Abstract
Objective
GDF5 and FRZB have been proposed as genetic loci conferring susceptibility to osteoarthritis (OA); however, the results of several studies investigating the association of OA with the rs143383 polymorphism of the GDF5 gene or the rs7775 and rs288326 polymorphisms of the FRZB gene have been conflicting or inconclusive. To examine these associations, we performed a large-scale meta-analysis of individual-level data.
Methods
Fourteen teams contributed data on polymorphisms and knee, hip, and hand OA. For rs143383, the total number of cases and controls, respectively, was 5,789 and 7,850 for hip OA, 5,085 and 8,135 for knee OA, and 4,040 and 4,792 for hand OA. For rs7775, the respective sample sizes were 4,352 and 10,843 for hip OA, 3,545 and 6,085 for knee OA, and 4,010 and 5,151 for hand OA, and for rs288326, they were 4,346 and 8,034 for hip OA, 3,595 and 6,106 for knee OA, and 3,982 and 5,152 for hand OA. For each individual study, sex-specific odds ratios (ORs) were calculated for each OA phenotype that had been investigated. The ORs for each phenotype were synthesized using both fixed-effects and random-effects models for allele-based effects, and also for haplotype effects for FRZB.
Results
A significant random-effects summary OR for knee OA was demonstrated for rs143383 (1.15 [95% confidence interval 1.09–1.22]) (P = 9.4 × 10−7), with no significant between-study heterogeneity. Estimates of effect sizes for hip and hand OA were similar, but a large between-study heterogeneity was observed, and statistical significance was borderline (for OA of the hip [P = 0.016]) or absent (for OA of the hand [P = 0.19]). Analyses for FRZB polymorphisms and haplotypes did not reveal any statistically significant signals, except for a borderline association of rs288326 with hip OA (P = 0.019).
Conclusion
Evidence of an association between the GDF5 rs143383 polymorphism and OA is substantially strong, but the genetic effects are consistent across different populations only for knee OA. Findings of this collaborative analysis do not support the notion that FRZB rs7775 or rs288326 has any sizable genetic effect on OA phenotypes.
Primary osteoarthritis (OA) is the most prevalent articular disorder and accounts for substantial morbidity and disability, particularly among the elderly, with a considerable health care burden in the developed countries. Its etiology is multifactorial. Many common genetic variants have been proposed as being associated with the risk of OA, albeit with inconsistent results among studies. Among them, 2 genes that have received extensive attention in the recent literature are the growth differentiation factor 5 gene (GDF5) and the Frizzled-related protein gene (FRZB). GDF5 has a role in the development and maintenance of bone and cartilage (1–5). FRZB is a key participant in the Wnt signaling pathway, which has been considered to be important in OA since it can influence chondrocyte differentiation and cartilage function (6–8).
With regard to GDF5, Miyamoto et al reported a strong association between the single-nucleotide polymorphism (SNP) rs143383 and hip and knee OA in Asian populations (9): the per–risk allele (T) summary odds ratio (OR) was 1.79 (P = 1.8 × 10−13) in 2 Japanese cohorts evaluated for associations with hip OA, and the ORs obtained in a Japanese and a Chinese cohort evaluated for associations with knee OA were 1.30 (P = 0.0021) and 1.54 (P = 2.8 × 10−4), respectively. Several studies of Caucasian populations have yielded conflicting results (10,11). Findings of an earlier meta-analysis (12) suggested that rs143383 is associated with a 1.21-fold increase in the risk of knee OA per risk allele copy, but significance levels were modest by current standards (P = 0.0004 in the per-allele model). A modest association was also observed for all cases combined (any joint involved [P = 0.006]); however, a large heterogeneity was observed (I2 = 85%).
Several studies have also investigated the relationship between OA and 2 polymorphisms in FRZB, rs7775 and rs288326 (7,13,14). Much like the findings with GDF5, studies have yielded conflicting results (13–17). A preliminary meta-analysis of published data showed no clear evidence of association between the rs7775 SNP and hip or knee OA (18). The original proposed association was stronger at the haplotype level (7), but published data did not enable evaluation of haplotype effects.
The inconclusive and conflicting results leave uncertainty with regard to the effects of these polymorphisms. Single studies are hampered by small sample size and lack of power (19), inconsistent definitions of OA phenotypes, difficulty in assessing haplotypes, lack of standardization, and potentially selective reporting in the published literature (20). Meta-analyses based on consortia may help overcome some of these limitations. Herein we report the results of a collaborative meta-analysis of individual-level data, from 14 teams, on the relationship of the GDF5 rs143383, FRZB rs7775, and FRZB rs288326 polymorphisms with OA phenotypes. We synthesized standardized data on hip, knee, and hand OA according to a common meta-analysis protocol.
METHODS
Study population
Groups from the Translational Research in Europe Applied Research in Osteoarthritis (TREAT-OA) consortium and other groups that were known to have genotyped the GDF5 and FRZB SNPs in their OA cohorts were invited to participate in the present study. Fourteen teams had generated such data (Table 1), 9 of which had already published these data in the literature (7,10–13,17,18); 1 team had published only some of the data (14), and 4 (Nottingham, Twins UK, Kujala, and deCODE) had not published any of the data. Thirteen teams studied populations of Caucasian descent (from Europe or the US), and 1 team studied a Japanese population (Table 1). Teams/studies are referred to below by the same designations that appear in Table 1, i.e., name of study or principal investigator.
Table 1.
Characteristics of the study populations included in the meta-analysis, and definition of phenotypes*
| Study or PI name | Country of origin | Study design | Female subjects, % | Age, mean ± SD years | Hip OA definition
|
Knee OA definition
|
Hand OA definition
|
||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| THR | Radiographic | TKR | Radiographic | Clinical | Radiographic | Clinical | |||||
| Loughlin (7) | UK | Case–control | 53 | NA | + | + | |||||
| Chapman (10) | UK | Case–control | 57 | 71.2 ± 7.8 (cases) | + | + | |||||
| Twins UK | UK | Twin study | 100 | 54.3 ± 7.8 | Croft grade ≥1 | K/L grade ≥2 | K/L grade >2 in >3 joints | ||||
| Chingford (10) | UK | Prospective cohort | 100 | 53.6 ± 6.0 | K/L grade ≥2 | K/L grade ≥2 | |||||
| Tsezou (11) | Greece | Case–control | 72 | 67.6 ± 8.4 | + | K/L grade ≥2 | |||||
| Ikegawa | Japan | Case–control | 70 | 57.8 ± 16.9 | K/L grade ≥2 | K/L grade ≥2 | |||||
| GARP (12) | The Netherlands | Family-based case– control study | 100 | 60.4 ± 7.5 | K/L grade ≥2 | K/L grade ≥2 | K/L grade ≥2 in >3 joints | ||||
| Gonzalez (10,17) | Spain | Case–control | 66 | 66.9 ± 8.3 | + | + | ACR | ||||
| deCODE | Iceland | Prospective cohort | 64 | 74.8 ± 11.9 | + | + | † | † | |||
| Rotterdam (18) | The Netherlands | Prospective cohort | 59 | 69.4 ± 9.1 | + | K/L grade ≥2 | K/L grade ≥2 | K/L grade ≥2 in 2 of 3 joint groups | |||
| Kujala | Finland | Family-based study (only index cases were used) | Hand OA 82, knee OA 76, controls 76 | Hand OA 62 ± 9, knee OA 67 ± 8, controls 58 ± 2 | + | Symptoms for TKR and K/L grade ≥3 | K/L grade ≥3 in bilateral DIP joints | Physician-diagnosed | |||
| Nottingham | UK | Case–control | GDF5 60.4, FRZB 56.9 | NA | ACR | ||||||
| SOF (14) | US | Prospective cohort | 100 | 78.5 ± 4.6 | JSN grade ≥3, summary grade ≥3, or definite osteophytes grade ≥2 and JSN grade ≥2 | ||||||
| Lories (16) | Belgium | Case–control | 100 | 76.7 ± 7.0 | + | ||||||
PI = principal investigator; OA = osteoarthritis; THR = total hip replacement; TKR = total knee replacement; NA = data not available; K/L = Kellgren/Lawrence; GARP = Genetics, Osteoarthritis, and Progression study; ACR = American College of Rheumatology; DIP = distal interphalangeal; SOF = Study of Osteoporotic Fractures; JSN = joint space narrowing.
