Abstract
Aneuploidy, defined as an abnormal number of chromosomes, is a hallmark of cancer. Paradoxically, aneuploidy generally has a negative impact on cell growth and fitness in nontransformed cells. In this work, we review recent progress in identifying how aneuploidy leads to genomic and chromosomal instability, how cells can adapt to the deleterious effects of aneuploidy, and how aneuploidy contributes to tumorigenesis in different genetic contexts. Finally, we also discuss how aneuploidy might be a target for anticancer therapies.
Aneuploidy is a common characteristic of cancer cells yet has a negative impact on normal cell physiology. Understanding the various molecular pathways that lead to aneuploidy is therefore critical.
As Horace famously wrote in his Odes, the “golden mean” is the secret to a happy, balanced life. Recent work, reviewed here, emphasizes the importance of this kind of balance for the genetics of human cells.
Maintaining a stable genome is critical for the preservation of genetic information during the life span of an organism. Despite mechanisms designed to ensure a diploid karyotype, errors can and do occur during chromosome segregation that result in the gain and loss of whole chromosomes. In vitro estimates suggest that normal, diploid cells missegregate a chromosome once every 100 cell divisions (Thompson and Compton 2008). The in vivo rate of chromosome missegregation is unknown, but could vary between different cell types. Even if the rate is low, an abnormal number of chromosomes, or aneuploidy, could have a significant impact on normal cell physiology, as well as tumorigenesis.
Aneuploidy, at the level of the organism, is detrimental and generally incompatible with life. In humans, only three aneuploidies—trisomy 13, 18, and 21—are viable, and only trisomy 21 is compatible with a life span beyond infancy (Hassold et al. 2007). Despite the deleterious consequences of aneuploidy in normal physiological contexts, an abnormal number of chromosomes is one of the hallmarks of cancer cells. Aneuploidy is found in ∼90% of solid tumors and >50% of blood cancers (Beroukhim et al. 2010; Mitelman et al. 2013). Whether aneuploidy is a cause or consequence of cell transformation is a frequent topic of debate. The challenge for establishing a causal relationship stems from the complexity of cancer cells, in which numerical chromosome abnormalities are rarely found in isolation but are usually accompanied by other genomic alterations, such as point mutations, translocations, and microsatellite instability. This complexity makes it difficult to define the initiating event(s) in tumorigenesis.
This review focuses on whole-chromosome aneuploidy, although it has been shown that the gain and loss of chromosome arms is also a common occurrence in cancer cells (Beroukhim et al. 2010; Mitelman et al. 2013). We review the molecular pathways leading to aneuploidy, the effects of aneuploidy on cellular physiology, and the links between aneuploidy and tumorigenesis. Finally, we also explore the exciting concept of targeting aneuploidy as a novel therapeutic approach in treating cancer.
CHROMOSOMAL INSTABILITY AND ANEUPLOIDY
Chromosomal instability (CIN) and aneuploidy are both common features of many cancers (Chandhok and Pellman 2009; Thompson et al. 2010; Thompson and Compton 2011b). However, it is important to note that CIN and aneuploidy are not synonymous. Aneuploidy refers to the state of the karyotype and denotes a chromosome number that deviates from the normal diploid number. CIN, on the other hand, refers to the rate of chromosome gains and losses, without taking into account the specific karyotype of a cell (Geigl et al. 2008). Consequently, not all aneuploid cells display CIN, with some aneuploid cells exhibiting a stable and unchanging karyotype. For example, individuals with trisomy 21, known as Down syndrome, exhibit an abnormal but stable karyotype that is defined by an extra copy of chromosome 21.
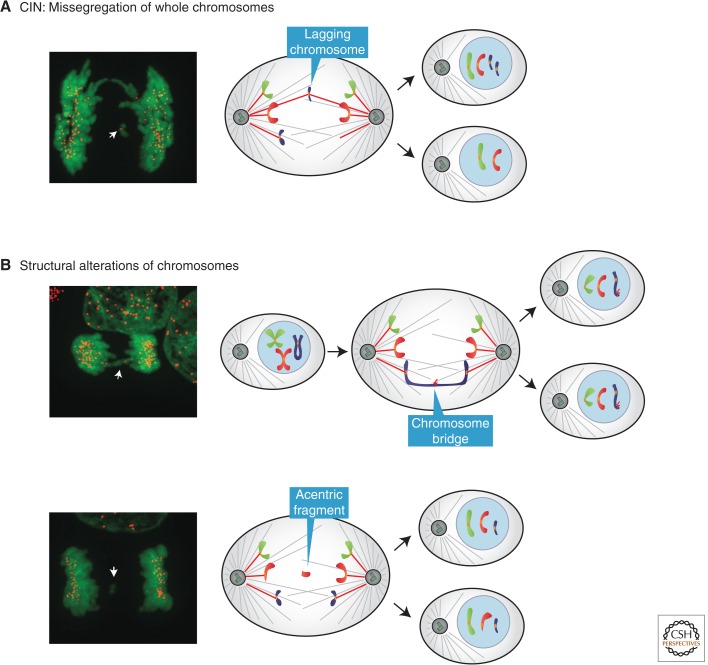
Having made the important distinction between CIN and aneuploidy, in light of recent literature it remains important to further clarify how we define and measure CIN. One can view “chromosomal instability” narrowly, as an elevated rate of missegregation of intact, whole chromosomes (Fig. 1A), or broadly, to include the missegregation of large “chunks” of chromosomes (Fig. 1B). This broader definition of CIN would include the generation of acentric chromosome fragments, which are broken chromosomes that lack centromeres and thus cannot attach to the mitotic spindle. CIN often also includes so-called chromosome bridges that result from incomplete DNA replication, telomere end-to-end fusions, or incompletely decatenated chromosomes (Terradas et al. 2010; Fenech et al. 2011). Although the resolution of chromosome bridges is incompletely understood, it is widely assumed that they are broken either during cytokinesis (Janssen et al. 2011) or in the subsequent cell cycle (McClintock 1941; De Lange 2005; Gisselsson 2008).
Figure 1.
Definition of chromosomal instability. (A) In this review, we refer to CIN as an elevated rate of whole-chromosome missegregation. CIN is often operationally defined as lagging, centromere-positive chromosomes at anaphase. Note that this operational definition cannot exclude the loss of the distal ends of chromosomes and thus does not completely exclude structural alterations to the chromosomes. (B) In some studies, CIN has been defined more broadly to include structural chromosome defects. These structural alterations include chromosome bridges or acentric chromosome fragments, which cannot attach to the spindle apparatus and are thus often partitioned into micronuclei. In this review, we adopt the narrow definition of CIN, excluding chromosome bridges and acentric chromosome fragments. (Fluorescence images from Ganem et al. 2009; reprinted, with permission, from the author.)
An illustration of the use of a broader definition of CIN is a recent study from Swanton and colleagues (Burrell et al. 2013). This paper suggested that the major cause of “CIN” is DNA replication stress. Because of the potential for replication stress to trigger DNA breaks and underreplicated chromosome segments, it is expected that replication stress would generate acentric chromosome fragments and chromosome bridges. It is also interesting to consider the possibility that replication stress, through novel mechanisms, would also affect the fidelity of mitosis and the distribution of intact chromosomes. However, in this study, when replication stress was induced by aphidicolin treatment, the frequency of missegregated whole chromosomes (lagging chromosomes; see below) was not significantly affected, whereas there was a large increase in chromosome bridges and acentric fragments. This supports the conventional view that replication stress mainly causes structural alterations of chromosomes, whereas the missegregation of intact chromosomes mainly results from mitotic errors. Because of this difference in the underlying mechanism, in this review we adopt the narrow definition of CIN as impacting the segregation of intact chromosomes.
We can in principle make a sharp distinction between the narrow and broad definitions of CIN, but in practice is it always clear? In fact, the measurement of CIN is not trivial, and it is limited to cultured cells because of technical constraints. CIN is the missegregation rate per generation. One classic way to assay for CIN is to use fluorescence in situ hybridization (FISH) of a clone of cells after a few generations, enabling an estimation of the rate of missegregation from the frequency of aneuploidy in the population (Lengauer et al. 1997; Thompson and Compton 2008). A more direct way is to perform FISH on pairs of cells immediately after mitosis (Cimini et al. 1999; Thompson and Compton 2008). This latter assay provides a direct per-generation measure of missegregation rates. These methods are valuable but have a caveat. Unless pairs of FISH probes detecting both chromosome arms are used, the methods cannot rigorously distinguish the missegregation of whole chromosomes from that of acentric chromosome fragments. Another useful common approach is to image anaphase cells to detect lagging chromosomes (Thompson and Compton 2008, 2011a; Bakhoum et al. 2009a; Ganem et al. 2009; Silkworth et al. 2009). Lagging chromosomes result from mitotic errors in which a chromatid is incorrectly attached to microtubules from opposite poles—termed merotelic attachments (Cimini et al. 2001; Cimini 2008; Gregan et al. 2011). Like anaphase FISH, it has the advantage of being a per-generation measurement and thus a “rate.” However, it is important to note that many lagging chromosomes are ultimately correctly segregated (Cimini et al. 2003, 2004; Gregan et al. 2011), so without extensive live-cell measurements, it is not possible to directly relate the frequency of lagging chromosomes to the rate of chromosome missegregation. Moreover, distinguishing lagging chromosomes (discrete chromosomes situated between the separating chromosome masses) from chromosome bridges (chromosomes stretched between the separating chromosome masses) requires high-quality images and careful analysis.
MOLECULAR MECHANISMS LEADING TO ANEUPLOIDY
Defects in multiple biological pathways can cause the missegregation of chromosomes. This includes abnormalities in the spindle assembly checkpoint (SAC), aberrant mitotic spindle geometry, abnormal microtubule–kinetochore attachments, and defects in sister chromatid cohesion. The persistence of any of these defects causes CIN.
Defects in the SAC
Defects in the SAC, which arrests cells with improper kinetochore–spindle attachments, can lead to CIN and aneuploidy. For proper chromosome segregation during mitosis, each sister chromatid must interact with microtubules originating from only one pole of the spindle. This configuration is known as biorientation. On a molecular level, the SAC inhibits the anaphase-promoting complex/cyclosome (APC/C), an E3 ubiquitin ligase required for mitotic exit and chromosome segregation (Primorac and Musacchio 2013). Inhibition of APC/C is mediated by the mitotic checkpoint complex, which is composed of the checkpoint proteins MAD2, BUBR1 and BUB3, and the APC/C coactivator CDC20. The SAC also requires several other proteins to function properly, including MAD1 and the BUB1 kinase (Musacchio and Salmon 2007).
Strong abrogation of the SAC in mice causes mitotic exit in the presence of nonbioriented chromosomes, widespread aneuploidy, and early embryonic lethality (Dobles et al. 2000; Kalitsis et al. 2000; Michel et al. 2001; Putkey et al. 2002). In humans, mutations in the SAC gene BUBR1 have been implicated in the pathogenesis of mosaic variegated aneuploidy, a disease characterized by constitutional mosaic aneuploidy and an increased predisposition to cancer (Hanks et al. 2004; Matsuura et al. 2006). More recently, mutations in the microtubule-interacting protein CEP57 were also identified as a cause of mosaic variegated aneuploidy, suggesting that that this rare clinical syndrome may be caused by defects in different proteins with diverse functions (Snape et al. 2011).
