Abstract
We studied the relationship between neuropeptide receptor transcript expression and current responses in the stomatogastric ganglion (STG) of the crab, Cancer borealis. We identified a transcript with high sequence similarity to crustacean cardioactive peptide (CCAP) receptors in insects and mammalian neuropeptide S receptors. This transcript was expressed throughout the nervous system, consistent with the role of CCAP in a range of different behaviors. In the STG, single-cell qPCR showed expression in only a subset of neurons. This subset had previously been shown to respond to CCAP with the activation of a modulator-activated inward current (IMI), with one exception. In the one cell type that showed expression but no IMI responses, we found CCAP modulation of synaptic currents. Expression levels within STG neuron types were fairly variable, but significantly different between some neuron types. We tested the magnitude and concentration dependence of IMI responses to CCAP application in two identified neurons, the lateral pyloric (LP) and the inferior cardiac (IC) neurons. LP had several-fold higher expression and showed larger current responses. It also was more sensitive to low CCAP concentrations and showed saturation at lower concentrations, as sigmoid fits showed smaller EC50 values and steeper slopes. In addition, occlusion experiments with proctolin, a different neuropeptide converging onto IMI, showed that saturating concentrations of CCAP activated all available IMI in LP, but only approximately two-thirds in IC, the neuron with lower receptor transcript expression. The implications of these findings for comodulation are discussed.
Keywords: crustacean cardioactive peptide, neuromodulation, proctolin, stomatogastric
Introduction
Neuromodulation underlies flexibility of neural circuit operation, and all circuits are likely under control of multiple neuromodulators at all times (Harris-Warrick, 2011; Jordan and Sławińska, 2011; Marder, 2012; Nadim and Bucher, 2014). Therefore, understanding circuit dynamics requires not just knowledge of connectivity and baseline intrinsic and synaptic dynamics, but detailed quantitative information about neuromodulatory effects on all circuit components (Harris-Warrick, 2011; Bargmann, 2012; Bargmann and Marder, 2013; Nadim and Bucher, 2014). A neuromodulator can target multiple aspects of excitability and synaptic function in a single neuron, and different neuromodulators can share intracellular targets. In addition, a neuromodulator can have different targets across different neuron types. These multiple patterns of divergence and convergence, and the fine-tuning of cellular and synaptic properties they provide, present a major challenge to the understanding of circuit dynamics (Nadim and Bucher, 2014).
Mapping neuromodulator effects quantitatively across circuit components may still only be an attainable goal in numerically simpler invertebrate circuits consisting of individually identifiable neurons, such as those found in the crustacean stomatogastric ganglion (STG) (Nusbaum and Beenhakker, 2002; Marder and Bucher, 2007). In the STG, the patterns of divergence and convergence of neuromodulator effects are fairly distinct between neuropeptides and biogenic amines (Marder et al., 2005; Marder and Bucher, 2007; Stein, 2009; Nadim and Bucher, 2014). Monoamines, such as dopamine, act on all neurons to modulate a cell-type specific subset of multiple voltage-gated ion channels and also affect many synapses (Harris-Warrick et al., 1998; Harris-Warrick and Johnson, 2010; Harris-Warrick, 2011). In contrast, neuropeptides appear to have only a limited number of subcellular targets, and each neuropeptide acts on only a specific subset of circuit neurons. A number of neuropeptides converge onto the same voltage-gated modulator-activated inward current, IMI (Golowasch and Marder, 1992; Swensen and Marder, 2000; DeLong et al., 2009), and there are only a few reports of neuropeptide actions on synapses (Thirumalai et al., 2006; Zhao et al., 2011) or other voltage-gated ion channels (Rodriguez et al., 2013).
The finding that each peptide acts only on a specific subset of circuit neurons serves as a qualitative explanation for the specific effects of each peptide on circuit activity (Swensen and Marder, 2001). However, it is not clear to which degree quantitative differences play a role, for example, manifest in differences in receptor expression levels and concentration dependence of effects. There are two main reasons for the lack of quantitative understanding. First, despite the fact that the majority of neuromodulators acting on the STG are neuropeptides (Li et al., 2003; Marder and Bucher, 2007), G-protein-coupled receptors (GPCRs) have only been identified for biogenic amines (Clark et al., 2004, 2008). Second, IMI responses to saturating concentrations of neuropeptide can be highly variable across individual animals (Goaillard et al., 2009).
Here we identify a putative neuropeptide receptor transcript with high sequence similarity to crustacean cardioactive peptide (CCAP) receptors in insects and show that expression is limited to the subset of STG neurons showing physiological responses. In two neuron types tested, differences in expression levels were consistent with differences in the magnitude and concentration dependence of modulatory effects.
Materials and Methods
Animals.
All experiments were performed on wild-caught adult male Jonah crabs, Cancer borealis, purchased from Fresh Lobster. Crabs were kept in aquaria supplied with artificial sea water (Instant Ocean), cooled to a temperature of 9°C–13°C. The animals were fed once a week until use. Before dissection, animals were anesthetized in ice for 30 min. For each experiment, individual tissues were dissected under chilled physiological saline, composed of (in mm) as follows: 440 NaCl, 11 KCl, 13 CaCl2, 26 MgCl2 (all from Fisher Scientific), and 10 HEPES (Sigma-Aldrich). The pH was adjusted to 7.45.
RNA isolation and cDNA construction.
Total RNA was isolated from tissues specific for each cDNA library application. To obtain the coding sequence of the C. borealis CCAP receptor (CbCCAPr), the brain, the two commissural ganglia (CoGs), the esophageal ganglion (OG), and the STG were pooled and used for RNA extraction. For analysis of CbCCAPr distribution, tissues were collected individually and included the brain, cardiac ganglion (CG), STG, OG, CoGs, thoracic ganglion (TG), the gastric mill muscle gm4, and the medial, dorsal muscle of the heart (HM). In both forms of RNA isolation, the TRI reagent protocol was used (Molecular Research Center). All tissues were placed directly into 360 μl (STG, OG, CoGs, CG, gm4), 500 μl (stomatogastric nervous system (STNS), brain, HM), or 750 μl (TG, pooled STNS, and brain) TRI reagent homogenization buffer. The tissue was completely homogenized and then stored overnight at −80°C. The following day, tissues were thawed and 250 μl TRI reagent was mixed into the homogenized TG or pooled STNS and brain samples. Samples were then processed according to the manufacturer's instructions and digested with DNase 1 (New England BioLab). Final RNA was suspended in 22, 30, or 50 μl DEPC-treated water, depending on tissue of origin. RNA concentration and quality were measured on a NanoDrop 2000c (Thermo Scientific).
Two forms of cDNA libraries were constructed. To clone CbCCAPr, the SMART method (Clontech) was used to develop cDNA compatible with rapid amplification of cDNA ends (RACE). Primers are listed in Table 1. Briefly, total RNA was incubated with TempSwitch and TRsa(T30) oligos and then reverse transcribed using SuperScript III RT (Invitrogen). PCR amplification of the cDNA library was then performed using SMART oligos and LA Taq polymerase reagents (Takara Bio). To evaluate the distribution of CbCCAPr throughout individual tissues of the animal, the standard SuperScript III RT protocol was used, using random hexamers and Oligo d(T)30 primers during reverse transcription. The amount of RNA used for reverse transcription was limited by the total RNA isolated from the smallest nervous tissues, the OG and STG, resulting in only ∼125 ng of RNA used from each tissue. Tissue-specific libraries were not amplified after reverse transcription, and any residual RNA was digested using RNase H (New England BioLabs). Both forms of cDNA were stored in aliquots at −20°C.
Table 1.
Primers used for cloning, distribution, and quantification of the CbCCAPra
| Primer | Sequence (5′ to 3′) |
|---|---|
| cDNA library development | |
| Template switch (TS) | AAGCAGTGGTATCAACGCAGAGTACGCrGrGrG |
| TRsa(T)30 | CGCAGTCGGTACT30 |
| CapPCR | AAG CAG TGG TAT CAA CGC AGA GTA |
| Cloning primers | |
| Lu4Cap | CGACGTGGACTATCCATGAACGCAAAGCAGTGGTATCAACGCAGAGTA |
| Lu4TRsa | CGACGTGGACTATCCATGAACGCACGCAGTCGGTACT30 |
| NsLu4 | TCGAGCGGCCGCCCGGGCAGGTCGACGTGGACTATCCATGAACGCA |
| CbCCAPr.5.F | CCTACGTCTTGGTGGCTCTC |
| CbCCAPr.8.R | TAATCTTGGCCTTTGGGATG |
| CbCCAPr.4.R | AAGGGGAAGGTGATGACGGTCACT |
| CbCCAPr.7.F | ATCGATTTCCCGAAGTTGTG |
| Tissue distribution PCR primers | |
| CbCCAPr.5.F | CCTACGTCTTGGTGGCTCTC |
| CbCCAPr.8.R | TAATCTTGGCCTTTGGGATG |
| Cbβtub.1.F | TCTGTGCTGGATGTAGTCCG |
| Cbβtub.3.R | AGAGTGGCGTTGTATGGCTC |
| qRT-PCR primers | |
| CbCCAPr.3.F | GGTGGCTCTGACTGTCTTCCTCTT |
| CbCCAPr.4.R | AAGGGGAAGGTGATGACGGTCACT |
aPrimers used for development of cDNA libraries suitable for RACE, cloning of CbCCAPr, tissue distribution, and qRT-PCR measurements. βtub, Beta-tubulin; F, forward; R, reverse.
Cloning the receptor transcript.
We first cloned and sequenced a partial cDNA fragment of CbCCAPr using an approach described previously (Schulz et al., 2006, 2007). Briefly, the receptor was amplified from a cDNA template derived from mixed nervous system tissue of C. borealis. We designed and used degenerate primer pairs based on conserved amino acid sequence compared across multiple invertebrate species. PCR products of predicted length were cloned into pGEM-T easy plasmid vector (Promega) and sequenced using traditional Sanger sequencing methods (University of Missouri DNA Core Facility). Sequences obtained were compared with orthologous sequences using BlastX (NCBI). With a putative fragment of CbCCAPr identified, gene-specific primers designed against the known fragment, and RACE oligo sequences (Table 1) were purchased from Integrated DNA Technologies. Touch-down and nested step-out PCRs to expand the amino- and carboxy termini of the fragment were performed using LA Taq polymerase reagents (Takara Bio), and products were cloned into pGEM-T vectors (Promega). Positive clones were sent to SeqWright for sequencing. Sequences comprising CbCCAPr were confirmed using BlastX (NCBI) and assembled using Lasergene SeqMan and SeqBuilder software (DNAstar). The assembled, putative coding sequence of CbCCAPr contains an open reading frame of 1005 bases, identified and translated into amino acids using the ExPASy translate tool (Swiss Institute of Bioinformatics).
Sequence alignment and comparison.
Multiple sequence alignment of CbCCAPr with other mammalian and arthropod Rhodopsin-like receptors was performed using MUSCLE (EMBL-EBI). Amino acid sequences were taken from the UniProtKB/Swiss-Prot database (EMBL-EBI). Accession numbers are listed in Table 2. ExPASy BoxShade server (SIB) was used to indicate conserved residues. The TMpred program (SIB) (Hofmann and Stoffel, 1993) was used to predict the transmembrane domains of CbCCAPr. Published transmembrane domains in the UniProtKB/Swiss-Prot databank were used for human and arthropod receptors. Statistical significance was assumed at e-values <0.001.
Table 2.
Accession numbers of select arthropod and human Rhodopsin-like receptors used in the multiple alignment shown in Figure 1a
| Receptor | Accession number |
|---|---|
| CbCCAPr | KM349850 |
| Apis mellifera CCAPr | XP_001122652.2 |
| Nasonia vitripennis CCAPr | XP_001602277.1 |
| T. castaneum CCAPr2 | NP_001076795.1 |
| T. castaneum CCAPr1 | NP_001076796.1 |
| Drosophila melanogaster CCAPr | NP_996297.3 |
| Culex quinquefasciatus CCAPr | XP_001847670.1 |
| Daphnia pulex CCAPr | EFX81678.1 |
| Homo sapiens NPSrA | NP_997055.1 |
| Homo sapiens NPSrB | NP_997056.1 |
| Homo sapiens V1Ar | NP_000697.1 |
| Homo sapiens V1Br | NP_000698.1 |
| Homo sapiens OXTr | NP_000907 |
aRhodopsin-like receptors: CCAPr; NPSrA, neuropeptide S receptor 1; NPSrB, neuropeptide S receptor isoforms B; V1Ar, vasopressin receptor 1A; V1Br, vasopressin receptor 1B; OXTr, oxytocin receptor.