Clinical and/or radiographic.
Outcomes assessed
Three outcomes, i.e., hip OA, knee OA, and hand OA, were addressed separately. We accepted phenotype definitions based on joint replacement, radiographic criteria, and clinical criteria. For radiographic criteria, we preferred the Kellgren/Lawrence (K/L) classification system (21), which is the most widely used scale for identifying and grading OA (grades 0–4, with 0 representing normal findings and 4 representing severe OA). A cutoff of K/L grade 2 was used to classify OA, unless the data had been generated with another cutoff and the definition could not be revisited. For clinical criteria, we preferred the American College of Rheumatology (ACR; formerly, the American Rheumatism Association) criteria (22–24) but also accepted other definitions that may have been preferred by local investigators, if information on ACR criteria was not available. The phenotype definitions used in each study are shown in Table 1.
Participants with >1 affected joint were considered cases for each of the analyses pertaining to OA phenotypes for which they had affected joints. Participants without any known affected joint were included as controls. If information on controls was available for only 1 joint and information on the other joints was missing, the participant was counted as a control only for the specific joint. Thus, for example, if an individual was considered as a control for hip OA and there was no recorded information on knee and hand OA, he or she was counted as a control only in the analysis of hip OA.
We also performed an analysis of associations with any OA regardless of site, but this was considered, from the outset of our study, a secondary exploratory analysis given the very large diversity in phenotype definitions. For this analysis, cases were subjects who had OA in at least 1 joint (hip, knee, or hand), and controls were those who had no OA documented in the joints assessed in each study, as per each study’s ascertainment procedures.
Genotyping procedures
Genotypes were determined by TaqMan analysis in the Rotterdam, Chingford, and Gonzalez studies, by other polymerase chain reaction methods in the Loughlin, Chapman, and Tsezou studies and the Study of Osteoporotic Fractures (SOF), by mass spectrometry in the Genetics, Osteoarthritis, and Progression (GARP) study and the Kujala study, and by pyrosequencing in the Lories study. The genotypes in the deCODE study were determined with the Centaurus platform. Twins UK study data were generated as part of ongoing genome-wide association studies using the Illumina platform. Invader assay, TaqMan, DNA fragment analysis, or direct sequencing were used in the Ikegama study. All teams performed random genotyping for quality controls, and the results were concordant.
Statistical analysis
Participants were stratified according to study and sex. Hardy-Weinberg equilibrium (HWE) was assessed in the control group in studies of unrelated cases and controls and in the whole cohort in population-based studies, by exact chi-square test. Deviation from HWE was considered nominally statistically significant at the P < 0.05 level.
For each phenotype of interest, allele-based natural logarithms of ORs and their respective standard errors were estimated across all strata. Specifically, an effect estimate (natural logarithm of the OR) and its standard error were estimated within each study separately for each sex. ORs correspond to the risk conferred by each copy of the proposed risk allele of each SNP; i.e., allele T for 143383, allele G for rs7775, and allele T for rs288326. In the deCODE study, the ORs were calculated as implemented in the NEMO software package (25). In studies that used familial designs (GARP, Twins UK) or had potential relatedness among the participants due to the structure of the population (deCODE), adjusted standard errors were computed to take into account the relatedness among the participants.
Between-study heterogeneity was tested using Cochran’s Q statistic, which is considered significant at P < 0.10. The extent of inconsistency across studies was quantified with the I2 statistic (26). We also computed the 95% confidence intervals (95% CIs) for I2 (27). The I2 ranges between 0 and 100%, and for operational purposes, values of 0–24%, 25–50%, 50–75%, and >75% are considered low, moderate, large, and very large, respectively (28). When there was very large or large (>50%) between-study heterogeneity, we used a simulation algorithm to evaluate how many studies had to be removed for the I2 to reach <25% (29).
The natural logarithms of the OR estimates were synthesized using fixed-effects models (30) and random-effects models (31). Fixed-effects models assume that the true genetic effect of the risk allele is constant among groups and the observed differences are due to chance. In random-effects models the risk allele effects for the individual studies are assumed to vary around some overall average effect, and they take into account the between-study heterogeneity (32). In the absence of heterogeneity, fixed and random effects coincide.
As a main analysis, we synthesized separately the ORs for hip, knee, and hand OA. We also synthesized information on all OA cases. Sex-specific subgroup analyses were also undertaken. Heterogeneity between the sex-specific summary effects was calculated using Cochran’s Q statistic. We also performed sensitivity analyses excluding studies in which there was nominally significant deviation from HWE. Furthermore, we undertook sensitivity analyses with only Caucasian-descent populations included.
For FRZB, we also performed an analysis of haplotypes. Haplotype reconstruction of rs7775 and rs288326 polymorphisms was performed using the population genotypic data separately for cases and controls, and separately for each sex stratum in each study. The possible haplotypes are CC, CT, GT, and GC (the first allele corresponds to the rs7775 and the second one to the rs288326 polymorphism). Haplotypes were inferred performing 1,000 iterations for 5 times with different seed for the random number generator. The frequency estimates were then checked to ensure consistency of the estimates across each run.
Nominal statistical significance is attributed using the traditional P < 0.05 level. However, this may be too lenient a criterion for claiming that an association is credible. Therefore, we also performed a calculation of the Bayes factors and credibility estimates for the observed associations. For each nominally statistically significant summary effect, we estimated the corresponding Bayes factor under a spike and smear prior, with average genetic effect, under the alternative corresponding to an OR of 1.2 (33,34). We also calculated the credibility of the association based on these Bayes factors under various assumptions for prior credibility (10% [strong prior based on biologic, functional, or other evidence], 0.1% [modest prior], or 0.0001% [agnostic prior]). Moreover, the nominally significant associations were graded according to the Venice criteria, which take into account the amount of data, replication consistency, and protection from bias (35). These indexes generate a composite assessment of strong, moderate, or weak credibility. Specifically, grades of A, B, or C are assigned for each of the above-mentioned criteria. An association with an assigned grade of C in any of the composites is considered as having weak credibility, while an association with an assigned grade of A in all 3 criteria is considered to have strong credibility.
All statistical analyses were performed with Stata 10 (StataCorp, College Station, TX). Haplotype reconstruction was performed using Phase 2.1 (36,37).
RESULTS
Data
Fourteen teams, 13 studying populations of Caucasian descent and 1 studying a Japanese population, contributed data to the collaborative analysis. As far as the authors are aware, all study teams that participate in TREAT-OA and that have genotype data available on the GDF5 and FRZB polymorphisms were included in this collaborative analysis. Eleven teams provided data on the GDF5 rs143383 SNP, and 10 teams contributed data on the FRZB rs7775 and rs288326 SNPs. Summary descriptions of the data on the different OA phenotypes (according to affected joints) are provided in Supplementary Table 1, available in the online version of this article at http://www3.interscience.wiley.com/journal/76509746/home.
