Abstract
Thoroughbred, a relatively recent horse breed, is best known for its use in horse racing. Although myostatin (MSTN) variants have been reported to be highly associated with horse racing performance, the trait is more likely to be polygenic in nature. The purpose of this study was to identify genetic variants strongly associated with racing performance by using estimated breeding value (EBV) for race time as a phenotype. We conducted a two-stage genome-wide association study to search for genetic variants associated with the EBV. In the first stage of genome-wide association study, a relatively large number of markers (~54,000 single-nucleotide polymorphisms, SNPs) were evaluated in a small number of samples (240 horses). In the second stage, a relatively small number of markers identified to have large effects (170 SNPs) were evaluated in a much larger number of samples (1,156 horses). We also validated the SNPs related to MSTN known to have large effects on racing performance and found significant associations in the stage two analysis, but not in stage one. We identified 28 significant SNPs related to 17 genes. Among these, six genes have a function related to myogenesis and five genes are involved in muscle maintenance. To our knowledge, these genes are newly reported for the genetic association with racing performance of Thoroughbreds. It complements a recent horse genome-wide association studies of racing performance that identified other SNPs and genes as the most significant variants. These results will help to expand our knowledge of the polygenic nature of racing performance in Thoroughbreds.
Keywords: Genome-wide Association Studies [GWAS], Thoroughbred, Racing Performance, Single Nucleotide Polymorphism, Estimated Breeding Value
INTRODUCTION
The Thoroughbred which is best known for horse racing is a relatively recent horse breed derived from a small number of Arabian stallions and native British mares in 17th and 18th century England (Hill et al., 2002). To measure horse racing performance, various phenotypic values are used including race time, best race time, rank, position rates, annual earnings, and earnings per start. In particular, race time for each race is the most direct measure of speed and hence, makes it a suitable quantitative measure for evaluating the genetics of racing performance (Moritsu et al., 1994; Oki et al., 1994). In a horse breeding study, race time showed moderate heritability in the range of 0.1 to 0.3 (Mota et al., 2005), with higher heritability for shorter distance race time. Previously, a study of 12,279 racehorses registered in the Korea Racing Authority (KRA), adjusted race time showed a 0.324 heritability (Park et al., 2011). However, as racing Thoroughbreds have multiple records for race time under different conditions and environmental factors, race time alone is not suitable as phenotypic value for genome-wide association studies (GWAS). But we can show racing performance of each Thoroughbred based on racing time by estimated breeding value (EBV). Originally, EBV is genetic value of complex traits which can help breeder choices. That value means how much each Thoroughbreds will pass a trait to its progeny, genetically. The EBV for racing time is single numerical value that can show racing performance of each Thoroughbred affected by only genetic effect. So we thought that EBV was suitable for GWAS as a phenotypic value.
A candidate gene approach to identify genetic variants associated with racing performance in Thoroughbreds revealed a single-nucleotide polymorphism (SNP; ECA18 g.66493737C/T) in the first intron of the equine myostatin gene (MSTN gene) (Hill et al., 2010c). Several GWAS have confirmed this finding that SNPs within or near the MSTN gene are strongly associated with racing performance (Binns et al., 2010; Hill et al., 2010c; Tozaki et al., 2010). Although MSTN variants have been reported to be highly associated with horse racing performance, this complex trait is more likely to be polygenic in nature. In the case of human athletic performance, more than 220 genes were reported to be associated with the phenotype (Bray et al., 2009). Similarly, we speculate that other SNPs not-related to MSTN could be associated with racing performance in Thoroughbreds.
To identify the genetic basis of horse racing performance, we used the EBV of race time as the phenotype for GWAS and conducted a joint-analysis of two-stage GWAS to search for significant genetic variants associated with race time. The EBV was used as the phenotype as it only considers the genetic component of phenotypic variance, increasing the statistical power of the analysis. In the first stage of GWAS, a relatively large number of markers were evaluated in a relatively small number of samples. In the second stage, a relatively small number of markers identified as having large effects in the first stage were evaluated in a relatively large number of samples. This joint analysis of two-stage GWAS has been shown to increase the power to detect genetic association (Skol et al., 2006; Skol et al., 2007). Using this approach, we identified 28 SNPs to be associated with the Thoroughbred racing performance. The SNPs were related to 17 genes including genes for myogenesis and muscle maintenance.
MATERIALS AND METHODS
Ethics and blood collection
Korea Racing Authority has established an animal experimentation ethics committee according to the Animals Protection Act 14 of Korea. This committee, titled Korea Racing Authority Institutional Animal Care and Use Committee (KRA IACUC) is composed of two external members and three internal members. One external member is a research veterinarian with experience in experimental animals (Veterinarian Act 2, paragraph 1, in Korea) and the other member is from an animal protection organization (Animals Protection Act 14, paragraph 2, in Korea). Three internal members are composed of the general manager (Chairman of KRA IACUC) and senior managers of the Equine Health and Welfare Section and the Disease Control and Prevention Section of the veterinary Center of KRA. KRA IACUC is under the auspices of the Equine Health and Welfare section of veterinary Center of KRA. The committee operates on a regular basis rather than approving each blood collection as blood collection of the race horses are performed routinely before every race. The KRA operates experimental procedures including drug testing and ethics problem according to international guidelines, which is guaranteed by an affiliate association of the Korean government (KRA Act, Article 44) and is a member of the Association of Official Racing Chemist (AORC). In addition, the owners of the horses in KRA have granted permission for blood extraction for research and development purposes (KRA Act, Article 11, 12, 36).
Genomic DNA of the Thoroughbreds was isolated from blood collected for drug testing, health care and horse bloodlines management by the KRA. Legally, 25 mL of blood, divided into three heparin tubes must be collected from the carotid artery of all race horses participating in the race 2 to 3 hours before the race. The samples are stored at KRA, and two samples are used in drug testing, while the third sample is stored for either DNA identification or for additional drug testing. After the race, urine is collected from horses with high standing in the race for primary drug testing. If prohibited drugs are discovered in the urine, the third sample of blood collected before the race is used for secondary drug testing. From an animal welfare point of view, drug testing protects the racehorse from use of prohibited drugs for enhanced racing performance.
The DNA information from the collected blood is used to preserve horse bloodlines and used in the genetic improvement of horse by genetic methods. With the development of genomics, horse breeding and selection strategies are moving towards the use of SNP information derived from genomic DNA. So KRA uses archived blood samples of racehorses that passed the dope test both before and after the race.
