Abstract
Polycomb-group (PcG) genes encode chromatin proteins involved in stable and heritable transcriptional silencing. PcG proteins participate in distinct multimeric complexes that deposit, or bind to, specific histone modifications (e.g., H3K27me3 and H2AK119ub1) to prevent gene activation and maintain repressed chromatin domains. PcG proteins are evolutionary conserved and play a role in processes ranging from vernalization and seed development in plants, over X-chromosome inactivation in mammals, to the maintenance of stem cell identity. PcG silencing is medically relevant as it is often observed in human disorders, including cancer, and tissue regeneration, which involve the reprogramming of PcG-controlled target genes.
Polycomb-group proteins are involved in transcriptional silencing and processes such as X-chromosome inactivation. They participate in multimeric complexes that deposit or bind to specific histone modifications (e.g., H3K27me3).
Overview
Organs of humans, animals, and plants are constructed from a large pool of distinct cell types, each performing a specialized physiological or structural function. With very few exceptions, all cell types contain the same genetic information encoded in their DNA. Thus, the distinctiveness of a given cell type is achieved through specific gene expression programs. As a consequence, cell lineages need to have these programs maintained during growth and cell division. This implies the existence of a memory system that ensures the faithful transmission of information (i.e., which gene is active or repressed) from mother to daughter cells. The existence of such a system is illustrated by the fact that cultured tissues of plants and animals usually maintain their differentiated characters even if grown in a foreign environment. By way of example, ivy plants regenerated after tissue culture produce the type of leaf corresponding to the phase of development from which the original tissue was taken (i.e., juvenile or adult leaf).
The major question to be addressed here and in Kingston and Tamkun (2014) concerns the molecular identity of factors contributing to the mechanism(s) of “cellular” or “transcriptional memory,” which maintains a determined state over many cell divisions. Genetic analyses in Drosophila melanogaster have identified regulators crucial in maintaining the morphology of individual body segments that are determined by the action of the HOX genes. In Drosophila males, the first thoracic segment has legs with sex combs. Legs on the second and third thoracic segment lack these structures. In the 1940s, Drosophila mutants were identified (Polycomb and extra sex combs) wherein males had sex combs on all legs. These morphological alterations reflect homeotic transformations of the second and third leg identities into the first leg identity. Subsequent molecular studies showed that these mutations did not affect the products of the HOX genes themselves, but rather the way HOX gene activity was spatially controlled. Throughout the years, a large number of similar regulatory genes were identified, and were classified into two antagonistic groups: the Polycomb (PcG) and Trithorax (TrxG) group. Whereas the PcG proteins are required to maintain the silenced state of developmental regulators such as the HOX genes, the TrxG proteins are generally involved in maintaining the active state of gene expression. Thus PcG and TrxG proteins embody the molecular components of cellular memory.
Proteins of both groups form large multimeric protein complexes that act on their target genes by modulating chromatin structure. In this article, we will focus on the molecular nature and function of two of the major Polycomb repressive complexes, PRC1 and PRC2. The molecular nature of the TrxG complexes will be described in Kingston and Tamkun (2014). In Drosophila, it was shown that transcription factors recruit PcG complexes to a DNA sequence called a PcG response element (PRE). Once recruited, they establish a silent chromatin state that can be inherited over many cell divisions. Members of PRC2 are highly conserved between plants and animals, whereas PRC1 proteins are less well conserved. This implies conservation, but also diversity, in the basic building blocks of the cellular memory system. In addition to the function of PcG complexes in the maintenance of cell types, they may also play important roles in stem cell plasticity and regeneration. Also, their deregulation can lead to neoplastic transformation and cancer. Thus, PcG proteins play a crucial role in many fundamental processes of normal development and disease in multicellular eukaryotes.
1. Introduction
All multicellular organisms start from a single cell, the zygote, which during development gives rise to a multitude of distinct cell types with specialized functions. This poses the problem of how, once determined, cell types can be maintained over many cell divisions occurring during growth phases.
1.1. The Concept of Cellular Memory
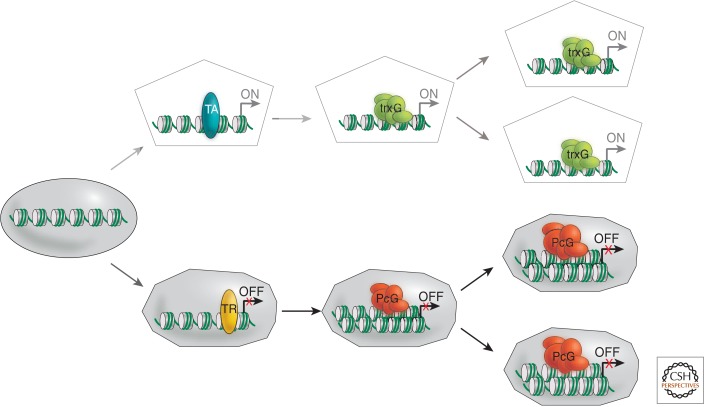
An adult animal has 200 to 300 structurally distinct cell types, whereas a plant has between 30 and 40. The identity and function of a given cell type is determined by its characteristic gene expression profile. During development and adult homeostasis, it is crucial to remember and faithfully reproduce this state after each cell division. This is particularly critical during the replication of genetic material (S-phase) and the separation of chromosomes during mitosis (M-phase; see Almouzni and Cedar 2014). These are recurring events at each cell cycle that interrupt gene expression processes. Thus, how can differential gene expression patterns be inherited from one cell generation to the next, as illustrated in Figure 1?
Figure 1.
The concept of cellular memory. Schematic illustration of the involvement of PcG and TrxG complexes in the determination of active and repressed states of gene expression and, thereby, cellular differentiation, which is maintained over many cell divisions. TA, transcriptional activator; TR, transcriptional repressor.
We know from experiments performed in the 1960s and 1970s that plant and animal tissues remember a determined state even after prolonged passage in culture (Hadorn 1968; Hackett et al. 1987). Hadorn and colleagues showed that imaginal disc cells found in Drosophila larvae have an intrinsic memory, allowing them to remember determined states that are fixed in early embryogenesis. Imaginal discs are clusters of epithelial cells set aside in the developing embryo as precursors for the formation of specific external structures and appendages during metamorphosis. For instance, of the two pairs of imaginal discs in the second thoracic segment, one forms a midleg and the other a wing (see Fig. 2 in Kingston and Tamkun 2014). Imaginal discs can be cultured by transplantation into the haemocoel of adult females, wherein they continue to proliferate, but do not differentiate. When transplanted back into a larva before metamorphosis, the disc will subsequently differentiate into the expected adult structures, even after successive passages in adult females. More recently, the PcG and TrxG proteins were shown to be required for the maintenance of the determined state of imaginal disc cells. Additionally, it was observed that in rare cases an imaginal disc could change its fate, a process called transdetermination. This process involves the down-regulation of PcG repression by the JNK signaling cascade in transdetermined cells (Lee et al. 2005). PcG mutants also have elevated frequencies of transdetermination, supporting a role for PcG proteins in maintaining imaginal disc cell fates (Katsuyama and Paro 2011). Thus, PcG proteins seem to play a crucial role in the maintenance and reprogramming of cellular fates during both normal development and regeneration.
1.2. The Genetic Identification of the Polycomb Group
In all metazoans, the anterior–posterior axis is specified through defined expression patterns of HOX genes (see Fig. 2 in Kingston and Tamkun 2014). During Drosophila embryogenesis, the activity of maternally (i.e., inherited through the oocyte) and zygotically produced transcription factors generates a specific combination of HOX expression patterns that define the morphology of each body segment. This segment-specific profile of HOX gene expression is maintained throughout the development of the fly, long after the early transcriptional regulators have disappeared. When the function of HOX genes was genetically characterized, many trans-acting regulators were isolated. Among the first, Polycomb (Pc) was identified and genetically analyzed by Pam and Ed Lewis (Lewis 1978). Heterozygous Pc mutant males have additional sex combs on the second and third legs. Homozygous mutants are embryonic lethal, showing a transformation of all cuticular segments toward the most posterior abdominal segment (Fig. 2C,D). These classical PcG phenotypes are caused by the ectopic expression of HOX genes. Thus, Pc and other genes with similar phenotypes were defined as repressors of HOX gene activity. Detailed analyses subsequently uncovered the fact that the PcG proteins are only required for the maintenance of HOX repression, rather than the position-specific establishment of HOX activity. This latter task is performed by the transcription factors encoded by the early acting segmentation genes. Based on their repressing or activating influence on HOX expression, these newly identified trans-acting regulators were divided into two antagonistic classes, the PcG and TrxG, respectively (Kennison 1995).
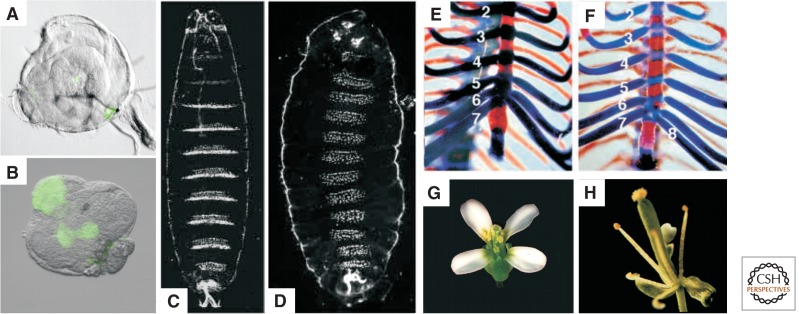
Figure 2.
Homeotic transformations in PcG mutants of various species. (A–D) Drosophila melanogaster, (E,F) Mus musculus, (G,H) Arabidopsis thaliana. (A,B) Leg imaginal discs undergoing a transdetermination event as indicated by the expression of the wing-specific gene vestigial (marked by green fluorescent protein [GFP]). (C,D) Cuticles of a wild-type (C) and a Su(z)12 mutant embryo (D). In the Su(z)12 mutant embryo, all abdominal, thoracic, and several head segments (not all visible in this focal plane) are homeotically transformed into copies of the eighth abdominal segment because of misexpression of the Abd-B gene in every segment. (E,F) Axial skeleton of newborn wild-type (E) and Ring1A-/- mice (F). Views of the thoracic regions of cleared skeletons show bone (red) and cartilage (blue). The mutant displays anterior transformation of the eighth thoracic vertebra as indicated by the presence of an eighth (1–8) vertebrosternal rib, instead of seven (1–7) as in the wild type. (G,H) Wild-type (G) and clf-2 mutant (H) flowers. The wild-type flower shows the normal arrangement of sepals, petals, stamens, and carpels. In the clf-2 flower, petals are absent or reduced in number. (A,B, Courtesy of N. Lee and R. Paro; C,D, reprinted, with permission, from Birve et al. 2001, © Company of Biologists Ltd; E,F, reprinted, with permission, from Lorente et al. 2000, © Company of Biologists Ltd; G,H, courtesy of J. Goodrich.)
The molecular isolation of Drosophila PcG genes made it possible to study the function of vertebrate orthologs in mice, which were subsequently also shown to be key regulators of HOX gene expression (van der Lugt et al. 1994; Core et al. 1997). In mammals, mutations in PcG genes typically lead to homeotic transformations of vertebrae (Fig. 2E,F). In addition, PcG genes play a crucial role in the control of cell proliferation, stem cell maintenance, and cancer (see Secs. 4.2 and 4.3).
In two other model organisms, namely, the worm Caenorhabditis elegans and the flowering plant Arabidopsis thaliana, the molecular characterization of mutants isolated in various genetic screens revealed the existence of other PcG protein orthologs in their genomes. In C. elegans, PcG members were identified in screens for maternal-effect sterile (mes) mutants and were shown to be involved in X-chromosome silencing in the hermaphrodite germline (Strome et al. 2014).
In Arabidopsis, PcG genes were identified in several genetic screens investigating distinct developmental processes (Hsieh et al. 2003). The first PcG gene in plants, CURLY LEAF (CLF), was identified as a mutant with homeotic transformations of floral organs (Goodrich et al. 1997). Mutations in the FERTILIZATION-INDEPENDENT SEED (FIS) class of genes were found in screens for mutants showing maternal-effect seed abortion (Grossniklaus et al. 1998), or allowing aspects of seed development to occur in the absence of fertilization (Luo et al. 1999; Ohad et al. 1999). Finally, PcG genes were identified in screens for flowering time mutants, for example, mutants that flower directly after germination (Yoshida et al. 2001) or that disrupt the vernalization response (Gendall et al. 2001)—the process rendering plants competent to flower after prolonged exposure to cold (discussed in detail in Baulcombe and Dean 2014).
The variety of processes regulated by PcG proteins illustrates the importance of maintaining the repressed state of key developmental regulators in different organisms. On the one hand, there is an amazing conservation of some biological functions from plants to mammals (e.g., the regulation of key developmental regulators such as homeotic genes or the involvement in the tight regulation of cell proliferation). On the other hand, PcG complexes appear to be versatile and dynamic molecular modules that have been used to control a large and diverse variety of developmental and cellular processes.
2. Establishing silencing marks on chromatin
PcG proteins are grouped into two major classes based on biochemical characterizations: members of the Polycomb repressive complex 1 or 2 (PRC1 and PRC2; Table 1). The two complexes are required for different steps in the repression of gene expression. PRC2 has histone modifying activity, namely, methylating H3K27 at genes targeted for silencing. PRC1 components can recognize and bind to this modification and induce appropriate structural changes in chromatin. Additionally, PRC1 is able to monoubiquitylate H2AK118/119 at target loci. Both complexes are widely conserved across metazoa (Whitcomb et al. 2007) and the plant kingdom (Köhler and Hennig 2010).
Table 1.
Core PcG proteins in model systems
| Drosophila melanogaster | Mus musculus | Arabidopsis thaliana | Caenorhabditis elegans | ||
|---|---|---|---|---|---|
| PcG DNA binding proteins | |||||
| PHO | Pleiohomeotic | Zinc finger | YY1 | ||
| PHOL | Pleiohomeotic-like | Zinc finger | |||
| PSQ | Pipsqueak | BTB-POZ domain | |||
| DSP1 | Dorsal switch protein 1 | HMG domain protein | HMGB2 | ||
| PRC2 core proteins | |||||
| ESC | Extra sex combs | WD 40 repeats | EED | FIE | MES-6 |
| E(Z) | Enhancer of zeste | SET domain | EZH1/ENX2 EZH2/ENX1 |
CLF MEA SWN |
MES-2 |
| SU(Z)12 | Suppressor of zeste 12 | Zinc finger VEFS box |
SU(Z)12 | FIS2 VRN2 EMF2 |
|
| p55 | p55 | Histone-binding domain | RBAP48 RBAP46 |
MSI1 (MSI2/3/4/5) |
|
| PRC1 core proteins | |||||
| PC | Polycomb | Chromodomain | CBX2/M33 CBX4/MPC2 CBX6 CBX7 CBX8/MPC3 |
||
| PH | Polyhomeotic | Zinc finger SAM/SPM domain |
EDR1/MPH1/RAE28 EDR2/MPH2 (EDR3) |
SOP-2 | |
| PSC | Posterior sex combs | Zinc finger HTH domain |
BMI1 MEL18/RNF110/ZFP144 |
AtBMI1A AtBMI1B AtBMI1C |
MIG-32 |
| SCE/dRING | Sex combs extra/dRing | RING zinc finger | RING1/RING1A RNF2/RING1B |
AtRING1A AtRING1B |
SPAT-3 |
2.1. Components and Evolutionary Conservation of PRC2
Several variants of PRC2 have been purified from Drosophila embryos, but all of these complexes contain four core proteins: the SET domain histone lysine methyltransferase Enhancer of zeste (E(Z)), the WD40 protein ESC, the histone binding protein p55, and Suppressor of zeste 12 (SU(Z)12; Table 1 and Fig. 3).