To evaluate the phylogenetic relationship of CbCCAPr with other Rhodopsin-like receptors, 61 protein sequences (accession numbers in Table 3) were aligned in MUSCLE, and phylogenetic analysis was conducted using MEGA software (version 6.0, ImageMagick Studio) (Tamura et al., 2013). A maximum likelihood consensus tree was inferred from 1000 bootstrap replicates, with the initial trees obtained by applying Neighbor-join and BioNJ algorithms to a matrix of pairwise distances estimated using a JTT model (Jones et al., 1992).
Table 3.
Accession numbers of the 61 protein sequences used in phylogenetic analysisa
| Receptor | Accession No. |
|---|---|
| CCAPr | |
| Apis mellifera CCAPr | XP_001122652.2 |
| Bombyx mori NPr A26 | BAG68425.1 |
| CbCCAPr | KM349850 |
| Daphnia pulex hypothetical CCAPr | EFX81678.1 |
| Drosophila melanogaster CCAPr | CCAPR_DROME |
| T. castaneum CCAPr1 | NP_001076796.1 |
| T. castaneum CCAPr2 | NP_001076795.1 |
| Neuropeptide S receptors | |
| Anopheles gambiae NPSr-like GPCR3 | AAS77205.1 |
| Bombyx mori NPr A30 | NP_001127746.1 |
| Homo sapiens NPSrA | NP_997055.1 |
| Homo sapiens NPSrB | NP_997056.1 |
| Mus musculus NPSr | NP_783609.1 |
| Vasopressin and oxytocin receptors | |
| Danio rerio OXTr | NP_001186299.1 |
| Danio rerio OXTr-like | NP_001186298.1 |
| Danio rerio VasotocinRx2 | XP_683692.1 |
| Daphnia pulex AVP/OXTr | E9HG37_DAPPU |
| Homo sapiens OXTr | NP_000907.2 |
| Homo sapiens V1Ar | NP_000697.1 |
| Homo sapiens V1Br | NP_000698.1 |
| Homo sapiens V2r | NP_000045.1 |
| Mus musculus OXTr | NP_001074616.1 |
| Mus musculus V1Ar | NP_058543.2 |
| Mus musculus V1Br | NP_036054.1 |
| Mus musculus V2r | NP_062277.1 |
| T. castaneum AVP/OXTr | B1NWV5_TRICA |
| Adipokinetic hormone/GNRHr | |
| Anopheles gambiae AKH/GNRHr | Q27J45_ANOGA |
| Danio rerio GNRHr2 | NP_001138451.1 |
| Danio rerio GNRHr4 | NP_001091663.1 |
| Drosophila melanogaster GNRHrA | NP_477387.1 |
| Drosophila melanogaster GNRHr2-A | NP_648571.1 |
| Homo sapiens GNRHr isoform 1 | NP_000397.1 |
| Mus musculus GNRHr | NP_034453.1 |
| T. castaneum AKH/GNRHr | Q1W7L1_TRICA |
| Gastrin/cholecystokinin receptors | |
| Anopheles gambiae CCKr-like | XP_001237203.1 |
| Danio rerio CCKrA-like | XP_697493.2 |
| Danio rerio CCKr-likeX1 | XP_002663361.2 |
| Drosophila melanogaster CCKr-like | NP_001097021.1 |
| Drosophila melanogaster CCKr-like17D3 | NP_001097023.1 |
| Homo sapiens CCKrA | NP_000721.1 |
| Homo sapiens Gastrin/CCKrB | NP_795344.1 |
| Mus musculus CCKrA | NP_033957.1 |
| Mus musculus CCKrB | NP_031653.1 |
| Orexin receptors | |
| Danio rerio ORXr2 | NP_001073337.1 |
| Harpegnathos saltator ORXr2 | E2BMY8_HARSA |
| Homo sapiens ORXr1 | NP_001516.2 |
| Mus musculus ORXr1 | NP_001156499.1 |
| Pyroglutamylated RFamide peptide receptors | |
| Danio rerio QRFPr | XP_001920042.3 |
| Homo sapiens QRFPr | NP_937822.2 |
| Mus musculus QRFPr | NP_937835.1 |
| Neuropeptide FF receptors | |
| Acromyrmex echinatior NPFFr2 | EGI57927.1 |
| Danio rerio NPFFr1 | NP_001082858.1 |
| Danio rerio NPFFr-like2 | NP_001165168.1 |
| Danio rerio NPFFr2 | XP_690069.5 |
| Drosophila melanogaster NPFFr-like SIFr | NP_001163674.1 |
| Homo sapiens NPFFr1 | NP_071429.1 |
| Homo sapiens NPFFr2 isoform1 | NP_004876.2 |
| Mus musculus NPFFr1 | NP_001170982.1 |
| Mus musculus NPFFr2 | NP_573455.2 |
| Frizzled receptors | |
| Drosophila melanogaster FRZ | FRIZ_DROME |
| Homo sapiens FRZ1 | NP_003496.1 |
| Mus musculus FRZ1 | NP_067432.2 |
aNPr, Neuropeptide receptor; NPSr, neuropeptide S receptor; OXTr, oxytocin receptor; AVP, arginine vasopressin; V1Ar, vasopressin receptor 1A; V1Br, vasopressin receptor 1B; V2r, vasopressin receptor 2; AKH, adipokinetic hormone; FRZ, frizzled receptor.
Tissue distribution of CbCCAPr transcript.
To evaluate the quality of individual tissue cDNAs, primers were designed against C. borealis β-tubulin (CbβTub, GenBank accession number HM157288.1; Table 1) using PCR. Bands of 250 base pairs (bp) were amplified (see Fig. 3B) from 30 cycles of 30 s at 94°C, 30 s at 60°C, and 1 min at 72°C. Primers designed to amplify a 458 bp fragment of CbCCAPr (Table 1) were then used for screening the expression of each tissue (see Fig. 3B) using 40 cycles of 30 s at 94°C, 30 s at 57°C, and 1 min at 72°C. For both CbβTub and CbCCAPr PCR amplification, control libraries developed without reverse transcriptase (No-RT) were run in parallel (data not shown), to exclude genomic DNA contamination and account for high cycle number. Products were visualized on 1.5%–1.8% ethidium bromide-stained agarose gels. Bands of appropriate sizes were randomly excised and sequenced to confirm product identity.
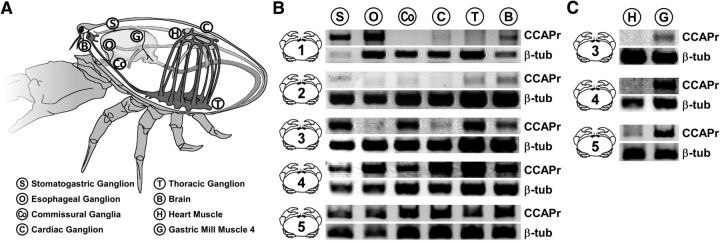
Figure 3.
CbCCAPr RNA is present throughout the nervous system and in a gastric mill muscle. A, Schematic of C. borealis anatomy, indicating the tissues harvested. B, C, Negative images of ethidium bromide-stained agarose gels from PCR amplification of CbCCAPr (top bands, 485 bp) and Cbβ-tubulin (bottom bands, 250 bp) from 5 animals.
CbCCAPr mRNA copy number quantification of individual STG neurons.
Individual neurons were identified physiologically with intracellular recordings using established criteria and harvested for single-cell quantitative PCR as previously described (Schulz et al., 2006). Total RNA was isolated using the RNeasy extraction kit (QIAGEN), reverse transcribed with SuperScript III, and then used as template for real-time PCR with the fluorescent reporter SYBR Green (SA Biosciences). Primers (Table 1) were designed specifically for real-time PCR detection of CbCCAPr transcripts using Primer3 software.
Electrophysiological recordings.
For electrophysiology experiments, the STNS was dissected from the stomach and transferred to a transparent Sylgard-lined (Dow Corning) dish in chilled physiological saline. A large petroleum jelly well was built around the STG for application of pharmacological agents. During all recordings, the preparation was continuously superfused with chilled saline (11°C–13°C). Intracellular recordings of neuron somata were obtained after desheathing the STG, using sharp glass microelectrodes filled with 0.6 M K2SO4 and 20 mm KCl (resistance: 10–30 ΩM). Intracellular recordings and voltage clamp were performed using Axoclamp 2B and 900A amplifiers (Molecular Devices). Electrode holders and headstages were mounted on mechanical (Leica) or motorized (Scientifica) micromanipulators. Traces were recorded using micro1401 mk2 digitizer boards (Cambridge Electronic Design) and Spike2 acquisition software (Cambridge Electronic Design, versions 7 and 8). Voltage and current traces were low-pass filtered in Spike2 to reduce noise as needed. Care was taken that filtering did not change the time course or amplitude of the signals of interest. Neurons were identified according to characteristic waveforms and by matching spike patterns to extracellular recordings from specific motor nerves. Extracellular recordings were obtained with stainless steel wire electrodes from petroleum jelly wells around motor nerves. Signals were amplified and filtered using A-M systems differential AC amplifiers (model 1700). All electrophysiological recordings were analyzed using custom programs written in the Spike2 script language.
Current measurements.
Two electrode voltage-clamp recordings were used to determine the effect of different concentrations of CCAP (Bachem) on synaptic currents and the modulator-activated inward current (IMI). Graded synaptic currents were recorded in response to depolarizing steps in a presynaptic neuron. Rhythmic activity and spiking were blocked by application of 100 nm TTX (Sigma-Aldrich). The postsynaptic neuron was held at −50 mV, and the presynaptic neuron was stepped from a holding potential of −60 mV. Short-term synaptic plasticity was tested with sets of five 0.5 s steps in the presynaptic neuron, at a frequency of 1 Hz. To test for postsynaptic effects, outward currents in response to 0.5 s puffs of 10 mm glutamate (l-glutamic acid monosodium salt, Sigma) onto the STG neuropil were measured in the postsynaptic neuron at a holding potential of −50 mV. Puffs were administered through a glass microelectrode with a broken tip, connected to a Toohey Spritzer (Toohey).
IMI was measured in two different ways. In some experiments, IV curves were obtained from difference currents measured in response to voltage ramps (at 90 mV/s) in control and CCAP (Golowasch and Marder, 1992; Goaillard et al., 2009). This was done in the presence of 100 nm TTX to block voltage-gated sodium channels, 1 μM picrotoxin to block inhibitory glutamatergic synapses, 200 μm CdCl2 to block L-type calcium currents, and 20 mm tetraethylammonium to block delayed rectifier and calcium-dependent potassium currents. All blockers were purchased from Sigma-Aldrich. IMI was obtained by subtracting the current response to the ramp in control saline from the response in CCAP, and current was subsequently plotted as a function of voltage. To test the concentration dependence of IMI, we held the membrane potential at −20 mV and applied CCAP in increasing concentrations from 100 pm to 1 μm. Each concentration was washed in for 6 min and washed out for 8–15 min. Blockers used in these experiments were the same as described for ramp protocols. Current amplitudes for each concentration were determined as the maximal difference to the holding current before CCAP application. In some experiments, the neuropeptide proctolin (Bachem) was applied in addition to CCAP to test for occlusion effects.
Statistical analysis.
All statistical analyses and data plots were generated in SigmaPlot (versions 11 and 12, Systat Software). Unless otherwise indicated, all data are presented as means ± SEM. Tests performed were t tests (paired or unpaired, as appropriate) and one-way or two-way ANOVA, for repeated measures when appropriate. Pairwise comparisons of normally distributed data following ANOVA used the Holm–Sidak method. For not normally distributed data, Kruskal-Wallis ANOVA was performed on ranks, and pairwise comparisons were performed using Dunn's method for unequal group sizes, and Tukey tests for equal group sizes.
