Abstract
Propionibacterium acnes is well-known as a human skin commensal but can also act as an invasive pathogen causing implant-associated infections. In order to resolve these types of P. acnes infections, the implants must be removed, due to the presence of an established biofilm that is recalcitrant to antibiotic therapy. In order to identify those P. acnes proteins produced in vivo during a biofilm infection, we established a rabbit model of implant-associated infection with this pathogen. P. acnes biofilms were anaerobically grown on dextran beads that were then inoculated into the left tibias of rabbits. At 4 weeks postinoculation, P. acnes infection was confirmed by radiograph, histology, culture, and PCR. In vivo-produced and immunogenic P. acnes proteins were detected on Western blot using serum samples from rabbits infected with P. acnes after these bacterial proteins were separated by two-dimensional gel electrophoresis. Those proteins that bound host antibodies were then isolated and identified by tandem mass spectrometry. Radiographs and histology demonstrated a disruption in the normal bone architecture and adherent biofilm communities in those animals with confirmed infections. A total of 24 immunogenic proteins were identified; 13 of these proteins were upregulated in both planktonic and biofilm modes, including an ABC transporter protein. We successfully adapted a rabbit model of implant-associated infection for P. acnes to identify P. acnes proteins produced during a chronic biofilm-mediated infection. Further studies are needed to evaluate the potential of these proteins for either a diagnostic test or a vaccine to prevent biofilm infections caused by P. acnes.
INTRODUCTION
Propionibacterium acnes is a Gram-positive facultative anaerobe that is a major colonizer and inhabitant of human skin (1) but that also causes invasive infections. Most P. acnes infections are chronic and include periodontitis, endodontic infections, endophthalmitis/keratitis, chronic rhinosinusitis, prostatitis, folliculitis associated with or without acne vulgaris, and implant-associated infections (2). With respect to implant-associated infections, P. acnes is frequently isolated from prosthetic shoulder joint infections, cerebrovascular device infections, fibrosis of breast implants, and infections of cardiovascular devices (2, 3). In all these diseases, biofilm formation is a major pathogenic strategy mediating the transition of P. acnes from a commensal to an opportunistic pathogen (2, 4, 5).
A biofilm is defined as a sessile community of microbial cells that (i) are attached to a substratum, interface, or each other; (ii) are embedded in a matrix of (at least partially self-produced) extracellular polymeric substances; and (iii) exhibit an altered phenotype with regard to growth, gene expression, and protein production compared to those in planktonic bacterial cells (6). The biofilm phenotype promotes the persistence of the infection, because microbes are embedded in the biofilm matrix and are recalcitrant to both antimicrobial treatment (7) and the host immune response (8). Compared to other bacterial species, there is a relative paucity of biofilm studies with P. acnes, since its pathogenic potential has only recently been recognized. Many putative virulence factors and strategies used against implant-associated infections caused by P. acnes are extrapolated from other pathogenic species, such as Staphylococcus aureus or Pseudomonas aeruginosa (2). A recent proteomic study analyzed the secretome of P. acnes during the planktonic mode of growth and identified several proteins that instigate the degradation of host tissue and inflammation (9). A comparison of the proteins produced in the biofilm mode of growth to those in the planktonic mode was beyond the scope of this study, but proteins produced in a biofilm mode are the critical phenotype in implant-associated infections. Biofilm bacteria presumably produce a different subset of proteins. Hence, we established a rabbit indwelling-device infection model caused by P. acnes to isolate serum for a biofilm immunoproteomic study of proteins that are produced in vivo. These proteins may be used in a vaccine formulation to prevent implant infections associated with P. acnes or as biomarkers for rapid diagnosis.
MATERIALS AND METHODS
Bacterial strains.
All in vitro biofilm experiments were performed using the P. acnes American Type Culture Collection (ATCC) strain ATCC 11827, since it has been used in multiple biofilm studies (10, 11). For the rabbit implant model, we used a pathogenic clinical strain, P. acnes RMA 13884, isolated from a bone biopsy sample from a patient with spinal osteomyelitis at the University of California at Los Angeles, Los Angeles, CA. Bacterial stock cultures were stored at −80°C in 20% glycerol. The strains were grown anaerobically at 37°C from stock cultures on brain heart infusion (BHI) agar plates (Difco) in a vinyl anaerobic gas chamber (Coy Laboratory Products, Grass Lake, MI).
Visualization of P. acnes ATCC 11827 and RMA 13884 in vitro biofilms on glass beads using scanning electron microscopy.
We anaerobically incubated the P. acnes strains ATCC 11827 and RMA 13884 with soda-lime solid-glass beads (Walter Stern, Inc., Port Washington, NY) for 2 days at 37°C under static conditions. After isolating the glass beads with the attached biofilm, we fixed the biofilm by incubating the beads in 100% ethanol for 10 min, washed them with distilled water (dH2O), and let the beads dry prior to performing scanning electron microscopy (SEM). SEM was performed using a Zeiss Supra 55VP field emission scanning electron microscope with a beam accelerating voltage of 1 kV and a working distance of 3 mm.
Multilocus sequence typing.
We characterized both P. acnes strains with the expanded multilocus sequence typing (MLST) (MLST8 genotyping) described in a study by McDowell et al. (12) (http://pubmlst.org/pacnes/) to identify six core housekeeping genes (aroE, atpD, guaA, lepA, sodA, and gmk) and two putative virulence genes (camp2 and tly). Gene fragments for the eight genes were amplified from purified P. acnes DNA, as previously described and sequenced (12–15).
Implant-associated infection model in rabbits.
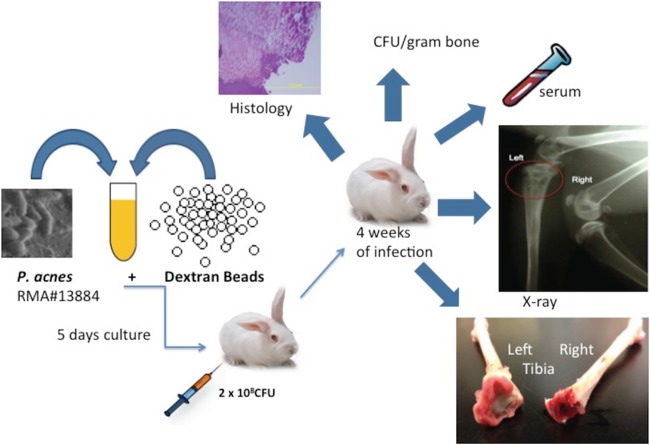
We used a rabbit tibial implant infection model that was adapted from Shirtliff et al. (16) and Mader and Shirtliff (17) for other bacterial species, including Streptococcus pyogenes (18), Staphylococcus epidermidis, and Acinetobacter baumannii (our unpublished data).
The inoculum was prepared by inoculating 10 ml of BHI broth (Difco) grown on BHI plates and incubating the culture for 5 days in the anaerobic gas chamber, subculturing into 10 ml of fresh BHI medium containing 10 mg/ml dextran beads (Cytodex microcarrier beads; Sigma), which represents the foreign implantable material, and incubating for an additional 5 days.
