Abstract
Embryo quality during the in vitro developmental period is of great clinical importance. Experimental genetic studies during this period have demonstrated the association between specific gene expression profiles and the production of healthy blastocysts. Although the quality of the oocyte may play a major role in embryo development, it has been well established that the post – fertilization period also has an important and crucial role in the determination of blastocyst quality. A variety of genes (such as OCT, SOX2, NANOG) and their related signaling pathways as well as transcription molecules (such as TGF-β, BMP) have been implicated in the pre- and post-implantation period. Furthermore, DNA methylation has been lately characterized as an epigenetic mark since it is one of the most important processes involved in the maintenance of genome stability. Physiological embryo development appears to depend upon the correct DNA methylation pattern. Due to the fact that soon after fertilization the zygote undergoes several morphogenetic and developmental events including activation of embryonic genome through the transition of the maternal genome, a diverse gene expression pattern may lead to clinically important conditions, such as apoptosis or the production of a chromosomically abnormal embryo. The present review focused on genes and their role during pre-implantation embryo development, giving emphasis on the various parameters that may alter gene expression or DNA methylation patterns. The pre-implantation embryos derived from in vitro culture systems (in vitro fertilization) and the possible effects on gene expression after the prolonged culture conditions are also discussed.
Keywords: Assisted reproduction, DNA methylation, Human embryology, Imprinting errors
INTRODUCTION
Methylation and de-methylation of DNA are epigenetic marks and have been characterized as one of the most important processes for maintenance of genome stability. Specifically, the DNA methylation during the fertilization phase and even more in the post-fertilization period have been characterized to be of major importance for embryo development, while during the same period many genes are implicated. Although most studies have been conducted on animal models, as it is unethical to perform experiments in human embryos, some solid conclusions can be drawn with similar if not equivalent observations to humans. Genes, such as OCT4, SOX2 and NANOG that have been characterized as pluripotency gene network, have been associated post-fertilization with the DNA methylation machinery, implying inter-dependency for proper embryo development. Apart from genes that participate in this crucial period, there are conditions that may influence the methylation machinery and this phenomenon is more profound during gamete manipulation and embryo culture in Assisted Reproduction Technologies (ART). External factors, such as temperature, light, culture media, hormonal stimulation protocol, demographic data of the patients and infertility causes may have an impact on the methylation machinery. Each factor may add a risk to the methylation process and all of them may result in an increased risk of aberrant DNA methylation. Example of the above assumption is the imprinting disorders that have been associated with ART procedures.
The present review will be focused on DNA methylation during post-fertilization, the genes that are involved during the developmental period, the conditions during ART procedures that influence embryo development, besides discussing the imprinting disorders and the association with DNA methylation.
1. THE METHYLATION – DE-METHYLATION CYCLE
The genome of both mammalian gametes is highly methylated. DNA methylation has been characterized as one of the most known epigenetic modification and occurs mainly at cytosines (5mC) in symmetrical dinucleotide CpG base pairs [1, 2]. It is important for stably repressing gene activity and for the establishment of differentially methylated regions (DMRs) at imprinted genes [3, 4]. Genomic imprinting is the allele-specific expression of several genes by both genomes and is responsible for normal embryo development [5, 6]. Upon fertilization, two highly specialized and methylated cells are combined in one cell, called zygote. The paternal genome is densely packaged while the remodeling of chromatin through removal of protamines and replacement by histones are the most remarkable transformation that take place in the cytoplasm of the oocyte [7]. At same time, the paternal pronucleus undergoes an acute and rapid loss of DNA methylation (de-methylation) [8-10]. This de-methylation occurs before paternal pronucleus begins DNA replication and is termed active de-methylation [10, 11]. The maternal pronucleus undergoes a replication-dependent loss of methylation and therefore this DNA de-methylation is in a step-wise manner, producing unequally methylated sister chromatids and is referred to as passive de-methylation [10-12]. Maternal de-methylation takes place until the morula developmental stage, while de novo genome methylation occurs after the fifth developmental cycle and coincides with the activation of embryonic genome and the initiation of the first differentiative events. The latter involves the establishment of the first two lineages, the inner cell mass (ICM) and the trophectoderm (TE). Although the blastomeres of a two - blastomere embryo are totipotent, it is likely that blastomeres located inside the morula are possible to become ICM, while those that are outside are more possible to result in TE. The ICM produce the embryo itself (all embryonic tissues), while the TE gives rise to the structure of the placenta. Interestingly, there are global differences in DNA methylation between lineages coming from TE, which are hypermethylated and those coming from ICM which are hypomethylated. Following implantation of the blastosyst (6-7 days post-fertilization), the embryo forms the hypoblast and the epiblast, where the latter at the 7-8 developmental day give rise to a small population of cells, the precursors of primordial germ cells (PGCs) [13]. These cells are highly methylated and by E8.5, the PGC progenitors start to migrate through the hindgut endoderm to the genital ridge arriving about E11.5 [14]. Upon arrival to the genital ridge, they undergo mitotic divisions and proliferate until E13.5. During this time of expansion and especially between E11.5-E12.5, the highly methylated PGCs undergo a rapid genome-wide loss of methylation also including the majority of parent-of-origin-specific DMRs of imprinted genes [15-18]. Soon after global methylation loss in the PGCs, the male and the female germline enters the mitotic and the meiotic arrest, respectively. Following sexual differentiation of PGCs, which coincides with the de-methylation process, it is necessary to lay down new imprints and therefore re-methylation takes place, while female and male PGCs reprogramming diverges. Reprogramming in germ cells is appropriate for resetting the imprints, so initial methylation of PGCs obliterates and re-methylation takes place to facilitate re-establishment in the germ line to ensure proper inheritance of imprints for the next generation. Re-methylation occurs earlier in the male germ line, at the pro-spermatogonia stage and before birth, while it is completed after birth and before the end of the pachytene phase of meiosis [19-21]. In contrast to male, female germ line starts to reprogram its genome post-natally, during oocyte growth and following pachytene phase of meiosis [22-24]. At the end of gametogenesis, both gametes acquire new imprints and are fully methylated. The differential epigenetic marking of the parental alleles that takes place during gametogenesis completes the genome methylation – de-methylation cycle.
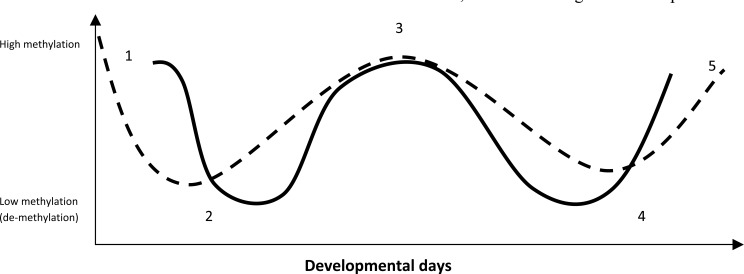
According to the methylation – de-methylation cycle described above and to the figure that has been produced according to the methylation reprogramming in the germ cells and in the developmental embryos in the respective developmental days, an interesting observation has been revealed, which highlights that the male PGCs are re-methylated earlier and in a more acute manner. Furthermore, the paternal pronucleus, following fertilization, is subjected to an acute de-methylation process and before the first DNA replication. In contrast, the female germ line is re-methylated in a much slower and gradual way in comparison to male germ line and also the maternal pronucleus is subjected to a gradual replication-dependent de-methylation. In other words, on one hand we have the acute re-methylation/de-methylation (for males) and on the other hand, the gradual re-methylation/de-methylation (for females) processes. It is very intriguing to know what makes this difference and for what reason two highly specified genomes that are coming from the same progenitors and are coming together behave so differently in this significant epigenetic process. Is it the genome itself? Does the Y or the X chromosome or specific sequences make the difference or there are some conditions that make this difference? Does evolution play any role in this process? In an attempt to approach the answers to these questions, we raised some speculations according to the way that the methylation – de-methylation cycle occurs in each genome: It is apparent from the schematic presentation (Fig. 1) that the male methylation – de-methylation cycle follows a “secondary immune-like response”, while the female cycle follows a rather “primary immune-like response”. Bearing in mind that the main difference between the two immune responses is the memory (the primary or the secondary contact with the antigen), we can speculate that due to the similarity of the male methylation – de-methylation pattern with the secondary immune-like response, the male genome and the subsequent methylation – de-methylation pattern seem to be a genome that during its life cycle undergoes genetic and epigenetic modifications at sequences that have been already methylated and de-methylated (probably in the previous generations) and these sequences are in the majority conserved from one generation to the next. On the contrary, the female mathylation – de-methylation pattern has similarities with the primary immune-like response and therefore the female genome consists of sequences that the majority of them have not methylated or de-methylated previously and are epigenetically modified for the first time. To sum up, the male genome seems to be genetically modified in the previous generations, while the female genome appears to be subjected to genetic modifications for the first time.
Fig. (1).
Primordial germ cells (PGCs) are highly methylated during the entry in the embryo (1). Afterwards the PGCs are migrating to the genital ridge and upon arrival to this point is has been observed a whole-genome demethylation (2). Soon after demethylation, re-methylation takes place in both male (black line) and female germ lines (black dotted line), so as new imprints to be established during gametogenesis (3). The two highly specified gametes are coming together (fertilization) and subsequently the paternal pronucleus is subjected to an acute active demethylation wave, while the female pronucleus undergoes a more step-by-step and passive de-methylation process (4). Finally, the implanted em-bryo/blastosyst, forming the ICM and the TE are hyper-methylated and hypo-methylated, respectively (5).
2. CONDITIONS AND GENES GOVERNING PRE- AND POST-IMPLANTATION EMBRYO DEVELOPMENT PERIOD
2.1. Paternal de-methylation
Following the molecular cascade of spermatozoon-oocyte interaction, fertilization in terms of formation of two pronuclei takes place [25]. Fertilization itself provokes a series of epigenetic changes among which de-methylation is one of the major waves of DNA methylation loss [26-28]. Before the S phase of the zygote, within about 4h post-fertilization, highly methylated sperm DNA is rapidly de-methylated by an active mechanism [29]. Bearing in mind that the parental genome is in different stage of the cell cycle carrying different DNA methylation patterns and chromatin organization, it follows a specific asymmetric loss of methylation that has been detected both by indirect immunofluorescence and by bisulphate sequencing of unique gene sequences and repeat families [30-32]. Today the exact mechanism of the active de-methylation has not yet been clarified. Nevertheless, a number of possible mechanisms have been proposed and all of them are intriguing but yet unclear which may function in the zygote. The first one involves the direct removal of the methyl group from the C-5 position in the heterocyclic ring, but this straight-forward form of de-methylation needs the presence of DNA demethylases the identity of which remains totally elusive. In mammalians, DNA glycosylases are unable to perform this reaction [33]. The second mechanism of active de-methylation is indirect and involves the function of some enzymes, such as deaminases. The deaminase is able to convert 5mC to thymidine (T) and therefore the resulting mismatch G:T would initiate the base excision repair (BER) pathway attracting a glycosylate to remove the T and replacing it by a cytocine (C). Candidate deaminases are members of the activation-induced deaminase (AID/ APOBEC) family, while candidate glycosylases include the methyl-CpG-binding domain (MBD4) and thymine DNA glycosylase (TDG) proteins in combination with the DNA damage response protein gadd45a [34, 35]. Although a more detailed biochemical mechanism will have to be clarified, there are reports demonstrating that gadd45a cannot demethylate the DNA [36, 37]. The third mechanism involves a newly identified base in mammals, hydroxymethylocytocine (hmC) [38, 39]. 5mC can be oxidized to 5hmC by the ten-eleven-translocation (TET) activity. 5hmC itself may be further oxidized by TETs to 5-formylcytosine (5fC) and 5-carboxylcytocine (5caC). The above derivatives can be excised by TDG or by single strand-selective monofunctional uracil DNA glycosylase 1 (SMUG1) (in case 5hmC deaminated to 5-hydromethyluracil, 5hmU) and MBD4, initiating in this way the BER pathway resulting in an unmodified C. Components of the BER family have been detected in the paternal pronucleus during the later phase of de-methylation, while inhibitors of the BER family demonstrate maintenance of DNA methylation levels [40, 41]. Among the TETs, TET3 is highly expressed in the zygote [42, 43], while its presence in both pronuclei at the time of fertilization underlies that also maternal pronuclei are subjected to some extent to active de-methylation [28]. Recently, AID was also suggested as an essential component of paternal de-methylation, while UNG2 and not TDG appear to be the initiating glycosylase for deamination. Furthermore, TET-mediated hydroxylation and de-methylation have been proved to occur prior to DNA synthesis [44]. Nevertheless, there is mounting evidence that TDG has a crucial role in de-methylation. Specifically, it has been observed that TDG interact with various transcription factors raising the possibility that TDG associates with gene transcription [45]. Gathering this information, it is likely that TDG, through the generation of 5fC and 5aC is the most validated pathway that explains completely active DNA de-methylation [46]. Recently, a role for the elongator complex in zygotic paternal genome de-methylation has been proposed [47]. This elongator complex possesses a lysine acetyl tranferase action regulating its transcriptional activity via acetylation of histone H3 [48]. The essential region for the structural integrity of the complex is a SAM domain, which is likely to play a role in directly removing 5mC [49]. Nevertheless, a significant amount of the 5mC derivatives remain refractory to the acute paternal de-methylation and are gradually dispersed during subsequent cell divisions [50, 51], raising the possibility that different pathways may activate or operate synergistically or in parallel to form a complex de-methylation network. We shall also bear in mind that a significant amount of 5hmC in the male pronucleus as well as 5mC in the female pronucleus undergo de-methylation in a passive-manner mechanism [43, 51, 52]. Conclusively, detailed genome-wide profiling experiments coupled with disruption of specific pathways may reveal the exact targets of these de-methylation pathways.
