh mulberry leaves under standard conditions.
Plasmid construction
Plasmids of Psw-peas-T-TALENs containing the cassettes of TALEN Repeat, FokI, and SV40 polyA were provided by View Solid Biotech (http://www.v-solid.com/). The TALEN cassette was moved to the transgenic plasmid PXL-BacII (kind gift from Prof. Malcolm Fraser in the University of Notre Dame) by digestion and ligation through NotI and HindIII restriction sites to generate intermediate plasmids PXLBacII -TALEN-L and PXLBacII-TALEN-R. Subsequently, the IE1 promoter was inserted into the HindIII site in the upstream region of TALEN repeat sequence to generate PXLBacII-IE1-TALEN-L-arm and PXLBacII -IE1-TALEN-R-arm. 3×P3-DsRed and 3×P3-EGFP were amplified from the plasmid pBac[3×P3/DsRed] and pBac[3×P3/EGFP] using primers F: 5′-TTATCGAATTCCTGCAGCCCGTACGCGTATCGATAAGCTT-3′ and R: 5′-GAGGTTTTTTAATTCGCTTCCCACAATGGTTAATTCG-3′ and inserted into NotI and SmaI site in multiple clone sites of the PXLBacII-IE1-TALEN-L-arm and PXLBacII-IE1-TALEN-R-arm, respectively, to generate PXLBacII-3×P3-DsRed-IE1-TALEN-R (pBac-DsxR) and PXLBacII-3×P3-EGFP-IE1-TALEN-L (pBac-DsxL).
Germ line transformation
DNA solutions containing pBac-DsxL or pBac-DsxR mixed with helper plasmids were microinjected into preblastoderm G0 embryos that then were incubated at 25 °C in a humidified chamber for 10–12 d until larval hatching (Tan et al., 2013). Larvae were reared on fresh mulberry leaves and putative transgenic G0 adults were mated to WT moths, and G1 progeny were scored for the presence of the marker gene using fluorescence microscopy (Nikon AZ100).
Mutagenesis analysis
Genomic DNA was extracted from B. mori larvae by using standard SDS lysis-phenol treatment after incubation with proteinase K, followed by RNase treatment and purification. PCR a mplification was carried out using 50ng genomic DNA as the template. Primers used for amplification of the target region were forward primer, 5′-GGAGACTGCACTATTTCAATGTT-3′ and reverse primer, 5′-CGTACGACGTGTCTATATTGCAT-3′, which were used to amplify a region of 608 base pairs (bp) in length that encompassed the target sites. PCR products were sub-cloned into pJET-1.2 vector (Fermentas) and sequenced.
mRNA detection of Vg and PBP genes
For Real-Time-PCR analysis, total RNA was extracted from silkworm larvae or cultured cells by using Trizol reagent (Invitrogen) and treated with RNase-free DNAse I (Ambion) followed the manufacturer’s protocol. cDNAs were synthesized using the Omniscript Reverse transcriptase kit (Qiagen), in a 20 µl reaction mixture containing 1 ug total RNA, followed the manufacturer’s instruction. RT-PCR reactions were carried out by using gene-specific primers (forward, 5′-GCCTCGATTTTCCAACTTCA -3′, reverse, 5′- CCATTCTGAAGCAACAGGAG -3′) for amplifying a 218 bp fragment of the BmVg gene; (forward, 5′-CATGGAGCCGATGAGACGAT-3′, reverse, 5′- TCATCGTTAGCTGGAGTGGACTT -3′) for amplifying an 80 bp fragment of the BmPBP gene. Another primer pair set (forward, 5′-TCAATCGGATCGCTATGACA-3′, reverse, 5′-ATGACGGGTCTTCTTGTTGG-3′) amplifies a 136 bp fragment from the B. mori ribosomal protein 49 (Bmrp49) as an internal control.
Courtship behavior analysis
Individual virgin females and males were separated at the pupal stage for morphological observation and courtship assays. Gross morphology of external genitals was investigated as reported previously (Suzuki et al., 2005). For courtship assays, males and females were collected at late pupal stage and aged individually for 3 days. Behavioral assays were performed at 25°C, 60% relative humidity under normal ambient light.
Supplementary Material
Acknowledgements
This work was supported by grants from the External Cooperation Program of BIC, Chinese Academy of Sciences (Grant No. GJHZ201305) and the National Science Foundation of China (31030060, 31272037 and 31372257). AAJ was supported in part by a grant from the NIH NIAID (AI29746).