Although these mouse models demonstrate that SAC defects can lead to tumorigenesis, mutations in SAC genes are quite rare in human cancers. In addition, there is not a clear correlation in these mice between the extent of CIN and the risk of developing cancer. Furthermore, many CIN cancer cell lines that were originally believed to have SAC defects were shown to have normal checkpoints (Tighe et al. 2001; Gascoigne and Taylor 2008).
Although mutations in SAC genes are infrequent in human cancer, alterations in the expression levels of SAC components are observed in a wide spectrum of tumors (Weaver and Cleveland 2006). Reduced levels of checkpoint components can weaken the SAC to an extent that results in aneuploidy but is still compatible with cell viability (Michel et al. 2001; Kops et al. 2004; Iwanaga et al. 2007; Weaver et al. 2007), which is often severely compromised by the complete loss of SAC components (Dobles et al. 2000; Kalitsis et al. 2000; Michel et al. 2001; Putkey et al. 2002). Multiple mechanisms, both transcriptional and posttranscriptional, have been reported to result in reduced levels of SAC proteins in tumors. For example, BRCA1 inactivation results in the decreased transcription of the MAD2 gene (Wang et al. 2004), the hypermethylation of the BUBR1 promoter results in reduced BUBR1 protein levels (Park et al. 2007), and the increased expression of breast cancer–specific gene 1 (BCSG1) stimulates the proteasome-mediated degradation of BUBR1 (Gupta et al. 2003).
The overexpression of SAC genes has also been reported in human tumors (Tanaka et al. 2001; Li et al. 2003; Hernando et al. 2004; Hayama et al. 2006) and can lead to aneuploidy and tumorigenesis in mouse models (Sotillo et al. 2007, 2010). This apparently counterintuitive observation can, in some cases, be explained by checkpoint-independent roles of SAC proteins in regulating chromosome segregation. For example, the inactivation of the RB or P53 tumor suppressors leads to the transcriptional up-regulation of MAD2 (Hernando et al. 2004; Schvartzman et al. 2011). MAD2 overexpression, in turn, increases the stability of kinetochore–microtubule attachments, which can lead to chromosome segregation errors (Kabeche and Compton 2012). Additionally, MAD2 overexpression can also lead to tetraploidy in mice (Sotillo et al. 2007), probably because a stronger SAC can lead to a sustained mitotic arrest, followed by mitotic escape in the absence of cytokinesis (Brito and Rieder 2006). Tetraploid cells have been shown to promote tumorigenesis in mice (Fujiwara et al. 2005) and can have multiple centrosomes, which increases the chance of forming merotelic attachments and developing aneuploidy (see below). Additionally, genes involved in SAC inactivation and mitotic progression, such as UBCH10 and CUEDC2, can lead to aneuploidy when overexpressed in cell lines and/or mouse models and are up-regulated in some human tumors (Reddy et al. 2007; van Ree et al. 2010; Gao et al. 2011; Xie et al. 2014). Taking into account these observations, it is likely that we are underestimating the frequency of alterations that impinge on the SAC in human tumors. Thus, the extent and role of SAC defects in human cancer will need to be clarified in future research.
Merotelic Attachments
Although the SAC delays anaphase onset in the absence of biorientation, there is a class of aberrant microtubule–kinetochore attachments, known as merotelic attachments, that are not detected by the SAC. Merotelic attachments occur when a single kinetochore attaches to microtubules that arise from both poles of the spindle. At the metaphase–anaphase transition, when the sister chromatids are physically separated to opposite poles, a chromosome attached to both of the poles is caught in a tug-of-war and can end up as a lagging chromosome (Cimini et al. 2001; Cimini 2008; Gregan et al. 2011). Most of the merotelic chromosomes in anaphase are correctly segregated (Cimini et al. 2003, 2004); however, a fraction of merotelic chromosomes lag at the spindle equator and can missegregate, generating aneuploid daughter cells (Cimini et al. 2001, 2003; Cimini 2008; Gregan et al. 2011).
Merotelic attachments are frequently observed in CIN cells (Thompson et al. 2010) and can be generated by different mechanisms, including centrosome amplification (Ganem et al. 2009) and hyperstabilized microtubule–kinetochore attachments (Bakhoum et al. 2009b). Centrosome amplification occurs frequently in vivo in cancer, is correlated with CIN, and can generate multipolar spindles in mitosis. However, multipolar cell divisions are rare because these multipolar spindles are often transient intermediates. Cancer cells with centrosome amplification usually cluster the extra centrosomes during mitosis to form a pseudobipolar spindle. This centrosome clustering allows the cells to divide with a bipolar spindle but results in an increased frequency of merotelic attachments (Ganem et al. 2009; Silkworth et al. 2009). Spindle multipolarity can also arise on spindle pole fragmentation, as after the depletion of the microtubule-associated protein TOGp or of the microtubule-, kinetochore-, and centrosome-associated proteins CLASP1 and -2 (Cassimeris and Morabito 2004; Logarinho et al. 2012).
Hyperstabilization of microtubule–kinetochore attachments can also cause merotelic attachments. During mitosis, erroneous attachments are normally corrected through mechanisms involving the destabilization of attachments by the Aurora B kinase (Andrews et al. 2004; Pinsky et al. 2006) and the microtubule depolymerases MCAK and KIF2B, which increase microtubule turnover at kinetochores (Bakhoum et al. 2009b). Consequently, the hyperstabilization of kinetochore–microtubule attachments results in inefficient correction of attachment errors, a higher rate of lagging chromosomes, and aneuploidy (Bakhoum et al. 2009b). Studies by Compton and coworkers have shown that CIN cell lines exhibit more stable kinetochore–microtubule attachments and lagging chromosomes than nontumor, diploid cells (Thompson and Compton 2008; Bakhoum et al. 2009a). Whether this observation extends to human cancers in vivo, however, remains untested and technically challenging to verify. It would also be important to understand the mechanism through which microtubules become hyperstabilized in cancer cells. Because of the deleterious effects of aneuploidy, it seems unlikely that cancer cells would be under direct selection for inaccurate chromosome segregation. Perhaps, however, microtubule stabilization is an obligatory accompaniment to some oncogenic mutations. In this case, the “benefit” of the growth-promoting mutation might outweigh the negative consequences that arise from the accompanying aneuploidy.
Cohesion Defects
The regulation of cohesion between sister chromatids is also required for the proper segregation of chromosomes. Specifically, the cohesin complex must be removed at the metaphase–anaphase transition (Musacchio and Salmon 2007; Nasmyth 2011). Sequencing of human homologs of budding yeast CIN genes in colorectal tumors has revealed mutations in four genes involved in chromosome cohesion, suggesting a link between cohesion and genome stability (Barber et al. 2008).
More recently, deletions or inactivating mutations in the STAG2 gene have been discovered in a number of different tumor types and human cancer cell lines, including glioblastoma, Ewing’s sarcoma, and acute myeloid leukemia (AML) (Walter et al. 2009; Rocquain et al. 2010; Solomon et al. 2011). STAG2 is the human ortholog of the yeast gene SCC3 and encodes a structural subunit of the cohesin complex, which physically holds together the sister chromatids. Inactivating STAG2 in non-CIN human cancer cell lines leads to decreased cohesion and aneuploidy (Solomon et al. 2011), which suggests a causal link between defects in chromosome cohesion, aneuploidy, and tumorigenesis.
However, STAG2 inactivation is not invariably linked to aneuploidy. Genome sequencing of 183 AML samples revealed mutations in STAG2 and the other cohesin genes, SMC3, SMC1A, and RAD21, in 19 samples, only one of which displayed CIN (Welch et al. 2012). Consequently, the mechanisms underlying the relationship between STAG2 inactivation and tumorigenesis might be tissue specific and more complex than expected, especially in light of the additional roles of STAG2 beyond chromatid cohesion, including transcriptional regulation (Dorsett 2011). However, even in tumors in which a STAG2 mutation drives tumorigenesis independently of aneuploidy, it would still be of interest to know whether the STAG2 mutation confers sensitivity to aneuploidy-targeting therapies.
It is unknown which of the different mechanisms leading to aneuploidy in vitro are most relevant in vivo. Animal models with defects in each of these pathways will provide useful information about their contribution to aneuploidy. Mice with mutations in SAC genes have been generated, but a defective mitotic checkpoint is not frequently observed in human cancers (Cahill et al. 1998; Myrie et al. 2000; Haruki et al. 2001; Gascoigne and Taylor 2008). Mouse models with extra centrosomes may be highly informative, given the frequent occurrence of multiple centrosomes in cancers. Overexpression of the kinase PLK4 drives centrosome amplification both in human cells in culture (Habedanck et al. 2005) and in a mouse model in which the restricted overexpression of Plk4 in the central nervous system caused microcephaly (Marthiens et al. 2013). Mice with inducible expression of Plk4 in different tissues would be an interesting model in which to study the effects of extra centrosomes on tumorigenesis in vivo. Regarding the role of merotelic attachments in the generation of aneuploidy, depletion of MCAK or KIF2B leads to the hyperstabilization of kinetochore–microtubule attachments (Bakhoum et al. 2009b), suggesting a strategy for animal models with increased merotely. Finally, given the unclear roles of cohesion defects in generating aneuploidy, mice with knockin of the cohesin gene mutations identified in human cancers could reveal whether aneuploidy develops in this genetic context, and if so, in what tissues and at what stage of tumor development.
CELLULAR EFFECTS OF ANEUPLOIDY
The deleterious effects of aneuploidy are well described at the level of the organism in a number of different species. Despite the challenges of investigating the consequences of aneuploidy on cell physiology, recent work has started to define the impact of aneuploidy on both budding yeast and mammalian cells.
Effects of Aneuploidy on Cell Fitness
Two groups have independently generated aneuploid yeast strains using different experimental methods (Torres et al. 2007; Pavelka et al. 2010). All of the aneuploid cells displayed growth defects and altered metabolism compared with the euploid, isogenic counterparts. The aneuploid yeast also showed a delay in the G1 phase of the cell cycle and increased glucose uptake. The deleterious consequences of aneuploidy are a result of imbalanced gene expression, as evidenced by the fact that artificial chromosomes containing human or mouse DNA do not have these effects on yeast (Torres et al. 2007).
The Amon laboratory also used mice with Robertsonian translocations to generate mouse embryonic fibroblast (MEF) cell lines with aneuploidy (trisomy) for specific chromosomes (Williams et al. 2008). The aneuploid MEFs exhibited impaired proliferation and metabolic abnormalities relative to the diploid MEFs. In general, the extent of growth inhibition correlated with the size of the extra chromosome (Torres et al. 2007; Williams et al. 2008). These results in MEFs are consistent with the observation that fibroblasts from human patients with trisomy 21 proliferate more slowly than diploid cells (Segal and McCoy 1974).