2.2. Maternal de-methylation
The maternal genome in oocytes is arrested at metaphase II (MII) with its 2C genome packaged with histones and upon fertilization, the maternal genome completes meiosis. Moreover, the maternal genome escapes 5mC loss in the fertilized oocyte and is passively de-methylated in subsequent divisions owing to the exclusion the DNA methyltrasferase 1, DNMT1 from the nucleus [10, 53]. DNMT1 is a member of the DNMT family, among which DNMT2, DNMT3a, DNMT3b and DNMTL have also been identified [54]. DNMTs can be divided into de novo methyltransferases that can methylate C to 5mC in unmethylated DNA and the maintenance DNMTs, which attach a methyl group to hemi-methylated DNA at the time of DNA replication [54, 55]. Except for the DNMT1, which is a maintenance methyltrasferase, all the other members of the DNMT family are de novo methyltrasferases. Experimental inactivation of DNMTs is thought to play major role in the development of pre-implantation embryos [56, 57]. Despite the excess levels of the DNMT1 isoform, DNMT1o, DNA methylation loss in the maternal pronucleus follows a passive manner due to the active exclusion of DNMT1o [51]. The exclusion of DNMT1 from the nucleus and the subsequent DNA replications result in de-methylation in a step-wise manner. Another mechanism that has been proposed for the escape of the maternal genome from rapid de-methylation is the exclusion of TET3 from the maternal pronucleus, because it has been observed that the maternal genome after fertilization contains 5mC. The molecular mechanism of exclusion of TET3 involves the recruitment of PGC7 to the H3K9me2 binding site, which in turn prevents TET3 to bind to the maternal genome [58].
2.3. Reprogramming in the Imprinted Regions of the Developing Embryos
Genomic imprinting is a typical example of parental asymmetry in the epigenome, while both epigenotypes are essential for normal embryo development under correct functional complementation of paternal and maternal genes (PEGs and MEGs). Today, approximately one hundred and fifty imprinted genes have been described so far in mammals. Many imprinted genes are located in clusters throughout the genome. These clusters typically contain at least one non-coding RNA region (ncRNA). These genes, in a cluster, are usually under the control of a discrete DNA element, known as an imprinting control region (ICR). One characteristic of ICRs is the profound difference in DNA methylation on the maternally- and paternally-derived sequences that are set up during gametogenesis, with one copy being highly methylated and the other unmethylated [61-63]. Interestingly, some ICRs are protected from both active and passive de-methylation during pre-implantation development. The same situation has been observed for intracisternal A particles (IAPs), long interspersed nuclear elements (LINEs) and satellite sequences. Today, it is still unclear which member of the DNMT family is responsible for the refractory situation of these sequences during global de-methylation. Nevertheless, the somatic form of the DMNT1, DMNT1s, is present during the first cell division, indicating that DMNT1s is mainly involved in the maintenance of methylation during this developmental period [64-66]. Although the DMNT1o isoform is difficult to be detected, it was suggested that it is localized in the nucleus [66]. Nevertheless, all reports indicate that both isoforms of DMNTs maintain imprinted methylation during the cleavage developmental period. It is likely that both isoforms co-operate and function in an inter-changeable manner, while the developmental time is crucial for their different actions. What protects paternally methylated ICRs from the active de-methylation and what directs the DMNTs to ICRs? It is important to note that not all imprinted genes are regulated in the same way and it is likely that cis and trans-acting factors are also involved in these procedures. CG-rich sequences, termed CG-islands are also found at the promoters and most of them are differentially methylated (DMRs) with the repressed allele being methylated and the active one being de-methylated [62, 63]. Nevertheless, there are imprinting regions in which monoallelic expression is independent of DNA methylation [67]. One defined factor that was found to be critical with a general role in DNA methylation maintenance is the PGC7/STELLA [68-70]. Zygotes coming from stella null oocytes showed premature DNA de-methylation of the maternal pronucleus, while PGC7 seems to be essential for the protection of maternal genome against tet3-dependent oxidation of 5mC [41]. The possible proposed mechanism is that PGC7 binds to the histone 3 di-methylated at lysine 9 (H3K9me2) [58], indicating that PGC7 is a maternal protein that protects imprinted regions of both parents from active de-methylation. A sequence-specific DNA binding factor, Zfp57, has also been implicated in the maintenance of DNA methylation. Zfp57 is a Kruppel-associated box-containing (KRAB) zinc finger protein, which belongs to a class of transcription factors that suppress transcription by recruiting KAP-1/TIF1β co-repressor complexes [71] and was identified during the study of human imprinted disorders of cases with 6q24 transient neonatal diabetes (TND) mellitus, which is caused by the hypomethylation of the imprinted promoter gene PLAGL1 [72]. Although the exact mechanism of the functioning of Zfp57 is not yet elucidated, it has been proposed as an “imprinted-specific” factor [67]. Specifically, Zfp57 binds to hemi-methylated sites recruiting the multifunctional Kap1 repressor protein ensuring in this way, the maintenance of the DNA methylation and restoration of the repressive histone modification that involves the tri-methylation of histone 3 at lysine 9 (H3K9me3) [73]. Zfp57, together with Kap1 form, is a complicated factor that is essential for the recruitment of the DNMT1 at the target sites, maintaining the DNA methylation pattern in this way. Besides the above complex, additional factors may be involved in the maintenance of DNA methylation among which an interesting class of proteins is the members of the nucleotide-binding oligomerization domain, leucine-rich repeat and pyrin domain containing protein (NLRP) family and TRIM28. The NLPRs seem to play some indirect roles in reproduction, while the NLRP7 gene has been implicated in recurrent molar pregnancies of diploid bi-parental origin [74, 75]. Although fertilization can be achieved, in these androgenetic-like pathology fetuses, from the molecular point of view, the maternal ICRs exert extensive loss of DNA methylation, while the paternal ICRs are unaffected. NLRP2 has also been found in a hypomethylated status in the ICRs of the imprinting KCNQ1 cluster gene [76]. TRIM28 is a maternal-derived protein [77] and it has been proposed to play a role in protection during passive de-methylation via the ZFP57 and TRIM28 interactions [78]. Today, it is difficult to conclude if all these factors have a role in the maintenance of methylation process, but it is likely that these proteins may be members of a more complicated complex that are derived mainly from the cytoplasm of the oocytes and may directly interact with other modyfing factors, like Ying Yang 1 (YY1) [79] or GSE. The latter is a maternal factor predominantly localized in the nucleus of cells exerting a significant reduction in the loss of 5mC and accumulation of 5hmC in the paternal pronucleus [80].
Although the factors that maintain DNA methylation in the imprinted regions have been a matter of intensive experimental focus, it is very intriguing to know what mechanisms involve the protection of the opposite allele to gain methylation. Besides the complicated methylation/de-methylation network, increasing evidence has proposed a role of histones in the protection of the opposite alleles. Briefly, open or active euchromatin is characterized by histone H3 lysine 4 tri-methylation (H3K4me3) and lysine 9 acetylation (H3K9ac), while silent or repressive heterochromatin is characterized by histone H3 lysine 9 (H3K9me3) and lysine 27 (H3K27me3) methylation. There are regions that have been identified with both activating and suppressing modifications (bivalent), with the possibility of one allele to be related to the euchromatin and the other to the heterochromatin state [81]. The acetylation of histones is catalyzed by histone acetyltransferases (HATs), while the removal of the acetyl group is achieved by histone deacetylases (HDACs). Both enzymes are components of the transcriptional co-activator/ repressor complexes and predominantly are bound to the core of the pluripotency factors OCT4, SOX2 and NANOG [82]. Besides, HDACs have been proposed to have a dual role during pre-implantation embryo development: firstly, to suppress the expression of genes that need to be inactive and secondly, to deacetylate active genes preventing excessive acetylation [83]. Although the mechanism might be more complicated, it is likely that protection of the unmethylated allele against the factors used for protecting the imprinted regions could be through generic molecules [67]. The identification of “transient” DMRs may be another component for the solution of the previous puzzle, which with combination with histone modifications may give rise to stable imprinting of the genes. Much of this evidence comes from the imprinted KCNQ1 gene region. On the paternal allele, the expression of the KCNQ1 silences all the subsequent genes in the domain, while in the maternal allele, the ICR methylation prevents expression of the KCNQ1, allowing the rest of the genes to be maternally expressed [84, 85]. The underlying mechanism in this situation is that the maternal alleles are activated by histone modifications and the paternal alleles by repressive marks in combination with the repressive Plycomb Group proteins [86, 87]. These suggestions indirectly indicate that histone modifications and histone-modifying enzymes, like Polycomb proteins, are critical for repressing genes in the KCNQ1 region [88]. Another example is that of the paternally Igf2 – maternally H19 locus [89]. The two genes share common sites of expression and share the same enhancer sequences located downstream of the H19 gene. The ICR domain [90] contains a binding site mainly for the zinc-finger insulator protein, CTCF, which binds to the maternal unmethylated allele [91] and blocks the enhancer to interact with the Igf2 promoter, thus resulting in the inactivation of the gene. Instead, maternal activity expresses the H19 gene. On the paternal allele, the ICR is methylated, and therefore, the CTCF cannot interact with the site, resulting in the expression of the Igf2 because the enhancers are unblocked and the promoters are active. We should also bear in mind that this mechanism is not always present in all imprinted genes [92].
2.4. Genes and Conditions Implicating in the Self-renewal and Pluripotency Network
DNA methylation is maintained at low levels in pluripotent cells. This hypothesis is based on the fact that while global DNA de-methylation is essential for the activation of the pluripotency network, pluripotent cells exhibit an uncoupling of DNA methylation pattern. It is likely that complete methylation loss results in genome instability especially during blastomere cleavage [93]. Furthermore, TET1/2 directly bound to the pluripotency network and especially to NANOG [94]. The pluripotent state is maintained by the interaction of the master transcriptional network, including OCT4 and NANOG with the methylation machinery. It appears that the expression of the pluripotency factors is accomplished by a stable hypo-methylated and not de-methylated genome.
OCT4 was discovered as a transcription factor playing a critical role in early embryo development. It is encoded by the gene POU5F1 and binds to an octamer motif of the ATGCAAAT sequence through the assistance of the POU domain [95]. OCT4, in combination with SOX2 and NANOG, constitutes a triad of transcription factors that are critical for self-renewal and pluripotency [96, 97]. The OCT4 gene contains three regulatory elements for the purpose of its transcription. Analysis of these elements involves a proximal enhancer (PE), a distal enhancer (DE) and a proximal promoter (PP) [97, 98]. According to the developmental stage, the two enhancers are differentially expressed. All these elements serve as a mask for various proteins and for DNA methylation. The DNA methylation status of these elements reflects the transcriptional status of the OCT4 gene. Therefore, the expression of the OCT4 is considered as a master regulator for the initiation and the maintenance of pluripotent cells during development since it has been found in unfertilized oocytes, early embryos and PGCs [99, 100]. It is interesting to note that precise levels of OCT4 must be sustained within a normal range in order for pluripotency to be maintained [101]. Furthermore, it was showed in transgenic ESC lines that elevated OCT4 levels differentiate to primitive endodermal and mesodermal lineages, while on the contrary, diminished OCT4 levels committed to trophectoderm lineage [101]. In order to achieve a precise level of OCT4, the chromatin state should be defined. In the undifferentiated status, the OCT4 is hypo-methylated and packaged with nucleosomes containing highly acetylated histone 3 (H3K9ac, H3K14ac) and di-methylated or tri-methylated histone H3 (H3K4me2, H3K4me3) [102]. The OCT4 locus however is subjected to a series of epigenetic modifications according to the transcriptional level. As far as DNA methylation is concerned, microRNAs (miR-290 through miR-295) and the Rbl2 transcriptional repressor, support the transcription of the DNMT3a and DNMT3b, which in turn methylate the DNA in the three regulatory elements of the OCT4 locus, promoting in this way the inactivation of the OCT4 gene [103]. Informatively, microRNAs are small non-coding RNAs playing an important role in post-transcription gene expression during animal development and regeneration [104-106]. Recently, two miRNA clusters, mir-290 and mir-302, are sequentially activated during development and the trajectory of any individual blastomere is strongly influenced by the combination of factors introduced [107]. The discovery of the Paf1 chromatin modifier shed some light on the efforts to uncover the OCT4 transcription puzzle [108]. Specifically, the Paf1C factor interacts with the RNA polymerase II in order to bind directly to the promoter and therefore, maintain an active or open chromatin structure. Recently, the structure of the human Paf1/Leo1 subcomplex was introduced within PAF1C complex. They form a tightly associated heterodimer through antiparallel beta-sheet interactions indicating that Leo1 binds to PAF1C through Paf1 and that the Ctr9 subunit is the key scaffold protein in assembling PAF1C. Furthermore, the Paf1/Leo1 heterodimer is necessary for its in vitro binding to histone H3, [109]. The best characterized histone modifier is the G9a methyltransferase, which induces either di- or tri-methylation of histone H3 at the PP of the OCT4 locus. These methylations recruit the heterochromatin protein 1 (HP1) and therefore trigger the heterochromatin formation, which silences the OCT4 expression. The whole process has been found to associate with the Cdk2ap1 factor which in turn interacts with the Mbd3 factor. The result of this combination is the acceleration of the process. Recently, G9a was found to colocalized with H3 lysine 9 monomethylation in a nuclear membrane-dependent manner during mouse embryo development [110]. Among the best characterized receptors that interact with the three regulatory elements of the OCT4 locus is the germ cell nuclear factor (GCNF). This protein binds to the PP and subsequently attracts both the methyltransferases DNMT3a and DNMT3b, suppressing in this way the OCT4 expression [111-113]. Alternatively, two other orphan receptors (they are referred to as receptors because the ligands have yet to be identified), of the chicken ovalbumin upstream promoter transcription factors (COUP-TFs) superfamily, COUP-TFI (EAR-3, NR2F1) and COUP-TFII (ARP-1, NR2F2) have been found to bind to the PP repressing the OCT4 expression [114], implying a major role of these receptors in various developmental processes [115]. Finally, phosphorylation within the homeobox region of OCT4 has been found to regulate its activity negatively by interrupting sequence-specific DNA binding [116].
