Abstract
A total of 59 lactic acid bacteria (LAB) strains were isolated from corn stover silage. According to phenotypic and chemotaxonomic characteristics, 16S ribosomal DNA (rDNA) sequences and recA gene polymerase chain reaction amplification, these LAB isolates were identified as five species: Lactobacillus (L.) plantarum subsp. plantarum, Pediococcus pentosaceus, Enterococcus mundtii, Weissella cibaria and Leuconostoc pseudomesenteroides, respectively. Those strains were also screened for antimicrobial activity using a dual-culture agar plate assay. Based on excluding the effects of organic acids and hydrogen peroxide, two L. plantarum subsp. plantarum strains ZZU 203 and 204, which strongly inhibited Salmonella enterica ATCC 43971T, Micrococcus luteus ATCC 4698T and Escherichia coli ATCC 11775T were selected for further research on sensitivity of the antimicrobial substance to heat, pH and protease. Cell-free culture supernatants of the two strains exhibited strong heat stability (60 min at 100°C), but the antimicrobial activity was eliminated after treatment at 121°C for 15 min. The antimicrobial substance remained active under acidic condition (pH 2.0 to 6.0), but became inactive under neutral and alkaline condition (pH 7.0 to 9.0). In addition, the antimicrobial activities of these two strains decreased remarkably after digestion by protease K. These results preliminarily suggest that the desirable antimicrobial activity of strains ZZU 203 and 204 is the result of the production of a bacteriocin-like substance, and these two strains with antimicrobial activity could be used as silage additives to inhibit proliferation of unwanted microorganism during ensiling and preserve nutrients of silage. The nature of the antimicrobial substances is being investigated in our laboratory.
Keywords: Antimicrobial Activity, Corn Stover Silage, Identification, Lactic Acid Bacteria
INTRODUCTION
Corn (Zea mays L.) is one of the high and stable-yield crops in the world, and although the stover still contains good nutrients suitable for livestock, 30% is generally incinerated in the corn field after corn cobs have been harvested. Finding a process to improve the nutritional value of corn stover has attracted increasing attention. Several processes have been developed that rely on heavy chemical treatments with ammonia and sodium hydroxide, but most of the processes were observed to reduce the palatability of stover to ruminants (Pang et al., 2011). Silage is a common preserved feed in many countries, including China. Epiphytic microorganisms, especially lactic acid bacteria (LAB), naturally present on forage crops are responsible for silage fermentation and also influence silage quality because LAB present in forage crops can convert sugar into lactic acid during the ensiling process (Muck et al., 1989; Cai et al., 1999), reduce the loss of nutrition by inhibiting the growth of undesirable microorganisms, and produce a special fragrance and flavour that are attractive to ruminant animals. Currently, corn stover silage has become more attractive due to its high nutritional value and good palatability and is rich in water-soluble carbohydrates which make it suitable for the LAB inhabit (Pang et al., 2011). For this reason we chose corn stover as research material for LAB isolation and selection.
An important objective of ensiling is to avoid the proliferation of harmful spoilage organisms (Tohno et al., 2012). The widespread use of antimicrobials as growth promoters in food for animals has been associated with the development of resistant bacteria (Karska-Wysocki et al., 2010). Recent scientific attention has been directed towards finding new ways to inhibit the colonisation of pathogenic bacteria. The consumption of probiotic cultures is a good way to exert a protective effect against pathogenic infections (Simova et al., 2008). Certain LAB strains can be used as probiotics for animals, not only because of the production of lactic acid, but other compounds, such as bacteriocins and bacteriocin-like substances (BLS) (Todorov et al., 2011) are produced. Different from antibiotics, bacteriocins produced by LAB are generally recognized as safe substances, they are ribosomal synthesized and become inactivated by digestive proteases (Riley, 2009; Hellal et al., 2012). Because of their potential use as non-toxic and safe additives for food preservation, bacteriocins from LAB have attracted increased attention (Hata et al., 2010). So choosing bacteriocin-producing LAB as inoculant will inhibit the activity of undesirable bacteria and enhance lactic acid fermentation to better preserve the ensiled crops (Weinberg et al., 2004). Compared to the ordinary bacteriocins produced by LAB, which are only active against closely related species, such as certain other Gram-positive bacteria (Cleveland et al., 2001), some bacteriocins and BLS may exhibit much broader antimicrobial activity. In other words, some of them can be active against both Gram-positive and certain Gram-negative bacteria (Todorov et al., 2011). In this research, we selected Salmonella enterica and Escherichia coli as Gram-negative indicator bacteria and Micrococcus luteus as Gram-positive indicator bacteria to identify the antimicrobial activity of LAB strains. Salmonella enterica is a common pathogenic bacteria to livestock, while the other two bacteria are also undesirable in silage, because their growth and proliferation not only consume nutriments, but may cause disease to animals (Sharma and Sihag, 2013; Chaudhari and Kariyawasam, 2014).
There are many reports about the characterization of LAB isolated from forage crops and grasses, e.g. corn, alfalfa, clover, sainfoin, timothy, orchardgrass, and so on (Pang et al., 2012; Tohno et al., 2012), and there has been much recent interest in researching LAB bacteriocins. A number of bacteriocins have been described for Lactobacillus (L.) plantarum isolated from such fermented products as milk, cheese, fermented cucumber, olives and dough (Todorov et al., 2011). However, adequate information regarding the characteristics and antimicrobial activity of natural LAB from corn stover silage is lacking. Therefore, the present study set out to screen, isolate and identify the LAB from corn stover silage in Henan province, China. The antimicrobial effects of these isolates and the sensitivity of the antimicrobial substance to heat, pH and protease were also subsequently evaluated. To the best of our knowledge, this is the first report describing the isolation of LAB with antimicrobial activity from corn stover silage, and investigating their characteristic for their further application in feed additives.
MATERIALS AND METHODS
Silages and bacterial isolates
Corn (cultivar: Zhengchaotian 2) stover was collected manually in Henan province, China, in September, 2011, and then samples were chopped to about 2 cm segments. Dry matter (DM) of the fresh corn stover material was 73.62%, and it contained 7.03% crude protein, 2.63% ether extract, 93.25% organic matter on a DM basis, which were measured based on the methods described by AOAC (1990). One hundred gram of the samples were packed into plastic film bags (Hiryu KN type; 18×26 cm; Asahikasei, Tokyo, Japan), and the bags were sealed with a vacuum sealer (BH 950; Panasonic, Osaka, Japan). Five small-scale silage sample bags were stored at room temperature (18°C to 26°C).
