Abstract
Tail-anchored (TA) proteins are a physiologically important class of membrane proteins targeted to the endoplasmic reticulum by the conserved GET pathway. During transit, their hydrophobic transmembrane domains (TMDs) are chaperoned by the cytosolic targeting factor Get3, but the molecular nature of the functional Get3-TA protein targeting complex remains unknown. We reconstituted the physiologic assembly pathway for a functional targeting complex and showed that it comprises a TA protein bound to a Get3 homodimer. Crystal structures of Get3 bound to different TA proteins showed an α-helical TMD occupying a hydrophobic groove that spans the Get3 homodimer. Our data elucidate the mechanism of TA protein recognition and shielding by Get3, and suggest general principles of hydrophobic domain chaperoning by cellular targeting factors.
Integral membrane proteins contain hydrophobic transmembrane domains (TMDs) that must be shielded from the cytosol until their insertion into the lipid bilayer. Whereas most eukaryotic membrane proteins are co-translationally targeted to the endoplasmic reticulum (ER) by the signal recognition particle (SRP) (1), tail-anchored (TA) membrane proteins are post-translationally targeted by the cytosolic factor, Get3 (2-7). This conserved ATPase changes conformation in a nucleotide-regulated manner (8-12) to bind TMDs in the cytosol and release them at its ER membrane receptor (6, 13-16).
Assembly of the Get3-TA targeting complex requires “pre-targeting” factors that mediate loading onto Get3 (17, 18). This pathway begins with TA protein in complex with the chaperone Sgt2. The Get4-Get5 scaffolding complex then recruits Sgt2 via Get5, while Get4 recruits ATP-bound Get3 (19). A hand-off reaction within this complex results in transfer of TA protein from Sgt2 to Get3. TA substrate-loaded Get3 then dissociates from Get4 (20-22), resulting in a targeting complex whose architecture and stoichiometry have been debated (8-12, 20-23).
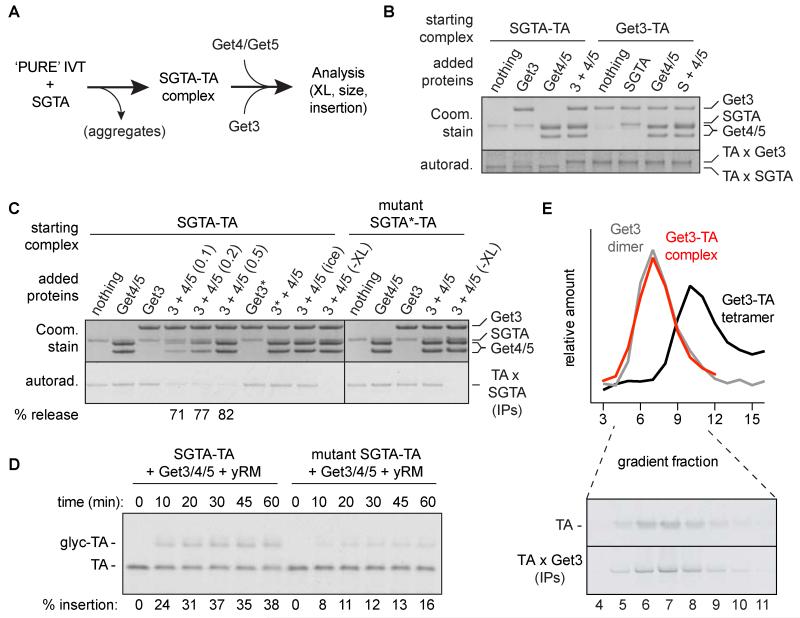
To define the physiologically relevant Get3 targeting complex, we recapitulated its assembly in vitro using purified recombinant factors at in vivo concentrations (Fig. 1A). Translation of radiolabeled TA protein in the presence of SGTA (the mammalian homolog of Sgt2) produced a stable complex detectable by chemical crosslinking (fig. S1). The TA protein remained associated with SGTA upon addition of either Get4-Get5 or Get3, but released efficiently when both factors were added (Fig. 1B). Correspondingly, Get3 efficiently acquired substrate from SGTA only when Get4-Get5 was present.
Fig. 1. Reconstitution of physiologic TA protein targeting complex assembly.
(A) Experimental strategy. (B) SGTA-TA or Get3-TA complexes (fig. S1 and S3) at 1 μM were incubated with 1 μM of the indicated proteins, followed by amine-reactive crosslinking. Reactions were analyzed by SDS-PAGE and Coomassie Blue staining to detect the input proteins (top) or autoradiography to detect the 35S-labeled TA protein crosslinks (bottom). (C) Reactions as in panel B were monitored by sulfhydryl-reactive crosslinking for TA protein release from SGTA (bottom). Reactions contained 0.5 μM of each factor, except lanes 4 and 5 which contained Get4-Get5 at 0.1 and 0.2 μM, respectively. Asterisks next to Get3 or SGTA indicate point mutants that disrupt interactions with Get4 or Get5, respectively. (D) Products of the indicated transfer reactions were incubated with yeast rough microsomes (yRM) and analyzed for insertion. (E) Sucrose gradient size analysis of Get3-TA complex formed by Get4-Get5-dependent loading from SGTA (red). Free, dimeric Get3 (grey) and E. coli-produced tetrameric Get3-TA substrate complex (black) are shown for comparison (fig. S5). Peak fractions containing substrate (red) were analyzed directly or after crosslinking and IP for Get3 to specifically detect Get3-TA complexes (bottom panel).