New Zealand White female rabbits (Charles Rivers Laboratory, Wilmington, MA), 8 weeks of age and weighing 1.5 to 2.0 kg, were anesthetized, and a bacterial infection was introduced according to a previously described method (18). In short, an 18-gauge needle was inserted into the intramedullary cavity of the left tibial metaphysis, and then 0.1 ml of a 5% (wt/vol) solution of morrhuate sodium (Eli Lilly, Indianapolis, IN), 0.1 ml of the P. acnes mixture (inoculum described above), and 0.2 ml of sterile saline were sequentially injected. The animals were returned to their cage, and serum samples were collected prior to infection and 28 days postinfection.
After 4 weeks, radiographs were obtained for both tibias, and the animals were sacrificed. Both the infected left and uninfected right tibias were removed, dissected so as to be free of all soft tissue, and processed for bacterial culture and histology. The right uninfected tibia was the negative control for all diagnostic methods. A small representative bone sample of the infected tibia was fixed in 4% paraformaldehyde for further imaging. The remaining bone fragments were suspended in 3 ml of sterile phosphate-buffered saline (PBS) per g of tissue and vortexed for 5 min. All samples were serially diluted in PBS, plated on BHI agar, and incubated in the anaerobic chamber at 37°C for 10 days for bacterial count determination. All samples were also plated on tryptic soy agar (Fluka Analytical) and incubated aerobically to exclude contamination. The limit of detection with this culture method was 10 CFU/g of bone. In clinical cases of P. acnes infection, cultures can often be negative even when histology shows inflammation and PCR results are clearly positive for P. acnes. This negative culture can be the result of inadequate sonication to liberate and culture the biofilm bacteria or escape detection through intracellular persistence at upwards of 8 months (19–21). Therefore, P. acnes-specific 16S rRNA gene PCR was performed on all samples as a secondary method to detect P. acnes in the tibial samples. In short, bacterial genomic DNA was extracted by boiling 1 ml of the tibial bone suspension (including bone marrow) for 10 min following several washes with PBS. The suspension was centrifuged at 8,000 rpm for 10 min. The P. acnes-specific 16S rRNA gene was amplified by using the specific primers PA-F (5′-GGGTTGTAAACCGCTTTCGCCT-3′) and PA-R (5′-GGCACACCCATCTCTGAGCAC-3′), and the conditions were as previously published (22). All PCR products were analyzed by electrophoresis.
Imaging of tibias.
The tibia fragments were formalin fixed, decalcified in EDTA, paraffin embedded, and sectioned. The tissue sections were stained with hematoxylin and eosin (H&E) to identify the infiltrating immune cells. An acute infection was defined by the predominance of polymorphonuclear neutrophils in the infiltrate, and chronic osteomyelitis was defined by the predominance of plasma cells, lymphocytes, and macrophages in the infiltrate, dead bone (necrosis), or bone sequester, and fibrosis of the remaining tissue (23). We classified an infection as subacute if the criteria for both the acute and chronic osteomyelitis stages were fulfilled. Gram staining (modified Brown-Brenn staining) was performed to visualize the bacteria. Based on the Gram stain findings, bacterial morphology was examined using the propidium iodide reagent from the LIVE/DEAD BacLight viability kit (Invitrogen, Carlsbad, CA) on adjacent tissue sections, according to the manufacturer's protocol. Since fixation in formalin kills most bacteria, we used only the BacLight stain that is able to penetrate dead cells (propidium iodide) to visualize the cells within the biofilm. The slides were examined with a Zeiss LSM 510 confocal laser scanning microscope (Carl Zeiss, Thornwood, NY).
Ethics statement.
All animal procedures were approved and performed under the supervision of the Institutional Care and Animal Use Committee at the University of Maryland, Baltimore, MD.
Growth conditions for in vitro planktonic and biofilm samples.
P. acnes strain ATCC 11827 was plated from the frozen stock onto BHI agar plates (Difco) and grown at 37°C under anaerobic conditions for 3 to 5 days. In order to prevent aberrant clonal isolate results that may be obtained by utilizing a single CFU, starter cultures were prepared by inoculating 10 ml of fresh BHI broth (Difco) with 8 to 10 single colonies of P. acnes (strain ATCC 11827) and incubated for 3 days at 37°C under anaerobic conditions.
Planktonic cultures were prepared by diluting the starter culture 1:100 in 50 ml of BHI and incubating for 3 days at 37°C under anaerobic conditions. On day 3, the planktonic cultures were centrifuged and resuspended in 500 μl of protein preservation solution (PBS supplemented with 0.02% sodium azide and cOmplete EDTA-free protease inhibitor tablets [Roche, Basel, Switzerland]). The bacterial suspensions were stored at −80°C until protein extraction.
In contrast, P. acnes biofilm samples were generated using 0.8-mm soda-lime solid-glass beads (Walter Stern, Inc., NY) as an abiotic surface for biofilm growth. The biofilm cultures were generated by diluting the starter culture 1:100 into 100 ml of fresh BHI containing sterile glass beads and incubating the cultures at 37°C under anaerobic conditions. The biofilm samples were harvested at various time points postinoculation for early (day 3), maturing (day 5), and fully mature (day 21) biofilms, in which fully mature biofilms showed no further increase in depth. To isolate the biofilm sample, the bacterial culture was drained out, and the glass beads were washed once with BHI broth. The glass beads were immersed in 30 ml of protein preservation solution and homogenized at 25,000 rpm for 4 min with a Polytron PT1200 homogenizer (Kinematica, Lucerne, Switzerland) to remove the biofilm from the surface of the beads. Similar to the planktonic cultures, the biofilm samples were concentrated in 500 μl of protein preservation solution and stored at −80°C. All samplings were performed in triplicate.
Preparation of membrane- and cell wall-associated protein extracts.
After thawing the frozen bacterial sample(s), 0.1-mm silica beads (BioSpec, Bartlesville, OK) were added to the cell suspension(s), and the cells were mechanically disrupted using a FastPrep instrument (Qbiogene, Irvine, CA) at 4 m/s for 45 s and then incubated on ice for 1 min. This cycle was repeated four more times. The bacterial lysates were centrifuged at 2,000 × g for 10 min at 4°C to remove large cell debris and beads. The supernatant was then separated into pellet (representing cell wall- and membrane-associated proteins) and cytosol (supernatant) fractions by centrifuging the sample at 15,000 × g and 4°C for 30 min (24). The pelleted fraction with the cell wall proteins from the planktonic or biofilm sample was resuspended, and the protein concentration was determined (25) using the Advanced protein assay reagent (Cytoskeleton, Inc., Denver, CO). Fifty-microgram aliquots of each protein sample were cleaned up and concentrated by tricarboxylic acid (TCA) precipitation. Subsequently, the 50-μg protein samples were used for the two-dimensional electrophoresis described below. The remainder of the supernatant was frozen at −80°C for future use.
