Abstract
The green lineage of chlorophyte algae and streptophytes form a large and diverse clade with multiple independent transitions to produce multicellular and/or macroscopically complex organization. In this review, I focus on two of the best-studied multicellular groups of green algae: charophytes and volvocines. Charophyte algae are the closest relatives of land plants and encompass the transition from unicellularity to simple multicellularity. Many of the innovations present in land plants have their roots in the cell and developmental biology of charophyte algae. Volvocine algae evolved an independent route to multicellularity that is captured by a graded series of increasing cell-type specialization and developmental complexity. The study of volvocine algae has provided unprecedented insights into the innovations required to achieve multicellularity.
Multicellularity requires several key innovations (e.g., cell–cell communication and adhesion). Two groups of green algae, charophytes and volvocines, provide unprecedented insights into the evolution of these innovations.
The transition from unicellular to multicellular organization is considered one of the major innovations in eukaryotic evolution (Szathmáry and Maynard-Smith 1995). Multicellular organization can be advantageous for several reasons. Foremost among these is the potential for cell-type specialization that enables more efficient use of scarce resources and can open up new adaptive niches (Stanley 1973; Szathmáry and Maynard-Smith 1995; Ispolatov et al. 2012). In addition, multicellularity allows organismal size to scale independently of cell size, thus freeing organisms from the constraints of individual cells and allowing them to evolve fundamentally new relationships with their physical and biological surroundings (Beardall et al. 2009). A third potential advantage of multicellularity and increased organismal size is escape from microscopic predators such as ciliates and rotifers that are limited by prey size (Bell 1985; Boraas et al. 1998). Reciprocally, increased size might also entail advantages in capturing more or larger prey.
There is some debate about how easy or difficult it has been for unicellular organisms to evolve multicellularity (Grosberg and Strathmann 2007). A conceptual division of multicellularity into two classes, simple and complex, has also been proposed (Knoll 2011). However, the argument that simple multicellularity is easier to achieve than complex multicellularity may be less compelling than it seems at first glance. In the ∼2-billion year history of eukaryotic life, “simple” multicellularity has arisen about two dozen times (Grosberg and Strathmann 2007; Knoll 2011; Adl et al. 2012). In contrast, complex multicellularity (defined by three-dimensional body plans and multiple cell types) has only arisen a few times with animals and plants being clear examples, and fungi, brown algae and red algae achieving somewhat lesser extents of complexity. In any case, because complex multicellularity almost certainly evolved from simple multicellularity, the odds of going from simple to complex multicellularity are quite high (approximately one in 12), whereas the initial transition from unicellular to simple multicellular seems far more difficult (one in thousands).
Achieving the benefits of multicellularity required evolutionary innovations. At minimum, multicellularity requires adhesive interactions between cells to maintain a coherent, physically connected form. Many multicellular organisms maintain not only adhesive connections between cells, but also physical bridges that allow the transfer of cytoplasmic materials directly between adjacent cells and mediate cell–cell communication. Diffusible extracellular molecules such as hormones are another means of signaling between cells that is common in multicellular organisms. In addition to adhesion and cell–cell communication, the overall size and shape of multicellular organisms typically follows a defined architecture or patterning that involves spatially coordinated growth and division among groups of cells. A final requirement for differentiated multicellular organization is regulation of reproductive potential. Reproduction in multicellular organisms is often confined to a subset of cells (germ cells or stem cells). This division of labor serves at least two purposes: First, by partitioning or restricting reproductive capacity to a subset of cells, genetic conflicts associated with a transition of fitness from individual cells to cooperating groups of cells can be mitigated (Michod and Roze 2001). Second, in multicellular organisms with indeterminate body plans such as land plants, vegetative stem cells play a critical role in integrating internal and external signals to regulate and establish overall organismal architecture (Smolarkiewicz and Dhonukshe 2013).