For imported stallions and retired racehorses of KRA that do not participate in races, 10 mL of blood was collected for DNA extraction. The collection of blood from these horses was approved as routine procedures for DNA information storage and horse bloodline management. Detailed records related to blood collection are outlined in Supplementary Table 2. The genotyping SNPs in this study was conducted for the dual purpose of academic achievement and horse preservation within the legal and ethical framework. All blood-collection was performed by KRA veterinarians.
Estimated breeding value of race time
To improve the accuracy of the EBV, we simplified the animal model by reducing the factors deemed unnecessary (racing year, racing type, and type of weight carried). A multiple-record animal model was used to estimate the genetic parameters as breeding value. The animal model used in this study is as follows:
where Y = the vector of observations, b = the vector of fixed effects, v = the vector of random effects, pe = the vector of permanent environmental effect: common environment, a = the vector of individual additive genetic effect, Z = relationship matrix and e = the vector of residual error. X, W1, W2, and Z are coefficient matrices for b, v, pe, and a, respectively.
Observations comprised a total of 262,326 race time records from 14,752 Thoroughbreds between 1994 and 2010 at Seoul Racecourse, Busan-Gyeongnam Racecourse of the KRA. The fixed effects used were racecourse, racing distance, country of foaling, sex, and age. Racing distances in the KRA were 1,000, 1,200, 1,300, 1,400, 1,600, 1,700, 1,800, 1,900, 2,000, 2,200, and 2,300 m. Countries of foaling were divided into Korea or others with sex divided into female, male, or gelding. Random effects were moisture, jockey, weight of handicap, and trainer. Different performances in a common environment represented the difference between the repeatability and heritability. Repeatability was explained by the permanent environment of the horse during the rearing period. The relationship matrix Z included all animal racers, non-racers, reproducers, and non-reproducers. The vector of the individual genetic effect, EBV, is the coefficients vector of Z matrix. Residual error, e, represent inexplicable factors such as temporary environmental effects (e.g., racing strategy, race condition, etc.).
All parameters were estimated using the ASREML program (Gilmour, 2000) facilitated by the derivative-free restricted maximum likelihood method for a single-trait animal model. Using this model, we calculated the EBV for race time.
Initial genome-wide scan: stage 1
DNA samples were obtained from a total of 240 Thoroughbreds registered in the KRA. The 180 racehorses and 60 stallions were genotyped for the initial genome-wide scan using EquineSNP50 Genotyping BeadChips (Illumina, San Diego, CA, USA). The chip includes 54,602 SNPs that are uniformly distributed on the 31 equine autosomes and X chromosome (average density of 1 SNP per approximately 43 kb) from the EquCab2 SNP database of the horse genome. All samples were genotyped in the National Instrumentation Center for Environmental Management (NICEM) at Seoul National University. We excluded SNPs with a missing rate of >0.05, minor allele frequency of <0.05, and Hardy-Weinberg equilibrium test p-value of <0.001. SNPs on the X chromosome were also excluded, retaining 41,371 autosomal SNPs for analysis. Association analysis of stage 1 was conducted on the basis of linear regression using the software PLINK (Purcell et al., 2007).
Joint analysis in replication study: stage 2
For stage 2 association analysis, 190 SNPs selected in stage 1 and the two SNPs (BIEC2-417495 and BIEC2-417274) related to MSTN, were included. For the replicate study, 916 Thoroughbreds with more than four race records in Korea were genotyped using the customized Equine BeadXpress SNP Chips (Illumina) at NICEM. A total of 172 SNPs were successfully genotyped on the Equine SNP chips. We applied the same quality control criteria as in stage 1 and retained 158 SNPs for further analysis. For the joint analysis, we combined the data from 158 SNPs from 240 Thoroughbreds from stage 1 and 916 Thoroughbreds from stage 2. The joint association analysis was based on linear regression implemented in PLINK. The same analyses were also conducted on the 2 SNPs related to MSTN gene. Haploview program was used to visualize several linkage disequilibrium (LD) blocks of interest regions.
RESULTS
Initial genome-wide scan of 240 Thoroughbred single-nucleotide polymorphisms: stage 1
In this GWAS, we compared the genotypes of Thoroughbreds with the EBV as a phenotypic value. We conducted the initial genome-wide scan in a population of 240 Thoroughbreds. The chromosome sorting is displayed as a Manhattan plot (Figure 1). Initial analysis of our data found 3,919 SNPs that were associated with the EBV for race time (unadjusted p-value, <0.05), though none of these SNPs exceeded the threshold of multiple tests (p<2.41 ×10−6, equivalent to p = 0.05 after Bonferroni correction). At this stage, speculation on the plausibility and biological significance of these candidate SNPs is not mature because of the inevitably high false-positive rate from several tens of thousands of tests are performed on the same data set. Replication of these findings in a larger population was required to identify significant SNPs associated with racing performance. We selected 190 SNPs that were evenly distributed on the equine chromosomes for the second stage.
Figure 1.
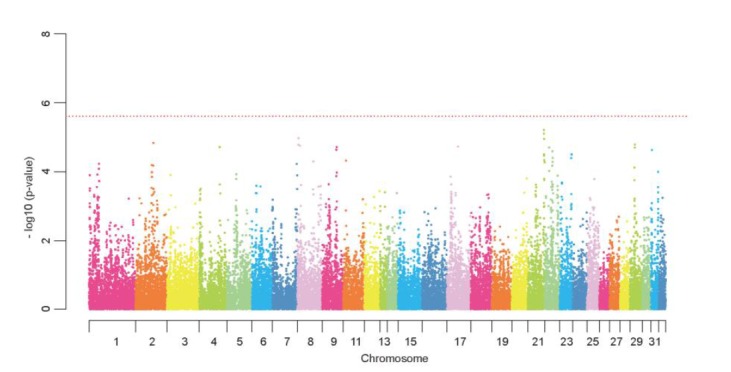
Manhattan plot of genome-wide association with the estimated breeding value for race time (Stage 1). In this plot, results are plotted as negative log-transformed p-values from the genotypic association test (observed −log 10 p-values by position); the red horizontal dotted line indicates p = 2.41×10−6 which means p = 0.05 after Bonferroni correction.
Joint analysis of 1,156 Thoroughbreds for replication study: stage 2
In the second stage, 916 additional samples were randomly selected from the Thoroughbred population of the KRA not included in stage one. To further test the association with the EBV, we genotyped 190 of the most associated SNPs from stage 1 in the 916 Thoroughbreds. After data quality control procedures, 158 SNPs were available for stage 2 for a total of 1,156 Thoroughbreds. These SNPs covered 119 distinct chromosomal regions defined by a maximal distance between two SNPs of <100 kb. Out of the 119 regions, 92 contained only one SNP, and 27 contained more than two SNPs. In joint analysis using the combined population data, 28 SNPs (17.7% of the 158 SNPs) achieved genome-wide significance criteria (p-value = 0.000632911, equivalent to p = 0.1 after Bonferroni correction) (Table 1).