Two-dimensional gel electrophoresis to visualize protein expression in the planktonic and biofilm phase and immunogenic protein identification.
Cell wall- and membrane-associated proteins were separated by their intrinsic charge and size using two-dimensional gel electrophoresis (2DGE), completed according to the principles of O'Farrell (26) and Brady et al. (27). After electrophoresis, the gels were fixed and stained with Sypro Ruby stain. Immunoproteomic studies/Western blot analysis to detect immunogens produced in the planktonic and biofilm modes of growth were conducted, as described previously, using serial serum samples from infected rabbits (27). The blots from three biofilm and planktonic protein samples were also performed with naive rabbit serum isolated prior to challenge to exclude the possibility that the rabbit already had circulating antibodies against P. acnes or antibodies that were cross-reactive with P. acnes proteins.
The blots were performed in triplicate for the planktonic samples isolated at one time point and the biofilm samples at different maturation stages and compared. Immunoreactive spots detected on two out of three Western blots were selected and aligned with the proteins on the corresponding 2DGE gel. These spots were excised and subjected to matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF MS), as described previously (27).
RESULTS
Strain characterization.
P. acnes ATCC 11827 used for the in vitro planktonic and biofilm sampling was characterized with the expanded MLST (13, 14, 28) as belonging to sequence type 103 (ST103) and clonal complex 1 (CC1) (lineage type IA1), and the clinical strain RMA 13884 used for the in vivo P. acnes infections was characterized as ST105 (IA1). This ST was previously found to link CC1 and CC4 into a much larger CC, with ST1 as the founder genotype and ST4 as the subfounder. We utilized both strains in this study, since antigen discovery using the serum infected by one strain of P. acnes being used to probe antigens produced by another well-characterized strain would strengthen our results of finding common immunogenic P. acnes proteins.
Since a recent study indicated that hemolytic P. acnes strains are more virulent than are nonhemolytic strains (29), we examined the hemolytic phenotypes of both our strains using Columbia agar with 5% sheep blood, and both strains were found to possess in vitro hemolytic activity. In addition, the two P. acnes strains demonstrated no relevant differences in protein expression in an investigation of the banding patterns at identical time points for multiple phases of growth (data not shown).
Biofilm formation confirmed by scanning electron microscopy.
SEM studies confirmed biofilm formation in both P. acnes strains (ATCC 11827 and RMA 13884) incubated with soda-lime solid-glass beads for 2 days at 37°C under static conditions. Figure 1 shows a young P. acnes (ATCC 11827 strain) biofilm with embedded cells within an exopolymeric matrix that appears similar to the glycocalyx of the biofilms of staphylococci.
FIG 1.
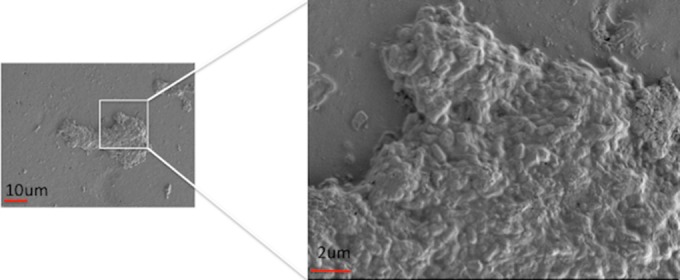
Scanning electron micrograph (SEM) of the P. acnes strain ATCC 11827 biofilm on solid glass beads (Walter Stern, Inc., Port Washington, NY) incubated for 2 days anaerobically at 37°C under static conditions (magnification, 2,000× [left panel] and 20,000× [right panel]), using a Zeiss Supra 55VP field emission scanning electron microscope with a beam accelerating voltage of 1 kV and a working distance of 3 mm.
Implant-associated infection model in rabbits.
We established a novel implant-associated infection model in rabbits to evaluate a P. acnes-mediated chronic infection and protein expression in vivo. We used rabbits because of their relative reproducibility and ease of diagnosis through presacrifice radiographs. In addition, this model has similar clinical and physiological properties of infection to those in humans and provides for ample serum samples to be obtained for subsequent Western blot (17). Based on previous literature with this rabbit model (17, 18), we chose 2 different infection doses. The rabbits infected with 6 × 106 CFU had slight macroscopic and radiological changes, but the tibial samples tested negative upon anaerobic culture and 16S rRNA PCR for P. acnes. Therefore, we surmised that this lower dose could be cleared by the host immune response in this model. In comparison, the macroscopic evaluation of the left tibia from rabbits infected with 2 × 108 CFU showed inflammation, periosteal elevation, and bone thickening (Fig. 2 and Table 1). Subsequent evaluation of the tibial samples from those rabbits confirmed P. acnes infection by bacterial culture and/or PCR testing. Histological sections of these infected rabbits showed either subacute or chronic osteomyelitis (histological classification adapted from that of Gaida et al. [23], defined in Materials and Methods), confirming the successful establishment of a chronic infection (Fig. 3). We further confirmed the presence of P. acnes and rod-shaped bacteria in histological sections with Gram stain and propidium iodide staining, respectively (Fig. 4).
FIG 2.
Schematic flow chart of the implant-associated osteomyelitis model in rabbits. After a 5-day incubation of the P. acnes strain RMA 13884 with dextran beads, rabbits were infected with a P. acnes inoculum in the left tibia. After 4 weeks of infection, blood serum was isolated, and X rays of both tibias were performed. The tibias were macroscopically and histologically examined, and the bacterial loads were quantified and reported as CFU per g of bone.
TABLE 1.
Characteristics of infected rabbits with P. acnes with two different bacterial inocula
| Measurement | Characteristics for rabbit: |
|||
|---|---|---|---|---|
| 1 | 2 | 3 | 4 | |
| Infective dose (total CFU) | 6 × 106 | 6 × 106 | 2 × 108 | 2 × 108 |
| Macroscopica | 1+ | 1+ | 2+ | 2+ |
| X rayb | 1+ | 1+ | 3+ | 3+ |
| CFU/g of bone | No growth | No growth | 4 × 107 | No growth |
| PCR specific for P. acnes | Negative | Negative | Positive | Positive |
| Histologyc | Acute | Normal | Subacute | Chronic |
Clinical criteria per eye: 0, normal; 1+, no bone involvement, soft tissue swelling at proximal tibial metaphysis; 2+, soft tissue abscess, <15% widening of proximal tibial metaphysis; 3+, >15% widening of proximal metaphysis.
Radiographic severity score: 0, normal; 1+, lytic changes around the needle stick, <5% disruption of the normal bone architecture; 2+, 5 to 15% disruption of normal bone architecture; 3+, 15 to 40% disruption of normal bone architecture; 4+, >40% disruption of normal bone architecture.