GDF5 rs143383
Eleven teams contributed cases and controls for study of the association of GDF5 rs143383 with OA in 1 or more affected joints (Supplementary Table 1). The allele contrast under analysis was T allele versus C allele. In 1 study (Gonzalez), genotype distributions were not consistent with HWE, in both sex-specific groups (P = 0.025 and P = 0.03 for men and women, respectively).
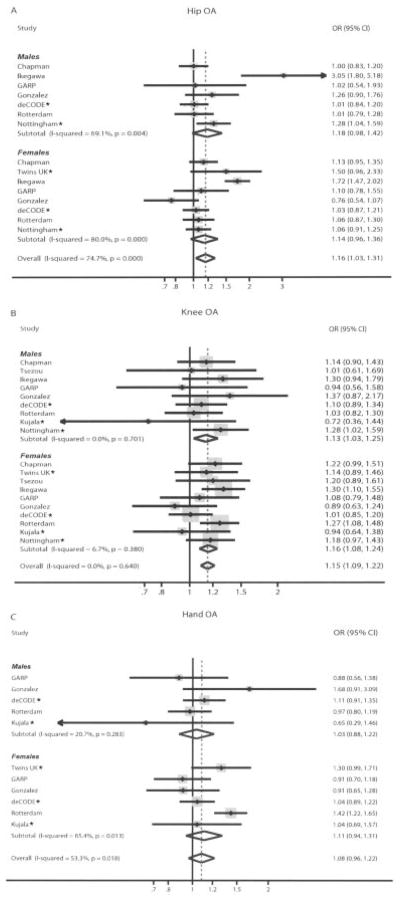
Hip OA
Eight teams contributed data for the analysis of GDF5 rs143383 and hip OA. There were data on 5,789 cases and 7,850 controls. The per-allele summary OR by random-effects analysis was nominally statistically significant (OR 1.16, 95% CI 1.03–1.31) (Table 2); however, very large heterogeneity was observed when all data were included in the analysis (I2 = 75%) (Figure 1A). When applying a simulation algorithm for sensitivity analysis of the heterogeneity, we found that the source of the variability was the Japanese study. Indeed, the I2 became 0% when only Caucasian subjects were considered, and the summary OR was 1.07 (95% CI 1.01–1.14) (P = 0.034). The summary effect remained nominally significant (P = 0.026) when studies with deviation from HWE and non-Caucasians were both removed from the analysis (OR 1.07 [95% CI 1.01–1.15]). The summary ORs were not significantly different between men and women (P = 0.35).
Table 2.
Fixed-effects and random-effects summary odds ratios and 95% confidence intervals for osteoarthritis risk with GDF5 rs143383 in various analyses*
| Analysis (no. of studies included) | Fixed-effects model
|
Random-effects model
|
I2, % (95% CI) | ||
|---|---|---|---|---|---|
| OR (95% CI) | P | OR (95% CI) | P | ||
| Hip OA | |||||
| Male subjects (7) | 1.10 (1.01–1.22) | 0.028 | 1.18 (0.98–1.42) | 0.074 | 69 (32–86) |
| Female subjects (8) | 1.17 (1.09–1.26) | 1.1 × 10−5 | 1.14 (0.97–1.36) | 0.12 | 80 (61–90) |
| All (8) | 1.15 (1.09–1.22) | 1.4 × 10−6 | 1.16 (1.03–1.31) | 0.016 | 75 (58–85) |
| Excluding Japanese subjects (7) | 1.07 (1.01–1.14) | 0.034 | 1.07 (1.01–1.14) | 0.034 | 0 (0–57) |
| Excluding studies without HWE (7) | 1.16 (1.10–1.23) | 5.9 × 10−7 | 1.19 (1.04–1.35) | 0.009 | 76 (58–86) |
| Excluding Japanese subjects and studies without HWE (6) | 1.07 (1.01–1.15) | 0.026 | 1.07 (1.01–1.15) | 0.026 | 0 (0–60) |
| Knee OA | |||||
| Male subjects (9) | 1.13 (1.03–1.25) | 0.011 | 1.13 (1.03–1.25) | 0.011 | 0 (0–65) |
| Female subjects (10) | 1.16 (1.08–1.24) | 2.5 × 10−5 | 1.16 (1.09–1.22) | 2.5 × 10−5 | 7 (0–65) |
| All (10) | 1.15 (1.09–1.22) | 9.4 × 10−7 | 1.15 (1.09–1.22) | 9.4 × 10−7 | 0 (0–49) |
| Excluding Japanese subjects (9) | 1.13 (1.06–1.20) | 9 × 10−5 | 1.13 (1.06–1.20) | 9 × 10−5 | 0 (0–51) |
| Excluding studies without HWE (9) | 1.16 (1.09–1.23) | 6.9 × 10−7 | 1.16 (1.09–1.23) | 6.9 × 10−7 | 0 (0–51) |
| Excluding Japanese subjects and studies without HWE (8) | 1.13 (1.07–1.22) | 7.3 × 10−5 | 1.13 (1.07–1.22) | 7.3 × 10−5 | 0 (0–54) |
| Hand OA | |||||
| Male subjects (5) | 1.04 (0.91–1.18) | 0.59 | 1.03 (0.88–1.22) | 0.69 | 21 (0–66) |
| Female subjects (6) | 1.16 (1.06–1.27) | 0.001 | 1.11 (0.94–1.31) | 0.22 | 65 (17–86) |
| All (6) | 1.12 (1.04–1.01) | 0.003 | 1.08 (0.96–1.22) | 0.19 | 53 (8–76) |
| Excluding studies without HWE (5) | 1.12 (1.04–1.21) | 0.003 | 1.09 (0.96–1.22) | 0.20 | 56 (8–79) |
OR = odds ratio; 95% CI = 95% confidence interval; OA = osteoarthritis; HWE = Hardy-Weinberg equilibrium.
Figure 1.
Forest plot of study-specific estimates and random-effects summary odds ratio (OR) estimates and 95% confidence intervals (95% CIs) for the association between the rs143383 polymorphism of the GDF5 gene and hip osteoarthritis (OA) (A), knee OA (B), and hand OA (C). Diamonds represent the point estimate (center of each diamond) and the 95% CIs (horizontal tips of each diamond). Dashed lines represent the summary OR estimate. Sizes of the shaded boxes represent the weight of each study. Asterisks indicate that the data are entirely new or expanded compared with what was included in a previous meta-analysis (12). GARP = Genetics, Osteoathritis, and Progression study.
Knee OA
Ten teams had available data for the analysis of GDF5 rs143383 and knee OA, contributing a total sample size of 5,085 cases and 8,135 controls. The random-effects summary OR was 1.15 (95% CI 1.09–1.22) (Table 2). No significant heterogeneity was detected (I2 = 0%) (Figure 1B). Sensitivity analysis with only Caucasian participants and excluding studies with deviation from HWE yielded almost identical results (OR 1.13 [95% CI 1.07–1.22]) (Table 2). The sex-specific effect estimates were 1.16 and 1.13 for women and men, respectively; no statistically significant heterogeneity between the 2 estimates was observed (P = 0.70).