Postsynaptic current as a function of presynaptic voltage and IMI as a function of the log of the CCAP concentration were fit with 3 parameter sigmoid functions of the following form: y = a/(1 + e−(x−x0)/b), where a is the maximal current (Imax), ×0 the voltage of half-activation (V1/2, synaptic currents) or the half-maximal effective concentration (EC50, IMI), and b the slope factor. In this form, the slope factor is not the same as the often used Hill slope (Prinz, 2010), and it is smaller at steeper slopes. Final figure mounting and editing were done in Canvas (version 11, ACD Systems).
Results
The putative CbCCAPr belongs to subfamily A6 within Rhodopsin-like GPCRs and is most closely related to other CCAP and neuropeptide S receptors
CCAP is a cyclic nonapeptide with mirror-image sequence similarity to vasopressin (Stangier et al., 1987). It mainly acts as a neurohormone in arthropods and plays important roles in the regulation of heart contractions (Stangier et al., 1987; Tublitz, 1989; Cruz-Bermúdez and Marder, 2007; Fort et al., 2007; Wasielewski and Skonieczna, 2008; Estévez-Lao et al., 2013; Lee et al., 2013); molting behavior (Ewer and Truman, 1996; Phlippen et al., 2000; Park et al., 2003); gut movements, digestion, and metabolism (Veelaert et al., 1997; Donini et al., 2002; Sakai et al., 2004; Sakai et al., 2006); and oviduct contractions (Donini et al., 2001). In the STG, it acts on a number of neurons and alters rhythmic motor activity (Weimann et al., 1997; Richards and Marder, 2000; Swensen and Marder, 2000, 2001; DeLong et al., 2009), as well as contraction properties of target muscles (Jorge-Rivera et al., 1998). GPCRs specific for CCAP have been identified in a number of insect species (Cazzamali et al., 2003; Belmont et al., 2006; Arakane et al., 2008; Li et al., 2011; Lee et al., 2013), but so far not in crustaceans. We identified a fragment of a putative receptor cloned from C. borealis with 66% identity to the Tribolium castaneum crustacean cardioactive peptide receptor 2 (TbCCAPr2) (Park et al., 2008; Li et al., 2011) and used cDNA libraries constructed from RNA isolated from the STNS and brain to assemble the partial coding sequence. The putative CbCCAPr (GenBank accession number: KM349850) comprises 332 amino acids and a seven-transmembrane domain region typical of GPCRs. Comparison of CbCCAPr to known proteins using BlastX (NCBI) analysis showed sequence homology to members of the Rhodopsin-like family of GPCRs, including other arthropod CCAP receptors and human vasopressin and neuropeptide S receptors.
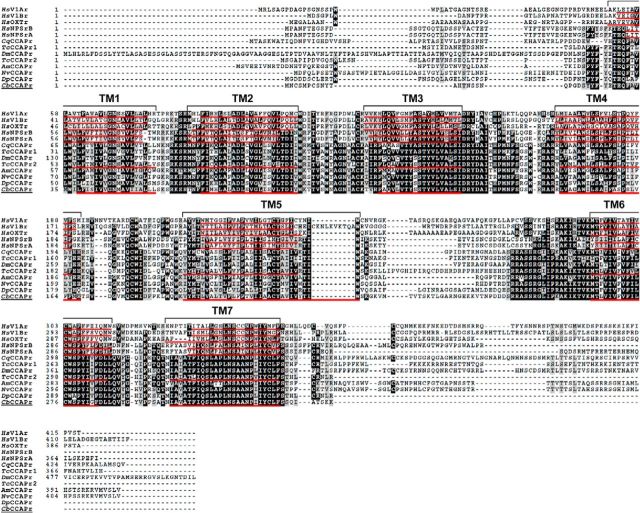
Multiple alignment of CbCCAPr with arthropod CCAP receptors and human vasopressin, oxytocin, and neuropeptide S (NPS) receptors revealed several regions of conserved residues (Fig. 1). Conserved residues occupy not only the transmembrane regions (TMs) but also intracellular and extracellular loops. Intracellular loop 3 showed the greatest diversity in residue compliment, aside from the carboxy- and amino-termini, which is likely due to specific residue motifs for coupling to G-proteins (Wess, 1997). Table 4 lists the percentage identity of amino acid residues between CbCCAPr and the full-length human and T. castaneum receptor sequences shown in the alignment in Figure 1. CbCCAPr is more similar to HsNPSrA than to HsV1Ar, with 41% sequence identity and 93% coverage compared with 33% and 89%, respectively, but most similar to the TcCCAPr2 (63% sequence identity and 96% coverage). Because the highest degree of similarity between GPCRs exists in the TM core structure (Bockaert and Pin, 1999), we also evaluated the relationships between receptor sequences excluding TMs. Without TMs, sequence identities dropped by 3%–9%, but e-values remained significant (Table 4).
Figure 1.
Multiple alignment of the CbCCAPr sequence with other arthropod CCAP receptor sequences and sequences for human vasopressin, oxytocin, and neuropeptide S receptors. Black represents identical amino acid residues. Gray represents similar residues. Transmembrane regions of CbCCAPr, human and T. castaneum receptors are underlined in red and labeled with brackets.
Table 4.
Percent amino acid identity shared between CbCCAPr and select human and arthropod receptors, with and without transmembrane domain regionsa
| CbCCAPr |
HsV1Ar |
HsNPSrA |
TcCCAPr |
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Whole |
(−) TM |
Whole |
(−) TM |
Whole |
(−) TM |
Whole |
(−) TM |
|||||||||
| ID | e-value | ID | e-value | ID | e-value | ID | e-value | ID | e-value | ID | e-value | ID | e-value | ID | e-value | |
| HsV1Ar | 33 | 1e-52 | 30 | 3e-06 | 29 | 6e-46 | 27 | 7e-09 | 33 | 2e-53 | 34 | 7e-05 | ||||
| HsV1Br | 33 | 3e-57 | 28 | 9e-09 | 54 | 2e-129 | 47 | 3e-53 | 31 | 1e-46 | 27 | 2e-09 | 34 | 6e-51 | 25 | 9e-06 |
| HsOXTr | 34 | 2e-52 | 28 | 1e-07 | 54 | 2e-118 | 47 | 1e-41 | 34 | 2e-46 | 30 | 8e-11 | 34 | 2e-49 | 38 | 3e-05 |
| HsNPSrA | 41 | 4e-81 | 36 | 3e-17 | 29 | 6e-46 | 27 | 7e-09 | 41 | 9e-86 | 33 | 6e-19 | ||||
| HsNPSrB | 41 | 2e-80 | 36 | 3e-17 | 29 | 2e-45 | 26 | 1e-08 | 99 | 0.0 | 99 | 7e-150 | 41 | 3e-85 | 33 | 1e-18 |
| TcCCAPr2 | 63 | 1e-151 | 54 | 7e-55 | 33 | 2e-53 | 34 | 7e-05 | 41 | 9e-86 | 33 | 6e-19 | ||||
aHsV1Ar, H. sapiens vasopressin receptor 1A; HsV1Br, H. sapiens vasopressin receptor 1B; HsOXTr, H. sapiens oxytocin receptor; HsNPSrA, H. sapiens neuropeptide S receptor 1; HsNPSrB, H. sapiens neuropeptide S receptor isoform B; TcCCAPr2, T. castaneum crustacean cardioactive peptide receptor 2; Whole, entire protein sequence; (−) TM, transmembrane domain regions removed; ID, sequence identity (in percent).
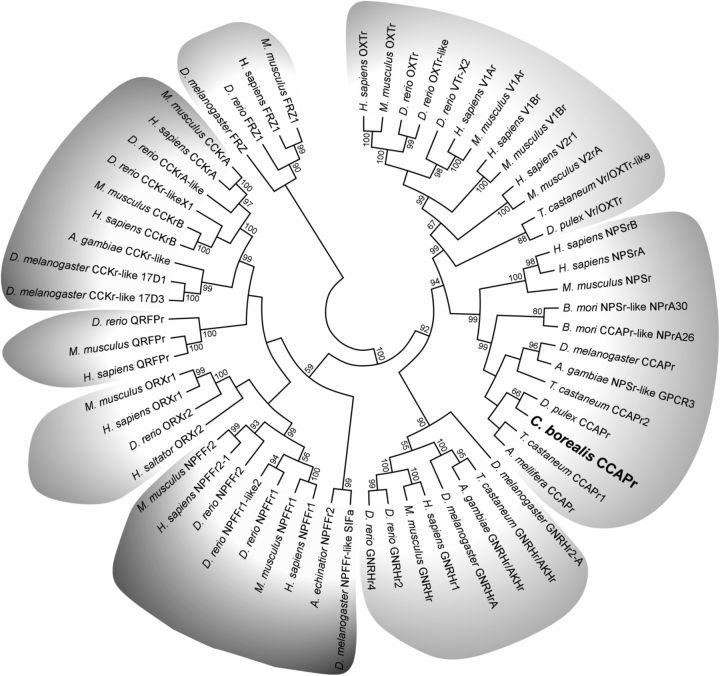
To better support the identity of CbCCAPr, we performed phylogenetic analysis. CCAP, vasopressin, and NPS receptors belong to the subfamily A6 of Rhodopsin-like GPCRs (Joost and Methner, 2002), which additionally include gonadotropin releasing hormone receptors (GNRHr), cholecystokinin receptors (CCKr), orexin receptors (ORXr), neuropeptide FF receptors (NPFFr), and pyroglutamylated receptors (QRFPr). Sequences representing each A6 receptor subclass were chosen from both vertebrate and invertebrate species. Frizzled GPCRs from vertebrates and Drosophila were used as the out-group. The maximum-likelihood consensus tree in Figure 2 shows CbCCAPr clusters with NPS and CCAP receptors and is most closely related to a hypothetical CCAPr predicted from the genome of the only other crustacean in the study, Daphnia pulex. The most closely related receptor group to CCAP and NPS receptors consists of vasopressin and oxytocin receptors. The group of CCAP, NPS, vasopressin, and oxytocin receptors is most closely related to GNRH receptors, consistent with a previous study (Pitti and Manoj, 2012). All other receptors cluster according to the receptor groups within subfamily A6, whereas Frizzled receptors are separate. The location of the CbCCAPr in the consensus tree strongly suggests that the putative sequence encodes for a GPCR with high similarity to arthropod CCAP receptors.
Figure 2.
Maximum-likelihood consensus tree of CbCCAPr and representatives of receptor types found in the A6 subfamily of Rhodopsin-like GPCRs. Sequences used for analysis include vasopressin receptors (V1Ar, V1Br, V2r), oxytocin receptors (OXTr), neuropeptide S receptors (NPSrA, NPSrB), gonadotropin releasing hormone receptors (GNRHr), neuropeptide FF receptors (NPFFr), orexin receptors (ORXr), pyroglutamylated RFamide peptide receptors (QRFPr), and cholecystokinin receptors (CCKr). Frizzled receptors (FRZ) were used as the out-group. Percentage bootstrap values of 1000 replicates are shown at the nodes; values <50% are hidden. Clades are shaded according to receptor type.
CbCCAPr is expressed in nervous tissues and the gastric mill gm4 muscle
Because of the widespread and diverse effects of CCAP on the nervous system and elsewhere, we investigated receptor mRNA expression throughout a range of different tissues. Five animals were used to extract RNA from six nervous tissues and two muscles (Fig. 3A): the thoracic ganglion, brain, cardiac ganglion, STG, CoGs, esophageal ganglion, heart muscle, and the gastric mill muscle gm4.
Figure 3B shows expression of CbCCAPr by each of the six nervous tissues, but at varying levels across individual animals. CbβTub is shown as a positive control. Libraries used as negative controls were generated in parallel and without reverse transcriptase (data not shown). The CoG sample in Animal 1 was the only one not showing expression. Figure 3C shows very weak or absent expression in the heart muscle but robust expression in gm4.