In order to comprehensively understand the change of microorganisms after ensiling, we investigated the numbers of 7 kinds of microorganisms: LAB, coliform bacteria, molds, yeasts, bacilli, aerobic bacteria and clostridia. Ten gram of fresh corn stover material were shaken well with 90 mL of sterilized distilled water, then a further tenfold serial dilutions, ranging from 10−2 to 10−5, were prepared and incubated anaerobically at 30°C for 48 h on lactobacilli de Man, Rogosa and Sharpe (MRS) agar (Difco Laboratories, Detroit, USA) before the numbers of LAB were measured by the plate count method. Coliform bacteria were cultivated on blue light broth agar (Nissui Ltd., Tokyo, Japan) incubated at 30°C for 48 h. Molds and yeasts were plated on potato dextrose agar incubated at 30°C for 24 h, and yeasts were distinguished from molds or bacteria by colony appearance and observation of cell morphology. Bacilli and aerobic bacteria were cultured onto nutrient agar under aerobic conditions for 24 h at 30°C. Clostridia were cultivated on clostridia count agar (Nissui Ltd., Tokyo, Japan) incubated anaerobically at 30°C for 48 h. Colonies were counted as viable numbers of microorganisms in colony forming units (CFU) per gram of fresh matter (FM). The isolation of LAB was determined by the Gram-stain appearance, catalase test and lactic acid productivity, and the pure cultures of LAB were grown on MRS agar at 30°C for 24 h, transferred to nutrient broth (Difco Laboratories, USA) with dimethyl sulfoxide at a ratio of 9:1, then stored as stock cultures at −80°C for further examination (Pang et al., 2011).
After 30 days ensiling, three bags at random were selected from those five silage bags for sampling, and a well-distributed 10 g of corn stover silage were taken out of each bag, respectively. Seven kinds of microorganisms in silage were measured according to the same methods as the fresh corn stover. Three replicate samples were used for microbial analysis for both the fresh corn stover and the silage materials, and the final results were expressed as mean values.
Morphological, physiological and biochemical tests
A total of 59 LAB strains were isolated from the silage samples, all of these were characterized phenotypically based on their morphology, production of gas from glucose according to Kozaki et al. (1992). Growth at different temperatures was observed in MRS broth after incubation at 5°C and 10°C for 14 days, respectively, and at 15°C, 20°C, 25°C, 30°C, 35°C, 40°C, 45°C, and 50°C for 7 days. Salt tolerance was determined in MRS broth with NaCl at 3.0 and 6.5% for 2 days. Growth of LAB at pH 3.0, 3.5, 4.0, 4.5, 5.0, 6.0, 7.0, 8.0, and 9.0 was determined in MRS broth after incubation at 30°C for 7 days. Triplicate preparations for each treatment and the final results were expressed as means.
Carbohydrate fermentation tests were carried out with the analytical profile index (API 50 CH) strips (bioMerieux, Tokyo, Japan) of 49 different compounds and one control, according to the manufacturer’s instructions, and reactions were determined after incubation at 30°C for 48 h.
16S rRNA gene sequencing
Fifty-nine LAB isolates grown in MRS broth for 8 h at 30°C were used for DNA extraction and purification, as described by Saitou and Miura (1963). The 16S rRNA gene sequence coding region was amplified by polymerase chain reaction (PCR) performed in a PCR thermal cycler (T-Gradient Thermoblock, Biometra Analytik GmbH, Gottingen, Germany). The PCR carried out using a TaKaRa 16S rDNA Bacterial Identification PCR Kit with the prokaryotic 16S rDNA universal primers 27F (5′-AGAGTTTGATCCTGGCTCAG-3′) and 1492R (5′-GGTTACCTTGTTACGACTT-3′). Sequence similarity searches were performed in the GenBank data library using the Basic Local Alignment Search Tool program. The sequence information was then imported into the CLUSTAL W software program for assembly and alignment. Phylogenetic trees were constructed by the neighbor-joining method (Saitou and Nei, 1987). Bacillus subtilis NCDO 1769T was used as an outgroup organism. The topologies of trees were evaluated by bootstrap analysis of the sequence data with molecular evolutionary genetics analysis 4 software (Tamura et al., 2007), based on 1,000 random resamplings (Eitan et al., 2006).
For further discrimination of strains in the L. plantarum group, a multiplex PCR assay was performed with the recA gene-based primers: paraF (5′-GTCACAGGCATTACGAA AAC-3′), pentF (5′-CAGTGGCGCGGTTGATATC-3′), planF (5′-CCGTTTATGCGGAACACCTA-3′), and pREV (5′-TCGGGATTACCAAACATCAC-3′). The PCR mixture and amplifications were performed as described by Torriani et al. (2001).
The nucleotide sequences for the 16S rDNA described in this report were deposited with the DNA Data Bank of Japan (DDBJ)/GenBank/European Molecular Biology Laboratory under accession nos. AB831177-AB831188 for the strains ZZU 204, ZZU 273, ZZU 274, ZZU 278, ZZU 283, ZZU 299, ZZU 203, ZZU 64, ZZU 205, ZZU 223, ZZU 170, and ZZU 50.
Detection inhibition of lactic acid bacteria
The selected LAB strains were inoculated into the tubes containing 1.5 mL MRS broth each and statically incubated at 30°C for 48 h. Cell free culture supernatants were obtained by centrifugation (10,000×g, 4°C, 20 min) of LAB cultures, and the supernatants were filtered through a 0.22 μm filter to remove residual cells, then, they were examined by the diameters of inhibition zones using agar diffusion assay method (Ennahar et al., 2000) with Salmonella enterica ATCC 43971T, Escherichia coli ATCC 11775T (Gram-negative bacteria) and Micrococcus luteus ATCC 4698T (Gram-positive bacteria) as indicator strains. Briefly, 300 μL of cell free supernatants were placed into wells (7.80 mm in diameter) on nutrient agar plates seeded with the above indicator strains. After incubation at 30°C for 12 h, the diameters of inhibition zones were recorded by vernier caliper (7-39792, measuring range: 0–150 mm, Wuhan Measuring Tool Enterprise, Wuhan, China).
In order to eliminate the organic acid function, the pH value of the cell free culture supernatants was adjusted to 6.0 with NaOH (2 mol/L) and the pH of MRS broth was also adjusted to the same value using lactic acid (2 mol/L).
The cell free culture supernatants (pH 6.0) were incubated at 37°C for 2 h with catalase at a final concentration of 3 mg/mL, and the same supernatants without catalase were used as control to eliminate the possible inhibitory action of hydrogen peroxide.
