Abstract
DNA has the ability to form a variety of secondary structures that can interfere with normal cellular processes, and many of these structures have been associated with neurological diseases and cancer. Secondary structure-forming sequences are often found at chromosomal fragile sites, which are hotspots for sister chromatid exchange, chromosomal translocations, and deletions. Structures formed at fragile sites can lead to instability by disrupting normal cellular processes such as DNA replication and transcription. The instability caused by disruption of replication and transcription can lead to DNA breakage, resulting in gene rearrangements and deletions that cause disease. In this review, we discuss the role of DNA secondary structure at fragile sites in human disease.
Keywords: Cancer, DNA fragility, DNA secondary structure, Fragile site, Genome instability, Neurological disease.
INTRODUCTION
DNA Secondary Structure in Human Disease
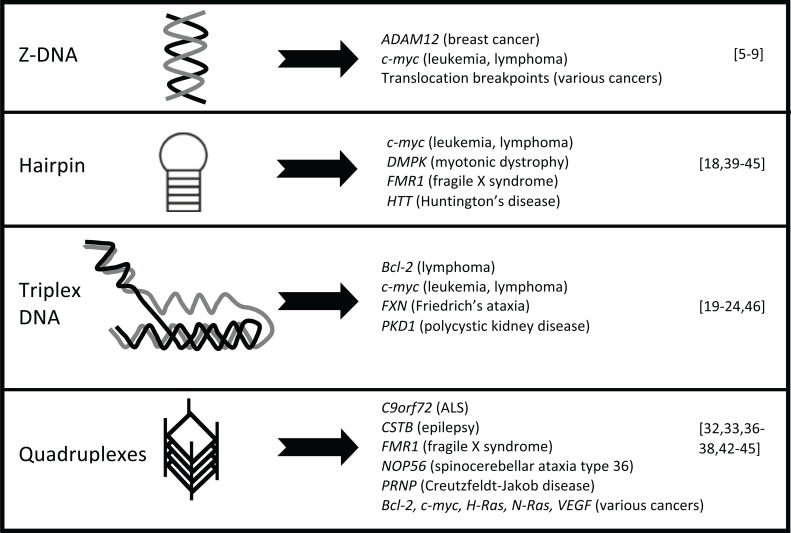
DNA is most commonly found as a right-handed double-helical structure known as B-DNA. However, as DNA undergoes dynamic processes, such as replication and transcription, B-DNA is able to take on alternative conformations including Z-DNA, hairpins, cruciforms, triplex DNA and quadruplexes, among others (Fig. 1) [1]. These structures are associated with a number of human diseases through their role in repetitive sequence amplification, genomic rearrangements, gene deletions and gene expression [2-4].
Fig. (1).
Diseases associated with secondary structure formation. a) Left-handed Z-DNA-forming sequences are found in the promoters of onco-genes and at DNA breakpoints in lymphoid tumors and B-cell precursor acute lymphoblastoid leukemia. b) Hairpin structures are found at genes involved in various cancers and many neurological diseases, including fragile X syndrome, myotonic dystrophy, and polyglutamine dis-eases such as Huntington’s disease. c) Triplex DNA is found at polypurine or polypyrimidine sequences, such as those found in cancer, Frie-drich’s ataxia, and autosomal dominant polycystic kidney disease. d) Quadruplex structures are associated with oncogene promoters in a num-ber of cancers and in neurological diseases, such as ALS/FTD, progressive myoclonus epilepsy type 1, fragile X syndrome, spinocerebellar ataxia type 36, and Creutzfeldt-Jakob disease.
Z-DNA is formed when purine bases within alternating purine-pyrimidine sequences adopt the syn conformation, allowing for left-handed helix formation. Bioinformatics studies have identified the co-localization of Z-DNA-forming sequences with sites of chromosomal breakage and translocations. GC and GT repeats capable of forming Z-DNA are commonly found in the promoters of oncogenes, such as in ADAM-12 and c-myc [5-7]. Additionally, translocation breakpoints in lymphoid tumors and B-cell precursor acute lymphoblastoid leukemia map to Z-DNA-forming sequences [8, 9].
Inverted repeats have the ability to form hairpin structures with palindromes as short as 14bp in vivo [10], and play a role in deletions, amplifications and translocations [4, 11-14]. Breakpoints in t(11;22) translocations found in parents of children with Emmanuel syndrome occur in AT-rich repeats at 11q23 and 22q11 [15-17]. A palindrome-mediated translocation, t(3:8)(p14.2;q24.1), is detected in renal carcinoma [18].
Polypurine or polypyrimidine sequences can form triplex DNA, and in sequences with mirror repeat symmetry, can form an intra-molecular triplex known as H-DNA [1]. These sequences localize with chromosomal breakpoint hotspots, such as those in Bcl-2 and c-myc [19-21], and sites of gene rearrangements [22-24]. The PKD1 gene, which is commonly mutated in autosomal dominant polycystic kidney disease, contains triplex DNA-forming sequences in the mutation cluster region [22].
Quadruplexes are formed in G-rich sequences by the Hoogsteen-bonding of four guanines, leading to a stack of guanine tetrads in a four-stranded structure [25]. High throughput sequencing of nascent DNA strands has shown that the majority of replication origins correspond to quadruplex motifs [26]. Quadruplex-forming sequences have been identified in mitotic and meiotic break sites and sites of instability [25]. These sequences are enriched at the telomere and in the promoters of many genes, including oncogenes such as Bcl-2, c-myc, c-kit, H-Ras, N-Ras and VEGF, though the propensity for structure-formation at these sites in vivo is still unknown [25]. In the absence of Pif1 helicase, quadruplex structures lead to deletions and rearrangements during replication [27, 28]. Quadruplex structures formed in telomere DNA are able to inhibit telomerase-dependent extension upon stabilization with structure-stabilizing compounds [29]. Genome-wide analysis of DNA breakpoints involved in cancer revealed that mutation breakpoints are enriched for quadruplex sequences, and that hypomethylation at these sites is associated with breakage [30]. Further, putative quadruplex-forming sequences are enriched at germline copy number variation breakpoints in neurodevelopmental diseases [31]. An expanded GGGGCC hexanucleotide repeat at C9orf72 can form quadruplex structures [32, 33], and is the most common genetic cause for familial amyotrophic lateral sclerosis-frontotemporal dementia (ALS-FTD) [34, 35]. Several other quadruplex-forming sequences have also been implicated in neurological diseases, including GGCCT repeats at NOP56 in spinocerebellar ataxia type 36 [36], and longer GC-rich repeats at CSTB and PRNP in progressive myoclonus epilepsy type 1 and Creutzfeldt-Jakob disease, respectively [37, 38].
Trinucleotide repeat expansion diseases, including fragile X syndrome, polyglutamine diseases, myotonic dystrophy, and Friedrich’s ataxia are manifested from trinucleotide repeats, which have been demonstrated to form various DNA secondary structures. Triplet repeats such as CAG in polyglutamine diseases [39], and CTG in myotonic dystrophy [40], are capable of forming hairpin structures that undergo expansion or deletion events in vivo [41]. The expanded CGG repeat at the FMR1 gene found in fragile X syndrome is able to form both hairpin and quadruplex structures, which block replication in vitro and in vivo [42-45]. The neurodegenerative disease Friedrich’s ataxia is caused by expansion of a GAA repeat in the FXN gene, and the expanded GAA repeat has been shown to form triplex structure [46].
Interestingly, trinucleotide repeat expansion diseases display a repeat-length threshold for disease severity. Repeat length is polymorphic but stable in normal individuals; however, expansion above the threshold leads to symptoms. Trinucleotide repeats in the normal range do not form highly stable DNA secondary structures, but the expanded repeats can lead to formation of structures too stable to be resolved, leading to disruption of cellular function and disease phenotype. Formation of DNA secondary structures is associated only with the repeats that can expand, suggesting that structure formation is important for repeat expansion [43]. In addition to repeat length, repeat purity also influences disease outcome, with repeat interruptions lessening the disease symptoms [47-49]. Interruptions of repeating sequences can severely diminish the ability of DNA to form stable secondary structure, suggesting that the ability to form structure is a key regulator of disease severity [43, 47-49].
Chromosomal Fragile Sites in Human Disease
Fragile sites are regions of the genome that are susceptible to breakage under conditions of replication stress [50]. They are defined cytogenetically as gaps or breaks in metaphase chromosomes, and can span up to several megabases in size. Fragile sites are classified based on their frequency in the population, as either common fragile sites, those present in all individuals, or rare fragile sites, which are only present in less than 5% of the population [51]. Expression of fragile sites is typically induced by treatment of cells with low-dose aphidicolin, an inhibitor of the replicative DNA polymerases α, δ and ε [52, 53]. However, fragile site breakage can also be induced by a number of chemotherapeutic and environmental chemicals [54], further demonstrating the importance in understanding the mechanism and consequences of breakage. While fragile sites are normally stable in cells, replication stress increases their susceptibility to deletions, rearrangements and sister chromatid exchanges [54, 55], perhaps due to the presence of sequences able to form stable secondary structures, including CGG repeats or AT-rich minisatellites.
Fragile site breakage is associated with a number of neurological diseases and cancer. Expanded CGG repeats were identified as the cause of disease at FRAXA, FRAXE, and FRA11B. FRAXA is expressed in patients with fragile X syndrome [56-58], and FRAXE is associated with a rare form of mental retardation [59]. Jacobsen syndrome is caused by deletion of the long arm of chromosome 11 due to breakage at FRA11B which contains an expanded CGG repeat located in the 5’-UTR of the CBL2 proto-oncogene [60]. In cells of patients with bipolar disorder, the rate of fragile site breakage is higher than in non-affected individuals [61]. Additionally, nearly half of genes linked to schizophrenia are located at fragile sites [62]. Cells from schizophrenia patients grown in the absence of folate display a higher frequency of fragile site breakage compared to cells from normal individuals [62]. Interestingly, schizophrenia and autism have been linked to folate metabolism genes [62], which can affect fragile site instability.