Effects of Aneuploidy on Transcription and Protein Composition
Recently, an innovative strategy based on the genetic insertion of the X-inactivation gene (XIST) has allowed the silencing of an extra chromosome 21 in induced pluripotent stem cells from Down syndrome patients (Jiang et al. 2013). Chromosome silencing results in an increase in the cell growth rate of 18%–34% compared with isogenic, nonsilenced cells. This result strongly suggests that transcription is required for aneuploidy to manifest its effects.
In budding yeast and mouse embryonic fibroblasts, aneuploid cells display a proportional increase in transcription of the genes on the extra chromosome (Torres et al. 2007; Williams et al. 2008). Different aneuploid yeast strains also show increased expression of common genes that are not located on the aneuploid chromosomes. In particular, these genes are involved in ribosome biogenesis and nucleic acid metabolism (Torres et al. 2007), a gene expression signature that is typical of the environmental stress response (Gasch et al. 2000) and observed when yeast grow under stress conditions or at slow rates. However, in a second study conducted with aneuploid yeast, the environmental stress response gene enrichment signature was less apparent and not correlated with either growth rate or number of aneuploid chromosomes (Pavelka et al. 2010). Although both studies suggest that aneuploidy can induce a general and non-chromosome-specific transcriptional response in yeast, the cause, extent, and nature of this response remain unclear. Further studies by Amon and coworkers have shown that stress response is an evolutionally widespread transcriptional consequence of aneuploidy, observed not only in budding yeast but also in fission yeast, plants, mice, and humans (Sheltzer et al. 2012).
The effects of aneuploidy on the proteome are even less clear. One study showed that aneuploid yeast strains do not show a correlation at the level of the proteome between gene copy number and protein level (Torres et al. 2007). Interestingly, many of the proteins whose levels do not correlate with gene number are part of multiprotein complexes. Consequently, one hypothesis suggests that aneuploid cells activate the chaperone and proteolytic pathways in an attempt to preserve protein complex stoichiometry. This increased protein production and degradation imposes an energetic burden on the cells, possibly causing the growth defects and higher energetic needs observed in the aneuploid strains. Proteotoxic stress could explain the increased sensitivity that some aneuploid cells show to inhibitors of protein synthesis, degradation, or folding (Torres et al. 2007; Tang et al. 2011).
The presence of dosage compensation at the level of the proteome in aneuploid yeast was not confirmed in a second study (Pavelka et al. 2010). The difference in these two studies may be related to technical differences in how the protein levels were measured or to yeast strain stability. Additional work has confirmed the activation of proteotoxic stress in aneuploid budding yeast (Oromendia et al. 2012). Disomic budding yeast strains show protein aggregates and reduced folding capacity of chaperones, corroborating the model of gene dosage compensation at the protein level in aneuploid cells. Moreover, a recent study in human cells further supports a model of proteome compensation to restore protein balance in trisomic and tetrasomic cells (Stingele et al. 2012). Storchova and coworkers found that approximately one-quarter of the proteins encoded on the extra chromosome did not scale with the DNA and mRNA content (Stingele et al. 2012). Again, these proteins were generally subunits of multiprotein complexes. The different aneuploid cells also shared common changes in gene expression. Specifically, pathways involved in DNA and RNA metabolism were down-regulated, whereas energy metabolic pathways, including autophagy, were up-regulated.
Overall, these studies suggest that aneuploid cells may share some adaptive cellular responses of dosage compensation at the level of the proteome, but the extent and mechanism of this compensation are the topic of active research.
Effects of Aneuploidy on Genome Stability
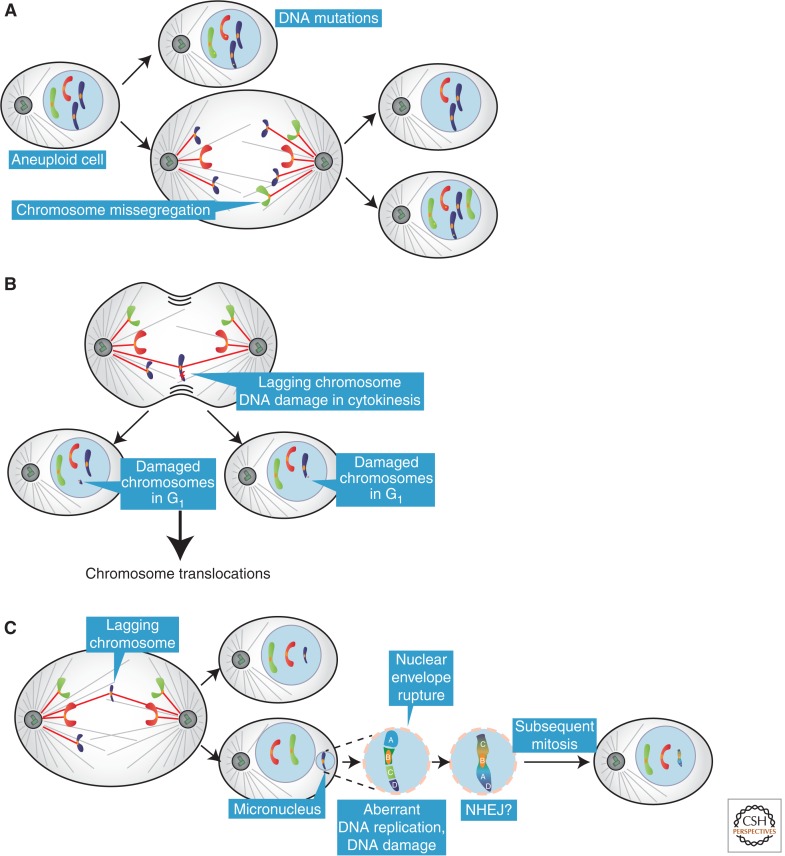
Recent work shows that aneuploidy itself can contribute to genome instability, likely through multiple mechanisms. Amon and coworkers found that the presence of a single extra chromosome in budding yeast increases the rate of point mutations, with effects ranging from twofold to sevenfold, and of mitotic recombination (Sheltzer et al. 2011). Aneuploid yeast strains also display an increase in the rate of chromosome missegregation, possibly generating a positive-feedback loop that sustains aneuploidy (Fig. 2A). The same finding was also reported in an independent study in which budding yeast strains with complex aneuploidies display CIN (Zhu et al. 2012). This increase in genomic instability in aneuploid strains is consistent with their higher sensitivity to genotoxins (Sheltzer et al. 2011).
Figure 2.
Effects of aneuploidy on genomic instability. (A) Aneuploid cells have an elevated rate of DNA mutations and chromosome missegregation compared with diploid cells. (B) A lagging chromosome in anaphase can be damaged if it is located in the proximity of the cleavage furrow. The damaged chromosome(s) can then generate chromosome translocations through nonhomologous end joining (NHEJ). (C) A lagging chromosome can be incorporated into a micronucleus if it does not reach the main chromatin masses. Chromosomes in micronuclei can accumulate damage and undergo fragmentation. The chromosome fragments in the micronucleus can be stitched together, likely through NHEJ, creating a highly rearranged derivative chromosome. In the subsequent mitosis, the nuclear envelope of the micronucleus can break down allowing the rearranged chromosome to be incorporated back into in the main genome.
A causal link between aneuploidy and CIN has also been described in human cells. Subclones of the colon cancer cell line HCT116 with experimentally induced aneuploidy show an increased frequency of chromosome missegregation in the absence of P53 (Thompson and Compton 2010). In addition to this direct effect of aneuploidy on genome instability, aneuploidy might theoretically lead to structural alterations of chromosomes because of imbalances in genes involved in DNA repair and replication (Duesberg et al. 2006).
Medema and coworkers showed that lagging chromosomes or chromosome bridges can be damaged if “trapped” in the proximity of the cleavage furrow at cytokinesis (Fig. 2B; Janssen et al. 2011). Furthermore, this damage can lead to unbalanced translocations in a process involving nonhomologous end joining (NHEJ). Although the DNA damage can be inhibited by blocking cytokinesis, it is currently unclear whether the chromosome breakage is caused by a physical insult resulting from cleavage furrow ingression or by endonucleases activated during cytokinesis.
Micronuclei are another major source of DNA damage resulting from errors in mitosis. Lagging chromosomes that are not incorporated into the main chromosome mass will be encapsulated into nuclear structures termed micronuclei (Terradas et al. 2010). Micronuclei are well-known features of human cancer cells that can result from mitotic errors or DNA damage. Micronuclei have many features of primary nuclei, but there has been controversy about their actual composition and functional properties. Studies differed on whether micronuclei replicate DNA, mount a normal DNA damage response, or assemble normal nuclear envelopes; moreover, the ultimate fate of chromosomes trapped within micronuclei and their contribution to the cancer genome was unclear.
Crasta et al. (2012) characterized the properties of newly generated micronuclei and tracked their fate over several cell division cycles. Newly formed micronuclei develop extensive DNA damage that accumulates after the initiation of DNA replication. Pulse labeling with bromodeoxyuridine showed that the density of DNA replication in micronuclei was reduced and replication was asynchronous, with many micronuclei still replicating DNA during the G2 phase. These data suggested that initiation of DNA replication had a role in generating DNA damage, but did not define the mechanism. Chromosome spreads and spectral karyotyping showed a pulverized appearance of chromosomes in micronucleated cells. By imaging, it was found that micronuclei can be reincorporated into the primary nucleus after mitosis. Thus, mutations acquired in micronuclei can be incorporated into the main genome.
Recently, Hetzer and coworkers have provided additional insight into the mechanism of DNA damage in micronuclei (Hatch et al. 2013). Earlier studies from this group had revealed that primary nuclei in cancer cells will undergo spontaneous rupture, manifest as spillage of nuclear contents into the cytoplasm (Vargas et al. 2012). For primary nuclei, these rupture events were almost always transient. However, in Hatch et al. (2013), it was observed that micronuclei also ruptured but in this case the rupture appeared to be irreversible. Notably, the populations of micronuclei displaying extensive DNA damage were almost always those that underwent rupture.
The recent study by Hetzer’s laboratory strongly suggests that rupture of micronuclei has a role in generating DNA damage, but the underlying mechanism has not been defined (Hatch et al. 2013). We have recently found that cells blocked from entering S phase by serum-starvation-displayed rupture at the frequency observed in the work by Hetzer and colleagues, but with little or no damage (A Spektor and D Pellman, unpubl.). This suggests that rupture of the nuclear envelope may be a necessary but not sufficient event for DNA damage to occur in micronuclei. One possible model for the damage is that the rupture of the nuclear envelope in micronuclei that are replicating their DNA may cause the sudden loss of replication proteins and the stalling of replication forks. Alternative but not mutually exclusive models can also be envisioned. For example, rupture of the nuclear envelope during S phase could expose the chromatin to cytosolic nucleases that cause DNA damage.
One idea that emerged from the Crasta et al. (2012) study is that DNA damage in micronuclei could explain a novel form of localized genome damage called chromothripsis (Stephens et al. 2011; Rausch et al. 2012). Discovered by cancer genome sequencing, chromothripsis appears to result from the shattering and random stitching together of individual chromosomes or chromosome arms. How extensive chromosome rearrangements can be highly localized is unclear; however, the physical partitioning of the affected chromosome into a micronucleus is an appealingly simple model.