After eliminating organic acid function and hydrogen peroxide, the cell free culture supernatants still retaining antimicrobial activity were selected out to determine the possible protein nature of the detected antimicrobial substances. The bacterial cell free culture supernatants (pH 6.0) were incubated at 37°C for 2 h with trypsin or protease K at a final concentration of 3 mg/mL individually, and those without enzyme treatment were used as control.
Sensitivity of the antimicrobial substance to heat and pH
Based on the research above, cell free culture supernatants of the LAB strains sensitive to protease were chosen for heat and pH stability tests. The thermal stability was tested by heating the cell free supernatants to 60°C, 80°C, and 100°C for 30 and 60 min and in an autoclave at 121°C for 15 min, then the activity determined as described before. The pH stability was determined by adjusting the cell free supernatant to pH 2.0 to 9.0 with sterile NaOH or lactic acid (both are 2 mol/L). After 2 h of incubation at 30°C, the samples were readjusted to pH 6.0 and the activity determined as described above.
RESULTS
Counts of microorganisms
The counts of microorganisms in fresh corn and 30 day silage are shown in Table 1. The counts of LAB increased from 1.4×105 CFU/g FM to 2.4×106 CFU/g FM after 30 days of ensiling, while other microorganisms were reduced by different degrees, especially molds and clostridia, which were not detected after ensiling. The counts of coliform bacteria were reduced from 1.2×105 to 1.1×103 CFU/g FM, and yeasts were reduced from 2.3×105 to 2.0×102 CFU/g FM. Three replicate samples were used for microbial analysis for both of the fresh corn stover and the silage materials, the final results were expressed as mean values.
Table 1.
Microbiological analysis of the fresh whole-plant corn and 30 days silage
| Sample | Counts (CFU/g of FM) of viable microorganisms | ||||||
|---|---|---|---|---|---|---|---|
|
| |||||||
| Lactic acid bacteria | Bacilli | Coliform bacteria | Aerobic bacteria | Molds | Yeasts | Clostridia | |
| Fresh whole-plant corn | 1.4×105 | 8.6×102 | 1.2×105 | 3.3×105 | 5.8×104 | 2.3×105 | 9.0×103 |
| 30 days corn stover silage | 2.4×106 | 4.2×102 | 1.1×103 | 2.0×103 | ND | 2.0×102 | ND |
CFU, colony forming unit; FM, fresh matter; ND, not detected.
Morphological, physiological and biochemical properties
A total of 59 isolates from corn silage were considered as LAB, based on the Gram-positive, reaction, negative catalase reaction, and production of lactic acid as the main fermentation product. The strains were divided into five groups (A to E) on the basis of morphological, physiological and biochemical properties. Strains in groups A (5 strains), C (5 strains), and E (5 strains) had the same growth temperature, salt tolerance, growth pH, and carbohydrate fermentation patterns. On the other hand, strains in groups B (10 strains) and D (34 strains) were separated into two and seven subgroups with different properties, respectively. There were total 12 subgroups and each representative strain from these subgroups is listed in Table 2 and 3. All of the isolates were Gram-positive and catalase-negative rods or cocci which could grow at pH ranging from 4.5 to 9.0 and at temperatures between 15°C and 40°C. The strains would not grow at pH levels of 3.0 or below and at temperatures of 50°C or above. Only strains in group A and group C could produce gas from glucose and form D (−) isomers of lactic acid, while other isolates could not. Most of the strains could grow in 3.0% (w/v) NaCl, except for ZZU 223. None of the strains produced acid from glycerol, D-arabinose, L-xylose, β-Methyl-xyloside, dulcitol, inositol, glycogene, xylitol, D-lyxose, D-fucose, L-arabitol, 2-ceto-gluconate, 5-ceto-gluconate. All of the strains produced acid from galactose, D-glucose, D-fructose, d-mannose, esculin, cellobiose, and saccharose. There was only one strain, ZZU 203, in group D that produced acids from L-sorbose. Strain ZZU 223 produced acids from the L-fucose, while strains in other groups did not. Strains in groups A and E did not produce acids from melezitose and D-raffinose, but the strains in other groups did. Strains in group A did not produce acids from L-arabinose and ribose, while strains in other cocci groups did. Strains in group C produced acids from α-methyl-D-glucoside, while strains in other groups did not. However, these strains did not produce acids from D-tagatose, while strains in other cocci groups did.
Table 2.
Characteristics of lactic acid bacteria strains isolated from silage samples
| Character | Group A | Group B | Group C | Group D | Group E | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
|
|
|
|
|
||||||||
| ZZU 50 | ZZU 64 | ZZU 223 | ZZU 170 | ZZU 203 | ZZU 204 | ZZU 273 | ZZU 274 | ZZU 278 | ZZU 283 | ZZU 299 | ZZU 205 | |
| Shape | Cocci | Cocci | Cocci | Cocci | Rod | Rod | Rod | Rod | Rod | Rod | Rod | Cocci |
| Gram stain | + | + | + | + | + | + | + | + | + | + | + | + |
| Catalase | − | − | − | − | − | − | − | − | − | − | − | − |
| Gas from glucose | + | − | − | + | − | − | − | − | − | − | − | − |
| Optical form of lactate | D(−) | DL | DL | D(−) | DL | DL | DL | DL | DL | DL | DL | L(−) |
| Fermentation type | Hetero | Homo | Homo | Hetero | Homo | Homo | Homo | Homo | Homo | Homo | Homo | Homo |
| Growth at temperature (°C) | ||||||||||||
| 5 | − | − | − | − | − | − | − | − | − | − | − | w |
| 10 | w | + | + | + | + | + | + | + | + | + | + | + |
| 15 | + | + | + | + | + | + | + | + | + | + | + | + |
| 20 | + | + | + | + | + | + | + | + | + | + | + | + |
| 25 | + | + | + | + | + | + | + | + | + | + | + | + |
| 30 | + | + | + | + | + | + | + | + | + | + | + | + |
| 35 | + | + | + | + | + | + | + | + | + | + | + | + |
| 40 | + | + | + | + | + | + | + | + | + | + | + | + |
| 45 | w | + | w | − | w | + | + | + | w | + | + | + |
| 50 | − | − | − | − | − | − | − | − | − | − | − | − |
| Growth in NaCl (w/v %) | ||||||||||||
| 3.0 | + | + | w | + | + | + | + | + | + | + | + | + |
| 6.5 | − | + | w | − | w | w | − | − | − | − | − | w |
| Growth at pH | ||||||||||||
| 3.0 | − | − | − | − | − | − | − | − | − | − | − | − |
| 3.5 | w | + | w | w | w | + | w | w | + | + | w | w |
| 4.0 | + | + | w | + | + | + | + | + | + | + | + | + |
| 4.5 | + | + | + | + | + | + | + | + | + | + | + | + |
| 5.0 | + | + | + | + | + | + | + | + | + | + | + | + |
| 6.0 | + | + | + | + | + | + | + | + | + | + | + | + |
| 7.0 | + | + | + | + | + | + | + | + | + | + | + | + |
| 8.0 | + | + | + | + | + | + | + | + | + | + | + | + |
| 9.0 | + | + | + | + | + | + | + | + | + | + | + | + |
+, positive; −, negative; w, weakly positive; Homo, homofermentative; Hetero, heterofermentative.