Over half of gene pairs involved in cancer-specific translocations contain at least one gene within a fragile site, suggesting a role for fragile site breakage in formation of gene rearrangements [63]. DNA breakage at fragile sites FRA3B and FRA16D can lead to deletions within tumor suppressor genes FHIT and WWOX, respectively, which are found in many cancers [64, 65]. Colorectal cancer patients show deletions within the PARK2 gene in FRA6E, and loss of PARK2 expression cooperates with APC mutations to cause colorectal cancer in mice [66]. Tumor cells of patients with oropharyngeal squamous cell carcinoma show decreased expression of six large genes, CTNNA3, DLG2, DMD, FHIT, NBEA and PARK2 (which are all located within fragile sites), in tumors, compared with matched normal samples [67]. Further, oncogenic viruses hepatitis B and human papillomavirus-16 preferentially integrate at fragile sites [68-72]. In addition to these associations between fragile sites and cancer, studies have revealed the direct involvement of fragile sites in oncogenesis. Treatment of cells with low doses of aphidicolin induces deletions within FHIT at the breakpoint cluster region of FRA3B, consistent with deletions seen in esophageal cancer, small-cell and non-small-cell lung cancers, and breast cancer [64]. Exposure of thyroid epithelial cells to fragile site-inducing conditions leads to formation of the RET/PTC rearrangement that causes papillary thyroid carcinoma [73]. Additionally, aphidicolin-induced breakpoints within the RET gene co-localize with the breakpoint cluster region found in papillary thyroid carcinoma patients with RET/PTC rearrangements [73].
These studies suggest that fragile site breakage can directly contribute to disease pathogenesis. Further understanding of the mechanisms for and consequences of fragile site breakage will be vital for prevention and/or treatment of fragile site-related disease.
The Role of Secondary Structures at Fragile Sites
The ability to form stable DNA secondary structure is one underlying feature of many fragile sites that appears to play a role in their instability. During DNA replication, as double-stranded DNA is unwound, such as at the Okazaki initiation zone, single-stranded DNA is exposed and stable secondary structures can form. Replication through trinucleotide repeat DNA demonstrates that instability increases when the structure-forming sequence is on the lagging strand template [40, 74-76]. Studies have shown that the CGG repeat at rare, folate-sensitive fragile sites can form hairpin and quadruplex structures, which were subsequently shown to perturb replication both in vitro and in vivo [42-45]. An AT-rich region within FRA16D was able to block replication in yeast [77], while the AT-rich FRA16B sequence was able to form secondary structure in vitro, which aligned with polymerase stall sites [74]. Additionally, FRA16B DNA showed instability during replication which is dependent on distance from the origin of replication and orientation of the sequence relative to the origin [74].
While over half of cancer-specific translocations contain at least one gene within a fragile site, many genes involved in cancer-specific copy number variations or translocations are not located within cytogenetically-defined fragile sites [63]. With the exception of a small number of fragile sites characterized at the molecular level [78], most fragile sites are identified by the appearance of gaps or breaks on metaphase chromosomes, and thus, can stretch up to several megabases in length. However, fragility is not present throughout the entire region, as evidenced by the difference between aphidicolin-induced breakage in RET and NCOA4, both located within FRA10G [73]. Conversely, non-fragile regions may contain “micro” fragile sites, which are too small to be observed cytogenetically. To better define fragile sites and to test whether the ability to form secondary structures correlates with DNA fragility, we analyzed DNA secondary structure-forming potential along chromosome 10 using the MFold program [79], which predicts the ability of DNA sequences to form multiple stem-loop structures, and estimates a free energy value for structure formation [80]. We found that aphidicolin-induced common fragile sites contain more secondary structure-forming segments, and these segments are clustered more densely than non-fragile DNA. Based on the secondary structure-forming ability, the previous cytogenetically-defined large fragile sites can be refined, and additional fragile sites in non-fragile regions identified. In in vitro re-duplexing assays, the RET intron 11 and PTEN exon 1 sequences have significantly more secondary structure formation than intron 7 of NCOA4 and exon 9 of PTEN, agreeing with the differential folding propensity predicted by the MFold program [79]. Further cell type-specific, genome-wide studies are needed to determine DNA breakage patterns at each of these sites, and to understand how each of these sites fit into the currently proposed models of fragility. These efforts are geared toward compiling a list of legitimate fragile sites for evaluation of disease susceptibility.
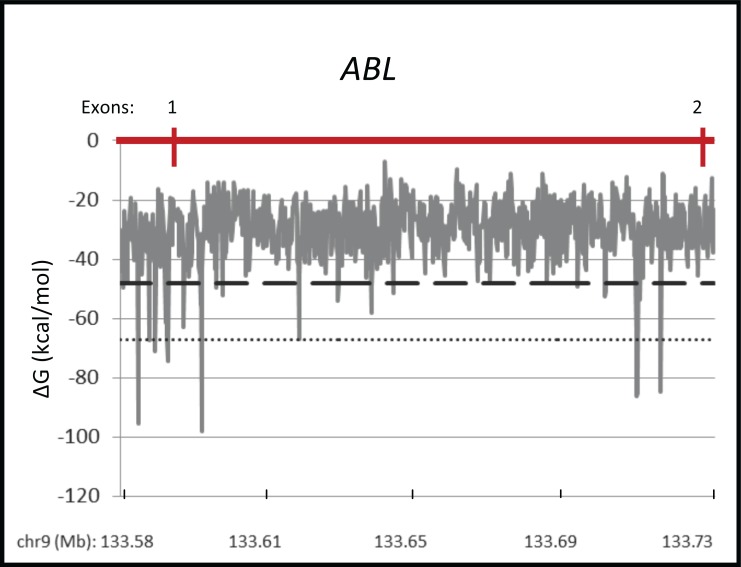
To further investigate the possibility of “micro” fragile sites that are undetected by cytogenetic methods, we analyzed the secondary structure-forming ability of the ABL gene, which is involved in the recurrent cancer-specific BCR-ABL translocation, and is not located within a cytogenetically-defined fragile site. Using the MFold program, we found that the breakpoint cluster region stretching from exon 1 to exon 2 of ABL shows the potential to form highly stable secondary structures (Fig. 2). Nearly fifty 300-nucleotide segments in the ABL breakpoint cluster region have folding free energy values within the same range as the most favorable 5% of the entire chromosome 9. Furthermore, two clusters of significant structure-forming segments contained 10 segments within the most favorable 1% of all free energy values measured on chromosome 9, suggesting that this region has the potential to form stable secondary structures that may play a role in breakage at ABL.
Fig. (2).
Secondary structure formation within the breakpoint cluster region of ABL. The MFold program predicted secondary structure formation in the breakpoint cluster region (exons 1-2, with 13 kb flanking sequence upstream) of the ABL gene (chr9: 133,576,165-133,729,624; hg19). Segments of 300 nt were analyzed with 150 nt shift windows along the gene sequence. Data is presented as predicted free energy of structure formation (ΔG) along the gene (mega-bases). Dashed and dotted lines, respectively, represent the threshold for the most favorable 5% and 1% of free energy values on chromo-some 9.
One limitation of the MFold program is that it is not able to assess the potential of DNA to form quadruplex structures, such as those able to be formed by expanded CGG repeats at fragile site FRAXA. Bioinformatics analysis of the human genome identified 375,000 potential quadruplex-forming sequences, but it is not known how many of these sites are able to actually form structures in vivo [81]. Several potential quadruplex-forming sequences identified were mapped to cytogenetically-defined fragile sites. Further genome-wide sequence analysis found that quadruplex-forming sequences were found to be enriched near sites of chromosomal rearrangement breakpoints, and were located within 70% of genes involved in lymphoid cancer rearrangements [82].
The GGGGCC hexanucleotide repeat expansion at C9orf72 in ALS-FTD [34, 35] is mapped within rare, folate-sensitive fragile site FRA9A, and common, BrdU-induced fragile site FRA9C, both cytogenetically located at 9p21. The hexanucleotide repeat has the ability to form quadruplex structure in vitro with as few as four repeats [32, 33]. The disease is associated with repeats that number from hundreds to thousands, while normal alleles contain between 2 and 25 repeats. The features of this repeat are very similar to CGG repeat-containing fragile sites, such as FRAXA, but more studies will be needed to identify the consequences of structure formation.
In addition to the secondary structure-forming ability, a genome-wide association study found that fragile sites are located away from centromeres, are enriched in Alu repeats, and are highly flexible [83]. Conversely, fragile sites were found to be negatively associated with CpG islands, H3K4 mono-methylation, and transcription start sites [83]. These studies will help to better define fragile sites by using molecular features possibly contributing to fragility. By more closely determining the constitution of a fragile site, we can begin to understand how breakage occurs, and identify ways to prevent or repair disease-causing fragility.
Mechanisms for Fragile Site Breakage
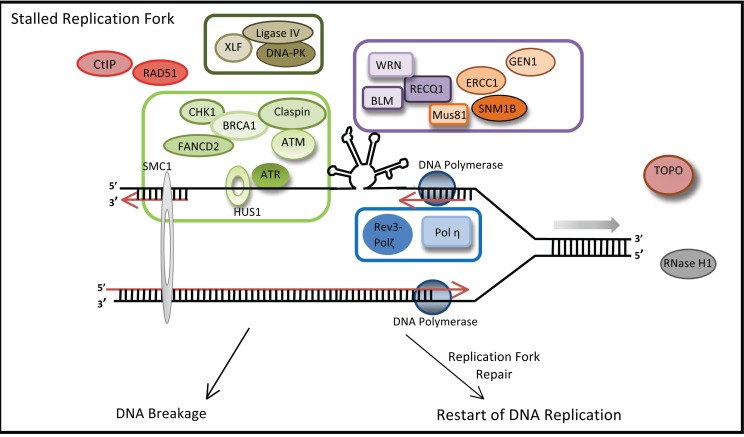
Most fragile sites studied to date are late-replicating, with replication further stalled in the presence of fragile-site inducing chemicals. Fragile site DNA is capable of stalling the replicative polymerase in vitro [74, 84], blocking replication fork progression [77] and inducing instability in yeast [76]. One working model of fragility postulates that during DNA replication, stress, such as low-dose aphidicolin, can cause uncoupling of the DNA helicase-topoisomerase complex from the replicative polymerase [55]. This uncoupling can lead to longer stretches of single-stranded DNA at the replication fork. Within fragile sites, the long single-stranded DNA has the ability to form a stable secondary structure, which can cause replication fork stall and collapse, resulting in DNA breakage. Loss of proteins involved in DNA repair and secondary structure resolution compounds the effects of sequence-specific fragility and increases instability (Fig. 3).