Contributor Information
Jun Xu, Email: xujun@sibs.ac.cn.
Yueqiang Wang, Email: yqwang@sippe.ac.cn.
Zhiqian Li, Email: zqli@sippe.ac.cn.
Lin Ling, Email: linglin1000@163.com.
Baosheng Zeng, Email: zengbaosheng@sibs.ac.cn.
Anthony A. James, Email: aajames@uci.edu.
Anjiang Tan, Email: tananjiang@sippe.ac.cn.
Yongping Huang, Email: yphuang@sibs.ac.cn.
References
- Anand A, Villella A, Ryner LC, Carlo T, Goodwin SF, Song HJ, et al. Molecular genetic dissection of the sex-specific and vital functions of the Drosophila melanogaster sex determination gene fruitless. Genetics. 2001;158:1569–1595. doi: 10.1093/genetics/158.4.1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auer TO, Duroure K, De Cian A, Concordet JP, Del Bene F. Highly efficient CRISPR/Cas9-mediated knock-in in zebrafish by homology-independent DNA repair. Genome Res. 2014;24:142–153. doi: 10.1101/gr.161638.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker BS. Sex in flies: the splice of life. Nature. 1989;340:521–524. doi: 10.1038/340521a0. [DOI] [PubMed] [Google Scholar]
- Duan J, Xu H, Ma S, Guo H, Wang F, Zhang L, et al. Ectopic expression of the male BmDSX affects formation of the chitin plate in female Bombyx mori. Mol Reprod Dev. 2014;81:240–247. doi: 10.1002/mrd.22290. [DOI] [PubMed] [Google Scholar]
- Garrett-Engele CM, Siegal ML, Manoli DS, Williams BC, Li H, Baker BS. intersex, a gene required for female sexual development in Drosophila, is expressed in both sexes and functions together with doublesex to regulate terminal differentiation. Development. 2002;129:4661–4675. doi: 10.1242/dev.129.20.4661. [DOI] [PubMed] [Google Scholar]
- Harrison DA. Sex determination: controlling the master. Curr Biol. 2007;17:R328–R330. doi: 10.1016/j.cub.2007.03.012. [DOI] [PubMed] [Google Scholar]
- Hockemeyer D, Wang H, Kiani S, Lai CS, Gao Q, Cassady JP, et al. Genetic engineering of human pluripotent cells using TALE nucleases. Nat Biotechnol. 2011;29:731–734. doi: 10.1038/nbt.1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang WY, Fu Y, Reyon D, Maeder ML, Tsai SQ, Sander JD, et al. Efficient genome editing in zebrafish using a CRISPR-Cas system. Nat Biotechnol. 2013;31:227–229. doi: 10.1038/nbt.2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito H, Fujitani K, Usui K, Shimizu-Nishikawa K, Tanaka S, Yamamoto D. Sexual orientation in Drosophila is altered by the satori mutation in the sex-determination gene fruitless that encodes a zinc finger protein with BTB domain. Proc Natl Acad Sci USA. 1996;93:9687–9692. doi: 10.1073/pnas.93.18.9687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito H, Sato K, Koganezawa M, Ote M, Matsumoto K, Hama C, et al. Fruitless recruits two antagonistic chromatin factors to establish single-neuron sexual dimorphism. Cell. 2012;149:1327–1338. doi: 10.1016/j.cell.2012.04.025. [DOI] [PubMed] [Google Scholar]
- Katsuyama T, Akmammedov A, Seimiya M, Hess SC, Sievers C, Paro R. An efficient strategy for TALEN-mediated genome engineering in Drosophila. Nucleic Acids Res. 2013;41:e163. doi: 10.1093/nar/gkt638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura K, Hachiya T, Koganezawa M, Tazawa T, Yamamoto D. Fruitless and doublesex coordinate to generate male-specific neurons that can initiate courtship. Neuron. 2008;59:759–769. doi: 10.1016/j.neuron.2008.06.007. [DOI] [PubMed] [Google Scholar]
- Kohatsu S, Koganezawa M, Yamamoto D. Female contact activates male-specific interneurons that trigger stereotypic courtship behavior in Drosophila. Neuron. 2011;69:498–508. doi: 10.1016/j.neuron.2010.12.017. [DOI] [PubMed] [Google Scholar]
