Abstract
Salmonella Typhimurium isolate D23580 represents a recently identified ST313 lineage of invasive non-typhoidal Salmonellae (iNTS). One of the differences between this lineage and other non-iNTS S. Typhimurium isolates is the presence of prophage BTP1. This prophage encodes a gtrC gene, implicated in O-antigen modification. GtrCBTP1 is essential for maintaining O-antigen length in isolate D23580, since a gtrBTP1 mutant yields a short O-antigen. This phenotype can be complemented by gtrCBTP1 or very closely related gtrC genes. The short O-antigen of the gtrBTP1 mutant was also compensated by deletion of the BTP1 phage tailspike gene in the D23580 chromosome. This tailspike protein has a putative endorhamnosidase domain and thus may mediate O-antigen cleavage. Expression of the gtrCBTP1 gene is, in contrast to expression of many other gtr operons, not subject to phase variation and transcriptional analysis suggests that gtrC is produced under a variety of conditions. Additionally, GtrCBTP1 expression is necessary and sufficient to provide protection against BTP1 phage infection of an otherwise susceptible strain. These data are consistent with a model in which GtrCBTP1 mediates modification of the BTP1 phage O-antigen receptor in lysogenic D23580, and thereby prevents superinfection by itself and other phage that uses the same O-antigen co-receptor.
Introduction
Salmonella enterica subspecies enterica (S. enterica) is responsible for 98% of human infections caused by Salmonella. Over 1500 serovars of S. enterica are recognized in the Kaufman–White scheme (Grimont and Weill, 2007), which uses differences in the O-antigen of the lipopolysaccharide (LPS) and flagellar H antigen to differentiate between isolates. Over 120 serovars are recognized as having the group O4 O-antigen (Grimont and Weill, 2007), which is characterized specifically by an abequose moiety linked to the mannose with a 1–3 linkage (Reeves, 1993). This is a subgroup of the O12 positive isolates that have an O-antigen subunit backbone consisting of mannose, rhamnose and galactose. Salmonella enterica subspecies enterica serovar Tyhimurium, hereafter referred to as S. Typhimurium, is one of these O4 (previously group B) serovars.
Salmonella Typhimurium is a non-host-restricted serovar typically associated with localized gastrointestinal infection in humans. However, there is an epidemic of invasive S. Typhimurium infections in Sub-Saharan Africa predominantly associated with the novel multilocus sequence type, ST313 (Kingsley et al., 2009; Okoro et al., 2012). It is hypothesized that the immunocompromised status of the population allowed evolution of these strains since it can be traced to follow the spread of HIV and is currently not commonly found outside of Africa (MacLennan et al., 2010; Okoro et al., 2012). Sequencing of a representative ST313 isolate, S. Typhimurium D23580, identified several pseudogenes compared with other S. Typhimurium isolates. Many of these are shared with the host-restricted S. Typhi (Holt et al., 2008), which could suggest that D23580 is becoming more host restricted. An additional differentiating feature is the distinct repertoire of five prophage-like elements in the D23580 genome (Kingsley et al., 2009). One of these is the BTP1 prophage, and it was recently determined that this prophage carries glycosyltransferase (gtr) genes that constitute a putative O-antigen modification operon (Davies et al., 2013).
The O-antigen-modifying gtr operons have been found in phage genomes, on prophages within Salmonella genomes and in the context of phage remnant sequences (Vernikos and Parkhill, 2006; Davies et al., 2013). The gtr operons described to date consist of three genes: gtrA, gtrB and gtrC. Based on biochemical analysis of P22 mediated O-antigen modification (Makela, 1973) and related gtr operons in Shigella flexneri, it is hypothesized that GtrA and GtrB add the glucose to a carrier lipid and flip it to the periplasm, where the GtrC protein performs the modification while the O-antigen side chain is being assembled (reviewed in Allison and Verma, 2000). Phylogenetic mapping of the GtrC proteins showed that there are at least 10 different ‘families’ of gtr operons in S. enterica genomes (Davies et al., 2013), and individual strains can encode for as many as four different gtr operons. It is thought that each family is responsible for a unique modification. Furthermore, the expression of many gtr operons is under the control of epigenetic phase variation (Broadbent et al., 2010a). Thus, the gtr repertoire of an isolate with four phase-varying gtr operons could result in up to 16 individual phenotypes within a population based just on the O-antigen composition.
Of the 10 GtrC families identified in Davies et al., the biochemical activity has been identified for only two, family 1 GtrC (serotype O1) and family 3 GtrC (serotype O122). Specifically, the phage P22 family 1 gtrC operon results in the addition of a glucose residue via an α1– > 6 linkage to the galactose of the O-antigen subunit and the O1 serotype (Fukazawa and Hartman, 1964; Makela, 1973; Van der Byl and Kropinski, 2000). Recently, the factor O122 modification was attributed to a gtr operon from family 3, which mediates addition of a glucose molecule to the galactose via an α1– > 4 linkage (Bogomolnaya et al., 2008). However, since the expression of both are under the control of phase variation, the associated factor O1 and O122 modifications will not always be detectable (Broadbent et al., 2010a). To date, the activity of the remaining families of GtrC proteins has not been elucidated.
There is evidence that the Gtr-mediated modification helps prevent superinfection by altering the O-antigen (co)-receptor recognized by the phage, which provides a rationale for the presence of these genes on phage genomes. The P22 tailspike protein (TSP) contains endorhamnosidase activity to cleave the O-antigen and better gain access to the bacterial surface. This is blocked by α1–6 glucosylation of the galactose in the O-antigen subunit (O1 serotype), which is mediated by the P22 gtr operon. The rhamnosidase activity is not blocked by the chromosomally encoded family 3 gtr operon that results in α1–4 glucosylation (O122 serotype). In contrast, phages 9NA and KB1 are able to cleave O-antigen containing the α1–6 glucosylation but not the α1–4 glucosylation, so there is some difference in the glycanase activity of different phages. P22 infection is also blocked by acetylation of the rhamnose (Wollin et al., 1987). More recently, the phase variable expression of the O122 modification was demonstrated to directly affect the ability of phage SPC35 to infect S. Typhimurium (Kim and Ryu, 2012).
Many gtr operons have been retained in the genome of Salmonella serovars despite the degradation of the surrounding phage sequences (Davies et al., 2013). This indicates that these operons may provide some additional, direct benefit for the bacterium. Indeed, in S. flexneri, glucosylation by the products encoded in the gtrV operon increases virulence of the strain by compacting the O-antigen layer to allow the type 3 secretion system better access to epithelial cells (West et al., 2005). In S. Typhimurium, the family 3 gtr-mediated glucosylation of the O-antigen is associated with an increase in the persistence of the bacteria in the intestine (Bogomolnaya et al., 2008).
The gtr operon encoded by BTP1 is classified as a family 2 gtr operon (Davies et al., 2013); this family is associated with invasive Salmonella serovars, such as S. Typhi and S. Paratyphi A. D23580 was the first S. Typhimurium isolate in which a family 2 gtr operon was identified, which suggests that the modification performed by this operon may have contributed to the invasive capacity of this invasive non-typhoidal Salmonellae (iNTS) isolate. In this study we examined the functionality and significance of the prophage-encoded gtrBTP1 present in the iNTS strain D23580.
Results
The gtrBTP1 operon is required for full length of O-antigen in iNTS strains with BTP1 prophage
To assess the role of the family 2 gtrBTP1 operon (STMMW_03911–03921), these two genes were deleted and introduced into the native BTP1 prophage in S. Typhimurium D23580 (sMV189), resulting in ΔgtrBTP1 (sMV386). The LPS was analyzed on a sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS–PAGE) gel to determine if a shift could be observed that would be indicative of addition of a sugar moiety (Davies et al., 2013). Unexpectedly, the O-antigen of this ΔgtrBTP1 strain was very short (Fig. 1A, lane 2). The O-antigen of the ΔgtrBTP1 strain differs from that of a mutant in the wzz gene, which is responsible for producing the long O-antigen chain length; this difference is especially evident in the range of 5–9 O-antigen subunits (Fig. 1A, lane 2 vs. 4). The ΔgtrBTP1 LPS phenotype also differed from a rough strain that lacks an O-antigen (data not shown). This indicates the gtrBTP1 mutation did not affect the core LPS and O-antigen biosynthesis pathways, and the reduced O-antigen length is due specifically to the absence of gtrBTP1. The short O-antigen phenotype was complemented by the cloned gtrBTP1 operon (sMV359) as well as by the family 2 gtr operon from S. Enteritidis PT4 (referred to as SEN-F2; sMV338); the GtrCSEN-F2 protein shares 77% identity with GtrCBTP1. However, complementation with the family 1 phage P22 gtr operon (sMV531) only partially restored the amount of long O-antigen (Fig. 1B).