The transfer reaction was rapid and unidirectional: once substrate released from SGTA, it did not re-bind (fig. S2). Likewise, substrate pre-loaded directly on Get3 (fig. S3) did not effectively transfer to SGTA (Fig. 1B). Structure-guided mutations disrupting either the SGTA-Get5 interaction [SGTA(C38S)] (24) or the Get4-Get3 interaction [Get3(E253R)] (20) abolished substrate release from SGTA (Fig. 1C). Targeting complex produced via Get4-Get5 supported TA protein insertion into yeast ER microsomes (Fig. 1D), while an identical reaction containing SGTA(C38S) showed reduced insertion (Fig. 1D). Thus, the recombinant assembly system requires all factors and interactions of the early GET pathway and produces insertion-competent Get3-TA protein targeting complex.
Three lines of evidence suggested that functional targeting complex assembled via pre-targeting factors consists of dimeric Get3 bound to TA protein. First, the targeting complex, containing a small (~10 kDa) TA protein, had the same native size as purified Get3 dimer and was clearly distinguishable from higher-order Get3 complexes (Fig. 1E). Such higher-order complexes, often seen when Get3 is co-expressed with TA protein in E. coli (fig. S5) (8, 22, 23), were not observed even when the loading reaction contained 10-fold excess Get3 (fig. S4A). Second, titration of Get3 into the loading reaction showed no evidence of cooperativity (fig. S4B), arguing against its higher-order assembly during targeting complex formation. Third, size-exclusion chromatography and multi-angle laser light scattering (SEC-MALLS) indicated that prior to loading, a single Get3 dimer is bound by two copies of the Get4-Get5 complex (fig S4C). Thus, TA protein is loaded onto dimeric Get3 to form a functional targeting complex.
To gain insight into how the TA protein is shielded by Get3 in this targeting complex, we sought to determine its structure. During physiologic targeting complex assembly, Get4 preferentially recruits and stabilizes ATP-bound Get3 (19, 20, 22). To mimic this during recombinant expression in E. coli, we biased Get3 to the ATP-bound state via the D57N hydrolysis mutant (10). Co-expression of this mutant with TA protein resulted in a targeting complex that was homogeneously dimeric for Get3 by SEC-MALLS (fig. S6A) and co-migrated with in vitro assembled targeting complex on sucrose gradients (fig. S6B).
To facilitate crystallization we generated a high-affinity synthetic antibody fragment (sAB) (25) that recognizes the closed (ATP-bound) conformation of Get3. Kinetic analysis revealed that this sAB binds with sub-nanomolar affinity to nucleotide-bound Get3, both in the presence and absence of TA protein (fig. S7). Thus, rather than inducing a large conformation change in Get3, the TA protein binds to a pre-organized conformation that closely resembles the closed (ATP-bound) state.
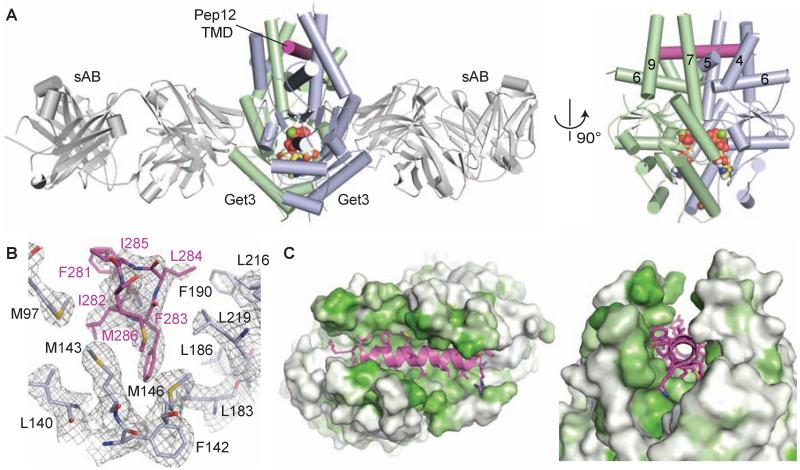
Using this sAB we crystallized Get3(D57N) in complex with the TMD of the yeast TA protein Pep12 (table S1). The structure reveals nucleotide-bound Get3 in a closed conformation with two sABs bound to equivalent sites on opposite faces of a Get3 homodimer (Fig. 2A); no higher-order Get3 oligomers are observed in the crystal (fig. S8). The closed conformation is nearly identical to that seen in previous Get3-ADP·AlF4− structures (~0.5 Å RMSD), in which two helical subdomains form a composite hydrophobic groove proposed to bind the TMDs of TA proteins (8, 10).
Fig. 2. The helical TMD of a TA substrate binds deep within the composite hydrophobic groove of dimeric Get3.