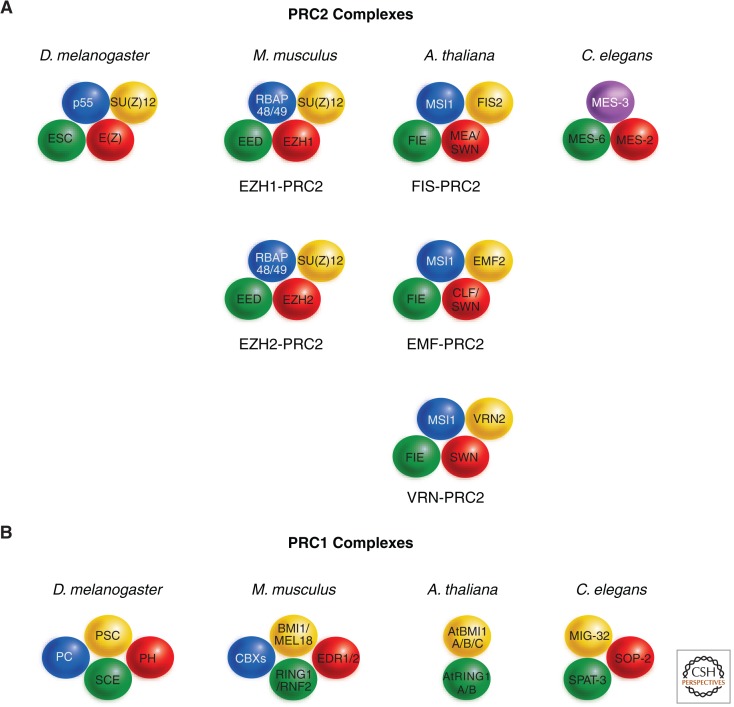
Figure 3.
Conserved PRC2 core complexes. The conserved core proteins of PRC2 (A) and PRC1 (B) complexes in Drosophila melanogaster, Mus musculus, Arabidopsis thaliana, and Caenorhabditis elegans are shown. (A) In the mouse, PRC2 variants containing EZH1 or EZH2 have distinct functions, whereas in Arabidopsis the ancestral complex has diversified into at least three variants with discrete functions during development. In C. elegans, the PRC2 core complex contains only three proteins, with MES-3 not having homology with any other identified PRC2 protein. Apart from these core proteins, several other proteins, which are not shown here, interact with PRC2. For instance, mammalian complexes can contain the histone lysine demethylase JARID2, the Zn-finger protein AEBP2, and various homologs of the Drosophila PCL protein (PCL1/2/3). Proteins that share the plant homeodomain (PHD)-domain with PCL, but are otherwise not closely related, are also associated with the VRN-PRC2 complex in Arabidopsis. Homologous proteins are indicated by the same color. (B) The core proteins of PRC1 are less conserved than those of PRC2 across the four species. In mammals, all genes encoding the PRC1 core subunits have been expanded (see Table 1), such that a variety of complexes with different isoform composition can be formed. In addition to the core components, several additional proteins can be found in PRC1 that are, however, less well characterized and are not shown. In plants, only homologs of Drosophila PSC and SCE have been identified; these are encoded by small gene families. Homologous proteins are indicated by the same color. (Based on Reyes and Grossniklaus 2003, Chanvivattana et al. 2004, and Margueron and Reinberg 2011.)
The E(z) gene encodes a 760-amino acid protein, containing a SET domain that exerts KMT (lysine methyltransferase) activity to histones. The SET domain is preceded by a CXC or Pre-SET domain (Tschiersch et al. 1994), which contains nine conserved cysteines that bind three zinc ions and is thought to stabilize the SET domain (see Fig. 1 in Cheng 2014). Such a structural role is supported by the fact that several temperature-sensitive E(z) alleles affect one of the conserved cysteines (Carrington and Jones 1996). In addition, E(Z) contains SANT domains implicated in histone binding, and a C5 domain required for the physical interaction with SU(Z)12. ESC is a short protein of 425 amino acids that contains five WD40 repeats, shown to form a β propeller structure. This serves as a platform for protein–protein interactions, hence, giving ESC a central role in PRC2 to physically interact with both E(z) and p55 in all model systems analyzed. The SU(Z)12 protein is 900 amino acids long and characterized by a C2H2-type zinc finger and a carboxy-terminal VEFS domain. The VEFS domain was identified as a conserved region between SU(Z)12 and its three homologs in plants: VERNALIZATION2 (VRN2), EMBRYONIC FLOWER2 (EMF2), and FIS2 (see Fig. 3). Several mutant Su(z)12 alleles alter this domain, showing that it is required for the interaction with the C5 domain of E(Z) (Chanvivattana et al. 2004; Yamamoto et al. 2004).
The p55 protein was not identified as a PcG member by genetic approaches, possibly because it takes part in a multitude of other protein complexes associated with chromatin (Hennig et al. 2005). The p55 protein was, however, identified biochemically as part of PRC2. It is 430 amino acids long and contains six WD40 repeats, which physically interact with ESC or its orthologs in mammals and plants (Tie et al. 2001; Köhler et al. 2003a).
In addition to the core PRC2 proteins, some variants of the complex contain the RPD3 histone deacetylase (HDAC), or the Polycomb-like (PCL) protein. The interaction with RPD3 is noteworthy as histone deacetylation is correlated with a repressed state of gene expression (see Seto and Yoshida 2014). The different compositions of PRC2 variants likely reflect both the dynamic changes that occur during development or cater for tissue-specific regulation. PRC2 is highly conserved in invertebrates, vertebrates, and plants (Fig. 3). In C. elegans, only homologs of E(Z) and ESC are present: MES-2 and MES-6. Together with a nonconserved protein, MES-3, they form a small complex of ∼230 kDa required to repress the X-chromosome and somatically active genes in the hermaphrodite germline (see Strome et al. 2014). In mammals and plants, all four core proteins of PRC2 are present. As in Drosophila, the mammalian complex is ∼600 kDa and is not only involved in regulating homeotic gene expression, but also in the control of cell proliferation, X-chromosome inactivation, and imprinted gene expression (for more detail, see Sec. 4; Barlowe and Bartolomei 2014; Brockdorff and Turner 2014).
In plants, several genes encoding PRC2 components have undergone duplications such that now they are present as small gene families. In Arabidopsis there is only one homolog of ESC, FERTILIZATION-INDEPENDENT ENDOSPERM (FIE), but three homologs of E(Z), three homologs of SU(Z)12, and five homologs of p55 (referred to as MSI1-5; Table 1). Varying combinations of these proteins form at least three distinct complexes that control specific developmental processes, namely, the FIS-PRC2, EMF-PRC2, and VRN-PRC2 complexes (Fig. 3).
The first of these complexes that was studied in detail is formed by members encoded by the FIS class genes, which play a crucial role in the control of cell proliferation in the seed (Grossniklaus et al. 2001). The FIS-PRC2 contains MEDEA (MEA), FIE, FIS2, and MSI1. Using chromatin immunoprecipitation (ChIP) against one of its components, the FIS-PRC2 was found to directly regulate MEA itself, as well as PHERES1 (PHE1) and FUSCA3 (FUS3), coding for transcription factors of the MADS-domain and B3-domain class, respectively (Köhler et al. 2003b; Baroux et al. 2006; Makarevich et al. 2006). Interestingly, the paternal allele of PHE1 is expressed at much higher levels than the maternal allele. This regulation of gene expression by genomic imprinting is under the control of the FIS-PRC2, which specifically represses the maternal allele (Köhler et al. 2005). Thus, as will be outlined in Section 4.1, the FIS-PRC2 shares with its mammalian counterpart functions in regulating cell proliferation as well as imprinted gene expression.
The EMF complex contains CLF and EMF2 (Chanvivattana et al. 2004). Mutations in either of them show weak homeotic transformations and an early flowering phenotype. The EMF-PRC2 is required to repress homeotic genes, whose combinatorial action determines the identity of floral organs (Goodrich et al. 1997). Thus, the EMF-PRC2 has a similar function in maintaining the repressed state of homeotic genes as PRC2 in Drosophila and vertebrates (Fig. 2). However, homeotic genes in plants do not encode homeodomain proteins, but rather transcription factors belonging to families containing the MADS-domain or the plant-specific AP2-domain. Strong emf2 mutants, however, have more severe phenotypes, and produce flowers directly after germination, bypassing the vegetative phase of development (Yoshida et al. 2001). Thus, the EMF-PRC2 plays a role both early in development, in which it prevents immediate flowering, and later during floral organogenesis (Chanvivattana et al. 2004). The EMF-PRC2 directly represses FLOWERING LOCUS T (FT) and SHOOTMERISTEMLESS (STM), which are both required for the transition to flowering, and the floral homeotic MADS-box gene AGAMOUS (AG), which together with the homeobox gene STM regulates the development floral organs (Fig. 4) (Schubert et al. 2006; Jiang et al. 2008). The FIS class proteins FIE and MSI1 have also been implicated in the control of homeotic gene expression (Figs. 3 and 4). Because mutations in both cause maternal-effect embryo lethality, this function was only revealed when partial loss-of-function alleles could be studied at later stages of development (Kinoshita et al. 2001; Hennig et al. 2003).
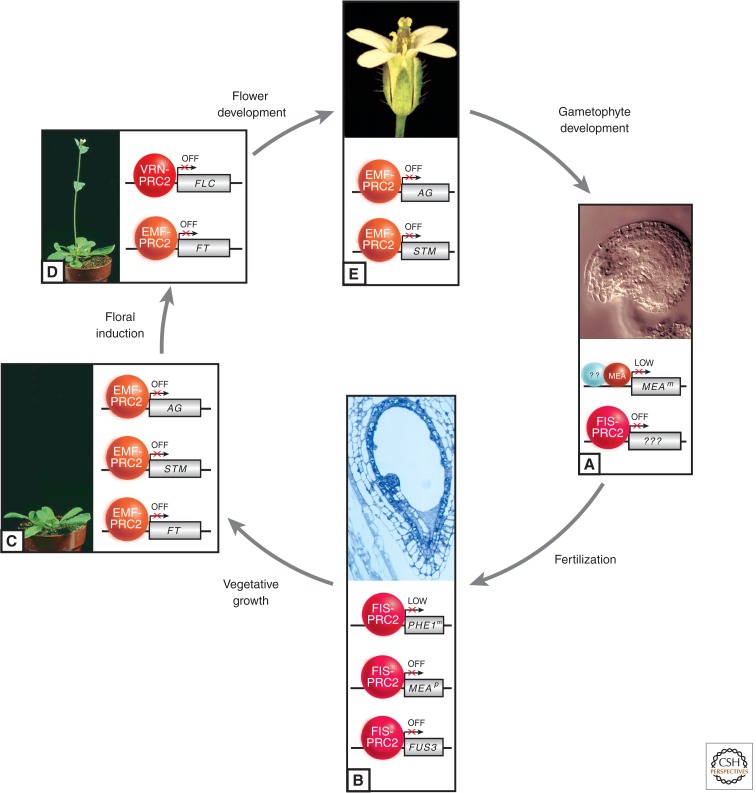
Figure 4.
Involvement of distinct PRC2 complexes at various stages of plant development. During the plant life cycle, distinct variants of PRC2 (see Fig. 3) control developmental progression. (A) A cleared wild-type ovule harboring the female gametophyte in its center is represented. The FIS-PRC2 represses unknown target genes that control proliferation of the central cell; consequently, in all fis class mutants, this cell proliferates in the absence of fertilization. Around fertilization, MEA is also required to maintain expression of the maternal MEA allele (MEAm) at a low level, but this activity is independent of other FIS-PRC2 components. (B) Section of a wild-type seed harboring embryo and endosperm, enclosed by the seed coat. After fertilization, the FIS-PRC2 is involved in the control of cell proliferation in embryo and endosperm. It maintains a low level of expression of the maternal PHE1m allele and is involved in keeping the paternal MEAp allele silent, although FIS-PRC2 only plays a minor part in its repression. Both parental alleles of FUS3 are repressed by the FIS-PRC2. (C) Wild-type plant before flowering. The EMF-PRC2 prevents flowering by repressing FT and directly represses the floral genes AG and STM. (D) Wild-type plant after bolting—that is, floral induction induced by appropriate photoperiod and/or vernalization. The former relieves repression by EMF-PRC2 of FT, a promoter of flowering, whereas the latter leads to repression of the floral repressor FLC, thus inducing flowering. The maintenance of FLC repression depends on the VRN-PRC2. (E) Wild-type Arabidopsis flower. During flower organogenesis, the EMF complex regulates floral homeotic genes, such as AG, which determine the identity of floral organs, and STM, which is involved in floral organ development. (A, Courtesy of J.M. Moore and U. Grossniklaus; B, courtesy of J.-P. Vielle-Calzada and U. Grossniklaus; C,D, courtesy of D. Weigel; E, reprinted, with permission, from Page and Grossniklaus 2002, © Macmillan.)
Finally, the VRN-PRC plays a key role in a well-known process called vernalization. This epigenetic regulation governs the timing of flowering in winter annuals, induced by extended periods of exposure to low temperatures, but the effect is only seen after many cell divisions (Fig. 4D; see Fig. 1 from Baulcombe and Dean 2014 for detail). A plant cell will remember that it was vernalized for many months, or even years, after the cold period. This cellular memory is even maintained through passages in cell culture, but not from one generation to the next (Sung and Amasino 2004). The VRN genes mediate the response to vernalization. VRN2 encodes a SU(Z)12 homolog (Gendall et al. 2001), which interacts with the plant E(Z) homologs CLF and SWINGER (SWN) in yeast two-hybrid assays (Chanvivattana et al. 2004). Floral induction is not only controlled by vernalization, but involves the perception of endogenous (developmental stage and age) as well as exogenous factors (day length, light conditions, temperature). Four pathways, two of which involve PcG factors, have been defined by genetic analyses (see Fig. 1B of Baulcombe and Dean 2014): (1) the autonomous pathway, which constitutively represses flowering presumably via PcG-mediated H3K27 methylation; (2) the vernalization pathway, which induces flowering in response to a prolonged exposure to cold temperature; (3) the photoperiod pathway, which accelerates flowering under long days; and (4) the gibberellin pathway, a phytohormone that promotes flowering. The flowering time gene FLOWERING LOCUS C (FLC), encoding a MADS-domain transcription factor, is a key integrator of the flowering response as a repressor of flowering. Although the initial repression of FLC during vernalization is independent of the VRN-PRC2, the maintenance of FLC repression requires VRN2 activity. Both the vernalization and the autonomous pathways reduce FLC expression, integrating diverse signals (Gendall et al. 2001; De Lucia et al. 2008; Jiang et al. 2008). The VRN-PRC2 contains the core subunits VRN2, SWN, FIE, and MSI1 (Fig. 3) and three associated PHD finger proteins (Wood et al., 2006; De Lucia et al. 2008). Interestingly, VRN2 interacts with the FLC locus independently of cold, and FLC silencing is triggered by the association of the VRN2-PRC2 with VRN5, a PHD finger protein that is cold-induced and shares limited similarity to PCL (see Sec. 2.2 and Fig. 2 from Baulcombe and Dean 2014). In summary, the regulation of flowering time involves both the VRN-PRC2 and EMF-PRC2, which regulate FLC and FT, respectively (Fig. 4).