Fig 1.
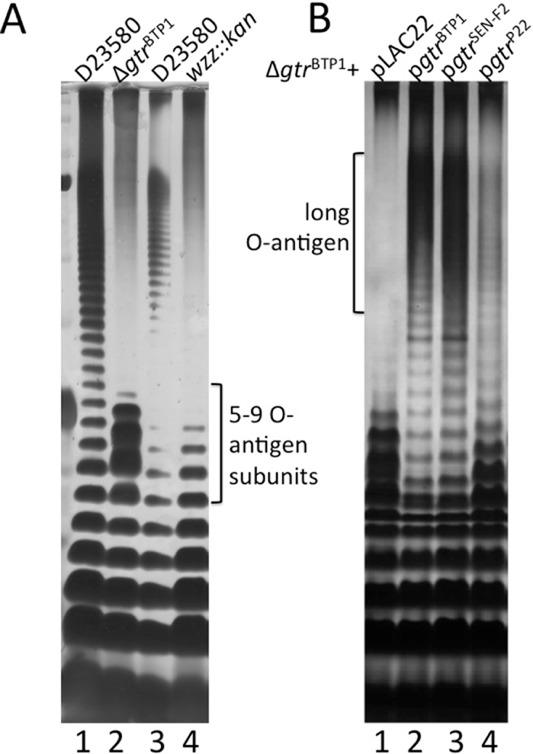
Deletion of the gtrBTP1 operon in S. Typhimurium D23580 affects O-antigen length. Crude LPS was run on a TSDS–PAGE gel and silver stained (see Experimental procedures section). Shown is LPS from stationary phase cultures of strain D23580 and its derivatives grown in LB; each rung represents a 1 unit difference in O-antigen length.A. The O-antigen phenotype of the ΔgtrBTP1 mutant does not resemble a wzz mutant. Lanes are as follows: (1) D23580; (2) ΔgtrBTP1; (3) D23580; (4) wzz::kan.B. Family 2 gtr operons complement the short O-antigen phenotype seen in ΔgtrBTP1. Lanes are as follows: (1) ΔgtrBTP1 + pLAC22; (2) ΔgtrBTP1 + pgtrBTP1; (3) ΔgtrBTP1 + pgtrSEN-F2; (4) ΔgtrBTP1 + pgtrP22. SEN-F2 = S. Enteritidis family 2 gtr operon.
GtrCBTP1 does not require GtrA/B for activity
Comparative genome analysis showed that family 2 gtr operons differed from other gtr operons due to the presence of a truncation of the gtrB gene (Davies et al., 2013). Analysis of specifically the gtrBTP1 family 2 operon identified additional differences. First, gtrBTP1 has a complete gtrB gene deletion, unlike the C-terminal gtrB truncation found in other family 2 gtr operons (Davies et al., 2013). Second, gtrABTP1 (referred to further as gtrA*) has a premature stop codon at nt 276 of the coding sequence, compared with one at nt 390 in the S. Typhi and S. Enteritidis family 2 gtr operons.
The mutations associated with the gtrBTP1 operon suggest that the GtrA/B proteins are no longer necessary for the activity of GtrC. Indeed, the ΔgtrBTP1 short O-antigen phenotype can be complemented by providing just GtrCBTP1 in trans (Fig. 2, lane 3). However, another possibility is that GtrA/B complementation occurs from one of the other chromosomal gtr operons. This is conceivable since the amino acid sequence identity of GtrA and GtrB between different gtr families is high, and the D23580 chromosome codes for family 3 and family 4 gtr operons in addition to gtrBTP1 (Davies et al., 2013). To address the necessity of GtrA/B for GtrCBTP1 functionality, a D23580 derivative with deletion of all three different gtr operons was constructed (‘basal’ D23580 strain) (sMV688). In this strain with an empty vector, the O-antigen is short (Fig. 2, lane 4). Expression of long O-antigen is restored by complementation with just GtrCBTP1 (Fig. 2, lane 5). This shows that GtrCBTP1-dependent O-antigen modification does not require any GtrA or GtrB proteins. Furthermore, this indicates a mechanistic difference between modification by family 2 GtrC proteins and GtrC proteins from families with conserved gtrAB genes.
Fig 2.
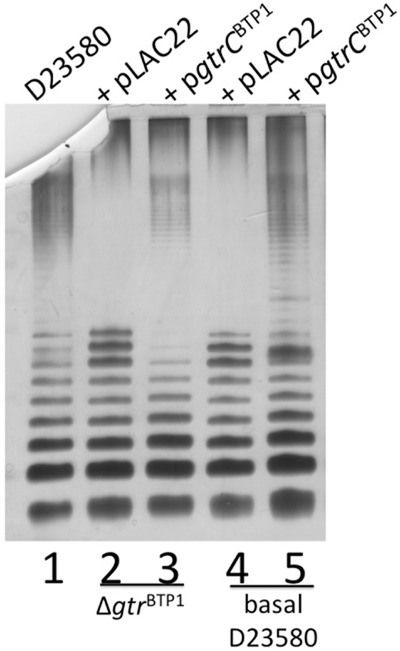
GtrA and GtrB are not required for biological activity of GtrCBTP1. LPS analysis details are as in legend for Fig. 1. Lanes are as follows: (1) D23580; (2) D23580 ΔgtrBTP1 + pLAC22; (3) D23580 ΔgtrBTP1 + pgtrCBTP1; (4) basal D23580 + pLAC22; (5) basal D23580 + pgtrCBTP1.
Identifying key features of the GtrCBTP1 protein
Alignment of the family 2 GtrC proteins with members of GtrC family 1 or 3, which have documented glucosylation activity, shows the former have a 33 amino acid conserved region toward the N-terminus that is not shared with the GtrC proteins from the other families (data not shown). BLAST analysis showed that this region aligned with the N-terminus of the S. Typhimurium O-antigen acetyltransferase OafA, and also contains the RXXR motif that is required for Oac-dependent acetylation of the O-antigen in S. flexneri (Thanweer and Verma, 2012) (Fig. 3A). A site-directed mutant in the RXXR motif was generated to determine if this region was also important for GtrCBTP1 activity; the R71A/R74A mutation of the GtrCBTP1 protein (sMV648) failed to complement the short O-antigen phenotype of the ΔgtrBTP1 strain (Fig. 3B, lane 4). This mutant GtrC protein is still expressed and can be detected in the membrane at levels similar to wild-type GtrC (Fig. 3C). These data show that the RXXR motif is required for GtrCBTP1 activity, in the same way as it is required for Oac acetylation in S. flexneri.
Fig 3.
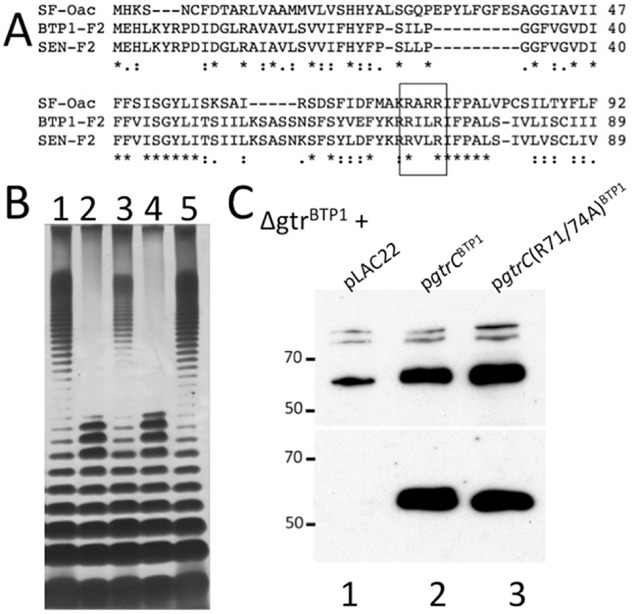
The RXXR motif is required for GtrCBTP1 biological activity.A. Family 2 GtrC proteins share homology with O-antigen acetyltransferase Oac from S. flexneri (SF). The RXXR motif found to be necessary for Oac activity is highlighted in the boxed region. Alignment was performed using TCOFFEE. SEN = S. Enteritidis; F2 = Family 2 GtrC.B. LPS analysis as in legend for Fig. 1. Lanes are as follows: (1) D23580; (2) ΔgtrBTP1 + pLAC22; (3) ΔgtrBTP1 + pgtrCBTP1; (4) ΔgtrBTP1 + pgtrC(R71/74A)BTP1; (5) D23580.C. Membrane localization of FLAG-tagged GtrCBTP1 and its R71A/R74A mutant derivative. Shown is a Western blot probed with anti-FLAG of WCL (top) and crude membrane extracts (bottom) made from D23580 ΔgtrBTP1 containing indicated vectors. Lanes: (1) pLAC22; (2) ΔgtrBTP1 + pgtrCBTP1; (3) ΔgtrBTP1 + pgtrC(R71/74A)BTP1.