CbCCAPr mRNA is expressed at varying levels across neuron types in the STG
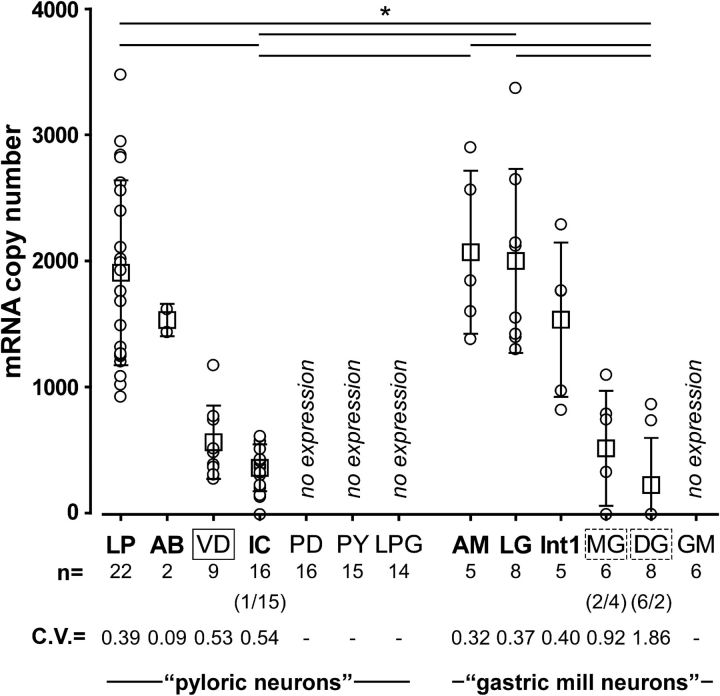
Figure 3B shows that CbCCAPr mRNA is expressed in the STG. We also determined the expression levels across different neuron types in the STG using single-cell qRT-PCR. In C. borealis, the STG contains 25 or 26 neurons, some of types that exist as a single copy and some of types that have multiple copies (Kilman and Marder, 1996). The majority of these neurons are members of two interacting central pattern-generating circuits controlling pyloric and gastric mill muscles of the stomach (Marder and Bucher, 2007). To varying degrees, most STG neurons take part in both the faster pyloric rhythm and the slower gastric mill rhythm (Weimann et al., 1991; Bucher et al., 2006), but for simplicity we separated the 13 neuron types into pyloric and gastric mill neurons, mostly based on the muscles that they control (Weimann et al., 1991). Figure 4 shows single measurements and means of mRNA copy numbers by cell type. There was substantial variability in expression levels within cell types (see coefficients of variation), and some cell types showed no expression at all. Among those that did show expression, mean levels were cell type-dependent and ranged from a few hundred copies to >2000. Pairwise comparisons revealed differences between the anterior median (AM) neuron and the dorsal gastric (DG) neurons, AM and the inferior cardiac (IC) neuron, the lateral gastric (LG) neuron and DG, LG and IC, the lateral pyloric (LP) and DG, and LP and IC. In some cell types with a low mean expression level, some individual samples showed no expression at all. This was true for IC in 1 of 16 samples, for the medial gastric (MG) neuron in 2 of 6 samples, and for DG in 6 of 8 samples.
Figure 4.
Single-cell qRT-PCR shows that CbCCAPr is expressed in a subset of STG neurons and at varying levels between cell types. For each cell type, mRNA copy numbers are plotted both for individual measurements (circles) and for means (squares). Cell types included all pyloric and gastric mill neurons (LP; AB; VD; IC; PY, pyloric constrictor neuron; LPG, lateral posterior gastric neuron; AM; Int1, interneuron 1; DG; GM). Cell types previously found to display physiological responses to CCAP are shown in bold. Solid line box around VD represents a mismatch between expression and physiological responsiveness. Dashed boxes around MG and DG represent ambiguity in expression and responsiveness. The number of individual cells measured for each cell type is given in the line beneath the cell type names. For IC, MG, and DG, the numbers in parentheses indicate the number of cells showing no expression/number of cells showing expression. The coefficients of variance are given to indicate variability of expression levels. Mean expression levels were cell type-dependent (one-way ANOVA on ranks, p < 0.001). Pairwise comparisons (Dunn's method) revealed differences as indicated by solid lines on top of the plot (p < 0.05 for each pair). *p < 0.05.
The effect of CCAP on intrinsic excitability has been tested for all STG neurons, in two different ways. Among pyloric neurons, voltage-clamp experiments revealed that CCAP activates IMI in LP and IC, but not in pyloric dilator (PD), PY, VD, and LPG (Swensen and Marder, 2001). IMI cannot be measured in voltage clamp from the anterior burster (AB) neuron, likely due to insufficient space clamp in soma recordings, but CCAP depolarizes synaptically isolated AB neurons in current-clamp recordings (Swensen and Marder, 2001). Among gastric neurons, only LG has been shown to activate IMI in response to CCAP application (DeLong et al., 2009). However, all other gastric mill neurons have been tested for depolarizing responses in current clamp (Kirby and Nusbaum, 2007). CCAP depolarized interneuron 1 (Int1) and AM, but not MG, DG, and the GM neurons. Cell types responsive to CCAP are shown in bold in Figure 4. Among pyloric neurons, CbCCAPr mRNA expression matches effects on intrinsic excitability, with the exception of the ventricular dilator (VD) neuron, which expresses at relatively low levels but in which IMI is not activated in response to CCAP. Among gastric mill neurons, CbCCAPr mRNA expression matches effects on intrinsic excitability unambiguously for AM, LG, and Int1, which show relatively high expression levels and also show physiological responses to CCAP, and GM, which neither shows expression nor responses. DG does not show depolarizing responses to CCAP. We found CbCCAPr mRNA expression in DG, but in only 2 of 8 samples. MG shows a somewhat ambiguous response to CCAP (Kirby and Nusbaum, 2007). A weak excitation is seen in low Ca2+ saline but ceases when responses in neurons that are electrically coupled to MG are suppressed. We found CbCCAPr mRNA expression in MG, but in only 4 of 6 samples. We conclude that there is some ambiguity in the cases of MG and DG, but that the only definitive mismatch between CbCCAPr mRNA expression and previously published physiological responses to CCAP exists for the VD neuron.
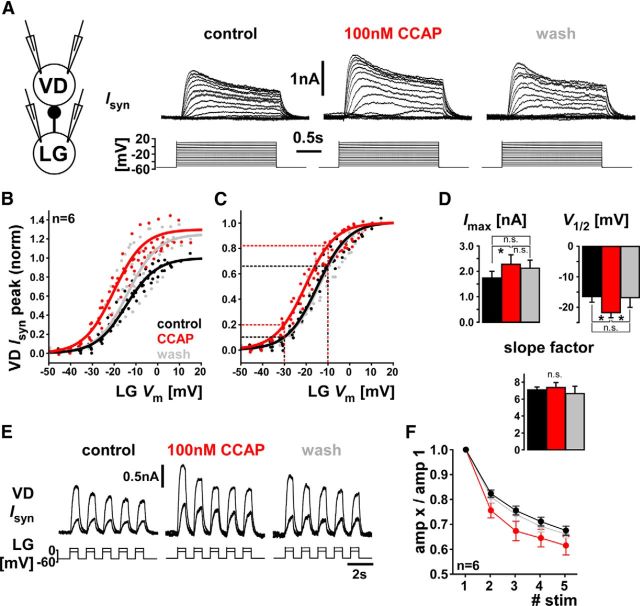
CCAP acts on the LG to VD synapse
CCAP does not activate IMI in the VD neuron, but expression of CbCCAPr mRNA suggests that CCAP may act on other subcellular targets. We therefore wanted to test whether CCAP acts on synaptic connections involving VD. Neuropeptide effects on synapses in the STG have not been studied extensively, but 2 cases involving other neuropeptides have been described (Thirumalai et al., 2006; Zhao et al., 2011). VD has no known chemical output synapses within the STG but receives graded inhibitory inputs from AB and LG through glutamatergic synapses (Nusbaum and Beenhakker, 2002). We used dual two-electrode voltage clamp to measure the LG to VD connection in control, 100 nm CCAP, and after a minimum of 15 min wash (Fig. 5A). Depolarizing voltage steps in LG elicited graded outward current responses in VD, with a transient and a sustained component typical for chemical synapses in the STG (Graubard et al., 1980; Manor et al., 1997; Zhao et al., 2011). The example traces in Figure 5A show an increase in Isyn in the presence of CCAP. We quantified the dependence of the peak synaptic current (Isyn peak) on presynaptic voltage. On average, CCAP increased Isyn peak by ∼30% (Fig. 5B) and shifted the dependence on presynaptic voltage (voltage of half activation, V1/2) by ∼−5 mV (Fig. 5C). Figure 5D shows that the change in Isyn peak was significant but did not wash. However, the shift in V1/2 was also significant, and did wash. The slope factor did not change. Even when only the change in V1/2 is considered (Fig. 5C), CCAP increased the synaptic current substantially at a given presynaptic voltage (25%–95% increase between −10 and −30 mV, dashed lines).
Figure 5.
CCAP modulates strength and dynamics of the LG to VD graded chemical synapse. A, Dual two-electrode voltage-clamp measurements of synaptic currents in VD (holding potential: −50 mV) in response to voltage steps in LG. Traces were averaged from 3 to 5 repeats of the same stimulation protocol. B, Cross-synaptic IV data for all three treatments from six experiments, normalized to Imax obtained from sigmoid fits to the control values in each experiment. Overlaid sigmoid curves were obtained by averaging sigmoidal fit parameters across experiments. C, The same cross-synaptic IV data as shown in B, but normalized to Imax obtained from sigmoid fits to values from each separate treatment. Dashed lines indicate the increase of synaptic currents at two different voltages that is solely due to the shift in V1/2. D, Statistical comparison of the sigmoidal fit parameters. One-way repeated-measures ANOVA showed significant differences across treatments for Imax (p < 0.05) and V1/2 (p < 0.05), but not the slope factor (p = 0.69). Asterisks indicate results from Holm–Sidak paired comparisons. E, VD neuron current responses to five 0.5 s steps at 1 Hz and at two different depolarization levels in the LG neuron. The synaptic current shows depression at both levels. Traces were averaged from 4 or 5 repeats of the same stimulation protocol. F, Plot of the mean response amplitudes, normalized to the first of the five responses. Two-way repeated-measures ANOVA showed a significant difference between treatments (p < 0.01), but no interaction between treatment and stimulus number (p = 0.37), meaning that the increase in depression was fairly uniform for stimuli 2 to 5. Holm–Sidak paired comparisons showed a significant difference between control and CCAP (p < 0.01) and CCAP and wash (p < 0.01), but not control and wash (p = 0.77). n.s., Not significant.
The effective strength of a synapse during repetitive activation depends critically on short-term synaptic dynamics (Nadim and Manor, 2000), and the graded synapses in the STG usually show substantial depression (Manor et al., 1997; Mamiya et al., 2003). We therefore also tested the effect of CCAP on synaptic dynamics. Figure 5E shows the VD current responses to repetitive stimulation at two different levels of LG depolarization in control, CCAP, and wash. We tested voltage steps of different amplitude because at STG synapses the sign of synaptic dynamics can depend on presynaptic voltage level (Zhao et al., 2011). For the range of LG voltages from −30 to 0 mV, the LG to VD synapse was always depressing (n = 5, with 2–5 voltage values per experiment; data not shown). We therefore only report responses to LG depolarization to −10 mV. Figure 5F shows that CCAP increased the amount of synaptic depression moderately (<10%) over the whole course of repeated stimulation.