Fig. (3).
Proteins involved in chromosomal fragility. During replication fork progression, replication stress can uncouple the helicase complex from the DNA polymerase, allowing for single-stranded DNA to form stable secondary structure that can cause fork stall and collapse. Proteins in-volved in the ATR pathway (light green box) are important for fragile site stability, as loss of expression leads to an increase in breakage. Sever-al proteins able to recognize and resolve DNA secondary structures (purple box) are also essential for fragility. Repair pathway proteins, such as those in NHEJ (dark green box) and translesion synthesis (blue box), also stabilize fragile sites..
Proteins Involved in DNA Fragility
Glover and colleagues have elegantly shown the critical role of ATR in maintenance of DNA fragility [85, 86]. ATR kinase responds to DNA damage and stalled and collapsed replication forks by activating a downstream pathway that leads to repair [87]. Reduction of ATR, and more particularly, its kinase activity leads to an increase in fragile site breakage, even in the absence of aphidicolin [85]. ATR is able to preferentially interact with fragile site FRA3B upon replication stress, with its kinase activity being necessary for this interaction [88]. Individuals with the genetic condition Seckel syndrome, caused by a mutation in the ATR gene resulting in hypomorphic expression, display an increase in fragile site breakage relative to unaffected individuals [86]. The kinase ATM is activated and forms foci with γH2AX upon aphidicolin exposure; however, loss of ATM alone does not lead to increased fragile site breakage [85, 89]. Loss of both ATM and ATR significantly increases fragile site breakage relative to ATR inhibition alone [89], suggesting that ATM is able to compensate for ATR inactivation to prevent further damage.
Proteins downstream of the ATR pathway, including BRCA1, CHK1, Claspin, FANCD2, HUS1, SMC1 and WRN, are all involved in stability of common fragile sites. Loss of expression of ATR substrates BRCA1 [90], CHK1 [91], and SMC1 [92] results in an increase of fragile site breakage. Proteins required for phosphorylation of ATR substrates such as HUS1 [93] and Claspin [94] are also important in stability of fragile sites. Cells with loss of FANCD2 expression or those from Fanconi anemia patients show increased fragile site breakage upon treatment with aphidicolin [95]. In addition to the ATR pathway, proteins from both homologous recombination (Rad51) and non-homologous end joining (DNA-PKcs, Ligase IV, and XLF) pathways are important in fragile site stability [96]. Translesion synthesis proteins pol η and pol ζ-Rev3 are essential for stability of fragile sites in human cells [97, 98]. Furthermore, in vitro synthesis of repetitive sequences within FRA16D and FRA3B suggest a role for DNA polymerase pol κ in fragile site replication [99]. These studies suggest that fragile sites are inherently recognized as DNA damage, and we postulate that their ability to form stable secondary structures not only licenses them to stall replication fork progression, but also to evade proper DNA repair, leading to DNA breaks or illegitimate repairs.
RPA-coated single-stranded DNA at sites of DNA damage is necessary for efficient TopBP1-dependent ATR activation, leading to downstream CHK1 phosphorylation [87, 100-102]. RPA binding requires roughly 30 nucleotides of single-stranded DNA to bind [103], while secondary structures can form on much shorter stretches [10]. DNA secondary structure formation at fragile sites may cause insufficient ATR pathway activation, due to a lack of RPA-bound DNA. Moreover, upon replication stress, excessive replication origin firing results in a depletion of free RPA in the nucleus, leaving single-stranded DNA at newly active origins exposed and prone to breakage [104, 105]. These studies suggest that a depletion of RPA, leading to stretches of unbound single-stranded DNA at fragile sites, can promote the formation of DNA secondary structures, and lead to breakage.
Interestingly, several proteins known to recognize DNA secondary structure also play a role in fragile site stability, including DNA helicases (BLM, RecQ1 and WRN), structure-specific endonucleases (CtIP, ERCC1 and MUS81), and an exonuclease (SNM1B). In cells of patients with Werner syndrome (deficient in WRN) or Bloom’s syndrome (deficient in BLM), fragile site breakage is increased following exposure to aphidicolin [84, 106]. Both BLM and WRN are able to resolve a variety of DNA secondary structures [107-110]. RecQ1, a helicase involved in repair of stalled replication forks, preferentially binds to fragile sites upon replication stress, and is necessary for fragile site stability [111]. ERCC1 and MUS81 endonucleases, which recognize DNA structures, interstrand crosslinks, and replication intermediates, promote fragile site breakage, possibly due to DNA cleavage at sites of stalled replication forks or secondary structures within fragile sites [112-115]. The noncatalytic partner of MUS81 is important, however, as MUS81-EME1 is essential for fragile site stability, while the MUS81-EME2 complex, which promotes replication fork cleavage and restart, is not [115]. Recently, the endonuclease activity of CtIP, involved in homologous recombination-mediated DNA repair, was shown to be required for repair of double-stranded DNA breaks at a secondary structure-forming AT-rich sequence in FRA16D [114, 116]. The SNM1B exonuclease recognizes interstrand crosslinks, and is necessary for resolution of replication fork stalling at fragile sites [117]. Finally, in transfection assays, the Holliday-junction resolvase GEN1 can generate breaks at palindromic AT-rich repeats [118].
DNA topoisomerases are a family of proteins responsible for resolving the supercoiling of DNA that occurs during replication and transcription [119]. Both topoisomerase I and IIα are also able to recognize and cleave DNA at sites of secondary structure formation [120-122]. Topoisomerases play multiple roles in fragile site stability, such as in the uncoupling of DNA polymerase and helicase/topoisomerase complex after replication perturbation [123], in resolving replication and transcription collision [124], and in yeast, mediating breakage at Mec1/ATR-sensitive fragile sites [125]. We showed that fragile site breakpoints within the RET gene were located near predicted topoisomerase I and/or IIα cleavage sites [126], and these sites displayed secondary structural features preferentially cleaved by both topoisomerase I and IIα. We further demonstrated that RET breakage was significantly decreased upon exposure to aphidicolin, in combination with topoisomerase catalytic inhibitors betulinic acid and merbarone. These data suggest that DNA topoisomerases I and II recognize secondary structures formed during replication of fragile sites, and initiate breakage at these sites.
The number and variety of proteins involved in fragile site stability suggests a complex mechanism involving many cellular processes, including DNA replication, transcription, DNA structure resolution, and DNA repair. These studies suggest that replication through fragile sites is a delicate process, with obstacles such as DNA secondary structure and/or replication stress leading to a perturbation at the replication fork. If any of the proteins involved in stabilizing the replication fork and repairing the damage are compromised, this stress can lead to fragile site breakage.
The Consequences of Replication and Transcription of Fragile Sites
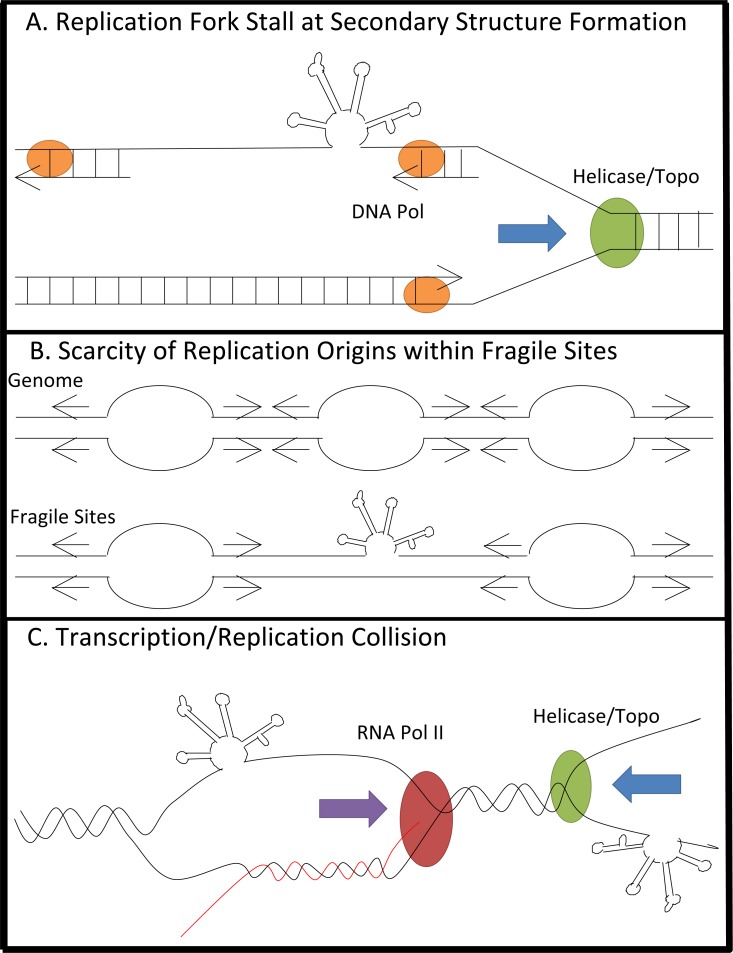
In addition to the replication-stalled model, paucity of replication initiation in fragile site regions [127, 128] and the presence of transcription-derived R-loops during DNA replication of fragile sites [124] are also suggested to be involved in the mechanism of fragility [129]. However, they are not mutually exclusive (Fig. 4).
Fig. (4).