ANEUPLOIDY-TOLERATING MUTATIONS
Although aneuploidy is usually detrimental for the viability of a cell and organism, it is an extremely common feature of cancer cells. This apparent paradox could be explained by aneuploidy being a secondary, or passenger, effect of cancer progression, occurring when cells have already acquired an uncontrolled growth phenotype. However, as discussed later in this review, a number of mouse models have shown that aneuploidy can directly promote cancer (Holland and Cleveland 2009). This suggests that cancer cells can adapt to aneuploidy to cope with the detrimental effects on proliferation and metabolism.
Amon and coworkers analyzed mutations occurring in aneuploid yeast strains that display higher proliferation rates after in vitro evolution (Torres et al. 2010). The investigators identified strain-specific alterations, as well as some shared mutations. One of these shared mutations was the inactivation of the deubiquitinating enzyme UBP6. UBP6 is associated with the proteasome and regulates proteasome activity. In the absence of UBP6, proteasomal degradation of substrates is accelerated (Hanna et al. 2006; Peth et al. 2009). UBP6 mutations in aneuploid yeast result in an attenuation of the imbalances in their protein composition, which leads to improved cell growth.
Polyploidization is another mechanism that may counteract the detrimental effects of aneuploidy. Multiple copies of the genome may buffer the presence of a single extra chromosome, attenuating protein imbalances. In support of this hypothesis, many cancers have a near-tetraploid karyotype (Carter et al. 2012; Mitelman et al. 2013). Recently, a computational analysis determined that whole-genome doubling is a frequent event in human cancers, with an incidence of >50% in some epithelial cancers (Carter et al. 2012) and with an overall frequency of 37%, as reported in the Cancer Genome Atlas Pan-Cancer data set (Zack et al. 2013). Tetraploidization often follows the occurrence of specific aneuploidies, again suggesting that it might be a mechanism to mitigate the deleterious effects of aneuploidy.
However, polyploidization may not be entirely beneficial. A recent study has reported that polyploid cancer cells in mice can trigger an immune response that limits tumor growth (Senovilla et al. 2012). This immunosurveillance mechanism appears to involve the exposure on the cell membrane of calreticulin, which is increased in polyploid cells. Why polyploid tumor cells would induce surface calreticulin is not yet clear. Whether this response is aneuploidy specific or polyploidy specific is also unclear. The former seems to be more likely because there are normal tissues that have a large fraction of polyploid cells, including hepatocytes (Gupta 2000), placental trophoblast giant cells (Ullah et al. 2009), megakaryocytes (Ravid et al. 2002), and myoblasts (Yaffe and Feldman 1965). This finding emphasizes that although polyploidization is advantageous for aneuploid cells in culture, its final outcome on cell fitness may depend on multiple factors.
Cells can also adapt to aneuploidy by suppressing pathways that are activated by aneuploidy and diminish cell fitness and growth. In cell culture, aneuploidy activates a P53-dependent response that delays the cell cycle or arrests cells in G1 (Li et al. 2010; Thompson and Compton 2010). Inactivation of P53 enables aneuploid cells to proliferate and expand, which is consistent with the co-occurrence of P53 inactivation and aneuploidy in several tumors (Tomasini et al. 2008). Activation of P53 by aneuploidy is accompanied by the production of reactive oxygen species (ROS) and the activation of ATM kinase (Li et al. 2010). The mechanism through which aneuploid cells generate ROS is not known, but it is tempting to speculate that ROS may accumulate because of the activation of metabolic pathways.
Additionally, some cell types can tolerate aneuploidy through mechanisms that are still unknown. For example, human and mouse hepatocytes and neurons display some levels of aneuploidy that do not appear to impair cellular function (Gupta 2000; Kingsbury et al. 2005; Yurov et al. 2007).
In summary, although aneuploidy is usually an adverse event that undermines cell fitness and physiology, it can be tolerated under specific circumstances.
ANEUPLOIDY AND CANCER
Aneuploidy as a Driver of Tumorigenesis
The cause-and-effect relationship between an abnormal chromosome number and tumorigenesis is complex. In the last decade, mouse models of aneuploidy have begun to shed light on the causal role of aneuploidy in cancer development. Most of the mouse models of aneuploidy were developed by inducing CIN through the manipulation of the levels of SAC proteins (Dobles et al. 2000; Kalitsis et al. 2000; Michel et al. 2001; Putkey et al. 2002; Baker et al. 2004; Iwanaga et al. 2007; Jeganathan et al. 2007; Perera et al. 2007). The complete knockout of any of these genes causes embryonic lethality in mice, but heterozygous deletions of the SAC genes or hypomorphic alleles result in viable animals. These mice display chronic CIN and aneuploidy. The extent of aneuploidy depends on the specific gene targeted and the residual level of protein. Importantly, most of these mice develop tumors at higher rates than isogenic controls, corroborating the hypothesis that CIN supports tumor formation. Examples include mice that are heterozygous for Mad1, Mad2, or Cenp-E, which develop benign lung tumors (Michel et al. 2001; Iwanaga et al. 2007; Weaver et al. 2007), and Bub1 hypomorphic mice, which develop a wide spectrum of lethal tumors (Jeganathan et al. 2007).
Although these mouse models have advanced our understanding of the role of aneuploidy in tumorigenesis, there are some important limitations. First, it is difficult to separate aneuploidy from CIN, making it impossible to study the effects of aneuploidy on cancer development in the absence of CIN, which is observed in some aneuploid tumors that are chromosomally stable (Lingle et al. 2002). Second, these models could also manifest phenotypes that are the result of unappreciated roles of the SAC proteins in tumorigenesis, independent of CIN and aneuploidy. Third, mutations in SAC genes are rare in human cancers (Cahill et al. 1998; Myrie et al. 2000; Haruki et al. 2001), calling into question the relevance of these models for the study of aneuploid human tumors. Moreover, the effects of CIN and aneuploidy on tumor development in mice are not always clear. For example, these mouse models often develop tumors very late, and some of them display a phenotype only on the chemical induction of carcinogenesis (Babu et al. 2003; Dai et al. 2004; Jeganathan et al. 2005; Kalitsis et al. 2005; Baker et al. 2006; Jeganathan et al. 2007). Furthermore, the levels of aneuploidy found in tissues do not directly correlate with the incidence of tumors (Babu et al. 2003; Baker et al. 2004, 2006; Dai et al. 2004; Jeganathan et al. 2005, 2007; Kalitsis et al. 2005). This lack of correlation might be a result of unrecognized functions of the mitotic checkpoint genes in suppressing tumorigenesis or to a complex balance between deleterious effects and growth-promoting effects in a specific tissue.
The overexpression of genes involved in the mitotic checkpoint can also lead to aneuploidy. Interestingly, higher levels of SAC proteins are observed more frequently in human cancers than their inactivation (Pérez de Castro et al. 2007). The overexpression of MAD2 and the kinetochore protein HEC1 is frequent in human cancers, with higher protein levels correlating with poorer prognosis (Tanaka et al. 2001; Li et al. 2003; Hernando et al. 2004; Hayama et al. 2006). Overexpression of HEC1 in vivo leads to aneuploidy and the onset of lung and liver tumors (Diaz-Rodriguez et al. 2008). Transgenic mice with conditional overexpression of MAD2 display a high degree of aneuploidy and develop a wide range of lethal tumors, in particular lung adenomas, hepatomas, and intestinal tumors (Sotillo et al. 2007, 2010). Recent work has shown that the overexpression of MAD2 in cultured human cells causes hyperstabilization of kinetochore–microtubule attachments (Kabeche and Compton 2012), possibly resulting in uncorrected merotelic attachments and chromosome missegregation. Importantly, the Mad2 mouse model showed that MAD2 overexpression is not required for tumor maintenance because turning off the Mad2 transgene after tumor formation did not have any effect on tumor progression and growth (Sotillo et al. 2007). This result suggests that a transformed phenotype initiated by CIN can be maintained without the initiating event. Additional experimental and clinical evidence supports the hypothesis that aneuploidy has a causal role in malignant transformation. Trisomy 21, or Down syndrome, is the most common whole-chromosome aneuploidy in humans and is the only trisomy that is compatible with survival to adult age (Hassold and Jacobs 1984). Individuals with Down syndrome display an increased incidence of acute lymphoblastic leukemia and acute myeloid leukemia (Satge et al. 1998), especially in childhood. Trisomy of chromosome 21 is also frequently observed as an acquired abnormality in hematological cancers, including a subtype of AML (Mitelman et al. 1990; Hama et al. 2008; Cheng et al. 2009). These observations suggest that chromosome 21 may contain oncogenes whose amplification drive the development of blood cancers.
Specific and recurrent chromosome gains and losses in cancer support the idea that the amplification of oncogenes or deletion of tumor suppressor genes through aneuploidy may drive tumorigenesis. For example, trisomy 8 is frequently found in acute leukemia and monosomy 7 is common in primary myelodysplasia and AML (Johnson and Cotter 1997; McKenna 2004; Paulsson and Johansson 2007). An extra copy of chromosome 7 bearing a mutated MET proto-oncogene is recurrent in human renal carcinomas (Fischer et al. 1998; Zhuang et al. 1998), and a mouse model of skin papilloma and squamous cell carcinoma displays frequent trisomy of chromosome 7 with a mutated HRAS allele (Bianchi et al. 1990).
Further evidence that aneuploidy promotes tumorigenesis through the loss of tumor suppressor genes is illustrated in a study using mice with reduced levels of BUB1 (Baker et al. 2009). Thirty percent or less of BUB1 levels led to increased tumor incidence in animals heterozygous for the tumor suppressor genes P53 (P53+/−) or APC (ApcMin/+). Bub1 hypomorphism caused higher levels of aneuploidy and, strikingly, loss of heterozygosity of the wild-type copy of the tumor suppressor gene in almost all the analyzed tumors. Interestingly, the vast majority of the tumors had two copies of the chromosome carrying the knockout (P53−) or mutated (ApcMin) allele of the tumor suppressor gene, indicating that loss of the wild-type allele was accompanied by the gain of its homologous chromosome. This suggests that there is a selective pressure for cells in retaining two copies of the chromosome containing the tumor suppressor, probably because of the presence of essential haploinsufficient genes. The effect of reduced levels of BUB1 in promoting tumorigenesis was not recapitulated in other genetic contexts, including heterozygosity of the tumor suppressor genes Rb and Pten. This highlights that the role of aneuploidy in promoting tumorigenesis is highly context-dependent.
Aneuploidy, in addition to creating imbalances in tumor suppressor genes and oncogenes, might also be beneficial for tumor development by creating karyotypes that are advantageous in specific environments. For example, a study in budding yeast showed that aneuploid strains grew more slowly than euploid strains in nonselective environments, but some of aneuploid strains grew more robustly under a variety of stress conditions (Pavelka et al. 2010). Similarly, aneuploidy might create heterogeneity in the tumor cell population, allowing some cells to develop a growth advantage in a specific microenvironment. Supporting this model, aneuploidy can promote tumor relapse in a mouse model. Expression of a mutant allele of K-ras in mice drives the formation of lung tumors, with K-RAS withdrawal resulting in tumor regression (Fischer et al. 2001). The concomitant overexpression of Mad2 with mutant K-ras does not affect tumor regression after K-RAS removal (Sotillo et al. 2010). However, the transient overexpression of Mad2 with K-ras gives rise to a higher frequency of tumor relapse after oncogene withdrawal (Sotillo et al. 2010). The relapsed tumors show high levels of aneuploidy, suggesting that the MAD2-induced CIN might have created genetic diversity, facilitating the emergence of resistant tumor cells.