Multiprotein repressive Polycomb group (PcG) in combination with the activating trithorax (TrxG) chromatin modyfing complexes has long been known to participate in the regulation of nuclear organization in order to activate or suppress their target-gene transcription, among which OCT4 is one of these genes [117]. The PcG complex is a chromatin regulator with its role restricted to establishing and maintaining the inactive status of genes during embryo development [118]. The Polycomb repressive complex 2 (PRC2) contains the embryonic ectoderm protein (EED), the suppressor of zeste 12 (SUZ12) and the enhancer of zeste 2 (EZH2), all of which function as histone methyltransferases providing either di or tri-methylation on histone H3 at K27 (H3K27me2/3) [119]. The other complex of the Polycomb family is PRC1 and its role is restricted to maintain genes in their silent status, which was originally achieved by the PRC2 [120]. Nucleosomes with tri-methylated histones (H3K27me3) attract the PRC1 complex through activation of the subunits RING1A and 1B, both of which belong to the E3 ligase family and perform monoubiquitination of H2A at the K119 [121]. Although a direct inter-relation between the two complexes has not yet been clarified, PRC1 has been found to interact with the major pluripotency gene network of OCT3, NANOG and SOX2 [122]. It is likely that both the Polycomb protein complexes are the major regulators for either differentiation or cell identity. Nevertheless, a relationship has been shown between PcGs and DNA methylation status [123]. Recently, it has been found that the protein JARID2 with histone demethylase activity is one of the components of the PRC2 complex [124-126]. It is worth mentioning that JARID2 has been reported to be an activator, as well as an inhibitor, of PRC2-mediated H3K27 methylation indicating a JARID-PRC2 duality [127]. JARID2 recruits PRC2 to target loci and with the interaction of MTF2 and esPRC2p48, stimulates synergistically the histone methyltransferase activity of PRC2. Taking all these mechanisms together, it appears that JARID contributes to the high levels of H3K27 methylation and gene activation or repression, even of these of plulipotency gene network, OCT4 and SOX2 [128].
In contrast to PcG complexes, the Trx group is involved in the transcriptional activity of the OCT4, SOX2 and KILF genes. The specific complex is implicated in post-transcriptional modifications, marking the open chromatin status, such as H3K4me3 [129]. Specifically, Wdr5, a key component of the TrxG, functions as a “presenter” of the H3K4 [130]. It was shown that Wdr5 interacts with H3K4me2 and mediates transition to the trimethylated state [131]. A complete loss of Wdr5 and thereby loss of the H3K4me3 mark, results in a lethal phenotype that bypasses self-renewal maintenance, indicating that Wdr5 directly acts through the loss of H3K4me3 [132]. Interconnectivity between the core transcriptional network (OCT4, SOX2, NANOG) and members of the trxG complex has been also proposed. OCT4 enhances basal Wdr5 through direct binding and transcriptional activation of its promoter. Afterwards, the DNA specificity conferred by OCT4 directs Wdr5 to genomic loci encoding self-renewal genes, such as OCT4 and NANOG, to re-establish a H3K4me3 chromatin signature. This elevated expression of H3K4me3 subsequently facilitates strong OCT4 occupancy to direct robust transcriptional activation [132]. Furthermore, Ino80, a chromatin remodeling ATPase, co-occupies pluripotency gene promoters with the master transcription factors, and its occupancy is dependent on OCT4 and Wdr5. Ino80 maintains a euchromatin state, thereby permitting RNA polymerase II for gene activation. These discoveries indicate a role for Ino80 in the expression of the pluripotency network and illustrate the coordination among chromatin remodeler, transcription factor and histone-modifying enzyme in the regulation of the pluripotent state [133]. Finally, histone acetyltransferase Mof appears to play an essential role in the maintenance of Embryonic Stem Cell (ESC) self-renewal and pluripotency, while the Mof deletion results in aberrant expression of the core transcription factors NANOG, OCT4, and SOX2, indicating that Mof is an integral component of the ESC core transcriptional network [134].
The achievement of reprogramming has drawn tremendous interest in the genetic field, using either the Yamanaka factors (OCT4, SOX2, KLF4 and c-MYC) [136]) or the Thomson factors (OCT4, SOX2, NANOG and LIN2) [137]. Both factors include the OCT4 and the SOX2 genes, but it is almost certain that other factors and family members facilitate the reprogramming process [138]. Nevertheless, OCT4 and NANOG have been recognized as master transcriptional organizers of ES cells [122, 139]. It seems that OCT4 and NANOG form a mutual interdependent transcriptional network with SOX2. OCT4 and SOX2 have been known to act synergistically to regulate their own transcription [140] as well as NANOG [141]. The pluripotency transcription factors form combinatorial complexes including OCT4-SOX2 [141], OCT4-NANOG [142] and NANOG-SALL4 [143], while recently it was demonstrated that the OCT4-NANOG-SALL4 network controls the developmental progress of the pre-implantation embryo [144].
During the exit of pluripotency and the beginning of the differentiation process, an acute decline in TET1/2 levels and an increase in DNMT3A/B enzymes have been observed [57], while it is has been demonstrated that at the 8-blastomere developmental stage, the DNMT1o is translocated into the nucleus of each blastomere [145]. During this transition, the nucleus of the majority of the cells seems to pass through an intermediate state of epigenetic priming that is characterized by high levels of DNMTs and TET enzymes [146]. Therefore, DNA methylation heterogeneity may result in transcriptional diversity, while it has been shown that DNA methylation dynamics may cause transcriptional changes and lineage choices in response to differentiated stimuli [147]. Several mechanisms have been proposed for creating DNA methylation heterogeneity [148], all of which very much depend on the balance between the expression of DNMTs and TETs [146]. Therefore, the maintenance of a hemi-methylated status and complete loss of DNA methylation may cause heterogenous methylation state at promoters, enhancers and other regulatory elements, modifying the transcriptional result in this way [148]. Besides, it has been demonstrated that most of the transcription factors display a methylation-sensitive sequence binding [149-151]. Primed cells are capable to diversify prior to lineage commitment through the mechanisms of gene transcription heterogeneity. This model allows cells to respond differently in various stimulations [148].
3. CONDITIONS THAT IMPACT ON DNA METHYLATION DURING PRE-IMPLANTATION EMBRYO DEVELOPMENT IN ASSISTED REPRODUCTIVE TECHNOLOGY (ART) CYCLES
Assisted reproductive technologies (ARTs) involve ovarian stimulation, gamete manipulation and embryo culture in order to improve fertility. Additionally, the ART itself encompasses the culture media and the embryo transfer procedure but not only these. It is obvious that during the various ART procedures, the gamete epigenomes are exposed to external stress factors that influence the establishment and the maintenance of genomic imprinting [152, 153], through alteration of the DNA methylation status (Table 1). Therefore, several reports have been published revealing the association of ART-conceived children and imprinting disorders [154, 155]. One of the issues that is relevant to the previous association is ovarian hyperstimulation. Taking into consideration that during oogenesis, acquisition of maternal DNA methylation begins in primary follicles and is completed in metaphase II (MII) oocytes, it is obvious that ovarian hyperstimulation may contribute in altering the methylation status, indicating superovulation as a potential imprinting disruptor. Hyperstimulation appears to impair imprinting acquisition of oocytes in a dose-dependent manner. It is possible that ovarian hyperstimulation may have adverse effects on the cytoplasmatic maternal factors (DPPA3, ZFP57, DMNT1) [156, 157]. In a recent study, hormonal induction resulted in a loss of maternal SNRPN, PEG3 and KCNQ1 methylation and a gain of maternal H19 methylation, with more embryos displaying altered imprinted methylation at high (10IU) compared with low (6.25IU) hormone treatment [157]. Furthermore, a subset of maternal transcripts is required for early developmental stages prior to embryonic gene activation [158, 159]. Does ovulation induction impact imprinting methylation maintenance since it is known that the oocyte defines the early methylation landscape? [160]. Although a recent study demonstrated that ovarian stimulation did not affect methylation status in the oocytes, it was shown in the same study that ovarian stimulation disrupts maternal-effects gene products essential for imprinting maintenance [161]. In a recent study, except for the dose of 0.075 IU/ml, the administration of increasing gonadotropin doses resulted in diverse methylation and de-methylation status of the H19, SNRPN, and PEG3 genes in MII oocytes. Furthermore, high gonadotropin concentrations caused hypo-methylation, while the genes that control spindle formation and cell-cycle were also downregulated [162]. Gathering these observations it is likely that ovulation induction has dual effects and functions via a dose-dependent manner, Firstly disrupting the imprinting acquisition during the growing phase of the oocyte and secondly, having a negative impact on maternal gene products that are required for imprinting maintenance.
Table 1.
The possible impact of various parameters of assisted reproductive technologies on imprinting disorders. Cumulatively all these factors may contribute to high risk of aberrant DNA methylation (the factors coming from ART and the relation to methylation have been discussed in the text in detail).
| Factors | Risk of Aberrant Methylation | References/comments |
|---|---|---|
| Age of woman | YES | Increased age |
| Age of man | MAYBE/NO | No reports |
| Ovarian stimulation | YES | [157, 162] |
| Semen sample | YES | [168-174]/decreased sperm parameters |
| Semen preparation | YES | No reports |
| Oocyte manipulation | YES | No reports |
| IVF insemination | NO | [185] |
| ICSI insemination | MAYBE/YES | [180] |
| Culture media | MAYBE/YES | [191-194] |
| Embryo | YES | [178, 179] |
| Temperature variations of incubators | MAYBE/YES | [177]/no actual reports |
| Causes of infertility | YES | [171, 206] |
There is growing evidence that the paternal epigenome plays a crucial role during embryo development. This role was confirmed by studies that analyzed sperm DNA methylation and histone modifications indicating the influence of paternal epigenome and transcriptome in early embryogenesis. Alterations in one of the above epigenetic marks have been shown to correlate with poor spermatogenesis, decreased fertility parameters, reduced fertilization potential and pregnancy outcome [163-165]. DNA methylation has been proved to contribute to male infertility as proved by experimental studies in knockout sperm mouse models for DNA methyltransferases [166]. Moreover, human sperm samples collected from IVF programs found that DNA methylation may influence embryo development and subsequently may associate with reduced pregnancy outcome, owing this result to the global sperm DNA hypo-methylation status [167]. A recent study attributes the production of poor embryo quality in the atypical distribution of retained histones in the sperm samples. Besides, poor embryo quality is often seen after ICSI techniques with round spermatids, because they carry alterations in the DNA methylation. The immature epigenetic landscape of these samples is unable to deposit proper methylation status in the embryo, resulting in poor embryo quality [168-170]. Some studies found increased percent of abberations in DNA methylation status in oligozoospermia samples, while similar aberrations were observed in severe, very severe olizoospermia and asheno-teratozoospermia samples [171-174]. High methylation of the Oct4 promoter was significantly more pronounced in embryos from couples where a male factor was the only known cause of infertility. Imprinting errors originating from sperm samples may be transmitted to fetuses through abnormal histone to protamine transition [175, 176]. It is worth mentioning that manipulations of sperm samples during the IVF procedure, such as various sperm preparation protocols (percoll or swim up), have not yet been done in order to examine the possible influence of these manipulations on sperm methylation landscape. Moreover, external environmental factors, like culture media, centrifugation conditions may influence the DNA methylation status of the sperm sample, while subtle temperature variations inside commercial IVF incubators, have been proved to impair embryo quality and probably fertilization ability of both gametes [177]. Given that a part of the sperm epigenome is essential for the early stages of embryo development, the subjection of the sperm sample to preparation protocols in combination with the culture media used, exposes the spermatozoa to diverse environmental conditions that may alter significant paternal epigenome or the sperm transcriptome and therefore may impair fertilization, embryo quality and even pregnancy outcome [163-165].
Besides, it is well known that embryo quality is strongly associated with IVF success. Even for those embryos that morphologically are of good quality, it is possible that they may possess aberrations in DNA methylation. These aberrations may be not visible and embryos may reach the blastocyst stage. Nevertheless, as mentioned above, these alterations may occur at the phase of lineage decision, or by the apoptosis process. Although a clear association between apoptosis and DNA methylation has not yet been defined, it is likely that alterations in DNA methylation precede the process of apoptosis. Apoptosis may be visibly determined by the presence of fragmentation (cellular parts without nucleus). Many embryos in IVF programs exhibit various amounts of fragmentation from the first divisions where DNA de-methylation occurs. Whether global incomplete DNA de-methylation induces the apoptosis/fragmentation process, is not known. Taking into consideration that both gametes and embryos are subjected to various manipulations, plus the fact that maternal factors and genes contribute to the developmental competence of the zygote, one can speculate that alterations in DNA methylation and apoptosis in terms of fragmentation, are procedures that associate exhibiting inter-dependency. Recently, it was found that ovarian stimulation induces elevated follicular fluid oocyte/cumulus free DNA concentrations and negative impact on embryo quality [178]. High apoptotic levels in the follicular fluid, exert through apoptosis negative effect on embryo quality and this negative effect started from the oocytes and proceeded to the zygote. It is possible that this negative effect includes aberrations in DNA methylation, although some new evidence has emerged [179]. Moreover, the OCT4-SALL4-NANOG network appears to control the developmental progression of the pre-implantation embryo, and alterations in DNA methylation status of these genes result in embryo developmental arrest, something which coincides with the presence of high fragmentation percent [144]. It is very much possible that upon dis-regulation of the gene network, the embryos exhibit high percent of fragmentation which is visible from the early stages of development.
Alterations in DNA methylation may derive either from the ICSI procedure during the insemination process or by the culture media during the post-zygotic divisions. Although the ICSI technique itself is not an invasive one, some reports indicate the association of the ICSI technique and aberrant DNA methylation during development [180]. An alteration of gene expression of ICSI-conceived pups was shown, suggesting that the major effect of ICSI at the neonatal stage reflects the consequences of a gene expression cascade starting from a change initially introduced in a small number of genes at the fertilization process [181]. Nevertheless, the ICSI-conceived mice were phenotypically normal [180]. Interestingly, ICSI produced a delay of two hours on active de-methylation of the paternal pronucleus attributed to the sperm DNA fragmentation and to the ICSI itself [182], while ICSI-derived gene expression changes were also observed at the blastocyst stage [183]. Given that male gene expression starts earlier in the zygote than in the female [184], the question that arises is whether ICSI exerts any impact on the paternally or on the maternally derived allele. It has been proposed that ICSI exhibits an impact on zygote gene activation but of paternally derived genomes [185], maybe due to direct impact on 5hmC [186] or to the alteration in calcium oscillation [187], because ICSI bypasses the acrosome reaction process.