2.2. The Chromatin-Modifying Activity of PRC2
How does PRC2 mediate its repressive effect? In Drosophila, mammals, and plants, the hallmark histone modification H3K27me3 is produced by PRC2 (Cao et al. 2002; Czermin et al. 2002). This modification is generally thought to be crucial for PcG silencing because its distribution matches the binding sites of PcG components determined by genome-wide ChIP studies (Schuettengruber et al. 2009; Kharchenko et al. 2010). The PRC2 core complex contains E(Z), which catalytically adds up to three methyl groups at the target lysine residue K27 of H3 through its SET-domain (Fig. 5A). However, E(Z) alone seems to be inactive and needs to associate with the other PRC2 subunits, ESC and SU(Z)12, to provide the necessary catalytic activity (Cao and Zhang 2004; Pasini et al. 2004; Nekrasov et al. 2005). Although the mechanistic basis of this enhancement remains elusive, the process is conserved in mammals. Several more proteins and subcomplexes related to PRC2 exist. For example, the ESC-like gene encodes a protein similar to ESC and is able to fully replace it in its absence (Wang et al. 2006; Kurzhals et al. 2008). Also, a PRC2 variant additionally containing PCL has been found to specifically enhance the last addition of a methyl group to generate H3K27me3 (Nekrasov et al. 2007). Without PCL, the bulk H3K27me3 is reduced in embryonic and larval tissues, leading to the derepression of several target genes. Interestingly, a similar function and complex has also been described for the PHD finger protein PHF1 in mammals (Cao et al. 2008; Sarma et al. 2008).
Figure 5.
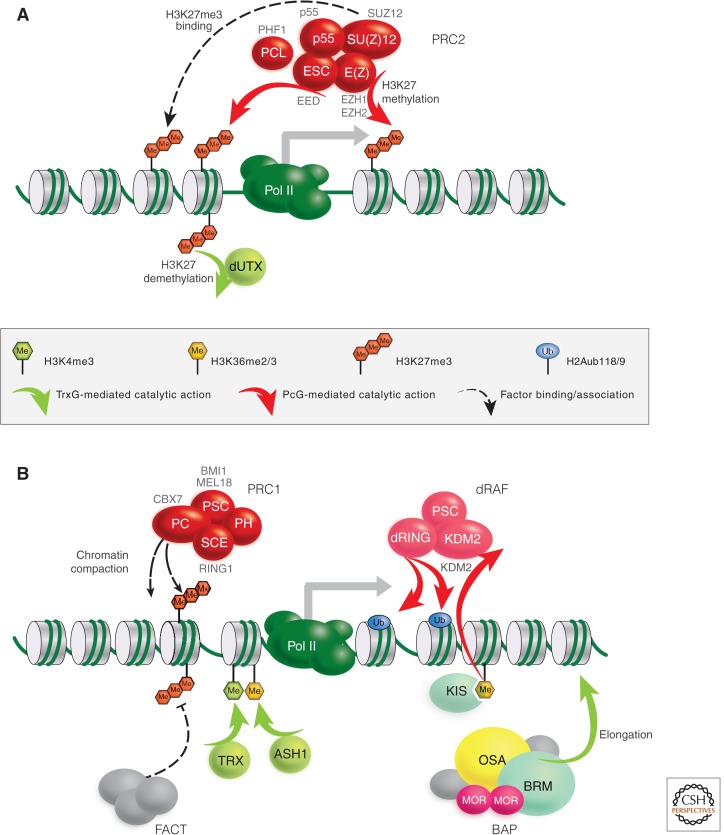
Schematic representation of the core PcG and TrxG protein complexes and their functions at promoters. Drosophila PcG proteins are depicted as red ovals with selected mammalian orthologs indicated in gray text. (A) Components and function of the PRC2 and counteracting activities of TrxG proteins (light green). (B) Components and functions of PRC1 and dRING-associated factor (dRAF) and the counteracting activities of the BAP SWI/SNF, facilitates chromatin transcription (FACT) remodeling complexes, and SET-domain histone KMTs TRX and ASH1. The TrxG protein Kismet-L is a member of the chromatin-helicase-DNA-binding (CHD) subfamily of chromatin-remodeling factors, stimulating elongation of Pol II. (Adapted from Enderle 2011.)
The mammalian PRC2 has also been found to counteract H3K4 methylation, an active histone modification, by recruiting the H3K4me3 demethylase RETINOBLASTOMA BINDING PROTEIN 2 (RBP2) to target genes (Pasini et al. 2008) and control transcriptional elongation via JARID 2 (Landeira et al. 2010). The biological implications of these variations and additional activities, however, are not yet fully understood. Moreover, even the molecular function of H3K27me3 is still a matter of debate. There is no evidence yet that H3K27 methylation may directly alter nucleosomal structure to repress target genes. Rather, it seems to provide a binding platform for other PcG proteins; the PRC1 complex binds, albeit weakly, to H3K27me3 through the chromodomain of its PC subunit, and also mammalian PRC2 itself has been shown to bind H3K27me3 via the Embryonic ectoderm development protein (EED; Fischle et al. 2003; Margueron et al. 2009). Interestingly, this interaction seems to trigger the lysine methyltransferase activity of E(Z), providing a self-reinforcing positive-feedback loop (as illustrated in Fig. 13 of Allis et al. 2014), potentially contributing to the heredity of the PcG/TrxG system (Margueron et al. 2009). The counteracting force of the TrxG has also been described in mammals: UTX/KDM6A, the mammalian ortholog of Drosophila dUTX showing some genetic characteristics of a TrxG member (Smith et al. 2008), is able to demethlyate H3K27me3 in vivo and in vitro (Agger et al. 2007; Lee et al. 2007). This reveals a direct antagonistic relationship between PcG and TrxG proteins in the modification of their target chromatin (Fig. 5A).
2.3. The Dynamic Function of PRC2 during Development
As pointed out in Sections 2.1 and 2.4, the PRC1 and PRC2 core complexes are associated with distinct factors that may play a role in recruiting PcG complexes to tissue-specific target loci or in modulating target gene activity. PcG complexes may even differ between target genes in the same cell, suggesting a highly dynamic behavior at different developmental stages. Studies performed in mammals and plants clearly show that PcG complexes have distinct memberships in specific tissues and their composition changes during cellular differentiation. Similar to the situation in plants described in Section 2.1, some of the genes encoding PRC2 subunits have been duplicated in mammals. For instance, PRC2 complexes containing either EZH1 or EZH2 are functionally distinct (Fig. 3). EZH1-containing PRC2 has weak KMT activity and is abundant in nondividing cells of adult organs, whereas EZH2 confers high KMT activity and is expressed strongly in proliferating cells (Margueron et al. 2008). Furthermore, different isoforms of EED, which are derived from the same messenger RNA but different translational start sites, are able to methylate H3K27 and H1K26 (Kuzmichev et al. 2005).
In Drosophila, PcG proteins maintain repressed states of homeotic genes, established during early embryogenesis, thereby fixing developmental decisions. Once the silent state of a PcG target has been fixed, it will often remain in that state for the remainder of an individual’s lifespan. In plants, a similar situation may occur with the VRN-PRC2; once vernalized, the target gene(s) will be permanently inactivated and only reset in the next generation (see Baulcombe and Dean 2014 for more detail). Other plant PRC2 variants, however, seem to respond quickly to developmental or environmental stimuli. For instance, one function of the FIS-PRC2 is to repress cell proliferation in the absence of fertilization. Upon fertilization, however, cell proliferation is rapidly induced, presumably through the derepression of PcG target genes. This indicates that PcG repression is the default state, which has to be overcome by some unknown mechanism to allow developmental progression to occur. Indeed, the major function of the various plant PRC2 variants seems to be the regulation of developmental transitions, such as those occurring at fertilization, during seed development, and after germination when the plant progresses from the juvenile to the adult, and finally the reproductive stage (reviewed in Holec and Berger 2012).
2.4. Components of the PRC1 Complex
The PRC1 core complex purified from Drosophila embryos contains stoichiometric amounts of PC, Polyhomeotic (PH), Posterior sex combs (PSC), and Sex combs extra (SCE/dRing 1) (Shao et al. 1999). Its mammalian counterpart comprises the same core components, but the genes encoding them have been amplified (Table 1). As mentioned before, the PC subunit is able to bind specifically to H3K27me3 in vitro. This does not necessarily mean that H3K27me3 is the primary recruiter of PRC1 (Fig. 5B) because it is experimentally challenging to distinguish recruitment from subsequent stabilization of local chromatin binding. However, experiments directly increasing the H3K27me3 levels in human cells clearly enhance PRC1 binding in vivo, demonstrating the importance of the PC subunit as a chromatin anchor point for the PRC1 (Fig. 5B) (Lee et al. 2007). Additionally, flies with a mutated H3K27 fail to repress transcription of PcG target genes depicting a similar phenotype as Polycomb mutants (Pengelly et al. 2013). The reconstituted core components of the mammalian PRC1 have been functionally tested on nucleosomal arrays and were found to inhibit chromatin remodeling by SWI/SNF and restrict access by RNA polymerase II (RNA Pol II) in vitro (Shao et al. 1999; King et al. 2002). Both the PRC1 subunits PSC and SU(Z)2 are crucial for chromatin accessibility, explaining the complete functional redundancy of PSC and SU(Z)2 (Lo et al. 2009). Another conserved hallmark function of PRC1 is its ability to monoubiquitinate H2A-K118/K119 (H2AK118/119ub1) by the E3 ubiquitin ligase SCE/dRing1 (Fig. 5B) (Wang et al. 2004b; Gutiérrez et al. 2012). This histone modification seems to be under a tight, dynamic control because PR-DUB, another PcG complex containing Calypso and Additional sex combs, actively removes this mark (Scheuermann et al. 2010). Moreover, there is cross talk with ubiquitination of histone H2B, extending the regulatory possibilities even further (reviewed in Weake and Workman 2008).
The function of H2AK118/119ub1 is not well understood, but somehow inhibits the recruitment of the FACT chromatin remodeling complex (Zhou et al. 2008). It was recently shown that the H2A ubiquitination activity of PRC1 is dispensable for target binding and its activity to compact chromatin at HOX loci, but is indispensable for efficient repression of target genes and thereby maintenance of embryonic stem (ES) cell identity (Endoh et al. 2012). The H2AK118/119ub1 mark in Drosophila is also set by the distinct and conserved dRAF complex (Fig. 5B) (Lagarou et al. 2008; Scheuermann et al. 2010). Interestingly, dRAF is able to demethylate H3K36 through its dKDM2 subunit, linking repression through H2A ubiquitination directly with the inhibition of transcriptional elongation by removing an activating mark.
The existence of a ubiquitously conserved PRC1 has been disputed because most of the core PRC1 subunits are not conserved in plants. However, although there is no clear PC homolog in plants, the Arabidopsis LIKE HETEROCHROMATIN PROTEIN1/TERMINAL FLOWER2 (LHP1/TFL2) protein serves as a functional counterpart to Drosophila PC. Like PC, LHP1/TFL2 binds H3K27me3 in vitro and is colocalized with this mark throughout the genome (Turck et al. 2007; Zhang et al. 2007). Also, lhp1/tfl2 mutants show some of the phenotypes typical of PcG mutants. Furthermore, there are multiple homologs of PSC and SCE/dRing (Table 1), and double mutants for Atbmi1a/1b or Atring1a/1b result in phenotypes similar to those observed in mutants affecting PRC2. Indeed, the AtBMI1 homologous proteins have been shown to mediate H2A monoubiquitination in vivo (Bratzel et al. 2010). The Arabidopsis PSC/BMI 1 and SCE/dRING homologs interact with each other, the chromodomain protein LHP1, and EMF1, a plant-specific nucleoprotein. Thus, plants have a PRC1-like complex that contains some PRC1 homologs, but also plant-specific factors. However, this PRC1-like activity only regulates a subset of the PRC2 targets as it was also reported for Drosophila (Gutiérrez et al. 2012). The PRC2 target gene AG, for instance, is not up-regulated in either the Atbmi1a/1b or the Atring1a/1b double mutant (Xu and Shen 2008; Bratzel et al. 2010).
In spite of the already dazzling variety in PcG functions found, there is still scope for a better functional understanding of the role that proteins, other than the core subunits, play. There is a large group of loosely associated subunits such as TBP-associated factors in PRC1. This interaction might indicate a role in inhibiting the assembly of the preinitiation complex of RNA Pol II (Dellino et al. 2004). Several other enzymatic functions also seem to contribute to PcG silencing; PRC1 members associate with HDAC1 (Huang et al. 2002), indicating that histone-deacetylation may play a role in PcG-silencing. Furthermore, the PcG gene super sex combs (sxc) encodes an enzyme that posttranslationally modifies PH and RNA Pol II with β-O-linked N-acetylglucosamine residues and is necessary for the repression of several HOX genes (Gambetta et al. 2009; Sinclair et al. 2009). Yet another interesting link is the possibility of a switch-like mechanism through the acetylation of H3K27 by the acetyltransferase cAMP response element binding protein-binding protein (CBP), antagonizing its methylation (Tie et al. 2009). Most interestingly, in mammalian stem cells, PcG-based promoters marked by H3K27me3 frequently become DNA methylated during differentiation, suggesting that Polycomb repression and de novo DNA methylation are linked (Mohn et al. 2008). A direct physical interaction of PRC2 components and the MET1 DNA methyltransferase was recently also found in plants, indicating an evolutionarily old interaction between these major epigenetic pathways (Schmidt et al. 2013).
2.5. The PcG Connection to Paused RNA Pol II Promoters
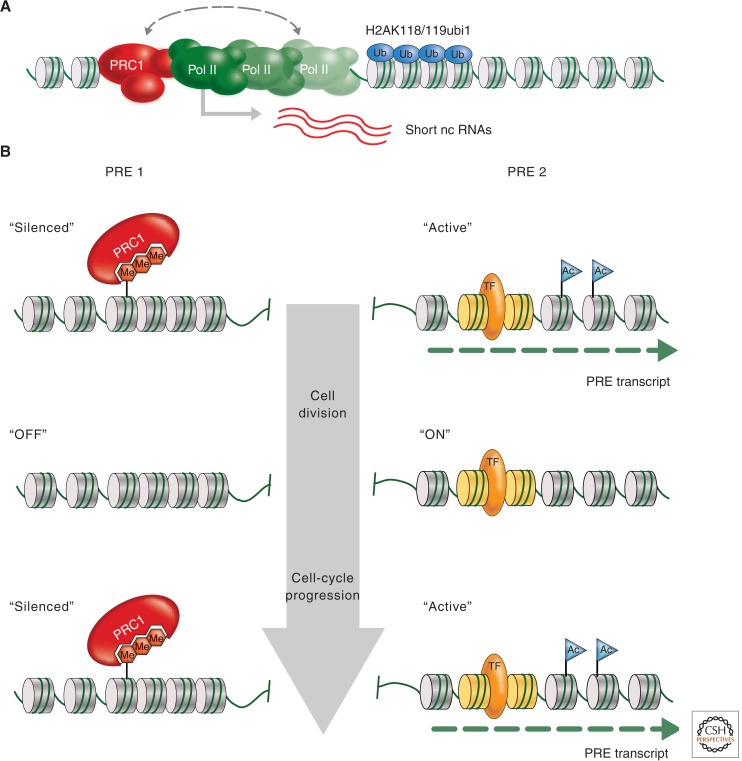
The mechanism by which PcG complexes interact with the promoter via binding to specific cis-regulatory elements (PREs, detailed in Sec. 3.1) to prevent transcription in Drosophila has become clearer in recent years. The anchoring of paused RNA Pol II complexes at promoters, preventing initiation, has been attributed to PRE–PRC1 interactions described by the use of reporter constructs (Dellino et al. 2004). In mouse ES cells, Ring1-mediated ubiquitination of H2A was found to restrain paused RNA Pol II at PcG target genes (Fig. 6A) (Stock et al. 2007). Genome-wide ChIP-Seq profiling of Drosophila tissue culture cells uncovered a strong overlap between PRC1-binding sites and promoters with paused RNA Pol II (Enderle et al. 2011). Indeed, this approach also found that many promoters of noncoding RNAs (ncRNAs) are targeted by PRC1. Among those, the promoters of primary transcripts for many micro RNAs stand out, suggesting that this important class of RNA regulators is under the control of the PcG system as well. Yet, the finding that paused Pol II at promoters is a major hallmark of PcG target genes indicates that there is a mechanistic link between PcG-mediated silencing and mechanisms of transcriptional elongation. In addition, PRC1 was shown to counteract remodeling of nucleosomes in vitro and to induce a compact chromatin structure. Thus, PRC1 potentially blocks the accessibility to DNA of transcription factors and other complexes required for transcription (Grau et al. 2011).