Table 1.
List of SNPs with a p-value of <0.000632911 (equivalent to p = 0.1 after Bonferroni correction) based on the linear regression model of stage 2 data (n = 1,156)
| Chr | BP | Neareat gene (Ensemble gene ID) |
SNP type | Minor allele | Major allele | Stage 1 | Stage 2 | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
|
|
|||||||||||
| p-value | MAF | SNP effect | p-value | MAF | SNP effect | |||||||
| BIEC2_906777 | 5 | 40747218 | - (ENSECAG00000019204) |
InterGenic | A | G | 1.63.E-04 | 0.356 | 0.278 | 8.38.E-05 | 0.330 | 0.141 |
| BIEC2_1026019 | 8 | 5906904 | INPP5J (ENSECAG00000006664) |
InterGenic | A | G | 1.64.E-05 | 0.159 | −0.4315 | 1.73.E-04 | 0.190 | −0.163 |
| BIEC2_1026200 | 8 | 6126833 | OSBP2 (ENSECAG00000015443) |
InterGenic | G | A | 1.06.E-05 | 0.107 | −0.5102 | 1.28.E-05 | 0.142 | −0.215 |
| BIEC2_1029757 | 8 | 11590300 | MVK (ENSECAG00000023067) |
InterGenic | C | A | 1.75.E-05 | 0.119 | −0.4715 | 8.85.E-06 | 0.137 | −0.219 |
| BIEC2_330101 | 16 | 14185461 | - | InterGenic | C | A | 1.39.E-04 | 0.254 | 0.3328 | 4.29.E-06 | 0.216 | 0.195 |
| BIEC2_330360 | 16 | 15550375 | - | InterGenic | A | G | 8.12.E-04 | 0.388 | 0.2536 | 2.00.E-06 | 0.371 | 0.167 |
| BIEC2_330495 | 16 | 16289366 | CNTN3 (ENSECAG00000013575) |
Genic | A | G | 3.81.E-04 | 0.298 | 0.2847 | 1.91.E-04 | 0.271 | 0.143 |
| BIEC2_330509 | 16 | 16334663 | InterGenic | G | A | 4.43.E-04 | 0.297 | 0.2818 | 8.79.E-05 | 0.274 | 0.150 | |
| BIEC2_330558 | 16 | 16948655 | PDZRN3 (ENSECAG00000014864) |
Genic | G | A | 6.02.E-04 | 0.377 | 0.2565 | 2.28.E-05 | 0.362 | 0.150 |
| BIEC2_330572 | 16 | 16993987 | Genic | A | G | 7.93.E-04 | 0.367 | 0.2484 | 4.53.E-05 | 0.358 | 0.145 | |
| BIEC2_330575 | 16 | 16999220 | Genic | A | G | 2.32.E-04 | 0.354 | 0.2734 | 1.73.E-04 | 0.339 | 0.134 | |
| BIEC2_330578 | 16 | 17001997 | Genic | G | A | 6.02.E-04 | 0.377 | 0.2565 | 1.92.E-05 | 0.363 | 0.152 | |
| BIEC2_330677 | 16 | 17421511 | PPP4R2 (ENSECAG00000000689) |
Genic | A | G | 6.13.E-04 | 0.377 | 0.2562 | 2.66.E-05 | 0.359 | 0.149 |
| BIEC2_330691 | 16 | 17546589 | GXYLT2 (ENSECAG00000008483) |
Genic | G | A | 7.05.E-04 | 0.371 | 0.2611 | 8.94.E-08 | 0.355 | 0.191 |
| BIEC2_330725 | 16 | 17763943 | SHQ1 (ENSECAG00000015673) |
InterGenic | G | A | 4.49.E-04 | 0.325 | 0.2802 | 9.40.E-07 | 0.316 | 0.180 |
| BIEC2_330739 | 16 | 17790340 | InterGenic | G | A | 6.71.E-04 | 0.315 | 0.2672 | 2.97.E-06 | 0.310 | 0.172 | |
| BIEC2_527753 | 20 | 29897398 | VARS2 (ENSECAG00000018202) |
Genic | A | G | 4.60.E-04 | 0.219 | 0.3011 | 1.32.E-05 | 0.172 | 0.195 |
| BIEC2_527879 | 20 | 30126579 | - (ENSECAG00000015285) |
InterGenic | A | G | 4.60.E-04 | 0.219 | 0.3011 | 6.86.E-06 | 0.172 | 0.202 |
| BIEC2_529755 | 20 | 32127869 | - (ENSECAG00000017401) |
InterGenic | A | C | 5.23.E-04 | 0.256 | 0.2969 | 3.80.E-05 | 0.210 | 0.171 |
| BIEC2_529760 | 20 | 32131071 | InterGenic | C | A | 2.39.E-04 | 0.296 | 0.2909 | 4.09.E-06 | 0.239 | 0.180 | |
| BIEC2_554645 | 21 | 18154976 | ARL15 (ENSECAG00000014970) |
Genic | A | G | 1.98.E-05 | 0.218 | −0.3696 | 7.44.E-06 | 0.213 | −0.188 |
| BIEC2_554739 | 21 | 18348602 | InterGenic | C | A | 9.53.E-05 | 0.265 | −0.3267 | 1.94.E-04 | 0.256 | −0.146 | |
| BIEC2_568963 | 21 | 47502588 | - | InterGenic | A | G | 4.70.E-04 | 0.471 | −0.2539 | 1.98.E-05 | 0.474 | −0.149 |
| BIEC2_569862 | 21 | 48967346 | CCT5 (ENSECAG00000019192) |
InterGenic | G | A | 2.59.E-04 | 0.373 | −0.2739 | 2.57.E-06 | 0.381 | −0.165 |
| BIEC2_570062 | 21 | 49384234 | Genic | A | C | 1.30.E-04 | 0.404 | 0.2819 | 2.12.E-04 | 0.386 | 0.127 | |
| BIEC2_570485 | 21 | 49894191 | TAS2R1 (ENSECAG00000005160) |
InterGenic | A | G | 3.40.E-04 | 0.215 | 0.3012 | 3.48.E-04 | 0.209 | 0.148 |
| BIEC2_732151 | 28 | 17405077 | - | InterGenic | A | C | 7.52.E-05 | 0.290 | 0.3151 | 4.42.E-04 | 0.274 | 0.134 |
| BIEC2_814518 | 30 | 3869336 | RGS7 (ENSECAG00000009422) |
InterGenic | A | G | 2.33.E-05 | 0.388 | 0.3152 | 5.79.E-04 | 0.390 | 0.119 |
BP, biological process; MAF, minor allele frequency; SNP, single-nucleotide polymorphism.