Hand OA
Six teams contributed data for the analysis of GDF5 rs143383 and hand OA, with 4,040 cases and 4,792 controls. A non–statistically significant summary OR (1.08 [95% CI 0.96–1.22]) with large heterogeneity was observed (Figure 1C). One group from 1 study (the female subjects in the Rotterdam study, in which a very large effect had been found) would have to be removed in order for any substantial reduction in heterogeneity to be achieved (I2 = 1%). The results were unaltered when sensitivity analysis excluding studies without HWE was performed (Table 2). The estimates did not differ beyond chance between men and women (P = 0.17).
All OA
Overall, the summary OR for association between GDF5 rs143383 and OA as assessed with random-effects models was not nominally statistically significant (OR 1.12 [95% CI 0.99–1.31]). Very large heterogeneity was observed (I2 = 80%, 95% CI 68–88%).
FRZB rs7775 and rs288326
Ten teams provided data for the study of FRZB rs7775 and rs288326 in OA. All subjects were of Caucasian origin. In 1 study (Chingford) there was a nominally significant deviation from HWE in rs288326 (P = 0.003 and P = 0.0001 for knee OA controls and hip OA controls, respectively). That study included only female subjects. The allele contrasts under study were G versus C for rs7775, and T versus C for rs288326.
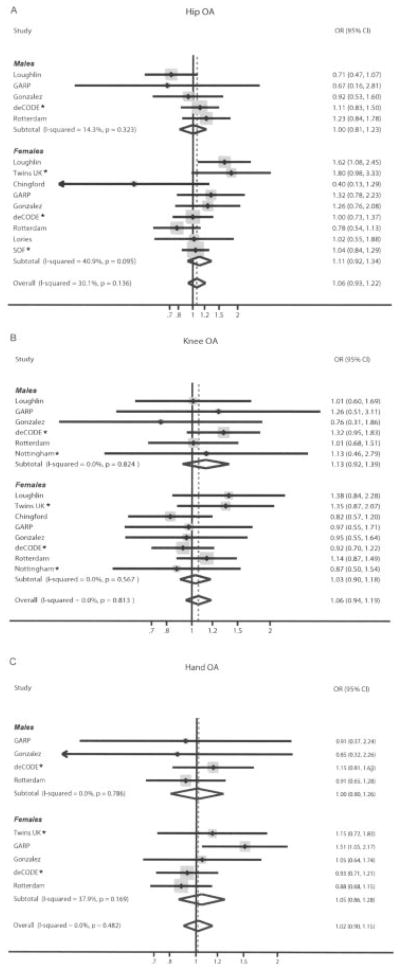
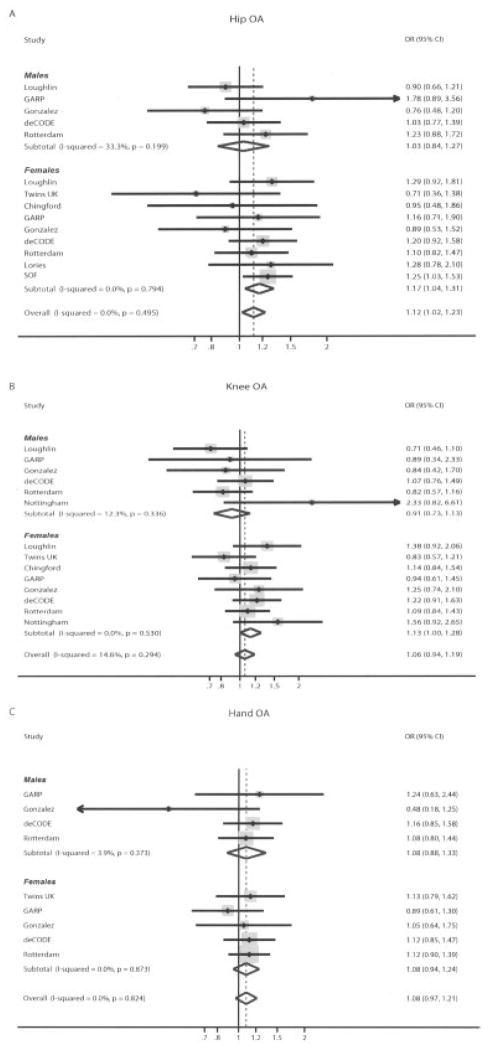
Hip OA
Overall 4,352 cases and 10,843 controls for rs7775 and 4,346 cases and 8,034 controls for rs288326 were available for the analyses of associations with hip OA. The summary effect was nominally statistically significant for rs288326. Specifically, the summary ORs assessed with random-effects models were 1.06 (95% CI 0.93–1.22) and 1.12 (95% CI 1.02–1.23) for rs7775 and rs288326, respectively (Table 3). There was moderate heterogeneity observed for rs7775 (I2 = 30%) and no significant heterogeneity for rs288326 (I2 = 0%) (Figures 2A and 3A). Sensitivity analysis excluding the 1 study with HWE deviation did not alter the calculated summary effect size for rs288326. Effects did not differ significantly between men and women (P = 0.52 and P = 0.19 for rs7775 and rs288326, respectively), but of note, the association with rs288326 reached nominal significance for women (Table 3).
Table 3.
Fixed-effects and random-effects summary odds ratios and 95% confidence intervals for osteoarthritis risk with FRZB rs7775 and rs288326 polymorphisms in various analyses*
| Analysis (no. of studies included) | Fixed-effects model
|
Random-effects model
|
I2, % (95% CI) | ||
|---|---|---|---|---|---|
| OR (95% CI) | P | OR (95% CI) | P | ||
| Hip OA | |||||
| rs7775 | |||||
| Male subjects (5) | 1.01 (0.84–1.21) | 0.95 | 1.00 (0.81–1.23) | 0.98 | 14 (0–82) |
| Female subjects (9) | 1.08 (0.95–1.24) | 0.22 | 1.11 (0.92–1.34) | 0.29 | 41 (0–73) |
| All (9) | 1.06 (0.95–1.18) | 0.30 | 1.06 (0.93–1.22) | 0.39 | 30 (0–63) |
| rs288326 | |||||
| Male subjects (5) | 1.02 (0.87–1.20) | 0.77 | 1.03 (0.84–1.27) | 0.77 | 33 (0–75) |
| Female subjects (9) | 1.17 (1.04–1.31) | 0.007 | 1.17 (1.04–1.31) | 0.007 | 0 (0–65) |
| All (9) | 1.12 (1.02–1.23) | 0.019 | 1.12 (1.02–1.23) | 0.019 | 0 (0–55) |
| Excluding studies without HWE (8) | 1.12 (1.02–1.23) | 0.017 | 1.12 (1.02–1.23) | 0.017 | 0 (0–57) |
| Knee OA | |||||
| rs7775 | |||||
| Male subjects (6) | 1.13 (0.92–1.39) | 0.26 | 1.13 (0.92–1.39) | 0.26 | 0 (0–75) |
| Female subjects (8) | 1.03 (0.90–1.18) | 0.67 | 1.03 (0.90–1.18) | 0.67 | 0 (0–68) |
| All (8) | 1.06 (0.95–1.19) | 0.32 | 1.06 (0.95–1.19) | 0.32 | 0 (0–55) |
| rs288326 | |||||
| Male subjects (6) | 0.91 (0.75–1.10) | 0.33 | 0.91 (0.73–1.13) | 0.39 | 12 (0–78) |
| Female subjects (8) | 1.13 (1.00–1.28) | 0.06 | 1.13 (1.00–1.28) | 0.06 | 0 (0–68) |
| All (8) | 1.06 (0.95–1.18) | 0.28 | 1.06 (0.94–1.19) | 0.34 | 15 (0–53) |
| Excluding studies without HWE (7) | 1.05 (0.94–1.18) | 0.40 | 1.05 (0.92–1.19) | 0.48 | 20 (0–58) |
| Hand OA | |||||
| rs7775 | |||||
| Male subjects (4) | 1.01 (0.80–1.26) | 0.96 | 1.01 (0.80–1.26) | 0.96 | 0 (0–85) |
| Female subjects (5) | 1.02 (0.88–1.19) | 0.77 | 1.05 (0.86–1.28) | 0.14 | 38 (0–77) |
| All (5) | 1.02 (0.90–1.15) | 0.79 | 1.02 (0.90–1.15) | 0.79 | 0 (0–65) |
| rs288326 | |||||
| Male subjects (4) | 1.08 (0.89–1.32) | 0.42 | 1.08 (0.88–1.33) | 0.45 | 4 (0–79) |
| Female subjects (5) | 1.08 (0.94–1.24) | 0.27 | 1.08 (0.94–1.24) | 0.27 | 0 (0–85) |
| All (5) | 1.08 (0.97–1.21) | 0.17 | 1.08 (0.97–1.21) | 0.17 | 0 (0–65) |
See Table 2 for definitions.