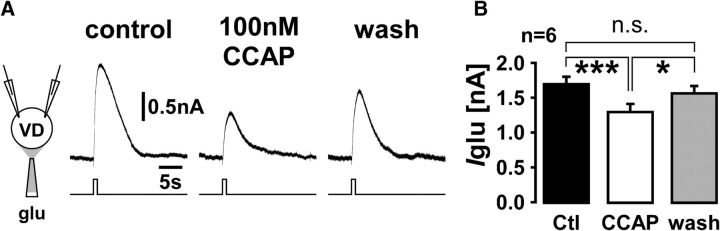
These results show that CCAP alters the LG to VD synaptic connection. We did not measure the AB to VD connection, the other synapse onto VD, but both LG (Kirby and Nusbaum, 2007; DeLong et al., 2009) and AB (Swensen and Marder, 2001) respond to CCAP in synaptic isolation, and both express CbCCAPr mRNA (Fig. 4). Consequently, the effects on synaptic strength and dynamics between LG and VD, and potentially AB and VD, could be either due to presynaptic or postsynaptic CCAP targets, or both. To test for unambiguously postsynaptic effects (i.e., to determine whether CCAP receptors on VD play a role in synaptic modulation), we also measured the currents in VD in response to glutamate, the transmitter used by both LG and AB. Figure 6A shows the multiple seconds long outward current responses to puffs of glutamate onto the STG neuropil in control, CCAP, and after washing for a minimum of 10 min. Surprisingly, CCAP reduced the amplitude of the response, on average by 24% (Fig. 6B). Therefore, postsynaptic effects of CCAP oppose the overall strengthening of the LG to VD synapse. We continuously monitored the input resistance (Rin) of VD in between glutamate puffs. Rin was similar in all conditions and not affected by CCAP (median value control: 11.0 MΩ, CCAP: 11.9 MΩ, wash: 12.7 MΩ; one-way ANOVA on ranks, p = 0.823). This suggests that the CCAP effect was solely due to changes in the activation of glutamate receptors. We conclude that CbCCAPr expression in VD contributes to synaptic modulation, which resolves the mismatch between receptor expression and previously published lack of VD responses to CCAP.
Figure 6.
CCAP reduces the current response to glutamate application in the VD neuron. A, VD current responses to 500 ms puff of 10 mm glutamate onto the STG neuropil in control saline (including 100 nm TTX), 100 nm CCAP, and after wash. Traces are averages from 4 to 8 repeats. B, Mean ± SEM responses from six experiments. Responses were significantly different between control saline (including 100 nm TTX) and CCAP, and CCAP and wash, but not between control and wash (one-way ANOVA for repeated measures, p < 0.01; Holm–Sidak paired comparisons). *p < 0.05. ***p < 0.001. n.s., Not significant.
Maximal amplitude and concentration dependence of CCAP elicited IMI differ between LP and IC
Despite substantial variability of CbCCAPr mRNA expression within cell types, mean transcript levels between some of the cell types were significantly different (Fig. 4). We therefore wanted to determine whether differences in mRNA expression levels between cell types correlate with differences in physiological responses, specifically the magnitude of IMI evoked by CCAP application. Because the effect of a given amount of IMI activation on a neuron's activity depends on the background of other ionic conductances, we chose to do this comparison between LP and IC neurons with very different expression levels but otherwise similar excitability. LP expresses CbCCAPr mRNA at a significantly higher level than IC, but both are pyloric neurons that fire in rebound from pacemaker inhibition during the same phase in the pyloric rhythm.
Even with similar receptor affinities, receptor and ion channel activation are only indirectly linked, and differences in the signaling pathway between cell types could yield different quantitative relationships between both. We therefore did not just compare maximal current responses but also tested different concentrations of CCAP. IMI is usually measured as the difference current obtained from imposing voltage ramps in control saline and in the presence of a neuromodulator (Golowasch and Marder, 1992). Across different individuals, CCAP-evoked IMI can be quite variable, both with regard to maximal current amplitude and voltage dependence (Goaillard et al., 2009). We found it easier to maintain stable voltage-clamp recordings and monitor current for the >2 h needed to apply different CCAP concentrations when a constant voltage was maintained. However, holding neurons at a single voltage would have yielded inaccurate measurements of concentration dependence had the voltage dependence changed at different concentrations within each experiment. Therefore, we first performed voltage ramp measurements in LP at three different CCAP concentrations and compared the resulting IV curves. Figure 7A shows an example of the raw IV curves obtained in control saline and 1 μm CCAP. Figure 7B shows IV plots of IMI, calculated as the difference between the raw IV functions at the three different CCAP concentrations. In all three experiments, we found that only the maximal amplitude of IMI (Fig. 7B, dashed line) changed at different concentrations, but not the voltage dependence. Therefore, changes in current obtained at a single voltage at different concentrations should only reflect the amount of channel activation and not changes in gating properties.
Figure 7.
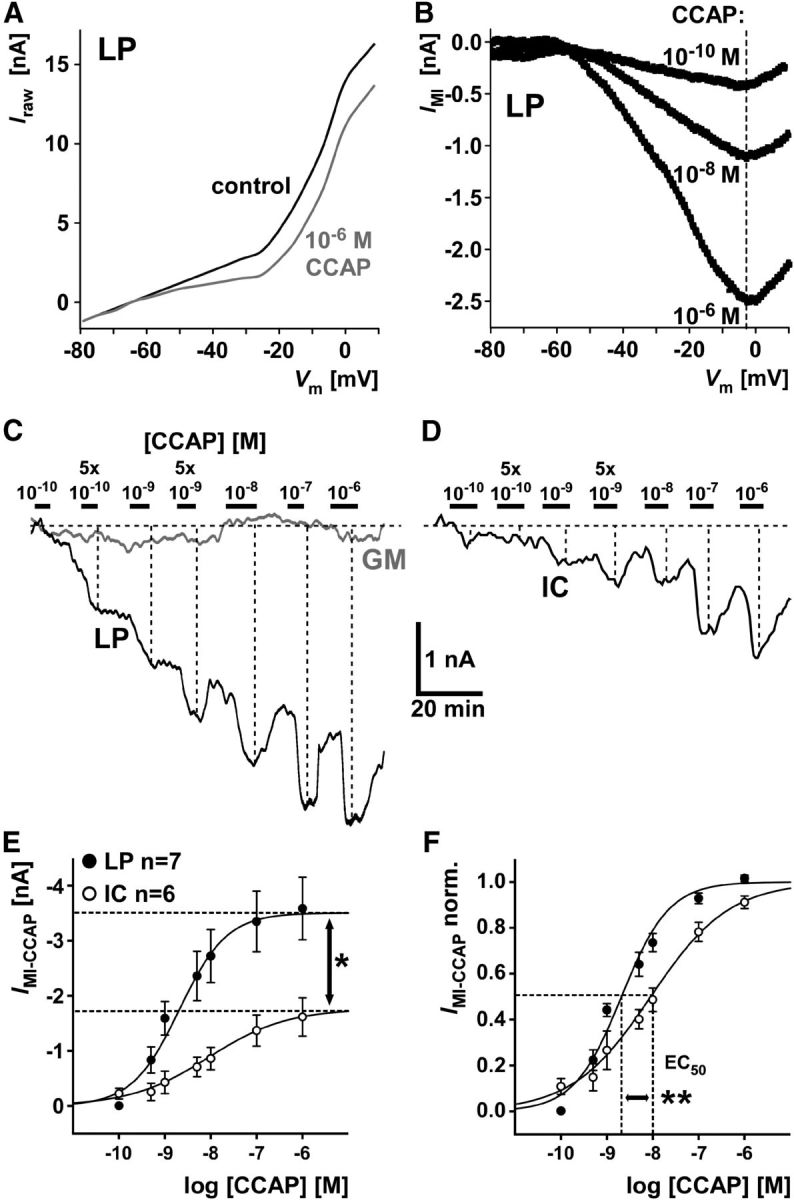
CCAP-elicited IMI in LP and IC differs in amplitude and concentration dependence. A, Raw currents as a function of voltage, measured from voltage ramps in control saline and CCAP in an LP neuron. B, IMI as a function of voltage, obtained as the difference between raw current–voltage relationships at three different CCAP concentrations. Dashed line indicates that the voltage at which the current peaked did not change with CCAP concentration. C, Filtered current traces from an LP and a GM neuron held at −20 mV, in response to application of CCAP at different concentrations. Dashed lines indicate that inward current responses increased with increasing CCAP concentration. D, Filtered current traces from an IC neuron, obtained in the same way as the traces shown in C. E, Mean current values at different CCAP concentration from measurements in LP and IC. Sigmoid fits were generated from parameters averaged across individual experiments. Maximum current values obtained from fits were significantly larger in LP than in IC (mean ± SEM: LP: 3.51 ± 0.55 nA; IC: 1.77 ± 0.37 nA; unpaired t test: *p < 0.05). F, Same data as in E, but normalized to the maximal current fit value in each experiment. Values for EC50 and the slope factor were significantly smaller in LP than in IC (mean ± SEM: EC50, LP: 2.07 ± 0.68 nm; IC: 9.02 ± 0.50 nm; unpaired t test: p < 0.01; slope factor, LP: 0.52 ± 0.04; IC: 0.86 ± 0.10; unpaired t test: p < 0.01). **p < 0.01.
To determine concentration dependence, we held the cells at −20 mV and measured the current amplitude evoked by different CCAP concentrations. Figure 7C shows filtered current traces simultaneously obtained from LP and GM at a holding potential of −20 mV and at different CCAP concentrations. GM is shown here as a control, as it does not express CbCCAPr. Figure 7D shows responses of IC from a different experiment. Most other voltage-gated currents were blocked (see Materials and Methods). Application of CCAP at increasing concentrations interspersed with incomplete washes yielded increasing inward current responses in LP and IC (dashed lines), but not in GM. Figure 7E shows mean IMI values in LP and IC as a function of CCAP concentration. Maximal current values were significantly larger in LP than in IC, matching the much higher CbCCAPr expression levels shown in Figure 4. Figure 7F shows the same data normalized to maximal current in each experiment. LP and IC IMI measurements also differed in concentration dependence, as both the EC50 and the slope factor in LP were significantly lower than in IC. A lower slope factor value means a steeper slope (see Materials and Methods). We therefore conclude that the higher expression level of CbCCAPr mRNA in LP is accompanied by stronger activation of IMI by CCAP, and a higher sensitivity to lower concentrations.
Saturating concentrations of CCAP activate IMI maximally in LP, but not IC
The difference in IMI responses between LP and IC at saturating CCAP concentrations could be solely due to different numbers of ion channels underlying IMI. In this case, both neuron types would express a sufficient number of CCAP receptors to activate all ion channels present. Saturation would be due to a limited number of ion channels, but LP would express more ion channels and therefore generate a larger current. Alternatively, saturation could occur because all receptors are activated at higher CCAP concentrations before all ion channels are activated. The larger response in LP would then be due to a larger percentage of ion channels opened when all receptors are activated. In this case, LP would show a larger response even if the number of ion channels underlying IMI was similar in both cell types. We wanted to distinguish between these two cases by using occlusion experiments. A number of different neuromodulators, neuropeptides in particular, can activate IMI in a given cell type. This convergence was established by showing that each neuromodulator activated a current with very similar properties and that the effects of each neuromodulator occlude each other (Swensen and Marder, 2000, 2001). In addition to CCAP, IMI is activated by the neuropeptide proctolin in both LP and IC (Swensen and Marder, 2001). In a subset of the experiments shown in Figure 7C–F, after measuring the current response in 1 μm CCAP, we added 1 μm proctolin to the bath solution. Figure 8A shows the concentration dependence of IMI, normalized to the fit maximum for CCAP alone in each experiment. Dashed lines indicate the fractional increase in IMI between 1 μm CCAP alone and 1 μm CCAP + 1 μm proctolin. Figure 8B shows the same data normalized to the response in 1 μm CCAP + 1 μm proctolin. The fraction of IMI activated by a saturating concentration of CCAP alone was significantly smaller in IC than in LP.
Figure 8.