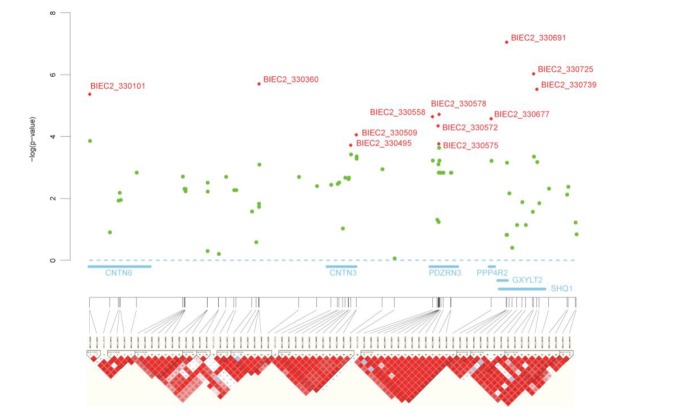
The most significant SNP (p = 8.941×10−8 in joint analysis), BIEC2_330691, identified by our two-stage GWAS was found on chromosome 16 with the other 11 most significant SNPs. Out of the 11 most significant SNPs, three were top-ranking and located in the sixth intron of the gene glucoside xylosyltransferase 2 (GXYLT2) (included in block 10 of Figure 3). Twelve of the top 27 SNPs were located together, (contained seven of 119 distinct chromosomal regions), spanning a 3.61 Mb region on chromosome 16 (chr16: 14.18 to 17.79 Mb). Those regions have not been previously reported as being related to the racing performance of Thoroughbreds (Figure 2 and Table 1). Four of these 12 significant SNPs were in PDZ domain-containing ring finger 3 (PDZRN3) genes and other four containing BIEC2_330691 was in a 0.37-Mb interval (chr16: 17.74 to 17.79 Mb). The LD calculations were performed within the 3.6-Mb region on chromosome 16 (chr16: 14.18 to 17.79 Mb). Thirteen discrete LD blocks were identified in the 3.6-Mb peak of association on chromosome 16 (Figure 3).
Figure 3.
Location of the association signal and pairwise linkage disequilibrium (LD) surrounding the most associated SNPs on chromosome 16:14.18–17.79 Mb. LD pattern is depicted using stage 1 data. Association signals are shown for all single-nucleotide polymorphisms (SNPs) genotyped in stage 1 samples (green circles, n = 240); significant SNPs in combined dataset of stage 2 (red diamond, n = 1,156). This region has six nearest genes related to significant SNPs. The most associated SNP, BIEC2-330691, lies in the gene GXYLT2 and are in small LD. Four SNPs lies in the gene PDZRN3 and are in almost complete LD. GXYLT2, glucoside xylosyltransferase 2; PDZRN3, PDZ domain-containing ring finger 3.
Figure 2.
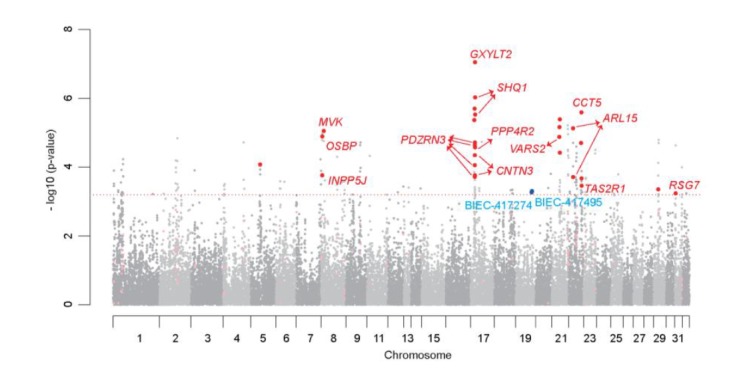
Manhattan plot of genome-wide association with the estimated breeding value for race time (Stage 1). In this plot, results are plotted as negative log-transformed p-values from the genotypic association test (observed −log 10 p-values by position); Result of stage 1 is shown in two different gray colors. Odd chromosome numbers are in dark grey, and even chromosome numbers in light grey. The red horizontal dotted line indicates stage 2 threshold. Single-nucleotide polymorphisms (SNPs) in stage 2 are shown in two red colors. Significant SNPs with a p-value less than threshold are in red, and others are in light red. Reported SNPs that associated with racing performance of Thoroughbred are in blue.
Six significant SNPs including the fourth most significant SNPs, BIEC2_569862, were located on chromosome 21. These SNPs were not located on a distinct chromosomal region but on two regions. One is chr21: 18.15 to 18.34 (2 SNPs) and another is chr21: 47.50 to 49.89 (4 SNPs). Four significant SNPs were on chromosome 20. Two SNPs were on one distinct chromosomal regions (chr20: 32.12 to 32.13, 2 SNPs) and other two were mostly near each other (chr20: 29.89 to 32.13). Three significant SNPs were dispersed in chromosome 3. Chromosome 5, 28, 30 each had one significant SNP.
Candidate genes associated with racing performance of Thoroughbreds
The transcriptional content of 28 significant SNPs was assessed using the EquCab2.0 assembly and annotation of the horse genome. The genes were annotated using the gene IDs from ENSEMBL genome browser EquCab2 (http://www.ensembl.org/). We collected the genes whose entire information was located within or near (±10 kb) the SNPs. Twenty-five SNPs associated with EBV for race time belonged to 17 genes (Ensemble Gene ID) which were mainly related to muscle terms (Table 1). Twenty SNPs of 25 SNPs were located in 13 protein-coding genes. Five of these identified genes contained more than one SNP. We found that 11 of the 13 genes associated with EBV were associated with muscle. Of the 11 genes related to muscle, six play a role in myogenesis. These six genes are GXYLT2, SHQ1 homolog, Saccharomyces cerevisiae (SHQ1), PDZRN3, ADP-ribosylation factor-like 15 (ARL15), oxysterol binding protein 2 (OSBP2), and mevalonate kinase (MVK).