Figure 2.
Forest plot of study-specific estimates and random-effects summary OR estimates and 95% CIs for the association between the rs7775 polymorphism of the FRZB gene and hip OA (A), knee OA (B), and hand OA (C). Diamonds represent the point estimate (center of each diamond) and the 95% CIs (horizontal tips of each diamond). Dashed lines represent the summary OR estimate. Sizes of the shaded boxes represent the weight of each study. Asterisks indicate that the data are entirely new or expanded compared with what was included in a previous meta-analysis (18). SOF = Study of Osteoporotic Fractures (see Figure 1 for other definitions).
Figure 3.
Forest plot of study-specific estimates and random-effects summary OR estimates and 95% CIs for the association between the rs288326 polymorphism of the FRZB gene and hip OA (A), knee OA (B), and hand OA (C). This polymorphism was not evaluated in a previous meta-analysis (18). Diamonds represent the point estimate (center of each diamond) and the 95% CIs (horizontal tips of each diamond). Dashed lines represent the summary OR estimate. Sizes of the shaded boxes represent the weight of each study. SOF = Study of Osteoporotic Fractures (see Figure 1 for other definitions).
Knee OA
Eight teams contributed 3,545 cases and 6,085 controls for rs7775 and 3,595 cases and 6,106 controls for rs288326, for the analyses of associations with knee OA. For both polymorphisms under study, the summary ORs computed with random-effects models were not statistically significant and were very close to the null (summary OR 1.06 [95% CI 0.95–1.19] and OR 1.06 [95% CI 0.94–1.19]) for rs7775 and rs288326, respectively) (Table 3). No statistically significant between-study heterogeneity was seen, with I2 = 0% and I2 = 15% for rs7775 and rs288336, respectively (Figures 2B and 3B). There was no significant difference between men and women for rs7775 (P = 0.48) or rs288326 (P = 0.064 [nonsignificant trend toward stronger effect in women]) (Table 3).
Hand OA
Overall, 4,010 cases and 5,151 controls for rs7775 and 3,982 cases and 5,152 controls for rs288326 were available from 5 teams, for the analyses of associations with hand OA. For both SNPs the summary ORs were not statistically significant and close to the null, with effect sizes of 1.02 and 1.08 for rs7775 and rs288326, respectively (Table 3). No significant heterogeneity was observed for either polymorphism (Figures 2C and 3C). There was no significant difference between men and women for rs7775 (P = 0.90) or rs288326 (P = 0.17).
All OA
The summary OR was not statistically significant for rs7775 or rs288326 when all OA cases were considered; by random-effects analysis, the summary ORs were 1.04 (95% CI 0.93–1.13) and 1.09 (95% CI 0.98–1.21), respectively. There was no between-study heterogeneity for either SNP (I2 = 80% [95% CI 0–58% and 0–42% for rs7775 and rs288326, respectively]).
Analysis of FRZB haplotypes
No evidence of association was found when FRZB haplotypes were considered. By random-effects analysis, in hip OA the summary ORs were 1.08 (95% CI 0.92–1.26) (P = 0.34) and 1.10 (95% CI 0.96–1.27) (P = 0.18) for the CT and GC haplotypes, respectively, with the CC haplotype as the reference. For knee OA, the summary ORs were 1.02 (95% CI 0.91–1.13) (P = 0.79) and 1.07 (95% CI 0.95–1.21) (P = 0.28) for the CT and GC haplotypes, respectively, with the CC haplotype as the reference. For the same comparisons, the summary ORs were 1.07 (95% CI 0.95–1.20) (P = 0.28) and 1.02 (95% CI 0.89–1.17) (P = 0.76) for hand OA.
The analysis of the GA haplotype that contains the rare alleles also showed no statistically significant effects, but data were sparse compared with data on the other haplotypes. Specifically, for hip OA the summary ORs were 0.96 (95% CI 0.41–2.24), 1.01 (95% CI 0.49–2.08), and 0.94 (95%CI 0.42–2.15) compared with the CT, CC, and GC haplotypes, respectively. The computed summary ORs for the same analysis for knee OA were 1.33 (95% CI 0.70–2.53), 1.31 (95% CI 0.68–2.54), and 1.26 (95% CI 0.60–2.62), respectively. For hand OA the computed results were 1.62 (95% CI 0.62–4.25), 1.63 (95% CI 0.65–4.10), and 1.63 (95% CI 0.62–4.27), respectively.
We also analyzed the haplotype consisting of the rare alleles versus all of the other 3 haplotypes combined. The summary OR using random-effects calculations was 0.99 (95% CI 0.48–2.07), 1.31 (95% CI 0.67–2.53), and 1.63 (95% CI 0.65–4.12) for hip, knee, and hand OA, respectively. When only women were considered, the summary OR was 1.70 (95% CI 0.82–3.50), 2.15 (95% CI 0.97–4.78), and 2.33 (95% CI 0.90–5.99) for hip, knee, and hand OA, respectively.
Bayes factors and credibility of significant signals
The estimated −log10 Bayes factor for the association between hip OA and GDF5 rs143383 was 0.60, and the credibility for the association to be true was 28%, 0.4%, and 0.0004% for the strong, modest, and agnostic priors, respectively. For knee OA and GDF5 rs143383, the estimated −log10 Bayes factor was 3.5, with the credibility for the association to be true being 99.7%, 76%, and 0.31% for the strong, modest, and agnostic priors, respectively. For the observed association between hip OA and FRZB rs288326, the estimated −log10 Bayes factor was 0.46 and the credibility for the association to be true was 22%, 0.29%, and 0.0003% for these 3 priors, respectively.
All 3 associations with nominal statistical significance are based on large-scale evidence (n >1,000 minor alleles in cases and controls combined; grade A per the Venice criteria for amount of evidence). The between-study heterogeneity was large for hip OA and GDF5 rs143383 (grade C per the Venice criteria), whereas no heterogeneity was observed for the knee OA and GDF5 rs143383 association or for the hip OA and FRZB rs288326 association (grade A per the Venice criteria for replication consistency). Excluding data from the first study on each polymorphism and OA and studies with HWE deviation would result in loss of the nominal significance for the hip OA and rs288326 association (OR 1.11, P = 0.052; grade C for protection from bias per the Venice criteria). Therefore, only the association of rs143383 would be graded as having strong credibility according to the Venice criteria.