Mechanisms for fragility during replication and transcription. A) Single-stranded DNA exposed during replication fork opening forms stable secondary structure, which can block replication and lead to fork collapse. B) Scarcity of replication origins within fragile sites causes replication to progress over long stretches of unstable DNA, which can slow the replicative polymerase. While non-fragile regions can overcome replication stress by firing additional replica-tion origins, fragile sites are unable to do so. C) Collision of replica-tion and transcription machinery at large genes leads to formation of DNA-RNA hybrids known as R-loops which are able to cause breakage. Although it has not yet been investigated, the potential of fragile site sequences to form highly stable secondary structures could participate in the latter two mechanisms as well. For example, the formation of R-loops promotes trinucleotide repeat instability. The ability of trinucleotide repeats to form stable secondary structures stabilizes the presence of R-loops by adopting a hairpin structure on the non-template DNA strand, thus favoring hybrid formation be-tween RNA transcripts and the DNA template strand [130-133].
Replication fork stalling was directly demonstrated at a common fragile site FRA16C (Fig. 4a) [127]. Under normal growth conditions, replication forks progress slowly and frequently stall at AT-rich regions of FRA16C. Upon replication stress, replication is further perturbed, and fork stalling is increased; however, unlike other regions of the genome, additional replication origins are not activated to compensate for replication fork stalling at FRA16C [127]. This suggests that replication machinery must travel through long stretches of structure-forming fragile site DNA sequences, without the ability to fire additional origins able to compensate for stalled or slowed replication forks, potentially leading to replication fork collapse and DNA breakage (Fig. 4a).
Debatisse and colleagues showed that at fragile sites FRA3B and FRA16D, the most commonly expressed fragile sites in human lymphoblastoid cells, lack of replication initiation causes fragility (Fig. 4b) [128]. Molecular combing showed that in lymphoblastoid cells, but not in fibroblasts, replication initiation events were excluded from FRA3B, meaning that replication of this region required convergence of replication forks originating from flanking regions. Compounding on this effect, origins in flanking regions fired in mid-S phase, and left little time to replicate a large origin-scarce region. This study suggests that a combination of replication origin scarcity and late replication completion are the cause of fragility, and that fragility occurs in a cell type-specific manner [128].
In the case of large genes, a model in which the collision between replication and transcription machineries leads to fragility is proposed (Fig. 4c) [124]. Transcription of large genes takes longer than one complete cell cycle, and can lead to collision of replication and transcription machinery resulting in formation of R-loops. Interestingly, RNase H1, which specifically degrades the RNA strands of R-loops, is able to suppress the instability of fragile sites containing long genes [124]. Although it has not yet been investigated, the potential of fragile site sequences to form highly stable secondary structures could participate in this mechanism as well. For example, the formation of R-loops promotes trinucleotide repeat instability. The ability of trinucleotide repeats to form stable secondary structures stabilizes the presence of R-loops by adopting a hairpin structure on the non-template DNA strand, thus favoring hybrid formation between RNA transcripts and the DNA template strand [130-133].
Recently, a new class of fragile sites was identified in mouse cells by genome-wide association with RPA [134]. These sites, termed early replicating fragile sites (ERFSs), co-localized with actively transcribed genes, and were enriched with BRCA1, (H2AX, and SMC5, indicating sites of replication fork collapse and DNA damage response activation. Interestingly, ERFSs were located at different genomic loci than late-replicating common fragile sites, but they were similarly prone to chromosomal breakage following ATR inhibition or oncogenic stress. The ERFSs identified in this study were bound by RPA, and replicated early in the cell cycle, while common fragile sites are typically late-replicating and not preferentially bound by RPA. We suggest that the lack of RPA binding at common fragile sites is due to the ability of these sites to form stable secondary structures. RPA-bound single-stranded DNA is an essential mediator of the ATR repair pathway [87, 100-102], but late replication at CFSs facilitates the formation of stable secondary structures which can diminish RPA binding and downstream ATR signaling, allowing for the structure to remain in place during replication and transcription, leading to fragility.
These models for the mechanism of fragile site breakage, though not mutually exclusive, appear to differ on a cell-to-cell basis based on a number of factors, including replication patterns, transcription levels, and protein expression. The relationship of the replication origins to the fragile site sequence, both in distance and orientation, is important in the stability of fragile sites, as a change in origin activation patterns is extensively linked to fragility in a cell type-specific manner [74, 76, 127-129, 135-139]. Further complication occurs when the variability in transcriptional activity across multiple cell types is considered, as collision of the replication and transcription machinery can also lead to instability [124]. The extensive interaction of proteins involved in a number of cellular processes, including replication, transcription and DNA repair, and how they negotiate DNA secondary structures, adds another layer of complexity [54, 140, 141]. All of these factors can vary from one cell to the next, making it clear that the cell types can be a major determinant in patterns of fragility.
ACKNOWLEDGEMENTS
This work was supported by the National Institutes of Health (RO1GM101192) to Y.-H. W..
ABBREVIATIONS
- ALS =
Amyotrophic lateral sclerosis
- FTD =
Frontotemporal dementia
- ERFS =
Early replicating fragile sites
CONCLUSION/PERSPECTIVES
Work in recent years has demonstrated the complexity of chromosomal fragility by suggesting several conflicting models to explain the mechanisms involved. These models suggest fragility to be cell type- and site-specific, but more work needs to be done to determine their prevalence. One shared feature of all fragile sites studied to date is their ability to form secondary structure. Upon closer examination of fragile sites, it is clear that hotspots of fragility coincide with sites where secondary structure is predicted to occur. Additionally, non-fragile regions display potential secondary structure-forming sites that coincide with disease-related DNA breakage, suggesting that the current cytogenetic definition of fragility is outdated. Further study on the prevalence of secondary structure throughout the genome may offer a clearer picture of chromosomal fragility.
CONFLICT OF INTEREST
The author(s) confirm that this article content has no conflict of interest.
REFERENCES
- 1.Choi J., Majima T. Conformational changes of non-B DNA. Chem. Soc. Rev. 2011;40(12):5893–5909. doi: 10.1039/c1cs15153c. [DOI] [PubMed] [Google Scholar]
- 2.Wells R.D. Non-B DNA conformations, mutagenesis and disease. Trends Biochem. Sci. 2007;32(6):271–278. doi: 10.1016/j.tibs.2007.04.003. [DOI] [PubMed] [Google Scholar]
- 3.Zhao J., Bacolla A., Wang G., Vasquez K.M. Non-B DNA structure-induced genetic instability and evolution. Cell. Mol. Life Sci. 2010;67(1):43–62. doi: 10.1007/s00018-009-0131-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vasquez K.M., Wang G. The yin and yang of repair mechanisms in DNA structure-induced genetic instability. Mutat. Res. 2013;743-744:118–131. doi: 10.1016/j.mrfmmm.2012.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ray B.K., Dhar S., Shakya A., Ray A. Z-DNA-forming silencer in the first exon regulates human ADAM-12 gene expression. Proc. Natl. Acad. Sci. USA. 2011;108(1):103–108. doi: 10.1073/pnas.1008831108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rimokh R., Rouault J.P., Wahbi K., Gadoux M., Lafage M., Archimbaud E., Charrin C., Gentilhomme O., Germain D., Samarut J., et al. A chromosome 12 coding region is juxtaposed to the MYC protooncogene locus in a t(8;12)(q24;q22) translocation in a case of B-cell chronic lymphocytic leukemia. Genes Chromosomes Cancer. 1991;3(1):24–36. doi: 10.1002/gcc.2870030106. [DOI] [PubMed] [Google Scholar]
- 7.Wölfl S., Wittig B., Rich A. Identification of transcriptionally induced Z-DNA segments in the human c-myc gene. Biochim. Biophys. Acta. 1995;1264(3):294–302. doi: 10.1016/0167-4781(95)00155-7. [DOI] [PubMed] [Google Scholar]