Aneuploidy as an Inhibitor of Tumorigenesis
The effects of aneuploidy on tumorigenesis seem to be highly context specific, as exemplified by a mouse model heterozygous for Cenp-E (Weaver et al. 2007). These mice have a modestly higher incidence of spontaneous lung tumors and lymphomas. At the same time, Cenp-E heterozygosity reduces the onset of carcinogen-induced tumors and increases the life span of mice devoid of the tumor suppressor P19ARF, despite high levels of aneuploidy. Similarly, ApcMin/+ mice, developed as a model for colon cancer, display an increased incidence of colon tumors on BubR1 haploinsufficiency. The same mice, however, have a reduction in the incidence of tumors of the small intestine (Rao et al. 2005).
Interestingly, individuals with Down syndrome have an increased risk of developing leukemia but a decreased risk of developing solid tumors (Satge et al. 1998; Korbel et al. 2009). The tumor suppressor effect of trisomy 21 in solid tumors may be caused by the overexpression of two genes located on chromosome 21, DSCR1 and DYRK1A, both of which block tumor angiogenesis by modulation of the calcineurin pathway (Baek et al. 2009; Korbel et al. 2009).
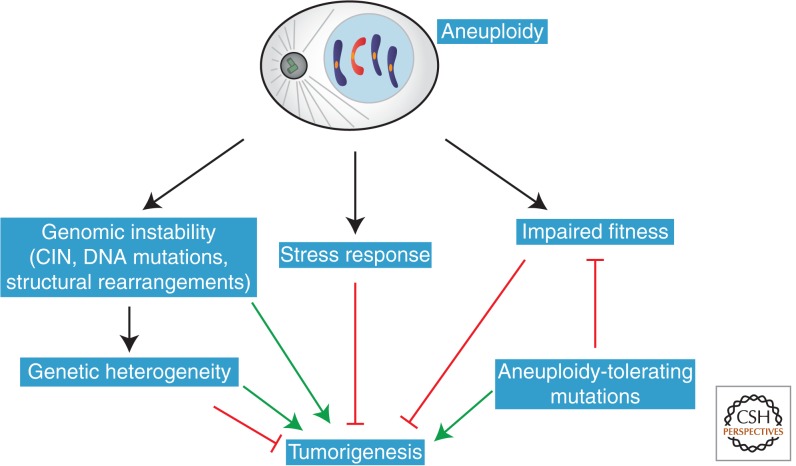
The inhibition of tumorigenesis by aneuploidy could also be explained by the reduced cellular fitness of aneuploid cells in the absence of aneuploidy-tolerating mutations. Aneuploidy could also generate genetic instability to an extent that is not compatible with the viability of specific tissues in defined genetic contexts. In summary, the effects of aneuploidy on tumor development are diverse and dependent on the specific tissue and the genetic background (Fig. 3).
Figure 3.
Effects of aneuploidy on tumorigenesis. Aneuploidy impairs cellular fitness, potentially preventing tumorigenesis in the absence of tolerating mutations. At the same time, aneuploidy also leads to genomic instability and genetic heterogeneity, which could provide a growth or survival advantage in a specific microenvironment.
ANEUPLOIDY AS A THERAPEUTIC TARGET
Targeting and killing aneuploid cells, but not diploid cells, is an attractive cancer therapy and can be approached from multiple directions. First, it may be possible to target both driver oncogenes and passenger genes on aneuploid chromosomes in a karyotype-specific type of therapeutic strategy. For example, the SLC19A1 gene, which codes for the reduced folate carrier that transports the antifolate drug methotrexate into cells, is located on chromosome 21, which is frequently gained in high-hyperdiploid pediatric acute lymphoblastic leukemia. Several studies have shown increased uptake and toxicity from methotrexate, attributed to these extra copies of the SLC19A1 gene, in the cells with extra copies of chromosome 21 (Zhang et al. 1998; Belkov et al. 1999).
Second, because aneuploid cells share some common features, including proteotoxic and metabolic stress, it may be possible to target aneuploidy itself in a more general strategy. This approach would not depend on the specific chromosomes gained or lost in a cancer cell. In fact, compounds that specifically inhibit the growth of aneuploid cells relative to diploid cells have been identified.
For example, aminoimidazole carboxamide ribonucleotide (AICAR), an energy stress–inducing compound that activates AMP-activated protein kinase, reduces the proliferation of trisomic mouse embryonic fibroblasts more than isogenic euploid cells (Tang et al. 2011). The aneuploidy-specific lethal effect of AICAR is synergistic with 17-allyamino-17-demethoxygeldanamycin (17-AAG), an inhibitor of the chaperone Hsp90. Importantly, the combined use of AICAR and 17-AAG showed increased lethality for human aneuploid cell lines with CIN compared with non-CIN cells (Tang et al. 2011). Both of these molecules are in clinical trials, although 17-AAG has not proven to be effective in cancer patients and is associated with toxic side effects (Heath et al. 2008; Solit et al. 2008; Gartner et al. 2012; Saif et al. 2013). In any case, these early results show that aneuploid cells can be selectively killed, even if the mechanisms underlying the lethality of these compounds are not fully understood.
Third, it is possible to selectively target CIN itself. As discussed earlier, CIN is often observed in cancer cells that have extra centrosomes, and many cancer cell lines cluster these extra centrosomes to generate a bipolar spindle (Ganem et al. 2009). An RNA interference screen was designed to identify genes that are required for centrosome clustering because preventing the clustering of centrosomes usually leads to multipolar mitoses and cell death (Kwon et al. 2008). One of the genes identified in the screen was the kinesin HSET, which is dispensable for the viability of normal cells but is essential for some cancer cells with supernumerary centrosomes. HSET is a feasible pharmacological target, as small-molecule inhibitors of kinesins have been developed and tested in clinical trials (Infante et al. 2012; Kantarjian et al. 2012).
Moreover, because several aneuploid human cancers have a near-tetraploid karyotype, tetraploidy itself might be an additional target for cancer therapy. In budding yeast it is well established that tetraploid cells display higher degrees of genome instability, including an elevated rate of chromosome missegregation and homologous recombination, possibly because of mismatches in the scaling of the mitotic spindle (Storchova et al. 2006). A genome-wide analysis in Saccharomyces cerevisiae validated the hypothesis of ploidy-specific lethality, identifying a small set of genes that are selectively required for the survival of tetraploid strains but not diploid strains. In particular, these genes are involved in homologous recombination, sister chromatid cohesion, and mitotic spindle function (Storchova et al. 2006). A similar systematic analysis is still missing in human cells, but the results could provide valuable hints of new targets in tetraploid cancer cells.
Overall, these examples support the concept that aneuploidy is an important target in cancer therapy. Further dissection of the molecular pathways that are selectively activated by aneuploidy could allow for the identification of additional therapeutic targets.
CONCLUDING REMARKS
Aneuploidy is generally a detrimental event at the level of the cell and organism, but it is also frequently observed in human tumors. This apparent paradox may be explained by the observation that aneuploid cells can acquire mutations to tolerate the negative effects of aneuploidy. Moreover, aneuploidy also confers a higher potential to evolve and adapt to selective conditions. Additionally, aneuploid cells display increased chromosome instability, DNA mutations, and genomic instability through mechanisms that include the acquisition of damaged lagging chromosomes and micronuclei. This generates cells with different karyotypes that may allow specific clones to expand in a tumor microenvironment with a unique selective pressure. At the same time, some karyotypes can also be counterselected in specific microenvironments, thereby explaining the context-specific effects of aneuploidy on tumorigenesis (Fig. 3).
Additional work is needed to establish whether the genetic instability observed in aneuploid cells directly promotes tumorigenesis. For example, can a cell with a damaged lagging chromosome or a micronucleus form a tumor when transplanted into a mouse? Moreover, the molecular pathways activated by aneuploidy that impair cell growth need to be dissected in more detail. The activation of a stress response in aneuploid cells in different species justifies the rationale for developing aneuploid-specific anticancer therapies. Understanding the mechanisms by which aneuploidy impairs cell growth, activates P53, and generates ROS will pave the way for the identification of therapeutic targets hitting aneuploid cells.