Table 3.
API 50 CH fermentation patterns of latic acid bacteria
| Substrate | Group A | Group B | Group C | Group D | Group E | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
|
|
|
|
|
||||||||
| ZZU 50 | ZZU 64 | ZZU 223 | ZZU 170 | ZZU 203 | ZZU 204 | ZZU 273 | ZZU 274 | ZZU 278 | ZZU 283 | ZZU 299 | ZZU 205 | |
| Erythritol | − | − | w | − | − | − | − | − | − | − | − | − |
| L-arabinose | w | + | + | + | + | + | + | + | + | + | + | + |
| Ribose | − | + | + | + | + | w | + | + | + | w | + | + |
| D-xylose | w | w | − | + | − | − | − | − | − | − | − | w |
| Adonitol | − | w | − | − | − | − | − | − | − | − | − | − |
| L-sorbose | − | − | − | − | + | − | − | − | − | − | − | − |
| Rhamnose | − | − | w | − | w | − | − | − | − | − | − | − |
| Mannitol | + | + | + | + | + | + | + | + | + | + | + | w |
| Sorbitol | − | + | + | − | + | + | + | + | + | + | + | w |
| α-Methyl-D-mannoside | − | − | − | − | + | − | + | + | + | + | + | − |
| α-Methyl-D-glucoside | − | − | − | + | − | − | − | − | − | − | − | − |
| N-Acetyl glucosamine | w | w | + | w | + | + | w | + | w | w | + | + |
| Amygdaline | + | w | + | − | + | + | + | + | + | + | + | w |
| Arbutine | + | w | + | − | + | + | + | + | + | + | + | w |
| Salicine | + | w | + | w | + | + | w | + | w | + | + | w |
| Maltose | w | + | + | + | + | + | + | + | + | + | + | + |
| Lactose | + | + | + | − | + | + | + | + | + | + | + | w |
| Melibiose | − | + | + | + | + | + | + | + | + | + | + | w |
| Trehalose | + | + | + | w | + | + | + | + | + | + | + | + |
| Inuline | − | − | − | − | − | − | − | − | − | − | w | − |
| Melezitose | − | + | + | + | + | + | + | + | + | + | + | − |
| D-raffinose | − | + | + | + | + | + | + | + | + | + | + | − |
| Starch | − | − | − | − | − | − | − | − | − | − | w | − |
| β-Gentiobiose | − | w | w | − | w | w | w | w | w | w | w | w |
| D-turanose | − | + | + | w | + | + | + | + | + | + | + | − |
| D-tagatose | + | + | + | − | − | − | − | + | + | − | − | + |
| L-ffucose | − | − | + | − | − | − | − | − | − | − | − | − |
| D-arabitol | − | w | − | − | − | − | − | − | − | − | − | − |
| Gluconate | − | w | − | − | − | − | w | w | w | w | w | − |
API, analytical profile index; +, positive; −, negative; w, weakly positive.
All strains produced acid from galactose, D-glucose, D-fructose, D-mannose, esculine, cellobiose, saccharose, but failed to produce acid from glycerol, D-arabinose, L-xylose, β-methyl-xyloside, dulcitol, inositol, glycogene, xylitol, D-lyxose, D-fucose, L-arabitol, 2-ceto-gluconate, 5-ceto-gluconate.
16S rRNA gene sequencing
The molecular classification of these isolated LAB strains was determined by 16S rDNA sequence analysis. Phylogenetic trees that were constructed using 12 representative strains, which were selected according to their morphological, physiological and biochemical properties, based on evolutionary distances determined by a neighbour-joining method are shown in Figure 1 and 2. The strain ZZU 50 in group A was placed in the Weissella cluster, with the species Weissella cibaria IMAU 10288T being the most closely related species, which was also supported by 99.7% sequence similarity. Strains in Group B were placed in the Pediococcus cluster and identified as Pediococcus pentosaceus with 16S rDNA sequence similarities above 99.8% with type strain DSM 20336T. Group C strain was placed in the cluster of the genus Leuconostoc in the phylogenetic tree and could be unquestionably identified as Leuconostoc pseudomesenteroides. Strains in Group D were grouped on the phylogenetic tree together with L. pentosus, L. plantarum subsp. plantarum, L. plantarum subsp. argentoratensis and L. paraplantarum, but they could not be identified on a species level based on 16S rRNA gene sequence analysis. Group E was placed in a cluster consisting of the genus Enterococcus (E.) and was assigned with confidence to E. mundtii.
Figure 1.
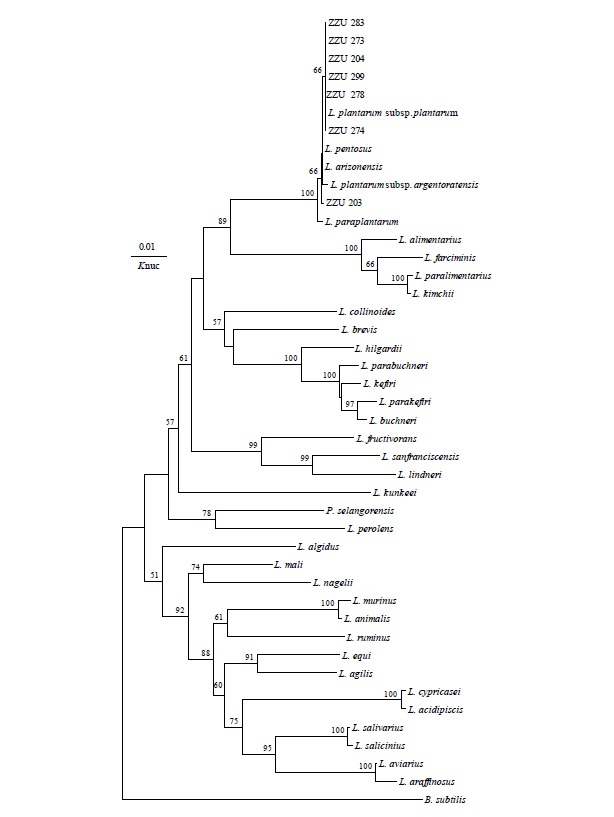
Phylogenetic tree showing the relative position of Lactobacillus plantarum as inferred by the neighbor-joining method with 16S rRNA gene sequences. Bootstrap values for a total of 1,000 replicates are shown at the nodes of the tree. Bacillus subtilis is used as an outgroup. The bar indicates 1% sequence divergence. Knuc, nucleotide substitution rates.