Other issues are the culture media, the days of culture and thereby the embryo transfer day. These issues seem to be very crucial since the reprogramming, gene expression and morphological changes take place during the first divisions. Given that human experiments are unethical in some countries and difficult to perform in some others, because there are no spare embryos to analyze or the embryos that are subjected to the analysis are of poor quality. Most studies are from mice which exhibit similar methylation patterns and imprinting methylation performance with humans [160, 188-190]. It has been demonstrated that the culture of embryos has negative effects on embryo development on both mouse and human embryos [191, 192], while some media have been proposed to maintain imprinting methylation [193, 194]. Nevertheless, accelerated developmental rates, which in most cases underlie excellent quality, have been associated with increased frequency of imprinting DNA methylation errors [195], proposing that faster development rates correlate with loss of genomic imprinting. In poor quality human embryos, aberrant methylation of H19 DMR was found [196], suggesting that if no other embryo transfer options are available, embryo transfer should not be performed because these embryos may exhibit increased percentage of imprinting errors. While another study came to the same conclusion, although morphologically similar quality day 3 embryos showed different methylation levels [197]. Moreover, it has been observed that changes in DNA methylation during the first divisions depend on gender, while the same study found that methylation levels were higher in the male bovine blastocysts compared to females, while DNA was more methylated in the 6-8 blastomere-stage female embryos [198]. Gathering all these observations it is obvious that suboptimal culture media may exert alterations in DNA imprinting methylation. Whether embryo transfer should be performed on day 2, 3 or at the blastocyst day according to human embryo transfer studies, it is better to transfer a blastocyst, but on the other hand, the risk of extended culture may cause DNA methylation aberrations. External effects that take place during fertilization and the subsequent cell divisions may induce long-lasting consequences on gene expression. These consequences maybe observed in the later stages of development, such as blastocyst stage. The conventional IVF technique appears to be a safe method, without emerging alterations in DNA methylation when embryos are cultured for only one day [185]. Culture media have been divided in two categories: the sequential, where media are defined according to the needs of the embryos and therefore different culture media are essential for fertilization through developing to 4-8 blastomeres and different media to blastocyst and the non-sequential, where the embryos are cultured in one medium allowing the embryo to use its own needs. To date, there is still a debate which media are more appropriate for embryo development. A disadvantage of the first media is that embryos are exposed to environmental stress (alter the optimal temperature due to the opening-closing of the incubators, light exposure ect) and consequently may create alterations in DNA methylation status. Recently, with the help of Embryoscope (monitoring of embryo development), two models of predicting excellent quality embryos [199, 200] in the very early stages of development (2-3 days post-fertilization) have been proposed. If this is the case then it is very much possible that embryos are not subjected to extended embryo culture, eliminating the risk of aberrations in DNA methylation.
Accumulating evidence indicates that the ambient environment of the developing embryos derived from ART may possess aberrations in DNA imprinting methylation. For that reason, many studies have focused on the association between DNA methylation errors and imprinting disorders in children born after ART [201]. The most prevalent imprinting disorders relative to ART concern the Bekwith-Wiedemann syndrome (BWS), the Silver-Russel syndrome (SRS) and the Angelman syndrome (AS) [202, 203]. The AS is caused by aberrant methylation of 2%-5% in the maternal allele, while BWS involves genetic or epigenetic alterations of the maternal allele with hyper-methylation on the H19 locus or hypo-methylation on LIT1. Finally, the SRS involves the hypo-methylation of H19 at the chromosome 11p15.5 [204]. Among the above imprinting disorders, DNA methylation errors are more prevalent in the SRS patients rather than in AS and BWS patients. Interestingly, when clinical features were compared between ART- and natural-conceived children, no significant differences were found [205] but it is possible that the resulting imprinting disorders derived from ART were due to the frequency and the extent of alterations in DNA methylation. The impact of ART on DNA imprinting methylation and vise versa cannot be evaluated because these patients differ mainly genetically and demographically. Moreover, couples seeking ART encounter different fertility problems, which may contribute variously to imprinting DNA methylation, something which was observed at least in oligozoospermia samples [171, 206]. Although an increase in imprinting disorders in children conceived through IVF and ICSI was reported, the heterogeneity in the types of fertility treatment, the imprinted regions studied and the methods of measurement reduce the ability to assess the full effect of ART on DNA methylation and imprinting [207]. DNA methylation, determined by pyrosequencing, in the imprinted gene, small nuclear ribonucleoprotein polypeptide N (SNRPN), was higher in children conceived by ICSI [208], indicating a role of ART in imprinting disorders [209]. Although small significant differences in several imprinting control regions were observed, these small DNA methylation changes did not reflect to the overall transcriptional levels of the genes adjacent to the ICRs (such as KCNQ1 and SNRPN), suggesting that methylation levels at imprinting control regions are not altered with ovulation induction or in in vitro fertilization [210], something which was supported by other reports [211, 212], indicating that defects in imprinting and genome-wide DNA methylation are not common in the in vitro fertilization population.
Conclusively, there is a surrounding suspicion between ART, infertility and aberrant DNA methylation in imprinting disorders. Whether the ART procedure itself, including manipulations of gametes, the culture media, the extended culture, the ovarian stimulation protocols or the infertility background of the couples remain to be elucidated, since there are reports with different conclusions, maybe due to the various genetic and demographic study cases. More studies therefore, are needed to ascertain if any association exists. The methylation – de-methylation cycle has been clarified in a great extent but some paths remain controversial and elusive. OCT4 gene has been proved from the majority of the reports to be the master gene of pluripotency, which in combination with the NANOG and SOX2 or SALL4 genes, creates a pluripotency network. Whether totipotency restoring is through epigenetic reprogramming, such as DNA de-methylation or histone modifications, is really a hot spot of epigenetics.
ACKNOWLEDGEMENTS
Declared none.
CONFLICT OF INTEREST
The author(s) confirm that this article content has no conflict of interest.
REFERENCES
- 1.Bird A.P., Wolffe A.P. Methylation-induced repression--belts, braces, and chromatin. Cell. 1999;99(5):451–454. doi: 10.1016/S0092-8674(00)81532-9. [DOI] [PubMed] [Google Scholar]
- 2.Jones P.A., Takai D. The role of DNA methylation in mammalian epigenetics. Science. 2001;293(5532):1068–1070. doi: 10.1126/science.1063852. [DOI] [PubMed] [Google Scholar]
- 3.Bird A. DNA methylation patterns and epigenetic memory. Genes Dev. 2002;16(1):6–21. doi: 10.1101/gad.947102. [DOI] [PubMed] [Google Scholar]
- 4.Edwards C.A., Ferguson-Smith A.C. Mechanisms regulating imprinted genes in clusters. Curr. Opin. Cell Biol. 2007;19(3):281–289. doi: 10.1016/j.ceb.2007.04.013. [DOI] [PubMed] [Google Scholar]
- 5.Surani M.A. Imprinting and the initiation of gene silencing in the germ line. Cell. 1998;93(3):309–312. doi: 10.1016/S0092-8674(00)81156-3. [DOI] [PubMed] [Google Scholar]
- 6.Tilghman S.M. The sins of the fathers and mothers: genomic imprinting in mammalian development. Cell. 1999;96(2):185–193. doi: 10.1016/S0092-8674(00)80559-0. [DOI] [PubMed] [Google Scholar]
- 7.McLay D.W., Clarke H.J. Remodelling the paternal chromatin at fertilization in mammals. Reproduction. 2003;125(5):625–633. doi: 10.1530/rep.0.1250625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mayer W., Niveleau A., Walter J., Fundele R., Haaf T. Demethylation of the zygotic paternal genome. Nature. 2000;403(6769):501–502. doi: 10.1038/35000656. [DOI] [PubMed] [Google Scholar]
- 9.Dean W., Santos F., Reik W. Epigenetic reprogramming in early mammalian development and following somatic nuclear transfer. Semin. Cell Dev. Biol. 2003;14(1):93–100. doi: 10.1016/S1084-9521(02)00141-6. [DOI] [PubMed] [Google Scholar]
- 10.Santos F., Hendrich B., Reik W., Dean W. Dynamic reprogramming of DNA methylation in the early mouse embryo. Dev. Biol. 2002;241(1):172–182. doi: 10.1006/dbio.2001.0501. [DOI] [PubMed] [Google Scholar]
- 11.Dean W., Santos F., Stojkovic M., Zakhartchenko V., Walter J., Wolf E., Reik W. Conservation of methylation reprogramming in mammalian development: aberrant reprogramming in cloned embryos. Proc. Natl. Acad. Sci. USA. 2001;98(24):13734–13738. doi: 10.1073/pnas.241522698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rougier N., Bourc’his D., Gomes D.M., Niveleau A., Plachot M., Pàldi A., Viegas-Péquignot E. Chromosome methylation patterns during mammalian preimplantation development. Genes Dev. 1998;12(14):2108–2113. doi: 10.1101/gad.12.14.2108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ginsburg M., Snow M.H., McLaren A. Primordial germ cells in the mouse embryo during gastrulation. Development. 1990;110(2):521–528. doi: 10.1242/dev.110.2.521. [DOI] [PubMed] [Google Scholar]
- 14.McLaren A. Primordial germ cells in the mouse. Dev. Biol. 2003;262(1):1–15. doi: 10.1016/S0012-1606(03)00214-8. [DOI] [PubMed] [Google Scholar]
- 15.Reik W., Walter J. Genomic imprinting: parental influence on the genome. Nat. Rev. Genet. 2001;2(1):21–32. doi: 10.1038/35047554. [DOI] [PubMed] [Google Scholar]
- 16.Hajkova P., Erhardt S., Lane N., Haaf T., El-Maarri O., Reik W., Walter J., Surani M.A. Epigenetic reprogramming in mouse primordial germ cells. Mech. Dev. 2002;117(1-2):15–23. doi: 10.1016/S0925-4773(02)00181-8. [DOI] [PubMed] [Google Scholar]
- 17.Li J.Y., Lees-Murdock D.J., Xu G.L., Walsh C.P. Timing of establishment of paternal methylation imprints in the mouse. Genomics. 2004;84(6):952–960. doi: 10.1016/j.ygeno.2004.08.012. [DOI] [PubMed] [Google Scholar]
- 18.Szabó P.E., Mann J.R., Pfeifer G.P. Methylation analysis by chemical DNA sequencing. Methods Mol. Biol. 2002;200:29–41. doi: 10.1385/1-59259-182-5:029. [DOI] [PubMed] [Google Scholar]
- 19.Davis T.L., Trasler J.M., Moss S.B., Yang G.J., Bartolomei M.S. Acquisition of the H19 methylation imprint occurs differentially on the parental alleles during spermatogenesis. Genomics. 1999;58(1):18–28. doi: 10.1006/geno.1999.5813. [DOI] [PubMed] [Google Scholar]
- 20.Davis T.L., Yang G.J., McCarrey J.R., Bartolomei M.S. The H19 methylation imprint is erased and re-established differentially on the parental alleles during male germ cell development. Hum. Mol. Genet. 2000;9(19):2885–2894. doi: 10.1093/hmg/9.19.2885. [DOI] [PubMed] [Google Scholar]
- 21.Lees-Murdock D.J., De Felici M., Walsh C.P. Methylation dynamics of repetitive DNA elements in the mouse germ cell lineage. Genomics. 2003;82(2):230–237. doi: 10.1016/S0888-7543(03)00105-8. [DOI] [PubMed] [Google Scholar]
- 22.Lucifero D., Mertineit C., Clarke H.J., Bestor T.H., Trasler J.M. Methylation dynamics of imprinted genes in mouse germ cells. Genomics. 2002;79(4):530–538. doi: 10.1006/geno.2002.6732. [DOI] [PubMed] [Google Scholar]
- 23.Lucifero D., Mann M.R., Bartolomei M.S., Trasler J.M. Gene-specific timing and epigenetic memory in oocyte imprinting. Hum. Mol. Genet. 2004;13(8):839–849. doi: 10.1093/hmg/ddh104. [DOI] [PubMed] [Google Scholar]
- 24.Obata Y., Kono T. Maternal primary imprinting is established at a specific time for each gene throughout oocyte growth. J. Biol. Chem. 2002;277(7):5285–5289. doi: 10.1074/jbc.M108586200. [DOI] [PubMed] [Google Scholar]