(A) Overview of dimeric S. cerevisiae Get3 bound to a truncated Pep12 TA substrate (magenta) and nucleotide (spheres), and sandwiched between two copies of an engineered sAB (grey). At right, a ‘side’ view of the complex is shown with sABs removed for clarity. (B) Details of the interaction between the Pep12 TMD C-terminus and a methionine-rich cluster at one end of the hydrophobic groove. Electron density is from a 2.05 Å 2Fo-Fc map contoured at 1.0σ. (C) Surface representations of the TA substrate binding site, colored from least (white) to most (green) hydrophobic.
As is typical for the fungal Get3 crystal structures, electron density is weakest within these dynamic helical subdomains. Nevertheless, unaccounted helical density was visible within the hydrophobic groove in early unbiased maps (fig. S9). After refinement, we assigned this density to the Pep12 TMD (Fig. 2B and fig. S9), excluding the possibility that it corresponds to flexible regions of Get3 folding into the groove.
The Pep12 TMD binds to Get3 at the bottom of the composite hydrophobic groove (Fig. 2 and fig. S9) where it spans the dimer interface and stabilizes the closed conformation of Get3. The most ordered interactions are found at the ends of the TMD, where bulky hydrophobic sidechains of the substrate contact groove residues including M97 (helix 4), L126 (helix 5), M143 and M146 (helix 6), L183, L186 and F190 (helix 7) and L216 and L219 (helix 9) (Fig. 2B). Consistent with their role in TMD binding, substitution of hydrophobic residues along helices 7 and 9 with polar or charged residues abolished Get3’s ability to induce TA protein release from SGTA (Fig. 3A).
Fig. 3. Dynamic shielding of the TMD.
(A) SGTA-TA complexes were prepared and subjected to transfer reactions with WT and mutant Get3 proteins as in Fig. 1C. LL-SS (L183S/L186S), LLL-SSS (L183S/L186S/L219S) and FL-DD (F190D/L216D) are hydrophobic groove mutants; E253R is a mutation that disrupts interaction with Get4. (B) ‘Top’ and ‘side’ views of Pep12 (magenta), Nyv1 (blue) and Sec22 (green) complexes superimposed on the free Get3 closed dimer structure (yellow; 2woj). Relative to free Get3, the end of helix 7 extends and begins to curve inward over the substrate. (C) WT or benzophenone-containing (at the indicated positions) Get3-TA complexes were prepared as in fig. S3 and the dimer peak was subjected to UV crosslinking. Uncrosslinked TA protein and its adducts to one or two Get3 proteins are indicated. (D) ‘Side’ views of the Get3 dimer, looking into the groove. In its transient empty state, Get3 is splayed apart, with two hydrophobic ‘half-sites’ occupied by the helix 8 region. ATP binding drives Get3 into a closed conformation which is captured by two copies of the Get4-Get5 complex. In this state, helix 8 is displaced, and the composite hydrophobic groove is now pre-organized for substrate binding. After substrate transfer from Sgt2, the targeting complex is released. The helix 8 region now dynamically shields the substrate during transit to the ER membrane.
The Pep12 TMD buries ~1,450 Å2 of hydrophobic surface area, distributed nearly evenly between the two Get3 subunits (Fig. 2C). This represents ~50% of the ordered hydrophobic surface area in the groove, and is significantly greater than in the SRP54-signal peptide interaction, where ~360 Å2 of hydrophobic surface area become buried upon binding (26, 27). The availability of such a large surface area likely explains how Get3 is able to accommodate hydrophobic sequences of differing lengths and composition.
Using the same strategies we also solved crystal structures of Get3(D57N) in complex with unrelated TMDs from Nyv1 and Sec22 (table S1). Density for these TMDs was less defined than for the Pep12 complex, but nevertheless sufficient to place helical TMDs (fig. S9). Like Pep12, these TMDs bind at the bottom of the hydrophobic groove, spanning the dimer interface (Fig. 3B). Thus, a single helix binding across the Get3 dimer represents the canonical mode of the Get3-TA substrate interaction.
Although much of the Get3 hydrophobic groove and substrate TMD are shielded in the targeting complexes, one surface of the TMD appears solvent exposed. Relative to previous closed Get3 structures, the groove in each substrate-bound complex is constricted at its apex where the ends of helix 7 curve inwards (Fig. 3B). Although the ‘TRC40-insert’, including helix 8, is poorly defined, we found by site-specific photocrosslinking that this region (and residues in helix 6 and 7) directly contact the TA substrate (Fig. 3C and fig. S10). Thus, helix 8 likely functions as a dynamic ‘lid’, protecting the TMD from aggregation, while still allowing substrate release after recruitment to Get1 (Fig. 3D) (13, 14).
Our biochemical and structural analyses define the functional targeting complex as a Get3 homodimer bound to a single TA protein. Although higher-order Get3 assembly has been postulated to promote ATP hydrolysis (22), this appears unnecessary since dimeric targeting complex was functional for TA protein insertion, indicating that it had hydrolyzed its ATP (Fig. 1D). Consistent with this, the catalytic machinery is organized for hydrolysis in the targeting complex structures (fig. S11). The higher-order Get3 oligomers that form during oxidative stress (28) are structurally and functionally distinct.