Footnotes
Editors: Mitsuhiro Yanagida, Anthony A. Hyman, and Jonathon Pines
Additional Perspectives on Mitosis available at www.cshperspectives.org
REFERENCES
- Andrews PD, Ovechkina Y, Morrice N, Wagenbach M, Duncan K, Wordeman L, Swedlow JR 2004. Aurora B regulates MCAK at the mitotic centromere. Dev Cell 6: 253–268. [DOI] [PubMed] [Google Scholar]
- Babu JR, Jeganathan KB, Baker DJ, Wu X, Kang-Decker N, van Deursen JM 2003. Rae1 is an essential mitotic checkpoint regulator that cooperates with Bub3 to prevent chromosome missegregation. J Cell Biol 160: 341–353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baek KH, Zaslavsky A, Lynch RC, Britt C, Okada Y, Siarey RJ, Lensch MW, Park IH, Yoon SS, Minami T, et al. 2009. Down’s syndrome suppression of tumour growth and the role of the calcineurin inhibitor DSCR1. Nature 459: 1126–1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker DJ, Jeganathan KB, Cameron JD, Thompson M, Juneja S, Kopecka A, Kumar R, Jenkins RB, de Groen PC, Roche P, et al. 2004. BubR1 insufficiency causes early onset of aging-associated phenotypes and infertility in mice. Nat Genet 36: 744–749. [DOI] [PubMed] [Google Scholar]
- Baker DJ, Jeganathan KB, Malureanu L, Perez-Terzic C, Terzic A, van Deursen JM 2006. Early aging–associated phenotypes in Bub3/Rae1 haploinsufficient mice. J Cell Biol 172: 529–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker DJ, Jin F, Jeganathan KB, van Deursen JM 2009. Whole chromosome instability caused by Bub1 insufficiency drives tumorigenesis through tumor suppressor gene loss of heterozygosity. Cancer Cell 16: 475–486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakhoum SF, Genovese G, Compton DA 2009a. Deviant kinetochore microtubule dynamics underlie chromosomal instability. Curr Biol 19: 1937–1942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakhoum SF, Thompson SL, Manning AL, Compton DA 2009b. Genome stability is ensured by temporal control of kinetochore-microtubule dynamics. Nat Cell Biol 11: 27–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barber TD, McManus K, Yuen KW, Reis M, Parmigiani G, Shen D, Barrett I, Nouhi Y, Spencer F, Markowitz S, et al. 2008. Chromatid cohesion defects may underlie chromosome instability in human colorectal cancers. Proc Natl Acad Sci 105: 3443–3448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belkov VM, Krynetski EY, Schuetz JD, Yanishevski Y, Masson E, Mathew S, Raimondi S, Pui CH, Relling MV, Evans WE 1999. Reduced folate carrier expression in acute lymphoblastic leukemia: A mechanism for ploidy but not lineage differences in methotrexate accumulation. Blood 93: 1643–1650. [PubMed] [Google Scholar]
- Beroukhim R, Mermel CH, Porter D, Wei G, Raychaudhuri S, Donovan J, Barretina J, Boehm JS, Dobson J, Urashima M, et al. 2010. The landscape of somatic copy-number alteration across human cancers. Nature 463: 899–905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bianchi AB, Aldaz CM, Conti CJ 1990. Nonrandom duplication of the chromosome bearing a mutated Ha-ras-1 allele in mouse skin tumors. Proc Natl Acad Sci 87: 6902–6906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brito DA, Rieder CL 2006. Mitotic checkpoint slippage in humans occurs via cyclin B destruction in the presence of an active checkpoint. Curr Biol 16: 1194–1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burrell RA, McClelland SE, Endesfelder D, Groth P, Weller MC, Shaikh N, Domingo E, Kanu N, Dewhurst SM, Gronroos E, et al. 2013. Replication stress links structural and numerical cancer chromosomal instability. Nature 494: 492–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cahill DP, Lengauer C, Yu J, Riggins GJ, Willson JK, Markowitz SD, Kinzler KW, Vogelstein B 1998. Mutations of mitotic checkpoint genes in human cancers. Nature 392: 300–303. [DOI] [PubMed] [Google Scholar]
- Carter SL, Cibulskis K, Helman E, McKenna A, Shen H, Zack T, Laird PW, Onofrio RC, Winckler W, Weir BA, et al. 2012. Absolute quantification of somatic DNA alterations in human cancer. Nat Biotechnol 30: 413–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cassimeris L, Morabito J 2004. TOGp, the human homolog of XMAP215/Dis1, is required for centrosome integrity, spindle pole organization, and bipolar spindle assembly. Mol Biol Cell 15: 1580–1590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandhok NS, Pellman D 2009. A little CIN may cost a lot: Revisiting aneuploidy and cancer. Curr Opin Genet Dev 19: 74–81. [DOI] [PubMed] [Google Scholar]
- Cheng Y, Wang H, Wang H, Chen Z, Jin J 2009. Trisomy 21 in patients with acute leukemia. Am J Hematol 84: 193–194. [DOI] [PubMed] [Google Scholar]
- Cimini D 2008. Merotelic kinetochore orientation, aneuploidy, and cancer. Biochim Biophys Acta 1786: 32–40. [DOI] [PubMed] [Google Scholar]
- Cimini D, Tanzarella C, Degrassi F 1999. Differences in malsegregation rates obtained by scoring ana-telophases or binucleate cells. Mutagenesis 14: 563–568. [DOI] [PubMed] [Google Scholar]
- Cimini D, Howell B, Maddox P, Khodjakov A, Degrassi F, Salmon ED 2001. Merotelic kinetochore orientation is a major mechanism of aneuploidy in mitotic mammalian tissue cells. J Cell Biol 153: 517–527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cimini D, Moree B, Canman JC, Salmon ED 2003. Merotelic kinetochore orientation occurs frequently during early mitosis in mammalian tissue cells and error correction is achieved by two different mechanisms. J Cell Sci 116: 4213–4225. [DOI] [PubMed] [Google Scholar]
- Cimini D, Cameron LA, Salmon ED 2004. Anaphase spindle mechanics prevent missegregation of merotelically oriented chromosomes. Curr Biol 14: 2149–2155. [DOI] [PubMed] [Google Scholar]
- Crasta K, Ganem NJ, Dagher R, Lantermann AB, Ivanova EV, Pan Y, Nezi L, Protopopov A, Chowdhury D, Pellman D 2012. DNA breaks and chromosome pulverization from errors in mitosis. Nature 482: 53–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai W, Wang Q, Liu T, Swamy M, Fang Y, Xie S, Mahmood R, Yang YM, Xu M, Rao CV 2004. Slippage of mitotic arrest and enhanced tumor development in mice with BubR1 haploinsufficiency. Cancer Res 64: 440–445. [DOI] [PubMed] [Google Scholar]
- De Lange T 2005. Telomere-related genome instability in cancer. Cold Spring Harb Symp Quant Biol 70: 197–204. [DOI] [PubMed] [Google Scholar]
- Diaz-Rodriguez E, Sotillo R, Schvartzman JM, Benezra R 2008. Hec1 overexpression hyperactivates the mitotic checkpoint and induces tumor formation in vivo. Proc Natl Acad Sci 105: 16719–16724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobles M, Liberal V, Scott ML, Benezra R, Sorger PK 2000. Chromosome missegregation and apoptosis in mice lacking the mitotic checkpoint protein Mad2. Cell 101: 635–645. [DOI] [PubMed] [Google Scholar]
- Dorsett D 2011. Cohesin: Genomic insights into controlling gene transcription and development. Curr Opin Genet Dev 21: 199–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duesberg P, Li R, Fabarius A, Hehlmann R 2006. Aneuploidy and cancer: From correlation to causation. Contrib Microbiol 13: 16–44. [DOI] [PubMed] [Google Scholar]
- Fenech M, Kirsch-Volders M, Natarajan AT, Surralles J, Crott JW, Parry J, Norppa H, Eastmond DA, Tucker JD, Thomas P 2011. Molecular mechanisms of micronucleus, nucleoplasmic bridge and nuclear bud formation in mammalian and human cells. Mutagenesis 26: 125–132. [DOI] [PubMed] [Google Scholar]
- Fischer J, Palmedo G, von Knobloch R, Bugert P, Prayer-Galetti T, Pagano F, Kovacs G 1998. Duplication and overexpression of the mutant allele of the MET proto-oncogene in multiple hereditary papillary renal cell tumours. Oncogene 17: 733–739. [DOI] [PubMed] [Google Scholar]
- Fischer C, Buthe J, Nollau P, Hollerbach S, Schulmann K, Schmiegel W, Wagener C, Tschentscher P 2001. Enrichment of mutant KRAS alleles in pancreatic juice by subtractive iterative polymerase chain reaction. Lab Invest 81: 827–831. [DOI] [PubMed] [Google Scholar]
- Fujiwara T, Bandi M, Nitta M, Ivanova EV, Bronson RT, Pellman D 2005. Cytokinesis failure generating tetraploids promotes tumorigenesis in p53-null cells. Nature 437: 1043–1047. [DOI] [PubMed] [Google Scholar]
- Ganem NJ, Godinho SA, Pellman D 2009. A mechanism linking extra centrosomes to chromosomal instability. Nature 460: 278–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao YF, Li T, Chang Y, Wang YB, Zhang WN, Li WH, He K, Mu R, Zhen C, Man JH, et al. 2011. Cdk1-phosphorylated CUEDC2 promotes spindle checkpoint inactivation and chromosomal instability. Nat Cell Biol 13: 924–933. [DOI] [PubMed] [Google Scholar]
- Gartner EM, Silverman P, Simon M, Flaherty L, Abrams J, Ivy P, Lorusso PM 2012. A phase II study of 17-allylamino-17-demethoxygeldanamycin in metastatic or locally advanced, unresectable breast cancer. Breast Cancer Res Treat 131: 933–937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasch AP, Spellman PT, Kao CM, Carmel-Harel O, Eisen MB, Storz G, Botstein D, Brown PO 2000. Genomic expression programs in the response of yeast cells to environmental changes. Mol Biol Cell 11: 4241–4257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gascoigne KE, Taylor SS 2008. Cancer cells display profound intra- and interline variation following prolonged exposure to antimitotic drugs. Cancer Cell 14: 111–122. [DOI] [PubMed] [Google Scholar]
- Geigl JB, Obenauf AC, Schwarzbraun T, Speicher MR 2008. Defining “chromosomal instability.” Trends Genet 24: 64–69. [DOI] [PubMed] [Google Scholar]
- Gisselsson D 2008. Classification of chromosome segregation errors in cancer. Chromosoma 117: 511–519. [DOI] [PubMed] [Google Scholar]
- Gregan J, Polakova S, Zhang L, Tolic-Norrelykke IM, Cimini D 2011. Merotelic kinetochore attachment: Causes and effects. Trends Cell Biol 21: 374–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta S 2000. Hepatic polyploidy and liver growth control. Semin Cancer Biol 10: 161–171. [DOI] [PubMed] [Google Scholar]
- Gupta A, Inaba S, Wong OK, Fang G, Liu J 2003. Breast cancer-specific gene 1 interacts with the mitotic checkpoint kinase BubR1. Oncogene 22: 7593–7599. [DOI] [PubMed] [Google Scholar]
- Habedanck R, Stierhof YD, Wilkinson CJ, Nigg EA 2005. The Polo kinase Plk4 functions in centriole duplication. Nat Cell Biol 7: 1140–1146. [DOI] [PubMed] [Google Scholar]
- Hama A, Yagasaki H, Takahashi Y, Nishio N, Muramatsu H, Yoshida N, Tanaka M, Hidaka H, Watanabe N, Yoshimi A, et al. 2008. Acute megakaryoblastic leukaemia (AMKL) in children: A comparison of AMKL with and without Down syndrome. Br J Haematol 140: 552–561. [DOI] [PubMed] [Google Scholar]
- Hanks S, Coleman K, Reid S, Plaja A, Firth H, Fitzpatrick D, Kidd A, Mehes K, Nash R, Robin N, et al. 2004. Constitutional aneuploidy and cancer predisposition caused by biallelic mutations in BUB1B. Nat Genet 36: 1159–1161. [DOI] [PubMed] [Google Scholar]