Acute osteomyelitis was defined by a predominance of polymorphonuclear neutrophils in the infiltrate; chronic osteomyelitis was defined as an infection with predominance of plasma cells, lymphocytes, and macrophages, dead bone (necrosis) or bone sequester, and fibrosis of the remaining tissue (23). We classified an infection as subacute if the criteria from acute and chronic were fulfilled.
FIG 3.
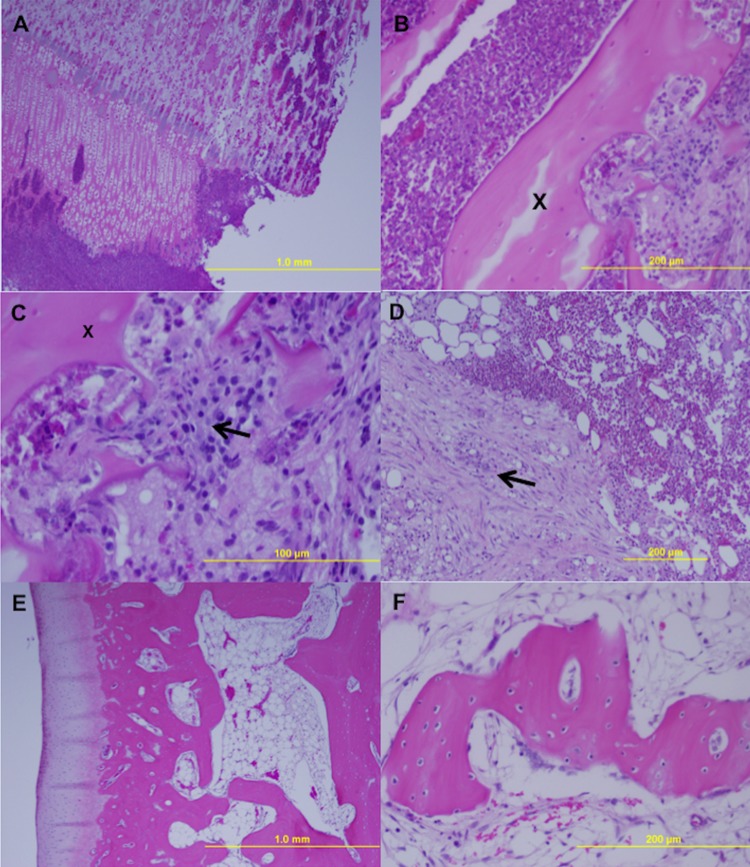
H&E-stained histologic sections from infected rabbit 3 (A to C) and rabbit 4 (D) showing subacute (rabbit 3) and more chronic (rabbit 4) osteomyelitis after challenge with P. acnes 2 × 108 CFU (classification adapted from that of Gaida et al. [23]). (A) Overview of inflammation of bone (cortex and medulla) and cartilage, with migration into the knee joint (4× magnification). (B and C) Inflammation with granulocytes with bone sequester (X) and osteoclasts (arrow) (magnification, 20× [B] and 40× [C]). (D) Chronic inflammation with fibrosis (arrow) of the medulla (10× magnification). (E and F) Noninfected tibia with an overview of the cartilage, bone cortex, and medulla as a control (magnification, 4× [E] and 40× [F]).
FIG 4.
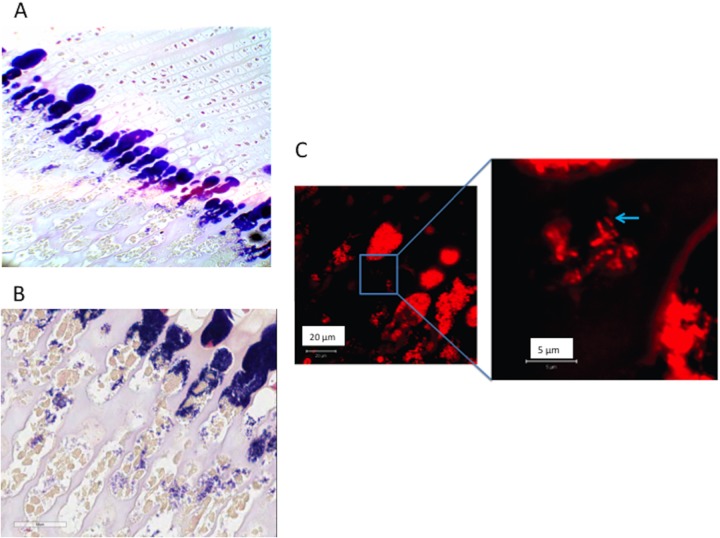
Formalin-fixed histology sections of osteomyelitis in rabbit 3 with positive Gram staining (blue) at the transition cartilage to bone (magnification, 20× [A] and 40× [B]) and confirmation of the rod shape of P. acnes (blue arrow) visualized with propidium iodide (LIVE/DEAD BacLight system) using confocal microscopy (C).
Identification of immunogenic proteins in P. acnes biofilm and/or planktonic growth phases.
We separated P. acnes proteins into cell wall/membrane and cytosol protein fractions. In order to find those proteins that would serve as a diagnostic or potential vaccine candidate, we targeted the cell wall/membrane protein fraction for further analysis with the Western blot technique using the immune serum from a culture-confirmed tibial infection in a rabbit (four weeks postchallenge). Immunogenic protein spots appearing on at least two blots were excised and identified by MALDI-TOF tandem mass spectrometry (MS/MS). We found 24 proteins that were immunogenic in either planktonic, biofilm, or both modes of growth. Table 2 lists the protein identities and provides the molecular mass, isoelectric point, protein function, and expression pattern (biofilm, planktonic, or both growth modes) of each immunogen. Representative sets of both the 2DGE gel and Western blot for a late-stage biofilm (21 days) and a planktonic sample (3 days) are shown in Fig. 5.
TABLE 2.