The structure of the Get3-TA substrate targeting complex illustrates a common strategy for binding to hydrophobic cargo. Like Get3, the signal sequence binding subunit of SRP (SRP54) captures substrates within a hydrophobic, methionine-rich groove presented on a helical scaffold (26, 27, 29). These scaffolds provide a large and intrinsically dynamic binding site that is not appreciably ordered by substrate capture. This likely confers the ability of Get3 and SRP54 to bind a variety of hydrophobic sequences—an essential property of both targeting systems. It will be of interest to determine whether these principles are shared by other TMD binding factors including SGTA and Bag6.
Materials and Methods
Preparation of individual proteins for functional analysis
Genes encoding full-length S. cerevisiae Get4, Get5 and Sgt2 were PCR amplified from genomic DNA. Sgt2 was subcloned into pET21c (Novagen) in-frame with a C-terminal 6xHis tag; Get5 was subcloned into a pCDF1b derivative (Novagen) modified to incorporate a tobacco etch virus (TEV) protease cleavage site between an N-terminal 6xHis tag and the polylinker; Get4 was subcloned into pET28 (Novagen) without modification. Full-length human SGTA was subcloned into pGEX6p1 with a 3C protease cleavage site between an N-terminal GST tag and the polylinker. Site-directed mutants were obtained by QuikChange mutagenesis (Stratagene) and verified by DNA sequencing.
Expression and purification of full-length Get3 (wild-type and mutants) was carried out as described previously (10). For photocrosslinking experiments, wild-type Get3 amber mutants prepared with a C-terminal 6xHis tag, were co-transformed with pEVOL-pBpF (30) into E. coli BL21(DE3) (Novagen). After the cells reached ~0.6 A600, 0.1 mM IPTG, 0.2% arabinose and 1 mM p-benzoylphenylalanine (BpF) (Bachem) were added, and the culture was grown for an additional 6 h at 25 °C. Purification was as above for Get3 except that 50 mM Hepes pH 7.5 was used as the buffer. Dimeric Get3-BpF mutants were purified by gel filtration and then used for photocrosslinking experiments.
Full-length Get4 and Get5 (wild-type and mutants) were co-expressed for 6 h at 25 °C in E. coli BL21(DE3)/pRIL, following induction with 0.1 mM IPTG. Cells were disrupted in buffer A (50 mM Tris, 500 mM NaCl, 10 mM imidazole, 5% glycerol, 5 mM β-mercaptoethanol, pH 7.5) with 1 mM PMSF and 0.02 mg/mL DNase using a high-pressure microfluidizer (Avestin). After clearing by centrifugation, the supernatant was batch-purified by nickel-affinity chromatography. Protein was eluted in buffer A containing 200 mM imidazole, and then dialyzed into 20 mM Tris, 200 mM NaCl, 2 mM DTT, pH 7.5. This was typically followed by gel filtration (Superdex 200 10/300 GL, GE Healthcare) in 20 mM Tris, 200 mM NaCl, 2 mM DTT, pH 7.5. Fractions were pooled and stored in aliquots at −80 °C. Protein concentrations were determined by Bradford (Bio-Rad).
Full-length human SGTA (wild-type and mutants) was expressed for 16 h at 16 °C in E. coli BL21(DE3), following induction with 0.2 mM IPTG. The GST-fusion was purified by standard methods and eluted with glutathione, followed by dialysis into 50 mM Hepes 7.4, 150 mM KOAc, 2 mM MgCl2, 1 mM DTT, and 10% Glycerol. After cleavage with 3C protease and subtraction using Glutathione resin, the protein was concentrated using Vivispin 10K cutoff filters and stored in aliquots at −80 °C.
Multi-angle laser light scattering
To obtain protein for size analysis (see fig. S5 and S6), dimeric targeting complexes were produced by co-expression in E. coli BL21(DE3)/pRIL (Novagen) after co-transformation with a plasmid encoding Get3(D57N), and a plasmid derived from the PURE system control plasmid (NEB) in which DHFR was replaced with full-length S. cerevisiae Sbh2 containing N-terminal Twin-strep and C-terminal opsin tags and a Pep12 TMD. Protein was expressed at RT for 4 h by induction with 0.1 mM IPTG after the cells reached an A600 of ~0.8. Cells were disrupted in buffer B (100 mM Tris, 150 mM NaCl, pH 8.0) and cleared by centrifugation. The supernatant was passed over Strep-Tactin agarose (IBA, Germany) three times. After washing with ten column volumes of buffer B, targeting complex was eluted with buffer B supplemented with 5 mM desthiobiotin (Novagen). These complexes were further purified by gel filtration in 10 mM Tris, 150 mM NaCl, pH 7.5. Fractions were pooled, concentrated, and stored in aliquots at −80 °C. Protein concentration was determined by Bradford assay (Bio-Rad).
Tetrameric targeting complexes were obtained similarly, except using wild-type Get3 and a truncated N-terminally 6xHis tagged Pep12262-288 substrate subcloned into pET28. Protein was expressed and purified as described below for the Get3(D57N)-TA substrate complexes.