Figure 6.
PRC1 at paused promoters and during cell division. (A) The PRC1 complex may repress target genes by stalling the elongation of RNA Pol II. This may be achieved by ubiquitination of histone H2A through the subunit SCE/dRING, compacting promoter proximal chromatin, or direct physical interaction with the transcriptional machinery (including the short RNAs produced by the paused RNA Pol II). (B) A possible model for how differential gene expression states can be inherited. The process of intergenic transcription places positive epigenetic marks (e.g., acetylated histone tails, histone variants) at PREs that control active genes (PRE 2). All other PREs are silenced by default (PRE 1). During DNA replication and mitosis, only the positive epigenetic signal needs to be transmitted to the daughter cells, ensuring that in the next interphase intergenic transcription is restarted at PRE 2 before default silencing is reestablished at all other PREs.
The stability of silencing complexes, as shown by anchoring via methylated histone tails, appears to be a major property of the long-term repressive function of PcG proteins. However, when analyzed in vivo at the cellular level, a remarkably dynamic behavior of the individual components is observed. PcG proteins cluster in PcG bodies, which vary in size and composition between cells (Bantignies and Cavalli 2011). Fluorescence recovery after photobleaching (FRAP) analyses of GFP-marked PC and PH proteins uncovered a very high rate of exchange between unbound proteins and their complexes at silenced target genes (Fonseca et al. 2012). These results suggest that long-term repression is primarily based on a chemical equilibrium between bound and unbound proteins rather than through high-affinity protection of DNA-binding sites. Additionally, a new method measuring nucleosomal turnover uncovered a rapid exchange over active gene bodies, epigenetic regulatory elements, and replication origins in Drosophila cells (Deal et al. 2010). Surprisingly, rapid nucleosome turnover is seen at many PcG- and TrxG-regulated elements. This finding questions whether PcG-associated histone marks can contribute to epigenetic stability. Indeed, it was recently shown that H3K4 methylation, a mark of active gene expression associated with the TrxG, is not essential; Drosophila cells completely lacking this histone mark show normal transcriptional activation in response to developmental signaling pathways (Hodl and Basler 2012).
2.6. Preventing Heritable Repression by Antisilencing
The binding of PRC1 complexes to PREs appears to be a default state, as many of the anchoring PcG components and DNA-binding proteins are expressed in all cells and transgenic constructs with PREs controlling reporter genes are globally silenced. The counteracting proteins of the TrxG do not, in fact, function as activators, but rather as antirepressors (Klymenko and Müller 2004; see Kingston and Tamkun 2014 and Fig. 7 therein). This antagonistic interplay of PcG and TrxG proteins seems to be conserved between animals and plants; for instance, several plant PRC2 targets such as the AG and FLC loci are similarly maintained in an active state through the activity of the homolog of Drosophila TRX, ATX1, which acts as a KMT specific to H3K4 (Alvarez-Venegas et al. 2003; Pien et al. 2008).
Figure 7.
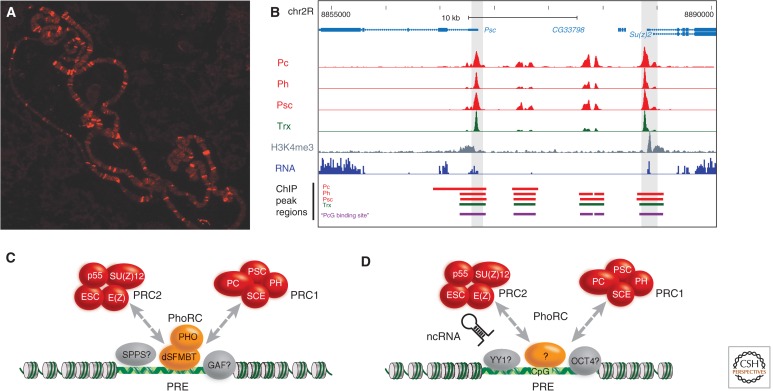
Chromosomal targeting of PRC1. (A) Immunostaining of Drosophila polytene chromosomes to visualize the distribution of the PC protein. (B) Genomic region encompassing the Drosophila PcG gene Psc and the Su(z)2 gene. The genome browser section shows the result of a ChIP-Seq and RNA-Seq analysis of Drosophila S2 tissue culture cells. The distributions of PRC1 components (red) and the TRX protein (green) are shown (data from Enderle et al. 2011). (C) In Drosophila, the PhoRC is a key player in chromatin targeting of PRC1 and PRC2, but a number of other transcription factors also contribute to target gene specificity. (D) In mouse and human, several different anchoring factors have been proposed. These include the Pho ortholog Ying and Yang 1 (YY1), transcription factors like Jarid 2 and Oct4, long ncRNAs, and the CpG content of the target sequence. (Adapted from Enderle 2011.)
To maintain active transcription of a PRE-controlled gene, the silencing at that PRE, thus, has to be prevented in a tissue- and stage-specific manner. In Drosophila, for example, the early cascade of transcription factors encoded by the segmentation genes controls the activation of HOX genes. Interestingly, these factors do not only induce transcription of the HOX genes, but also of intergenic ncRNAs that are transcribed through the associated PREs often found upstream or downstream. It was shown that transcription through PREs is required to prevent silencing and to maintain the active state of a reporter gene using transgenic constructs (Schmitt et al. 2005). The process of transcription most probably remodels PRE chromatin to generate an active state that is characterized, for instance, by a lack of repressive histone methylation and the presence of histone acetylation. Thus, although the DNA binding proteins will attract PRC1 to this particular activated PRE, the histone environment will not allow anchoring of PC via H3K27me3, and no stable silencing would be established. Because silencing is induced by default in the PcG system, epigenetic inheritance of a differential gene expression pattern only requires the transmission of the active PRE state during DNA replication and mitosis (Fig. 6B). How this is achieved at the molecular level and which epigenetic mark(s) are responsible for maintaining an active PRE state is still an open question. It has been suggested that particular TrxG factors might act as “bookmarks” during epigenetic bottleneck stages, like DNA replication and mitosis, to mark a gene for continued expression (Blobel et al. 2009). Hence, finding the molecular constitution of chromatin components that self-template during DNA replication and carry over the signals for activity to the daughter cells may be key for advancing our understanding of epigenetic inheritance.
3. Targeting PcG complexes to silenced genes
3.1. PcG Response Elements
A striking feature of the PRC1 and PRC2 core complexes is that they do not contain any obvious DNA sequence-binding activity, raising the question of how they might be targeted. Although PRC1 is bound to chromatin through its affinity for H3K27me3 and a consistent hierarchical recruitment has been shown at the bxd genomic region in Drosophila (Wang et al. 2004a), H3K27me3 alone is not sufficient to explain the targeting of the complex. First of all, PcG-binding sites are generally devoid of histones and a place of rapid nucleosome turnover (Mito et al. 2007; Deal et al. 2010). Furthermore, the generally broad distribution of H3K27me3 does not fit the localized binding of PRC1 (Fig. 7B) (Schuettengruber et al. 2009; Enderle et al. 2011). Also, removal of H3K27me3 does not lead to the immediate displacement of PRC1. Indeed, there are examples of PRC1 binding to sites without any apparent H3K27 methylation (Schoeftner et al. 2006; Tavares et al. 2012). Overall, H3K27me3 may contribute to several different low-affinity steps working together in recruitment or, more interestingly, allowing PRC1 to reach and modify histones distant to its initial binding site in a local domain.
The best characterized binding sites for PRC1 and PRC2 proteins were originally identified at the bithorax complex and subsequently termed PREs (Simon et al. 1993). PREs are thought to act as intergenic cis-regulatory elements, controlling gene expression by looping to the promoter regions of their target HOX genes. But mostly, PREs have been characterized by their ability to confer PcG silencing on reporter genes. PREs frequently contain binding sites for the zinc finger DNA-binding proteins Pleiohomeotic (PHO) and the related PHO-like (PHOL) protein, and these sites are essential for transgene and endogenous silencing functions (Fig. 7C). PHO forms a heterodimeric complex together with the Scm-related gene containing four mbt domains (SFMBT) protein, termed the Pho Repressive Complex (PhoRC) (Klymenko et al. 2006). The genome-wide distribution of PhoRC confirms its central role in the recruitment of PcG proteins: 45% of PHO-binding sites in larval and embryonic tissue are cooccupied by both PRC1 and PRC2. At the same time, the majority of PH-binding sites in embryos are enriched for PHO (Schuettengruber et al. 2009). Conversely, these data also show that PhoRC binding cannot be the only factor for targeting PRC1 because many loci bind PcG proteins without PhoRC being present. This is also reflected in PHO-binding sites being necessary, but not sufficient, for recruitment of PRC1 and PRC2. Several other proteins with DNA-binding abilities have been found as physical or genetic interactors of PcG proteins. Among them are Pipsqueak (PSQ) as subunit of the CHRASCH complex, Dorsal switch protein 1 (DSP1), Grainyhead (GRH), GAGA factor (GAF), and Sp1-like factor for pairing sensitive-silencing (SPSS), which is an Sp1/Klf protein family member (Fig. 7C). Despite this diversity, many PcG-binding sites do not contain any binding sites for the aforementioned transcription factors. Consistently, prediction algorithms based on their consensus binding sites are only able to predict a fraction of the many PRC1- and PRC2-binding sites found in a single cell type (Ringrose et al. 2003; Schwartz et al. 2006).
In contrast to Drosophila, PREs are ill-defined in plant and mammalian genomes, and only a few sequences have been described that, at least partially, fulfill the criteria for PRE function. According to Schwartz and Pirrotta (2008), the minimum criteria include (1) PREs attract H3K27me3, (2) they should form a new binding site for PcG proteins when inserted at a new location within the genome, and (3) they confer PcG-based repression to a reporter gene. Although no PREs have fulfilled all three criteria in plants, some sequences at well-studied PcG targets likely function as PREs. For instance, a transgene including sequences overlapping the promoter and parts of the coding sequence of the AG locus, a target of the EMF-PRC2, renders the reporter gene responsive to CLF, leading to H3K27me3 deposition (Schubert et al. 2006). More recently, the promoter of LEAFY COTYLEDON2 (LEC2), which is regulated by EMF-PRC2, was shown to contain a repressive LEC2 element (RLE), which is sufficient to trigger H3K27me3 deposition and silencing of a reporter gene in seedlings (Berger et al. 2011). Although the RLE is close to a CT-rich cis-regulatory element with similarities to the GAGA-box of Drosophila PREs, a possible function of the BASIC PENTACYSTEINE (BCP) proteins binding these CT-rich sequences in recruiting PRC2 has not yet been shown.
DNA sequences from the FLC and MEA loci can similarly confer PcG-dependent silencing to reporter genes, although in the case of MEA the FIS-PRC2 plays only a minor part in the repression of the paternal allele (Sheldon et al. 2002; Wöhrmann et al. 2012). For AG, FLC, or MEA, no DNA-binding factors are currently known that recruit PRC2 variants to these loci. Thus, other factors, such as a specific chromatin structure or long ncRNAs (lncRNAs), may be involved in PRC2 recruitment. Recent studies have, however, shown that DNA-binding proteins do play a role in PRC2 recruitment at the WUSCHEL (WUS) locus, whose repression is crucial for the appropriate termination of floral meristems. WUS is a target of the EMF-PRC2, and H3K27me3 levels at the WUS locus are reduced similarly in ag, clf, and swn mutants, which act in the same genetic pathway. As H3K27me3 levels at WUS increase rapidly after the experimental induction of AG, the MADS-domain protein AG likely plays a role in recruiting PcG proteins to this locus.
In the mouse and human genome, PcG proteins predominantly occupy regions around gene promoters (Boyer et al. 2006; Lee et al. 2006; Ku et al. 2008). Two intergenic PREs, however, were identified in mammalian genomes (Sing et al. 2009; Woo et al. 2010). An intergenic region of 1.8 kb, termed D11.12, was bound by PcG proteins in the human HOX-D complex. The other intergenic PRE, a 3-kb “PRE-kr” at the MafB/Kreisler locus in mouse, also recruited PcG proteins and was able to regulate its expression pattern. PcG protein recruitment is necessary for their potential to exert gene silencing. Importantly, the D11.12 element is able to maintain repression of a luciferase transgene throughout cell differentiation, providing the first example of a mammalian PRE sequence. Interestingly, both elements contain binding sites for the mammalian homolog of the Drosophila PhoRC, which contains SFMBT and YY1 (Fig. 7D). Still, binding of YY1 only accounts for a fraction of the genome-wide PRC2 sites in ES cells (Squazzo et al. 2006). Although there is substantial overlap with the pluripotency factors NANOG, OCT4, and SOX2, these three proteins have not been copurified with PcG complexes yet (Boyer et al. 2006; Lee et al. 2006). Also, the recent identification of the mammalian GAF homolog may provide new insights into PcG recruitment by transcription factors in the future (Matharu et al. 2010).
A surprising aspect was found in ES cells, in which nearly all PRC2 binding sites are found at CpG islands or other highly GC-enriched sequences (Ku et al. 2008). Indeed, GC-rich DNA from bacterial genomes is able to initiate recruitment of PRC2 (Mendenhall et al. 2010). This is especially interesting because Mixed lineage leukemia (MML), the mammalian TRX homolog, also has a preference for CpG dinucleotides, revealing a shared discriminant for targeting. Other interesting recruitment factors are specific lncRNAs, which are discussed in Section 3.2.
3.2. PcG Proteins Bind to Noncoding RNAs
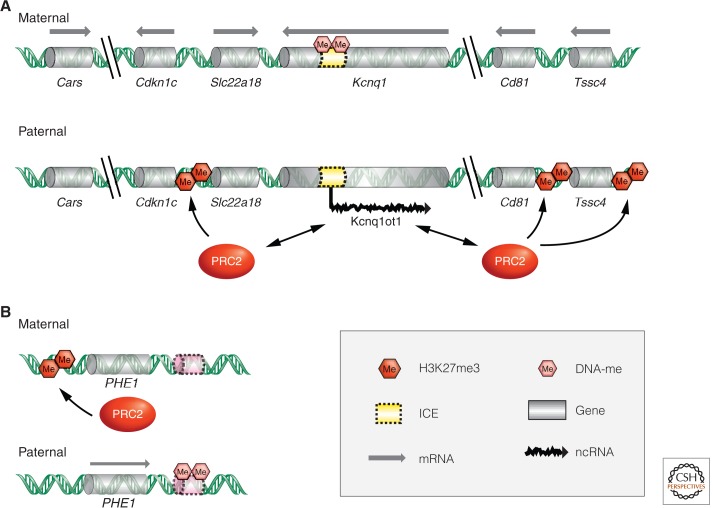
Several ncRNAs have been proposed to recruit PcG proteins in mammals. The most prominent example may be HOTAIR, a 2.2-kb ncRNA from the human HOX-C cluster, acting in trans to mediate gene repression (described in Rinn 2014; also Rinn et al. 2007). HOTAIR is one of many transcripts identified in the intergenic regions of human HOX clusters and its depletion leads to the loss of H3K27me3 in a large genomic region of HOX-D. Indeed, HOTAIR interacts with PRC2 components in vitro, suggesting that it may also recruit KMT activity to the HOX-D complex (Rinn et al. 2007). Another interaction between PRC2 components and an lncRNA is observed at the paternally imprinted Kcnq1 locus in mouse (Fig. 8) (Wu and Bernstein 2008). Similar to HOTAIR, the 91-kb-long primary transcript of Kcnq1 overlapping transcript 1 (Kcnq1ot1), coimmunoprecipitates with EZH2 and SUZ12, and additionally with the H3K9-specific KMT G9a (Kanduri et al. 2006; Pandey et al. 2008). The paternally transcribed ncRNA may facilitate silencing by recruitment of methyltransferases in cis, leading to the inactivation and compaction of genes at the locus (Terranova et al. 2008).