- 25.Anifandis G., Messini C., Dafopoulos K., Sotiriou S., Messinis I. Molecular and cellular mechanisms of sperm-oocyte interactions opinions relative to in vitro fertilization (IVF). Int. J. Mol. Sci. 2014;15(7):12972–12997. doi: 10.3390/ijms150712972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kobayashi H., Sakurai T., Imai M., Takahashi N., Fukuda A., Yayoi O., Sato S., Nakabayashi K., Hata K., Sotomaru Y., Suzuki Y., Kono T. Contribution of intragenic DNA methylation in mouse gametic DNA methylomes to establish oocyte-specific heritable marks. PLoS Genet. 2012;8(1):e1002440. doi: 10.1371/journal.pgen.1002440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Smith Z.D., Meissner A. DNA methylation: roles in mammalian development. Nat. Rev. Genet. 2013;14(3):204–220. doi: 10.1038/nrg3354. [DOI] [PubMed] [Google Scholar]
- 28.Wang L., Zhang J., Duan J., Gao X., Zhu W., Lu X., Yang L., Zhang J., Li G., Ci W., Li W., Zhou Q., Aluru N., Tang F., He C., Huang X., Liu J. Programming and inheritance of parental DNA methylomes in mammals. Cell. 2014;157(4):979–991. doi: 10.1016/j.cell.2014.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Oswald J., Engemann S., Lane N., Mayer W., Olek A., Fundele R., Dean W., Reik W., Walter J. Active demethylation of the paternal genome in the mouse zygote. Curr. Biol. 2000;10(8):475–478. doi: 10.1016/S0960-9822(00)00448-6. [DOI] [PubMed] [Google Scholar]
- 30.Beaujean N., Hartshorne G., Cavilla J., Taylor J., Gardner J., Wilmut I., Meehan R., Young L. Non-conservation of mammalian preimplantation methylation dynamics. Curr. Biol. 2004;14(7):R266–R267. doi: 10.1016/j.cub.2004.03.019. [DOI] [PubMed] [Google Scholar]
- 31.Fulka H., Mrazek M., Tepla O., Fulka J., Jr DNA methylation pattern in human zygotes and developing embryos. Reproduction. 2004;128(6):703–708. doi: 10.1530/rep.1.00217. [DOI] [PubMed] [Google Scholar]
- 32.Lane N., Dean W., Erhardt S., Hajkova P., Surani A., Walter J., Reik W. Resistance of IAPs to methylation reprogramming may provide a mechanism for epigenetic inheritance in the mouse. Genesis. 2003;35(2):88–93. doi: 10.1002/gene.10168. [DOI] [PubMed] [Google Scholar]
- 33.Cortázar D., Kunz C., Saito Y., Steinacher R., Schär P. The enigmatic thymine DNA glycosylase. DNA Repair (Amst.) 2007;6(4):489–504. doi: 10.1016/j.dnarep.2006.10.013. [DOI] [PubMed] [Google Scholar]
- 34.Rai K., Huggins I.J., James S.R., Karpf A.R., Jones D.A., Cairns B.R. DNA demethylation in zebrafish involves the coupling of a deaminase, a glycosylase, and gadd45. Cell. 2008;135(7):1201–1212. doi: 10.1016/j.cell.2008.11.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhu J.K. Active DNA demethylation mediated by DNA glycosylases. Annu. Rev. Genet. 2009;43:143–166. doi: 10.1146/annurev-genet-102108-134205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jin S.G., Guo C., Pfeifer G.P. GADD45A does not promote DNA demethylation. PLoS Genet. 2008;4(3):e1000013. doi: 10.1371/journal.pgen.1000013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Engel N., Tront J.S., Erinle T., Nguyen N., Latham K.E., Sapienza C., Hoffman B., Liebermann D.A. Conserved DNA methylation in Gadd45a(-/-) mice. Epigenetics. 2009;4(2):98–99. doi: 10.4161/epi.4.2.7858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kriaucionis S., Heintz N. The nuclear DNA base 5-hydroxymethylcytosine is present in Purkinje neurons and the brain. Science. 2009;324(5929):929–930. doi: 10.1126/science.1169786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tahiliani M., Koh K.P., Shen Y., Pastor W.A., Bandukwala H., Brudno Y., Agarwal S., Iyer L.M., Liu D.R., Aravind L., Rao A. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. 2009;324(5929):930–935. doi: 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hajkova P., Jeffries S.J., Lee C., Miller N., Jackson S.P., Surani M.A. Genome-wide reprogramming in the mouse germ line entails the base excision repair pathway. Science. 2010;329(5987):78–82. doi: 10.1126/science.1187945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wossidlo M., Arand J., Sebastiano V., Lepikhov K., Boiani M., Reinhardt R., Schöler H., Walter J. Dynamic link of DNA demethylation, DNA strand breaks and repair in mouse zygotes. EMBO J. 2010;29(11):1877–1888. doi: 10.1038/emboj.2010.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gu T.P., Guo F., Yang H., Wu H.P., Xu G.F., Liu W., Xie Z.G., Shi L., He X., Jin S.G., Iqbal K., Shi Y.G., Deng Z., Szabó P.E., Pfeifer G.P., Li J., Xu G.L. The role of Tet3 DNA dioxygenase in epigenetic reprogramming by oocytes. Nature. 2011;477(7366):606–610. doi: 10.1038/nature10443. [DOI] [PubMed] [Google Scholar]
- 43.Iqbal K., Jin S.G., Pfeifer G.P., Szabó P.E. Reprogramming of the paternal genome upon fertilization involves genome-wide oxidation of 5-methylcytosine. Proc. Natl. Acad. Sci. USA. 2011;108(9):3642–3647. doi: 10.1073/pnas.1014033108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Santos F., Peat J., Burgess H., Rada C., Reik W., Dean W. Active demethylation in mouse zygotes involves cytosine deamination and base excision repair. Epigenetics Chromatin. 2013;6(1):39. doi: 10.1186/1756-8935-6-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dalton S.R., Bellacosa A. DNA demethylation by TDG. Epigenomics. 2012;4(4):459–467. doi: 10.2217/epi.12.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kohli R.M., Zhang Y. TET enzymes, TDG and the dynamics of DNA demethylation. Nature. 2013;502(7472):472–479. doi: 10.1038/nature12750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Okada Y., Yamagata K., Hong K., Wakayama T., Zhang Y. A role for the elongator complex in zygotic paternal genome demethylation. Nature. 2010;463(7280):554–558. doi: 10.1038/nature08732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Creppe C., Buschbeck M. Elongator: an ancestral complex driving transcription and migration through protein acetylation. 2011. [DOI] [PMC free article] [PubMed]
- 49.Wu S.C., Zhang Y. Active DNA demethylation: many roads lead to Rome. Nat. Rev. Mol. Cell Biol. 2010;11(9):607–620. doi: 10.1038/nrm2950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Inoue A., Zhang Y. Replication-dependent loss of 5-hydroxymethylcytosine in mouse preimplantation embryos. Science. 2011;334(6053):194. doi: 10.1126/science.1212483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Inoue A., Shen L., Dai Q., He C., Zhang Y. Generation and replication-dependent dilution of 5fC and 5caC during mouse preimplantation development. Cell Res. 2011;21(12):1670–1676. doi: 10.1038/cr.2011.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhang P., Su L., Wang Z., Zhang S., Guan J., Chen Y., Yin Y., Gao F., Tang B., Li Z. The involvement of 5-hydroxymethylcytosine in active DNA demethylation in mice. Biol. Reprod. 2012;86(4):104. doi: 10.1095/biolreprod.111.096073. [DOI] [PubMed] [Google Scholar]
- 53.Howell C.Y., Bestor T.H., Ding F., Latham K.E., Mertineit C., Trasler J.M., Chaillet J.R. Genomic imprinting disrupted by a maternal effect mutation in the Dnmt1 gene. Cell. 2001;104(6):829–838. doi: 10.1016/S0092-8674(01)00280-X. [DOI] [PubMed] [Google Scholar]
- 54.Bestor T.H. The DNA methyltransferases of mammals. Hum. Mol. Genet. 2000;9(16):2395–2402. doi: 10.1093/hmg/9.16.2395. [DOI] [PubMed] [Google Scholar]
- 55.Margot J.B., Ehrenhofer-Murray A.E., Leonhardt H. Interactions within the mammalian DNA methyltransferase family. 2003. [DOI] [PMC free article] [PubMed]
- 56.Hirasawa R., Sasaki H. Dynamic transition of Dnmt3b expression in mouse pre- and early post-implantation embryos. Gene Expr. Patterns. 2009;9(1):27–30. doi: 10.1016/j.gep.2008.09.002. [DOI] [PubMed] [Google Scholar]
- 57.Oda M., Oxley D., Dean W., Reik W. Regulation of lineage specific DNA hypomethylation in mouse trophectoderm. PLoS One. 2013;8(6):e68846. doi: 10.1371/journal.pone.0068846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Nakamura T., Liu Y.J., Nakashima H., Umehara H., Inoue K., Matoba S., Tachibana M., Ogura A., Shinkai Y., Nakano T. PGC7 binds histone H3K9me2 to protect against conversion of 5mC to 5hmC in early embryos. Nature. 2012;486(7403):415–419. doi: 10.1038/nature11093. [DOI] [PubMed] [Google Scholar]
- 59.Fitzpatrick G.V., Soloway P.D., Higgins M.J. Regional loss of imprinting and growth deficiency in mice with a targeted deletion of KvDMR1. Nat. Genet. 2002;32(3):426–431. doi: 10.1038/ng988. [DOI] [PubMed] [Google Scholar]
- 60.Lin S.P., Youngson N., Takada S., Seitz H., Reik W., Paulsen M., Cavaille J., Ferguson-Smith A.C. Asymmetric regulation of imprinting on the maternal and paternal chromosomes at the Dlk1-Gtl2 imprinted cluster on mouse chromosome 12. Nat. Genet. 2003;35(1):97–102. doi: 10.1038/ng1233. [DOI] [PubMed] [Google Scholar]
- 61.Williamson C.M., Turner M.D., Ball S.T., Nottingham W.T., Glenister P., Fray M., Tymowska-Lalanne Z., Plagge A., Powles-Glover N., Kelsey G., Maconochie M., Peters J. Identification of an imprinting control region affecting the expression of all transcripts in the Gnas cluster. Nat. Genet. 2006;38(3):350–355. doi: 10.1038/ng1731. [DOI] [PubMed] [Google Scholar]
- 62.Smallwood S.A., Kelsey G. De novo DNA methylation: a germ cell perspective. Trends Genet. 2012;28(1):33–42. doi: 10.1016/j.tig.2011.09.004. [DOI] [PubMed] [Google Scholar]
- 63.Tomizawa S., Kobayashi H., Watanabe T., Andrews S., Hata K., Kelsey G., Sasaki H. Dynamic stage-specific changes in imprinted differentially methylated regions during early mammalian development and prevalence of non-CpG methylation in oocytes. Development. 2011;138(5):811–820. doi: 10.1242/dev.061416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Cirio M.C., Ratnam S., Ding F., Reinhart B., Navara C., Chaillet J.R. Preimplantation expression of the somatic form of Dnmt1 suggests a role in the inheritance of genomic imprints. 2008. [DOI] [PMC free article] [PubMed]
- 65.Hirasawa R., Chiba H., Kaneda M., Tajima S., Li E., Jaenisch R., Sasaki H. Maternal and zygotic Dnmt1 are necessary and sufficient for the maintenance of DNA methylation imprints during preimplantation development. Genes Dev. 2008;22(12):1607–1616. doi: 10.1101/gad.1667008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kurihara Y., Kawamura Y., Uchijima Y., Amamo T., Kobayashi H., Asano T., Kurihara H. Maintenance of genomic methylation patterns during preimplantation development requires the somatic form of DNA methyltransferase 1. Dev. Biol. 2008;313(1):335–346. doi: 10.1016/j.ydbio.2007.10.033. [DOI] [PubMed] [Google Scholar]
- 67.Hanna C.W., Kelsey G. The specification of imprints in mammals. Heredity (Edinb) 2014;113(2):176–183. doi: 10.1038/hdy.2014.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Nakamura T., Arai Y., Umehara H., Masuhara M., Kimura T., Taniguchi H., Sekimoto T., Ikawa M., Yoneda Y., Okabe M., Tanaka S., Shiota K., Nakano T. PGC7/Stella protects against DNA demethylation in early embryogenesis. Nat. Cell Biol. 2007;9(1):64–71. doi: 10.1038/ncb1519. [DOI] [PubMed] [Google Scholar]
- 69.Saitou M., Barton S.C., Surani M.A. A molecular programme for the specification of germ cell fate in mice. Nature. 2002;418(6895):293–300. doi: 10.1038/nature00927. [DOI] [PubMed] [Google Scholar]
- 70.Payer B., Saitou M., Barton S.C., Thresher R., Dixon J.P., Zahn D., Colledge W.H., Carlton M.B., Nakano T., Surani M.A. Stella is a maternal effect gene required for normal early development in mice. Curr. Biol. 2003;13(23):2110–2117. doi: 10.1016/j.cub.2003.11.026. [DOI] [PubMed] [Google Scholar]
- 71.Abrink M., Ortiz J.A., Mark C., Sanchez C., Looman C., Hellman L., Chambon P., Losson R. Conserved interaction between distinct Krüppel-associated box domains and the transcriptional intermediary factor 1 beta. Proc. Natl. Acad. Sci. USA. 2001;98(4):1422–1426. doi: 10.1073/pnas.041616998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Mackay D.J., Callaway J.L., Marks S.M., White H.E., Acerini C.L., Boonen S.E., Dayanikli P., Firth H.V., Goodship J.A., Haemers A.P., Hahnemann J.M., Kordonouri O., Masoud A.F., Oestergaard E., Storr J., Ellard S., Hattersley A.T., Robinson D.O., Temple I.K. Hypomethylation of multiple imprinted loci in individuals with transient neonatal diabetes is associated with mutations in ZFP57. Nat. Genet. 2008;40(8):949–951. doi: 10.1038/ng.187. [DOI] [PubMed] [Google Scholar]