GXYLT2 encodes the 37.4-kDa proteoglycan core protein, glucoside xylosyltransferase 2. Proteoglycans, one of the macromolecule groups in the extracellular matrix, impact the regulation of muscle cell proliferation and differentiation during myogenesis (Velleman et al., 2012). The fact that the SNP with the most significant p-value was located in the intron of GXYLT2 suggests that this gene is a likely to be associated with racing performance (Figure 3). SHQ1 encodes an assembly factor required for the assembly of telomerase RNPs (Grozdanov et al., 2009). O’Connor et al. (2009) showed that the telomerase activity in muscle stem cells is retained in old and age-specific telomere shortening and is not detected in the old differentiated muscle fibers in other mammals (O’Connor et al., 2009). In addition, the second and fifth most significant SNPs, BIEC2_330725 and BIEC2_330739, respectively were located near the SHQ1 gene (Figure 3). Ko (2006) suggested that PDZRN3 plays an crucial role in the differentiation of myoblasts into myotubes by acting of myogenin (Ko et al., 2006). PDZRN3 contains four significant SNPs in the 3.6-Mb region on chromosome 16 (Figure 3). Two significant SNPs were found in the ARL15 gene on chromosome 21, which encodes the ADP-ribosylation factor-like 15. ADP-ribosylation factor is reported to be critical regulator of myoblast fusion (Bach et al., 2010) (Supplementary Figure 1). OSBP2 encodes oxysterol binding protein 2 and is highly regulated during transitional-phase post-differentiation induction during myogenic differentiation (Szustakowski et al., 2006) (Supplementary Figure 4). Therefore, before oxyterol operation, mevalonate kinase (encoded by MVK gene) which is an intermediate substance of the oxysterol synthesis pathway may be important in myogenesis (Liscurn, 2002) (Supplementary Figure 5).
Five other genes identified in this study are related to muscle maintenance. The five genes include contactin 3 (CNTN3); Chaperonin Containing TCP1, Subunit 5 (CCT5); valyl-tRNA synthetase 2, mitochondrial (VARS2) and inositol polyphosphate-5-phosphatase J (INPP5J), Protein Phosphatase 4, Regulatory Subunit 2 (PPP4R2). The CNTN3 encodes contactin 1 protein, which is a member of the immunoglobulin superfamily. Jelinsky (2010) defined a set of tendon-selective genes present in both adult rat and human tendons that contained CNTN3 (Jelinsky et al., 2010). Tendon connects muscle to bone, so plays a crucial role in muscle functions. Two SNPs were located in the CNTN3 gene on chromosome 16 (Figure 3). Two SNPs were also found in the CCT5 gene on chromosome 21. TCP-1 Ring Complex encoded CCT is known to play a synergistic role in the process of actin folding (Kim et al., 2008). VARS2 encodes valyl-tRNA synthetase 2. Phosphorylation of aminoacyl-tRNA synthetases could play a role in the regulation of protein synthesis in a similar manner as insulin regulates muscle (Kimball et al., 1994). INPP5J encodes inositol polyphosphate 5-phosphatase J; its role as a second messenger in signal transduction has been well established in many cell types involved in skeletal muscle signaling (Moschella et al., 1995). PPP4R2 is important partner of survival of motor neuron (SMN) protein which affect modulation of skeletal muscle (Bosio et al., 2012).
Comparison of previously reported polymorphisms associated with racing performance
Hill (2010) and Binns (2010) previously reported a peak of association with best race distance on chromosome 18, and each identified the most important SNPs as BIEC2-417495 (Hill et al., 2010d) and BIEC2-417274 (Binns et al., 2010), respectively. These two SNPs did not exceed the threshold genome-wide significance criteria (p-value = 0.000632911, equivalent to p = 0.1 after Bonferroni correction) in stage 1 of this study. However, we tested the relationship of these SNPs with EBV once more for comparison with our result in the second stage. Two reported SNPs reached the threshold in the joint analysis of this study (Figure 2 and Table 1). Moreover, LD blocks across a 1.7-Mb region on chromosomes in a study by Hill (2010) were almost identical to the LD blocks using our stage 1 data (Hill et al., 2010c) (Figure 4).
Figure 4.

Location of the association signal and pairwise linkage disequilibrium (LD) surrounding two reported single-nucleotide polymorphisms (SNPs) related to gene myostatin. LD pattern is depicted using stage 1 data (n = 240). Association signals of two SNPs are shown in combined dataset of stage 2 (red diamond, n = 1,156).
Evaluation of single-nucleotide polymorphisms effects on estimated breeding value
Using the results of the linear regression model, we wanted to identify 28 SNP effects in this study. To evaluate the effects of 28 significant SNPs and compared them with the two SNPs of MSTN, we made 30 plots for the effect allele score with EBV (Figures 5 to 6, Supplementary Figures 7 to 10). For each subject in this population, each minor allele was scored for the EBV. Each box plot shows the relationship between the effect allele number and EBV. Each allele had either a positive or negative effect on the racing performance of Thoroughbreds. In chromosome 16, all 12 SNPs had a positive effect on racing performance (Figure 5). In other chromosomes, nine out of 15 SNPs had positive effects while the remaining seven had negative effects. (Supplementary Figures 7 to 10). Both SNPs related to MSTN showed a negative effect on the EBV (Figure 6).
Figure 5.
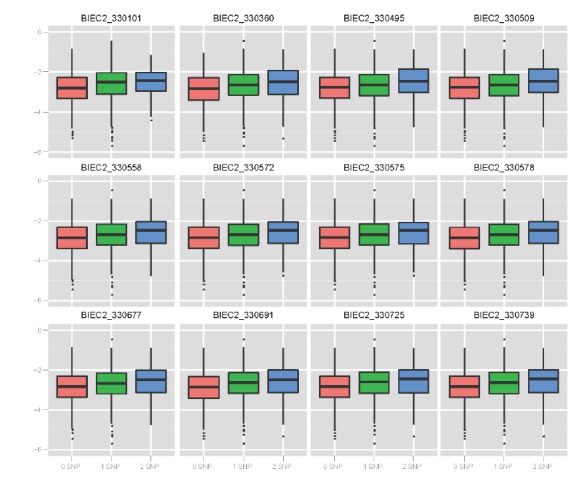
Boxplots show cumulative effect for estimated breeding value of the effect allele number of significant 12 single-nucleotide polymorphisms (SNPs) on chromosome 16. All 12 SNPs on chromosome 16 have positive effects.
Figure 6.

Boxplots show cumulative effect for estimated breeding value of the effect allele number of significant 2 single-nucleotide polymorphisms (SNPs) near myostatin. These two SNPs have negative effects.
DISCUSSION
To identify SNPs underlying the racing performance in Thoroughbred, we applied a two-stage GWAS to search for genetic variants associated with the EBV for race time. We present the results of a joint-analysis of two-stage GWAS involving 1,156 Thoroughbreds. In the first stage of our GWAS, a relatively large number of markers were evaluated in a relatively small number of samples. In the second stage, a relatively small number of markers with large effects were evaluated in a much larger number of samples. In data set of the first and second stage were combined in a joint analysis to increase the power to detect genetic associations (Skol et al., 2006; Skol et al., 2007). Using this approach, we identified 28 SNPs associated with racing performance. The SNPs were in genes related to myogenesis and muscle maintenance that have not been previously reported.