Full-length Get3(D57N)-Get4-Get5 complexes were obtained by incubating Get3(D57N) (containing an N-terminal 6xHis tag) with 2 mM ATP and 2 mM MgCl2. After incubating for 15 minutes, Get4-Get5 (with an N-terminal 6xHis tag on Get5) was added to give final protein concentrations of 60 μM Get3(D57N), and 40 μM Get4-Get5. Samples were incubated for an additional 30 minutes and then analyzed immediately.
The absolute molecular masses of targeting complexes were measured by static multi-angle laser light scattering (MALLS), essentially as described (14). Samples were injected onto a Superdex 200 10/300 GL gel-filtration column (GE Healthcare) equilibrated with 20 mM Tris, 150 mM NaCl, pH 7.5 (targeting complexes) or 20 mM Hepes, 150 mM NaCl, 0.5 mM ATP, 0.5 mM MgCl2, pH 7.5 [Get3(D57N)/4/5 and Get4-Get5 complexes]. The purification system was coupled to an online, static, light scattering detector (Dawn HELEOS II, Wyatt Technology), a refractive-index detector (Optilab rEX, Wyatt Technology) and a ultraviolet-light detector (UPC-900, GE Healthcare). Absolute weight-averaged molar masses were calculated using the ASTRA software (Wyatt Technology).
Tail-anchored substrate transfer reaction
A previously described native human Sec61β construct (7) was modified to contain the TMD from VAMP, followed by a C-terminal opsin tag (fig. S1). This ORF was subcloned in place of DHFR in the control T7-driven plasmid for in vitro transcription and translation in the PURE system (NEB). Yeast Get3 antibody was as described previously (10). The SGTA antibody was generated against a synthetic C-terminal peptide conjugated to KLH. The SGTA sequence used was CRSRRPSASNDDQQE, with the extra cysteine added at the N-terminus for KLH conjugation.
Chaperone-TA complexes were obtained by supplementing the PURE translation system (NEB) with plasmid encoding the VAMP TMD-containing substrate, 35S-methionine and 25 μM of purified Sgt2, SGTA or Get3. After incubating for 90 min at 37 °C, reactions were diluted with ice cold assay buffer (50 mM Hepes pH 7.4, 125 mM KOAc, 2 mM MgCl2), and separated at 4 °C through a 5-25% sucrose gradient (55,000 rpm/5 h in a TLS55 rotor); fractions containing the soluble complexes (see fig. S1 and S3) were pooled and either used immediately or frozen in liquid nitrogen and stored at −80 °C.
Substrate transfer reactions were carried out in 50 mM Hepes pH 7.4, 125 mM KOAc, 4 mM MgCl2 and 1 mM ATP and subjected to amine-reactive, sulfhydryl-reactive, or UV crosslinking. Amine-reactive crosslinking used 250 μM disuccinimidyl suberate (Pierce) at 22° C for 30 min. Sulfhydryl-reactive crosslinking used 200 μM bismaleimidohexane (Pierce) for 30 min on ice. Photo-crosslinking via BpF with UV was for 15 min on ice using a 365 nm longwave UV spot lamp (UVP) placed 10 cm from the sample. All crosslinking reactions were terminated by addition of excess SDS-PAGE buffer, followed optionally by immunoprecipitation (with anti-SGTA or anti-Get3 antibodies), separation by SDS-PAGE on 12% Tris-Tricine gels, Coomassie blue staining and autoradiography.
Size analysis of the Get3-TA substrate complexes formed by Get4-Get5-dependent loading from SGTA was performed using high-resolution 5-25% sucrose gradients (55,000 rpm/5 h in a TLS55 rotor); gradient fractions were analyzed by SDS-PAGE and quantified by phosphorimaging. Free, dimeric Get3 and E. coli-produced tetrameric Get3-TA substrate complexes (described above) were used as molecular weight standards; samples were analyzed by Coomassie staining and quantified by densitometry.
Insertion activity was analyzed by incubating TA substrate complexes (as indicated in the Figure legends) with yeast rough microsomes, prepared from wild-type yeast essentially as described previously (14). Insertion was monitored by TA protein glycosylation and quantified by phosphorimaging.
Phage display
Gel filtration purified Get3(D57N) (130 μM in 20 mM Hepes, 150 mM NaCl, pH 8.0) was preincubated with 2 mM ATP and 2 mM MgCl2 for 30 minutes, followed by addition of a 5-fold molar excess of biotinylation reagent (NHS-SS-PEG4-Biotin) (Thermo Scientific). Biotinylation was carried out for 1 h at 25 °C and quenched with 2 mM Tris pH 8.0. After desalting on a PD10 column, Get3(D57N) dimers were purified by SEC and concentrated. The extent of biotinylation and efficiency of antigen capture were tested by pulldown with Streptavidin MagneSphere particles. To obtain sABs selective to the closed conformation of Get3, solutions were supplemented with 2 mM ATP and 2 mM MgCl2 throughout the selection process. Phage display (using a synthetic antibody phage library provided by S. Koide) and initial clone testing was performed as described previously (25). Conformational specificity was confirmed in single point competitive ELISA prior to gel filtration analysis and kinetic analysis by SPR (see below).