Figure 8.
Interplay of PcG-mediated repression and DNA methylation regulates genomic imprinting in plants and mammals. (A) Regulation of genomic imprinting at the Kcnq1 domain on distal chromosome 7. The imprinting control element (ICE) is maternally methylated and prevents the transcription of the lncRNA Kcnq1ot1 from the maternal chromosome. The paternally expressed Kcnq1ot1 associates with chromatin and recruits chromatin modifying complexes, such as PRC2, to mediate and maintain transcriptional silencing of several paternal, protein-coding alleles. (B) In Arabidopsis seeds, the paternally expressed PHE1 gene is maternally repressed by the action of PRC2. A cis-regulatory element (shaded pink) downstream of the PHE1 gene must be methylated for paternal expression, but demethylated for maternal repression.
Another cis-acting ncRNA that is a crucial component for inactivating one of the X chromosomes in female mammals is the 17-kb-long X inactive-specific transcript (Xist). It contains a 28-bp repeat element that interacts with EZH2 in vitro and in vivo (Zhao et al. 2010). This element folds into a double stem-loop structure and is necessary for X inactivation (covered in more detail in Brockdorff and Turner 2014). Recently, similar stem-loop structures have been identified in a class of short promoter-proximal ncRNAs, which are transcribed from H3K27me3-marked genes (Kanhere et al. 2010). The small ncRNAs interact with PRC2 in vitro through secondary structures, revealing PRC2-bound and folded ncRNAs as a repeating theme in mammalian cells. However, details of the recruiting mechanism and a common RNA motif have yet to be described. Also, RNA-interacting PcG proteins are not restricted to members of the PRC2 complex. A recent example is ANRIL, an ncRNA at the murine Ink4b/Arf/Ink4a locus. This transcript has been shown to specifically associate with PRC1 members through the chromodomain of CBX7 (Yap et al. 2010). A local competition between ANRIL and H3K27me3 for the binding of CBX7 may therefore remove PRC1 from chromatin, leading to the derepression of the Ink4b/Arf/Ink4a locus.
An interaction between lncRNAs also seems to play a role in the regulation of the Arabidopsis FLC locus and, hence, vernalization (see Baulcombe and Dean 2014 for details). Expression of the sense ncRNA COLDAIR and the antisense ncRNA COOLAIR are induced by cold, and COLDAIR was shown to physically interact with CLF, indicating a possible role in VRN-PRC2 recruitment (Swiezewski et al. 2009). However, FLC transgenes without the COLDAIR promoter respond to cold, and also antisense COOLAIR is not required for vernalization-induced repression of FLC, such that the functional requirements for these ncRNAs are not clear (Sheldon et al. 2002; Helliwell et al. 2011). However, as the expression and processing of the COOLAIR ncRNA is affected by different genotypes and environments, and these correlate with changes in chromatin marks (reviewed in Ietswaart et al. 2012), it is possible that ncRNAs also recruit PRC2 components to target loci as they do in mammals.
4. PcG repression in development and disease
4.1. From Gene to Chromosome Repression
Mutations in members of the murine PRC1 complex show homeotic transformations of the axial skeleton. This can cause the appearance of additional vertebrae as a consequence of the derepression of HOX genes (Fig. 2E,F) (Core et al. 1997). In addition, the mutant mice display severe combined immunodeficiencies caused by a lack of proliferative responses of hematopoietic cells (Raaphorst 2005). The role of PcG proteins has been particularly well studied in blood cells, in line with the fact that most blood cell lineages are characterized by their well-described cell-type-specific transcription programs. Lineage commitment and restriction need to be faithfully maintained through cell division. In PcG knockout mice, B- and T-cell precursor populations are produced normally, indicating that the establishment of lineage-specific gene expression patterns does not depend on PcG proteins. These proteins do, however, contribute to the irreversibility of the lineage choice, rather than the decision to follow a particular developmental pathway.
PcG proteins play a major role in controlling proliferation as well as the control of HOX genes, whose expression patterns characterize different blood cell lineages. The Bmi1 gene, an ortholog of Drosophila Psc belonging to the PRC1 group, was initially identified as an oncogene that, in collaboration with myc, induces murine lymphomagenesis (van Lohuizen et al. 1991). The Bmi1 protein controls the cell-cycle regulators p16INK4a and p19ARF (Jacobs et al. 1999). Both Bmi1 and the related protein Mel-18 are negative regulators of the Ink4c-Arf locus required for normal lymphoid proliferation control. Misregulation of this important cell-cycle checkpoint affects apoptosis and senescence in mice.
Mammalian PcG proteins are also associated with X-chromosome inactivation as mentioned in Section 3.2 (see also Brockdorff and Turner 2014). The inactivation of one X chromosome in XX female cells is accompanied by a series of chromatin modifications that involve PcG proteins. In particular, components of the PRC2 complex, like the ESC homolog EED, or the E(Z) homolog ENX1 (Table 1), play a major role in the establishment of histone marks associated with transcriptional silencing. Transient association of PRC2 with the X chromosome, coated by Xist RNA, is accompanied by H3K27 methylation. In contrast, eed mutant mouse embryos show no recruitment of the ENX1 KMT, and consequently no H3K27me3 is observed. However, the absence of these PRC2 components does not lead to a complete derepression of the entire inactive X chromosome; rather, some cells display sporadic reexpression of X-linked genes and an increase in epigenetic marks associated with an active state (H3K9ac and H3K4me3). This is likely because other, partially redundant, epigenetic mechanisms are in place to ensure the maintenance of one inactive X chromosome.
Recruitment of PRC2 to the inactive X chromosome is dependent on Xist RNA. As association of PRC2 to the inactive X is only transient, it appears that the complex is only required to set epigenetic marks (i.e., H3K27me3) for the maintenance of silencing. Currently, it is not known whether the PRC1 complex directly recognizes these marks. PRC1 is involved in the permanent silencing of the inactive X chromosome. The PRC2 component EED is required to recruit the PRC1 components MPH1 and MPH2, whereas RING1b, which can ubiquitinate H2A, is recruited independently of EED (Schoeftner et al. 2006). Thus, Xist RNA can recruit PRC1 components in both PRC2-dependent and -independent ways. In the absence of PRC2, Xist-dependent PRC1 recruitment is sufficient for PcG-based X-chromosome inactivation, which is further consolidated and maintained by DNA methylation.
PRC2 is involved in X-chromosome inactivation both in the embryo, in which an X chromosome is chosen at random for inactivation, and in extraembryonic tissues, wherein the paternally inherited X chromosome is systematically inactivated (imprinted X-chromosome inactivation). In addition, it was found that PRC2 is involved in the regulation of some autosomal imprinted genes. PRC2-mediated repression is, thus, a silencing mechanism that operates in addition to DNA methylation in the regulation of imprinted gene expression. By way of example, an analysis of 14 imprinted loci from six distinct imprinting clusters showed that four of these were biallelically expressed in eed mutant mice (Mager et al. 2003). At the Kcnq1 imprinting cluster (Fig. 8), for instance, the predominantly maternally expressed genes Cdk1, Cd81, and Tssc4 become biallelically expressed in ezh2 mutant mice (Terranova et al. 2008). As similar results were also observed in mutants deficient for Ring1b, both PRC1 and PRC2 appear to be involved in regulating the monoallelic expression of some imprinted genes. Furthermore, it was shown that EZH2 is required for the association of the lncRNA Kcnq1ot1 along the Kcnq1 imprinting cluster (Terranova et al. 2008), confirming a link between PRC2 and ncRNAs in the regulation of imprinted genes (Fig. 8). Interestingly, all loci that lost imprinted expression were normally repressed when paternally inherited, whereas none of the maternally repressed loci were affected. As there appears to be a cross talk between PcG-based repression and gene silencing by DNA methylation, it is possible that PRC2 complex plays a role in the regulation of these imprinted genes via DNA methylation (see Li and Zhang 2014).
An involvement of PRC2 in the regulation of imprinted gene expression has also been reported in Arabidopsis, in which the PHE1 locus is expressed at much higher levels from the paternal allele (Köhler et al. 2005). In mutants affecting the E(z) homolog MEA, the maternal PHE1 allele is specifically derepressed. MEA also regulates its own imprinted expression, noted by the strong repression of maternal MEA early in reproductive development in a mea mutant background. This effect, however, is independent of the other components of the FIS-PRC2 (Fig. 4) (Baroux et al. 2006). In contrast, later in development the FIS-PRC2 contributes to the stable repression of the paternal MEA allele (Baroux et al. 2006; Gehring et al. 2006; Jullien et al. 2006). In this latter case, the FIS-PRC2 is involved in the silencing of a paternally repressed imprinted allele similar to the situation in mammals. But MEA also has a role in keeping expression of the maternal PHE1 and MEA alleles at low levels. Similar to the situation in mammals, regulation of imprinted expression at the PHE1 and MEA loci involves both PRC2 and DNA methylation (Fig. 8). Although DNA methylation at the locus was suggested to regulate higher-order chromatin structure rather than directly distinguishing maternal and paternal alleles (Wöhrmann et al. 2012), both these epigenetic pathways seem to work together at the PHE1 locus (Makarevich et al. 2008). The silencing of the maternal PHE1 allele depends on a cis-regulatory region, which is differentially methylated. This regulatory element downstream of PHE1 is methylated on the expressed paternal allele, but must not be methylated to mediate PRC2-dependent repression of the maternal allele (Fig. 8).
As PRC2 components are present in plants, invertebrates, and mammals, PRC2 represents an ancient molecular module suitable for gene repression that was already present in the unicellular ancestor of plants and animals, before the evolution of multicellularity. It was recently shown that DNA methylation at both the MEA and PHE1 locus is affected in mea mutants, and PRC2 components directly interact with the DNA methyltransferase MET1, as found for the mammalian counterparts (Schmidt et al. 2012). Thus, although the interaction of these two major epigenetic pathways in the control of gene expression may have an ancient evolutionary origin, they were independently recruited for the regulation of imprinted genes in plants and mammals, the two lineages in which genomic imprinting evolved (Raissig et al. 2011).
4.2. Consequences of Aberrant Transcriptional Activation
The finding that Bmi1 misregulation causes malignant lymphomas in mice raises the question of whether human BMI1 (a PRC1 component) itself contributes to the development of cancer in a similar fashion. There is now accumulating evidence that altered PcG gene expression is widespread in human malignant lymphomas (Shih et al. 2012). For instance, the level of BMI1 overexpression in B-cell lymphomas correlates with the degree of malignancy, suggesting that PRC1 components do play a role in the development of human cancers. However, the target genes of BMI1 in human cells appear to be different from those of mouse lymphocytes, as no obvious down-regulation of p16INK4a could be correlated with the overexpression of the oncogenes.
PcG gene overexpression is not only observed in hematological malignancies, but is also found in solid tumors, including meduloblastomas, and tumors originating from liver, colon, breast, lung, penis, and prostate (Fig. 9). The high expression of a PRC2 marker, EZH2, is often found in early stages of highly proliferative lung carcinomas. This suggests that the well-known cascade of PRC2 initiation and PRC1 maintenance might also accompany the development of a tumor cell lineage (for a review, see Sauvageau and Sauvageau 2010).
Figure 9.
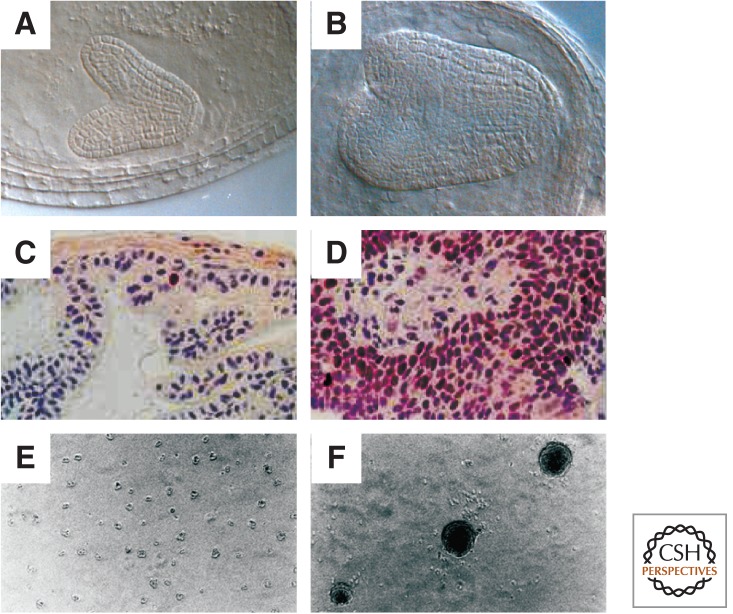
PRC2 regulates cell proliferation in mammals and plants. (A,B) Plant embryos derived from wild-type and mea mutant egg cells. MEA encodes a protein of the FIS-PRC2 and regulates cell proliferation. The mea embryo (B) is much larger than the corresponding wild-type embryo (A) at the same stage of development (late heart stage). Mutant embryos develop slower and have approximately twice the number of cell layers. (C,D) Normal and cancerous prostate epithelium of mice. In the cancerous epithelium, Ezh2 expression is highly increased (labeled with an anti-Ezh2 antibody). Thus, both loss of E(Z) function in plants and overexpression of E(Z) function in mice can lead to defects in cell proliferation. (E,F) Control and RING1 overexpressing rat 1a fibroblast cells. Overexpression of RING1 leads to anchorage-independent growth in soft agar, typical of neoplastically transformed cells. (A,B, Courtesy of J.-P. Vielle-Calzada and U. Grossniklaus; C,D, reprinted, with permission, from Kuzmichev et al. 2005, © National Academy of Sciences; E,F, reprinted, with permission, from Satijn and Otte 1999, © American Society for Microbiology.)
Interestingly, PRC2 components also play a crucial role in the control of cell proliferation in Arabidopsis. Although aberrant growth does not lead to cancer and death in plants, a strict control of cell proliferation is essential for normal development. In mutants of the fis class, the two fertilization products of flowering plants, the embryo and endosperm, overproliferate and the resulting seeds abort (Grossniklaus et al. 2001; Hsieh et al. 2003). Effects on cell proliferation are also observed in double mutants of clf and swn, two of the three plant E(z) homologs. Such plants undergo normal seed development, but produce a mass of proliferating, undifferentiated tissue (callus) rather than a differentiated shoot after germination (Chanvivattana et al. 2004).
Although it is currently not known how exactly PRC2 controls cell proliferation in plants, it is likely to involve interactions with RBR, the plant homolog of the Retinoblastoma (Rb) protein (Ebel et al. 2004; Mosquna et al. 2004). Mutants of the fis class do not only show proliferation defects during seed development after fertilization, but the FIS genes are also required to prevent proliferation of the endosperm in the absence of fertilization. This latter aspect of the phenotype is shared with rbr mutants and can be explained by the fact that RBR regulates the expression of genes encoding PRC2 components and MET1 (Johnston et al. 2008). Remarkably, the Rb pathway also regulates the mammalian Ezh2 and Eed genes encoding PRC2 subunits (Bracken et al. 2003), illustrating conserved regulatory networks between plants and animals.
4.3. Maintaining Stem Cell Fate
PcG regulation plays a very early role during oogenesis in the mouse for the formation of totipotent cell identities in the progeny (Posfai et al. 2012). Genetic ablation of the PRC1 components RING1 and RNF2 results in loss of chromatin-bound PRC1 in oocytes, induction of massive transcriptional misregulation during oocyte growth, and a developmental arrest at the two-cell stage of embryogenesis. These results indicate that PRC1 functions during oogenesis to specify maternal contributions in the cytoplasm as well as on maternal chromosomes, both of which contribute to the developmental competence of preimplantation embryos. Indeed, cultured mouse embryonic stem cells were used very effectively to study the role of PcG proteins in many aspects of cell proliferation and differentiation. The introduction of new technologies like ChIP-Seq allowed the correlation of PRC1/PRC2 components with many epigenetic marks, genetic regulatory elements of the murine genome, and the identification of functions related to ES cell pluripotency and plasticity during embryonic development (Boyer et al. 2006). Bivalent chromatin domains, characterized by the coexistence of the active H3K4me3 and repressive H3K27me3 histone marks, are resolved during differentiation (Mikkelsen et al. 2007). The cellular memory system takes a leading role in this process. Permanently repressed genes are tagged by the PcG system, together with DNA methylation, to establish stable silencing marks. Conversely, the TrxG system reiterates the H3K4me3 mark to keep the corresponding differentiation genes active.