Various racing traits such as race time, best race time, rank, starting position, and annual earnings are used to measure racing performance. Out of these, race time of each race is the most direct measure of speed and a suitable quantitative measure for evaluating the racing performance of horses (Moritsu et al., 1994). However, racing performance is a complex phenotype affected by a variety of different factors such as management of Thoroughbreds, climate, geographical region, age, and reproductive status. In addition, management of exercise conditioning and nutrition has been shown to account for about 65% of racing capacity in the development of elite Thoroughbreds. Even considering these factors, previous studies show that race time had a heritability in the range of 0.1 to 0.3. Also, a significant proportion of variation in athletic ability has been shown to be heritable (Gaffney and Cunningham, 1988).
In animal breeding, EBV is used to rank breeding stock for selection as it only considers the genetic effect on phenotype and predicts the genetic value of an individual based on the phenotypes measured in their relatives. Breeding value is the sum of gene effects of an animal as measured by the performance of its progeny. To exclude other effects and increase the power of the analysis, we calculated a composite phenotype for genetic merit (i.e., EBV) of race time. Each Thoroughbred racehorse has multiple records for race time and each record was achieved under different conditions and environment factors. This makes using race time as a phenotypic value for GWAS difficult. As EBV is a single numerical prediction value that indicates how each Thoroughbred contributes to its progeny, it is suitable as a phenotype for GWAS. However, with the use of EBV as a phenotype in GWAS, inflation becomes a problem.
To investigate the population stratification in stage 1 data, we calculated the lambda value by the statistical package R. The lambda value of stage 1 data was 1.38, which is much higher than that of other studies. The inflation of significant association signals most likely resulted from the relatedness of the horses studied, which contain a massively structured population and high LD. In addition, because EBV comprises only genetic factors, the inflation value increases more than expected. However, inflation is a normal phenomenon in animal GWASs and not a problem for detecting significant SNPs. We excluded relatedness from race time as much as possible by using the EBV and conducted stringent multiple testing (Bonferroni correction) to reduce false-positive errors. Moreover, we performed association analysis using a combined dataset in stage 2 to detecting significant SNPs.
We carried out a two-stage GWAS to search for common variants associated with EBV for race time. GWAS is a promising method to discover common genetic variants that could explain diseases or economic phenotypes of animals and plants. Due to the high cost of genotyping, often a two stage design is used, in which a portion of the sample is genotyped on a large number of markers in stage 1 and a certain number of these markers are followed-up by genotyping the remaining samples in stage 2. Compared with one-stage designs that genotype all samples on all markers, well-constructed two-stage association designs maintain power and reduce cost (Thomas et al., 2004). Most often, two-stage design is used to include a replication study in the second stage to define the findings that reach statistical thresholds. We used an alternative strategy, in which the second stage analysis was a joint analysis that combines the data from both stages. This increases the power to detect genetic association (Skol et al., 2006). Skol (2006) reported that joint analysis is far more powerful than replication-based analysis (Skol et al., 2006) and recommended this strategy for two-stage GWAS studies that genotype a large portion of the samples in stage 1 and genotype a large portion of SNPs in stage 2. In this study, the number of selected markers in stage 2 was smaller than expected. However, as Thoroughbreds have stronger and larger LD than other species because of selective breeding over the three centuries, a small portion of the markers still retained power in the joint analysis stage. This indicated that a low number of markers could explain large regions of the genome. Our study was sufficiently powered to reliably detect SNPs moderately associated with the racing performance of Thoroughbreds.
Twenty-eight loci of the studied clearly achieved a genome-wide level of significance. This contrasts with GWASs in other phenotypes linked to racing performance such as racing distance, where the most associated SNP related to MSTN had a p-value of <1.61×10−9 (Hill et al., 2010c). The most significant SNP in our study had a P-value of <8.9×10−8, and 28 SNPs exceeded the threshold after multiple testing. However, no overlap was observed between the results of this study and those of previously published horse GWAS studies (Gu et al., 2010; Hill et al., 2010a,b,c). In previous studies, rare mutations with large effects on the racing performance of Thoroughbreds have been identified in genes such as MSTN. Several possible explanations exist for this discrepancy in the results. Tens of thousands of tests performed in GWASs increases the likelihood that associated SNPs in the initial genome-wide scan represent false-positives arising by chance. Another possible explanation for this lack of overlap may be that EBV is a more directly related to phenotype, which would significantly expand the power of GWASs. The identification of multiple SNPs involved in disparate biological pathways supports this notion.
In humans, numerous GWAS and candidate gene studies have revealed more than 220 gene loci (Bray et al., 2009) that influence athletic performance. This hints at the fact that racing performance has the potential to be a polygenic in nature. However in Thoroughbreds only a small number of racing performance-associated sequence variants have been reported (Gu et al., 2010; Hill et al., 2010a,b,c).
In this study, SNPs were identified in genes with diverse functions related to those previously identified as being selected in the Thoroughbred. Gu (2009) indicated that genomic regions containing genes responsible for other biological functions such as insulin signaling, fatty acid metabolism, steroid metabolic processes, and muscle strength have been selected for during the development of the Thoroughbred (Gu et al., 2009). INPP5J in chromosome 8 revealed that these enzymes regulate insulin signaling (Ooms et al., 2009). In addition, VARS2 could play a role in the regulation of protein synthesis such as that in muscle by insulin. MVK, encoding mevalonate kinase, is a part of the mevalonate pathway, which is involved in steroid biosynthesis (Goldstein and Brown, 1990). A GWAS revealed variants in ARL15 that influence adiponectin levels involved in fatty acid breakdown (Richards et al., 2009).