Identities and characteristics of immunogenic antigens of P. acnes, listed by expression profile and molecular mass
| Protein no. | Full name of proteina | Protein nameb | Best protein accession no.a | Functiona | pIc | Molecular mass (kDa) | Expression (biofilm, planktonic, or both) |
|---|---|---|---|---|---|---|---|
| 1 | Methylmalonyl-CoA mutase | MutA | E4DBY8 | Lactate fermentation to propionate and acetate | 4.698 | 69.111 | Both |
| 2 | Chaperone protein DnaK | DnaK | W4TZS5 | Heat shock protein, ATP binding | 4.476 | 59.03 | Both |
| 3 | 60-kDa chaperonin 1 | GroL1 | P0CY97 | Heat shock protein, protein misfolding, ATP binding | 4.613 | 56.840 | Both |
| 4 | Pyruvate kinase | PykA | G5EZ02 | Glycolysis | 5.01 | 51.091 | Both |
| 5 | Phosphoglycerate kinase | Pgk1 | Q6A9J3 | Carbohydrate degradation, glycolysis | 4.752 | 42.218 | Both |
| 6 | DivIVA domain protein | DivIVA | E4D8W2 | Cell division | 5.016 | 40,124 | Both |
| 7 | Glyceraldehyde-3-phosphate dehydrogenase | GAPDH | G7U8Y4 | Glucose metabolic process | 5.485 | 35.95 | Both |
| 8 | Malate dehydrogenase | Mdh | Q6A6Z5 | TCA cycle | 4.831 | 34.686 | Both |
| 9 | Carbamate kinase | ArcC | D4HCA4 | Arginine metabolism | 4.641 | 31.977 | Both |
| 10 | Translation elongation factor | Tef | W4TZY5 | Protein biosynthesis | 4.599 | 29,648 | Both |
| 11 | ABC transporter, ATP-binding protein | Abc1 | D4HAH2 | ATB binding | 6.18 | 26.002 | Both |
| 12 | 2,3-Bisphosphoglycerate-dependent phosphoglycerate mutase | BgmA | Q6AAU8 | Glycolysis | 5.981 | 27.991 | Both |
| 13 | Methylmalonyl-CoA epimerase | Mce | D4HDR2 | Amino acid metabolism, isomerase | 5.593 | 16.84 | Both |
| 14 | Translation elongation factor G, partial | FusA | F9NNW3 | Protein biosynthesis | 4.789 | 76.605 | Biofilm |
| 15 | Succinate dehydrogenase or fumarate reductase | SdhA | E4E2R8 | Oxidoreductase in carbohydrate metabolism, TCA cycle | 5.898 | 75.667 | Biofilm |
| 16 | Oxaloacetate decarboxylase | OxdC | G7U7Q6 | Glycolysis, oxaloacetate metabolism | 5.23 | 54,690 | Biofilm |
| 17 | Putative tryptophan 2,3-dioxygenase | Ptd1 | E4D811 | Aromatic amino acid family metabolic process | 5.011 | 50.782 | Biofilm |
| 18 | Fumarate hydratase class II | FumC | G7U975 | TCA cycle, fumarate metabolic process | 5.512 | 50.689 | Biofilm |
| 19 | Elongation factor | Tuf | Q6A6L7 | Protein biosynthesis | 5.214 | 44.14 | Biofilm |
| 20 | RecA protein | RecA | Q5IV56 | DNA repair | 5.258 | 37.153 | Biofilm |
| 21 | Fructose-biphosphate aldolase class II | FbaA | E4D532 | TCA cycle, fumarate metabolic process | 5.037 | 36,771 | Biofilm |
| 22 | Succinyl-CoA ligase | SucD | E4D6H5 | Ligase (e.g., ATP binding) | 5.708 | 30.566 | Biofilm |
| 23 | Enolase | Eno | Q6AAB8 | Glycolysis (cell attachment) | 4.418 | 45.53 | Planktonic |
| 24 | Pyridoxal 5′-phosphate lyase | PdxS | G8V9H3 | Pyridoxal phosphate biosynthetic process (vitamin B6) | 4.788 | 31.530 | Planktonic |
Identification and function of the proteins were obtained from the UniProt database of P. acnes complete proteome (ID 1.81) (www.uniprot.org). CoA, coenzyme A; TCA cycle, tricarboxylic acid cycle.
The protein names were used for labeling the protein spots in Fig. 5.
Average values derived from isoelectric point calculator (http://isoelectric.ovh.org).
FIG 5.
Western blot (A and C) and corresponding 2DGE gels (B and D) with identification of immunogenic proteins of P. acnes in a biofilm of 21 days (A and B) and in a planktonic phase of 3 days (C and D). For the Western blot, rabbit serum after 4 weeks of infection was used to identify immunogenic proteins. pH levels are indicated at the bottom and the molecular weights (MW) of the proteins (in thousands) in the middle of the panels. These protein spots were visually identified on Sypro-stained gels and excised for MALDI-TOF MS analysis. The abbreviated protein names are explained in Table 2. Note that some spots listed in Table 2 are not present in these figures, as not all spots were visible in every sample at every time point. UP, uncharacterized protein.
DISCUSSION
P. acnes has not historically been associated with indwelling medical device infections, since cultures would often require incubation periods of up to 13 days (30). In addition, the presence of P. acnes in a biopsy specimen has often been interpreted as a contaminant in many clinical laboratories. However, improved preparatory procedures, including the sonication of explanted implants and tissue biopsy specimens for bacterial culture, as well as molecular methods, such as specific PCR, have increased the diagnosis rates for P. acnes (2, 31). Even with these advances, diagnosis during early onset of the infection is still difficult, because patient complaints of an infected shoulder arthroplasty are limited to pain and stiffness only (32), thereby allowing the infection to develop into a mature biofilm infection. These types of infections are recalcitrant to antibiotics alone and require extensive surgical debridement combined with antibiotic therapy for infection resolution. In the present study, we successfully developed a rabbit model for chronic P. acnes indwelling medical device infections and generated convalescent-phase serum for further immunoproteomic studies. In the subsequent immunoproteomic analyses of planktonic- and biofilm-derived samples, we identified a total of 23 immunogenic cell wall proteins and found that 13 proteins were upregulated in both the planktonic and biofilm modes of growth.
Current animal models for P. acnes bone infections associated with an implant are limited to a hematogenous infection model of total knee arthroplasties (33), an implant-associated osteomyelitis model in rabbits (34), and a bone implant model in BALB/c mice (35). These studies examined the capability of P. acnes to establish chronic infections but did not evaluate the in vivo protein expression of P. acnes and/or the host humoral response. We chose to develop a comparatively easy and representative model of musculoskeletal infection and used this model to examine the protein expression of P. acnes during a chronic infection. Our model was a rabbit osteomyelitis model established for S. aureus by Mader and Shirtliff (17) and was successfully adapted for A. baumannii (our unpublished data) and S. pyogenes (18) by using an intramedullary injection of P. acnes-infected dextran beads to provide an abiotic surface for colonization and biofilm growth.
While other bacterial species (e.g., S. aureus [17]) require a low bacterial inoculum, 108 CFU was needed for P. acnes to establish a chronic infection, most likely due to the reduced virulence associated with this bacterial species and the resulting indolent infection that it produces. This observation is also similar to the findings of the two nonhematogenous studies that required a high inoculum of 107 to 108 to achieve chronic infection (34, 35).
The clinical strain RMA 13884 in this study was chosen since (i) it was relevant and recently isolated from a clinical case of spinal osteomyelitis with hardware, and (ii) it matched the 1A lineage with the P. acnes laboratory strain from the ATCC that has been used in multiple in vitro studies (10, 11). Although some P. acnes strains are more frequently associated with implant-associated infections (e.g., 1B isolates) (12, 36, 37), the 1A lineage types used in this study are also capable of causing these chronic infections, as shown by the isolation of a 1A lineage strain from an infected patient and its ability to induce a chronic infection in this study. Future studies will include a comparison of the infectious doses between the arrays of lineages but that were beyond the scope of the present study.