DISCUSSION
In the present study, which is, to our knowledge, by far the largest study of genetics in OA undertaken to date, we found strong support for the notion that the GDF5 rs143383 polymorphism is a determinant of risk of OA. The evidence is stronger and most consistent for knee OA, but the magnitude of the effect seems considerably smaller than originally proposed. Our meta-analyses used new or expanded data on 4 large studies compared with a previous meta-analysis of GDF5 (12) and new or expanded data on 4 large studies compared with a previous meta-analysis of FRZB (18), and the amount of available data is now more than doubled for all phenotypes and polymorphisms. One of the new data sets (deCODE) is the largest single study conducted on these polymorphisms. Also, in the previous meta-analysis of FRZB, only 1 variant was addressed, haplotype analyses were not performed, and data on hand OA were not included. Our results yield far more robust statistical support for the association of GDF5 rs143383 with OA, for which previous statistical support was more tenuous, even though the absolute magnitude of the effect (odds ratio) demonstrated in our meta-analysis is smaller than previously thought. For FRZB rs7775 and rs288326, the addition of more data demonstrates more conclusively that there is no overall effect, and excludes modest overall effects (that would be pertinent to both women and men) that could not have been excluded based on results of the previous meta-analysis.
Even for the strongest association signal we observed, that between GDF5 rs143383 and knee OA, the effect size was very small, with an OR of only 1.15 overall (1.13 excluding the Japanese study and studies with deviation from HWE). This is much smaller than the effect of GDF5 originally proposed in the primary publication that addressed populations of Asian descent (9). Diversity between populations of different racial descents may reflect different linkage disequilibrium patterns and may even indicate that this marker is not necessarily the true or only culprit. Nevertheless, Miyamoto et al found functional evidence for a role of the rs143383 polymorphism (9), and based on the findings of some previous studies, a functional role of GDF5 in the pathogenesis of OA is plausible. In humans, GDF5 is present in adult articular cartilage and also stimulates proteoglycan synthesis in articular cartilage explants (38). In addition, mutations in GDF5 have been implicated in disorders of skeletal development, such as various forms of chondrodysplasia, synphalangism, and type C brachydactyly (9,39). Findings in a mouse study by Masuya et al further support the notion that GDF5 has a critical role in joint formation and OA development (40).
The more prominent effect sizes reported in earlier studies may have been inflated (41), and the large number of samples now analyzed provides an estimate of genetic effect in knee OA that is probably more realistic. Despite the very large sample size we accrued, our findings are not conclusive with regard to an association between GDF5 rs143383 and OA of other joints. Small effects with ORs in the range of 1.1–1.15 are certainly plausible for these sites as well, but the evidence remains weak. These results highlight the need to perform extremely large studies with very careful definitions and measurements in order to understand the effects of common genetic variants on OA outcomes (42), and are consistent with experience regarding the magnitude of genetic effects that is currently obtained with high-throughput platform testing for various diseases (43). According to data in the HuGE Navigator (44) as of December 2, 2008, there are at least 95 genes that have been proposed to be associated with OA. However, the studies performed have almost always been too small for the results to be conclusive, or even considerably suggestive, for validation of such modest effects.
The ORs for FRZB gene polymorphisms rs7775 and rs288326 were very close to unity. In the first study from which a role of these polymorphisms was proposed, Loughlin et al (7) observed that the risk allele for rs7775 was responsible for a 1.5-fold increased risk of total hip replacement. Min and colleagues (13) also found a strong association of rs7775 with generalized OA. We observed only a nominally statistically significant association of rs288326 with hip OA; however, the potential association of rs288326 with hip OA in women should not be disregarded. One may argue that the anatomy predisposing to hip OA may differ in men and women, and Wnt signaling may affect anatomy. A possible sex-specific association may require further documentation, acknowledging that robust documentation of significantly different effects between men and women would require extremely large sample sizes (45).
Nevertheless, the overall epidemiologic credibility of this association is currently weak, despite the supporting biology. Effects did not differ significantly across studies, and the largest estimate of between-study inconsistency in the FRZB data was obseved for rs7775 and hip OA, but it was still not large (I2 = 30%). It should be acknowledged that the Loughlin study and the SOF investigated subjects with severe radiographic hip OA, defined by findings equivalent to a K/L grade of ≥3 or total hip replacement, with these cases of severe hip OA compared with controls with no radiographic hip OA (K/L grade of 0 [SOF] or no signs or symptoms of arthritis or joint disease [Loughlin study]). The phenotype definitions of radiographic hip OA and the severity of the disease varied across other studies included in the present meta-analysis. Additional studies with greater standardization of the definition of severe OA in cases and more rigorous ascertainment of lack of even mild disease in controls may be useful.
Very large collaborative studies would also be needed for better understanding of the heterogeneity of genetic effects across different populations and settings. For example, no significant heterogeneity was observed for the association of GDF5 rs143383 with knee OA; however, the results must be interpreted with caution since the upper confidence interval of I2 was 49%. A large degree of heterogeneity was observed in many of the other GDF5 analyses. Part of this heterogeneity was attenuated when the only study of GDF5 in an Asian population was excluded from the analysis.
In addition, similar to the above-described situation with regard to studies of FRZB, different criteria were used for definition of cases in different studies of GDF5, and this phenotypic heterogeneity may have led to heterogeneity in the magnitude of the genetic effects. Most of the participating study teams defined OA radiographically, using the K/L classification criteria. The K/L classification provides a composite estimate of osteophytes and joint space narrowing, and the lack of focus on joint space narrowing has been considered to be a flaw of that system (46) and might be the reason many different descriptions/definitions have been used for determination of K/L grade (47). Another important issue is that differences in radiographic assessment protocols across centers might have also introduced heterogeneity. In particular, differences in the position of the knee in which the radiographs were obtained yield different K/L grades (46). All of these issues underscore the fact that further consensus is needed in order to achieve standardization of phenotype definitions in future studies of OA.
Supplementary Material
Acknowledgments
The TreatOA project is supported through Coordination Theme 1 (Health) of the European Union 7th Framework Programme (grant HEALTH-F2-2008-00). The GARP study is supported by Leiden University Medical Center, the Dutch Arthritis Association, and Pfizer, Groton, Connecticut. The genotypic work was supported by the Netherlands Organization of Scientific Research (grants MW 904-61-095, 911-03-016, 917 66344, and 911-03-012), Leiden University Medical Center, and the Center of Medical System Biology, Leiden, The Netherlands. The Study of Osteoporotic Fractures is supported by the NIH (National Institute on Aging grants AG-05407, AR-35582, AG-05394, AR-35584, AR-35583, R01-AG-005407, R01-AG-027576-22, 2 R01-AG-005394-22A1, and 2 R01-AG-027574-22A1). Dr. Lane’s work was supported by NIH grant K24-AR-04884.
We thank Dr. Jeanine Houwing Duistermaat (Department of Medical Statistics, Leiden University Medical Center) for statistical support and Dr. Steffan D. Bos (Department of Molecular Epidemiology, Leiden University Medical Center) for assistance with data acquisition. We are grateful for the valuable contributions of Hafdis T. Helgadottir and the other personnel at the core facilities of deCODE Genetics.
Footnotes
Drs. Halldorsson, Jonsdottir, and Stefansson own stock and/or stock options in deCODE Genetics.
AUTHOR CONTRIBUTIONS
Dr. Ioannidis had full access to all of the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
Study design. Loughlin, Ingvarsson, Jonsson, Kloppenburg, Lane, Näkki, Nevitt, Spector, Valdes, Ioannidis.