sAB production
sABs were subcloned into the expression vector RH2.2 (gift from S. Sidhu) using Hind III and SalI restriction sites. Sequence confirmed clones were transformed into E. coli BL21 (DE3)/pRIL (Novagen) and sABs were expressed for 24 h at 25 °C using autoinduction in a LEX fermentor system with air flow rate of 2 L/min. Cells were disrupted in lysis buffer containing 50 mM Tris, 500 mM NaCl, and 0.05% Triton X-100, pH 8.0 using a high pressure microfluidizer (Avestin). Lysate was cleared by centrifugation and loaded onto HiTrap MabSelect SuRe 5 mL column, equilibrated with buffer containing 50 mM Tris, 500 mM NaCl pH 8.0. Column was washed with 10 volumes of equilibration buffer and then protein was eluted with 0.1 M acetic acid. Fractions containing protein were directly loaded onto ion exchange Resource S 1 mL column. Column was washed with buffer containing 50 mM sodium acetate pH 5.0 at 5 mL/min. sABs were eluted with a linear gradient 0-50% of buffer containing 50 mM sodium acetate, 2 M NaCl, pH 5.0. Pure sABs were dialyzed against buffer containing 20 mM Tris, 150 mM NaCl, pH 7.5.
Surface plasmon resonance
Interaction analyses were performed at 20 °C using a BIACORE 3000 (GE Healthcare). SEC purified, 6xHis tagged Get3(D57N) and Get3(D57N)-Pep12262-288 complex (with an N-terminal 6xHis tag on the TA substrate only) were immobilized on an NTA sensor chip. For the analysis, running buffer contained: 10 mM Hepes, 150 mM NaCl, pH 7.4, 0.05% Tween 20 supplemented with 1 mM MgCl2 and +/− 1 mM nucleotide (ATP or ADP). 6xHis-tagged Get3(D57N) and Get3(D57N)-Pep12262-288 complex were captured by injecting 5 μL of 30 nM protein solution at a flow rate of 5 μL/min. Up to three blanks were injected to ensure stability of the surface before analyte injections were started. For each assay, two-fold dilution series of sAB (clone ID 47E1_2) starting at 10 nM were injected over the NTA chip surface at a flow rate of 30 μL/min to minimize mass transport effects for 150 s. The resulting response unit change was measured for 300 s after the injection finished. Following each sample injection, the NTA chip surface was regenerated with 50 μL of 5 M GdmHCl, 100 mM EDTA, 2% Tween 20 solution at a flow rate of 50 μL/min. All conditions were tested at 7 different sAB concentrations, and each concentration was tested in triplicate. Injections were randomized to avoid systematic errors. Data processing and kinetic analysis were performed using in Scrubber 2 program (BioLogic software). All sensorgrams were double referenced using blank channel and buffer injections. For the determination of kinetic rate constants, all data sets were fit to a simple 1:1 interaction model using nonlinear regression analysis.
Preparation of Get3(D57N)-TA substrate complexes for crystallization
The gene encoding native, full-length S. cerevisiae Get3(D57N) was subcloned into pCDF1b (Novagen). Truncated TA substrates corresponding to Pep12262-288, Sec22184-214 and Nyv1225-251, modified to contain an N-terminal 6xHis tag, were subcloned into pET28. TA substrate and Get3(D57N) plasmids were co-transformed into E. coli BL21(DE3)/pRIL (Novagen), and expression was carried out at room temperature for 6 h following induction with 0.1 mM IPTG after the cells reached an A600 of ~0.6. After resuspending in buffer A supplemented with 1 mM PMSF, cells were disrupted using a microfluidizer (Avestin). After clearing by centrifugation, the supernatant was batch-purified by nickel-affinity chromatography. Protein was eluted in buffer A containing 200 mM imidazole, dialyzed into 20 mM Tris, 200 mM NaCl, 2 mM DTT, pH 7.5 and followed by gel filtration (Superdex 200 10/300 GL, GE Healthcare) in 20 mM Tris, 150 mM NaCl, pH 7.5. Fractions corresponding to dimeric Get3(D57N)-TA substrate complex were pooled, concentrated and stored in aliquots at −80 °C. Protein concentrations were determined by Bradford assay (Bio-Rad).
Purified Get3(D57N)-TA substrate complexes were incubated for 30 minutes with 2 mM ATP and 2 mM MgCl2 (Sec22, Nyv1) or with 2 mM ADP and 2 mM MgCl2 (Pep12). Note that to conserve reagents (especially the sAB), we did not prepare all TMD-nucleotide combinations, under the assumption that we would be able to fully exchange nucleotide on the comparatively slow timescale of crystallization. Next, the complex was incubated for 30 min with a 1.2 molar excess of purified sAB (clone ID 47E1_2), and then spun down at 13,000 rpm for 10 minutes at 4 °C. Finally, the complex was separated from excess sAB by gel filtration. Fractions were pooled, concentrated to ~8-10 mg/mL in 10 mM Tris, 150 mM NaCl, pH 7.5, and stored in aliquots at −80 °C.