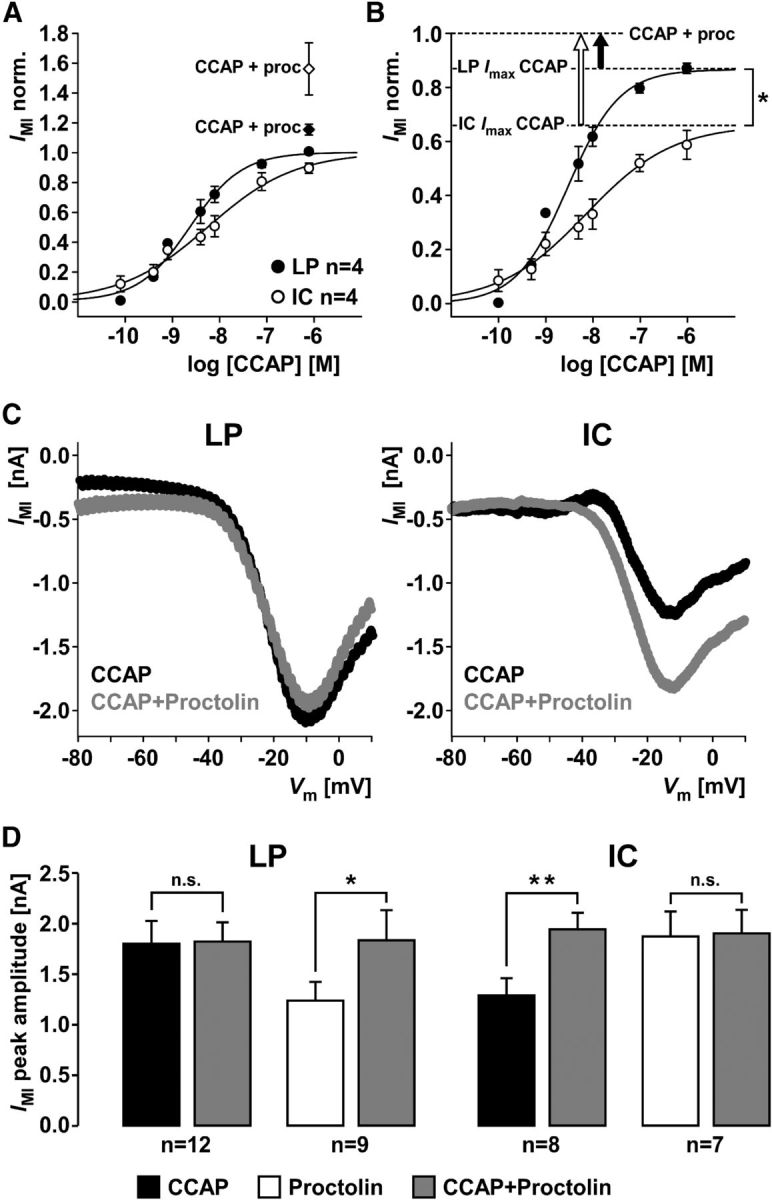
Differences in occlusion of IMI activation by CCAP and proctolin between LP and IC. A, Mean current values at different CCAP concentrations from measurements at −20 mV in LP and IC, normalized to the maximal current fit value in each experiment. After the concentration series of CCAP, a mix of 1 μm CCAP and 1 μm proctolin was added. Addition of proctolin yielded larger currents (diamonds) than saturating CCAP concentrations alone (circles). B, Same data from the CCAP concentration series as in A, but normalized to the responses to the mix of CCAP and proctolin. Addition of proctolin yielded a significantly larger increase in current in IC than in LP (unpaired t test, p < 0.05). C, Example IV curves generated from the difference of responses to voltage ramps in control saline, 1 μm CCAP, and 1 μm CCAP + 1 μm proctolin. Peak current amplitude did not change in LP but increased in IC. D, Mean peak current amplitudes across experiments, measured from IV curves as shown in C. Current amplitudes in LP did not change between CCAP alone and CCAP + proctolin (paired t test, p = 0.89). In contrast, current amplitudes in IC significantly increased between CCAP alone and CCAP + proctolin (paired t test, p < 0.01). When proctolin was applied alone first, adding CCAP significantly increased current amplitudes in LP (paired t test, p < 0.05), but not IC (paired t test, p = 0.81). *p < 0.05. **p < 0.01. n.s., Not significant.
An important caveat in interpreting these data is that, after the repeated CCAP applications in these experiments, nonmaximal IMI responses to saturating CCAP concentrations could be mostly due to receptor desensitization. The difference between LP and IC could thus reflect differences in desensitization rather than differences in the quantities of receptors or ion channels. We therefore performed separate occlusion experiments, with minimal numbers of repeated applications of only the saturating concentrations. In this case, we used voltage ramps and compared peak currents from IV curves. Figure 8C shows example IV curves from single experiments in which 1 μm CCAP was first applied alone, and then together with 1 μm proctolin. The addition of proctolin had little effect on the IV curve in LP but increased the peak current in IC. We also performed experiments in which proctolin was applied alone first, and then together with CCAP. Figure 8D shows bar plots of the mean peak current amplitudes measured in these experiments. Whereas proctolin responses after repeated CCAP applications at different concentrations still caused a small increase in IMI in LP (Fig. 8A,B), possibly due to receptor desensitization, the ramp measurements in only saturating concentrations showed complete occlusion of proctolin by CCAP. In contrast, IC responses increased significantly when proctolin was added. The reverse was true when proctolin was applied first, with only LP showing a significant increase in peak currents when CCAP was added.
We conclude that the complete occlusion seen in LP when applying CCAP first, and in IC when applying proctolin first, means that coapplication of CCAP and proctolin activates all available IMI in both cells. In LP, this means that the CCAP response saturates because all available target ion channels are activated and addition of a converging neuromodulator cannot increase current responses. In IC, the CCAP response saturates because all available CCAP receptors are occupied, but receptors for other neuromodulators can activate additional target ion channels. Saturating CCAP concentrations therefore only activate two-thirds (66%) of all available IMI in IC. We did not test whether 1 μm proctolin is a saturating concentration in LP, but the complete occlusion of CCAP responses by proctolin in IC suggests that proctolin activated all available ion channels. When response amplitudes from coapplication of both modulators were pooled, independent of the sequence of application, mean values for LP and IC did not differ (LP: 1.83 ± 0.16 nA SEM; IC: 1.93 ± 0.13 nA SEM; unpaired t test: p = 0.67). Therefore, the smaller CCAP responses in IC are consistent with a smaller number of CCAP receptors, which matches the significantly lower CbCCAPr mRNA expression shown in Figure 4.
Discussion
Circuit function is dependent on neuromodulation, a term used for a variety of types of neuronal communication beyond fast neurotransmission (Katz, 1999; Bucher and Marder, 2013), but most commonly for diffuse release of transmitters acting on GPCRs (Hille, 1992). Because GPCR signaling involves pathways shared between different receptors and with multiple intracellular targets, the patterns of activation across different neurons are complex (Harris-Warrick and Johnson, 2010; Bucher and Marder, 2013; Nadim and Bucher, 2014). Even when detailed quantitative information about circuit-wide neuromodulator targets has been gathered, for example, about dopamine effects on STG neurons (Harris-Warrick et al., 1998; Harris-Warrick and Johnson, 2010), the complexity arising from effects on many circuit components has so far precluded comprehensive quantitative models of circuit modulation. We studied quantitative aspects of neuropeptide neuromodulation because neuropeptide effects in the STG appear to be less divergent than amine effects. We show that both receptor expression levels and ion channel activation through these receptors can vary substantially across neuron types.
The identity and distribution of CbCCAPr
We identified a transcript with high similarity to arthropod CCAP receptors. It is a putative receptor because we did not deorphanize it (e.g., by heterologous protein expression and pharmacology) (Civelli et al., 2013). However, its identity is strongly supported by sequence similarity and expression pattern. Phylogenetic analysis showed a close relationship to arthropod CCAP and mammalian NPS receptors. Sequence comparison was consistent with previous studies of insect CCAP receptor homology, which placed them in subfamily A6 of Rhodopsin-like GPCRs (Joost and Methner, 2002), together with vasopressin and NPS receptors (Park et al., 2002; Li et al., 2011; Pitti and Manoj, 2012). Some insect species have two CCAP receptors, but this is likely due to gene duplication in insect lineages (Li et al., 2011), and we found no evidence of multiple receptor genes.
The transcript was expressed in all nerve tissues tested, consistent with the diverse behaviors under control of CCAP. Expression in the gm4 muscle is consistent with the finding that CCAP enhances gm4 contraction amplitude (Jorge-Rivera et al., 1998). The absence of expression in the heart muscle is surprising, as CCAP was originally named for its effect on heart rate (Stangier et al., 1987). However, crustacean hearts are neurogenic (Cooke, 2002), and CCAP has multiple targets in the regulation of heart contractions, including the cardiac ganglion (Cruz-Bermúdez and Marder, 2007; Fort et al., 2007). Our finding may mean that CCAP mainly acts presynaptically to the muscle in C. borealis.
Strong support for receptor identity comes from the expression pattern in the STG. The CbCCAPr transcript was differentially expressed across different neuron types and found in all neurons previously shown to respond to CCAP (Swensen and Marder, 2000, 2001; Kirby and Nusbaum, 2007; DeLong et al., 2009). In most cases, neurons lacking CCAP modulation of excitability did not express CbCCAPr. VD was the only cell type that clearly showed expression despite not displaying CCAP effects on excitability. We resolved this mismatch by showing a change in postsynaptic responses to exogenously applied transmitter.
CCAP modulation of the LG to VD synapse
Neuropeptides alter both excitability and synaptic function in many systems (Taghert and Nitabach, 2012; van den Pol, 2012). However, there are only two reports of neuropeptide effects on synapses in the STG. In the lobster, red pigment concentrating hormone strengthens the synapse from LP to the PD neurons (Thirumalai et al., 2006). In C. borealis, the same synapse is modulated by proctolin (Zhao et al., 2011). Our description of CCAP modulation at the LG to VD synapse is novel in two ways. First, it was previously unknown that a peptide can act on STG neurons without also activating IMI. Effects of red pigment concentrating hormone on excitability in H. americanus have not been studied, but in C. borealis, both LP and PD show IMI responses to proctolin (Swensen and Marder, 2001). The only other identified target of peptide modulation is a transient inward current activated by pyrokinin in LG (Rodriguez et al., 2013). This current is also activated in parallel to IMI. Our expression results confirm that neuropeptide receptors are not present in all neurons, but the selective effect on synaptic current in VD means that IMI activation cannot serve as a proxy for receptor expression. This has to be taken into account for future attempts at circuit-wide mapping of neuropeptide effects.
The second novel aspect is that CCAP modulation at the LG to VD synapse has both presynaptic and postsynaptic components, which has not been tested for other neuropeptides. It is particularly intriguing that presynaptic and postsynaptic effects had opposing signs. As the postsynaptic current responses were clearly reduced by CCAP, the overall strengthening of the synapse can only be explained by a dominant presynaptic enhancement. Presynaptic enhancement and postsynaptic reduction of responses at STG synapses have also been shown for dopamine (Cleland and Selverston, 1997; Johnson and Harris-Warrick, 1997), and Harris-Warrick and Johnson (2010) argue that functionally opposing modulatory mechanisms may stabilize the modulatory state of a network and prevent overmodulation.
Quantitative differences in expression levels and IMI responses and their implications for comodulation
We found stronger IMI responses and higher sensitivity to CCAP in LP compared with IC. Overall, low activation thresholds and EC50 values for IMI in both cells are consistent with the fact that CCAP acts exclusively as a neurohormone in the STG (Christie et al., 1995; Li et al., 2003; Chen et al., 2009) and affects circuit output at low concentrations (Weimann et al., 1997). Similar EC50 values have been found in binding assays for insect CCAP receptors (Lee et al., 2013). It is intriguing that, in addition to the stronger responsiveness of LP, we also found higher CbCCAPr transcript expression. There is no a priori reason to assume that IMI responses scale with mRNA expression level. The correlation between mRNA and protein abundance is often poor (Maier et al., 2009), and protein and mRNA expression levels are not necessarily at the same ratio in each cell type. In addition, different concentration dependence can be due to differences in receptor affinities (Baker and Hill, 2007). Furthermore, for a given number of receptors activated, there could be quantitative differences in second messenger signaling and activation of target ion channels (Hill, 2006). Stronger support for a correlation between transcript expression levels and IMI responses would come from testing whether they covary across all cell types in the STG, but this exceeded the scope of our study.
Even without the corroborating evidence of different transcript levels, the finding remains that both the magnitude and concentration dependence of IMI activation were different between the two cell types tested. In addition, occlusion experiments showed that saturating concentrations of CCAP activate the totally available IMI in LP, but only approximately two-thirds in IC. These findings have important implications for comodulation. Neuropeptides are often released in conjunction with classical transmitters or other peptides (Hökfelt et al., 2000; Merighi, 2002; Salio et al., 2006; van den Pol, 2012). In the STG, a multitude of neuromodulators are present as neurohormones or released into the neuropil from descending neurons containing multiple modulators (Nusbaum et al., 2001; Marder and Bucher, 2007). Figure 8B illustrates that, at a given concentration, the percentage of totally available IMI activated can be substantially different between the two cell types. Therefore, nonsaturating concentrations of different neuropeptides present at the same time can have quantitatively very different effects across cell types.