Figure 2.
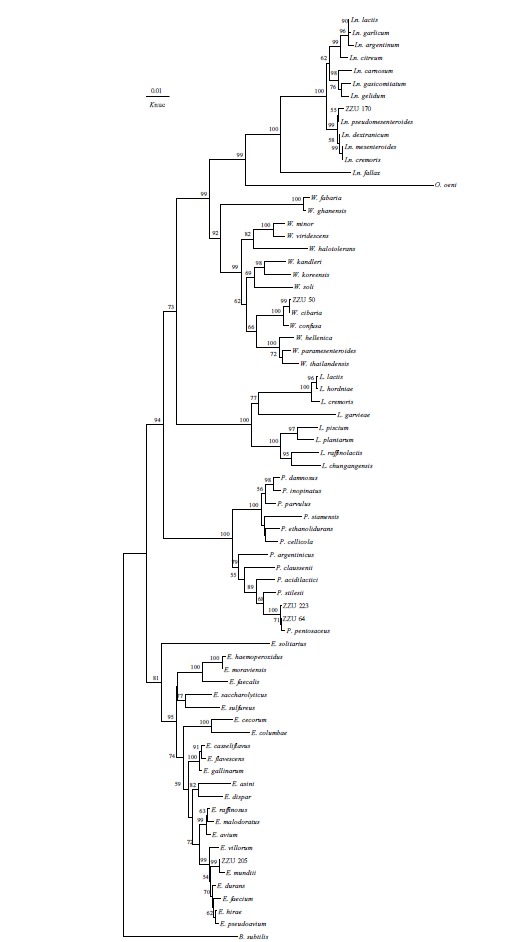
Phylogenetic tree showing the relative position of Leuconostoc pseudomesenteroides, Weissella cibaria, Pediococcus pentosaceus and Enterococcus mundtii species as inferred by the neighbor-joining method with 16S rRNA gene sequences. Bacillus subtilis is used as an outgroup. The bar indicates 1% sequence divergence. Ln., Leuconostocs; L., Lactococcus; E., Enterococcus; P., Pediococcus and W., Weissella.
Amplification products obtained from the recA gene multiplex assay
Amplification products obtained from the recA gene are shown in Figure 3. Group D strains and type strain L. plantarum subsp. plantarum JCM 1149T produced 318 bp recA gene amplification products. Type strain L. paraplantarum JCM 12533T and L. pentosus JCM 1558T produced 107 bp and 218 bp, respectively. L. plantarum subsp. argentoratensis JCM 16169T produced two distinct PCR products of 318 and 120 bp, while the negative control L. casei JCM 16167T did not produce any amplicons. Thus, strains in group D were clearly identified as L. plantarum subsp. plantarum.
Figure 3.

Amplification products obtained from the recA multiplex assay. Lane M contained a 2-kb PLUS DNA ladder (Wako Pure Chemical Industries, Ltd., Tokyo, Japan). Lanes 1, 2, 3, 4, and 5, PCR amplification products from L. casei JCM 16167T (negative control), L. paraplantarum JCM 12533T, L. pentosus JCM 1558T, L. plantarum subsp. plantarum JCM 1149T, L. plantarum subsp. argentoratensis JCM 16169T respectively; Lanes 6, 7, 8, 9, 10, 11, and 12 PCR amplification products from ZZU 203, ZZU204, ZZU273, ZZU274, ZZU278, ZZU 283, and ZZU 299, respectively. PCR, polymerase chain reaction.
In conclusion, the LAB species in this study were finally classified as Weissella cibaria (8.5%), Pediococcus pentosaceus (16.9%), Leuconostoc pseudomesenteroides (8.5%), L. plantarum subsp. plantarum (57.6%) and E. mundtii (8.5%).
Antibacterial activities
A total of 59 LAB isolates were investigated, of which 56 strains showed distinct antimicrobial activity against Salmonella enterica ATCC 43971T, Micrococcus luteus ATCC 4698T and Escherichia coli ATCC 11775T indicators. There were 39 strains with inhibitory activity against both Gram-positive bacteria, such as Micrococcus luteus ATCC 4698T, and Gram-negative bacteria, such as Salmonella enterica ATCC 43971T and Escherichia coli ATCC 11775T. The antibacterial activities of the representative strains (diameter of inhibition zone is more than 8.00 mm) are shown in Table 4.
Table 4.
Antimicrobial activity of the representative strains
| Strain number | Antimicrobial activity | ||
|---|---|---|---|
|
| |||
| Salmonella enterica ATCC 43971T | Escherichia coli ATCC 11775T | Micrococcus luteus ATCC 4698T | |
| ZZU 50 | ++ | +++ | +++ |
| ZZU 64 | + | + | ++ |
| ZZU 170 | + | + | + |
| ZZU 203 | +++ | +++ | +++ |
| ZZU 204 | ++ | +++ | ++++ |
| ZZU 205 | + | ++ | + |
| ZZU 223 | + | +++ | + |
| ZZU 273 | ++ | +++ | +++ |
| ZZU 274 | +++ | ++ | +++ |
| ZZU 278 | +++ | +++ | +++ |
| ZZU 283 | +++ | + | +++ |
| ZZU 299 | +++ | + | +++ |
After eliminating organic acid function, the MRS broth had no antimicrobial activity and only cell free supernatants of strain ZZU 203 and 204 still exhibited distinct inhibitory activity against all the indicators (Table 5). Further, after catalase treatment, the antimicrobial activity was still the same as the control. After treatment by protease K, the antimicrobial activity of cell free supernatants of the two strains disappeared. After treatment with trypsin the antimicrobial activity of strain ZZU 203 disappeared, while ZZU 204 still exhibited residual antimicrobial activity.
Table 5.