- Kondo S, Ueda R. Highly improved gene targeting by germline-specific Cas9 expression in Drosophila. Genetics. 2013;195:715–721. doi: 10.1534/genetics.113.156737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li T, Liu B, Spalding MH, Weeks DP, Yang B. High-efficiency TALEN-based gene editing produces disease-resistant rice. Nat Biotechnol. 2012;30:390–392. doi: 10.1038/nbt.2199. [DOI] [PubMed] [Google Scholar]
- Ren X, Sun J, Housden BE, Hu Y, Roesel C, Lin S, et al. Optimized gene editing technology for Drosophila melanogaster using germ line-specific Cas9. Proc Natl Acad Sci USA. 2013;110:19012–19017. doi: 10.1073/pnas.1318481110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sajwan S, Takasu Y, Tamura T, Uchino K, Sezutsu H, Zurovec M. Efficient disruption of endogenous Bombyx gene by TAL effector nucleases. Insect Biochem Mol Biol. 2013;43:17–23. doi: 10.1016/j.ibmb.2012.10.011. [DOI] [PubMed] [Google Scholar]
- Steinmann-Zwicky M, Amrein H, Nöthiger R. Genetic control of sex determination in Drosophila. Adv Genet. 1990;27:189–237. doi: 10.1016/s0065-2660(08)60026-7. [DOI] [PubMed] [Google Scholar]
- Suzuki MG, Funaguma S, Kanda T, Tamura T, Shimada T. Analysis of the biological functions of a doublesex homologue in Bombyx mori. Dev Genes Evol. 2003;213:345–354. doi: 10.1007/s00427-003-0334-8. [DOI] [PubMed] [Google Scholar]
- Suzuki MG, Funaguma S, Kanda T, Tamura T, Shimada T. Role of the male BmDSX protein in the sexual differentiation of Bombyx mori. Evol Dev. 2005;7:58–68. doi: 10.1111/j.1525-142X.2005.05007.x. [DOI] [PubMed] [Google Scholar]
- Takasu Y, Kobayashi I, Beumer K, Uchino K, Sezutsu H, Sajwan S, et al. Targeted mutagenesis in the silkworm Bombyx mori using zinc finger nuclease mRNA injection. Insect Biochem Mol Biol. 2010;40:759–765. doi: 10.1016/j.ibmb.2010.07.012. [DOI] [PubMed] [Google Scholar]
- Tamura T, Thibert C, Royer C, Kanda T, Abraham E, Kamba M, et al. Germline transformation of the silkworm Bombyx mori L. using a piggyBac transposon-derived vector. Nat Biotechnol. 2000;18:81–84. doi: 10.1038/71978. [DOI] [PubMed] [Google Scholar]
- Tan A, Fu G, Jin L, Guo Q, Li Z, Niu B, et al. Transgene-based, female-specific lethality system for genetic sexing of the silkworm, Bombyx mori. Proc Natl Acad Sci USA. 2013;111:6766–6770. doi: 10.1073/pnas.1221700110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treen N, Yoshida K, Sakuma T, Sasaki H, Kawai N, Yamamoto T, et al. Tissue-specific and ubiquitous gene knockouts by TALEN electroporation provide new approaches to investigating gene function in Ciona. Development. 2014;141:481–487. doi: 10.1242/dev.099572. [DOI] [PubMed] [Google Scholar]
- Urnov FD, Miller JC, Lee YL, Beausejour CM, Rock JM, Augustus S, et al. Highly efficient endogenous human gene correction using designed zinc-finger nucleases. Nature. 2005;435:646–651. doi: 10.1038/nature03556. [DOI] [PubMed] [Google Scholar]
- Vincent S, Perkins LA, Perrimon N. Doublesex surprises. Cell. 2001;106:399–402. doi: 10.1016/s0092-8674(01)00468-8. [DOI] [PubMed] [Google Scholar]
- Wang Y, Li Z, Xu J, Zeng B, Ling L, You L, et al. The CRISPR/Cas system mediates efficient genome engineering in Bombyx mori. Cell Res. 2013;23:1414–1416. doi: 10.1038/cr.2013.146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterbury JA, Jackson LL, Schedl P. Analysis of the doublesex female protein in Drosophila melanogaster: role on sexual differentiation and behaviorand dependence on intersex. Genetics. 1999;152:1653–1667. doi: 10.1093/genetics/152.4.1653. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.