Crystallization and data collection
All crystals of the S. cerevisiae Get3(D57N)-TA complexes with sAB were grown at room temperature by hanging drop vapor diffusion. Initial high-throughput screening was done with either ATP or ADP added in excess to the protein solution. Although we obtained crystals in both nucleotide conditions, the most promising were with ATP for Pep12 and Nyv1, and ADP for Sec22; these were subsequently optimized. Micro-crystals from screening experiments were crushed and used to seed experiments using varying concentrations of PEG 3350 and succinic acid pH 7.0. Optimization led to the production of single crystals.
Crystals of Get3(D57N)-Pep12262-288 were obtained by mixing equal volumes of a protein solution containing 2 mM ATP and 2 mM MgCl2 with a reservoir solution containing 16% PEG 3350 and 25 mM succinic acid pH 7.0. Crystals were cryoprotected in mother liquor supplemented with 20% ethylene glycol, 2 mM ATP and 2 mM MgCl2, and flash frozen in liquid nitrogen.
Crystals of the S. cerevisiae Get3(D57N)-Nyv1225-251 complex with sAB were grown by mixing equal volumes of protein solution containing 2 mM ATP and 2 mM MgCl2 with 15% PEG 3350, 25 mM succinic acid pH 7.0. Crystals were cryoprotected in mother liquor supplemented with 10% PEG 400, 2 mM ATP and 2 mM MgCl2, and flash frozen in liquid nitrogen.
Crystals of the S. cerevisiae Get3(D57N)-Sec22184-214 complex with sAB were obtained by mixing equal volumes of a protein solution containing 2 mM ADP and 2 mM MgCl2 with a reservoir solution containing 13% PEG 3350, 25 mM succinic acid pH 7.0. Crystals were briefly soaked in mother liquor supplemented with 20% ethylene glycol, 2 mM ADP, and 2 mM MgCl2, and flash frozen in liquid nitrogen.
The presence of all protein components (including the TMD) was established by analyzing washed crystals by SDS-PAGE (fig. S12). All diffraction data were collected from single crystals at 100 K at APS beamline 24ID-C (lambda=0.9795 Å) on a PILATUS 6MF pixel-array detector. Data were processed using the Xia2 (31) pipeline to XDS (32); data collection and processing statistics are listed in table S1.
Structure determination and refinement
The structure of the Pep12262-288 complex was determined to a resolution of 2.05 Å by molecular replacement with PHASER (33), using the closed dimer form of S. cerevisiae Get3 (PDB 2woj; with the helical subdomain trimmed) (10), and a sAB (PDB 3pgf, with the complementarity determining regions omitted) (34), used as search models. Unbiased electron density maps, calculated after manual building and refinement of Get3 and the sAB, revealed clear positive Fo-Fc difference density for the helical Pep12 TMD (fig. S9). After initial placement of the TMD, iterative refinement and model building in PHENIX (35) and COOT (36) allowed us to assign its sequence (Fig. 2). The final model contains one Get3 homodimer, one Pep12 TMD, two sAB complexes, one zinc atom, two magnesium atoms, two molecules each of ADP and ATP (with average occupancies of 0.36 and 0.64 respectively) and 928 water molecules.
The structures of the Nyv1225-251 (determined to 2.35 Å) and Sec22184-214 (determined to 2.75 Å) complexes were obtained by molecular replacement as described above, except that the refined model for the sAB (from the Pep12 complex) was used along with trimmed 2woj dimer as the search models. Weak difference density was visible within the groove in unbiased electron density maps (fig. S9). After placement of a helical TMD, additional refinement and model building confirmed the presence of the TMDs; however, because the density was weak, we modeled the TMDs as poly-alanine helices and did not define their orientation. The final Nyv1 model contains two Get3 homodimers, two Nyv1 TMDs, four sAB complexes, two zinc atoms, four magnesium atoms, four ATP molecules and 808 waters; The final Sec22 model contains four Get3 homodimers, four Sec22 TMDs, eight sAB complexes, four zinc atoms, eight magnesium atoms, eight molecules each of ADP and ATP (with average occupancies of 0.45 and 0.55 respectively), and 425 waters.
Refinement and validation statistics are listed in table S1. Structure figures were generated using PyMOL (http://www.pymol.org/).
Over the course of this project we screened the diffraction properties of hundreds of crystals and collected a series of datasets on different Pep12, Nyv1 and Sec22 crystals; these diffracted to varying resolutions, and often possessed different space groups and cell dimensions. The best of these datasets gave rise to electron density maps that showed convincing helical TMD density in the hydrophobic groove; others, while largely identical in structure, were apparently less ordered in the groove, making assignment of the TA substrate difficult. However, there was no obvious correlation between nucleotide state and the quality of electron density in the groove. Importantly, the Nyv1 complex (space group P1; ATP only), Pep12 (space group P212121; mixture of ATP/ADP) and Sec22 (space group P1; mixture of ATP/ADP) are remarkably similar—both in terms of overall structure and in the active site details (see fig. S11), despite the different crystal forms and nucleotide composition. Thus, the structures we report do not appear to be sensitive to ATP vs. ADP.