- 8.Boehm T., Mengle-Gaw L., Kees U.R., Spurr N., Lavenir I., Forster A., Rabbitts T.H. Alternating purine-pyrimidine tracts may promote chromosomal translocations seen in a variety of human lymphoid tumours. EMBO J. 1989;8(9):2621–2631. doi: 10.1002/j.1460-2075.1989.tb08402.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sinclair P.B., Parker H., An Q., Rand V., Ensor H., Harrison C.J., Strefford J.C. Analysis of a breakpoint cluster reveals insight into the mechanism of intrachromosomal amplification in a lymphoid malignancy. Hum. Mol. Genet. 2011;20(13):2591–2602. doi: 10.1093/hmg/ddr159. [DOI] [PubMed] [Google Scholar]
- 10.Nag D.K., Petes T.D. Seven-base-pair inverted repeats in DNA form stable hairpins in vivo in Saccharomyces cerevisiae. Genetics. 1991;129(3):669–673. doi: 10.1093/genetics/129.3.669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Akgün E., Zahn J., Baumes S., Brown G., Liang F., Romanienko P.J., Lewis S., Jasin M. Palindrome resolution and recombination in the mammalian germ line. Mol. Cell. Biol. 1997;17(9):5559–5570. doi: 10.1128/mcb.17.9.5559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhou Z.H., Akgūn E., Jasin M. Repeat expansion by homologous recombination in the mouse germ line at palindromic sequences. Proc. Natl. Acad. Sci. USA. 2001;98(15):8326–8333. doi: 10.1073/pnas.151008498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tanaka H., Tapscott S.J., Trask B.J., Yao M.C. Short inverted repeats initiate gene amplification through the formation of a large DNA palindrome in mammalian cells. Proc. Natl. Acad. Sci. USA. 2002;99(13):8772–8777. doi: 10.1073/pnas.132275999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cunningham L.A., Coté A.G., Cam-Ozdemir C., Lewis S.M. Rapid, stabilizing palindrome rearrangements in somatic cells by the center-break mechanism. Mol. Cell. Biol. 2003;23(23):8740–8750. doi: 10.1128/MCB.23.23.8740-8750.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kurahashi H., Shaikh T.H., Hu P., Roe B.A., Emanuel B.S., Budarf M.L. Regions of genomic instability on 22q11 and 11q23 as the etiology for the recurrent constitutional t(11;22) Hum. Mol. Genet. 2000;9(11):1665–1670. doi: 10.1093/hmg/9.11.1665. [DOI] [PubMed] [Google Scholar]
- 16.Edelmann L., Spiteri E., Koren K., Pulijaal V., Bialer M.G., Shanske A., Goldberg R., Morrow B.E. AT-rich palindromes mediate the constitutional t(11;22) translocation. Am. J. Hum. Genet. 2001;68(1):1–13. doi: 10.1086/316952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kurahashi H., Emanuel B.S. Long AT-rich palindromes and the constitutional t(11;22) breakpoint. Hum. Mol. Genet. 2001;10(23):2605–2617. doi: 10.1093/hmg/10.23.2605. [DOI] [PubMed] [Google Scholar]
- 18.Kato T., Franconi C.P., Sheridan M.B., Hacker A.M., Inagakai H., Glover T.W., Arlt M.F., Drabkin H.A., Gemmill R.M., Kurahashi H., Emanuel B.S. Analysis of the t(3;8) of hereditary renal cell carcinoma: a palindrome-mediated translocation. Cancer Genet. 2014;207(4):133–140. doi: 10.1016/j.cancergen.2014.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Raghavan S.C., Swanson P.C., Wu X., Hsieh C.L., Lieber M.R. A non-B-DNA structure at the Bcl-2 major breakpoint region is cleaved by the RAG complex. Nature. 2004;428(6978):88–93. doi: 10.1038/nature02355. [DOI] [PubMed] [Google Scholar]
- 20.Raghavan S.C., Chastain P., Lee J.S., Hegde B.G., Houston S., Langen R., Hsieh C.L., Haworth I.S., Lieber M.R. Evidence for a triplex DNA conformation at the bcl-2 major breakpoint region of the t(14;18) translocation. J. Biol. Chem. 2005;280(24):22749–22760. doi: 10.1074/jbc.M502952200. [DOI] [PubMed] [Google Scholar]
- 21.Belotserkovskii B.P., De Silva E., Tornaletti S., Wang G., Vasquez K.M., Hanawalt P.C. A triplex-forming sequence from the human c-MYC promoter interferes with DNA transcription. J. Biol. Chem. 2007;282(44):32433–32441. doi: 10.1074/jbc.M704618200. [DOI] [PubMed] [Google Scholar]
- 22.Bacolla A., Jaworski A., Larson J.E., Jakupciak J.P., Chuzhanova N., Abeysinghe S.S., O’Connell C.D., Cooper D.N., Wells R.D. Breakpoints of gross deletions coincide with non-B DNA conformations. Proc. Natl. Acad. Sci. USA. 2004;101(39):14162–14167. doi: 10.1073/pnas.0405974101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bacolla A., Wojciechowska M., Kosmider B., Larson J.E., Wells R.D. The involvement of non-B DNA structures in gross chromosomal rearrangements. DNA Repair (Amst.) 2006;5(9-10):1161–1170. doi: 10.1016/j.dnarep.2006.05.032. [DOI] [PubMed] [Google Scholar]
- 24.Bacolla A., Wells R.D. Non-B DNA conformations as determinants of mutagenesis and human disease. Mol. Carcinog. 2009;48(4):273–285. doi: 10.1002/mc.20507. [DOI] [PubMed] [Google Scholar]
- 25.Wu Y., Brosh R.M., Jr G-quadruplex nucleic acids and human disease. FEBS J. 2010;277(17):3470–3488. doi: 10.1111/j.1742-4658.2010.07760.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Besnard E., Babled A., Lapasset L., Milhavet O., Parrinello H., Dantec C., Marin J.M., Lemaitre J.M. Unraveling cell type-specific and reprogrammable human replication origin signatures associated with G-quadruplex consensus motifs. Nat. Struct. Mol. Biol. 2012;19(8):837–844. doi: 10.1038/nsmb.2339. [DOI] [PubMed] [Google Scholar]
- 27.Lopes J., Piazza A., Bermejo R., Kriegsman B., Colosio A., Teulade-Fichou M.P., Foiani M., Nicolas A. G-quadruplex-induced instability during leading-strand replication. EMBO J. 2011;30(19):4033–4046. doi: 10.1038/emboj.2011.316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Paeschke K., Capra J.A., Zakian V.A. DNA replication through G-quadruplex motifs is promoted by the Saccharomyces cerevisiae Pif1 DNA helicase. Cell. 2011;145(5):678–691. doi: 10.1016/j.cell.2011.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zahler A.M., Williamson J.R., Cech T.R., Prescott D.M. Inhibition of telomerase by G-quartet DNA structures. Nature. 1991;350(6320):718–720. doi: 10.1038/350718a0. [DOI] [PubMed] [Google Scholar]
- 30.De S., Michor F. DNA secondary structures and epigenetic determinants of cancer genome evolution. Nat. Struct. Mol. Biol. 2011;18(8):950–955. doi: 10.1038/nsmb.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bose P., Hermetz K.E., Conneely K.N., Rudd M.K. Tandem repeats and G-rich sequences are enriched at human CNV breakpoints. PLoS One. 2014;9(7):e101607. doi: 10.1371/journal.pone.0101607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Reddy K., Zamiri B., Stanley S.Y., Macgregor R.B., Jr, Pearson C.E. The disease-associated r(GGGGCC)n repeat from the C9orf72 gene forms tract length-dependent uni- and multimolecular RNA G-quadruplex structures. J. Biol. Chem. 2013;288(14):9860–9866. doi: 10.1074/jbc.C113.452532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Haeusler A.R., Donnelly C.J., Periz G., Simko E.A., Shaw P.G., Kim M.S., Maragakis N.J., Troncoso J.C., Pandey A., Sattler R., Rothstein J.D., Wang J. C9orf72 nucleotide repeat structures initiate molecular cascades of disease. Nature. 2014;507(7491):195–200. doi: 10.1038/nature13124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.DeJesus-Hernandez M., Mackenzie I.R., Boeve B.F., Boxer A.L., Baker M., Rutherford N.J., Nicholson A.M., Finch N.A., Flynn H., Adamson J., Kouri N., Wojtas A., Sengdy P., Hsiung G.Y., Karydas A., Seeley W.W., Josephs K.A., Coppola G., Geschwind D.H., Wszolek Z.K., Feldman H., Knopman D.S., Petersen R.C., Miller B.L., Dickson D.W., Boylan K.B., Graff-Radford N.R., Rademakers R. Expanded GGGGCC hexanucleotide repeat in noncoding region of C9ORF72 causes chromosome 9p-linked FTD and ALS. Neuron. 2011;72(2):245–256. doi: 10.1016/j.neuron.2011.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Renton A.E., Majounie E., Waite A., Simón-Sánchez J., Rollinson S., Gibbs J.R., Schymick J.C., Laaksovirta H., van Swieten J.C., Myllykangas L., Kalimo H., Paetau A., Abramzon Y., Remes A.M., Kaganovich A., Scholz S.W., Duckworth J., Ding J., Harmer D.W., Hernandez D.G., Johnson J.O., Mok K., Ryten M., Trabzuni D., Guerreiro R.J., Orrell R.W., Neal J., Murray A., Pearson J., Jansen I.E., Sondervan D., Seelaar H., Blake D., Young K., Halliwell N., Callister J.B., Toulson G., Richardson A., Gerhard A., Snowden J., Mann D., Neary D., Nalls M.A., Peuralinna T., Jansson L., Isoviita V.M., Kaivorinne A.L., Hölttä-Vuori M., Ikonen E., Sulkava R., Benatar M., Wuu J., Chiò A., Restagno G., Borghero G., Sabatelli M., Heckerman D., Rogaeva E., Zinman L., Rothstein J.D., Sendtner M., Drepper C., Eichler E.E., Alkan C., Abdullaev Z., Pack S.D., Dutra A., Pak E., Hardy J., Singleton A., Williams N.M., Heutink P., Pickering-Brown S., Morris H.R., Tienari P.J., Traynor B.J., ITALSGEN Consortium A hexanucleotide repeat expansion in C9ORF72 is the cause of chromosome 9p21-linked ALS-FTD. Neuron. 2011;72(2):257–268. doi: 10.1016/j.neuron.2011.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kobayashi H., Abe K., Matsuura T., Ikeda Y., Hitomi T., Akechi Y., Habu T., Liu W., Okuda H., Koizumi A. Expansion of intronic GGCCTG hexanucleotide repeat in NOP56 causes SCA36, a type of spinocerebellar ataxia accompanied by motor neuron involvement. Am. J. Hum. Genet. 2011;89(1):121–130. doi: 10.1016/j.ajhg.2011.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Borel C., Migliavacca E., Letourneau A., Gagnebin M., Béna F., Sailani M.R., Dermitzakis E.T., Sharp A.J., Antonarakis S.E. Tandem repeat sequence variation as causative cis-eQTLs for protein-coding gene expression variation: the case of CSTB. Hum. Mutat. 2012;33(8):1302–1309. doi: 10.1002/humu.22115. [DOI] [PubMed] [Google Scholar]