Most of the immunogenic proteins identified in this study (Table 2) are known to have a metabolic function in bacteria. This phenomenon of proteins with metabolic functions having antigenic potential has been observed in other immunoproteomic studies with P. acnes (9, 38) and other Gram-positive pathogenic strains, such as S. aureus (27) and S. pyogenes (18). We focused our immunoproteomic analysis on cell wall- and membrane-associated proteins of P. acnes, since previous studies have shown that these proteins may be able to be used as novel diagnostic biomarkers or vaccine candidates (39). Our study might exclude secreted and/or cytosolic proteins (9) that are also important for biofilm pathogenesis and for the development of vaccine candidates. One protein of particular interest to our laboratory was an ABC transporter protein (accession no. D4HAH2, Table 2). We previously identified an ABC transporter in S. aureus that was highly immunogenic in infected rabbits (27, 39). Furthermore, patients with wound infections, bacteremia and sepsis, pneumonia, arthritis, urinary tract infections, catheter-related bloodstream infections, and peritonitis due to S. aureus have a similar response, with high IgG levels against the ABC transporter (40, 41).
The secretome for planktonic P. acnes was already published by Holland et al. (9). This study found that secreted proteins, such as glycoside hydrolases, esterases, and proteases, were mainly involved in degradation, but it also identified unique proteins, such as Christie-Atkins-Munch-Petersen (CAMP) factors, glyceraldehyde-3-phosphate dehydrogenase (GAPDH), and hypothetical proteins. Our immunoproteomic study identified GAPDH as an immunogenic protein as well, which suggests that GAPDH is associated with the cell wall, in addition to being secreted into the extracellular milieu. This observation confirms the findings in studies of other Gram-positive bacteria, such as streptococci (18, 42); in these, GAPDH is described as a multifunctional protein, which is anchorless, found on the surface of many Gram-positive pathogens, and it contributes to adhesion and virulence. Based on its ability to perform dual or multiple mechanistically distinct functions, it belongs to a group of moonlighting proteins (43). We found other moonlighting proteins in the cell wall fraction, such as elongation factors, 10-kDa chaperonin, and enzymes involved in glycolysis, pyruvate metabolism, and the tricarboxylic acid cycle (Table 2).
Of note, we found the protein enolase to be upregulated in all stages of in vitro planktonic and biofilm growth, but we did not detect immunoreactive spots corresponding to this protein on the biofilm blots, which has to be interpreted as a false-negative result. The enolase protein converts 2-phosphoglycerate to phosphoenolpyruvate during glycolysis but might also mediate bacterial attachment to the abiotic and biotic surfaces. In Streptococcus pneumoniae, the protein enolase serves as a pneumococcal plasminogen receptor as a surface-displaced protein and is therefore a key factor in attachment (44). To date, there are no studies describing the function of enolase in P. acnes.
The results of our immunoproteomic study are robust, since we focused only on immunoreactive spots and identified the proteins that were seen in at least two of three 2DGE gels. Since the protein concentration of samples excised from the 2DGE gel significantly influences the quality of MALDI spectra (45), we were not able to identify low-abundance protein spots (labeled as unknown proteins [UP] in Fig. 5). The P. acnes strains we used were not sequenced yet; therefore, some of the unidentified proteins by MALDI spectra might also be strain specific. Another limitation of our experimental design is the use of rabbit instead of human serum to investigate the humoral immune response. Differential protein expression of P. acnes in a host or differences in the immune responses might influence whether a vaccine candidate in one host will have efficacy against P. acnes in a second host species. However, an ethical agreement for the use of human serum is very strict, and the Helsinki Declaration demands preliminary results to be determined in animal studies first (46), which we present in this study. Therefore, further studies with human serum are now possible to conduct in order to verify our study results. Another limitation of the study is that the number of rabbits used was relatively low, and only two isolates of a single clonal type of P. acnes were used. Although this study demonstrates that the rabbit osteomyelitis model can be used to study P. acnes indwelling medical device infections, care must be taken to draw conclusions until an expanded study using more animals and strains can be performed.
The rates of indwelling medical device infections associated with P. acnes have long been underestimated. Due to the problematic diagnosis and indolent nature of these infections, they often progress to a full mature biofilm infection that relies on surgical intervention for resolution. Therefore, the early diagnosis and/or prevention of these infections will minimize surgical procedures and improve clinical outcomes. In addition, while a number of studies have evaluated the potential for active and passive vaccines to prevent the chronic skin disease acne vulgaris, which is also caused by P. acnes (47–49), no subsequent studies have evaluated the efficacy of this vaccine(s) against implant-associated infections. To identify effective vaccine candidates for P. acnes implant-associated infections, it is necessary to find key P. acnes virulence factors and/or proteins that are produced in vivo, recognized by the host immune system, and immunogenic. In addition, both planktonic and biofilm immunogenic cell wall proteins have to be taken into account, since targeting both phenotypes might be crucial to the generation of an effective vaccine against P. acnes-mediated implant infections.
Taken together, we found 13 upregulated proteins of P. acnes mainly associated with metabolic functions in both planktonic and biofilm growth stages, which need to be further analyzed as vaccine candidates within immunization protection studies or as biomarkers in diagnostic tests for implant-associated infections.
ACKNOWLEDGMENTS
We thank Ellie J. C. Goldstein for providing the P. acnes clinical strains used in this study and A. McDowell for helping with the MLST identification.
We have no commercial or other association that might pose a conflict of interest.
This work was supported by the National Institute of Allergy and Infectious Diseases, National Institutes of Health grant (R01 AI69568-01A2), a 3-year fellowship grant by the Swiss National Science Foundation (SNF) (Switzerland) (grant PBZHP3_141483), and a grant from the Swiss Foundation for Medical-Biological Grants (SSMBS) (Switzerland) (grant P3MP3_148362/1).