Acquisition of data. Chapman, Meulenbelt, Loughlin, Carr, M. Doherty, S. Doherty, Gómez-Reino, Gonzalez, Hauksson, Hofman, Hart, Ikegawa, Jiang, Jonsdottir, Jonsson, Kerkhof, Lane, Li, Lories, van Meurs Näkki, Nevitt, Rodriguez-Lopez, Shi, Slagboom, Stefansson, Tsezou, Wallis, Watson, Uitterlinden, Valdes.
Analysis and interpretation of data. Evangelou, Meulenbelt, Karassa, Gómez-Reino, Gonzalez, Hauksson, Jonsdottir, Jonsson, Kloppenburg, Li, van Meurs, Näkki, Rodriguez-Lopez, Slagboom, Ioannidis.
Manuscript preparation. Evangelou, Meulenbelt, Karassa, Loughlin, Hauksson, Ingvarsson, Jonsson, Kerkhof, Kloppenburg, Lane, Li, Lories, Näkki, Slagboom, Valdes, Ioannidis.
Statistical analysis. Evangelou, Meulenbelt, Halldorsson, Hauksson, Lane, Näkki, Ioannidis.
Sample collection. Ingvarsson.
References
- 1.Luyten FP. Cartilage-derived morphogenetic protein-1. Int J Biochem Cell Biol. 1997;29:1241–4. doi: 10.1016/s1357-2725(97)00025-3. [DOI] [PubMed] [Google Scholar]
- 2.Francis-West PH, Abdelfattah A, Chen P, Allen C, Parish J, Ladher R, et al. Mechanisms of GDF-5 action during skeletal development. Development. 1999;126:1305–15. doi: 10.1242/dev.126.6.1305. [DOI] [PubMed] [Google Scholar]
- 3.Edwards CJ, Francis-West PH. Bone morphogenetic proteins in the development and healing of synovial joints. Semin Arthritis Rheum. 2001;31:33–42. doi: 10.1053/sarh.2001.24875. [DOI] [PubMed] [Google Scholar]
- 4.Mikic B. Multiple effects of GDF-5 deficiency on skeletal tissues: implications for therapeutic bioengineering. Ann Biomed Eng. 2004;32:466–76. doi: 10.1023/b:abme.0000017549.57126.51. [DOI] [PubMed] [Google Scholar]
- 5.Nickel J, Kotzsch A, Sebald W, Mueller TD. A single residue of GDF-5 defines binding specificity to BMP receptor IB. J Mol Biol. 2005;349:933–47. doi: 10.1016/j.jmb.2005.04.015. [DOI] [PubMed] [Google Scholar]
- 6.Enomoto-Iwamoto M, Kitagaki J, Koyama E, Tamamura Y, Wu C, Kanatani N, et al. The Wnt antagonist Frzb-1 regulates chondrocyte maturation and long bone development during limb skeletogenesis. Dev Biol. 2002;251:142–56. doi: 10.1006/dbio.2002.0802. [DOI] [PubMed] [Google Scholar]
- 7.Loughlin J, Dowling B, Chapman K, Marcelline L, Mustafa Z, Southam L, et al. Functional variants within the secreted frizzled-related protein 3 gene are associated with hip osteoarthritis in females. Proc Natl Acad Sci U S A. 2004;101:9757–62. doi: 10.1073/pnas.0403456101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lories RJ, Peeters J, Bakker A, Tylzanowski P, Derese I, Schrooten J, et al. Articular cartilage and biomechanical properties of the long bones in Frzb-knockout mice. Arthritis Rheum. 2007;56:4095–103. doi: 10.1002/art.23137. [DOI] [PubMed] [Google Scholar]
- 9.Miyamoto Y, Mabuchi A, Shi D, Kubo T, Takatori Y, Saito S, et al. A functional polymorphism in the 5′ UTR of GDF5 is associated with susceptibility to osteoarthritis. Nat Genet. 2007;39:529–33. doi: 10.1038/2005. [DOI] [PubMed] [Google Scholar]
- 10.Southam L, Rodriguez-Lopez J, Wilkins JM, Pombo-Suarez M, Snelling S, Gomez-Reino JJ, et al. An SNP in the 5′-UTR of GDF5 is associated with osteoarthritis susceptibility in Europeans and with in vivo differences in allelic expression in articular cartilage. Hum Mol Genet. 2007;16:2226–32. doi: 10.1093/hmg/ddm174. [DOI] [PubMed] [Google Scholar]
- 11.Tsezou A, Satra M, Oikonomou P, Bargiotas K, Malizos KN. The growth differentiation factor 5 (GDF5) core promoter polymorphism is not associated with knee osteoarthritis in the Greek population. J Orthop Res. 2008;26:136–40. doi: 10.1002/jor.20464. [DOI] [PubMed] [Google Scholar]
- 12.Chapman K, Takahashi A, Meulenbelt I, Watson C, Rodriguez-Lopez J, Egli R, et al. A meta-analysis of European and Asian cohorts reveals a global role of a functional SNP in the 5′ UTR of GDF5 with osteoarthritis susceptibility. Hum Mol Genet. 2008;17:1497–504. doi: 10.1093/hmg/ddn038. [DOI] [PubMed] [Google Scholar]
- 13.Min JL, Meulenbelt I, Riyazi N, Kloppenburg M, Houwing-Duistermaat JJ, Seymour AB, et al. Association of the Frizzled-related protein gene with symptomatic osteoarthritis at multiple sites. Arthritis Rheum. 2005;52:1077–80. doi: 10.1002/art.20993. [DOI] [PubMed] [Google Scholar]
- 14.Lane NE, Lian K, Nevitt MC, Zmuda JM, Lui L, Li J, et al. Frizzled-related protein variants are risk factors for hip osteoarthritis. Arthritis Rheum. 2006;54:1246–54. doi: 10.1002/art.21673. [DOI] [PubMed] [Google Scholar]
- 15.Valdes AM, Loughlin J, van Oene M, Chapman K, Surdulescu GL, Doherty M, et al. Sex and ethnic differences in the association of ASPN, CALM1, COL2A1, COMP, and FRZB with genetic susceptibility to osteoarthritis of the knee [published erratum appears in Arthritis Rheum 2009;60:178] Arthritis Rheum. 2007;56:137–46. doi: 10.1002/art.22301. [DOI] [PubMed] [Google Scholar]
- 16.Lories RJ, Boonen S, Peeters J, de Vlam K, Luyten FP. Evidence for a differential association of the Arg200Trp single-nucleotide polymorphism in FRZB with hip osteoarthritis and osteoporosis [letter] Rheumatology (Oxford) 2006;45:113–4. doi: 10.1093/rheumatology/kei148. [DOI] [PubMed] [Google Scholar]
- 17.Rodriguez-Lopez J, Pombo-Suarez M, Liz M, Gomez-Reino JJ, Gonzalez A. Further evidence of the role of frizzled-related protein gene polymorphisms in osteoarthritis. Ann Rheum Dis. 2007;66:1052–5. doi: 10.1136/ard.2006.065938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kerkhof JM, Uitterlinden AG, Valdes AM, Hart DJ, Rivadeneira F, Jhamai M, et al. Radiographic osteoarthritis at three joint sites and FRZB, LRP5, and LRP6 polymorphisms in two population-based cohorts. Osteoarthritis Cartilage. 2008;16:1141–9. doi: 10.1016/j.joca.2008.02.007. [DOI] [PubMed] [Google Scholar]
- 19.Ioannidis JP, Trikalinos TA, Khoury MJ. Implications of small effect sizes of individual genetic variants on the design and interpretation of genetic association studies of complex diseases. Am J Epidemiol. 2006;164:609–14. doi: 10.1093/aje/kwj259. [DOI] [PubMed] [Google Scholar]