Variability of CbCCAPr transcript expression levels and IMI responses
In addition to varying CbCCAPr transcript expression across tissues and cell types, we found interindividual differences. Variability in expression in different tissues may be partially due to the fact that CCAP is an important regulator of molting behavior (Ewer and Truman, 1996; Phlippen et al., 2000; Park et al., 2003), and we used wild-caught animals that were likely at different stages in the molt cycle. Within STG cell types, mRNA copy numbers varied substantially. In the STG, transcript expression levels for K+ channel genes vary to a similar degree, well correlated with differences in the magnitude of K+ currents, a proxy for protein expression (Schulz et al., 2006). Therefore, there is no reason to assume that the variability in CbCCAPr transcripts was due to experimental error. Expression levels of different ion channels covary in a cell type-specific manner (Schulz et al., 2007), suggesting that cellular and synaptic parameters are coregulated to produce robust circuit output from varying underlying parameter values (Prinz et al., 2004; Marder and Goaillard, 2006; Golowasch, 2014; Marder et al., 2014). CCAP elicited IMI in LP varies substantially (Goaillard et al., 2009), but it was not clear whether this is due to ion channels or receptors. Concentration dependence was relatively consistent across individuals in our study, but variability in expression also suggests that responsiveness to neuromodulators is another free parameter in regulating consistent circuit function. This raises the question whether expression of neuromodulator receptors is coregulated with other parameters determining excitability, particularly IMI. However, currently, the molecular identity of the ion channels carrying IMI is unknown.
Footnotes
This work was supported by National Institute of Neurological Disorders and Stroke Grants NS058825 and NS083319 to D.B. We thank Drs. Jorge Golowasch and Farzan Nadim for helpful discussions; and Dr. Andrea Kohn for guidance with receptor cloning.
The authors declare no competing financial interests.
References
- Arakane Y, Li B, Muthukrishnan S, Beeman RW, Kramer KJ, Park Y. Functional analysis of four neuropeptides, EH, ETH, CCAP and bursicon, and their receptors in adult ecdysis behavior of the red flour beetle, Tribolium castaneum. Mech Dev. 2008;125:984–995. doi: 10.1016/j.mod.2008.09.002. [DOI] [PubMed] [Google Scholar]
- Baker JG, Hill SJ. Multiple GPCR conformations and signalling pathways: implications for antagonist affinity estimates. Trends Pharmacol Sci. 2007;28:374–381. doi: 10.1016/j.tips.2007.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bargmann CI. Beyond the connectome: how neuromodulators shape neural circuits. Bioessays. 2012;34:458–465. doi: 10.1002/bies.201100185. [DOI] [PubMed] [Google Scholar]
- Bargmann CI, Marder E. From the connectome to brain function. Nat Methods. 2013;10:483–490. doi: 10.1038/nmeth.2451. [DOI] [PubMed] [Google Scholar]
- Belmont M, Cazzamali G, Williamson M, Hauser F, Grimmelikhuijzen CJ. Identification of four evolutionarily related G protein-coupled receptors from the malaria mosquito Anopheles gambiae. Biochem Biophys Res Commun. 2006;344:160–165. doi: 10.1016/j.bbrc.2006.03.117. [DOI] [PubMed] [Google Scholar]
- Bockaert J, Pin JP. Molecular tinkering of G protein-coupled receptors: an evolutionary success. EMBO J. 1999;18:1723–1729. doi: 10.1093/emboj/18.7.1723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bucher D, Marder E. SnapShot: neuromodulation. Cell. 2013;155:482–482.e1. doi: 10.1016/j.cell.2013.09.047. [DOI] [PubMed] [Google Scholar]
- Bucher D, Taylor AL, Marder E. Central pattern generating neurons simultaneously express fast and slow rhythmic activities in the stomatogastric ganglion. J Neurophysiol. 2006;95:3617–3632. doi: 10.1152/jn.00004.2006. [DOI] [PubMed] [Google Scholar]
- Cazzamali G, Hauser F, Kobberup S, Williamson M, Grimmelikhuijzen CJ. Molecular identification of a Drosophila G protein-coupled receptor specific for crustacean cardioactive peptide. Biochem Biophys Res Commun. 2003;303:146–152. doi: 10.1016/S0006-291X(03)00302-4. [DOI] [PubMed] [Google Scholar]
- Chen R, Ma M, Hui L, Zhang J, Li L. Measurement of neuropeptides in crustacean hemolymph via MALDI mass spectrometry. J Am Soc Mass Spectrom. 2009;20:708–718. doi: 10.1016/j.jasms.2008.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christie AE, Skiebe P, Marder E. Matrix of neuromodulators in neurosecretory structures of the crab Cancer borealis. J Exp Biol. 1995;198:2431–2439. doi: 10.1242/jeb.198.12.2431. [DOI] [PubMed] [Google Scholar]
- Civelli O, Reinscheid RK, Zhang Y, Wang Z, Fredriksson R, Schiöth HB. G protein-coupled receptor deorphanizations. Annu Rev Pharmacol Toxicol. 2013;53:127–146. doi: 10.1146/annurev-pharmtox-010611-134548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark MC, Dever TE, Dever JJ, Xu P, Rehder V, Sosa MA, Baro DJ. Arthropod 5-HT2 receptors: a neurohormonal receptor in decapod crustaceans that displays agonist independent activity resulting from an evolutionary alteration to the DRY motif. J Neurosci. 2004;24:3421–3435. doi: 10.1523/JNEUROSCI.0062-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark MC, Khan R, Baro DJ. Crustacean dopamine receptors: localization and G protein coupling in the stomatogastric ganglion. J Neurochem. 2008;104:1006–1019. doi: 10.1111/j.1471-4159.2007.05029.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleland TA, Selverston AI. Dopaminergic modulation of inhibitory glutamate receptors in the lobster stomatogastric ganglion. J Neurophysiol. 1997;78:3450–3452. doi: 10.1152/jn.1997.78.6.3450. [DOI] [PubMed] [Google Scholar]
- Cooke IM. Reliable, responsive pacemaking and pattern generation with minimal cell numbers: the crustacean cardiac ganglion. Biol Bull. 2002;202:108–136. doi: 10.2307/1543649. [DOI] [PubMed] [Google Scholar]
- Cruz-Bermúdez ND, Marder E. Multiple modulators act on the cardiac ganglion of the crab, Cancer borealis. J Exp Biol. 2007;210:2873–2884. doi: 10.1242/jeb.002949. [DOI] [PubMed] [Google Scholar]
- DeLong ND, Kirby MS, Blitz DM, Nusbaum MP. Parallel regulation of a modulator-activated current via distinct dynamics underlies comodulation of motor circuit output. J Neurosci. 2009;29:12355–12367. doi: 10.1523/JNEUROSCI.3079-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donini A, Agricola H, Lange AB. Crustacean cardioactive peptide is a modulator of oviduct contractions in Locusta migratoria. J Insect Physiol. 2001;47:277–285. doi: 10.1016/S0022-1910(00)00112-8. [DOI] [PubMed] [Google Scholar]
- Donini A, Ngo C, Lange AB. Evidence for crustacean cardioactive peptide-like innervation of the gut in Locusta migratoria. Peptides. 2002;23:1915–1923. doi: 10.1016/S0196-9781(02)00174-2. [DOI] [PubMed] [Google Scholar]
- Estévez-Lao TY, Boyce DS, Honegger HW, Hillyer JF. Cardioacceleratory function of the neurohormone CCAP in the mosquito Anopheles gambiae. J Exp Biol. 2013;216:601–613. doi: 10.1242/jeb.077164. [DOI] [PubMed] [Google Scholar]
- Ewer J, Truman JW. Increases in cyclic 3′, 5′-guanosine monophosphate (cGMP) occur at ecdysis in an evolutionarily conserved crustacean cardioactive peptide-immunoreactive insect neuronal network. J Comp Neurol. 1996;370:330–341. doi: 10.1002/(SICI)1096-9861(19960701)370:3<330::AID-CNE4>3.3.CO%3B2-5. [DOI] [PubMed] [Google Scholar]
- Fort TJ, García-Crescioni K, Agricola HJ, Brezina V, Miller MW. Regulation of the crab heartbeat by crustacean cardioactive peptide (CCAP): central and peripheral actions. J Neurophysiol. 2007;97:3407–3420. doi: 10.1152/jn.00939.2006. [DOI] [PubMed] [Google Scholar]
- Goaillard JM, Taylor AL, Schulz DJ, Marder E. Functional consequences of animal-to-animal variation in circuit parameters. Nat Neurosci. 2009;12:1424–1430. doi: 10.1038/nn.2404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golowasch J. Ionic current variability and functional stability in the nervous system. BioScience. 2014;64:570–580. doi: 10.1093/biosci/biu070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golowasch J, Marder E. Proctolin activates an inward current whose voltage dependence is modified by extracellular Ca2+ J Neurosci. 1992;12:810–817. doi: 10.1523/JNEUROSCI.12-03-00810.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graubard K, Raper JA, Hartline DK. Graded synaptic transmission between spiking neurons. Proc Natl Acad Sci U S A. 1980;77:3733–3735. doi: 10.1073/pnas.77.6.3733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris-Warrick RM. Neuromodulation and flexibility in Central Pattern Generator networks. Curr Opin Neurobiol. 2011;21:685–692. doi: 10.1016/j.conb.2011.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris-Warrick RM, Johnson BR. Checks and balances in neuromodulation. Front Behav Neurosci. 2010;4:pii47. doi: 10.3389/fnbeh.2010.00047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris-Warrick RM, Johnson BR, Peck JH, Kloppenburg P, Ayali A, Skarbinski J. Distributed effects of dopamine modulation in the crustacean pyloric network. Ann N Y Acad Sci. 1998;860:155–167. doi: 10.1111/j.1749-6632.1998.tb09046.x. [DOI] [PubMed] [Google Scholar]
- Hill SJ. G-protein-coupled receptors: past, present and future. Br J Pharmacol. 2006;147(Suppl 1):S27–S37. doi: 10.1038/sj.bjp.0706455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hille B. G protein-coupled mechanisms and nervous signaling. Neuron. 1992;9:187–195. doi: 10.1016/0896-6273(92)90158-A. [DOI] [PubMed] [Google Scholar]
- Hofmann K, Stoffel W. TMbase: a database of membrane spanning proteins segments. Biol Chem Hoppe Seyler. 1993;374:166. [Google Scholar]
- Hökfelt T, Broberger C, Xu ZQ, Sergeyev V, Ubink R, Diez M. Neuropeptides: an overview. Neuropharmacology. 2000;39:1337–1356. doi: 10.1016/S0028-3908(00)00010-1. [DOI] [PubMed] [Google Scholar]
- Johnson BR, Harris-Warrick RM. Amine modulation of glutamate responses from pyloric motor neurons in lobster stomatogastric ganglion. J Neurophysiol. 1997;78:3210–3221. doi: 10.1152/jn.1997.78.6.3210. [DOI] [PubMed] [Google Scholar]
- Jones DT, Taylor WR, Thornton JM. The rapid generation of mutation data matrices from protein sequences. Comput Appl Biosci. 1992;8:275–282. doi: 10.1093/bioinformatics/8.3.275. [DOI] [PubMed] [Google Scholar]
- Joost P, Methner A. Phylogenetic analysis of 277 human G-protein-coupled receptors as a tool for the prediction of orphan receptor ligands. Genome Biol. 2002;3:RESEARCH0063. doi: 10.1186/gb-2002-3-11-research0063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan LM, Sławińska U. Modulation of rhythmic movement: control of coordination. Prog Brain Res. 2011;188:181–195. doi: 10.1016/B978-0-444-53825-3.00017-6. [DOI] [PubMed] [Google Scholar]
- Jorge-Rivera JC, Sen K, Birmingham JT, Abbott LF, Marder E. Temporal dynamics of convergent modulation at a crustacean neuromuscular junction. J Neurophysiol. 1998;80:2559–2570. doi: 10.1152/jn.1998.80.5.2559. [DOI] [PubMed] [Google Scholar]