GtrCBTP1 distinguishes itself from S. flexneri Oac, first, by being in the context of a (remnant) gtr operon. Second, topological analysis demonstrated that Oac has 10 transmembrane domains and a C-terminal cytoplasmic domain (Thanweer and Verma, 2012), whereas GtrCBTP1 is predicted to have 11 transmembrane domains and a 273 amino acid long C-terminus in the periplasm (Fig. 4A). To determine the localization of the RXXR motif and the C-terminal domain, GtrCBTP1 protein topology was analyzed with PhoA protein fusions. The PhoA enzyme is only active in the periplasm, so the location of loops between transmembrane domains can be determined by assessing the absence or presence of PhoA enzymatic activity in strains containing these fusions. Five different fusions were created (vectors pMV406-410) and PhoA activity was investigated in both Escherichia coli and S. Typhimurium. A fusion at amino acid 37, at the end of the first predicted transmembrane domain, was PhoA-positive, whereas fusions at amino acids 56 and 76 were PhoA-negative; the latter two amino acids flank the RXXR loop, indicating those essential amino acids are located in the cytoplasm (Fig. 4B). In agreement with the topology prediction, PhoA fusions at amino acids 370 (at the beginning of the predicted C-terminal periplasmic region) and 640 (at the C-terminal end) resulted in PhoA-positive colonies, indicating periplasmic localization. GtrCBTP1 activity furthermore requires at least the final 238 amino acids of the predicted 273 amino acid periplasmic C-terminus, as the O-antigen of a strain with gtrCBTP1 truncated at amino acid 402 (sMV548) resembles that of the ΔgtrBTP1 strain (sMV386) (Fig. 4C). These data are consistent with the long C-terminus of GtrCBTP1 being located in the periplasm and having an essential role for the biological function of the protein.
Fig 4.
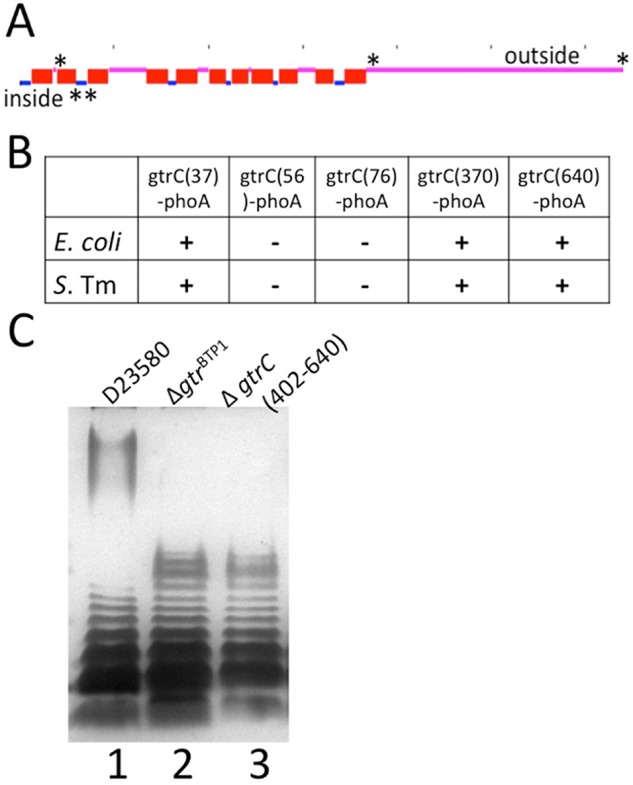
Membrane topology of GtrCBTP1 and requirement for the C-terminal periplasmic domain for activity.A. Locations of the GtrCBTP1–PhoA fusions along the TMHMM (Moller et al., 2001) topology prediction are given with an asterisk (*). Dashes along top indicate 100aa increments.B. Phenotype of GtrCBTP1–PhoA fusion in both E. coli and S. Typhimurium (S. Tm) strain 4/74. (−) indicates white color and (+) blue color. The location of the amino acid fusion is given in the vector name along the top row.C. The C-terminus of GtrCBTP1 is required for activity. LPS analysis as in the legend for Fig. 1. Lanes are as follows: (1) D23580; (2) ΔgtrBTP1; (3) gtrC(Δ402–640)BTP1.
Taken together, the data described earlier suggest that the family 2 gtr operon encodes an O-antigen acetyltransferase. First, there is no requirement for GtrAB proteins that are necessary for glucose modification, and second, the RXXR motif is essential, similar to the role of this motif in S. flexneri O-antigen acetyltransferase Oac.
gtrCBTP1 expression is not phase variable
Expression of most gtr operons across the gtr families, except family 4, is under the control of phase variation associated with a signature sequence in the regulatory region (Broadbent et al., 2010a; Davies et al., 2013). The sequence upstream of gtrA* in BTP1, however, contains a 365 nucleotide sequence that disrupts this signature sequence. To assess whether the insertion disrupts phase variation, a strain carrying a fusion of this region from gtrBTP1 to lacZ was analyzed. This strain gave rise to colonies with a uniform Lac+ (ON) phenotype (data not shown), reflecting a lack of phase variation.
Two transcriptional start sites (TSS) of gtrBTP1 were identified in strain D23580 by differential RNA-seq analysis. One TSS was upstream of gtrA*, and the second upstream of gtrCBTP1 (Fig. 5A). The former is the same TSS previously identified for the gtrLT2_I operon (STM0559-7) in S. Typhimurium strain LT2 that is phase variable (Broadbent et al., 2010b). However, as shown earlier, in strain D23580, transcription from this promoter region is not controlled by phase variation. The second TSS is in a region created by the gtrB deletion in D23580 and presumably would not be present in other gtr operons. Transcriptomic data from RNA-seq experiments (Fig. 5B) indicate that both gtrA* and gtrCBTP1 are usually expressed, and therefore a degree of O-antigen modification is likely to occur, under a wide range of growth conditions. These ranged from standard laboratory growth conditions to those designed to mimic in vivo infections (Kroger et al., 2013; see Experimental procedures section for further details). Future work is needed to determine the relative contribution of the two promoters for gtrCBTP1 expression.
Fig 5.
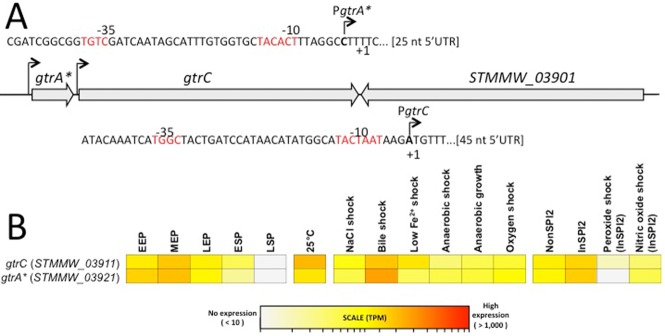
Genetic organisation and transcriptional profiling of the gtrBTP1 locus. The gtrA*, gtrC and STMMW_03901 (TSP) genes are encoded within the phage BTP1 locus in S. Typhimurium strain D23580 (Kingsley et al., 2009).A. The locations of the transcription start sites (TSS) of gtrC (STMMW_03911) and gtrA* (STMMW_3921) are shown, and the predicted -10/-35 promoter motifs and the initiating nucleotide of both gtr transcripts are highlighted in bold. The nucleotide position of the gtrC (STMMW_03911) gene is 406441–408363 (negative strand), the gtrA* (STMMW_3921) gene is 408435–408710 (negative strand) and the STMMW_03901 gene is 404416–406401 (positive strand) on the D23580 chromosome.B. Absolute gene expression of the gtrBTP1 locus. The heat map shows the levels of gene expression of the gtrBTP1 genes as TPM values, derived as described (Kroger et al., 2013).
The ΔgtrBTP1 effect on O-antigen length requires the BTP1 phage tailspike protein
Analysis of the sequence adjacent to the gtrBTP1 operon identified gene STMMW_03901, which is convergently transcribed (see Fig. 5A). STMMW_03901 encodes for a TSP with 70% identity to that of the well-studied Salmonella phage P22 protein. The phage P22 TSP has endorhamnosidase activity that allows cleavage of the O-antigen. Specific amino acids important for this enzymatic activity of P22 TSP (namely, Glu 359, Asp 390 and Asp 392) are present in BTP1 TSP (Baxa et al., 1996; Steinbacher et al., 1996). Thus, the shorter O-antigen of the ΔgtrBTP1 strain could be due to BTP1 TSP-dependent cleavage of full length O-antigen.