Antimicrobial activity of ZZU 203 and 204 by several treatments
| Indicator strains | Antimicrobial activity | |||||||
|---|---|---|---|---|---|---|---|---|
|
| ||||||||
| pH 6.0 | Catalase | Proteinase K | Trypsin | |||||
|
|
|
|
|
|||||
| ZZU 203 | ZZU 204 | ZZU 203 | ZZU 204 | ZZU 203 | ZZU 204 | ZZU 203 | ZZU 204 | |
| Salmonella enterica ATCC 43971T | + | ++ | + | ++ | − | − | − | − |
| Escherichia coli ATCC 11775T | ++ | ++ | ++ | ++ | − | − | − | + |
| Micrococcus luteus ATCC 4698T | +++ | ++ | +++ | ++ | − | − | − | + |
+, Diameter of inhibition zone: 8.00 to 12.00 mm; ++, 12.00 to 16.00 mm; +++, 16.00 to 20.00 mm; ++++, more than 20.00 mm; −, no inhibition zone; the diameter of inhibition zone including that of Oxford cup (7.80 mm).
Table 6 presents the residual antimicrobial activity of cell free supernatants from two strains ZZU 203 and 204 after thermal treatments at 60°C, 80°C, and 100°C for 30 min and 60 min. The activities remained stable for 60 min at 100°C but decreased beyond that temperature. In addition, the activity completely disappeared after treatment at 121°C for 15 min. The residual activities of the antimicrobial substances at variable pH from 2.0 to 9.0 are also showed in Table 6. The antimicrobial substance retained activity between 2.0 and 6.0, however, it was completely inactive above pH 7.0.
Table 6.
Effects of temperature and pH on the antimicrobial activity of ZZU 203 and 204
| Treatment | Antimicrobial activity | ||||||
|---|---|---|---|---|---|---|---|
|
| |||||||
| Salmonella enterica ATCC 43971T | Escherichia coli ATCC 11775T | Micrococcus luteus ATCC 4698T | |||||
|
|
|
|
|||||
| ZZU 203 | ZZU 204 | ZZU 203 | ZZU 204 | ZZU 203 | ZZU 204 | ||
| Heat (°C) | 60, 30 min | +++ | ++ | ++ | ++ | ++++ | ++++ |
| 60, 60 min | +++ | ++ | ++ | ++ | +++ | ++++ | |
| 80, 30 min | +++ | ++ | + | ++ | +++ | +++ | |
| 80, 60 min | +++ | ++ | + | ++ | +++ | +++ | |
| 100, 30 min | ++ | ++ | + | ++ | +++ | +++ | |
| 100, 60 min | ++ | ++ | + | ++ | +++ | +++ | |
| 121, 15 min | − | − | − | − | − | − | |
| pH | 2.0 | +++ | +++ | +++ | +++ | +++ | +++ |
| 3.0 | ++ | +++ | ++ | ++ | +++ | +++ | |
| 4.0 | ++ | ++ | ++ | ++ | +++ | +++ | |
| 5.0 | ++ | ++ | + | + | +++ | ++ | |
| 6.0 | + | ++ | + | + | ++ | ++ | |
| 7.0 | − | − | − | − | − | − | |
| 8.0 | − | − | − | − | − | − | |
| 9.0 | − | − | − | − | − | − | |
+, Diameter of inhibition zone: 8.00 to 12.00 mm; ++, 12.00 to 16.00 mm; +++, 16.00 to 20.00 mm; ++++, more than 20.00 mm; −, no inhibition zone; the diameter of inhibition zone including that of Oxford cup (7.80 mm).
DISCUSSION
Corn stover is an increasingly important source of animal feed in China (Pang et al., 2011) and the preservation of fresh corn stover as silage, which depends on the production of sufficient acid to inhibit the activity of undesirable microorganisms under anaerobic conditions, is widespread around the world, including in China. Epiphytic microorganisms, especially LAB naturally present on forage crops, are responsible for silage fermentation and also influence silage quality (Lin et al., 1992; Cai and Kumai 1994; Cai et al., 1997; Pang et al., 2011). The counts and characteristics of LAB have become significant factors in predicting the adequacy of silage fermentation and in determining whether to apply silage bacteria inoculants (Cai and Kumai, 1994; Cai et al., 1997; Cai et al., 1999). In this study, 1.4×105 CFU/g FM LAB were observed in fresh corn stover, which suggested that the counts of LAB were abundant and the lactic acid fermentation to preserve the nutrient of the silage was guaranteed.
Fermentation of sugar by heterofermentative LAB will produce acetic acid, alcohol and CO2 specifically during ensiling. The formation of a large amount of acetic acid in the silage is not an advantage because such formation consumes energy. As the formation of acetic acid always entails the formation of CO2, a significantly larger amount of energy is lost in this process than during the formation of lactic acid. Fermentation of sugar by homofermentative LAB transforms sugar into lactic acid at a ratio of almost 1:1. This rapid fermentation saves nutrients, thus preserving a large amount of carbohydrates, which would be lost if the silage were left untreated or treated with acetic acid producing bacteria. In contrast with the lactic acid-producing cocci, lactobacilli play a more important role in promoting lactic acid fermentation over long periods of time (Pang et al., 2011).
It was observed that most of the isolates in this study were lactic acid-producing rods (57.6%) and all of them belonged to L. plantarum subsp. plantarum. The high counts of L. plantarum subsp. plantarum might indicate that this species plays an important role in fermentation processes by effectively promoting lactic acid fermentation. Similar results were reported by Ennahar et al. (2003) where L. plantarum subsp. plantarum was the most frequently isolated microorganism in paddy rice silage. These results are in agreement with those of previous studies (Lin et al., 1992; Cai et al., 1998; Pang et al., 2011), which have reported similar LAB species composition in corn stover in regions of America, Japan and China.