- Hanna J, Hathaway NA, Tone Y, Crosas B, Elsasser S, Kirkpatrick DS, Leggett DS, Gygi SP, King RW, Finley D 2006. Deubiquitinating enzyme Ubp6 functions noncatalytically to delay proteasomal degradation. Cell 127: 99–111. [DOI] [PubMed] [Google Scholar]
- Haruki N, Saito H, Harano T, Nomoto S, Takahashi T, Osada H, Fujii Y, Takahashi T 2001. Molecular analysis of the mitotic checkpoint genes BUB1, BUBR1 and BUB3 in human lung cancers. Cancer Lett 162: 201–205. [DOI] [PubMed] [Google Scholar]
- Hassold TJ, Jacobs PA 1984. Trisomy in man. Annu Rev Genet 18: 69–97. [DOI] [PubMed] [Google Scholar]
- Hassold T, Hall H, Hunt P 2007. The origin of human aneuploidy: Where we have been, where we are going. Hum Mol Genet 16: R203–R208. [DOI] [PubMed] [Google Scholar]
- Hatch EM, Fischer AH, Deerinck TJ, Hetzer MW 2013. Catastrophic nuclear envelope collapse in cancer cell micronuclei. Cell 154: 47–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayama S, Daigo Y, Kato T, Ishikawa N, Yamabuki T, Miyamoto M, Ito T, Tsuchiya E, Kondo S, Nakamura Y 2006. Activation of CDCA1-KNTC2, members of centromere protein complex, involved in pulmonary carcinogenesis. Cancer Res 66: 10339–10348. [DOI] [PubMed] [Google Scholar]
- Heath EI, Hillman DW, Vaishampayan U, Sheng S, Sarkar F, Harper F, Gaskins M, Pitot HC, Tan W, Ivy SP, et al. 2008. A phase II trial of 17-allylamino-17-demethoxygeldanamycin in patients with hormone-refractory metastatic prostate cancer. Clin Cancer Res 14: 7940–7946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernando E, Nahle Z, Juan G, Diaz-Rodriguez E, Alaminos M, Hemann M, Michel L, Mittal V, Gerald W, Benezra R, et al. 2004. Rb inactivation promotes genomic instability by uncoupling cell cycle progression from mitotic control. Nature 430: 797–802. [DOI] [PubMed] [Google Scholar]
- Holland AJ, Cleveland DW 2009. Boveri revisited: Chromosomal instability, aneuploidy and tumorigenesis. Nat Rev Mol Cell Biol 10: 478–487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Infante JR, Kurzrock R, Spratlin J, Burris HA, Eckhardt SG, Li J, Wu K, Skolnik JM, Hylander-Gans L, Osmukhina A, et al. 2012. A Phase I study to assess the safety, tolerability, and pharmacokinetics of AZD4877, an intravenous Eg5 inhibitor in patients with advanced solid tumors. Cancer Chemother Pharmacol 69: 165–172. [DOI] [PubMed] [Google Scholar]
- Iwanaga Y, Chi YH, Miyazato A, Sheleg S, Haller K, Peloponese JM Jr, Li Y, Ward JM, Benezra R, Jeang KT 2007. Heterozygous deletion of mitotic arrest-deficient protein 1 (MAD1) increases the incidence of tumors in mice. Cancer Res 67: 160–166. [DOI] [PubMed] [Google Scholar]
- Janssen A, van der Burg M, Szuhai K, Kops GJ, Medema RH 2011. Chromosome segregation errors as a cause of DNA damage and structural chromosome aberrations. Science 333: 1895–1898. [DOI] [PubMed] [Google Scholar]
- Jeganathan KB, Malureanu L, van Deursen JM 2005. The Rae1-Nup98 complex prevents aneuploidy by inhibiting securin degradation. Nature 438: 1036–1039. [DOI] [PubMed] [Google Scholar]
- Jeganathan K, Malureanu L, Baker DJ, Abraham SC, van Deursen JM 2007. Bub1 mediates cell death in response to chromosome missegregation and acts to suppress spontaneous tumorigenesis. J Cell Biol 179: 255–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang J, Jing Y, Cost GJ, Chiang JC, Kolpa HJ, Cotton AM, Carone DM, Carone BR, Shivak DA, Guschin DY, et al. 2013. Translating dosage compensation to trisomy 21. Nature 500: 296–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson E, Cotter FE 1997. Monosomy 7 and 7q− associated with myeloid malignancy. Blood Rev 11: 46–55. [DOI] [PubMed] [Google Scholar]
- Kabeche L, Compton DA 2012. Checkpoint-independent stabilization of kinetochore-microtubule attachments by Mad2 in human cells. Curr Biol 22: 638–644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalitsis P, Earle E, Fowler KJ, Choo KH 2000. Bub3 gene disruption in mice reveals essential mitotic spindle checkpoint function during early embryogenesis. Genes Dev 14: 2277–2282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalitsis P, Fowler KJ, Griffiths B, Earle E, Chow CW, Jamsen K, Choo KH 2005. Increased chromosome instability but not cancer predisposition in haploinsufficient Bub3 mice. Genes Chromosomes Cancer 44: 29–36. [DOI] [PubMed] [Google Scholar]
- Kantarjian HM, Padmanabhan S, Stock W, Tallman MS, Curt GA, Li J, Osmukhina A, Wu K, Huszar D, Borthukar G, et al. 2012. Phase I/II multicenter study to assess the safety, tolerability, pharmacokinetics and pharmacodynamics of AZD4877 in patients with refractory acute myeloid leukemia. Invest New Drugs 30: 1107–1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kingsbury MA, Friedman B, McConnell MJ, Rehen SK, Yang AH, Kaushal D, Chun J 2005. Aneuploid neurons are functionally active and integrated into brain circuitry. Proc Natl Acad Sci 102: 6143–6147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kops GJ, Foltz DR, Cleveland DW 2004. Lethality to human cancer cells through massive chromosome loss by inhibition of the mitotic checkpoint. Proc Natl Acad Sci 101: 8699–8704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korbel JO, Tirosh-Wagner T, Urban AE, Chen XN, Kasowski M, Dai L, Grubert F, Erdman C, Gao MC, Lange K, et al. 2009. The genetic architecture of Down syndrome phenotypes revealed by high-resolution analysis of human segmental trisomies. Proc Natl Acad Sci 106: 12031–12036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon M, Godinho SA, Chandhok NS, Ganem NJ, Azioune A, Thery M, Pellman D 2008. Mechanisms to suppress multipolar divisions in cancer cells with extra centrosomes. Genes Dev 22: 2189–2203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lengauer C, Kinzler KW, Vogelstein B 1997. Genetic instability in colorectal cancers. Nature 386: 623–627. [DOI] [PubMed] [Google Scholar]
- Li GQ, Li H, Zhang HF 2003. Mad2 and p53 expression profiles in colorectal cancer and its clinical significance. World J Gastroenterol 9: 1972–1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Fang X, Baker DJ, Guo L, Gao X, Wei Z, Han S, van Deursen JM, Zhang P 2010. The ATM-p53 pathway suppresses aneuploidy-induced tumorigenesis. Proc Natl Acad Sci 107: 14188–14193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lingle WL, Barrett SL, Negron VC, D’Assoro AB, Boeneman K, Liu W, Whitehead CM, Reynolds C, Salisbury JL 2002. Centrosome amplification drives chromosomal instability in breast tumor development. Proc Natl Acad Sci 99: 1978–1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logarinho E, Maffini S, Barisic M, Marques A, Toso A, Meraldi P, Maiato H 2012. CLASPs prevent irreversible multipolarity by ensuring spindle-pole resistance to traction forces during chromosome alignment. Nat Cell Biol 14: 295–303. [DOI] [PubMed] [Google Scholar]
- Marthiens V, Rujano MA, Pennetier C, Tessier S, Paul-Gilloteaux P, Basto R 2013. Centrosome amplification causes microcephaly. Nat Cell Biol 15: 731–740. [DOI] [PubMed] [Google Scholar]
- Matsuura S, Matsumoto Y, Morishima K, Izumi H, Matsumoto H, Ito E, Tsutsui K, Kobayashi J, Tauchi H, Kajiwara Y, et al. 2006. Monoallelic BUB1B mutations and defective mitotic-spindle checkpoint in seven families with premature chromatid separation (PCS) syndrome. Am J Med Genet A 140: 358–367. [DOI] [PubMed] [Google Scholar]
- McClintock B 1941. The stability of broken ends of chromosomes in Zea Mays. Genetics 26: 234–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenna RW 2004. Myelodysplasia and myeloproliferative disorders in children. Am J Clin Pathol 122 Suppl: S58–S69. [DOI] [PubMed] [Google Scholar]
- Michel LS, Liberal V, Chatterjee A, Kirchwegger R, Pasche B, Gerald W, Dobles M, Sorger PK, Murty VV, Benezra R 2001. MAD2 haplo-insufficiency causes premature anaphase and chromosome instability in mammalian cells. Nature 409: 355–359. [DOI] [PubMed] [Google Scholar]
- Mitelman F, Heim S, Mandahl N 1990. Trisomy 21 in neoplastic cells. Am J Med Genet 7 Suppl: 262–266. [DOI] [PubMed] [Google Scholar]
- Mitelman F, Johansson B, Mertens F (eds.). 2013. Mitelman Database of Chromosome Aberrations and Gene Fusions in Cancer, cgap.nci.nih.gov/Chromosomes/Mitelman. [Google Scholar]
- Musacchio A, Salmon ED 2007. The spindle-assembly checkpoint in space and time. Nat Rev Mol Cell Biol 8: 379–393. [DOI] [PubMed] [Google Scholar]
- Myrie KA, Percy MJ, Azim JN, Neeley CK, Petty EM 2000. Mutation and expression analysis of human BUB1 and BUB1B in aneuploid breast cancer cell lines. Cancer Lett 152: 193–199. [DOI] [PubMed] [Google Scholar]
- Nasmyth K 2011. Cohesin: A catenase with separate entry and exit gates? Nature Cell Biol 13: 1170–1177. [DOI] [PubMed] [Google Scholar]
- Oromendia AB, Dodgson SE, Amon A 2012. Aneuploidy causes proteotoxic stress in yeast. Genes Dev 26: 2696–2708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park HY, Jeon YK, Shin HJ, Kim IJ, Kang HC, Jeong SJ, Chung DH, Lee CW 2007. Differential promoter methylation may be a key molecular mechanism in regulating BubR1 expression in cancer cells. Exp Mol Med 39: 195–204. [DOI] [PubMed] [Google Scholar]
- Paulsson K, Johansson B 2007. Trisomy 8 as the sole chromosomal aberration in acute myeloid leukemia and myelodysplastic syndromes. Pathol Biol (Paris) 55: 37–48. [DOI] [PubMed] [Google Scholar]
- Pavelka N, Rancati G, Zhu J, Bradford WD, Saraf A, Florens L, Sanderson BW, Hattem GL, Li R 2010. Aneuploidy confers quantitative proteome changes and phenotypic variation in budding yeast. Nature 468: 321–325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perera D, Tilston V, Hopwood JA, Barchi M, Boot-Handford RP, Taylor SS 2007. Bub1 maintains centromeric cohesion by activation of the spindle checkpoint. Dev Cell 13: 566–579. [DOI] [PubMed] [Google Scholar]
- Pérez de Castro I, de Cárcer G, Malumbres M 2007. A census of mitotic cancer genes: New insights into tumor cell biology and cancer therapy. Carcinogenesis 28: 899–912. [DOI] [PubMed] [Google Scholar]
- Peth A, Besche HC, Goldberg AL 2009. Ubiquitinated proteins activate the proteasome by binding to Usp14/Ubp6, which causes 20S gate opening. Mol Cell 36: 794–804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinsky BA, Kung C, Shokat KM, Biggins S 2006. The Ipl1-Aurora protein kinase activates the spindle checkpoint by creating unattached kinetochores. Nat Cell Biol 8: 78–83. [DOI] [PubMed] [Google Scholar]
- Primorac I, Musacchio A 2013. Panta rhei: The APC/C at steady state. J Cell Biol 201: 177–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putkey FR, Cramer T, Morphew MK, Silk AD, Johnson RS, McIntosh JR, Cleveland DW 2002. Unstable kinetochore-microtubule capture and chromosomal instability following deletion of CENP-E. Dev Cell 3: 351–365. [DOI] [PubMed] [Google Scholar]