- 73.Liu Y., Toh H., Sasaki H., Zhang X., Cheng X. An atomic model of Zfp57 recognition of CpG methylation within a specific DNA sequence. Genes Dev. 2012;26(21):2374–2379. doi: 10.1101/gad.202200.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Tian X., Pascal G., Monget P. Evolution and functional divergence of NLRP genes in mammalian reproductive systems. BMC Evol. Biol. 2009;9:202. doi: 10.1186/1471-2148-9-202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Murdoch S., Djuric U., Mazhar B., Seoud M., Khan R., Kuick R., Bagga R., Kircheisen R., Ao A., Ratti B., Hanash S., Rouleau G.A., Slim R. Mutations in NALP7 cause recurrent hydatidiform moles and reproductive wastage in humans. Nat. Genet. 2006;38(3):300–302. doi: 10.1038/ng1740. [DOI] [PubMed] [Google Scholar]
- 76.Meyer E., Lim D., Pasha S., Tee L.J., Rahman F., Yates J.R., Woods C.G., Reik W., Maher E.R. Germline mutation in NLRP2 (NALP2) in a familial imprinting disorder (Beckwith-Wiedemann Syndrome). PLoS Genet. 2009;5(3):e1000423. doi: 10.1371/journal.pgen.1000423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Messerschmidt D.M., de Vries W., Ito M., Solter D., Ferguson-Smith A., Knowles B.B. Trim28 is required for epigenetic stability during mouse oocyte to embryo transition. Science. 2012;335(6075):1499–1502. doi: 10.1126/science.1216154. [DOI] [PubMed] [Google Scholar]
- 78.Zuo X., Sheng J., Lau H.T., McDonald C.M., Andrade M., Cullen D.E., Bell F.T., Iacovino M., Kyba M., Xu G., Li X. Zinc finger protein ZFP57 requires its co-factor to recruit DNA methyltransferases and maintains DNA methylation imprint in embryonic stem cells via its transcriptional repression domain. J. Biol. Chem. 2012;287(3):2107–2118. doi: 10.1074/jbc.M111.322644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Mahadevan S., Wen S., Wan Y.W., Peng H.H., Otta S., Liu Z., Iacovino M., Mahen E.M., Kyba M., Sadikovic B., Van den Veyver I.B. NLRP7 affects trophoblast lineage differentiation, binds to overexpressed YY1 and alters CpG methylation. Hum. Mol. Genet. 2014;23(3):706–716. doi: 10.1093/hmg/ddt457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Hatanaka Y., Shimizu N., Nishikawa S., Tokoro M., Shin S.W., Nishihara T., Amano T., Anzai M., Kato H., Mitani T., Hosoi Y., Kishigami S., Matsumoto K. GSE is a maternal factor involved in active DNA demethylation in zygotes. PLoS One. 2013;8(4):e60205. doi: 10.1371/journal.pone.0060205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Bernstein B.E., Mikkelsen T.S., Xie X., Kamal M., Huebert D.J., Cuff J., Fry B., Meissner A., Wernig M., Plath K., Jaenisch R., Wagschal A., Feil R., Schreiber S.L., Lander E.S. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125(2):315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- 82.Kidder B.L., Palmer S. HDAC1 regulates pluripotency and lineage specific transcriptional networks in embryonic and trophoblast stem cells. Nucleic Acids Res. 2012;40(7):2925–2939. doi: 10.1093/nar/gkr1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kraushaar D.C., Zhao K. The epigenomics of embryonic stem cell differentiation. Int. J. Biol. Sci. 2013;9(10):1134–1144. doi: 10.7150/ijbs.7998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Lee M.P., DeBaun M.R., Mitsuya K., Galonek H.L., Brandenburg S., Oshimura M., Feinberg A.P. Loss of imprinting of a paternally expressed transcript, with antisense orientation to KVLQT1, occurs frequently in Beckwith-Wiedemann syndrome and is independent of insulin-like growth factor II imprinting. Proc. Natl. Acad. Sci. USA. 1999;96(9):5203–5208. doi: 10.1073/pnas.96.9.5203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Mancini-Dinardo D., Steele S.J., Levorse J.M., Ingram R.S., Tilghman S.M. Elongation of the Kcnq1ot1 transcript is required for genomic imprinting of neighboring genes. Genes Dev. 2006;20(10):1268–1282. doi: 10.1101/gad.1416906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Medvedev S.P., Pokushalov E.A., Zakian S.M. Epigenetics of pluripotent cells. Acta Naturae. 2012;4(4):28–46. [PMC free article] [PubMed] [Google Scholar]
- 87.Tollervey J.R., Lunyak V.V. Epigenetics: judge, jury and executioner of stem cell fate. Epigenetics. 2012;7(8):823–840. doi: 10.4161/epi.21141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Terranova R., Yokobayashi S., Stadler M.B., Otte A.P., van Lohuizen M., Orkin S.H., Peters A.H. Polycomb group proteins Ezh2 and Rnf2 direct genomic contraction and imprinted repression in early mouse embryos. Dev. Cell. 2008;15(5):668–679. doi: 10.1016/j.devcel.2008.08.015. [DOI] [PubMed] [Google Scholar]
- 89.Bartolomei M.S., Zemei S., Tilghman S.M. Parental imprinting of the mouse H19 gene. 1991. [DOI] [PubMed]
- 90.Tremblay K.D., Duran K.L., Bartolomei M.S. A 5′ 2-kilobase-pair region of the imprinted mouse H19 gene exhibits exclusive paternal methylation throughout development. Mol. Cell. Biol. 1997;17(8):4322–4329. doi: 10.1128/mcb.17.8.4322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Bell A.C., Felsenfeld G. Methylation of a CTCF-dependent boundary controls imprinted expression of the Igf2 gene. Nature. 2000;405(6785):482–485. doi: 10.1038/35013100. [DOI] [PubMed] [Google Scholar]
- 92.da Rocha S.T., Edwards C.A., Ito M., Ogata T., Ferguson-Smith A.C. Genomic imprinting at the mammalian Dlk1-Dio3 domain. Trends Genet. 2008;24(6):306–316. doi: 10.1016/j.tig.2008.03.011. [DOI] [PubMed] [Google Scholar]
- 93.Smith Z.D., Meissner A. DNA methylation: roles in mammalian development. Nat. Rev. Genet. 2013;14(3):204–220. doi: 10.1038/nrg3354. [DOI] [PubMed] [Google Scholar]
- 94.Costa Y., Ding J., Theunissen T.W., Faiola F., Hore T.A., Shliaha P.V., Fidalgo M., Saunders A., Lawrence M., Dietmann S., Das S., Levasseur D.N., Li Z., Xu M., Reik W., Silva J.C., Wang J. NANOG-dependent function of TET1 and TET2 in establishment of pluripotency. Nature. 2013;495(7441):370–374. doi: 10.1038/nature11925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Tantin D. Oct transcription factors in development and stem cells: insights and mechanisms. Development. 2013;140(14):2857–2866. doi: 10.1242/dev.095927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Boiani M., Schöler H.R. Regulatory networks in embryo-derived pluripotent stem cells. Nat. Rev. Mol. Cell Biol. 2005;6(11):872–884. doi: 10.1038/nrm1744. [DOI] [PubMed] [Google Scholar]
- 97.Niwa H. Open conformation chromatin and pluripotency. Genes Dev. 2007;21(21):2671–2676. doi: 10.1101/gad.1615707. [DOI] [PubMed] [Google Scholar]
- 98.Yeom Y.I., Fuhrmann G., Ovitt C.E., Brehm A., Ohbo K., Gross M., Hübner K., Schöler H.R. Germline regulatory element of Oct-4 specific for the totipotent cycle of embryonal cells. Development. 1996;122(3):881–894. doi: 10.1242/dev.122.3.881. [DOI] [PubMed] [Google Scholar]
- 99.Evans M.J., Kaufman M.H. Establishment in culture of pluripotential cells from mouse embryos. Nature. 1981;292(5819):154–156. doi: 10.1038/292154a0. [DOI] [PubMed] [Google Scholar]
- 100.Martin G.R. Isolation of a pluripotent cell line from early mouse embryos cultured in medium conditioned by teratocarcinoma stem cells. Proc. Natl. Acad. Sci. USA. 1981;78(12):7634–7638. doi: 10.1073/pnas.78.12.7634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Niwa H., Miyazaki J., Smith A.G. Quantitative expression of Oct-3/4 defines differentiation, dedifferentiation or self-renewal of ES cells. Nat. Genet. 2000;24(4):372–376. doi: 10.1038/74199. [DOI] [PubMed] [Google Scholar]
- 102.Feldman N., Gerson A., Fang J., Li E., Zhang Y., Shinkai Y., Cedar H., Bergman Y. G9a-mediated irreversible epigenetic inactivation of Oct-3/4 during early embryogenesis. Nat. Cell Biol. 2006;8(2):188–194. doi: 10.1038/ncb1353. [DOI] [PubMed] [Google Scholar]
- 103.Sinkkonen L., Hugenschmidt T., Berninger P., Gaidatzis D., Mohn F., Artus-Revel C.G., Zavolan M., Svoboda P., Filipowicz W. MicroRNAs control de novo DNA methylation through regulation of transcriptional repressors in mouse embryonic stem cells. Nat. Struct. Mol. Biol. 2008;15(3):259–267. doi: 10.1038/nsmb.1391. [DOI] [PubMed] [Google Scholar]
- 104.Bartel D.P. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136(2):215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Yang Z., Wu J. MicroRNAs and regenerative medicine. DNA Cell Biol. 2007;26(4):257–264. doi: 10.1089/dna.2006.0548. [DOI] [PubMed] [Google Scholar]
- 106.Stefani G., Slack F.J. Small non-coding RNAs in animal development. Nat. Rev. Mol. Cell Biol. 2008;9(3):219–230. doi: 10.1038/nrm2347. [DOI] [PubMed] [Google Scholar]
- 107.Parchem R.J., Ye J., Judson R.L., LaRussa M.F., Krishnakumar R., Blelloch A., Oldham M.C., Blelloch R. Two miRNA clusters reveal alternative paths in late-stage reprogramming. Cell Stem Cell. 2014;14(5):617–631. doi: 10.1016/j.stem.2014.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Ding L., Paszkowski-Rogacz M., Nitzsche A., Slabicki M.M., Heninger A.K., de Vries I., Kittler R., Junqueira M., Shevchenko A., Schulz H., Hubner N., Doss M.X., Sachinidis A., Hescheler J., Iacone R., Anastassiadis K., Stewart A.F., Pisabarro M.T., Caldarelli A., Poser I., Theis M., Buchholz F. A genome-scale RNAi screen for Oct4 modulators defines a role of the Paf1 complex for embryonic stem cell identity. Cell Stem Cell. 2009;4(5):403–415. doi: 10.1016/j.stem.2009.03.009. [DOI] [PubMed] [Google Scholar]
- 109.Chu X., Qin X., Xu H., Li L., Wang Z., Li F., Xie X., Zhou H., Shen Y., Long J. Structural insights into Paf1 complex assembly and histone binding. Nucleic Acids Res. 2013;41(22):10619–10629. doi: 10.1093/nar/gkt819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Li B., Tang N., Chen S., Li X., Huang X., Wang X., Sun F. G9a co-localized with histone H3 lysine 9 monomethylation but not dimethylation in a nuclear membrane-dependent manner during mouse preimplantation embryo development. J. Assist. Reprod. Genet. 2013;30(3):441–448. doi: 10.1007/s10815-012-9911-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Gu P., LeMenuet D., Chung A.C., Mancini M., Wheeler D.A., Cooney A.J. Orphan nuclear receptor GCNF is required for the repression of pluripotency genes during retinoic acid-induced embryonic stem cell differentiation. Mol. Cell. Biol. 2005;25(19):8507–8519. doi: 10.1128/MCB.25.19.8507-8519.2005. [DOI] [PMC free article] [PubMed] [Google Scholar] [Research Misconduct Found]
- 112.Sato N., Kondo M., Arai K. The orphan nuclear receptor GCNF recruits DNA methyltransferase for Oct-3/4 silencing. Biochem. Biophys. Res. Commun. 2006;344(3):845–851. doi: 10.1016/j.bbrc.2006.04.007. [DOI] [PubMed] [Google Scholar]
- 113.Wang Q., Cooney A.J. Revisiting the role of GCNF in embryonic development. Semin. Cell Dev. Biol. 2013;24(10-12):679–686. doi: 10.1016/j.semcdb.2013.08.003. [DOI] [PubMed] [Google Scholar]
- 114.Ben-Shushan E., Sharir H., Pikarsky E., Bergman Y. A dynamic balance between ARP-1/COUP-TFII, EAR-3/COUP-TFI, and retinoic acid receptor:retinoid X receptor heterodimers regulates Oct-3/4 expression in embryonal carcinoma cells. Mol. Cell. Biol. 1995;15(2):1034–1048. doi: 10.1128/mcb.15.2.1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Lin F.J., Qin J., Tang K., Tsai S.Y., Tsai M.J. Coup d’Etat: an orphan takes control. Endocr. Rev. 2011;32(3):404–421. doi: 10.1210/er.2010-0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Brumbaugh J., Hou Z., Russell J.D., Howden S.E., Yu P., Ledvina A.R., Coon J.J., Thomson J.A. Phosphorylation regulates human OCT4. Proc. Natl. Acad. Sci. USA. 2012;109(19):7162–7168. doi: 10.1073/pnas.1203874109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Schuettengruber B., Chourrout D., Vervoort M., Leblanc B., Cavalli G. Genome regulation by polycomb and trithorax proteins. Cell. 2007;128(4):735–745. doi: 10.1016/j.cell.2007.02.009. [DOI] [PubMed] [Google Scholar]
- 118.Ringrose L., Ehret H., Paro R. Distinct contributions of histone H3 lysine 9 and 27 methylation to locus-specific stability of polycomb complexes. Mol. Cell. 2004;16(4):641–653. doi: 10.1016/j.molcel.2004.10.015. [DOI] [PubMed] [Google Scholar]
- 119.Cao R., Wang L., Wang H., Xia L., Erdjument-Bromage H., Tempst P., Jones R.S., Zhang Y. Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science. 2002;298(5595):1039–1043. doi: 10.1126/science.1076997. [DOI] [PubMed] [Google Scholar]
- 120.Elderkin S., Maertens G.N., Endoh M., Mallery D.L., Morrice N., Koseki H., Peters G., Brockdorff N., Hiom K. A phosphorylated form of Mel-18 targets the Ring1B histone H2A ubiquitin ligase to chromatin. Mol. Cell. 2007;28(1):107–120. doi: 10.1016/j.molcel.2007.08.009. [DOI] [PubMed] [Google Scholar]
- 121.Jones A., Wang H. Polycomb repressive complex 2 in embryonic stem cells: an overview. Protein Cell. 2010;1(12):1056–1062. doi: 10.1007/s13238-010-0142-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Boyer L.A., Mathur D., Jaenisch R. Molecular control of pluripotency. Curr. Opin. Genet. Dev. 2006;16(5):455–462. doi: 10.1016/j.gde.2006.08.009. [DOI] [PubMed] [Google Scholar]