Bacteriocins often exhibit a narrow killing spectrum and inhibit only bacteria that are closely related to those from which they have been formed. For instance, bacteriocins from Gram-positive bacteria are generally not effective against Gram-negative bacteria (Smaoui et al., 2010). Only a few bacteria have been observed that may exhibit broad-spectrum activity against a wide range of microorganisms, including Gram-positive and Gram-negative bacteria (Benitez et al., 2011). Todorov et al. (2011) reported that a cell-free supernatant and semi-purified bacteriocin, Bacteriocin ST16Pa, of L. plantarum subsp. plantarum, a strain isolated from papaya, presented a large spectrum of activity against both certain Gram-positive bacteria and Gram-negative bacteria, such as Escherichia coli. In this paper, LAB strains isolated from the corn stover silage were tested for their ability to inhibit the growth of the following indicator microorganisms: pathogenic bacteria including one Gram-positive (Micrococcus luteus ATCC 4698T) and two Gram-negative bacteria (Salmonella enterica ATCC 43971T and Escherichia coli ATCC 11775T). Moreover, two LAB strains of these tested strains also exhibited strong antimicrobial activity after excluding the effects of organic acids and hydrogen peroxide by neutralizing pH and eliminating hydrogen peroxide from the cell-free culture supernatants. However, all of activities disappeared completely when the cell-free supernatant was treated with proteinase K, which indicated that the antimicrobial activity may attributed to the production of a proteinaceous substance. However, the antimicrobial activity of strain ZZU 203 disappeared, while ZZU 204 still exhibited residual antimicrobial activity after treatment with trypsin. This finding may indicate that the substance with strong antimicrobial activity is sensitive to trypsin, but trypsin does not completely suppress the antimicrobial activity. This result may be observed because the two types of proteases have different cutting sites. Proteinase K is a broad-spectrum serine protease, and the predominant site of cleavage is the peptide bond adjacent to the carboxyl group of aliphatic and aromatic amino acids with blocked alpha amino groups (Kraus et al., 1976), whereas trypsin cleaves peptide chains primarily at the carboxyl side of the amino acids lysine or arginine (Rawlings and Barrett, 1994).
Yang et al. (2012) reported that BLS produced by Streptococcus thermophilus (ASR-1) and Lactococcus casei (JFR-5) was thermally stable at 80°C and 100°C for 60 and 90 min but completely lost its anti-listerial activities after exposure to 121°C for 15 min. In this study, the activity of the inhibitory compounds was not significantly altered by boiling for 60 min, which indicates that the antimicrobial activity of the culture supernatant is heat-resistant. However, the inhibitory activities of ZZU 203 and 204 were completely suppressed by autoclaving at 121°C for 15 min. This finding indicated that the substance with strong antimicrobial activity is thermally stable at 100°C but is destroyed at 121°C.
Due to a modification of ionisation and/or a partial denaturation of the molecule, the bacteriocin-like compounds of the strains were highly sensitive to the variations of the medium pH. Indeed, these compounds are more active at acid pH than at neutral or alkaline pH. Magnusson and Schnürer (2001) found that the antifungal proteinaceous compound produced by the Lactobacillus. coryniformis subsp. coryniformis Si3 strain was stable at pH between 3.0 and 4.5, but rapidly decreased between 4.5 and 6.0, completely disappeared above pH 6.0. Based on our research, the cell-free supernatants of strains ZZU 203 and 204 retained antimicrobial activity after incubation at pH 2.0 to 6.0 and lost activity after treatment at pH 7.0 to 9.0. Bacteriocins generated by LAB strains are generally stable at acidic or neutral pH, indicating that these substances are well-adapted to the environment produced by these bacteria. From a technological point of view, the high pH stability observed in the low pH range is an important factor, enabling their use in silages.
The different species and characteristic of epiphytic LAB can change and influence silage quality, and the population of epiphytic LAB are not always sufficiently large or of suitable composition to promote efficient fermentation under farm conditions. We hope that this work can lay a foundation for preparing an improved LAB inoculant for silage. The two strains ZZU 203 and 204 can potentially be used as silage additives. Investigation of the nature of the antimicrobial substances for silage preparation and trials is underway in our laboratory.
ACKNOWLEDGMENTS
This work was supported by Foundation of Henan Science and Technology Committee (Grant No. 132300410054, 132300410123 and 132300413205), National Natural Science Foundation of China (Grant No. 41101244) and an Exploration, Torch-plan Project grant (112102310069) from Henan Province, a grant from the Special Fund for Agro-Scientific Research in the Public Interest of China (201103007).
REFERENCES
- AOAC. Official Methods of Analysis. 15th edn. Association of Official Analytical Chemists; Arlington, VA, USA: 1990. [Google Scholar]
- Benitez LB, Caumo K, Brandelli A, Rott MB. Bacteriocin-like substance from Bacillus amyloliquefaciens shows remarkable inhibition of Acanthamoeba polyphaga. Parasitol Res. 2011;108:687–691. doi: 10.1007/s00436-010-2114-5. [DOI] [PubMed] [Google Scholar]
- Cai Y, Kumai S. The proportion of lactate isomers in farm silage and the influence of inoculation with lactic acid bacteria on the proportion of L-lactate in silage. Jpn J Zootech Sci. 1994;65:788–795. [Google Scholar]
- Cai Y, Ohmomo S, Ogawa M, Kumai S. Effect of NaCl-tolerant lactic acid bacteria and NaCl on the fermentation characteristics and aerobic stability of silage. J Appl Microbiol. 1997;83:307–313. doi: 10.1046/j.1365-2672.1997.00229.x. [DOI] [PubMed] [Google Scholar]
- Cai Y, Benno Y, Ogawa M, Ohmomo S, Kumai S, Nakase K. Influence of Lactobacillus spp. from an inoculant and of Weissella and Leuconostoc spp. from forage crops on silage. Appl Environ Microbiol. 1998;64:2982–2987. doi: 10.1128/aem.64.8.2982-2987.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai Y. Identification and characterization of Enterococcus species isolated from forage crops and their influence on silage fermentation. J Dairy Sci. 1999;82:2466–2471. doi: 10.3168/jds.S0022-0302(99)75498-6. [DOI] [PubMed] [Google Scholar]