An outstanding question in the evolution of multicellularity is its early origins. It is increasingly clear that the path to multicellular evolution is dependent on the genetic and cellular “toolkits” available in unicellular ancestors, as well as on the specific circumstances and environment in which multicellularity arose (Rokas 2008; Knoll 2011; Bowman 2013; Niklas and Newman 2013; Smolarkiewicz and Dhonukshe 2013). By examining the origins of multicellularity in diverse lineages, it can be determined whether commonalities exist that transcend lineage-specific solutions to achieve multicellular organization.
UNIQUE ASPECTS OF MULTICELLULARITY IN PLANTS AND GREEN ALGAE
Plants and animals are interesting focal groups because they arose from unicellular ancestors that appear to have had very different propensities or success rates in experimenting with multicellular organization. The closest unicellular ancestors to animals, the choanoflagellates, show a limited degree of colonial or multicellular organization outside of their one big hit in the metazoan lineage (Dayel et al. 2011; Alegado and King 2014). The streptophytes (charophyte algae + embryophytes [land plants]) and their sister group, the chlorophytes (green algae), have repeatedly generated multicellular taxa as well as macroscopic unicellular forms that show many of the traits that are typically considered hallmarks of multicellularity (Fig. 1) (De Clerck et al. 2012; Leliaert et al. 2012). This diversity in the green lineage represents a potential treasure trove of information on the evolution of multicellular development. A second reason to focus on plants and algae involves the fundamental constraint that a cell wall places on the ability of cells to move and reorganize spatially. This constraint, which is not present in metazoans, has shaped the means by which multicellular organization arose in the green lineage.
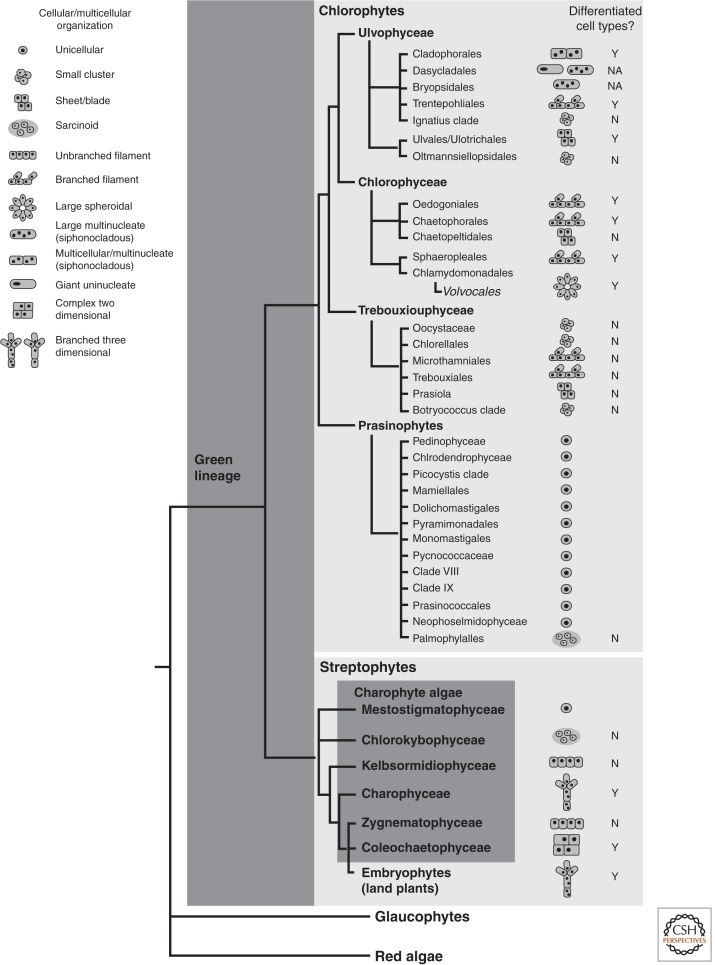
Figure 1.