- 38.Mead S., Webb T.E., Campbell T.A., Beck J., Linehan J.M., Rutherfoord S., Joiner S., Wadsworth J.D., Heckmann J., Wroe S., Doey L., King A., Collinge J. Inherited prion disease with 5-OPRI: phenotype modification by repeat length and codon 129. Neurology. 2007;69(8):730–738. doi: 10.1212/01.wnl.0000267642.41594.9d. [DOI] [PubMed] [Google Scholar]
- 39.Fan H.C., Ho L.I., Chi C.S., Chen S.J., Peng G.S., Chan T.M., Lin S.Z., Harn H.J. Polyglutamine (PolyQ) diseases: genetics to treatments. Cell Transplant. 2014;23(4-5):441–458. doi: 10.3727/096368914X678454. [DOI] [PubMed] [Google Scholar]
- 40.Cleary J.D., Nichol K., Wang Y.H., Pearson C.E. Evidence of cis-acting factors in replication-mediated trinucleotide repeat instability in primate cells. Nat. Genet. 2002;31(1):37–46. doi: 10.1038/ng870. [DOI] [PubMed] [Google Scholar]
- 41.Lahue R.S., Slater D.L. DNA repair and trinucleotide repeat instability. Front. Biosci. 2003;8:s653–s665. doi: 10.2741/1107. [DOI] [PubMed] [Google Scholar]
- 42.Fry M., Loeb L.A. The fragile X syndrome d(CGG)n nucleotide repeats form a stable tetrahelical structure. Proc. Natl. Acad. Sci. USA. 1994;91(11):4950–4954. doi: 10.1073/pnas.91.11.4950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gacy A.M., Goellner G., Juranić N., Macura S., McMurray C.T. Trinucleotide repeats that expand in human disease form hairpin structures in vitro. Cell. 1995;81(4):533–540. doi: 10.1016/0092-8674(95)90074-8. [DOI] [PubMed] [Google Scholar]
- 44.Usdin K., Woodford K.J. CGG repeats associated with DNA instability and chromosome fragility form structures that block DNA synthesis in vitro. Nucleic Acids Res. 1995;23(20):4202–4209. doi: 10.1093/nar/23.20.4202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Samadashwily G.M., Raca G., Mirkin S.M. Trinucleotide repeats affect DNA replication in vivo. Nat. Genet. 1997;17(3):298–304. doi: 10.1038/ng1197-298. [DOI] [PubMed] [Google Scholar]
- 46.Rajeswari M.R. DNA triplex structures in neurodegenerative disorder, Friedreich’s ataxia. J. Biosci. 2012;37(3):519–532. doi: 10.1007/s12038-012-9219-1. [DOI] [PubMed] [Google Scholar]
- 47.Jarem D.A., Huckaby L.V., Delaney S. AGG interruptions in (CGG)(n) DNA repeat tracts modulate the structure and thermodynamics of non-B conformations in vitro. Biochemistry. 2010;49(32):6826–6837. doi: 10.1021/bi1007782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pearson C.E., Eichler E.E., Lorenzetti D., Kramer S.F., Zoghbi H.Y., Nelson D.L., Sinden R.R. Interruptions in the triplet repeats of SCA1 and FRAXA reduce the propensity and complexity of slipped strand DNA (S-DNA) formation. Biochemistry. 1998;37(8):2701–2708. doi: 10.1021/bi972546c. [DOI] [PubMed] [Google Scholar]
- 49.Weisman-Shomer P., Cohen E., Fry M. Interruption of the fragile X syndrome expanded sequence d(CGG)(n) by interspersed d(AGG) trinucleotides diminishes the formation and stability of d(CGG)(n) tetrahelical structures. Nucleic Acids Res. 2000;28(7):1535–1541. doi: 10.1093/nar/28.7.1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Richards R.I. Fragile and unstable chromosomes in cancer: causes and consequences. Trends Genet. 2001;17(6):339–345. doi: 10.1016/S0168-9525(01)02303-4. [DOI] [PubMed] [Google Scholar]
- 51.Glover T.W. Common fragile sites. Cancer Lett. 2006;232(1):4–12. doi: 10.1016/j.canlet.2005.08.032. [DOI] [PubMed] [Google Scholar]
- 52.Cheng C.H., Kuchta R.D. DNA polymerase epsilon: aphidicolin inhibition and the relationship between polymerase and exonuclease activity. Biochemistry. 1993;32(33):8568–8574. doi: 10.1021/bi00084a025. [DOI] [PubMed] [Google Scholar]
- 53.Glover T.W., Berger C., Coyle J., Echo B. DNA polymerase alpha inhibition by aphidicolin induces gaps and breaks at common fragile sites in human chromosomes. Hum. Genet. 1984;67(2):136–142. doi: 10.1007/BF00272988. [DOI] [PubMed] [Google Scholar]
- 54.Dillon L.W., Burrow A.A., Wang Y.H. DNA instability at chromosomal fragile sites in cancer. Curr. Genomics. 2010;11(5):326–337. doi: 10.2174/138920210791616699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Durkin S.G., Glover T.W. Chromosome fragile sites. Annu. Rev. Genet. 2007;41:169–192. doi: 10.1146/annurev.genet.41.042007.165900. [DOI] [PubMed] [Google Scholar]
- 56.Kremer E.J., Pritchard M., Lynch M., Yu S., Holman K., Baker E., Warren S.T., Schlessinger D., Sutherland G.R., Richards R.I. Mapping of DNA instability at the fragile X to a trinucleotide repeat sequence p(CCG)n. Science. 1991;252(5013):1711–1714. doi: 10.1126/science.1675488. [DOI] [PubMed] [Google Scholar]
- 57.Pieretti M., Zhang F.P., Fu Y.H., Warren S.T., Oostra B.A., Caskey C.T., Nelson D.L. Absence of expression of the FMR-1 gene in fragile X syndrome. Cell. 1991;66(4):817–822. doi: 10.1016/0092-8674(91)90125-I. [DOI] [PubMed] [Google Scholar]
- 58.Verkerk A.J., Pieretti M., Sutcliffe J.S., Fu Y.H., Kuhl D.P., Pizzuti A., Reiner O., Richards S., Victoria M.F., Zhang F.P., et al. Identification of a gene (FMR-1) containing a CGG repeat coincident with a breakpoint cluster region exhibiting length variation in fragile X syndrome. Cell. 1991;65(5):905–914. doi: 10.1016/0092-8674(91)90397-H. [DOI] [PubMed] [Google Scholar]
- 59.Sofola O.A., Jin P., Botas J., Nelson D.L. Argonaute-2-dependent rescue of a Drosophila model of FXTAS by FRAXE premutation repeat. Hum. Mol. Genet. 2007;16(19):2326–2332. doi: 10.1093/hmg/ddm186. [DOI] [PubMed] [Google Scholar]
- 60.Jones C., Müllenbach R., Grossfeld P., Auer R., Favier R., Chien K., James M., Tunnacliffe A., Cotter F. Co-localisation of CCG repeats and chromosome deletion breakpoints in Jacobsen syndrome: evidence for a common mechanism of chromosome breakage. Hum. Mol. Genet. 2000;9(8):1201–1208. doi: 10.1093/hmg/9.8.1201. [DOI] [PubMed] [Google Scholar]
- 61.Demirhan O., Tastemir D., Sertdemir Y. The expression of folate sensitive fragile sites in patients with bipolar disorder. Yonsei Med. J. 2009;50(1):137–141. doi: 10.3349/ymj.2009.50.1.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Smith C.L., Bolton A., Nguyen G. Genomic and epigenomic instability, fragile sites, schizophrenia and autism. Curr. Genomics. 2010;11(6):447–469. doi: 10.2174/138920210793176001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Burrow A.A., Williams L.E., Pierce L.C., Wang Y.H. Over half of breakpoints in gene pairs involved in cancer-specific recurrent translocations are mapped to human chromosomal fragile sites. BMC Genomics. 2009;10:59. doi: 10.1186/1471-2164-10-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Arlt M.F., Durkin S.G., Ragland R.L., Glover T.W. Common fragile sites as targets for chromosome rearrangements. DNA Repair (Amst.) 2006;5(9-10):1126–1135. doi: 10.1016/j.dnarep.2006.05.010. [DOI] [PubMed] [Google Scholar]
- 65.Ohta M., Inoue H., Cotticelli M.G., Kastury K., Baffa R., Palazzo J., Siprashvili Z., Mori M., McCue P., Druck T., Croce C.M., Huebner K. The FHIT gene, spanning the chromosome 3p14.2 fragile site and renal carcinoma-associated t(3;8) breakpoint, is abnormal in digestive tract cancers. Cell. 1996;84(4):587–597. doi: 10.1016/S0092-8674(00)81034-X. [DOI] [PubMed] [Google Scholar]
- 66.Poulogiannis G., McIntyre R.E., Dimitriadi M., Apps J.R., Wilson C.H., Ichimura K., Luo F., Cantley L.C., Wyllie A.H., Adams D.J., Arends M.J. PARK2 deletions occur frequently in sporadic colorectal cancer and accelerate adenoma development in Apc mutant mice. Proc. Natl. Acad. Sci. USA. 2010;107(34):15145–15150. doi: 10.1073/pnas.1009941107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Gao G., Kasperbauer J.L., Tombers N.M., Wang V., Mayer K., Smith D.I. A selected group of large common fragile site genes have decreased expression in oropharyngeal squamous cell carcinomas. Genes Chromosomes Cancer. 2014;53(5):392–401. doi: 10.1002/gcc.22150. [DOI] [PubMed] [Google Scholar]
- 68.Feitelson M.A., Lee J. Hepatitis B virus integration, fragile sites, and hepatocarcinogenesis. Cancer Lett. 2007;252(2):157–170. doi: 10.1016/j.canlet.2006.11.010. [DOI] [PubMed] [Google Scholar]
- 69.Jiang S., Yang Z., Li W., Li X., Wang Y., Zhang J., Xu C., Chen P.J., Hou J., McCrae M.A., Chen X., Zhuang H., Lu F. Re-evaluation of the carcinogenic significance of hepatitis B virus integration in hepatocarcinogenesis. PLoS One. 2012;7(9):e40363. doi: 10.1371/journal.pone.0040363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ragin C.C., Reshmi S.C., Gollin S.M. Mapping and analysis of HPV16 integration sites in a head and neck cancer cell line. Int. J. Cancer. 2004;110(5):701–709. doi: 10.1002/ijc.20193. [DOI] [PubMed] [Google Scholar]
- 71.Dall K.L., Scarpini C.G., Roberts I., Winder D.M., Stanley M.A., Muralidhar B., Herdman M.T., Pett M.R., Coleman N. Characterization of naturally occurring HPV16 integration sites isolated from cervical keratinocytes under noncompetitive conditions. Cancer Res. 2008;68(20):8249–8259. doi: 10.1158/0008-5472.CAN-08-1741. [DOI] [PubMed] [Google Scholar]