Stem cells play an ever-increasing role in medicine. Their potential to provide progenitors for the healing of damaged tissue places them into a well treasured tool box of regenerative medicine. Not surprisingly, it is in the very well characterized blood cell lineage wherein we know most about the identity and location of stem cells. Hematopoietic stem cells (HSCs) maintain the pool of blood cells by self-renewing as well as producing daughter cells that differentiate into the lymphoid, myeloid, and erythroid lineages. The stem cell niche in the adult bone marrow provides the cells with specific external signals to maintain their fate. On the other hand, cell intrinsic cues for the maintenance of the “stemness” state seem to rely on the PcG system.
Mouse mutants affecting PRC1 genes (e.g., bmi1, mel-18, mph1/rae28, and m33; see Table 1) suffer from various defects in the hematopoietic system, such as hyperplasia in spleen and thymus, reduction in B and T cells, and an impaired proliferative response of lymphoid precursors to cytokines. The requirements for Bmi1 and Mel18 in stem cell self-renewal during different stages of development suggest a changing pool of target genes between embryonic and adult stem cells.
The PcG system is also required for neural stem cells (NSCs) as indicated by the neuronal defects observed in bmi1 mouse mutants (Bruggeman et al. 2005; Zencak et al. 2005). In particular, these mice are depleted of cerebral NSCs postnatally, indicating an in vivo requirement for Bmi1 in NSC renewal. It appears that embryonic NSC maintenance is thus under a different PcG network control than adult NSC self-renewal, similar to the regulation of the hematopoietic system.
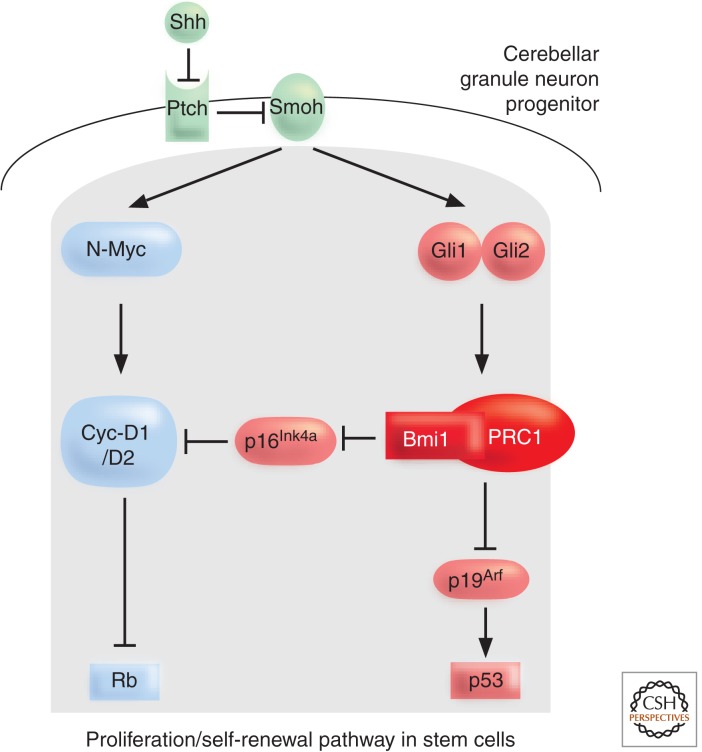
External signals like the Sonic hedgehog (Shh) signaling cascade modulate the Bmi1 response in NSCs and ensure a proliferative/self-renewal capacity (Leung et al. 2004). The identification of these external cues controlling PcG repression came through the analysis of the development of cerebellar granule neuron progenitors (CGNPs). A postnatal wave of proliferation is induced by the signaling factor Shh, secreted by the Purkinje cells. The Shh signal branches to control N-Myc and Bmi1 levels (Fig. 10). Thus, Bmi1-deficient CGNPs have a defective proliferative response upon Shh stimulation. The Shh signal is able to control proliferation of these stem cells ultimately by modulating both the downstream Rb pathway (via N-myc and Bmi1/p16INK4a) and p53 pathway (via Bmi1/p19ARF). This mechanism explains why hyperactivation of Shh signaling leads to the development of medulloblastomas. HSCs are regulated by a similar Indian hedgehog-controlled pathway. And, in NSCs, expression of the Hoxd8, Hoxd9, and Hoxc9 loci is under the control of Bmi1. The appropriate HOX expression profile confers the necessary stem cell fate.
Figure 10.
Sonic Hedgehog signaling maintains proliferation/self-renewal of cerebellar progenitor cells. The Shh signaling cascade regulates both the Rb pathway (which can be bound by the PRC2 RbAp48 protein) as well as the p53 pathway via Bmi1 control of the p16/p19 proliferation checkpoint. Inhibition of Smoothened (Smoh) by the Shh receptor Patched (Ptch) results in downstream signaling in the nucleus. One part of the signal induces N-Myc, Cyclin D1, and Cyclin D2, whereas the other part activates Bmi1 via the Gli effectors. (Adapted, with permission, from Valk-Lingbeek et al. 2004, © Elsevier.)
Indeed, as stem cells represent a defined and committed cellular state, it is not surprising that the PcG system maintains this particular fate in a mitotically heritable fashion. In the future, it will be interesting to identify the pool of targets of the PcG system in the different adult stem cell populations, and to learn how to influence the maintenance system to allow for the controlled reprogramming of stem cell fates. At the moment, little is known about the role of PcG genes in stem cell maintenance in plants. However, the reprogramming of plant cells, which are totipotent and have the potential to form a complete new organism under appropriate conditions, involves PcG regulation. Indeed, plants lacking the E(z) homologs CLF and SWN produce a mass of undifferentiated cells after germination, suggesting that PcG genes are required to maintain a differentiated state (Chanvivattana et al. 2004). Interestingly, the same PcG genes are required for the in vitro reprogramming of differentiated leaf cells into totipotent callus cells, possibly because PRC2 is required to repress leaf differentiation genes during this reprogramming process (He et al. 2012). In floral meristems, PRC2 plays a role to repress WUS (see Sec. 3.1), which itself is required for stem cell maintenance (Liu et al. 2011). Thus, although the molecular mechanisms differ greatly between animals and plants, PRC2 has been recruited to regulate cell stem identity and cell differentiation in both lineages.
5. Conclusion and outlook
It has been remarkable to follow the development of our understanding of PcG epigenetic regulation, starting with the initial genetic identification of a Drosophila mutant possessing additional sex combs on the second and third leg. This eventually led to the discovery of a new class of regulators found to be required for fundamental epigenetic processes such as vernalization in plants and silencing of the mammalian X chromosome. Control of genetic information is highly influenced by chromatin structure and composition of histones in their various modified forms. The proteins of the PcG are directly involved in generating epigenetic marks, for instance, H3K27me3 and H2AK118/119ub1, as a consequence of developmental decisions. The same group “reads” (i.e., shows high affinity to) these epigenetic marks through the action of the PRC1 proteins, and translates them into a stable, transcriptionally repressed state. In the model organism Drosophila, we have a relatively clear picture of how PcG complexes are anchored at PREs, for a defined group of target genes that are subject to long-term repression. However, to date very few PREs have been identified in other organisms. Although the basic functions of PcG proteins remain the same, it is not well understood how they are targeted to their site of action. Additionally, we need to get a better understanding of how an apparently dynamic group of proteins can impose a stable state of transcriptional repression through a chemical equilibrium.
The other major question in PcG research focuses on the heritability of the repressed state, the very essence of epigenetics. What is the identity of the molecular marks required to transmit a state of gene expression through DNA replication and mitosis? Do both active and repressed states need corresponding epigenetic marks, which are transmitted to daughter cells, or is only one sufficient, whereas the other represents the default state? The mechanism by which PcG proteins impose silencing on transcription during the interphase of the cell cycle has become increasingly clear. In the future, the focus of research will be on how the information regarding a state of gene expression endures the DNA replication process and is faithfully transmitted to the daughter cells following mitosis.
ACKNOWLEDGMENTS
R.P. thanks Daniel Enderle for help with the manuscript and design of some of the figures; U.G. is grateful to Michael Raissig and Heike Wöhrmann for their support in preparing figures, and to Hanspeter Schöb for help with the bibliography. Work on PcG-mediated silencing mechanisms in R.P.'s group is supported by the ETH Zürich and the Swiss National Science Foundation, and in U.G.'s laboratory by the University of Zürich, the Swiss National Science Foundation, and the European Research Council.
Footnotes
Editors: C. David Allis, Marie-Laure Caparros, Thomas Jenuwein, and Danny Reinberg
Additional Perspectives on Epigenetics available at www.cshperspectives.org
REFERENCES
*Reference is also in this collection.
- Agger K, Cloos PA, Christensen J, Pasini D, Rose S, Rappsilber J, Issaeva I, Canaani E, Salcini AE, Helin K 2007. UTX and JMJD3 are histone H3K27 demethylases involved in HOX gene regulation and development. Nature 449: 731–734. [DOI] [PubMed] [Google Scholar]
- *.Allis CD, Jenuwein T, Reinberg D 2014. Overview and concepts. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a018739. [DOI] [Google Scholar]
- *.Almouzni G, Cedar H 2014. Maintenance of epigenetic information. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a019372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez-Venegas R, Pien S, Sadder M, Witmer X, Grossniklaus U, Avramova Z 2003. ATX-1, an Arabidopsis homolog of trithorax, activates flower homeotic genes. Curr Biol 13: 627–637. [DOI] [PubMed] [Google Scholar]
- Bantignies F, Cavalli G 2011. Polycomb group proteins: Repression in 3D. Trends Genet 27: 454–474. [DOI] [PubMed] [Google Scholar]
- *.Barlow DP, Bartolomei MS 2014. Genomic imprinting in mammals. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a018382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baroux C, Gagliardini V, Page DR, Grossniklaus U 2006. Dynamic regulatory interactions of Polycomb group genes: MEDEA autoregulation is required for imprinted gene expression in Arabidopsis. Genes Dev 20: 1081–1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Baulcombe DC, Dean C 2014. Epigenetic regulation in plant responses to the environment. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a019471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger N, Dubreucq B, Roudier F, Dubos C, Lepiniec L 2011. Transcriptional regulation of Arabidopsis LEAFY COTYLEDON2 involves RLE, a cis-element that regulates trimethylation of histone H3 at lysine-27. Plant Cell 23: 4065–4078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birve A, Sengupta AK, Beuchle D, Larsson J, Kennison JA, Rasmuson-Lestander A, Müller J 2001. Su(z)12, a novel Drosophila Polycomb group gene that is conserved in vertebrates and plants. Development 128: 3371–3379. [DOI] [PubMed] [Google Scholar]
- Blobel GA, Kadauke S, Wang E, Lau AW, Zuber J, Chou MM, Vakoc CR 2009. A reconfigured pattern of MLL occupancy within mitotic chromatin promotes rapid transcriptional reactivation following mitotic exit. Mol Cell 36: 970–983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyer LA, Plath K, Zeitlinger J, Brambrink T, Medeiros LA, Lee TI, Levine SS, Wernig M, Tajonar A, Ray MK, et al. 2006. Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature 441: 349–353. [DOI] [PubMed] [Google Scholar]
- Bracken AP, Pasini D, Capra M, Prosperini E, Colli E, Helin K 2003. EZH2 is downstream of the pRB-E2F pathway, essential for proliferation and amplified in cancer. Embo J 22: 5323–5335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bratzel F, López-Torrejón G, Koch M, Del Pozo JC, Calonje M 2010. Keeping cell identity in Arabidopsis requires PRC1 RING-finger homologs that catalyze H2A monoubiquitination. Curr Biol 20: 1853–1859. [DOI] [PubMed] [Google Scholar]
- *.Brockdorff N, Turner B 2014. Dosage compensation in mammals. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a019406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruggeman SW, Valk-Lingbeek ME, van der Stoop PP, Jacobs JJ, Kieboom K, Tanger E, Hulsman D, Leung C, Arsenijevic Y, Marino S, et al. 2005. Ink4a and Arf differentially affect cell proliferation and neural stem cell self-renewal in Bmi1-deficient mice. Genes Dev 19: 1438–1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao R, Zhang Y 2004. SUZ12 is required for both the histone methyltransferase activity and the silencing function of the EED-EZH2 complex. Mol Cell 15: 57–67. [DOI] [PubMed] [Google Scholar]
- Cao R, Wang L, Wang H, Xia L, Erdjument Bromage H, Tempst P, Jones RS, Zhang Y 2002. Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science 298: 1039–1043. [DOI] [PubMed] [Google Scholar]
- Cao R, Wang H, He J, Erdjument-Bromage H, Tempst P, Zhang Y 2008. Role of hPHF1 in H3K27 methylation and Hox gene silencing. Mol Cell Biol 28: 1862–1872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrington EA, Jones RS 1996. The Drosophila Enhancer of zeste gene encodes a chromosomal protein: Examination of wild-type and mutant protein distribution. Development 122: 4073–4083. [DOI] [PubMed] [Google Scholar]
- Chanvivattana Y, Bishopp A, Schubert D, Stock C, Moon YH, Sung ZR, Goodrich J 2004. Interaction of Polycomb-group proteins controlling flowering in Arabidopsis. Development 131: 5263–5276. [DOI] [PubMed] [Google Scholar]
- *.Cheng X 2014. Structural and functional coordination of DNA and histone methylation. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a018747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Core N, Bel S, Gaunt SJ, Aurrand Lions M, Pearce J, Fisher A, Djabali M 1997. Altered cellular proliferation and mesoderm patterning in Polycomb-M33-deficient mice. Development 124: 721–729. [DOI] [PubMed] [Google Scholar]
- Czermin B, Melfi R, McCabe D, Seitz V, Imhof A, Pirrotta V 2002. Drosophila Enhancer of Zeste/ESC complexes have a histone H3 methyltransferase activity that marks chromosomal Polycomb sites. Cell 111: 185–196. [DOI] [PubMed] [Google Scholar]
- Deal RB, Henikoff JG, Henikoff S 2010. Genome-wide kinetics of nucleosome turnover determined by metabolic labeling of histones. Science 328: 1161–1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dellino GI, Schwartz YB, Farkas G, McCabe D, Elgin SC, Pirrotta V 2004. Polycomb silencing blocks transcription initiation. Mol Cell 13: 887–893. [DOI] [PubMed] [Google Scholar]
- De Lucia F, Crevillen P, Jones AM, Greb T, Dean C 2008. A PHD-Polycomb repressive complex 2 triggers the epigenetic silencing of FLC during vernalization. Proc Natl Acad Sci 105: 16831–16836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebel C, Mariconti L, Gruissem W 2004. Plant retinoblastoma homologues control nuclear proliferation in the female gametophyte. Nature 429: 776–780. [DOI] [PubMed] [Google Scholar]
- Enderle D 2011. Non-coding RNAs at intergenic Polycomb group binding sites. PhD thesis University of Heidelberg, Heidelberg. [Google Scholar]