- 123.Viré E., Brenner C., Deplus R., Blanchon L., Fraga M., Didelot C., Morey L., Van Eynde A., Bernard D., Vanderwinden J.M., Bollen M., Esteller M., Di Croce L., de Launoit Y., Fuks F. The Polycomb group protein EZH2 directly controls DNA methylation. Nature. 2006;439(7078):871–874. doi: 10.1038/nature04431. [DOI] [PubMed] [Google Scholar]
- 124.Li G., Margueron R., Ku M., Chambon P., Bernstein B.E., Reinberg D. Jarid2 and PRC2, partners in regulating gene expression. Genes Dev. 2010;24(4):368–380. doi: 10.1101/gad.1886410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Peng J.C., Valouev A., Swigut T., Zhang J., Zhao Y., Sidow A., Wysocka J. Jarid2/Jumonji coordinates control of PRC2 enzymatic activity and target gene occupancy in pluripotent cells. Cell. 2009;139(7):1290–1302. doi: 10.1016/j.cell.2009.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Shen X., Kim W., Fujiwara Y., Simon M.D., Liu Y., Mysliwiec M.R., Yuan G.C., Lee Y., Orkin S.H. Jumonji modulates polycomb activity and self-renewal versus differentiation of stem cells. Cell. 2009;139(7):1303–1314. doi: 10.1016/j.cell.2009.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Herz H.M., Shilatifard A. The JARID2-PRC2 duality. Genes Dev. 2010;24(9):857–861. doi: 10.1101/gad.1921610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Zhang Z., Jones A., Sun C.W., Li C., Chang C.W., Joo H.Y., Dai Q., Mysliwiec M.R., Wu L.C., Guo Y., Yang W., Liu K., Pawlik K.M., Erdjument-Bromage H., Tempst P., Lee Y., Min J., Townes T.M., Wang H. PRC2 complexes with JARID2, MTF2, and esPRC2p48 in ES cells to modulate ES cell pluripotency and somatic cell reprogramming. Stem Cells. 2011;29(2):229–240. doi: 10.1002/stem.578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Ringrose L., Ehret H., Paro R. Distinct contributions of histone H3 lysine 9 and 27 methylation to locus-specific stability of polycomb complexes. Mol. Cell. 2004;16(4):641–653. doi: 10.1016/j.molcel.2004.10.015. [DOI] [PubMed] [Google Scholar]
- 130.Dou Y., Milne T.A., Ruthenburg A.J., Lee S., Lee J.W., Verdine G.L., Allis C.D., Roeder R.G. Regulation of MLL1 H3K4 methyltransferase activity by its core components. Nat. Struct. Mol. Biol. 2006;13(8):713–719. doi: 10.1038/nsmb1128. [DOI] [PubMed] [Google Scholar]
- 131.Wysocka J., Swigut T., Milne T.A., Dou Y., Zhang X., Burlingame A.L., Roeder R.G., Brivanlou A.H., Allis C.D. WDR5 associates with histone H3 methylated at K4 and is essential for H3 K4 methylation and vertebrate development. Cell. 2005;121(6):859–872. doi: 10.1016/j.cell.2005.03.036. [DOI] [PubMed] [Google Scholar]
- 132.Ang Y.S., Gaspar-Maia A., Lemischka I.R., Bernstein E. Stem cells and reprogramming: breaking the epigenetic barrier? Trends Pharmacol. Sci. 2011;32(7):394–401. doi: 10.1016/j.tips.2011.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Wang L., Du Y., Ward J.M., Shimbo T., Lackford B., Zheng X., Miao Y.L., Zhou B., Han L., Fargo D.C., Jothi R., Williams C.J., Wade P.A., Hu G. INO80 facilitates pluripotency gene activation in embryonic stem cell self-renewal, reprogramming, and blastocyst development. Cell Stem Cell. 2014;14(5):575–591. doi: 10.1016/j.stem.2014.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Li X., Li L., Pandey R., Byun J.S., Gardner K., Qin Z., Dou Y. The histone acetyltransferase MOF is a key regulator of the embryonic stem cell core transcriptional network. Cell Stem Cell. 2012;11(2):163–178. doi: 10.1016/j.stem.2012.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Sawan C., Hernandez-Vargas H., Murr R., Lopez F., Vaissière T., Ghantous A.Y., Cuenin C., Imbert J., Wang Z.Q., Ren B., Herceg Z. Histone acetyltransferase cofactor Trrap maintains self-renewal and restricts differentiation of embryonic stem cells. 2013. [DOI] [PubMed]
- 136.Takahashi K., Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126(4):663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 137.Yu J., Vodyanik M.A., Smuga-Otto K., Antosiewicz-Bourget J., Frane J.L., Tian S., Nie J., Jonsdottir G.A., Ruotti V., Stewart R., Slukvin I.I., Thomson J.A. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318(5858):1917–1920. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- 138.Feng B., Jiang J., Kraus P., Ng J.H., Heng J.C., Chan Y.S., Yaw L.P., Zhang W., Loh Y.H., Han J., Vega V.B., Cacheux-Rataboul V., Lim B., Lufkin T., Ng H.H. Reprogramming of fibroblasts into induced pluripotent stem cells with orphan nuclear receptor Esrrb. Nat. Cell Biol. 2009;11(2):197–203. doi: 10.1038/ncb1827. [DOI] [PubMed] [Google Scholar]
- 139.Kim J., Chu J., Shen X., Wang J., Orkin S.H. An extended transcriptional network for pluripotency of embryonic stem cells. Cell. 2008;132(6):1049–1061. doi: 10.1016/j.cell.2008.02.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Chew J.L., Loh Y.H., Zhang W., Chen X., Tam W.L., Yeap L.S., Li P., Ang Y.S., Lim B., Robson P., Ng H.H. Reciprocal transcriptional regulation of Pou5f1 and Sox2 via the Oct4/Sox2 complex in embryonic stem cells. Mol. Cell. Biol. 2005;25(14):6031–6046. doi: 10.1128/MCB.25.14.6031-6046.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Rodda D.J., Chew J.L., Lim L.H., Loh Y.H., Wang B., Ng H.H., Robson P. Transcriptional regulation of nanog by OCT4 and SOX2. J. Biol. Chem. 2005;280(26):24731–24737. doi: 10.1074/jbc.M502573200. [DOI] [PubMed] [Google Scholar]
- 142.Zhang L., Rayner S., Katoku-Kikyo N., Romanova L., Kikyo N. Successful co-immunoprecipitation of Oct4 and Nanog using cross-linking. Biochem. Biophys. Res. Commun. 2007;361(3):611–614. doi: 10.1016/j.bbrc.2007.07.089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Wu Q., Chen X., Zhang J., Loh Y.H., Low T.Y., Zhang W., Zhang W., Sze S.K., Lim B., Ng H.H. Sall4 interacts with Nanog and co-occupies Nanog genomic sites in embryonic stem cells. J. Biol. Chem. 2006;281(34):24090–24094. doi: 10.1074/jbc.C600122200. [DOI] [PubMed] [Google Scholar]
- 144.Tan M.H., Au K.F., Leong D.E., Foygel K., Wong W.H., Yao M.W. An Oct4-Sall4-Nanog network controls developmental progression in the pre-implantation mouse embryo. Mol. Syst. Biol. 2013;9:632. doi: 10.1038/msb.2012.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Ratnam S., Mertineit C., Ding F., Howell C.Y., Clarke H.J., Bestor T.H., Chaillet J.R., Trasler J.M. Dynamics of Dnmt1 methyltransferase expression and intracellular localization during oogenesis and preimplantation development. Dev. Biol. 2002;245(2):304–314. doi: 10.1006/dbio.2002.0628. [DOI] [PubMed] [Google Scholar]
- 146.Ficz G., Reik W. Reprogramming by cell fusion: boosted by Tets. Mol. Cell. 2013;49(6):1017–1018. doi: 10.1016/j.molcel.2013.03.014. [DOI] [PubMed] [Google Scholar]
- 147.Challen G.A., Sun D., Jeong M., Luo M., Jelinek J., Berg J.S., Bock C., Vasanthakumar A., Gu H., Xi Y., Liang S., Lu Y., Darlington G.J., Meissner A., Issa J.P., Godley L.A., Li W., Goodell M.A. Dnmt3a is essential for hematopoietic stem cell differentiation. Nat. Genet. 2012;44(1):23–31. doi: 10.1038/ng.1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Lee H.J., Hore T.A., Reik W. Reprogramming the methylome: erasing memory and creating diversity. Cell Stem Cell. 2014;14(6):710–719. doi: 10.1016/j.stem.2014.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Hu S., Wan J., Su Y., Song Q., Zeng Y., Nguyen H.N., Shin J., Cox E., Rho H.S., Woodard C., Xia S., Liu S., Lyu H., Ming G.L., Wade H., Song H., Qian J., Zhu H. DNA methylation presents distinct binding sites for human transcription factors. eLife. 2013;2:e00726. doi: 10.7554/eLife.00726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Iurlaro M., Ficz G., Oxley D., Raiber E.A., Bachman M., Booth M.J., Andrews S., Balasubramanian S., Reik W. A screen for hydroxymethylcytosine and formylcytosine binding proteins suggests functions in transcription and chromatin regulation. Genome Biol. 2013;14(10):R119. doi: 10.1186/gb-2013-14-10-r119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Spruijt C.G., Baymaz H.I., Vermeulen M. Identifying specific protein-DNA interactions using SILAC-based quantitative proteomics. Methods Mol. Biol. 2013;977:137–157. doi: 10.1007/978-1-62703-284-1_11. [DOI] [PubMed] [Google Scholar]
- 152.DeBaun M.R., Niemitz E.L., Feinberg A.P. Association of in vitro fertilization with Beckwith-Wiedemann syndrome and epigenetic alterations of LIT1 and H19. Am. J. Hum. Genet. 2003;72(1):156–160. doi: 10.1086/346031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Maher E.R., Afnan M., Barratt C.L. Epigenetic risks related to assisted reproductive technologies: epigenetics, imprinting, ART and icebergs? Hum. Reprod. 2003;18(12):2508–2511. doi: 10.1093/humrep/deg486. [DOI] [PubMed] [Google Scholar]
- 154.Le Bouc Y., Rossignol S., Azzi S., Steunou V., Netchine I., Gicquel C. Epigenetics, genomic imprinting and assisted reproductive technology. Ann. Endocrinol. (Paris) 2010;71(3):237–238. doi: 10.1016/j.ando.2010.02.004. [DOI] [PubMed] [Google Scholar]
- 155.Bowdin S., Allen C., Kirby G., Brueton L., Afnan M., Barratt C., Kirkman-Brown J., Harrison R., Maher E.R., Reardon W. A survey of assisted reproductive technology births and imprinting disorders. Hum. Reprod. 2007;22(12):3237–3240. doi: 10.1093/humrep/dem268. [DOI] [PubMed] [Google Scholar]
- 156.Denomme M.M., Mann M.R. Maternal control of genomic imprint maintenance. Reprod. Biomed. Online. 2013;27(6):629–636. doi: 10.1016/j.rbmo.2013.06.004. [DOI] [PubMed] [Google Scholar]
- 157.Market Velker BA, Denomme MM, Mann MR. Loss of genomic imprinting in mouse embryos with fast rates of preimplantation development in culture. Biol. Reprod. 2012;5(86):143–16. doi: 10.1095/biolreprod.111.096602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Sirard M.A. Factors affecting oocyte and embryo transcriptomes. Reprod. Domest. Anim. 2012;47(Suppl. 4):148–155. doi: 10.1111/j.1439-0531.2012.02069.x. [DOI] [PubMed] [Google Scholar]
- 159.Bruce A.W. Generating different genetic expression patterns in the early embryo: insights from the mouse model. Reprod. Biomed. Online. 2013;27(6):586–592. doi: 10.1016/j.rbmo.2013.03.024. [DOI] [PubMed] [Google Scholar]
- 160.Smith Z.D., Chan M.M., Mikkelsen T.S., Gu H., Gnirke A., Regev A., Meissner A. A unique regulatory phase of DNA methylation in the early mammalian embryo. Nature. 2012;484(7394):339–344. doi: 10.1038/nature10960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Denomme M.M., Zhang L., Mann M.R. Embryonic imprinting perturbations do not originate from superovulation-induced defects in DNA methylation acquisition. Fertil. Steril. 2011;96(3):734–738.e2. doi: 10.1016/j.fertnstert.2011.06.055. [DOI] [PubMed] [Google Scholar]
- 162.Lu C.L., Wang T.R., Yan L.Y., Xia X., Zhu X.H., Li R., Zhao H.C., Yan J., Yin T.L., Jin H.Y., Zhang Y., Zhang W.X., Feng H.L., Qiao J. Gonadotropin-mediated dynamic alterations during bovine oocyte maturation in vitro. Biol. Reprod. 2014;91(2):44. doi: 10.1095/biolreprod.114.117945. [DOI] [PubMed] [Google Scholar]
- 163.Aoki V.W., Liu L., Jones K.P., Hatasaka H.H., Gibson M., Peterson C.M., Carrell D.T. Sperm protamine 1/protamine 2 ratios are related to in vitro fertilization pregnancy rates and predictive of fertilization ability. Fertil. Steril. 2006;86(5):1408–1415. doi: 10.1016/j.fertnstert.2006.04.024. [DOI] [PubMed] [Google Scholar]
- 164.Oakes C.C., La Salle S., Smiraglia D.J., Robaire B., Trasler J.M. Developmental acquisition of genome-wide DNA methylation occurs prior to meiosis in male germ cells. Dev. Biol. 2007;307(2):368–379. doi: 10.1016/j.ydbio.2007.05.002. [DOI] [PubMed] [Google Scholar]
- 165.Glaser S., Lubitz S., Loveland K.L., Ohbo K., Robb L., Schwenk F., Seibler J., Roellig D., Kranz A., Anastassiadis K., Stewart A.F. The histone 3 lysine 4 methyltransferase, Mll2, is only required briefly in development and spermatogenesis. Epigenetics Chromatin. 2009;2(1):5. doi: 10.1186/1756-8935-2-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.La Salle S., Oakes C.C., Neaga O.R., Bourc’his D., Bestor T.H., Trasler J.M. Loss of spermatogonia and wide-spread DNA methylation defects in newborn male mice deficient in DNMT3L. BMC Dev. Biol. 2007;7:104. doi: 10.1186/1471-213X-7-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Benchaib M., Braun V., Ressnikof D., Lornage J., Durand P., Niveleau A., Guérin J.F. Influence of global sperm DNA methylation on IVF results. Hum. Reprod. 2005;20(3):768–773. doi: 10.1093/humrep/deh684. [DOI] [PubMed] [Google Scholar]