This hypothesis was tested by deleting the TSP gene in the D23580 ΔgtrBTP1 mutant (Fig. 6). When the ΔgtrBTP1 mutation is combined with the TSP deletion, the same full length O-antigen is seen as in the wild-type strain (Fig. 6, lane 3). This correlation between O-antigen length, depending on the presence of gtrCBTP1 and the TSP with its associated rhamnosidase activity, held true for two other iNTS strains (5597 and A38596) that are phylogenetic relatives of D23580 and harbor the BTP1 prophage (Kingsley et al., 2009; Okoro et al., 2012) (Fig. 6, lanes 4–9). These data lead us to conclude that in wild-type D23580, GtrCBTP1 acts to protect the O-antigen from cleavage by the phage BTP1 tailspike-encoded rhamnosidase activity.
Fig 6.
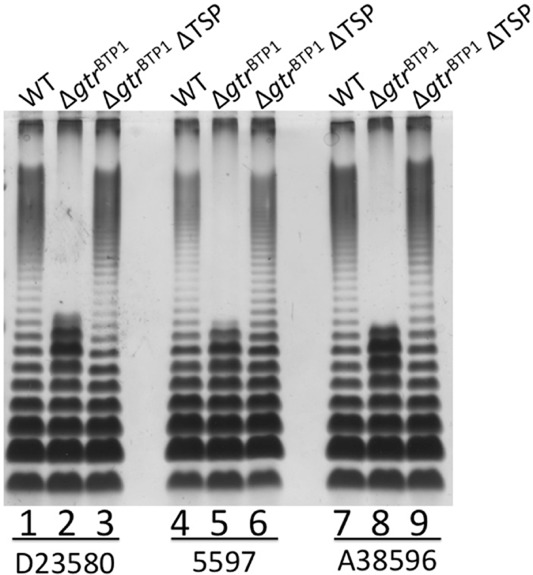
O-antigen length in iNTS strains with the BTP1 prophage is defined by a combination of the TSP rhamnosidase and GtrCBTP1 activity. LPS analysis as in legend for Fig. 1. Lanes are as follows: (1) D23580; (2) D23580 ΔgtrBTP1; (3) D23580 ΔgtrBTP1ΔTSP; (4) iNTS strain 5597; (5) 5597 ΔgtrBTP1; (6) 5597 ΔgtrBTP1ΔTSP; (7) iNTS strain A38596; (8) A38596 ΔgtrBTP1; (9) A38596 ΔgtrBTP1ΔTSP. WT = wild-type strain.
Significance of GtrCBTP1 in phage–bacteria interactions
The findings described earlier are consistent with a model in which prophage-encoded GtrCBTP1 mediates O-antigen modification, possibly via acetylation. Modification of a receptor is a common phage strategy to protect a lysogen from superinfection by phages that use the same (co)-receptor, in this case the O-antigen. Phage P22 is known to bind O-antigen as a receptor via its TSP and the BTP1 TSP sequence is homologous to that of P22 (Steinbacher et al., 1997), suggesting that BTP1 may also use the O-antigen as a (co)-receptor. Furthermore, our data indicate that unmodified, full-length O-antigen on the cell surface can be cleaved by the BTP1 phage TSP similar to P22 (Steinbacher et al., 1997), which can facilitate phage penetration.
In this model, the short O-antigen of the ΔgtrBTP1 strain is a result of BTP1 phage-dependent O-antigen cleavage and, therefore, free phage BTP1 particles should be present in cultures of strain D23580. Indeed, plaques are observed upon spotting supernatant of a culture of strain D23580 on a lawn of ‘basal’ LT2, which is a mutant that lacks both native gtr operons (Fig. 7A). No plaques were observed with the supernatant of a D23580 mutant that has the BTP1 TSP gene deleted from the prophage, indicating these plaques are derived from BTP1 phage and not from other prophage present in this strain (Fig. 7A) (Kingsley et al., 2009). Furthermore, culture supernatants from the D23580-related iNTS strains A130 and A38596 also showed plaque formation, indicative of free BTP1 phage (data not shown).
Fig 7.
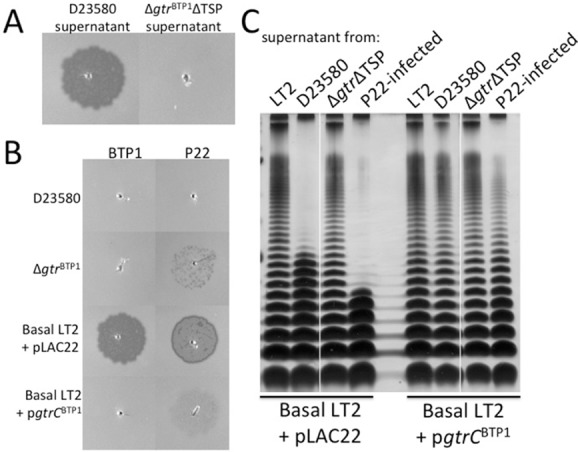
GtrCBTP1 O-antigen modification prevents infection by both P22 and BTP1 and inhibits cleavage of the O-antigen by the BTP1 phage.A. Supernatant from D23580 cultures, containing BTP1 phage, or the ΔgtrΔTSP mutant were spotted on lawns of basal LT2 + pLAC22.B. Supernatant from cultures contacting BTP1 or P22 phage were spotted at equal titer (about 3.5 × 105 pfu ml−1) on lawns of wild-type D23580, the ΔgtrBTP1 mutant, basal LT2 + pLAC22 or basal LT2 + pgtrCBTP1.C. Cultures of basal LT2 containing either empty vector or pgtrCBTP1 were incubated with supernatant of cultures from strains (as indicated along top of gel) to investigate O-antigen cleavage from the surface. LPS analysis performed as described in Fig. 1.
To examine whether the GtrCBTP1 O-antigen modification affects phage susceptibility, spot titer tests were carried out with the BTP1- and P22-containing supernatants. Plaque formation by these phages was tested against the basal LT2 strain either with or without GtrCBTP1, and against wild-type D23580 and the ΔgtrBTP1 mutant. As expected, no plaque formation occurs on a D23580 lawn, consistent with an active phage repressor system. Interestingly, some P22 plaques did form on the ΔgtrBTP1 mutant, suggesting that P22 is a heteroimmune phage (right column, Fig. 7B). Importantly, GtrCBTP1 activity blocked infection by both phages BTP1 and P22 (Fig. 7B). Specifically, BTP1 and P22 formed plaques on the basal LT2 strain containing only empty vector (Fig. 7B). This plaque formation, however, is absent when spotting BTP1 phage on this strain containing pgtrCBTP1 (sMV816). Similarly, P22 infection of LT2 is strongly inhibited by pgtrCBTP1 (Fig. 7B). P22 also does not infect wild-type D23580 but can infect the ΔgtrBTP1 D23580 mutant. These data illustrate the protective nature of the modification of the O-antigen by GtrCBTP1.
Importantly, both the BTP1 and P22 phages mediated cleavage of the full length O-antigen present on basal LT2 (Fig. 7C, lanes 2 and 4). When the GtrCBTP1 protein was present, O-antigen was not cleaved from the surface of the bacteria and full length LPS was maintained (Fig. 7C, lanes 6 and 8). No O-antigen cleavage was observed with supernatant from the ΔgtrBTP1ΔTSP strain that lacks both gtrBTP1 and TSP (Fig. 7C, lanes 3 and 7). Thus, GtrCBTP1 activity protects the O-antigen from BTP1 TSP mediated O-antigen cleavage.
Another role of phage receptor modification could be the prevention of readsorption of phage released by lysogenic kin. To address whether GtrCBTP1-mediated O-antigen modification can prevent readsorption, phage titers from wild-type D23580 and the ΔgtrBTP1 mutant were compared (Fig 8A). Phage concentrations produced by overnight cultures of D23580 were 5.8 × 107 pfu/ml, but the ΔgtrBTP1 mutant only produced 1.4 × 106 pful/ml. The lower titers in the absence of GtrCBTP1 could be due to phage particles rebinding to the surface of the bacteria that express an un-modified O-antigen. To further test adsorption directly, BTP1 phage-containing supernatant was incubated with the LT2 strain that lacks O-antigen modification, and with this strain expressing GtrCBTP1 (Fig. 8B). Adsorption with the unmodified strain resulted in a greater decrease in titer than adsorption with the strain expressing modified O-antigen. This is consistent with the conclusion that modification can reduce readsorption of phage. Less adsorption to the LT2 strain with modified O-antigen was also observed with P22 phage, consistent with the lower level of infection observed on strains with GtrCBTP1-mediated O-antigen modification (Fig. 7). Together, our data support a model in which BTP1 phage-encoded GtrC modifies the BTP1 O-antigen (co)-receptor to reduce superinfection by heteroimmune phages that use the same O-antigen moiety as BTP1 as co-receptor, and to limit readsorption by kin.