- Chaudhari AA, Kariyawasam S. An experimental infection model for Escherichia coli egg peritonitis in layer chickens. Avian Dis. 2014;58:25–33. doi: 10.1637/10536-032213-Reg.1. [DOI] [PubMed] [Google Scholar]
- Cleveland J, Montville TJ, Nes IF, Chikindas ML. Bacteriocins: Safe, natural antimicrobials for food preservation. Int J Food Microbiol. 2001;71:1–20. doi: 10.1016/s0168-1605(01)00560-8. [DOI] [PubMed] [Google Scholar]
- Eitan BD, Shapiro OH, Siboni N, Kushmaro A. Advantage of using inosine at the 3′ Termini of 16S rRNA gene universal primers for the study of microbial diversity. Appl Environ Microbiol. 2006;72:6902–6906. doi: 10.1128/AEM.00849-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ennahar S, Sashihara T, Sonomoto K, Ishizaki A. Class IIa bacteriocins: Biosynthesis, structure and activity. FEMS Microbiol Rev. 2000;24:85–106. doi: 10.1111/j.1574-6976.2000.tb00534.x. [DOI] [PubMed] [Google Scholar]
- Ennahar S, Cai Y, Fujita Y. Phylogenetic diversity of Lactic acid bacteria associated with paddy rice silage as determined by 16S ribosomal DNA analysis. Appl Environ Microbiol. 2003;69:444–451. doi: 10.1128/AEM.69.1.444-451.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hata T, Tanaka R, Ohmomo S. Isolation and characterization of plantaricin ASM1: A new bacteriocin produced by Lactobacillus plantarum A-1. Int J Food Microbiol. 2010;137:94–99. doi: 10.1016/j.ijfoodmicro.2009.10.021. [DOI] [PubMed] [Google Scholar]
- Hellal A, Amrouche L, Ferhat Z, Laraba F. Characterization of bacteriocin from Lactococcus isolated from traditional Algerian dairy products. Ann Microbiol. 2012;62:177–185. [Google Scholar]
- Karska-Wysocki B, Bazo M, Smoragiewicz W. Antibacterial activity of Lactobacillus acidophilus and Lactobacillus casei against methicillin-resistant Staphylococcus aureus (MRSA) Microbiol Res. 2010;165:674–686. doi: 10.1016/j.micres.2009.11.008. [DOI] [PubMed] [Google Scholar]
- Kozaki M, Uchimura T, Okada S. Experimental Manual for Lactic Acid Bacteria. Asakurasyoten; Tokyo, Japan: 1992. [Google Scholar]
- Kraus E, Kiltz HH, Femfert UF. The specificity of proteinase K against oxidized insulin B chain. Hoppe Seylers Z Physiol Chem. 1976;357:233–237. [PubMed] [Google Scholar]
- Lin C, Bolsen KK, Brent BE, Fung DYC. Epiphytic lactic acid bacteria succession during the pre-ensiling and ensiling periods of alfalfa and maize. J Appl Bacteriol. 1992;73:375–387. [Google Scholar]
- Magnusson J, Schnürer J. Lactobacillus coryniformis subsp. coryniformis strain Si3 produces a broad-spectrum proteinaceous antifungal compound. Appl Environ Microbiol. 2001;67:1–5. doi: 10.1128/AEM.67.1.1-5.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muck RE. Initial bacterial numbers on lucerne prior to ensiling. Grass Forage Sci. 1989;44:19–25. [Google Scholar]
- Pang H, Zhang M, Qin G, Tan Z, Li Z, Wang Y, Cai Y. Identification of lactic acid bacteria isolated from corn stovers. Anim Sci J. 2011;82:642–653. doi: 10.1111/j.1740-0929.2011.00894.x. [DOI] [PubMed] [Google Scholar]
- Pang H, Tan Z, Qin G, Wang Y, Li Z, Jin Q, Cai Y. Phenotypic and phylogenetic analysis of lactic acid bacteria isolated from forage crops and grasses in the Tibetan Plateau. J Microbiol. 2012;50:63–71. doi: 10.1007/s12275-012-1284-5. [DOI] [PubMed] [Google Scholar]
- Rawlings ND, Barrett AJ. Families of serine peptidases. Method Enzymol. 1994;244:19–61. doi: 10.1016/0076-6879(94)44004-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riley MA. Bacteriocins, biology, ecology, and evolution. In: Moselio S, editor. Encyclopedia of Microbiology. 3rd Ed. Academic Press; Massachusetts, USA: 2009. pp. 32–44. [Google Scholar]
- Saitou H, Miura KI. Preparation of transforming deoxyribonucleic acid by phenol treatment. Biochim Biophys Acta. 1963;72:619–629. [PubMed] [Google Scholar]
- Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- Sharma P, Sihag RC. Pathogenicity test of bacterial and fungal fish pathogens in Cirrihinus mrigala infected with EUS disease. Pak J Biol Sci. 2013;16:1204–1207. doi: 10.3923/pjbs.2013.1204.1207. [DOI] [PubMed] [Google Scholar]
- Simova ED, Beshkova DM, Angelov MP, Dimitrov ZhP. Bacteriocin production by strain Lactobacillus delbrueckii ssp. bulgaricus BB18 during continuous prefermentation of yogurt starter culture and subsequent batch coagulation of milk. J Ind Microbiol Biotechnol. 2008;35:559–567. doi: 10.1007/s10295-008-0317-x. [DOI] [PubMed] [Google Scholar]
- Smaoui S, Elleuch L, Bejar W, Karray-Rebai I, Ayadi I, Jaouadi B, Mathieu F, Chouayekh H, Bejar S, Mellouli L. Inhibition of Fungi and Gram-Negative Bacteria by Bacteriocin BacTN635 Produced by Lactobacillus plantarum sp. TN635. Appl Biochem Biotechnol. 2010;162:1132–1146. doi: 10.1007/s12010-009-8821-7. [DOI] [PubMed] [Google Scholar]
- Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- Tohno M, Kobayashi H, Nomura M, Kitahara M, Ohkuma M, Uegaki R, Cai Y. Genotypic and phenotypic characterization of lactic acid bacteria isolated from Italian ryegrass silage. Anim Sci J. 2012;83:111–120. doi: 10.1111/j.1740-0929.2011.00923.x. [DOI] [PubMed] [Google Scholar]
- Todorov SD, Prévost H, Lebois M, Dousset X, LeBlanc JG, Franco BDGM. Bacteriocinogenic Lactobacillus plantarum ST16Pa isolated from papaya (Carica papaya) - From isolation to application: Characterization of a bacteriocin. Food Res Int. 2011;44:1351–1363. [Google Scholar]
- Torriani S, Felis GE, Dellaglio F. Differentiation of Lactobacillus plantarum, L. pentosus, and L. paraplantarum by rec A gene sequence analysis and multiplex PCR assay with rec A gene-derived primers. Appl Environ Microbiol. 2001;67:3450–3454. doi: 10.1128/AEM.67.8.3450-3454.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinberg ZG, Muck RE, Weimer PJ, Chen Y, Gamburg M. Lactic acid bacteria used in inoculants for silage as probiotics for ruminants. Appl Biochem Biotechnol. 2004;118:1–9. doi: 10.1385/abab:118:1-3:001. [DOI] [PubMed] [Google Scholar]
- Yang E, Fan L, Jiang Y, Doucette C, Fillmore S. Antimicrobial activity of bacteriocin-producing lactic acid bacteria isolated from cheeses and yogurts. AMB Express. 2012;2:48. doi: 10.1186/2191-0855-2-48. [DOI] [PMC free article] [PubMed] [Google Scholar]