- 168.Hammoud S.S., Nix D.A., Hammoud A.O., Gibson M., Cairns B.R., Carrell D.T. Genome-wide analysis identifies changes in histone retention and epigenetic modifications at developmental and imprinted gene loci in the sperm of infertile men. Hum. Reprod. 2011;26(9):2558–2569. doi: 10.1093/humrep/der192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Kishigami S., Van Thuan N., Hikichi T., Ohta H., Wakayama S., Mizutani E., Wakayama T. Epigenetic abnormalities of the mouse paternal zygotic genome associated with microinsemination of round spermatids. Dev. Biol. 2006;289(1):195–205. doi: 10.1016/j.ydbio.2005.10.026. [DOI] [PubMed] [Google Scholar]
- 170.Saitou M., Kagiwada S., Kurimoto K. Epigenetic reprogramming in mouse pre-implantation development and primordial germ cells. Development. 2012;139(1):15–31. doi: 10.1242/dev.050849. [DOI] [PubMed] [Google Scholar]
- 171.Kobayashi H., Sato A., Otsu E., Hiura H., Tomatsu C., Utsunomiya T., Sasaki H., Yaegashi N., Arima T. Aberrant DNA methylation of imprinted loci in sperm from oligospermic patients. Hum. Mol. Genet. 2007;16(21):2542–2551. doi: 10.1093/hmg/ddm187. [DOI] [PubMed] [Google Scholar]
- 172.Marques C.J., Costa P., Vaz B., Carvalho F., Fernandes S., Barros A., Sousa M. Abnormal methylation of imprinted genes in human sperm is associated with oligozoospermia. Mol. Hum. Reprod. 2008;14(2):67–74. doi: 10.1093/molehr/gam093. [DOI] [PubMed] [Google Scholar]
- 173.Sato A., Hiura H., Okae H., Miyauchi N., Abe Y., Utsunomiya T., Yaegashi N., Arima T. Assessing loss of imprint methylation in sperm from subfertile men using novel methylation polymerase chain reaction Luminex analysis. Fertil. Steril. 2011;95(1):129–134, 134.e1-134.e4. doi: 10.1016/j.fertnstert.2010.06.076. [DOI] [PubMed] [Google Scholar]
- 174.Boissonnas C.C., Abdalaoui H.E., Haelewyn V., Fauque P., Dupont J.M., Gut I., Vaiman D., Jouannet P., Tost J., Jammes H. Specific epigenetic alterations of IGF2-H19 locus in spermatozoa from infertile men. Eur. J. Hum. Genet. 2010;18(1):73–80. doi: 10.1038/ejhg.2009.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Al-Khtib M., Blachère T., Guérin J.F., Lefèvre A. Methylation profile of the promoters of Nanog and Oct4 in ICSI human embryos. Hum. Reprod. 2012;27(10):2948–2954. doi: 10.1093/humrep/des284. [DOI] [PubMed] [Google Scholar]
- 176.Carell T. Catalysis and regulation. Curr. Opin. Struct. Biol. 2012;22(6):689–690. doi: 10.1016/j.sbi.2012.10.006. [DOI] [PubMed] [Google Scholar]
- 177.Anifandis G. Temperature variations inside commercial IVF incubators. J. Assist. Reprod. Genet. 2013;30(12):1587–1588. doi: 10.1007/s10815-013-0138-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Dimopoulou M, Anifandis G, Messini CI, Dafopoulos K, Kouris S, Sotiriou S, Satra M, Vamvakopoulos N, Messinis IE. Follicular fluid oocyte/cumulus-free DNA concentrations as a potential biomolecular marker of embryo quality and IVF outcome. Biomed. Res. Int. 2014;2014:289306. doi: 10.1155/2014/289306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Ju S., Rui R., Lu Q., Lin P., Guo H. Analysis of apoptosis and methyltransferase mRNA expression in porcine cloned embryos cultured in vitro. J. Assist. Reprod. Genet. 2010;27(1):49–59. doi: 10.1007/s10815-009-9378-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.de Waal E., Yamazaki Y., Ingale P., Bartolomei M.S., Yanagimachi R., McCarrey J.R. Gonadotropin stimulation contributes to an increased incidence of epimutations in ICSI-derived mice. Hum. Mol. Genet. 2012;21(20):4460–4472. doi: 10.1093/hmg/dds287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Kohda T., Ishino F. Embryo manipulation via assisted reproductive technology and epigenetic asymmetry in mammalian early development. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2013;368(1609):20120353. doi: 10.1098/rstb.2012.0353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Fernández-Gonzalez R., Moreira P.N., Pérez-Crespo M., Sánchez-Martín M., Ramirez M.A., Pericuesta E., Bilbao A., Bermejo-Alvarez P., de Dios Hourcade J., de Fonseca F.R., Gutiérrez-Adán A. Long-term effects of mouse intracytoplasmic sperm injection with DNA-fragmented sperm on health and behavior of adult offspring. Biol. Reprod. 2008;78(4):761–772. doi: 10.1095/biolreprod.107.065623. [DOI] [PubMed] [Google Scholar]
- 183.Giritharan G., Li M.W., Di Sebastiano F., Esteban F.J., Horcajadas J.A., Lloyd K.C., Donjacour A., Maltepe E., Rinaudo P.F. Effect of ICSI on gene expression and development of mouse preimplantation embryos. Hum. Reprod. 2010;25(12):3012–3024. doi: 10.1093/humrep/deq266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Aoki F., Worrad D.M., Schultz R.M. Regulation of transcriptional activity during the first and second cell cycles in the preimplantation mouse embryo. Dev. Biol. 1997;181(2):296–307. doi: 10.1006/dbio.1996.8466. [DOI] [PubMed] [Google Scholar]
- 185.Kohda T., Kishigami S., Kaneko-Ishino T., Wakayama T., Ishino F. Gene expression profile normalization in cloned mice by trichostatin A treatment. Cell. Reprogram. 2012;14(1):45–55. doi: 10.1089/cell.2011.0062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Yoshizawa Y., Kato M., Hirabayashi M., Hochi S. Impaired active demethylation of the paternal genome in pronuclear-stage rat zygotes produced by in vitro fertilization or intracytoplasmic sperm injection. Mol. Reprod. Dev. 2010;77(1):69–75. doi: 10.1002/mrd.21109. [DOI] [PubMed] [Google Scholar]
- 187.Ozil J.P., Banrezes B., Tóth S., Pan H., Schultz R.M. Ca2+ oscillatory pattern in fertilized mouse eggs affects gene expression and development to term. Dev. Biol. 2006;300(2):534–544. doi: 10.1016/j.ydbio.2006.08.041. [DOI] [PubMed] [Google Scholar]
- 188.Borgel J., Guibert S., Li Y., Chiba H., Schübeler D., Sasaki H., Forné T., Weber M. Targets and dynamics of promoter DNA methylation during early mouse development. Nat. Genet. 2010;42(12):1093–1100. doi: 10.1038/ng.708. [DOI] [PubMed] [Google Scholar]
- 189.El Hajj N., Trapphoff T., Linke M., May A., Hansmann T., Kuhtz J., Reifenberg K., Heinzmann J., Niemann H., Daser A., Eichenlaub-Ritter U., Zechner U., Haaf T. Limiting dilution bisulfite (pyro)sequencing reveals parent-specific methylation patterns in single early mouse embryos and bovine oocytes. Epigenetics. 2011;6(10):1176–1188. doi: 10.4161/epi.6.10.17202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Heinzmann J., Hansmann T., Herrmann D., Wrenzycki C., Zechner U., Haaf T., Niemann H. Epigenetic profile of developmentally important genes in bovine oocytes. Mol. Reprod. Dev. 2011;78(3):188–201. doi: 10.1002/mrd.21281. [DOI] [PubMed] [Google Scholar]
- 191.Khosla S., Dean W., Reik W., Feil R. Culture of preimplantation embryos and its long-term effects on gene expression and phenotype. Hum. Reprod. Update. 2001;7(4):419–427. doi: 10.1093/humupd/7.4.419. [DOI] [PubMed] [Google Scholar]
- 192.Chen S.L., Shi X.Y., Zheng H.Y., Wu F.R., Luo C. Aberrant DNA methylation of imprinted H19 gene in human preimplantation embryos. Fertil. Steril. 2010;94(6):2356–2358, 2358.e1. doi: 10.1016/j.fertnstert.2010.01.120. [DOI] [PubMed] [Google Scholar]
- 193.Fauque P., Jouannet P., Lesaffre C., Ripoche M.A., Dandolo L., Vaiman D., Jammes H. Assisted Reproductive Technology affects developmental kinetics, H19 Imprinting Control Region methylation and H19 gene expression in individual mouse embryos. BMC Dev. Biol. 2007;7:116. doi: 10.1186/1471-213X-7-116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Market-Velker B.A., Fernandes A.D., Mann M.R. Side-by-side comparison of five commercial media systems in a mouse model: suboptimal in vitro culture interferes with imprint maintenance. Biol. Reprod. 2010;83(6):938–950. doi: 10.1095/biolreprod.110.085480. [DOI] [PubMed] [Google Scholar]
- 195.Market Velker BA, Denomme MM, Mann MR. Loss of genomic imprinting in mouse embryos with fast rates of preimplantation development in culture. Biol. Reprod. 2012;86(5): 1–16. doi: 10.1095/biolreprod.111.096602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Ibala-Romdhane S., Al-Khtib M., Khoueiry R., Blachère T., Guérin J.F., Lefèvre A. Analysis of H19 methylation in control and abnormal human embryos, sperm and oocytes. Eur. J. Hum. Genet. 2011;19(11):1138–1143. doi: 10.1038/ejhg.2011.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Shi X., Chen S., Zheng H., Wang L., Wu Y. Abnormal DNA Methylation of Imprinted Loci in Human Preimplantation Embryos. Reprod. Sci. 2014;21(8):978–983. doi: 10.1177/1933719113519173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Dobbs K.B., Rodriguez M., Sudano M.J., Ortega M.S., Hansen P.J. Dynamics of DNA methylation during early development of the preimplantation bovine embryo. PLoS One. 2013;8(6):e66230. doi: 10.1371/journal.pone.0066230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Meseguer M., Herrero J., Tejera A., Hilligsøe K.M., Ramsing N.B., Remohí J. The use of morphokinetics as a predictor of embryo implantation. Hum. Reprod. 2011;26(10):2658–2671. doi: 10.1093/humrep/der256. [DOI] [PubMed] [Google Scholar]
- 200.Wong C.C., Loewke K.E., Bossert N.L., Behr B., De Jonge C.J., Baer T.M., Reijo Pera R.A. Non-invasive imaging of human embryos before embryonic genome activation predicts development to the blastocyst stage. Nat. Biotechnol. 2010;28(10):1115–1121. doi: 10.1038/nbt.1686. [DOI] [PubMed] [Google Scholar]
- 201.Chiba H., Hiura H., Okae H., Miyauchi N., Sato F., Sato A., Arima T. DNA methylation errors in imprinting disorders and assisted reproductive technology. Pediatr. Int. 2013;55(5):542–549. doi: 10.1111/ped.12185. [DOI] [PubMed] [Google Scholar]
- 202.Gosden R., Trasler J., Lucifero D., Faddy M. Rare congenital disorders, imprinted genes, and assisted reproductive technology. Lancet. 2003;361(9373):1975–1977. doi: 10.1016/S0140-6736(03)13592-1. [DOI] [PubMed] [Google Scholar]
- 203.Maher E.R. Imprinting and assisted reproductive technology. Hum. Mol. Genet. 2005;14(Spec No 1):R133–R138. doi: 10.1093/hmg/ddi107. [DOI] [PubMed] [Google Scholar]
- 204.Bliek J., Terhal P., van den Bogaard M.J., Maas S., Hamel B., Salieb-Beugelaar G., Simon M., Letteboer T., van der Smagt J., Kroes H., Mannens M. Hypomethylation of the H19 gene causes not only Silver-Russell syndrome (SRS) but also isolated asymmetry or an SRS-like phenotype. Am. J. Hum. Genet. 2006;78(4):604–614. doi: 10.1086/502981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Hiura H., Okae H., Miyauchi N., Sato F., Sato A., Van De Pette M., John R.M., Kagami M., Nakai K., Soejima H., Ogata T., Arima T. Characterization of DNA methylation errors in patients with imprinting disorders conceived by assisted reproduction technologies. Hum. Reprod. 2012;27(8):2541–2548. doi: 10.1093/humrep/des197. [DOI] [PubMed] [Google Scholar]
- 206.Marques C.J., Carvalho F., Sousa M., Barros A. Genomic imprinting in disruptive spermatogenesis. Lancet. 2004;363(9422):1700–1702. doi: 10.1016/S0140-6736(04)16256-9. [DOI] [PubMed] [Google Scholar]
- 207.Lazaraviciute G., Kauser M., Bhattacharya S., Haggarty P., Bhattacharya S. A systematic review and meta-analysis of DNA methylation levels and imprinting disorders in children conceived by IVF/ICSI compared with children conceived spontaneously. Hum. Reprod. Update. 2014;20(6):840–852. doi: 10.1093/humupd/dmu033. [DOI] [PubMed] [Google Scholar]
- 208.Whitelaw N., Bhattacharya S., Hoad G., Horgan G.W., Hamilton M., Haggarty P. Epigenetic status in the offspring of spontaneous and assisted conception. Hum. Reprod. 2014;29(7):1452–1458. doi: 10.1093/humrep/deu094. [DOI] [PubMed] [Google Scholar]
- 209.Eroglu A., Layman L.C. Role of ART in imprinting disorders. Semin. Reprod. Med. 2012;30(2):92–104. doi: 10.1055/s-0032-1307417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Rancourt R.C., Harris H.R., Michels K.B. Methylation levels at imprinting control regions are not altered with ovulation induction or in vitro fertilization in a birth cohort. Hum. Reprod. 2012;27(7):2208–2216. doi: 10.1093/humrep/des151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Oliver V.F., Miles H.L., Cutfield W.S., Hofman P.L., Ludgate J.L., Morison I.M. Defects in imprinting and genome-wide DNA methylation are not common in the in vitro fertilization population. Fertil. Steril. 2012;97(1):147–53.e7. doi: 10.1016/j.fertnstert.2011.10.027. [DOI] [PubMed] [Google Scholar]
- 212.Feng C., Tian S., Zhang Y., He J., Zhu X.M., Zhang D., Sheng J.Z., Huang H.F. General imprinting status is stable in assisted reproduction-conceived offspring. Fertil. Steril. 2011;96(6):1417–1423.e9. doi: 10.1016/j.fertnstert.2011.09.033. [DOI] [PubMed] [Google Scholar]