Fig 8.
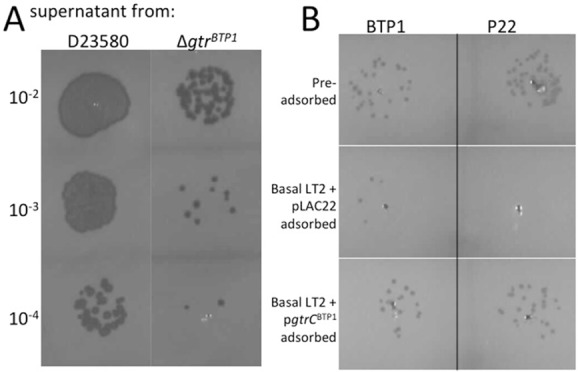
Modification by GtrCBTP1 prevents readsorption of phage.A. BTP1 titer is reduced in a strain with a deletion of gtrBTP1. Supernatants containing BTP1 phage, obtained from a D23580 culture or the ΔgtrBTP1 mutant, were spotted on lawns of basal LT2. Dilutions from the original supernatant are given.B. BTP1 and P22 phage titers are affected by adsorption with strains having or lacking GtrCBTP1 O-antigen modification. BTP1 and P22 phage were incubated for 5 minutes with the indicated strains (top row shows phage titer pre-adsorption). Bacteria and phage bound to their surface were removed by pelleting and the free, unadsorbed phage numbers determined by spotting the supernatant on a lawn of basal LT2. The same dilution of phage for all samples is shown.
Discussion
The BTP1 prophage is one of the distinguishing genetic features of the iNTS S. Typhimurium strain D23580 (Kingsley et al., 2009). Previously, a Salmonella-wide genome analysis for the occurrence of O-antigen modifying gtr operons determined that this prophage encoded an uncharacterized gtr operon (Davies et al., 2013). Here we analyzed the operon and its significance for this strain. This gtr operon has one full-length protein: the GtrCBTP1 protein. We determined that GtrCBTP1 has hallmarks consistent with acetyltransferase activity. Expression of GtrCBTP1 occurs under a wide range of growth conditions that are relevant to infection, and, unlike many other gtr operons, expression of gtrBTP1 is not subject to phase variation. We show that the GtrCBTP1-mediated modification plays an important role in the bacterium–phage interaction by protecting the bacterium from infection by both BTP1 and the related P22 phage.
Previously characterized gtrABC operons mediate the glucosylation of the O-antigen, but have different receiving moieties and linkages (Allison and Verma, 2000). In contrast, our data suggest that the gtrBTP1 locus mediates acetylation of the O-antigen, and we propose that GtrCBTP1 is the protein responsible for the acetylation of the rhamnose residue in the O-2 and O-3 positions that was recently identified in biochemical analysis of the O-antigen of strain D23580 (Micoli et al., 2014). First, GtrCBTP1 contains an acetyltransferase domain from the acyl_transf_3 superfamily within its first 360 amino acids. Second, we showed that a RXXR protein motif that is essential for the Oac acetyltransferase activity in S. flexneri (Thanweer and Verma, 2012) is present in GtrCBTP1 and also is essential for its activity (Fig. 3A and B). GtrA and GtrB have roles in transferring a glucose moiety across the inner membrane; the absence of gtrB and truncation of gtrA, and the fact they are not required for this modification (Fig. 2), provides further evidence that the activity of GtrCBTP1 does not involve glucosylation.
All the data are consistent with the conclusion that the family 2 GtrCBTP1 mediates acetylation of the rhamnose in the O-antigen. Based on high amino acid identity among the GtrC proteins of this family (Davies et al., 2013) and the ability of the S. Enteritidis family 2 gtr operon to complement the ΔgtrBTP1 short O-antigen phenotype, it is highly likely that all family 2 GtrC proteins have acetyltransferase activity. In support of this conclusion, analysis of the O-antigen of defined strains of S. Typhi confirmed that acetylation of the rhamnose requires the S. Typhi family 2 gtr operon (manuscript in preparation). This S. Typhi GtrC has 77% identity with GtrCBTP1 and 99% identity with the S. Enteritidis family 2 GtrC protein, which complements the short O-antigen phenotype of our ΔgtrBTP1 mutant. Thus there is strong support for the conclusion that all family 2 GtrC proteins carry out acetylation of the rhamnose in the O-antigen of these strains.
GtrCBTP1 and other O-antigen acetyltransferases, such as Oac in S. flexneri and OafA in S. Typhimurium (Slauch et al., 1996; Thanweer et al., 2008; Thanweer and Verma, 2012), share some predicted structural features. As noted earlier, all three acetyltransferase proteins contain the RXXR motif, which is necessary for the O-antigen modification by Oac and GtrCBTP1. A cytoplasmic location of this motif has been experimentally confirmed for Oac (Thanweer et al., 2008) and GtrCBTP1 (Fig. 4), and is predicted for OafA (Thanweer and Verma, 2012). The three acetyltransferases are also membrane proteins with multiple membrane spanning regions, ranging from 9 to 11 transmembrane domains. One of the significant differences between GtrCBTP1, OafA and Oac is the length and location of the C-terminal soluble domain. GtrCBTP1 and OafA contain a large (270 aa for GtrCBTP1, predicted 255 aa for OafA) periplasmic tail that is necessary for activity (Fig. 4C and Hauser et al., 2011). In contrast, Oac has a relatively short (22 aa), cytoplasmically located C-terminal end. The role and relevance of the location of the C-termini remain to be elucidated, but it suggests that Salmonella GtrC acetyltransferase enzymes have an additional activity or different mechanisms of activity than Oac from S. flexneri.
For these O-antigen acetyltranferases, it is unclear from where the acetyl moiety is derived, and where or when the moiety is transferred to the growing O-antigen chain. Given the requirement for both a cytoplasmic and a periplasmic domain, and the presence of multiple transmembrane domains, one possibility is that family 2 GtrC proteins function as a channel for the acetyl moiety in addition to carrying out the enzymatic transfer reaction. In this model, the RXXR cytoplasmic loop might be involved in binding the acetyl group for transfer to the periplasm. Alternatively, modification of the O-antigen subunit could occur in the cytoplasm as has been proposed for OafA (Slauch et al., 1996). The C-terminal domain may mediate necessary protein–protein interactions, potentially with the O-antigen assembly complex. Further work on the structure/function relationship of Gtr proteins is needed to elucidate these and many other aspects of gtr-mediated O-antigen modification.
Acetylation of the O-antigen, and specifically of the rhamnose moiety, has been previously described in context of lysogenic conversion by phages A3 and A4 (Wollin et al., 1987). This modification inhibited P22 adsorption and it was inferred that TSP-mediated O-antigen cleavage was inhibited (Wollin et al., 1987). Similarly, our data indicate that P22 and BTP1 phage-mediated infection is inhibited by GtrCBTP1 activity, likely because the acetyl modification blocks the TSP endorhamnosidase activity. This was evident from the short O-antigen phenotype in the absence of GtrCBTP1, and the dependency on the TSP for this phenotype. Effective protection against phage infection may also be facilitated by a high level of acetylation, with an estimated 80% of O-antigen subunits containing the modification in D23580 (Micoli et al., 2014). There are two contributing factors to this high degree of modification. First, GtrCBTP1 expression was not subject to phase variation, and thus all cells in a population will have modified O-antigen. This is in contrast to gtrP22-mediated glucosylation, for which the mixed population generated by phase variation was directly shown to affect the dynamics of interactions of a population of cells with phage (Kim and Ryu, 2012). Second, the fact that gtrA*C appears to be expressed in a wide range of growth conditions suggests that the O-antigen will usually be in the modified state (Fig. 5B). Thus, a benefit of the BTP1 prophage for the bacterium is that it confers effective protection on the entire bacterial population against infection by BTP1 and both homo- and heteroimmune phages that use the same O-antigen moiety as co-receptor.
There are several aspects of the BTP1 phage that could potentially contribute to the virulence of iNTS BTP1 lysogens. One is the high titer of BTP1 prophage (an average of 7 × 107 pfu ml−1) we found in overnight cultures, even in the absence of phage-inducing conditions. Phage-associated lysis could produce bacterial debris, and this, in turn, could affect the development of the immune response to the iNTS infection. Additionally, the BTP1-encoded family 2 gtrBTP1 operon is not under the control of phase variation, whereas other family 2 operons are phase variable, including those from S. Enteritidis and S. Typhi and the majority of gtr operons (Broadbent et al., 2010a). Thus, D23580 does not have the option for phase-variable immune evasion from antibodies developed to the acetylated O-antigen, which could be significant. However, whether acetyl modification affects the course of infection has yet to be determined. The family 2 gtr operons are associated with many invasive serovars of S. enterica, and it will be important to determine whether, as we propose, O-antigen acetylation contributes to the typical course of infection, including systemic disease, as reported for the iNTS isolates.