- 20.Kavvoura FK, Liberopoulos G, Ioannidis JP. Selection in reported epidemiological risks: an empirical assessment. PLoS Med. 2007;4:e79. doi: 10.1371/journal.pmed.0040079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kellgren JH, Lawrence JS. Radiological assessment of osteoarthrosis. Ann Rheum Dis. 1957;16:494–502. doi: 10.1136/ard.16.4.494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Altman R, Alarcon G, Appelrouth D, Bloch D, Borenstein D, Brandt K, et al. The American College of Rheumatology criteria for the classification and reporting of osteoarthritis of the hand. Arthritis Rheum. 1990;33:1601–10. doi: 10.1002/art.1780331101. [DOI] [PubMed] [Google Scholar]
- 23.Altman R, Alarcon G, Appelrouth D, Bloch D, Borenstein D, Brandt K, et al. The American College of Rheumatology criteria for the classification and reporting of osteoarthritis of the hip. Arthritis Rheum. 1991;34:505–14. doi: 10.1002/art.1780340502. [DOI] [PubMed] [Google Scholar]
- 24.Altman R, Asch E, Bloch D, Bole G, Borenstein D, Brandt K, et al. Development of criteria for the classification and reporting of osteoarthritis: classification of osteoarthritis of the knee. Arthritis Rheum. 1986;29:1039–49. doi: 10.1002/art.1780290816. [DOI] [PubMed] [Google Scholar]
- 25.Gretarsdottir S, Thorleifsson G, Reynisdottir ST, Manolescu A, Jonsdottir S, Jonsdottir T, et al. The gene encoding phospho-diesterase 4D confers risk of ischemic stroke. Nat Genet. 2003;35:131–8. doi: 10.1038/ng1245. [DOI] [PubMed] [Google Scholar]
- 26.Higgins JP, Thompson SG. Quantifying heterogeneity in a meta-analysis. Stat Med. 2002;21:1539–58. doi: 10.1002/sim.1186. [DOI] [PubMed] [Google Scholar]
- 27.Ioannidis JP, Patsopoulos NA, Evangelou E. Uncertainty in heterogeneity estimates in meta-analyses. BMJ. 2007;335:914–6. doi: 10.1136/bmj.39343.408449.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Higgins JP, Thompson SG, Deeks JJ, Altman DG. Measuring inconsistency in meta-analyses. BMJ. 2003;327:557–60. doi: 10.1136/bmj.327.7414.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Patsopoulos NA, Evangelou E, Ioannidis JP. Sensitivity of between-study heterogeneity in meta-analysis: proposed metrics and empirical evaluation. Int J Epidemiol. 2008;37:1148–57. doi: 10.1093/ije/dyn065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lau J, Ioannidis JP, Schmid CH. Quantitative synthesis in systematic reviews. Ann Intern Med. 1997;127:820–6. doi: 10.7326/0003-4819-127-9-199711010-00008. [DOI] [PubMed] [Google Scholar]
- 31.DerSimonian R, Laird N. Meta-analysis in clinical trials. Control Clin Trials. 1986;7:177–88. doi: 10.1016/0197-2456(86)90046-2. [DOI] [PubMed] [Google Scholar]
- 32.Kavvoura FK, Ioannidis JP. Methods for meta-analysis in genetic association studies: a review of their potential and pitfalls. Hum Genet. 2008;123:1–14. doi: 10.1007/s00439-007-0445-9. [DOI] [PubMed] [Google Scholar]
- 33.Ioannidis JP. Calibration of credibility of agnostic genome-wide associations. Am J Med Genet B Neuropsychiatr Genet. 2008;147B:964–72. doi: 10.1002/ajmg.b.30721. [DOI] [PubMed] [Google Scholar]
- 34.Ioannidis JP. Effect of formal statistical significance on the credibility of observational associations. Am J Epidemiol. 2008;168:374–83. doi: 10.1093/aje/kwn156. [DOI] [PubMed] [Google Scholar]
- 35.Ioannidis JP, Boffetta P, Little J, O’Brien TR, Uitterlinden AG, Vineis P, et al. Assessment of cumulative evidence on genetic associations: interim guidelines. Int J Epidemiol. 2008;37:120–32. doi: 10.1093/ije/dym159. [DOI] [PubMed] [Google Scholar]
- 36.Stephens M, Smith NJ, Donnelly P. A new statistical method for haplotype reconstruction from population data. Am J Hum Genet. 2001;68:978–89. doi: 10.1086/319501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stephens M, Scheet P. Accounting for decay of linkage disequilibrium in haplotype inference and missing-data imputation. Am J Hum Genet. 2005;76:449–62. doi: 10.1086/428594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Erlacher L, McCartney J, Piek E, ten Dijke P, Yanagishita M, Oppermann H, et al. Cartilage-derived morphogenetic proteins and osteogenic protein-1 differentially regulate osteogenesis. J Bone Miner Res. 1998;13:383–92. doi: 10.1359/jbmr.1998.13.3.383. [DOI] [PubMed] [Google Scholar]
- 39.Savarirayan R, White SM, Goodman FR, Graham JM, Jr, Delatycki MB, Lachman RS, et al. Broad phenotypic spectrum caused by an identical heterozygous CDMP-1 mutation in three unrelated families. Am J Med Genet A. 2003;117A:136–42. doi: 10.1002/ajmg.a.10924. [DOI] [PubMed] [Google Scholar]
- 40.Masuya H, Nishida K, Furuichi T, Toki H, Nishimura G, Kawabata H, et al. A novel dominant-negative mutation in Gdf5 generated by ENU mutagenesis impairs joint formation and causes osteoarthritis in mice. Hum Mol Genet. 2007;16:2366–75. doi: 10.1093/hmg/ddm195. [DOI] [PubMed] [Google Scholar]
- 41.Ioannidis JP. Why most discovered true associations are inflated. Epidemiology. 2008;19:640–8. doi: 10.1097/EDE.0b013e31818131e7. [DOI] [PubMed] [Google Scholar]
- 42.Burton PR, Hansell AL, Fortier I, Manolio TA, Khoury MJ, Little J, et al. Size matters: just how big is BIG? Quantifying realistic sample size requirements for human genome epidemiology. Int J Epidemiol. 2009;38:263–73. doi: 10.1093/ije/dyn147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Manolio TA, Brooks LD, Collins FS. A HapMap harvest of insights into the genetics of common disease. J Clin Invest. 2008;118:1590–605. doi: 10.1172/JCI34772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yu W, Gwinn M, Clyne M, Yesupriya A, Khoury MJ. A navigator for human genome epidemiology. Nat Genet. 2008;40:124–5. doi: 10.1038/ng0208-124. [DOI] [PubMed] [Google Scholar]
- 45.Patsopoulos NA, Tatsioni A, Ioannidis JP. Claims of sex differences: an empirical assessment in genetic associations. JAMA. 2007;298:880–93. doi: 10.1001/jama.298.8.880. [DOI] [PubMed] [Google Scholar]
- 46.Hart DJ, Spector TD. The classification and assessment of osteoarthritis. Baillieres Clin Rheumatol. 1995;9:407–32. doi: 10.1016/s0950-3579(05)80198-0. [DOI] [PubMed] [Google Scholar]
- 47.Schiphof D, Boers M, Bierma-Zeinstra SM. Differences in descriptions of Kellgren and Lawrence grades of knee osteoarthritis. Ann Rheum Dis. 2008;67:1034–6. doi: 10.1136/ard.2007.079020. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.