Supplementary Material
Materials and Methods
Figs. S1 to S12
Table S1
References (30-37)
Acknowledgements
We thank S. Shao for help with assay development; S. Koide for the phage library; S. Sidhu for the sAB expression vector; M. Kivlen for plasmids; F. Bezanilla, E. Perozo and J. Piccirilli for instrumentation; members of the Keenan, Hegde and Kossiakoff labs for support; and the NE-CAT (24-ID-C) beamline staff at APS for technical assistance. NE-CAT is supported by NIH Grant P41 GM103403 and U.S. DOE contract DE-AC02-06CH11357. Additional support was from the U.K. MRC (MC_UP_A022_1007 to R.S.H.), the NIH (U01 GM094588 and U54 GM087519 to A.A.K.; R01 GM086487 to R.J.K.) and the Chicago Biomedical Consortium (to A.A.K. and R.J.K.). The Protein Data Bank accession codes are: 4XTR (Pep12), 4XVU (Nyv1) and 4XWO (Sec22).
References and Notes
- 1.Akopian D, et al. Annu Rev Biochem. 2013;82:693–721. doi: 10.1146/annurev-biochem-072711-164732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chartron JW, et al. Curr Opin Struct Biol. 2012;22:217–224. doi: 10.1016/j.sbi.2012.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Denic V, et al. Cold Spring Harb Perspect Biol. 2013;5:a013334. doi: 10.1101/cshperspect.a013334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hegde RS, et al. Nat Rev Mol Cell Biol. 2011;12:787–798. doi: 10.1038/nrm3226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Favaloro V, et al. J Cell Sci. 2008;121:1832–1840. doi: 10.1242/jcs.020321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schuldiner M, et al. Cell. 2008;134:634–645. doi: 10.1016/j.cell.2008.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stefanovic S, et al. Cell. 2007;128:1147–1159. doi: 10.1016/j.cell.2007.01.036. [DOI] [PubMed] [Google Scholar]
- 8.Bozkurt G, et al. Proc Natl Acad Sci USA. 2009;106:21131–21136. doi: 10.1073/pnas.0910223106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hu J, et al. PLoS ONE. 2009;4:e8061. doi: 10.1371/journal.pone.0008061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mateja A, et al. Nature. 2009;461:361–366. doi: 10.1038/nature08319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Suloway CJ, et al. Proc Natl Acad Sci U S A. 2009;106:14849–14854. doi: 10.1073/pnas.0907522106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yamagata A, et al. Genes Cells. 2010;15:29–41. doi: 10.1111/j.1365-2443.2009.01362.x. [DOI] [PubMed] [Google Scholar]
- 13.Stefer S, et al. Science. 2011;333:758–762. doi: 10.1126/science.1207125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mariappan M, et al. Nature. 2011;477:61–66. doi: 10.1038/nature10362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang F, et al. Nature. 2014;512:441–444. doi: 10.1038/nature13471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang F, et al. Mol Cell. 2011;43:738–750. doi: 10.1016/j.molcel.2011.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang F, et al. Mol Cell. 2010;40:159–171. doi: 10.1016/j.molcel.2010.08.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mariappan M, et al. Nature. 2010;466:1120–1124. doi: 10.1038/nature09296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chartron JW, et al. Proc Natl Acad Sci USA. 2010;107:12127–12132. doi: 10.1073/pnas.1006036107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gristick HB, et al. Nat Struct Mol Biol. 2014;21:437–442. doi: 10.1038/nsmb.2813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rome ME, et al. Proc Natl Acad Sci U S A. 2014;111:E4929–4935. doi: 10.1073/pnas.1411284111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rome ME, et al. Proc Natl Acad Sci U S A. 2013;110:7666–7671. doi: 10.1073/pnas.1222054110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Suloway CJ, et al. EMBO J. 2012;31:707–719. doi: 10.1038/emboj.2011.433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chartron JW, et al. Cell Rep. 2012;2:1620–1632. doi: 10.1016/j.celrep.2012.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Paduch M, et al. Methods. 2013;60:3–14. doi: 10.1016/j.ymeth.2012.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hainzl T, et al. Nat Struct Mol Biol. 2011;18:389–391. doi: 10.1038/nsmb.1994. [DOI] [PubMed] [Google Scholar]
- 27.Janda CY, et al. Nature. 2010;465:507–510. doi: 10.1038/nature08870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Voth W, et al. Mol Cell. 2014;56:116–127. doi: 10.1016/j.molcel.2014.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Keenan RJ, et al. Cell. 1998;94:181–191. doi: 10.1016/s0092-8674(00)81418-x. [DOI] [PubMed] [Google Scholar]
- 30.Chin JW, et al. Proc Natl Acad Sci U S A. 2002;99:11020–11024. doi: 10.1073/pnas.172226299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Winter G. J Appl Crystallogr. 2010;43:186–190. [Google Scholar]
- 32.Kabsch W. Acta Crystallogr D Biol Crystallogr. 2010;66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McCoy AJ, et al. J Appl Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rizk SS, et al. Nat Struct Mol Biol. 2011;18:437–442. doi: 10.1038/nsmb.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Adams PD, et al. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Emsley P, et al. Acta Crystallogr D Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pedrazzini E, et al. J Cell Biol. 2000;148:899–914. doi: 10.1083/jcb.148.5.899. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Materials and Methods
Figs. S1 to S12
Table S1
References (30-37)