- 72.Matovina M., Sabol I., Grubisić G., Gasperov N.M., Grce M. Identification of human papillomavirus type 16 integration sites in high-grade precancerous cervical lesions. Gynecol. Oncol. 2009;113(1):120–127. doi: 10.1016/j.ygyno.2008.12.004. [DOI] [PubMed] [Google Scholar]
- 73.Gandhi M., Dillon L.W., Pramanik S., Nikiforov Y.E., Wang Y.H. DNA breaks at fragile sites generate oncogenic RET/PTC rearrangements in human thyroid cells. Oncogene. 2010;29(15):2272–2280. doi: 10.1038/onc.2009.502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Burrow A.A., Marullo A., Holder L.R., Wang Y.H. Secondary structure formation and DNA instability at fragile site FRA16B. Nucleic Acids Res. 2010;38(9):2865–2877. doi: 10.1093/nar/gkp1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Mirkin S.M. DNA structures, repeat expansions and human hereditary disorders. Curr. Opin. Struct. Biol. 2006;16(3):351–358. doi: 10.1016/j.sbi.2006.05.004. [DOI] [PubMed] [Google Scholar]
- 76.Balakumaran B.S., Freudenreich C.H., Zakian V.A. CGG/CCG repeats exhibit orientation-dependent instability and orientation-independent fragility in Saccharomyces cerevisiae. Hum. Mol. Genet. 2000;9(1):93–100. doi: 10.1093/hmg/9.1.93. [DOI] [PubMed] [Google Scholar]
- 77.Zhang H., Freudenreich C.H. An AT-rich sequence in human common fragile site FRA16D causes fork stalling and chromosome breakage in S. cerevisiae. Mol. Cell. 2007;27(3):367–379. doi: 10.1016/j.molcel.2007.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Savelyeva L., Brueckner L.M. Molecular characterization of common fragile sites as a strategy to discover cancer susceptibility genes. Cell. Mol. Life Sci. 2014;71(23):4561–4575. doi: 10.1007/s00018-014-1723-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Dillon L.W., Pierce L.C., Ng M.C., Wang Y.H. Role of DNA secondary structures in fragile site breakage along human chromosome 10. Hum. Mol. Genet. 2013;22(7):1443–1456. doi: 10.1093/hmg/dds561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31(13):3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Huppert J.L., Balasubramanian S. G-quadruplexes in promoters throughout the human genome. Nucleic Acids Res. 2007;35(2):406–413. doi: 10.1093/nar/gkl1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Katapadi V.K., Nambiar M., Raghavan S.C. Potential G-quadruplex formation at breakpoint regions of chromosomal translocations in cancer may explain their fragility. Genomics. 2012;100(2):72–80. doi: 10.1016/j.ygeno.2012.05.008. [DOI] [PubMed] [Google Scholar]
- 83.Fungtammasan A., Walsh E., Chiaromonte F., Eckert K.A., Makova K.D. A genome-wide analysis of common fragile sites: what features determine chromosomal instability in the human genome? Genome Res. 2012;22(6):993–1005. doi: 10.1101/gr.134395.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Shah S.N., Opresko P.L., Meng X., Lee M.Y., Eckert K.A. DNA structure and the Werner protein modulate human DNA polymerase delta-dependent replication dynamics within the common fragile site FRA16D. Nucleic Acids Res. 2010;38(4):1149–1162. doi: 10.1093/nar/gkp1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Casper A.M., Nghiem P., Arlt M.F., Glover T.W. ATR regulates fragile site stability. Cell. 2002;111(6):779–789. doi: 10.1016/S0092-8674(02)01113-3. [DOI] [PubMed] [Google Scholar]
- 86.Casper A.M., Durkin S.G., Arlt M.F., Glover T.W. Chromosomal instability at common fragile sites in Seckel syndrome. Am. J. Hum. Genet. 2004;75(4):654–660. doi: 10.1086/422701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Cimprich K.A., Cortez D. ATR: an essential regulator of genome integrity. Nat. Rev. Mol. Cell Biol. 2008;9(8):616–627. doi: 10.1038/nrm2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Wan C., Kulkarni A., Wang Y.H. ATR preferentially interacts with common fragile site FRA3B and the binding requires its kinase activity in response to aphidicolin treatment. Mutat. Res. 2010;686(1-2):39–46. doi: 10.1016/j.mrfmmm.2009.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Ozeri-Galai E., Schwartz M., Rahat A., Kerem B. Interplay between ATM and ATR in the regulation of common fragile site stability. Oncogene. 2008;27(15):2109–2117. doi: 10.1038/sj.onc.1210849. [DOI] [PubMed] [Google Scholar]
- 90.Arlt M.F., Xu B., Durkin S.G., Casper A.M., Kastan M.B., Glover T.W. BRCA1 is required for common-fragile-site stability via its G2/M checkpoint function. Mol. Cell. Biol. 2004;24(15):6701–6709. doi: 10.1128/MCB.24.15.6701-6709.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Durkin S.G., Arlt M.F., Howlett N.G., Glover T.W. Depletion of CHK1, but not CHK2, induces chromosomal instability and breaks at common fragile sites. Oncogene. 2006;25(32):4381–4388. doi: 10.1038/sj.onc.1209466. [DOI] [PubMed] [Google Scholar]
- 92.Musio A., Montagna C., Mariani T., Tilenni M., Focarelli M.L., Brait L., Indino E., Benedetti P.A., Chessa L., Albertini A., Ried T., Vezzoni P. SMC1 involvement in fragile site expression. Hum. Mol. Genet. 2005;14(4):525–533. doi: 10.1093/hmg/ddi049. [DOI] [PubMed] [Google Scholar]
- 93.Zhu M., Weiss R.S. Increased common fragile site expression, cell proliferation defects, and apoptosis following conditional inactivation of mouse Hus1 in primary cultured cells. Mol. Biol. Cell. 2007;18(3):1044–1055. doi: 10.1091/mbc.E06-10-0957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Focarelli M.L., Soza S., Mannini L., Paulis M., Montecucco A., Musio A. Claspin inhibition leads to fragile site expression. Genes Chromosomes Cancer. 2009;48(12):1083–1090. doi: 10.1002/gcc.20710. [DOI] [PubMed] [Google Scholar]
- 95.Howlett N.G., Taniguchi T., Durkin S.G., D’Andrea A.D., Glover T.W. The Fanconi anemia pathway is required for the DNA replication stress response and for the regulation of common fragile site stability. Hum. Mol. Genet. 2005;14(5):693–701. doi: 10.1093/hmg/ddi065. [DOI] [PubMed] [Google Scholar]
- 96.Schwartz M., Zlotorynski E., Goldberg M., Ozeri E., Rahat A., le Sage C., Chen B.P., Chen D.J., Agami R., Kerem B. Homologous recombination and nonhomologous end-joining repair pathways regulate fragile site stability. Genes Dev. 2005;19(22):2715–2726. doi: 10.1101/gad.340905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Rey L., Sidorova J.M., Puget N., Boudsocq F., Biard D.S., Monnat R.J., Jr, Cazaux C., Hoffmann J.S. Human DNA polymerase eta is required for common fragile site stability during unperturbed DNA replication. Mol. Cell. Biol. 2009;29(12):3344–3354. doi: 10.1128/MCB.00115-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Bhat A., Andersen P.L., Qin Z., Xiao W. Rev3, the catalytic subunit of Polζ, is required for maintaining fragile site stability in human cells. Nucleic Acids Res. 2013;41(4):2328–2339. doi: 10.1093/nar/gks1442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Walsh E., Wang X., Lee M.Y., Eckert K.A. Mechanism of replicative DNA polymerase delta pausing and a potential role for DNA polymerase kappa in common fragile site replication. J. Mol. Biol. 2013;425(2):232–243. doi: 10.1016/j.jmb.2012.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Costanzo V., Shechter D., Lupardus P.J., Cimprich K.A., Gottesman M., Gautier J. An ATR- and Cdc7-dependent DNA damage checkpoint that inhibits initiation of DNA replication. Mol. Cell. 2003;11(1):203–213. doi: 10.1016/S1097-2765(02)00799-2. [DOI] [PubMed] [Google Scholar]
- 101.Zou L., Elledge S.J. Sensing DNA damage through ATRIP recognition of RPA-ssDNA complexes. Science. 2003;300(5625):1542–1548. doi: 10.1126/science.1083430. [DOI] [PubMed] [Google Scholar]
- 102.Choi J.H., Lindsey-Boltz L.A., Kemp M., Mason A.C., Wold M.S., Sancar A. Reconstitution of RPA-covered single-stranded DNA-activated ATR-Chk1 signaling. Proc. Natl. Acad. Sci. USA. 2010;107(31):13660–13665. doi: 10.1073/pnas.1007856107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Kim C., Paulus B.F., Wold M.S. Interactions of human replication protein A with oligonucleotides. Biochemistry. 1994;33(47):14197–14206. doi: 10.1021/bi00251a031. [DOI] [PubMed] [Google Scholar]
- 104.Fernandez-Capetillo O., Nussenzweig A. Naked replication forks break apRPArt. Cell. 2013;155(5):979–980. doi: 10.1016/j.cell.2013.10.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Toledo L.I., Altmeyer M., Rask M.B., Lukas C., Larsen D.H., Povlsen L.K., Bekker-Jensen S., Mailand N., Bartek J., Lukas J. ATR prohibits replication catastrophe by preventing global exhaustion of RPA. Cell. 2013;155(5):1088–1103. doi: 10.1016/j.cell.2013.10.043. [DOI] [PubMed] [Google Scholar]
- 106.Pirzio L.M., Pichierri P., Bignami M., Franchitto A. Werner syndrome helicase activity is essential in maintaining fragile site stability. J. Cell Biol. 2008;180(2):305–314. doi: 10.1083/jcb.200705126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Pellicioli A., Muzi-Falconi M. A blooming resolvase at chromosomal fragile sites. Nat. Cell Biol. 2013;15(8):883–885. doi: 10.1038/ncb2812. [DOI] [PubMed] [Google Scholar]