Experimental procedures
Bacterial strains and culture conditions
Strains are listed in Table 1. Strains were maintained in either Luria broth or on Luria–Bertani (LB) plates at 37°C. Strains harboring temperature-sensitive vectors were grown at 30°C. For selection of mutants or maintenance of vectors, the following antibiotic concentrations were used: tetracycline (15 μg ml−1), ampicillin (100 μg ml−1), chloramphenicol (34 μg ml−1 for vectors or 8 μg ml−1 for chromosomal inserts) or kanamycin (30 μg ml−1).
Table 1.
Strains used in this study
| Strain name in text | Strain number | Relevant genotype | Plasmid | Source |
|---|---|---|---|---|
| Escherichia coli isolates | ||||
| E. coli–gtrC(37)BTP1–phoA | MV1557 | NEB5a | pMV406 | This study |
| E. coli–gtrC(56)BTP1–phoA | MV1558 | NEB5a | pMV407 | This study |
| E. coli–gtrC(76)BTP1–phoA | MV1559 | NEB5a | pMV408 | This study |
| E. coli–gtrC(370)BTP1–phoA | MV1560 | NEB5a | pMV409 | This study |
| E. coli–gtrC(640)BTP1–phoA | MV1561 | NEB5a | pMV410 | This study |
| Salmonella Typhimurium isolates | ||||
| D23580 | sMV189 | Wild-type | Kingsley et al., 2009 | |
| gtrABTP1–lacZ | sMV523 | sMV77att::pMV349 | This study | |
| ΔgtrBTP1 | sMV386 | D23580 ΔgtrBTP1 (STMMW_03921-STMMW_03911) | This study | |
| Δwzz | sMV664 | D23580 wzz::kan (STMMW_21101) | This study | |
| ΔgtrBTP1 + pLAC22 | sMV610 | D23580 ΔgtrBTP1 | pLAC22 | This study |
| ΔgtrBTP1 + pgtrBTP | sMV569 | D23580 ΔgtrBTP1 | pMV359 | This study |
| ΔgtrBTP1 + pgtrSEN-F2 | sMV571 | D23580 ΔgtrBTP1 | pMV338 | This study |
| ΔgtrBTP1 + pgtrP22 | sMV531 | D23580 ΔgtrBTP1 | pMV333 | This study |
| ΔgtrBTP1 ΔTSP | sMV800 | D23580 ΔgtrBTP1 ΔTSP | This study | |
| ΔgtrBTP1 + pgtrCBTP1 | sMV647 | D23580 ΔgtrBTP1 | pMV390 | This study |
| ΔgtrBTP1 + pgtrC (R71/74A)BTP1 | sMV648 | D23580 ΔgtrBTP1 | pMV392 | This study |
| gtrC(Δ402–640)BTP1 | sMV548 | D23580 gtrC(402–640)BTP1::kan | This study | |
| S. Tm-gtrC(37)BTP1–phoA | sMV697 | S. Typhimurium 4/74 | pMV406 | This study |
| S. Tm-gtrC(56)BTP1–phoA | sMV698 | S. Typhimurium 4/74 | pMV407 | This study |
| S. Tm-gtrC(76)BTP1–phoA | sMV699 | S. Typhimurium 4/74 | pMV408 | This study |
| S. Tm-gtrC(370)BTP1–phoA | sMV700 | S. Typhimurium 4/74 | pMV409 | This study |
| S. Tm-gtrC(640)BTP1–phoA | sMV701 | S. Typhimurium 4/74 | pMV410 | This study |
| Basal D23580 | sMV688 | D23580 ΔoafA; ΔgtrBTP1; ΔgtrFam3; ΔgtrFam4 | This study | |
| Basal D23580 + pLAC22 | sMV691 | D23580 ΔoafA; ΔgtrBTP1; ΔgtrFam3; ΔgtrFam4 | pLAC22 | This study |
| Basal D2350 + pgtrCBTP1 | sMV689 | D23580 ΔoafA; ΔgtrBTP1; ΔgtrFam3; ΔgtrFam4 | pMV390 | This study |
| Basal LT2 + pLAC22 | sMV815 | LT2 ΔoafA; gtrFam3::tet; ΔgtrFam4::kan | pMVLAC22 | This study |
| Basal LT2 + pgtrBTP1 | sMV816 | LT2 ΔoafA; gtrFam3::tet; ΔgtrFam4::kan | pMV390 | This study |
| 5597 | sMV762 | iNTS, lineage I | Kingsley et al., 2009 | |
| 5597ΔgtrBTP1 | sMV796 | 5597 gtrBTP1::tet | This study | |
| 5597ΔgtrBTP1ΔTSP | sMV801 | 5597 gtrBTP1::tet;TSP::kan | This study | |
| A38596 | sMV765 | iNTS, lineage II | Kingsley et al., 2009 | |
| A38596ΔgtrBTP1 | sMV792 | A38596 A38596 gtrBTP1::tet | This study | |
| A38596ΔgtrBTP1ΔTSP | sMV802 | A38596 gtrBTP1::tet; TSP::kan | This study | |
| A130 | sMV761 | iNTS, lineage I | Kingsley et al., 2009 |
Molecular biology techniques
Standard molecular biology techniques were used (Sambrook et al., 1987). DNA was obtained using either genomic DNA isolated as described (Ausubel et al., 1989) or Qiagen miniprep kit for plasmids. PCRs were performed using either Merck's KOD Hot Start (Billerica, MA, USA) or Promega's GoTaq Flexi polymerases (Madison, WI, USA). Restriction enzymes and T4 ligase for cloning were purchased from NEB (Ipswich, MA, USA). All vectors and plasmids generated for this study are listed in Table 2. The site-directed mutagenesis was performed on constructs in the pLAC22 vector (Warren et al., 2000) with primers oMV954 (GAATTTTACAAGAGAGCAATACTAGCAATATTCCCAGCT) and oMV955 (AGCTGGGAATATTGCTAGTATTGCTCTCTTGTAAAATTC) using Agilent's QuikChange Lightning kit (Santa Clara, CA, USA). Bold regions indicate codons changed to introduce the mutations.
Table 2.
Plasmids used in this study
| Parent plasmid | Description; gene cloned with oligonucleotides used | Source | |
|---|---|---|---|
| pCP20-TcR | pCP20 | TcR cassette inserted into SmaI site of pCP20 [oMV466; oMV467]a | This study |
| pMV349 | pMV243 | BTP1 regulatory region–lacZ fusion [oMV413; oMV875] | This study |
| pMV333 | pLAC22 | gtrABC(P22) [oMV776; oMV778] | Broadbent et al., 2010a |
| pMV338 | pLAC22 | gtrABC(S. Enteritidis family 2) [oMV776; oMV780] | This study |
| pMV359 | pLAC22 | gtrABC(BTP1) [oMV897; oMV898] | This study |
| pMV390 | pLAC22 | BTP1 GtrC-FLAG [oMV898; oMV958/959] | This study |
| pMV392 | pLAC22 | BTP1 GtrC (R54A/R57A)-FLAG; [oMV954; oMV955] | This study |
| pMV406 | pSK4158 | gtrC(BTP1)–phoA fusion at amino acid 37 [oMV993; oMV1003] | This study |
| pMV407 | pSK4158 | gtrC(BTP1)–phoA fusion at amino acid 56 [oMV994; oMV1003] | This study |
| pMV408 | pSK4158 | gtrC (BTP1)–phoA fusion at amino acid 76 [oMV995; oMV1003] | This study |
| pMV409 | pSK4158 | gtrC (BTP1)–phoA fusion at amino acid 370 [oMV996; oMV1003] | This study |
| pMV410 | pSK4158 | gtrC (BTP1)–phoA fusion at amino acid 640 [oMV997; oMV1003] | This study |
Oligos are given in Supplemental Table S1.
Constructon of lacZ reporter fusions and detection of LacZ activity
The CRIM system was used to insert gtr promoter–lacZ fusions into the attB site of S. Typhimurium LT2 (Haldimann and Wanner, 2001) using vector pMV243 (Broadbent et al., 2010a). Strains containing inserts were streaked on minimal M9 media agar plates containing 40 μg ml−1 X-gal (5-bromo-4-chloro-3-indolyl-ß-d-galactoside) (Melford; Ipswich, Suffolk, UK).