REFERENCES
- 1.Grice EA, Kong HH, Conlan S, Deming CB, Davis J, Young AC, NISC Comparative Sequencing Program, Bouffard GG, Blakesley RW, Murray PR, Green ED, Turner ML, Segre JA. 2009. Topographical and temporal diversity of the human skin microbiome. Science 324:1190–1192. doi: 10.1126/science.1171700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Achermann Y, Goldstein EJ, Coenye T, Shirtliff ME. 2014. Propionibacterium acnes: from commensal to opportunistic biofilm-associated implant pathogen. Clin Microbiol Rev 27:419–440. doi: 10.1128/CMR.00092-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Portillo ME, Corvec S, Borens O, Trampuz A. 2013. Propionibacterium acnes: an underestimated pathogen in implant-associated infections. Biomed Res Int 2013:804391. doi: 10.1155/2013/804391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bayston R, Ashraf W, Barker-Davies R, Tucker E, Clement R, Clayton J, Freeman BJ, Nuradeen B. 2007. Biofilm formation by Propionibacterium acnes on biomaterials in vitro and in vivo: impact on diagnosis and treatment. J Biomed Mater Res A 81:705–709. doi: 10.1002/jbm.a.31145. [DOI] [PubMed] [Google Scholar]
- 5.Ramage G, Tunney MM, Patrick S, Gorman SP, Nixon JR. 2003. Formation of Propionibacterium acnes biofilms on orthopaedic biomaterials and their susceptibility to antimicrobials. Biomaterials 24:3221–3227. doi: 10.1016/S0142-9612(03)00173-X. [DOI] [PubMed] [Google Scholar]
- 6.Archer NK, Mazaitis MJ, Costerton JW, Leid JG, Powers ME, Shirtliff ME. 2011. Staphylococcus aureus biofilms: properties, regulation, and roles in human disease. Virulence 2:445–459. doi: 10.4161/viru.2.5.17724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ceri H, Olson ME, Stremick C, Read RR, Morck D, Buret A. 1999. The Calgary Biofilm Device: new technology for rapid determination of antibiotic susceptibilities of bacterial biofilms. J Clin Microbiol 37:1771–1776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Høiby N, Ciofu O, Johansen HK, Song ZJ, Moser C, Jensen PØ, Molin S, Givskov M, Tolker-Nielsen T, Bjarnsholt T. 2011. The clinical impact of bacterial biofilms. Int J Oral Sci 3:55–65. doi: 10.4248/IJOS11026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Holland C, Mak TN, Zimny-Arndt U, Schmid M, Meyer TF, Jungblut PR, Brüggemann H. 2010. Proteomic identification of secreted proteins of Propionibacterium acnes. BMC Microbiol 10:230. doi: 10.1186/1471-2180-10-230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Furustrand Tafin U, Corvec S, Betrisey B, Zimmerli W, Trampuz A. 2012. Role of rifampin against Propionibacterium acnes biofilm in vitro and in an experimental foreign-body infection model. Antimicrob Agents Chemother 56:1885–1891. doi: 10.1128/AAC.05552-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Furustrand Tafin U, Trampuz A, Corvec S. 2013. In vitro emergence of rifampicin resistance in Propionibacterium acnes and molecular characterization of mutations in the rpoB gene. J Antimicrob Chemother 68:523–528. doi: 10.1093/jac/dks428. [DOI] [PubMed] [Google Scholar]
- 12.McDowell A, Barnard E, Nagy I, Gao A, Tomida S, Li H, Eady A, Cove J, Nord CE, Patrick S. 2012. An expanded multilocus sequence typing scheme for Propionibacterium acnes: investigation of ‘pathogenic,’ ‘commensal’ and antibiotic resistant strains. PLoS One 7:e41480. doi: 10.1371/journal.pone.0041480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McDowell A, Gao A, Barnard E, Fink C, Murray PI, Dowson CG, Nagy I, Lambert PA, Patrick S. 2011. A novel multilocus sequence typing scheme for the opportunistic pathogen Propionibacterium acnes and characterization of type I cell surface-associated antigens. Microbiology 157:1990–2003. doi: 10.1099/mic.0.049676-0. [DOI] [PubMed] [Google Scholar]
- 14.McDowell A, Valanne S, Ramage G, Tunney MM, Glenn JV, McLorinan GC, Bhatia A, Maisonneuve JF, Lodes M, Persing DH, Patrick S. 2005. Propionibacterium acnes types I and II represent phylogenetically distinct groups. J Clin Microbiol 43:326–334. doi: 10.1128/JCM.43.1.326-334.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Valanne S, McDowell A, Ramage G, Tunney MM, Einarsson GG, O'Hagan S, Wisdom GB, Fairley D, Bhatia A, Maisonneuve JF, Lodes M, Persing DH, Patrick S. 2005. CAMP factor homologues in Propionibacterium acnes: a new protein family differentially expressed by types I and II. Microbiology 151:1369–1379. doi: 10.1099/mic.0.27788-0. [DOI] [PubMed] [Google Scholar]
- 16.Shirtliff ME, Calhoun JH, Mader JT. 2002. Experimental osteomyelitis treatment with antibiotic-impregnated hydroxyapatite. Clin Orthop Relat Res (401):239–247. [DOI] [PubMed] [Google Scholar]
- 17.Mader JT, Shirtliff ME. 1999. The rabbit model of bacterial osteomyelitis of the tibia, p 581–591. In Zak O, Sande M (ed), Handbook of animal models of infection. Academic Press Ltd., London, England. [Google Scholar]
- 18.Freiberg JA, McIver KS, Shirtliff ME. 2014. In vivo expression of Streptococcus pyogenes immunogenic proteins during tibial foreign body infection. Infect Immun 82:3891–3899. doi: 10.1128/IAI.01831-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fischer N, Mak TN, Shinohara DB, Sfanos KS, Meyer TF, Brüggemann H. 2013. Deciphering the intracellular fate of Propionibacterium acnes in macrophages. Biomed Res Int 2013:603046. doi: 10.1155/2013/603046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Webster GF, Leyden JJ, Musson RA, Douglas SD. 1985. Susceptibility of Propionibacterium acnes to killing and degradation by human neutrophils and monocytes in vitro. Infect Immun 49:116–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Csukas Z, Banizs B, Rozgonyi F. 2004. Studies on the cytotoxic effects of Propionibacterium acnes strains isolated from cornea. Microb Pathog 36:171–174. doi: 10.1016/j.micpath.2003.09.002. [DOI] [PubMed] [Google Scholar]
- 22.Sfanos KS, Isaacs WB. 2008. An evaluation of PCR primer sets used for detection of Propionibacterium acnes in prostate tissue samples. Prostate 68:1492–1495. doi: 10.1002/pros.20820. [DOI] [PubMed] [Google Scholar]
- 23.Gaida M, Mayer B, Stegmaier S, Schirmacher P, Wagner C, Hänsch G. 2012. Polymorphonuclear neutrophils in osteomyelitis: link to osteoclast generation and bone resorption. Eur J Inflamm 10:413–426. [Google Scholar]
- 24.Webster GF, Indrisano JP, Leyden JJ. 1985. Antibody titers to Propionibacterium acnes cell wall carbohydrate in nodulocystic acne patients. J Investig Dermatol 84:496–500. doi: 10.1111/1523-1747.ep12273462. [DOI] [PubMed] [Google Scholar]
- 25.Bradford MM. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 26.O'Farrell PH. 1975. High resolution two-dimensional electrophoresis of proteins. J Biol Chem 250:4007–4021. [PMC free article] [PubMed] [Google Scholar]