- 108.Pichierri P., Rosselli F., Franchitto A. Werner’s syndrome protein is phosphorylated in an ATR/ATM-dependent manner following replication arrest and DNA damage induced during the S phase of the cell cycle. Oncogene. 2003;22(10):1491–1500. doi: 10.1038/sj.onc.1206169. [DOI] [PubMed] [Google Scholar]
- 109.Bacolla A., Wang G., Jain A., Chuzhanova N.A., Cer R.Z., Collins J.R., Cooper D.N., Bohr V.A., Vasquez K.M. Non-B DNA-forming sequences and WRN deficiency independently increase the frequency of base substitution in human cells. J. Biol. Chem. 2011;286(12):10017–10026. doi: 10.1074/jbc.M110.176636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Kamath-Loeb A.S., Shen J.C., Schmitt M.W., Loeb L.A. The Werner syndrome exonuclease facilitates DNA degradation and high fidelity DNA polymerization by human DNA polymerase δ. J. Biol. Chem. 2012;287(15):12480–12490. doi: 10.1074/jbc.M111.332577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Lu X., Parvathaneni S., Hara T., Lal A., Sharma S. Replication stress induces specific enrichment of RECQ1 at common fragile sites FRA3B and FRA16D. Mol. Cancer. 2013;12(1):29. doi: 10.1186/1476-4598-12-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Naim V., Wilhelm T., Debatisse M., Rosselli F. ERCC1 and MUS81-EME1 promote sister chromatid separation by processing late replication intermediates at common fragile sites during mitosis. Nat. Cell Biol. 2013;15(8):1008–1015. doi: 10.1038/ncb2793. [DOI] [PubMed] [Google Scholar]
- 113.Ying S., Minocherhomji S., Chan K.L., Palmai-Pallag T., Chu W.K., Wass T., Mankouri H.W., Liu Y., Hickson I.D. MUS81 promotes common fragile site expression. Nat. Cell Biol. 2013;15(8):1001–1007. doi: 10.1038/ncb2773. [DOI] [PubMed] [Google Scholar]
- 114.Minocherhomji S., Hickson I.D. Structure-specific endonucleases: guardians of fragile site stability. Trends Cell Biol. 2014;24(5):321–327. doi: 10.1016/j.tcb.2013.11.007. [DOI] [PubMed] [Google Scholar]
- 115.Pepe A., West S.C. MUS81-EME2 promotes replication fork restart. Cell Reports. 2014;7(4):1048–1055. doi: 10.1016/j.celrep.2014.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Wang H., Li Y., Truong L.N., Shi L.Z., Hwang P.Y., He J., Do J., Cho M.J., Li H., Negrete A., Shiloach J., Berns M.W., Shen B., Chen L., Wu X. CtIP maintains stability at common fragile sites and inverted repeats by end resection-independent endonuclease activity. Mol. Cell. 2014;54(6):1012–1021. doi: 10.1016/j.molcel.2014.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Mason J.M., Das I., Arlt M., Patel N., Kraftson S., Glover T.W., Sekiguchi J.M. The SNM1B/APOLLO DNA nuclease functions in resolution of replication stress and maintenance of common fragile site stability. Hum. Mol. Genet. 2013;22(24):4901–4913. doi: 10.1093/hmg/ddt340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Inagaki H., Ohye T., Kogo H., Tsutsumi M., Kato T., Tong M., Emanuel B.S., Kurahashi H. Two sequential cleavage reactions on cruciform DNA structures cause palindrome-mediated chromosomal translocations. Nat. Commun. 2013;4:1592. doi: 10.1038/ncomms2595. [DOI] [PubMed] [Google Scholar]
- 119.Vos S.M., Tretter E.M., Schmidt B.H., Berger J.M. All tangled up: how cells direct, manage and exploit topoisomerase function. Nat. Rev. Mol. Cell Biol. 2011;12(12):827–841. doi: 10.1038/nrm3228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Been M.D., Champoux J.J. Breakage of single-stranded DNA by eukaryotic type 1 topoisomerase occurs only at regions with the potential for base-pairing. J. Mol. Biol. 1984;180(3):515–531. doi: 10.1016/0022-2836(84)90025-1. [DOI] [PubMed] [Google Scholar]
- 121.Froelich-Ammon S.J., Gale K.C., Osheroff N. Site-specific cleavage of a DNA hairpin by topoisomerase II. DNA secondary structure as a determinant of enzyme recognition/cleavage. J. Biol. Chem. 1994;269(10):7719–7725. [PubMed] [Google Scholar]
- 122.Jonstrup A.T., Thomsen T., Wang Y., Knudsen B.R., Koch J., Andersen A.H. Hairpin structures formed by alpha satellite DNA of human centromeres are cleaved by human topoisomerase IIalpha. Nucleic Acids Res. 2008;36(19):6165–6174. doi: 10.1093/nar/gkn640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Arlt M.F., Glover T.W. Inhibition of topoisomerase I prevents chromosome breakage at common fragile sites. DNA Repair (Amst.) 2010;9(6):678–689. doi: 10.1016/j.dnarep.2010.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Helmrich A., Ballarino M., Tora L. Collisions between replication and transcription complexes cause common fragile site instability at the longest human genes. Mol. Cell. 2011;44(6):966–977. doi: 10.1016/j.molcel.2011.10.013. [DOI] [PubMed] [Google Scholar]
- 125.Hashash N., Johnson A.L., Cha R.S. Topoisomerase II- and condensin-dependent breakage of MEC1ATR-sensitive fragile sites occurs independently of spindle tension, anaphase, or cytokinesis. PLoS Genet. 2012;8(10):e1002978. doi: 10.1371/journal.pgen.1002978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Dillon L.W., Pierce L.C., Lehman C.E., Nikiforov Y.E., Wang Y.H. DNA topoisomerases participate in fragility of the oncogene RET. PLoS One. 2013;8(9):e75741. doi: 10.1371/journal.pone.0075741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Ozeri-Galai E., Lebofsky R., Rahat A., Bester A.C., Bensimon A., Kerem B. Failure of origin activation in response to fork stalling leads to chromosomal instability at fragile sites. Mol. Cell. 2011;43(1):122–131. doi: 10.1016/j.molcel.2011.05.019. [DOI] [PubMed] [Google Scholar]
- 128.Letessier A., Millot G.A., Koundrioukoff S., Lachagès A.M., Vogt N., Hansen R.S., Malfoy B., Brison O., Debatisse M. Cell-type-specific replication initiation programs set fragility of the FRA3B fragile site. Nature. 2011;470(7332):120–123. doi: 10.1038/nature09745. [DOI] [PubMed] [Google Scholar]
- 129.Debatisse M., Le Tallec B., Letessier A., Dutrillaux B., Brison O. Common fragile sites: mechanisms of instability revisited. Trends Genet. 2012;28(1):22–32. doi: 10.1016/j.tig.2011.10.003. [DOI] [PubMed] [Google Scholar]
- 130.Grabczyk E., Mancuso M., Sammarco M.C. A persistent RNA.DNA hybrid formed by transcription of the Friedreich ataxia triplet repeat in live bacteria, and by T7 RNAP in vitro. Nucleic Acids Res. 2007;35(16):5351–5359. doi: 10.1093/nar/gkm589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Lin Y., Dent S.Y., Wilson J.H., Wells R.D., Napierala M. R loops stimulate genetic instability of CTG.CAG repeats. Proc. Natl. Acad. Sci. USA. 2010;107(2):692–697. doi: 10.1073/pnas.0909740107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Reddy K., Tam M., Bowater R.P., Barber M., Tomlinson M., Nichol Edamura K., Wang Y.H., Pearson C.E. Determinants of R-loop formation at convergent bidirectionally transcribed trinucleotide repeats. Nucleic Acids Res. 2011;39(5):1749–1762. doi: 10.1093/nar/gkq935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Lin Y., Wilson J.H. Transcription-induced DNA toxicity at trinucleotide repeats: double bubble is trouble. Cell Cycle. 2011;10(4):611–618. doi: 10.4161/cc.10.4.14729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Barlow J.H., Faryabi R.B., Callén E., Wong N., Malhowski A., Chen H.T., Gutierrez-Cruz G., Sun H.W., McKinnon P., Wright G., Casellas R., Robbiani D.F., Staudt L., Fernandez-Capetillo O., Nussenzweig A. Identification of early replicating fragile sites that contribute to genome instability. Cell. 2013;152(3):620–632. doi: 10.1016/j.cell.2013.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Le Tallec B., Koundrioukoff S., Wilhelm T., Letessier A., Brison O., Debatisse M. Updating the mechanisms of common fragile site instability: how to reconcile the different views? Cell. Mol. Life Sci. 2014;71(23):4489–4494. doi: 10.1007/s00018-014-1720-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Mirkin E.V., Mirkin S.M. To switch or not to switch: at the origin of repeat expansion disease. Mol. Cell. 2014;53(1):1–3. doi: 10.1016/j.molcel.2013.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Ozeri-Galai E., Tur-Sinai M., Bester A.C., Kerem B. Interplay between genetic and epigenetic factors governs common fragile site instability in cancer. Cell. Mol. Life Sci. 2014;71(23):4495–4506. doi: 10.1007/s00018-014-1719-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Coquelle A., Rozier L., Dutrillaux B., Debatisse M. Induction of multiple double-strand breaks within an hsr by meganucleaseI-SceI expression or fragile site activation leads to formation of double minutes and other chromosomal rearrangements. Oncogene. 2002;21(50):7671–7679. doi: 10.1038/sj.onc.1205880. [DOI] [PubMed] [Google Scholar]
- 139.Debatisse M., Coquelle A., Toledo F., Buttin G. Gene amplification mechanisms: the role of fragile sites. Recent Results Cancer Res. 1998;154:216–226. doi: 10.1007/978-3-642-46870-4_13. [DOI] [PubMed] [Google Scholar]
- 140.Franchitto A., Pichierri P. Replication fork recovery and regulation of common fragile sites stability. Cell. Mol. Life Sci. 2014;71(23):4507–4517. doi: 10.1007/s00018-014-1718-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Franchitto A. Genome instability at common fragile sites, searching for the cause of their instability. 2013. [DOI] [PMC free article] [PubMed]