- Enderle D, Beisel C, Stadler MB, Gerstung M, Athri P, Paro R 2011. Polycomb preferentially targets stalled promoters of coding and noncoding transcripts. Genome Res 21: 216–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endoh M, Endo TA, Endoh T, Isono K, Sharif J, Ohara O, Toyoda T, Ito T, Eskeland R, Bickmore WA, et al. 2012. Histone H2A mono-ubiquitination is a crucial step to mediate PRC1-dependent repression of developmental genes to maintain ES cell identity. PLoS Genet 8: e1002774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischle W, Wang Y, Jacobs SA, Kim Y, Allis CD, Khorasanizadeh S 2003. Molecular basis for the discrimination of repressive methyl-lysine marks in histone H3 by Polycomb and HP1 chromodomains. Genes Dev 17: 1870–1881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fonseca JP, Steffen PA, Müller S, Lu J, Sawicka A, Seiser C, Ringrose L 2012. In vivo Polycomb kinetics and mitotic chromatin binding distinguish stem cells from differentiated cells. Genes Dev 26: 857–871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gambetta M, Oktaba K, Müller J 2009. Essential role of the glycosyltransferase sxc/Ogt in Polycomb repression. Science 325: 93–96. [DOI] [PubMed] [Google Scholar]
- Gehring M, Huh JH, Hsieh T-F, Penterman J, Choi Y, Harada JJ, Goldberg RB, Fischer RL 2006. DEMETER DNA glycosylase establishes MEDEA Polycomb gene self-imprinting by allele-specific demethylation. Cell 124: 495–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gendall AR, Levy YY, Wilson A, Dean C 2001. The VERNALIZATION 2 gene mediates the epigenetic regulation of vernalization in Arabidopsis. Cell 107: 525–535. [DOI] [PubMed] [Google Scholar]
- Goodrich J, Puangsomlee P, Martin M, Long D, Meyerowitz EM, Coupland G 1997. A Polycomb-group gene regulates homeotic gene expression in Arabidopsis. Nature 386: 44–51. [DOI] [PubMed] [Google Scholar]
- Grau DJ, Chapman BA, Garlick JD, Borowsky M, Francis NJ, Kingston RE 2011. Compaction of chromatin by diverse Polycomb group proteins requires localized regions of high charge. Genes Dev 25: 2210–2221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grossniklaus U, Vielle Calzada JP, Hoeppner MA, Gagliano WB 1998. Maternal control of embryogenesis by MEDEA, a Polycomb group gene in Arabidopsis. Science 280: 446–450. [DOI] [PubMed] [Google Scholar]
- Grossniklaus U, Spillane C, Page DR, Köhler C 2001. Genomic imprinting and seed development: Endosperm formation with and without sex. Curr Opin Plant Biol 4: 21–27. [DOI] [PubMed] [Google Scholar]
- Gutiérrez L, Oktaba K, Scheuermann JC, Gambetta MC, Ly-Hartig N, Müller J 2012. The role of the histone H2A ubiquitinase Sce in Polycomb repression. Development 139: 117–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hackett WP, Cordero RE, Sinivasan C 1987. Apical meristem characteristics and activity in relation to juvenility in Hedera. Butterworth, London, UK. [Google Scholar]
- Hadorn E 1968. Transdetermination in cells. Sci Am 219: 110–114. [DOI] [PubMed] [Google Scholar]
- He C, Chen X, Huang H, Xu L 2012. Reprogramming of H3K27me3 is critical for acquisition of pluripotency from cultured Arabidopsis tissues. PLoS Genet 8: e1002911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helliwell CA, Robertson M, Finnegan EJ, Buzas DM, Dennis ES 2011. Vernalization-repression of Arabidopsis FLC requires promoter sequences but not antisense transcripts. PloS One 6: e21513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hennig L, Taranto P, Walser M, Schonrock N, Gruissem W 2003. Arabidopsis MSI1 is required for epigenetic maintenance of reproductive development. Development 130: 2555–2565. [DOI] [PubMed] [Google Scholar]
- Hennig L, Bouveret R, Gruissem W 2005. MSI1-like proteins: An escort service for chromatin assembly and remodeling complexes. Trends Cell Biol 15: 295–302. [DOI] [PubMed] [Google Scholar]
- Hodl M, Basler K 2012. Transcription in the absence of histone H3.2 and H3K4 methylation. Curr Biol 22: 1–5. [DOI] [PubMed] [Google Scholar]
- Holec S, Berger F 2012. Polycomb group complexes mediate developmental transitions in plants. Plant Phys 158: 35–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh TF, Hakim O, Ohad N, Fischer RL 2003. From flour to flower: How Polycomb group proteins influence multiple aspects of plant development. Trends Plant Sci 8: 439–445. [DOI] [PubMed] [Google Scholar]
- Huang DH, Chang YL, Yang CC, Pan IC, King B 2002. pipsqueak encodes a factor essential for sequence-specific targeting of a Polycomb group protein complex. Mol Cell Biol 22: 6261–6271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ietswaart R, Wu Z, Dean C 2012. Flowering time control: Another window to the connection between antisense RNA and chromatin. Trends Genet 28: 445–53. [DOI] [PubMed] [Google Scholar]
- Jacobs JJ, Scheijen B, Voncken JW, Kieboom K, Berns A, van Lohuizen M 1999. Bmi-1 collaborates with c-Myc in tumorigenesis by inhibiting c-Myc-induced apoptosis via INK4a/ARF. Genes Dev 13: 2678–2690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang D, Wang Y, He Y 2008. Repression of FLOWERING LOCUS C and FLOWERING LOCUS T by the Arabidopsis Polycomb repressive complex 2 components. PLoS One 3: e3404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston AJ, Matveeva E, Kirioukhova O, Grossniklaus U, Gruissem W 2008. A dynamic reciprocal RBR-PRC2 regulatory circuit controls Arabidopsis gametophyte development. Curr Biol 18: 1680–1686. [DOI] [PubMed] [Google Scholar]
- Jullien PE, Katz A, Oliva M, Ohad N, Berger F 2006. Polycomb group complexes self-regulate imprinting of the Polycomb group gene MEDEA in Arabidopsis. Curr Biol 16: 486–492. [DOI] [PubMed] [Google Scholar]
- Kanduri C, Thakur N, Pandey RR 2006. The length of the transcript encoded from the Kcnq1ot1 antisense promoter determines the degree of silencing. EMBO J 25: 2096–2106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanhere A, Viiri K, Araújo CC, Rasaiyaah J, Bouwman RD, Whyte WA, Pereira CF, Brookes E, Walker K, Bell GW, et al. 2010. Short RNAs are transcribed from repressed Polycomb target genes and interact with Polycomb repressive complex-2. Mol Cell 38: 675–688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katsuyama T, Paro R 2011. Epigenetic reprogramming during tissue regeneration. FEBS Lett 585: 1617–1624. [DOI] [PubMed] [Google Scholar]
- Kennison JA 1995. The Polycomb and trithorax group proteins of Drosophila: Trans-regulators of homeotic gene function. Annu Rev Genet 29: 289–303. [DOI] [PubMed] [Google Scholar]
- Kharchenko PV, Alekseyenko AA, Schwartz YB, Minoda A, Riddle NC, Ernst J, Sabo PJ, Larschan E, Gorchakov AA, Gu T, et al. 2010. Comprehensive analysis of the chromatin landscape in Drosophila melanogaster. Nature 471: 480–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King IF, Francis NJ, Kingston RE 2002. Native and recombinant Polycomb group complexes establish a selective block to template accessibility to repress transcription in vitro. Mol Cell Biol 22: 7919–7928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Kingston R, Tamkun J 2014. Transcriptional regulation by trithorax group proteins. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a019349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinoshita T, Harada JJ, Goldberg RB, Fischer RL 2001. Polycomb repression of flowering during early plant development. Proc Natl Acad Sci 98: 14156–14161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klymenko T, Müller J 2004. The histone methyltransferases Trithorax and Ash1 prevent transcriptional silencing by Polycomb group proteins. EMBO Rep 5: 373–377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klymenko T, Papp B, Fischle W, Kocher T, Schelder M, Fritsch C, Wild B, Wilm M, Müller J 2006. A Polycomb group protein complex with sequence-specific DNA-binding and selective methyl-lysine-binding activities. Genes Dev 20: 1110–1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köhler C, Hennig L 2010. Regulation of cell identity by plant Polycomb and trithorax group proteins. Curr Opin Genet Dev 20: 541–547. [DOI] [PubMed] [Google Scholar]
- Köhler C, Hennig L, Bouveret R, Gheyselinck J, Grossniklaus U, Gruissem W 2003a. Arabidopsis MSI1 is a component of the MEA/FIE Polycomb group complex and required for seed development. EMBO J 22: 4804–4814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köhler C, Hennig L, Spillane C, Pien S, Gruissem W, Grossniklaus U 2003b. The Polycomb-group protein MEDEA regulates seed development by controlling expression of the MADS-box gene PHERES1. Genes Dev 17: 1540–1553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köhler C, Page DR, Gagliardini V, Grossniklaus U 2005. The Arabidopsis thaliana MEDEA Polycomb group protein controls expression of PHERES1 by parental imprinting. Nat Genet 37: 28–30. [DOI] [PubMed] [Google Scholar]
- Ku M, Koche RP, Rheinbay E, Mendenhall EM, Endoh M, Mikkelsen TS, Presser A, Nusbaum C, Xie X, Chi AS, et al. 2008. Genomewide analysis of PRC1 and PRC2 occupancy identifies two classes of bivalent domains. PLoS Genet 4: e1000242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurzhals RL, Tie F, Stratton CA, Harte PJ 2008. Drosophila ESC-like can substitute for ESC and becomes required for Polycomb silencing if ESC is absent. Dev Biol 313: 293–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuzmichev A, Margueron R, Vaquero A, Preissner TS, Scher M, Kirmizis A, Ouyang X, Brockdorff N, Abate-Shen C, Farnham P, Reinberg D 2005. Composition and histone substrates of Polycomb repressive group complexes change during cellular differentiation. Proc Natl Acad Sci 102: 1859–1864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagarou A, Mohd-Sarip A, Moshkin YM, Chalkley GE, Bezstarosti K, Demmers JA, Verrijzer CP 2008. dKDM2 couples histone H2A ubiquitylation to histone H3 demethylation during Polycomb group silencing. Genes Dev 22: 2799–2810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landeira D, Sauer S, Poot R, Dvorkina M, Mazzarella L, Jørgensen HF, Pereira CF, Leleu M, Piccolo FM, Spivakov M, et al. 2010. Jarid2 is a PRC2 component in embryonic stem cells required for multi-lineage differentiation and recruitment of PRC1 and RNA Polymerase II to developmental regulators. Nature 12: 618–624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee N, Maurange C, Ringrose L, Paro R 2005. Suppression of Polycomb group proteins by JNK signalling induces transdetermination in Drosophila imaginal discs. Nature 438: 234–237. [DOI] [PubMed] [Google Scholar]
- Lee TI, Jenner RG, Boyer LA, Guenther MG, Levine SS, Kumar RM, Chevalier B, Johnstone SE, Cole MF, Isono K, et al. 2006. Control of developmental regulators by Polycomb in human embryonic stem cells. Cell 125: 301–313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee MG, Villa R, Trojer P, Norman J, Yan K-P, Reinberg D, Di Croce L, Shiekhattar R 2007. Demethylation of H3K27 regulates Polycomb recruitment and H2A ubiquitination. Science 318: 447–450. [DOI] [PubMed] [Google Scholar]
- Leung C, Lingbeek M, Shakhova O, Liu J, Tanger E, Saremaslani P, van Lohuizen M, Marino S 2004. Bmi1 is essential for cerebellar development and is overexpressed in human medulloblastomas. Nature 428: 337–341. [DOI] [PubMed] [Google Scholar]
- Lewis EB 1978. A gene complex controlling segmentation in Drosophila. Nature 276: 565–570. [DOI] [PubMed] [Google Scholar]
- *.Li E, Zhang Y 2014. DNA methylation in mammals. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a019133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Kim YJ, Müller R, Yumul RE, Liu C, Pan Y, Cao X, Goodrich J, Chen X 2011. AGAMOUS terminates floral stem cell maintenance in Arabidopsis by directly repressing WUSCHEL through recruitment of Polycomb Group proteins. Plant Cell 23: 3654–3670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo SM, Ahuja NK, Francis NJ 2009. Polycomb group protein Suppressor 2 of zeste is a functional homolog of Posterior Sex Combs. Mol Cell Biol 29: 515–525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorente MD, Marcos-Gutierrez C, Perez C, Schoorlemmer J, Ramirez A, Magin T, Vidal M 2000. Loss- and gain-of-function mutations show a Polycomb group function for Ring1A in mice. Development 127: 5093–5100. [DOI] [PubMed] [Google Scholar]
- Luo M, Bilodeau P, Koltunow A, Dennis ES, Peacock WJ, Chaudhury AM 1999. Genes controlling fertilization-independent seed development in Arabidopsis thaliana. Proc Natl Acad Sci 96: 296–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mager J, Montgomery ND, de Villena FP, Magnuson T 2003. Genome imprinting regulated by the mouse Polycomb group protein Eed. Nat Genet 33: 502–507. [DOI] [PubMed] [Google Scholar]
- Makarevich G, Leroy O, Akinci U, Schubert D, Clarenz O, Goodrich J, Grossniklaus U, Köhler C 2006. Different Polycomb group complexes regulate common target genes in Arabidopsis. EMBO Rep 7: 947–952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarevich G, Villar CB, Erilova A, Köhler C 2008. Mechanism of PHERES1 imprinting in Arabidopsis. J Cell Sci 121: 906–912. [DOI] [PubMed] [Google Scholar]
- Margueron R, Reinberg D 2011. The Polycomb complex PRC2 and its mark in life. Nature 469: 343–349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margueron R, Li G, Sarma K, Blais A, Zavadil J, Woodcock CL, Dynlacht BD, Reinberg D 2008. Ezh1 and Ezh2 maintain repressive chromatin through different mechanisms. Mol Cell 32: 503–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margueron R, Justin N, Ohno K, Sharpe ML, Son J, Drury WJ 3rd, Voigt P, Martin SR, Taylor WR, De Marco V, et al. 2009. Role of the Polycomb protein EED in the propagation of repressive histone marks. Nature 461: 762–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matharu NK, Hussain T, Sankaranarayanan R, Mishra RK 2010. Vertebrate homologue of Drosophila GAGA factor. J. Mol Biol 400: 434–447. [DOI] [PubMed] [Google Scholar]
- Mendenhall EM, Koche RP, Truong T, Zhou VW, Issac B, Chi AS, Ku M, Bernstein BE 2010. GC-rich sequence elements recruit PRC2 in mammalian ES cells. PLoS Genet 6: e1001244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikkelsen TS, Ku M, Jaffe DB, Issac B, Lieberman E, Giannoukos G, Alvarez P, Brockman W, Kim T-K, Koche RP, et al. 2007. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature 448: 553–560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mito Y, Henikoff JG, Henikoff S 2007. Histone replacement marks the boundaries of cis-regulatory domains. Science 315: 1408–1411. [DOI] [PubMed] [Google Scholar]
- Mohn F, Weber M, Rebhan M, Roloff TC, Richter J, Stadler MB, Bibel M, Schübeler D 2008. Lineage-specific Polycomb targets and de novo DNA methylation define restriction and potential of neuronal progenitors. Mol Cell 30: 755–766. [DOI] [PubMed] [Google Scholar]
- Mosquna A, Katz A, Shochat S, Grafi G, Ohad N 2004. Interaction of FIE, a Polycomb protein, with pRb: A possible mechanism regulating endosperm development. Mol Genet Genomics 271: 651–657. [DOI] [PubMed] [Google Scholar]