- Rao CV, Yang YM, Swamy MV, Liu T, Fang Y, Mahmood R, Jhanwar-Uniyal M, Dai W 2005. Colonic tumorigenesis in BubR1+/−ApcMin/+ compound mutant mice is linked to premature separation of sister chromatids and enhanced genomic instability. Proc Natl Acad Sci 102: 4365–4370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rausch T, Jones DT, Zapatka M, Stutz AM, Zichner T, Weischenfeldt J, Jager N, Remke M, Shih D, Northcott PA, et al. 2012. Genome sequencing of pediatric medulloblastoma links catastrophic DNA rearrangements with TP53 mutations. Cell 148: 59–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravid K, Lu J, Zimmet JM, Jones MR 2002. Roads to polyploidy: The megakaryocyte example. J Cell Physiol 190: 7–20. [DOI] [PubMed] [Google Scholar]
- Reddy SK, Rape M, Margansky WA, Kirschner MW 2007. Ubiquitination by the anaphase-promoting complex drives spindle checkpoint inactivation. Nature 446: 921–925. [DOI] [PubMed] [Google Scholar]
- Rocquain J, Gelsi-Boyer V, Adelaide J, Murati A, Carbuccia N, Vey N, Birnbaum D, Mozziconacci MJ, Chaffanet M 2010. Alteration of cohesin genes in myeloid diseases. Am J Hematol 85: 717–719. [DOI] [PubMed] [Google Scholar]
- Saif MW, Erlichman C, Dragovich T, Mendelson D, Toft D, Burrows F, Storgard C, Von Hoff D 2013. Open-label, dose-escalation, safety, pharmacokinetic, and pharmacodynamic study of intravenously administered CNF 1010 (17-[allylamino]-17-demethoxygeldanamycin [17-AAG]) in patients with solid tumors. Cancer Chemother Pharmacol 71: 1345–1355. [DOI] [PubMed] [Google Scholar]
- Satge D, Sommelet D, Geneix A, Nishi M, Malet P, Vekemans M 1998. A tumor profile in Down syndrome. Am J Med Genet 78: 207–216. [PubMed] [Google Scholar]
- Schvartzman JM, Duijf PH, Sotillo R, Coker C, Benezra R 2011. Mad2 is a critical mediator of the chromosome instability observed upon Rb and p53 pathway inhibition. Cancer Cell 19: 701–714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segal DJ, McCoy EE 1974. Studies on Down's syndrome in tissue culture: I. Growth rates and protein contents of fibroblast cultures. J Cell Physiol 83: 85–90. [DOI] [PubMed] [Google Scholar]
- Senovilla L, Vitale I, Martins I, Tailler M, Pailleret C, Michaud M, Galluzzi L, Adjemian S, Kepp O, Niso-Santano M, et al. 2012. An immunosurveillance mechanism controls cancer cell ploidy. Science 337: 1678–1684. [DOI] [PubMed] [Google Scholar]
- Sheltzer JM, Blank HM, Pfau SJ, Tange Y, George BM, Humpton TJ, Brito IL, Hiraoka Y, Niwa O, Amon A 2011. Aneuploidy drives genomic instability in yeast. Science 333: 1026–1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheltzer JM, Torres EM, Dunham MJ, Amon A 2012. Transcriptional consequences of aneuploidy. Proc Natl Acad Sci 109: 12644–12649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silkworth WT, Nardi IK, Scholl LM, Cimini D 2009. Multipolar spindle pole coalescence is a major source of kinetochore mis-attachment and chromosome missegregation in cancer cells. PloS ONE 4: e6564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snape K, Hanks S, Ruark E, Barros-Nunez P, Elliott A, Murray A, Lane AH, Shannon N, Callier P, Chitayat D, et al. 2011. Mutations in CEP57 cause mosaic variegated aneuploidy syndrome. Nat Genet 43: 527–529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solit DB, Osman I, Polsky D, Panageas KS, Daud A, Goydos JS, Teitcher J, Wolchok JD, Germino FJ, Krown SE, et al. 2008. Phase II trial of 17-allylamino-17-demethoxygeldanamycin in patients with metastatic melanoma. Clin Cancer Res 14: 8302–8307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solomon DA, Kim T, Diaz-Martinez LA, Fair J, Elkahloun AG, Harris BT, Toretsky JA, Rosenberg SA, Shukla N, Ladanyi M, et al. 2011. Mutational inactivation of STAG2 causes aneuploidy in human cancer. Science 333: 1039–1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sotillo R, Hernando E, Diaz-Rodriguez E, Teruya-Feldstein J, Cordon-Cardo C, Lowe SW, Benezra R 2007. Mad2 overexpression promotes aneuploidy and tumorigenesis in mice. Cancer Cell 11: 9–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sotillo R, Schvartzman JM, Socci ND, Benezra R 2010. Mad2-induced chromosome instability leads to lung tumour relapse after oncogene withdrawal. Nature 464: 436–440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens PJ, Greenman CD, Fu B, Yang F, Bignell GR, Mudie LJ, Pleasance ED, Lau KW, Beare D, Stebbings LA, et al. 2011. Massive genomic rearrangement acquired in a single catastrophic event during cancer development. Cell 144: 27–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stingele S, Stoehr G, Peplowska K, Cox J, Mann M, Storchova Z 2012. Global analysis of genome, transcriptome and proteome reveals the response to aneuploidy in human cells. Mol Syst Biol 8: 608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storchova Z, Breneman A, Cande J, Dunn J, Burbank K, O’Toole E, Pellman D 2006. Genome-wide genetic analysis of polyploidy in yeast. Nature 443: 541–547. [DOI] [PubMed] [Google Scholar]
- Tanaka K, Nishioka J, Kato K, Nakamura A, Mouri T, Miki C, Kusunoki M, Nobori T 2001. Mitotic checkpoint protein hsMAD2 as a marker predicting liver metastasis of human gastric cancers. Jpn J Cancer Res 92: 952–958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang YC, Williams BR, Siegel JJ, Amon A 2011. Identification of aneuploidy-selective antiproliferation compounds. Cell 144: 499–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terradas M, Martin M, Tusell L, Genesca A 2010. Genetic activities in micronuclei: Is the DNA entrapped in micronuclei lost for the cell? Mutat Res 705: 60–67. [DOI] [PubMed] [Google Scholar]
- Thompson SL, Compton DA 2008. Examining the link between chromosomal instability and aneuploidy in human cells. J Cell Biol 180: 665–672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson SL, Compton DA 2010. Proliferation of aneuploid human cells is limited by a p53-dependent mechanism. J Cell Biol 188: 369–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson SL, Compton DA 2011a. Chromosome missegregation in human cells arises through specific types of kinetochore-microtubule attachment errors. Proc Natl Acad Sci 108: 17974–17978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson SL, Compton DA 2011b. Chromosomes and cancer cells. Chromosome Res 19: 433–444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson SL, Bakhoum SF, Compton DA 2010. Mechanisms of chromosomal instability. Curr Biol 20: R285–R295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tighe A, Johnson VL, Albertella M, Taylor SS 2001. Aneuploid colon cancer cells have a robust spindle checkpoint. EMBO Rep 2: 609–614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomasini R, Mak TW, Melino G 2008. The impact of p53 and p73 on aneuploidy and cancer. Trends Cell Biol 18: 244–252. [DOI] [PubMed] [Google Scholar]
- Torres EM, Sokolsky T, Tucker CM, Chan LY, Boselli M, Dunham MJ, Amon A 2007. Effects of aneuploidy on cellular physiology and cell division in haploid yeast. Science 317: 916–924. [DOI] [PubMed] [Google Scholar]
- Torres EM, Dephoure N, Panneerselvam A, Tucker CM, Whittaker CA, Gygi SP, Dunham MJ, Amon A 2010. Identification of aneuploidy-tolerating mutations. Cell 143: 71–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ullah Z, Lee CY, Lilly MA, DePamphilis ML 2009. Developmentally programmed endoreduplication in animals. Cell Cycle 8: 1501–1509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Ree JH, Jeganathan KB, Malureanu L, van Deursen JM 2010. Overexpression of the E2 ubiquitin-conjugating enzyme UbcH10 causes chromosome missegregation and tumor formation. J Cell Biol 188: 83–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vargas JD, Hatch EM, Anderson DJ, Hetzer MW 2012. Transient nuclear envelope rupturing during interphase in human cancer cells. Nucleus 3: 88–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter MJ, Payton JE, Ries RE, Shannon WD, Deshmukh H, Zhao Y, Baty J, Heath S, Westervelt P, Watson MA, et al. 2009. Acquired copy number alterations in adult acute myeloid leukemia genomes. Proc Natl Acad Sci 106: 12950–12955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang RH, Yu H, Deng CX 2004. A requirement for breast-cancer-associated gene 1 (BRCA1) in the spindle checkpoint. Proc Natl Acad Sci 101: 17108–17113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver BA, Cleveland DW 2006. Does aneuploidy cause cancer? Curr Opin Cell Biol 18: 658–667. [DOI] [PubMed] [Google Scholar]
- Weaver BA, Silk AD, Montagna C, Verdier-Pinard P, Cleveland DW 2007. Aneuploidy acts both oncogenically and as a tumor suppressor. Cancer Cell 11: 25–36. [DOI] [PubMed] [Google Scholar]
- Welch JS, Ley TJ, Link DC, Miller CA, Larson DE, Koboldt DC, Wartman LD, Lamprecht TL, Liu F, Xia J, et al. 2012. The origin and evolution of mutations in acute myeloid leukemia. Cell 150: 264–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams BR, Prabhu VR, Hunter KE, Glazier CM, Whittaker CA, Housman DE, Amon A 2008. Aneuploidy affects proliferation and spontaneous immortalization in mammalian cells. Science 322: 703–709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie C, Powell C, Yao M, Wu J, Dong Q 2014. Ubiquitin-conjugating enzyme E2C: A potential cancer biomarker. Int J Biochem Cell Biol 47: 113–117. [DOI] [PubMed] [Google Scholar]
- Yaffe D, Feldman M 1965. The formation of hybrid multinucleated muscle fibers from myoblasts of different genetic origin. Dev Biol 11: 300–317. [DOI] [PubMed] [Google Scholar]
- Yurov YB, Iourov IY, Vorsanova SG, Liehr T, Kolotii AD, Kutsev SI, Pellestor F, Beresheva AK, Demidova IA, Kravets VS, et al. 2007. Aneuploidy and confined chromosomal mosaicism in the developing human brain. PloS ONE 2: e558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zack TI, Schumacher SE, Carter SL, Cherniack AD, Saksena G, Tabak B, Lawrence MS, Zhang CZ, Wala J, Mermel CH, et al. 2013. Pan-cancer patterns of somatic copy number alteration. Nat Genet 45: 1134–1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Taub JW, Williamson M, Wong SC, Hukku B, Pullen J, Ravindranath Y, Matherly LH 1998. Reduced folate carrier gene expression in childhood acute lymphoblastic leukemia: Relationship to immunophenotype and ploidy. Clin Cancer Res 4: 2169–2177. [PubMed] [Google Scholar]
- Zhu J, Pavelka N, Bradford WD, Rancati G, Li R 2012. Karyotypic determinants of chromosome instability in aneuploid budding yeast. PLoS Genet 8: e1002719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuang Z, Park WS, Pack S, Schmidt L, Vortmeyer AO, Pak E, Pham T, Weil RJ, Candidus S, Lubensky IA, et al. 1998. Trisomy 7-harbouring non-random duplication of the mutant MET allele in hereditary papillary renal carcinomas. Nat Genet 20: 66–69. [DOI] [PubMed] [Google Scholar]