Due to the a priori hypothesis of the candidate gene approach, we did not perform direct experimental evaluation of genes with racing performance-associated SNPs (Jorgensen et al., 2009). The results of this study are important in that they reveal many SNPs that are associated with racing performance. Assuming that the significant variants identified in this study are truly associated with the EBV for race time, we investigated the LD block that includes the genes identified in this study. Next, assuming that LD is significant with racing performance of Thoroughbreds, we speculated that these genes are strong candidates for racing performance of Thoroughbreds. However, these biological hypotheses should be interpreted cautiously as simply the genes that are closest to the significant SNPs. In our study significant SNPs may affect the expression of cis genes up to 10 kb away or act in trans to alter gene expression on other chromosomes (Myers et al., 2007). Also the SNPs could alter the function or tissue-specific expression of a previously unidentified microRNA or genetic element. Therefore we used two indirect evaluations for our study. The first was the addition of two reported SNPs related to MSTN in the joint analysis. Based on previous candidate gene approach, SNPs, BIEC2-417495, and BIEC2-417274, near the equine MSTN gene were found to have a highly significant association with racing performance in Thoroughbreds (Binns et al., 2010; Hill et al., 2010d). Several MSTN variants are likely to be responsible for large amounts of the phenotypic variance. In domestic animals’ traits, few quantitative trait loci have been shown to have large effects on the phenotype. For example, most of the morphological traits across domestic dog breeds and the double-muscling gene in cattle are based on few SNPs. In addition, several GWASs confirmed that SNPs within or near the MSTN gene are strongly associated with racing performance in Thoroughbreds (Binns et al., 2010; Hill et al., 2010c; Tozaki et al., 2010; Tozaki et al., 2011). This region contained BIEC2-417495, BIEC2-417274, and the top SNP (g.66493737C>T) associated with optimum race distance according to Hill (2010) (Hill et al., 2010c). These SNPs are related to MSTN, which is a member of the transforming growth factor β family expressed in skeletal muscle and acts as a negative regulator of the proliferation and differentiation of myocytes (Hill et al., 2010c).
However racing performance is more likely to be polygenic in nature. In this study all p-values of the 28 selected SNPs were more significant than the two previously identified SNPs related to MSTN. Therefore we consider that the 28 SNPs of this study are as important as or more so than those two reported SNPs. The second indirect evaluation was a display of relationships between EBV and effect allele number. We noted that the two SNPs related to MSTN have a negative effect. Thoroughbreds with one or two of these SNPs could produce about 0.5-second faster progeny than those with no SNPs (Figure 5). The 28 SNPs identified in this study had similar or steeper slopes for racing performance than MSTN SNPs. Thus, we speculated that the 28 SNPs identified in this study influences the racing capacity of Thoroughbreds.
The two-stage GWAS of this study conducted in a large population of Thoroughbreds suggested the presence of 17 protein coding genes related to muscle based on 28 SNPs associated with EBV for race time. Our results strongly support a major involvement of myogenesis in the genetic predisposition for high race performance and suggest several genes as genetic factors for muscle maintenance. These candidate genes may provide insights into the genetic secrets underlying the racing performance of Thoroughbreds.
Supplementary Data
REFERENCES
- Bach A-S, Enjalbert S, Comunale F, Bodin S, Vitale N, Charrasse S, Gauthier-Rouvière C. ADP-ribosylation factor 6 regulates mammalian myoblast fusion through phospholipase D1 and phosphatidylinositol 4,5-bisphosphate signaling pathways. Mol Biol Cell. 2010;21:2412–2424. doi: 10.1091/mbc.E09-12-1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binns M, Boehler DA, Lambert DH. Identification of the myostatin locus (MSTN) as having a major effect on optimum racing distance in the Thoroughbred horse in the USA. Anim Genet. 2010;41:154–158. doi: 10.1111/j.1365-2052.2010.02126.x. [DOI] [PubMed] [Google Scholar]
- Bosio Y, Berto G, Camera P, Bianchi F, Ambrogio C, Claus P, Di Cunto F. PPP4R2 regulates neuronal cell differentiation and survival, functionally cooperating with SMN. Eur J Cell Biol. 2012;91:662–674. doi: 10.1016/j.ejcb.2012.03.002. [DOI] [PubMed] [Google Scholar]
- Bray MS, Hagberg JM, Pérusse L, Rankinen T, Roth SM, Wolfarth B, Bouchard C. The human gene map for performance and health-related fitness phenotypes: the 2006–2007 update. Med Sci Sports Exerc. 2009;41:34–72. doi: 10.1249/mss.0b013e3181844179. [DOI] [PubMed] [Google Scholar]
- Gaffney B, Cunningham EP. Estimation of genetic trend in racing performance of thoroughbred horses. Nature. 1988;332:722–724. doi: 10.1038/332722a0. [DOI] [PubMed] [Google Scholar]
- Goldstein JL, Brown MS. Regulation of the mevalonate pathway. Nature. 1990;343:425–430. doi: 10.1038/343425a0. [DOI] [PubMed] [Google Scholar]
- Grozdanov PN, Roy S, Kittur N, Meier UT. SHQ1 is required prior to NAF1 for assembly of H/ACA small nucleolar and telomerase RNPs. RNA. 2009;15:1188–1197. doi: 10.1261/rna.1532109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu J, MacHugh D, McGivney B, Park S, Katz L, Hill E. Association of sequence variants in CKM (creatine kinase, muscle) and COX4I2 (cytochrome c oxidase, subunit 4, isoform 2) genes with racing performance in Thoroughbred horses. Equine Vet J. 2010;42:569–575. doi: 10.1111/j.2042-3306.2010.00181.x. [DOI] [PubMed] [Google Scholar]
- Gu J, Orr N, Park SD, Katz LM, Sulimova G, MacHugh DE, Hill EW. A genome scan for positive selection in thoroughbred horses. PloS one. 2009;4(6):e5767. doi: 10.1371/journal.pone.0005767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill EW, Bradley DG, Al-Barody M, Ertugrul O, Splan RK, Zakharov I, Cunningham EP. History and integrity of thoroughbred dam lines revealed in equine mtDNA variation. Anim Genet. 2002;33:287–294. doi: 10.1046/j.1365-2052.2002.00870.x. [DOI] [PubMed] [Google Scholar]