- Nekrasov M, Wild B, Müller J 2005. Nucleosome binding and histone methyltransferase activity of Drosophila PRC2. EMBO Rep 6: 348–353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nekrasov M, Klymenko T, Fraterman S, Papp B, Oktaba K, Kocher T, Cohen A, Stunnenberg HG, Wilm M, Müller J 2007. Pcl-PRC2 is needed to generate high levels of H3-K27 trimethylation at Polycomb target genes. EMBO J 26: 4078–4088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohad N, Yadegari R, Margossian L, Hannon M, Michaeli D, Harada JJ, Goldberg RB, Fischer RL 1999. Mutations in FIE, a WD polycomb group gene, allow endosperm development without fertilization. Plant Cell 11: 407–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey RR, Mondal T, Mohammad F, Enroth S, Redrup L, Komorowski J, Nagano T, Mancini-Dinardo D, Kanduri C 2008. Kcnq1ot1 antisense noncoding RNA mediates lineage-specific transcriptional silencing through chromatin-level regulation. Mol Cell 32: 232–246. [DOI] [PubMed] [Google Scholar]
- Pasini D, Bracken AP, Jensen MR, Denchi EL, Helin K 2004. Suz12 is essential for mouse development and for EZH2 histone methyltransferase activity. EMBO J 23: 4061–4071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasini D, Hansen KH, Christensen J, Agger K, Cloos PA, Helin K 2008. Coordinated regulation of transcriptional repression by the RBP2 H3K4 demethylase and Polycomb-Repressive Complex 2. Genes Dev 22: 1345–1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pengelly AR, Copur Ö, Jäckle H, Herzig A, Müller J 2013. A histone mutant reproduces the phenotype caused by loss of histone-modifying factor Polycomb. Science 339: 698–699. [DOI] [PubMed] [Google Scholar]
- Pien S, Fleury D, Mylne JS, Crevillen P, Inze D, Avramova Z, Dean C, Grossniklaus U 2008. ARABIDOPSIS TRITHORAX1 dynamically regulates FLOWERING LOCUS C activation via histone 3 lysine 4 trimethylation. Plant Cell 20: 580–588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posfai E, Kunzmann R, Brochard V, Salvaing J, Cabuy E, Roloff TC, Liu Z, Tardat M, van Lohuizen M, Vidal M, et al. 2012. Polycomb function during oogenesis is required for mouse embryonic development. Genes Dev 26: 920–932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raaphorst FM 2005. Deregulated expression of Polycomb-group oncogenes in human malignant lymphomas and epithelial tumors. Hum Mol Genet 14 Spec No 1: R93–R100. [DOI] [PubMed] [Google Scholar]
- Raissing RT, Baroux C, Grossniklaus U 2011. Regulation and flexibility of genomic imprinting during seed development. Plant Cell 23: 16–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes JC, Grossniklaus U 2003. Diverse functions of Polycomb group proteins during plant development. Semin Cell Dev Biol 14: 77–84. [DOI] [PubMed] [Google Scholar]
- Ringrose L, Rehmsmeier M, Dura JM, Paro R 2003. Genome-wide prediction of Polycomb/Trithorax response elements in Drosophila melanogaster. Dev Cell 5: 759–771. [DOI] [PubMed] [Google Scholar]
- *.Rinn JL 2014. lncRNAs: Linking RNA to chromatin. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a018614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinn JL, Kertesz M, Wang JK, Squazzo SL, Xu X, Brugmann SA, Goodnough LH, Helms JA, Farnham PJ, Segal E, et al. 2007. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell 129: 1311–1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarma K, Margueron R, Ivanov A, Pirrotta V, Reinberg D 2008. Ezh2 requires PHF1 to efficiently catalyze H3 lysine 27 trimethylation in vivo. Mol Cell Biol 28: 2718–2731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauvageau M, Sauvageau G 2010. Polycomb group proteins: Multi-faceted regulators of somatic stem cells and cancer. Cell Stem Cell 7: 299–313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheuermann JC, Gaytán de Ayala Alonso A, Oktaba K, Ly-Hartig N, McGinty RK, Fraterman S, Wilm M, Muir TW, Müller J 2010. Histone H2A deubiquitinase activity of the Polycomb repressive complex PR-DUB. Nature 465: 243–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt A, Wöhrmann HJ, Raissig MT, Arand J, Gheyselinck J, Gagliardini V, Heichinger C, Walter J, Grossniklaus U 2013. The Polycomb group protein MEDEA and the DNA methyltransferase MET1 interact to repress autonomous endosperm development in Arabidopsis. Plant J 73: 776–787. [DOI] [PubMed] [Google Scholar]
- Schmitt S, Prestel M, Paro R 2005. Intergenic transcription through a Polycomb group response element counteracts silencing. Genes Dev 19: 697–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoeftner S, Sengupta AK, Kubicek S, Mechtler K, Spahn L, Koseki H, Jenuwein T, Wutz A 2006. Recruitment of PRC1 function at the initiation of X inactivation independent of PRC2 and silencing. EMBO J 25: 3110–3122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schubert D, Primavesi L, Bishopp A, Roberts G, Doonan J, Jenuwein T, Goodrich J 2006. Silencing by plant Polycomb-group genes requires dispersed trimethylation of histone H3 at lysine 27. EMBO J 25: 4638–4649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuettengruber B, Ganapathi M, Leblanc B, Portoso M, Jaschek R, Tolhuis B, van Lohuizen M, Tanay A, Cavalli G 2009. Functional anatomy of Polycomb and trithorax chromatin landscapes in Drosophila embryos. PLoS Biol 7: e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz YB, Pirrotta V 2008. Polycomb complexes and epigenetic states. Curr Opin Cell Biol 20: 266–273. [DOI] [PubMed] [Google Scholar]
- Schwartz YB, Kahn TG, Nix DA, Li XY, Bourgon R, Biggin M, Pirrotta V 2006. Genome-wide analysis of Polycomb targets in Drosophila melanogaster. Nat Genet 38: 700–705. [DOI] [PubMed] [Google Scholar]
- *.Seto E, Yoshida M 2014. Erasers of histone acetylation: The histone deacetylase enzymes. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a018713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao Z, Raible F, Mollaaghababa R, Guyon JR, Wu CT, Bender W, Kingston RE 1999. Stabilization of chromatin structure by PRC1, a Polycomb complex. Cell 98: 37–46. [DOI] [PubMed] [Google Scholar]
- Sheldon CC, Conn AB, Dennis ES, Peacock WJ 2002. Different regulatory regions are required for the vernalization-induced repression of FLOWERING LOCUS C and for the epigenetic maintenance of repression. Plant Cell 14: 2527–2537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shih AH, Abdel-Wahab O, Patel JP, Levine RL 2012. The role of mutations in epigenetic regulators in myeloid malignancies. Nat Rev Can 12: 599–612. [DOI] [PubMed] [Google Scholar]
- Simon J, Chiang A, Bender W, Shimell MJ, O’Connor M 1993. Elements of the Drosophila bithorax complex that mediate repression by Polycomb group products. Dev Biol 158: 131–144. [DOI] [PubMed] [Google Scholar]
- Sinclair DA, Syrzycka M, Macauley MS, Rastgardani T, Komljenovic I, Vocadlo DJ, Brock HW, Honda BM 2009. Drosophila O-GlcNAc transferase (OGT) is encoded by the Polycomb group (PcG) gene, super sex combs (sxc). Proc Natl Acad Sci 106: 13427–13432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sing A, Pannell D, Karaiskakis A, Sturgeon K, Djabali M, Ellis J, Lipshitz HD, Cordes SP 2009. A vertebrate Polycomb response element governs segmentation of the posterior hindbrain. Cell 138: 885–897. [DOI] [PubMed] [Google Scholar]
- Smith ER, Lee MG, Winter B, Droz NM, Eissenberg JC, Shiekhattar R, Shilatifard A 2008. Drosophila UTX is a histone H3 Lys27 demethylase that colocalizes with the elongating form of RNA polymerase II. Mol Cell Biol 28: 1041–1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Squazzo SL, O’Geen H, Komashko VM, Krig SR, Jin VX, Jang SW, Margueron R, Reinberg D, Green R, Farnham PJ 2006. Suz12 binds to silenced regions of the genome in a cell-type-specific manner. Genome Res 16: 890–900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stock JK, Giadrossi S, Casanova M, Brookes E, Vidal M, Koseki H, Brockdorff N, Fisher AG, Pombo A 2007. Ring1-mediated ubiquitination of H2A restrains poised RNA polymerase II at bivalent genes in mouse ES cells. Nat Cell Biol 9: 1428–1435. [DOI] [PubMed] [Google Scholar]
- *.Strome S, Kelly WG, Ercan S, Lieb JD 2014. Regulation of the X chromosomes in C. elegans. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a018366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung S, Amasino RM 2004. Vernalization in Arabidopsis thaliana is mediated by the PHD finger protein VIN3. Nature 427: 159–164. [DOI] [PubMed] [Google Scholar]
- Swiezewski S, Liu F, Magusin A, Dean C 2009. Cold-induced silencing by long antisense transcripts of an Arabidopsis Polycomb target. Nature 462: 799–802. [DOI] [PubMed] [Google Scholar]
- Tavares L, Dimitrova E, Oxley D, Webster J, Poot R, Demmers J, Bezstarosti K, Taylor S, Ura H, Koide H, et al. 2012. RYBP-PRC1 complexes mediate H2A ubiquitylation at Polycomb target sites independently of PRC2 and H3K27me3. Cell 148: 664–678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terranova R, Yokobayashi S, Stadler MB, Otte AP, van Lohuizen M, Orkin SH, Peters AH 2008. Polycomb group proteins Ezh2 and Rnf2 direct genomic contraction and imprinted repression in early mouse embryos. Dev Cell 15: 668–679. [DOI] [PubMed] [Google Scholar]
- Tie F, Furuyama T, Prasad Sinha J, Jane E, Harte PJ 2001. The Drosophila Polycomb group proteins ESC and E(Z) are present in a complex containing the histone-binding protein p55 and the histone deacetylase RPD3. Development 128: 275–286. [DOI] [PubMed] [Google Scholar]
- Tie F, Banerjee R, Stratton CA, Prasad-Sinha J, Stepanik V, Zlobin A, Diaz MO, Scacheri PC, Harte PJ 2009. CBP-mediated acetylation of histone H3 lysine 27 antagonizes Drosophila Polycomb silencing. Development 136: 3131–3141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tschiersch B, Hofmann A, Krauss V, Dorn R, Korge G, Reuter G 1994. The protein encoded by the Drosophila position-effect variegation suppressor gene Su(var)3-9 combines domains of antagonistic regulators of homeotic gene complexes. EMBO J 13: 3822–3831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turck F, Roudier F, Farrona S, Martin-Magniette M-L, Guillaume E, Buisine N, Gagnot S, Martienssen RA, Coupland G, Colot V 2007. Arabidopsis TFL2/LHP1 specifically associates with genes marked by trimethylation of histone H3 lysine 27. PLoS Genet 3: e86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valk-Lingbeek ME, Bruggeman SW, van Lohuizen M 2004. Stem cells and cancer; the Polycomb connection. Cell 118: 409–418. [DOI] [PubMed] [Google Scholar]
- van der Lugt NM, Domen J, Linders K, van Roon M, Robanus Maandag E, te Riele H, van der Valk M, Deschamps J, Sofroniew M, van Lohuizen M, et al. 1994. Posterior transformation, neurological abnormalities, and severe hematopoietic defects in mice with a targeted deletion of the bmi-1 proto-oncogene. Genes Dev 8: 757–769. [DOI] [PubMed] [Google Scholar]
- van Lohuizen M, Frasch M, Wientjens E, Berns A 1991. Sequence similarity between the mammalian bmi-1 protooncogene and the Drosophila regulatory genes Psc and Su(z)2. Nature 353: 353–355. [DOI] [PubMed] [Google Scholar]
- Wang L, Brown JL, Cao R, Zhang Y, Kassis JA, Jones RS 2004a. Hierarchical recruitment of Polycomb group silencing complexes. Mol Cell 14: 637–646. [DOI] [PubMed] [Google Scholar]
- Wang H, Wang L, Erdjument-Bromage H, Vidal M, Tempst P, Jones RS, Zhang Y 2004b. Role of histone H2A ubiquitination in Polycomb silencing. Nature 431: 873–878. [DOI] [PubMed] [Google Scholar]
- Wang L, Jahren N, Vargas ML, Andersen EF, Benes J, Zhang J, Miller EL, Jones RS, Simon JA 2006. Alternative ESC and ESC-like subunits of a Polycomb group histone methyltransferase complex are differentially deployed during Drosophila development. Mol Cell Biol 26: 2637–2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weake VM, Workman JL 2008. Histone ubiquitination: Triggering gene activity. Mol Cell 29: 653–663. [DOI] [PubMed] [Google Scholar]
- Whitcomb SJ, Basu A, Allis CD, Bernstein E 2007. Polycomb group proteins: An evolutionary perspective. Trends Genet 23: 494–502. [DOI] [PubMed] [Google Scholar]
- Wöhrmann HJ, Gagliardini V, Raissig MT, Wehrle W, Arand J, Schmidt A, Tierling S, Page DR, Schöb H, Walter J, et al. 2012. Identification of a DNA methylation-independent imprinting control region at the Arabidopsis MEDEA locus. Genes Dev 26: 1837–1850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo CJ, Kharchenko PV, Daheron L, Park PJ, Kingston RE 2010. A region of the human HOXD cluster that confers Polycomb-group responsiveness. Cell 140: 99–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood CC, Robertson M, Tanner G, Peacock WJ, Dennis ES, Helliwell CA 2006. The Arabidopsis thaliana vernalization response requires a Polycomb-like protein complex that also includes VERNALIZATION INSENSITIVE-3. Proc Natl Acad Sci 103: 14631–14636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu HA, Bernstein E 2008. Partners in imprinting: Noncoding RNA and Polycomb group proteins. Dev Cell 15: 637–638. [DOI] [PubMed] [Google Scholar]
- Xu L, Shen W-H 2008. Polycomb silencing of KNOX genes confines shoot stem cell niches in Arabidopsis. Curr Biol 18: 1966–1971. [DOI] [PubMed] [Google Scholar]
- Yamamoto K, Sonoda M, Inokuchi J, Shirasawa S, Sasazuki T 2004. Polycomb group suppressor of zeste 12 links heterochromatin protein 1α and enhancer of zeste 2. J Biol Chem 279: 401–406. [DOI] [PubMed] [Google Scholar]
- Yap KL, Li S, Muñoz-Cabello AM, Raguz S, Zeng L, Mujtaba S, Gil J, Walsh MJ, Zhou M-M 2010. Molecular interplay of the noncoding RNA ANRIL and methylated histone H3 lysine 27 by Polycomb CBX7 in transcriptional silencing of INK4a. Mol Cell 38: 662–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida N, Yanai Y, Chen L, Kato Y, Hiratsuka J, Miwa T, Sung ZR, Takahashi S 2001. EMBRYONIC FLOWER2, a novel Polycomb group protein homolog, mediates shoot development and flowering in Arabidopsis. Plant Cell 13: 2471–2481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zencak D, Lingbeek M, Kostic C, Tekaya M, Tanger E, Hornfeld D, Jaquet M, Munier FL, Schorderet DF, van Lohuizen M, et al. 2005. Bmi1 loss produces an increase in astroglial cells and a decrease in neural stem cell population and proliferation. J Neurosci 25: 5774–5783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Germann S, Blus BJ, Khorasanizadeh S, Gaudin V, Jacobsen SE 2007. The Arabidopsis LHP1 protein colocalizes with histone H3 Lys27 trimethylation. Nat Struct Mol Biol 14: 869–871. [DOI] [PubMed] [Google Scholar]
- Zhao J, Ohsumi TK, Kung JT, Ogawa Y, Grau DJ, Sarma K, Song JJ, Kingston RE, Borowsky M, Lee JT 2010. Genome-wide identification of Polycomb-associated RNAs by RIP-seq. Mol Cell 40: 939–953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou W, Zhu P, Wang J, Pascual G, Ohgi KA, Lozach J, Glass CK, Rosenfeld MG 2008. Histone H2A monoubiquitination represses transcription by inhibiting RNA polymerase II transcriptional elongation. Mol Cell 29: 69–80. [DOI] [PMC free article] [PubMed] [Google Scholar]