- Katz PS, editor. Beyond neurotransmission: neuromodulation and its importance for information processing. Oxford: UP; 1999. [Google Scholar]
- Kilman VL, Marder E. Ultrastructure of the stomatogastric ganglion neuropil of the crab, Cancer borealis. J Comp Neurol. 1996;374:362–375. doi: 10.1002/(SICI)1096-9861(19961021)374:3<362::AID-CNE5>3.0.CO%3B2-%23. [DOI] [PubMed] [Google Scholar]
- Kirby MS, Nusbaum MP. Peptide hormone modulation of a neuronally modulated motor circuit. J Neurophysiol. 2007;98:3206–3220. doi: 10.1152/jn.00795.2006. [DOI] [PubMed] [Google Scholar]
- Lee D, Vanden Broeck J, Lange AB. Identification and expression of the CCAP receptor in the Chagas' disease vector, Rhodnius prolixus, and its involvement in cardiac control. PLoS One. 2013;8:e68897. doi: 10.1371/journal.pone.0068897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Beeman RW, Park Y. Functions of duplicated genes encoding CCAP receptors in the red flour beetle, Tribolium castaneum. J Insect Physiol. 2011;57:1190–1197. doi: 10.1016/j.jinsphys.2011.05.011. [DOI] [PubMed] [Google Scholar]
- Li L, Kelley WP, Billimoria CP, Christie AE, Pulver SR, Sweedler JV, Marder E. Mass spectrometric investigation of the neuropeptide complement and release in the pericardial organs of the crab, Cancer borealis. J Neurochem. 2003;87:642–656. doi: 10.1046/j.1471-4159.2003.02031.x. [DOI] [PubMed] [Google Scholar]
- Maier T, Güell M, Serrano L. Correlation of mRNA and protein in complex biological samples. FEBS Lett. 2009;583:3966–3973. doi: 10.1016/j.febslet.2009.10.036. [DOI] [PubMed] [Google Scholar]
- Mamiya A, Manor Y, Nadim F. Short-term dynamics of a mixed chemical and electrical synapse in a rhythmic network. J Neurosci. 2003;23:9557–9564. doi: 10.1523/JNEUROSCI.23-29-09557.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manor Y, Nadim F, Abbott LF, Marder E. Temporal dynamics of graded synaptic transmission in the lobster stomatogastric ganglion. J Neurosci. 1997;17:5610–5621. doi: 10.1523/JNEUROSCI.17-14-05610.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marder E. Neuromodulation of neuronal circuits: back to the future. Neuron. 2012;76:1–11. doi: 10.1016/j.neuron.2012.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marder E, Bucher D. Understanding circuit dynamics using the stomatogastric nervous system of lobsters and crabs. Annu Rev Physiol. 2007;69:291–316. doi: 10.1146/annurev.physiol.69.031905.161516. [DOI] [PubMed] [Google Scholar]
- Marder E, Goaillard JM. Variability, compensation and homeostasis in neuron and network function. Nat Rev Neurosci. 2006;7:563–574. doi: 10.1038/nrn1949. [DOI] [PubMed] [Google Scholar]
- Marder E, Bucher D, Schulz DJ, Taylor AL. Invertebrate central pattern generation moves along. Curr Biol. 2005;15:R685–R699. doi: 10.1016/j.cub.2005.08.022. [DOI] [PubMed] [Google Scholar]
- Marder E, Goeritz ML, Otopalik AG. Robust circuit rhythms in small circuits arise from variable circuit components and mechanisms. Curr Opin Neurobiol. 2014;31:156–163. doi: 10.1016/j.conb.2014.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merighi A. Costorage and coexistence of neuropeptides in the mammalian CNS. Prog Neurobiol. 2002;66:161–190. doi: 10.1016/S0301-0082(01)00031-4. [DOI] [PubMed] [Google Scholar]
- Nadim F, Bucher D. Neuromodulation of neurons and synapses. Curr Opin Neurobiol. 2014;29:48–56. doi: 10.1016/j.conb.2014.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadim F, Manor Y. The role of short-term synaptic dynamics in motor control. Curr Opin Neurobiol. 2000;10:683–690. doi: 10.1016/S0959-4388(00)00159-8. [DOI] [PubMed] [Google Scholar]
- Nusbaum MP, Beenhakker MP. A small-systems approach to motor pattern generation. Nature. 2002;417:343–350. doi: 10.1038/417343a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nusbaum MP, Blitz DM, Swensen AM, Wood D, Marder E. The roles of co-transmission in neural network modulation. Trends Neurosci. 2001;24:146–154. doi: 10.1016/S0166-2236(00)01723-9. [DOI] [PubMed] [Google Scholar]
- Park JH, Schroeder AJ, Helfrich-Förster C, Jackson FR, Ewer J. Targeted ablation of CCAP neuropeptide-containing neurons of Drosophila causes specific defects in execution and circadian timing of ecdysis behavior. Development. 2003;130:2645–2656. doi: 10.1242/dev.00503. [DOI] [PubMed] [Google Scholar]
- Park Y, Kim YJ, Adams ME. Identification of G protein-coupled receptors for Drosophila PRXamide peptides, CCAP, corazonin, and AKH supports a theory of ligand-receptor coevolution. Proc Natl Acad Sci U S A. 2002;99:11423–11428. doi: 10.1073/pnas.162276199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park Y, Aikins J, Wang LJ, Beeman RW, Oppert B, Lord JC, Brown SJ, Lorenzen MD, Richards S, Weinstock GM, Gibbs RA. Analysis of transcriptome data in the red flour beetle, Tribolium castaneum. Insect Biochem Mol Biol. 2008;38:380–386. doi: 10.1016/j.ibmb.2007.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phlippen MK, Webster SG, Chung JS, Dircksen H. Ecdysis of decapod crustaceans is associated with a dramatic release of crustacean cardioactive peptide into the haemolymph. J Exp Biol. 2000;203:521–536. doi: 10.1242/jeb.203.3.521. [DOI] [PubMed] [Google Scholar]
- Pitti T, Manoj N. Molecular evolution of the neuropeptide S receptor. PLoS One. 2012;7:e34046. doi: 10.1371/journal.pone.0034046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prinz AA, Bucher D, Marder E. Similar network activity from disparate circuit parameters. Nat Neurosci. 2004;7:1345–1352. doi: 10.1038/nn1352. [DOI] [PubMed] [Google Scholar]
- Prinz H. Hill coefficients, dose-response curves and allosteric mechanisms. J Chem Biol. 2010;3:37–44. doi: 10.1007/s12154-009-0029-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards KS, Marder E. The actions of crustacean cardioactive peptide on adult and developing stomatogastric ganglion motor patterns. J Neurobiol. 2000;44:31–44. doi: 10.1002/1097-4695(200007)44:1<31::AID-NEU4>3.0.CO%3B2-F. [DOI] [PubMed] [Google Scholar]
- Rodriguez JC, Blitz DM, Nusbaum MP. Convergent rhythm generation from divergent cellular mechanisms. J Neurosci. 2013;33:18047–18064. doi: 10.1523/JNEUROSCI.3217-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakai T, Satake H, Minakata H, Takeda M. Characterization of crustacean cardioactive peptide as a novel insect midgut factor: isolation, localization, and stimulation of alpha-amylase activity and gut contraction. Endocrinology. 2004;145:5671–5678. doi: 10.1210/en.2004-0722. [DOI] [PubMed] [Google Scholar]
- Sakai T, Satake H, Takeda M. Nutrient-induced alpha-amylase and protease activity is regulated by crustacean cardioactive peptide (CCAP) in the cockroach midgut. Peptides. 2006;27:2157–2164. doi: 10.1016/j.peptides.2006.04.009. [DOI] [PubMed] [Google Scholar]
- Salio C, Lossi L, Ferrini F, Merighi A. Neuropeptides as synaptic transmitters. Cell Tissue Res. 2006;326:583–598. doi: 10.1007/s00441-006-0268-3. [DOI] [PubMed] [Google Scholar]
- Schulz DJ, Goaillard JM, Marder E. Variable channel expression in identified single and electrically coupled neurons in different animals. Nat Neurosci. 2006;9:356–362. doi: 10.1038/nn1639. [DOI] [PubMed] [Google Scholar]
- Schulz DJ, Goaillard JM, Marder EE. Quantitative expression profiling of identified neurons reveals cell-specific constraints on highly variable levels of gene expression. Proc Natl Acad Sci U S A. 2007;104:13187–13191. doi: 10.1073/pnas.0705827104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stangier J, Hilbich C, Beyreuther K, Keller R. Unusual cardioactive peptide (CCAP) from pericardial organs of the shore crab Carcinus maenas. Proc Natl Acad Sci U S A. 1987;84:575–579. doi: 10.1073/pnas.84.2.575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein W. Modulation of stomatogastric rhythms. J Comp Physiol A Neuroethol Sens Neural Behav Physiol. 2009;195:989–1009. doi: 10.1007/s00359-009-0483-y. [DOI] [PubMed] [Google Scholar]
- Swensen AM, Marder E. Multiple peptides converge to activate the same voltage-dependent current in a central pattern-generating circuit. J Neurosci. 2000;20:6752–6759. doi: 10.1523/JNEUROSCI.20-18-06752.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swensen AM, Marder E. Modulators with convergent cellular actions elicit distinct circuit outputs. J Neurosci. 2001;21:4050–4058. doi: 10.1523/JNEUROSCI.21-11-04050.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taghert PH, Nitabach MN. Peptide neuromodulation in invertebrate model systems. Neuron. 2012;76:82–97. doi: 10.1016/j.neuron.2012.08.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 2013;30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thirumalai V, Prinz AA, Johnson CD, Marder E. Red pigment concentrating hormone strongly enhances the strength of the feedback to the pyloric rhythm oscillator but has little effect on pyloric rhythm period. J Neurophysiol. 2006;95:1762–1770. doi: 10.1152/jn.00764.2005. [DOI] [PubMed] [Google Scholar]
- Tublitz N. Insect cardioactive peptides: neurohormonal regulation of cardiac activity by two cardioacceleratory peptides during flight in the tobacco hawkmoth, Manduca sexta. J Exp Biol. 1989;142:31–48. doi: 10.1242/jeb.142.1.31. [DOI] [PubMed] [Google Scholar]
- van den Pol AN. Neuropeptide transmission in brain circuits. Neuron. 2012;76:98–115. doi: 10.1016/j.neuron.2012.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veelaert D, Passier P, Devreese B, Vanden Broeck J, Van Beeumen J, Vullings HG, Diederen JH, Schoofs L, De Loof A. Isolation and characterization of an adipokinetic hormone release-inducing factor in locusts: the crustacean cardioactive peptide. Endocrinology. 1997;138:138–142. doi: 10.1210/endo.138.1.4855. [DOI] [PubMed] [Google Scholar]
- Wasielewski O, Skonieczna M. Pleiotropic effects of the neuropeptides CCAP and myosuppressin in the beetle, Tenebrio molitor L. J Comp Physiol B. 2008;178:877–885. doi: 10.1007/s00360-008-0276-6. [DOI] [PubMed] [Google Scholar]
- Weimann JM, Meyrand P, Marder E. Neurons that form multiple pattern generators: identification and multiple activity patterns of gastric/pyloric neurons in the crab stomatogastric system. J Neurophysiol. 1991;65:111–122. doi: 10.1152/jn.1991.65.1.111. [DOI] [PubMed] [Google Scholar]
- Weimann JM, Skiebe P, Heinzel HG, Soto C, Kopell N, Jorge-Rivera JC, Marder E. Modulation of oscillator interactions in the crab stomatogastric ganglion by crustacean cardioactive peptide. J Neurosci. 1997;17:1748–1760. doi: 10.1523/JNEUROSCI.17-05-01748.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wess J. G-protein-coupled receptors: molecular mechanisms involved in receptor activation and selectivity of G-protein recognition. FASEB J. 1997;11:346–354. [PubMed] [Google Scholar]
- Zhao S, Sheibanie AF, Oh M, Rabbah P, Nadim F. Peptide neuromodulation of synaptic dynamics in an oscillatory network. J Neurosci. 2011;31:13991–14004. doi: 10.1523/JNEUROSCI.3624-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]