- Hill EW, Eivers SS, McGivney BA, Fonseca RG, Gu J, Smith NA, Browne JA, MacHugh DE, Katz LM. Moderate and high intensity sprint exercise induce differential responses in COX4I2 and PDK4 gene expression in Thoroughbred horse skeletal muscle. Equine Vet J. 2010a;42:576–581. doi: 10.1111/j.2042-3306.2010.00206.x. [DOI] [PubMed] [Google Scholar]
- Hill EW, Gu J, McGivney BA, MacHugh DE. Targets of selection in the Thoroughbred genome contain exercise-relevant gene SNPs associated with elite racecourse performance. Anim Genet. 2010b;41:56–63. doi: 10.1111/j.1365-2052.2010.02104.x. [DOI] [PubMed] [Google Scholar]
- Hill EW, Gu J, Eivers SS, Fonseca RG, McGivney BA, Govindarajan P, Orr N, Katz LM, MacHugh D. A sequence polymorphism in MSTN predicts sprinting ability and racing stamina in thoroughbred horses. PLoS One. 2010c;5(1):e8645. doi: 10.1371/journal.pone.0008645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill EW, McGivney BA, Gu J, Whiston R, MacHugh DE. A genome-wide SNP-association study confirms a sequence variant (g. 66493737C> T) in the equine myostatin (MSTN) gene as the most powerful predictor of optimum racing distance for Thoroughbred racehorses. BMC Genomics. 2010d;11:552. doi: 10.1186/1471-2164-11-552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jelinsky SA, Archambault J, Li L, Seeherman H. Tendon-selective genes identified from rat and human musculoskeletal tissues. J Orthop Res. 2010;28:289–297. doi: 10.1002/jor.20999. [DOI] [PubMed] [Google Scholar]
- Jorgensen TJ, Ruczinski I, Kessing B, Smith MW, Shugart YY, Alberg AJ. Hypothesis-driven candidate gene association studies: practical design and analytical considerations. Am J Epidemiol. 2009;170:986–993. doi: 10.1093/aje/kwp242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Löwe T, Hoppe T. Protein quality control gets muscle into shape. Trends Cell Biol. 2008;18:264–272. doi: 10.1016/j.tcb.2008.03.007. [DOI] [PubMed] [Google Scholar]
- Kimball SR, Vary TC, Jefferson LS. Regulation of protein synthesis by insulin. Ann Rev Physiol. 1994;56:321–348. doi: 10.1146/annurev.ph.56.030194.001541. [DOI] [PubMed] [Google Scholar]
- Ko J-A, Kimura Y, Matsuura K, Yamamoto H, Gondo T, Inui M. PDZRN3 (LNX3, SEMCAP3) is required for the differentiation of C2C12 myoblasts into myotubes. J Cell Sci. 2006;119:5106–5113. doi: 10.1242/jcs.03290. [DOI] [PubMed] [Google Scholar]
- Liscurn L. Cholesterol biosynthesis. New Comprehensive Biochemistry. 2002;36:409–431. [Google Scholar]
- Moritsu Y, Funakoshi H, Ichikawa S. Genetic evaluation of sires and environmental factors influencing best racing times of Thoroughbred horses in Japan. J Equine Sci. 1994;5:53–58. [Google Scholar]
- Moschella MC, Watras J, Jayaraman T, Marks AR. Inositol 1, 4, 5-trisphosphate receptor in skeletal muscle: differential expression in myofibres. J Muscle Res Cell Motil. 1995;16:390–400. doi: 10.1007/BF00114504. [DOI] [PubMed] [Google Scholar]
- Mota MDS, Abrahão AR, Oliveira HN. Genetic and environmental parameters for racing time at different distances in Brazilian Thoroughbreds. J Anim Breed Genet. 2005;122:393–399. doi: 10.1111/j.1439-0388.2005.00551.x. [DOI] [PubMed] [Google Scholar]
- Myers AJ, Gibbs JR, Webster JA, Rohrer K, Zhao A, Marlowe L, Kaleem M, Leung D, Bryden L, Nath P, et al. A survey of genetic human cortical gene expression. Nat Genet. 2007;39:1494–1499. doi: 10.1038/ng.2007.16. [DOI] [PubMed] [Google Scholar]
- O’Connor MS, Carlson ME, Conboy IM. Differentiation rather than aging of muscle stem cells abolishes their telomerase activity. Biotechnol Prog. 2009;25:1130–1137. doi: 10.1002/btpr.223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oki H, Sasaki Y, Willham RL. Genetics of racing performance in the Japanese Thoroughbred horse. J Anim Breed, Genet. 1994;111:128–137. doi: 10.1111/j.1439-0388.1994.tb00445.x. [DOI] [PubMed] [Google Scholar]
- Ooms L, Horan K, Rahman P, Seaton G, Gurung R, Kethesparan D, Mitchell C. The role of the inositol polyphosphate 5-phosphatases in cellular function and human disease. Biochem J. 2009;419:29–49. doi: 10.1042/BJ20081673. [DOI] [PubMed] [Google Scholar]
- Park J-E, Lee J-R, Oh S, Lee JW, Oh H-S, Kim H. Principal components analysis applied to genetic evaluation of racing performance of Thoroughbred race horses in Korea. Livest Sci. 2011;135:293–299. [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MAR, Bender D, Maller J, Sklar P, De Bakker PIW, Daly MJ, Sham PC. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards JB, Waterworth D, O’Rahilly S, Hivert M-F, Loos RJF, Perry JRB, Tanaka T, Timpson NJ, Semple RK, Soranzo N, et al. A genome-wide association study reveals variants in ARL15 that influence adiponectin levels. PLoS Genet. 2009;5(12):e1000768. doi: 10.1371/journal.pgen.1000768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skol AD, Scott LJ, Abecasis GR, Boehnke M. Joint analysis is more efficient than replication-based analysis for two-stage genome-wide association studies. Nat Genet. 2006;38:209–213. doi: 10.1038/ng1706. [DOI] [PubMed] [Google Scholar]
- Skol AD, Scott LJ, Abecasis GR, Boehnke M. Optimal designs for two-stage genome-wide association studies. Genet Epidemiol. 2007;31:776–788. doi: 10.1002/gepi.20240. [DOI] [PubMed] [Google Scholar]
- Szustakowski JD, Lee J-H, Marrese CA, Kosinski PA, Nirmala N, Kemp DM. Identification of novel pathway regulation during myogenic differentiation. Genomics. 2006;87:129–138. doi: 10.1016/j.ygeno.2005.08.009. [DOI] [PubMed] [Google Scholar]
- Thomas D, Xie R, Gebregziabher M. Two-Stage sampling designs for gene association studies. Genet Epidemiol. 2004;27:401–414. doi: 10.1002/gepi.20047. [DOI] [PubMed] [Google Scholar]
- Tozaki T, Hill EW, Hirota K, Kakoi H, Gawahara H, Miyake T, Sugita S, Hasegawa T, Ishida N, Nakano Y, Kurosawa M. A cohort study of racing performance in Japanese Thoroughbred racehorses using genome information on ECA18. Anim Genet. 2012;43:42–52. doi: 10.1111/j.1365-2052.2011.02201.x. [DOI] [PubMed] [Google Scholar]
- Tozaki T, Miyake T, Kakoi H, Gawahara H, Sugita S, Hasegawa T, Ishida N, Hirota K, Nakano Y. A genome-wide association study for racing performances in Thoroughbreds clarifies a candidate region near the MSTN gene. Anim Genet. 2010;41:28–35. doi: 10.1111/j.1365-2052.2010.02095.x. [DOI] [PubMed] [Google Scholar]
- Velleman SG, Shin J, Li X, Song Y. Review: The skeletal muscle extracellular matrix: Possible roles in the regulation of muscle development and growth. Can J Anim Sci. 2012;92:1–10. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.