Generation of mutant strains
Allelic replacement was used to introduce mutations to the chromosome (Datsenko and Wanner, 2000). Antibiotic resistance cassettes were obtained from Tn10 for Tc-resistance (TcR) and pKD13 for KmR. To generate multiple mutations in a single strain, these resistance cassettes were occasionally removed (as indicated in Table 1) before the next mutation was introduced. Since the pKD13 KmR cassette is flanked by flippase recognition target (FRT) sites, the introduction of pCP20 (Cherepanov and Wackernagel, 1995) or its pCP20-TcR derivative allowed for the expression of Flp recombinase to produce unmarked mutant strains. When the Tn10 TcR cassette was used, lambda-red recombination was used to introduce an oligonucleotide homologous to the region surrounding the TcR cassette. This combined with counter-selection (Bochner et al., 1980) produced scarless mutations of the targeted genes. For example, to obtain strain ΔgtrBTP1 (sMV386), which contains a deletion of the BTP1 prophage-encoded gtrA* and the gtrC genes in D23580, the sequence from 101 nt upstream of the start codon of the pseudogene STMMW_03921 (gtrA*) to 15 nt downstream of the stop codon of STMMW_03911 (gtrCBTP1) was first replaced with a tetracycline cassette (amplified using oMV754/oMV755) (Table S1). This sequence was, in turn, replaced with oMV758 using the counter-selection method described. These genes are located on the phage genome between the phage TSP (TSP gene, STMMW_03901) and the phage–host genome junction.
LPS extraction and visualization
Crude LPS extracts were prepared as described (Davies et al., 2013). After running the samples on TSDS–PAGE (Tricine-SDS–PAGE) gels, two methods were used for visualization: either silver stain as described previously (Davies et al., 2013) or Western blot. For Western blots, the samples were transferred to polyvinylidene fluoride (PVDF) and blocked with phosphate buffered saline with 0.1% Tween-20 (PBS-T) and 5% milk. The O4 serum from Statens Serum Institute (Denmark) was used as primary antibody followed by goat anti-rabbit IgG-HRP from Sigma (A0545). Millipore's Luminato Western HRP substrate was used for detection.
Membrane preparation and FLAG visualization
Vectors encoding for FLAG-tagged versions of the GtrCBTP1 protein were expressed in ΔgtrBTP1. Mid-exponential phase cultures were induced with 100 μM ITPG and expression allowed to occur for 30 minutes. Cells were then pelleted. Whole cell lysate (WCL) samples were immediately boiled in SDS buffer (0.1 m Tris–HCl, pH 6.8, 4% β-mercaptoethanol, 4% SDS, 20% glycerol). For crude membrane preparations, the samples were resuspended in 1 ml PBS and subjected to a freeze/thaw cycle (−80°C/RT) three times. Unbroken cells were pelleted by centrifugation for 2 min (16,060× g) and the supernatant transferred to a new Eppendorf. Membranes were pelleted by centrifugation at 4°C for 1 hour (16,060× g). The pellet was then resuspended in SDS buffer and boiled for analysis by Western.
For protein detection, SDS–PAGE gels were used and then transferred to PVDF. Blots were blocked with 5% milk PBS-T and the primary antibody was Sigma's Monoclonal ANTI-FLAG M2 antibody produced in mouse (F3615). Secondary antibody was goat anti-mouse IgG-HRP (Sigma; A0168).
Determining topology of BTP1 GtrC protein
The gtrC–phoA fusion vectors were introduced into E. coli NEB5a and S. Typhimurium 4/74. After verification by colony PCR using primers oMV1003 (CATGAAGCTTATTAATGCAGCTGGCACGAC) and oMV1006 (CGTTGGGTGATCTTTTTCGT) that strains contained the appropriate vectors, colonies were patched onto LB agar plates containing chloramphenicol and X-Phos (5-bromo-4-chloro-3-indolyl-phosphate, 40 μg ml−1; Melford) for detection of the blue color change.
RNA-seq
RNA-seq was performed on S. Typhimurium strain D23580 cultured in 16 environmental conditions: early exponential phase, mid-exponential phase, late exponential phase, early stationary phase, late stationary phase, 25°C, NaCl shock, Bile shock, low Fe2+ shock, anaerobic shock, anaerobic growth, oxygen shock, nonSPI2, InSPI2, peroxide shock (InSPI2) and nitric oxide shock (InSPI2) as described (Kroger et al., 2012; 2013,). Sequence reads were mapped to the S. Typhimurium D23580 genome (Genbank Accession FN424405). The BTP1 gtrA* and gtrC TSSs were identified in D23580 grown to ESP by dRNA-seq (Sharma et al., 2010). Transcript per million (TPM) values for absolute levels of gtrA* and gtrC gene expression were derived as described previously (Kroger et al., 2013).
Preparation of phage-containing supernatants
BTP1 phage was prepared by pelleting overnight cultures of D23580, removing the supernatant and treating with a few drops of chloroform to kill any remaining bacteria. This produced phage titers with an average of 7 × 107 pfu ml−1. To generate P22 supernatant for the experiments, a phage lysate (50 μl at 104 pfu ml−1 of wild-type P22) was added to a 10 ml culture of exponential phase basal LT2 and growth allowed to continue overnight. Bacteria were again pelleted, the supernatant removed and treated with chloroform. This produced a supernatant containing 3.5 × 1010 pfu ml−1.
Top agar phage assays
To create the lawn of bacteria, 100 μl of an overnight culture was mixed with 3.5 ml of molten 0.7% agar, poured onto the top of an LB agar plate and allowed to dry. Five microliters of phage-containing supernatants or dilutions thereof were spotted onto the bacterial lawns. The agar was touched with the pipette tip to indicate where drops were placed. The drops were allowed to dry before incubation overnight at 37°C.
Verifying O-antigen cleavage by BTP1 phage
To investigate O-antigen cleavage, phage-susceptible strains were grown to overnight, pelleted and then resuspended in the phage-containing supernatants containing chloramphenicol (100 μg ml−1) to prevent further bacterial growth and lysis from the phage. Samples were incubated with the phage for 1 hour at 37°C before bacteria were pelleted and LPS prepared as described.
Adsorption assay
BTP1 and P22 were diluted to a titer of 3.5 × 107 pfu ml−1. Bacterial cells (approximately 3 × 108 cfu ml−1) were resuspended in the diluted phage (multiplicity of infection of 1:10) and incubated at room temperature for 5 minutes. Bacteria were then pelleted and the supernatant was removed. Dilutions of the supernatant were used in the spot titer assay on a lawn of basal LT2 to determine number of phage remaining in the supernatant.
Acknowledgments
This work was supported by the Wellcome Trust grant [WT094333MA]. We would like to thank Dr. Robert Heyderman for providing the D23580 strain, Dr. Robert Kingsley for providing the other iNTS isolates and Dr. Abram Aertsen for the gift of the wild-type P22 phage lysate. We would also like to thank Tom Taynton for generating the pMV333 vector and Sarah Broadbent for creating strain sMV523.
Supporting information
Additional supporting information may be found in the online version of this article at the publisher's web-site.