- 27.Brady RA, Leid JG, Camper AK, Costerton JW, Shirtliff ME. 2006. Identification of Staphylococcus aureus proteins recognized by the antibody-mediated immune response to a biofilm infection. Infect Immun 74:3415–3426. doi: 10.1128/IAI.00392-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McDowell A, Perry AL, Lambert PA, Patrick S. 2008. A new phylogenetic group of Propionibacterium acnes. J Med Microbiol 57:218–224. doi: 10.1099/jmm.0.47489-0. [DOI] [PubMed] [Google Scholar]
- 29.Nodzo SR, Hohman DW, Crane JK, Duquin TR. 2014. Hemolysis as a clinical marker for Propionibacterium acnes orthopedic infection. Am J Orthop 43:E93–E97. [PubMed] [Google Scholar]
- 30.Butler-Wu SM, Burns EM, Pottinger PS, Magaret AS, Rakeman JL, Matsen FA III, Cookson BT. 2011. Optimization of periprosthetic culture for diagnosis of Propionibacterium acnes prosthetic joint infection. J Clin Microbiol 49:2490–2495. doi: 10.1128/JCM.00450-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Trampuz A, Piper KE, Jacobson MJ, Hanssen AD, Unni KK, Osmon DR, Mandrekar JN, Cockerill FR, Steckelberg JM, Greenleaf JF, Patel R. 2007. Sonication of removed hip and knee prostheses for diagnosis of infection. N Engl J Med 357:654–663. doi: 10.1056/NEJMoa061588. [DOI] [PubMed] [Google Scholar]
- 32.Wang B, Toye B, Desjardins M, Lapner P, Lee C. 2013. A 7-year retrospective review from 2005 to 2011 of Propionibacterium acnes shoulder infections in Ottawa, Ontario, Canada. Diagn Microbiol Infect Dis 75:195–199. doi: 10.1016/j.diagmicrobio.2012.10.018. [DOI] [PubMed] [Google Scholar]
- 33.Blomgren G. 1981. Hematogenous infection of total joint replacement. An experimental study in the rabbit. Acta Orthop Scand Suppl 187:1–64. [DOI] [PubMed] [Google Scholar]
- 34.Gahukamble AD, McDowell A, Post V, Salavarrieta Varela J, Rochford ET, Richards RG, Patrick S, Moriarty TF. 2014. Propionibacterium acnes and Staphylococcus lugdunensis cause pyogenic osteomyelitis in an intramedullary nail model in rabbits. J Clin Microbiol 52:1595–1606. doi: 10.1128/JCM.03197-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shiono Y, Ishii K, Nagai S, Kakinuma H, Sasaki A, Aiawa M, Okada Y, Toyama Z, Matsumoto M. 2014. Propionibacterium acnes cause delayed surgical site infection only in the presence of implant. Orthop Res Soc (ORS) Annu Meet 15 to 18 March 2014, New Orleans, LA. [Google Scholar]
- 36.Sampedro MF, Piper KE, McDowell A, Patrick S, Mandrekar JN, Rouse MS, Steckelberg JM, Patel R. 2009. Species of Propionibacterium and Propionibacterium acnes phylotypes associated with orthopedic implants. Diagn Microbiol Infect Dis 64:138–145. doi: 10.1016/j.diagmicrobio.2009.01.024. [DOI] [PubMed] [Google Scholar]
- 37.McDowell A, Nagy I, Magyari M, Barnard E, Patrick S. 2013. The opportunistic pathogen Propionibacterium acnes: insights into typing, human disease, clonal diversification and CAMP factor evolution. PLoS One 8:e70897. doi: 10.1371/journal.pone.0070897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li L, Sun C, Yang F, Yang S, Feng X, Gu J, Han W, Langford PR, Lei L. 2013. Identification of proteins of Propionibacterium acnes for use as vaccine candidates to prevent infection by the pig pathogen Actinobacillus pleuropneumoniae. Vaccine 31:5269–5275. doi: 10.1016/j.vaccine.2013.08.054. [DOI] [PubMed] [Google Scholar]
- 39.Brady RA, O'May GA, Leid JG, Costerton JW, Shirtliff ME. 2011. Resolution of Staphylococcus aureus biofilm infection using vaccination and antibiotic treatment. Infect Immun 79:1797–1803. doi: 10.1128/IAI.00451-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dryla A, Prustomersky S, Gelbmann D, Hanner M, Bettinger E, Kocsis B, Kustos T, Henics T, Meinke A, Nagy E. 2005. Comparison of antibody repertoires against Staphylococcus aureus in healthy individuals and in acutely infected patients. Clin Diagn Lab Immunol 12:387–398. doi: 10.1128/CDLI.12.3.387-398.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.den Reijer PM, Lemmens-den Toom N, Kant S, Snijders SV, Boelens H, Tavakol M, Verkaik NJ, van Belkum A, Verbrugh HA, van Wamel WJ. 2013. Characterization of the humoral immune response during Staphylococcus aureus bacteremia and global gene expression by Staphylococcus aureus in human blood. PLoS One 8:e53391. doi: 10.1371/journal.pone.0053391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bergmann S, Rohde M, Hammerschmidt S. 2004. Glyceraldehyde-3-phosphate dehydrogenase of Streptococcus pneumoniae is a surface-displayed plasminogen-binding protein. Infect Immun 72:2416–2419. doi: 10.1128/IAI.72.4.2416-2419.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Henderson B, Martin A. 2011. Bacterial virulence in the moonlight: multitasking bacterial moonlighting proteins are virulence determinants in infectious disease. Infect Immun 79:3476–3491. doi: 10.1128/IAI.00179-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bergmann S, Schoenen H, Hammerschmidt S. 2013. The interaction between bacterial enolase and plasminogen promotes adherence of Streptococcus pneumoniae to epithelial and endothelial cells. Int J Med Microbiol 303:452–462. doi: 10.1016/j.ijmm.2013.06.002. [DOI] [PubMed] [Google Scholar]
- 45.Drevinek M, Dresler J, Klimentova J, Pisa L, Hubalek M. 2012. Evaluation of sample preparation methods for MALDI-TOF MS identification of highly dangerous bacteria. Lett Appl Microbiol 55:40–46. doi: 10.1111/j.1472-765X.2012.03255.x. [DOI] [PubMed] [Google Scholar]
- 46.American Society for Clinical Investigation. 1967. Declaration of Helsinki: recommendations guiding doctors in clinical research. J Clin Invest 46:1140. doi: 10.1172/JCI105595. [DOI] [Google Scholar]
- 47.Nakatsuji T, Liu YT, Huang CP, Zouboulis CC, Gallo RL, Huang CM. 2008. Vaccination targeting a surface sialidase of P. acnes: implication for new treatment of acne vulgaris. PLoS One 3:e1551. doi: 10.1371/journal.pone.0001551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liu PF, Nakatsuji T, Zhu W, Gallo RL, Huang CM. 2011. Passive immunoprotection targeting a secreted CAMP factor of Propionibacterium acnes as a novel immunotherapeutic for acne vulgaris. Vaccine 29:3230–3238. doi: 10.1016/j.vaccine.2011.02.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nakatsuji T, Tang DC, Zhang L, Gallo RL, Huang CM. 2011. Propionibacterium acnes CAMP factor and host acid sphingomyelinase contribute to bacterial virulence: potential targets for inflammatory acne treatment. PLoS One 6:e14797. doi: 10.1371/journal.pone.0014797. [DOI] [PMC free article] [PubMed] [Google Scholar]