Supporting information
References
- Allison GE, Verma NK. Serotype-converting bacteriophages and O-antigen modification in Shigella flexneri. Trends Microbiol. 2000;8:17–23. doi: 10.1016/s0966-842x(99)01646-7. [DOI] [PubMed] [Google Scholar]
- Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, et al. Current Protocols in Molecular Biology. New York, NY: Wiley Interscience; 1989. [Google Scholar]
- Baxa U, Steinbacher S, Miller S, Weintraub A, Huber R, Seckler R. Interactions of phage P22 tails with their cellular receptor, Salmonella O-antigen polysaccharide. Biophys J. 1996;71:2040–2048. doi: 10.1016/S0006-3495(96)79402-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bochner BR, Huang HC, Schieven GL, Ames BN. Positive selection for loss of tetracycline resistance. J Bacteriol. 1980;143:926–933. doi: 10.1128/jb.143.2.926-933.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogomolnaya LM, Santiviago CA, Yang HJ, Baumler AJ, Andrews-Polymenis HL. ‘Form variation’ of the O12 antigen is critical for persistence of Salmonella Typhimurium in the murine intestine. Mol Microbiol. 2008;70:1105–1119. doi: 10.1111/j.1365-2958.2008.06461.x. [DOI] [PubMed] [Google Scholar]
- Broadbent SE, Davies MR, van der Woude MW. Phase variation controls expression of Salmonella lipopolysaccharide modification genes by a DNA methylation-dependent mechanism. Mol Microbiol. 2010a;77:337–353. doi: 10.1111/j.1365-2958.2010.07203.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broadbent SE, van der Woude M, Aziz N. Accurate and simple sizing of primer extension products using a non-radioactive approach facilitates identification of transcription initiation sites. J Microbiol Methods. 2010b;81:256–258. doi: 10.1016/j.mimet.2010.03.021. [DOI] [PubMed] [Google Scholar]
- Cherepanov PP, Wackernagel W. Gene disruption in Escherichia coli: TcR and KmR cassettes with the option of Flp-catalyzed excision of the antibiotic-resistance determinant. Gene. 1995;158:9–14. doi: 10.1016/0378-1119(95)00193-a. [DOI] [PubMed] [Google Scholar]
- Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci USA. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies MR, Broadbent SE, Harris SR, Thomson NR, van der Woude MW. Horizontally acquired glycosyltransferase operons drive salmonellae lipopolysaccharide diversity. PLoS Genet. 2013;9:e1003568. doi: 10.1371/journal.pgen.1003568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukazawa Y, Hartman PE. A P22 bacteriophage mutant defective in antigen conversion. Virology. 1964;23:279–283. doi: 10.1016/0042-6822(64)90296-x. [DOI] [PubMed] [Google Scholar]
- Grimont PAD, Weill F-X. Institut Pasteur, Paris, France: WHO Collaborating Centre for Reference and Research on Salmonella. 9th edn. Paris: Institut Pasteur; 2007. Antigenic formulae of the Salmonella serovars. [Google Scholar]
- Haldimann A, Wanner BL. Conditional-replication, integration, excision, and retrieval plasmid-host systems for gene structure-function studies of bacteria. J Bacteriol. 2001;183:6384–6393. doi: 10.1128/JB.183.21.6384-6393.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauser E, Junker E, Helmuth R, Malorny B. Different mutations in the oafA gene lead to loss of O5-antigen expression in Salmonella enterica serovar Typhimurium. J Appl Microbiol. 2011;110:248–253. doi: 10.1111/j.1365-2672.2010.04877.x. [DOI] [PubMed] [Google Scholar]
- Holt KE, Parkhill J, Mazzoni CJ, Roumagnac P, Weill FX, Goodhead I, et al. High-throughput sequencing provides insights into genome variation and evolution in Salmonella Typhi. Nat Genet. 2008;40:987–993. doi: 10.1038/ng.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M, Ryu S. Spontaneous and transient defence against bacteriophage by phase-variable glucosylation of O-antigen in Salmonella enterica serovar Typhimurium. Mol Microbiol. 2012;86:411–425. doi: 10.1111/j.1365-2958.2012.08202.x. [DOI] [PubMed] [Google Scholar]
- Kingsley RA, Msefula CL, Thomson NR, Kariuki S, Holt KE, Gordon MA, et al. Epidemic multiple drug resistant Salmonella Typhimurium causing invasive disease in sub-Saharan Africa have a distinct genotype. Genome Res. 2009;19:2279–2287. doi: 10.1101/gr.091017.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroger C, Dillon SC, Cameron AD, Papenfort K, Sivasankaran SK, Hokamp K, et al. The transcriptional landscape and small RNAs of Salmonella enterica serovar Typhimurium. Proc Natl Acad Sci USA. 2012;109:E1277–E1286. doi: 10.1073/pnas.1201061109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroger C, Colgan A, Srikumar S, Handler K, Sivasankaran SK, Hammarlof DL, et al. An infection-relevant transcriptomic compendium for Salmonella enterica Serovar Typhimurium. Cell Host Microbe. 2013;14:683–695. doi: 10.1016/j.chom.2013.11.010. [DOI] [PubMed] [Google Scholar]
- MacLennan CA, Gilchrist JJ, Gordon MA, Cunningham AF, Cobbold M, Goodall M, et al. Dysregulated humoral immunity to nontyphoidal Salmonella in HIV-infected African adults. Science. 2010;328:508–512. doi: 10.1126/science.1180346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makela PH. Glucosylation of lipopolysaccharide in Salmonella: mutants negative for O antigen factor 1221. J Bacteriol. 1973;116:847–856. doi: 10.1128/jb.116.2.847-856.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Micoli F, Ravenscroft N, Cescutti P, Stefanetti G, Londero S, Rondini S, Maclennan CA. Structural analysis of O-polysaccharide chains extracted from different Salmonella Typhimurium strains. Carbohydr Res. 2014;385:1–8. doi: 10.1016/j.carres.2013.12.003. [DOI] [PubMed] [Google Scholar]
- Moller S, Croning MD, Apweiler R. Evaluation of methods for the prediction of membrane spanning regions. Bioinformatics. 2001;17:646–653. doi: 10.1093/bioinformatics/17.7.646. [DOI] [PubMed] [Google Scholar]
- Okoro CK, Kingsley RA, Connor TR, Harris SR, Parry CM, Al-Mashhadani MN, et al. Intracontinental spread of human invasive Salmonella Typhimurium pathovariants in sub-Saharan Africa. Nat Genet. 2012;44:1215–1221. doi: 10.1038/ng.2423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeves P. Evolution of Salmonella O antigen variation by interspecific gene transfer on a large scale. Trends Genet. 1993;9:17–22. doi: 10.1016/0168-9525(93)90067-R. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch E, Maniatis T. Molecular Cloning: A Laboratory Manual. Plain View: Cold Spring Harbor Laboratory Press; 1987. [Google Scholar]
- Sharma CM, Hoffmann S, Darfeuille F, Reignier J, Findeiss S, Sittka A, et al. The primary transcriptome of the major human pathogen Helicobacter pylori. Nature. 2010;464:250–255. doi: 10.1038/nature08756. [DOI] [PubMed] [Google Scholar]
- Slauch JM, Lee AA, Mahan MJ, Mekalanos JJ. Molecular characterization of the oafA locus responsible for acetylation of Salmonella Typhimurium O-antigen: oafA is a member of a family of integral membrane trans-acylases. J Bacteriol. 1996;178:5904–5909. doi: 10.1128/jb.178.20.5904-5909.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinbacher S, Baxa U, Miller S, Weintraub A, Seckler R, Huber R. Crystal structure of phage P22 tailspike protein complexed with Salmonella sp. O-antigen receptors. Proc Natl Acad Sci USA. 1996;93:10584–10588. doi: 10.1073/pnas.93.20.10584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinbacher S, Miller S, Baxa U, Budisa N, Weintraub A, Seckler R, Huber R. Phage P22 tailspike protein: crystal structure of the head-binding domain at 2.3 A, fully refined structure of the endorhamnosidase at 1.56 A resolution, and the molecular basis of O-antigen recognition and cleavage. J Mol Biol. 1997;267:865–880. doi: 10.1006/jmbi.1997.0922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thanweer F, Verma NK. Identification of critical residues of the serotype modifying O-acetyltransferase of Shigella flexneri. BMC Biochem. 2012;13:13. doi: 10.1186/1471-2091-13-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thanweer F, Tahiliani V, Korres H, Verma NK. Topology and identification of critical residues of the O-acetyltransferase of serotype-converting bacteriophage, SF6, of Shigella flexneri. Biochem Biophys Res Commun. 2008;375:581–585. doi: 10.1016/j.bbrc.2008.08.069. [DOI] [PubMed] [Google Scholar]
- Van der Byl C, Kropinski AM. Sequence of the genome of Salmonella bacteriophage P22. J Bacteriol. 2000;182:6472–6481. doi: 10.1128/jb.182.22.6472-6481.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vernikos GS, Parkhill J. Interpolated variable order motifs for identification of horizontally acquired DNA: revisiting the Salmonella pathogenicity islands. Bioinformatics. 2006;22:2196–2203. doi: 10.1093/bioinformatics/btl369. [DOI] [PubMed] [Google Scholar]
- Warren JW, Walker JR, Roth JR, Altman E. Construction and characterization of a highly regulable expression vector, pLAC11, and its multipurpose derivatives, pLAC22 and pLAC33. Plasmid. 2000;44:138–151. doi: 10.1006/plas.2000.1477. [DOI] [PubMed] [Google Scholar]
- West NP, Sansonetti P, Mounier J, Exley RM, Parsot C, Guadagnini S, et al. Optimization of virulence functions through glucosylation of Shigella LPS. Science. 2005;307:1313–1317. doi: 10.1126/science.1108472. [DOI] [PubMed] [Google Scholar]
- Wollin R, Stocker BA, Lindberg AA. Lysogenic conversion of Salmonella typhimurium bacteriophages A3 and A4 consists of O-acetylation of rhamnose of the repeating unit of the O-antigenic polysaccharide chain. J Bacteriol. 1987;169:1003–1009. doi: 10.1128/jb.169.3.1003-1009.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting information