Multicellularity and morphological complexity in the green lineage. Simplified cladogram based on data in Leliaert et al. (2012) and De Clerck et al. (2012) showing relationships among the green lineage (chlorophytes and streptophytes) and their sister groups, red algae and glaucophytes, which together comprise the Archaeplastida. The major classes in the green lineage are in bold and, in some cases, further expanded into subgroups. Note that the prasinophytes are a paraphyletic grouping of the most primitive chlorophyte algae. The streptophytes are divided into the charophyte algae (a paraphyletic grade) and the embryophytes (land plants). To the right are depicted the most complex type of cellular/multicellular organization in each clade and whether cell type differentiation occurs in multicellular forms. NA, not applicable for giant unicellular forms.
Cellular Diversity in the Green Lineage
Among unicellular green algae are found spectacular amounts of cellular diversification (Lewis and McCourt 2004; Leliaert et al. 2012). The ancestral species in the green lineage were likely to be small unicellular marine biflagellates that originated around 1 billion years ago (De Clerck et al. 2012), and this form is still prevalent among modern aquatic algae. A well-studied example of a unicellular biflagellate is Chlamydomonas reinhardtii whose highly stereotyped cell architecture is described later in this review. Figure 1 shows a simplified phylogenetic tree depicting the major lineages of green algae and their land plant relatives discussed in this review.
Reductions in cell size and cellular complexity have occurred multiple times among green lineage resulting in small coccoid species such as Ostreococcus that have lost flagella and probably underwent genomic reduction and streamlining (Derelle et al. 2006). On the other hand, some of the largest and most complex single cells on earth are found among green algae, including species such as Acetabularia whose wine-cup-shaped cells are measured in centimeters (Mine et al. 2008). Additional cellular diversity in microalgae includes different cell shapes such as crescents, rectangles, ovoids, elongated spines, and flattened discs. Some species have no cell wall, whereas walled species use a variety of wall polymers including cellulose, pectins, chitin-like molecules, or hydroxyproline-rich glycoproteins. Besides sugar-polymer-based walls, some species cover themselves with hard calcified scales (Popper et al. 2011; Domozych et al. 2012; Leliaert et al. 2012).
Cell division in green organisms can occur through binary fission, but a form of division called multiple fission (or palintomy) is also common in chlorophyte algae (Pickett-Heaps 1975; Šetlík and Zachleder 1984; Graham et al. 2009). In multiple fission cell cycles, mother cells divide more than once in rapid succession to produce 2n daughters (2,4,8,16, …) within a single mother cell wall. Multiple fission has been exploited by green algae as a means of forming multicellular colonies, most notably in the volvocine algae that are discussed below. Different mechanisms of cytokinesis have also evolved in the green lineage including a specialized microtubule structure called the phycoplast, as well as an analogous structure, the phragmoplast, which is used in streptophytes (Pickett-Heaps 1976). The former mechanism involves membrane furrowing guided by microtubules that are parallel to the plane of division, whereas the latter involves formation of new cross-walls and plasma membrane at the overlap region between parallel microtubules emanating from each daughter nucleus after mitosis.
Chlorophyte and charophyte algae generally have haploid dominant life cycles with 1n gametophyte cells that can reproduce vegetatively (i.e., asexually). In gametophyte or haploid dominant life cycles, growth and proliferation are generally restricted to the haploid (1n) phase, whereas the diploid (2n) phase is a zygotic spore. Nonetheless, the Ulvophytes (a predominantly multicellular order of chlorophyte algae) contain genera with a significant diploid sporophytic stage. This derived state of sporophytic growth and development also evolved independently in the embryophytes (land plants) in which the gametophyte stage was concomitantly reduced and the sporophyte dominant life cycle predominates (Niklas and Kutschera 2009).
Many species also have a sexual cycle in which at least one of the gametes is a unicellular biflagellate. Anisogamy and oogamy (sperm and egg-mating systems) evolved more than once in different chlorophyte subgroups from isogamous mating systems seen in most unicelluar species. Although aflagellate gametes are found in some charophyte algae such as Zygnematales that mate via a specialized process called conjugation (Graham et al. 2009; Sekimoto et al. 2012), the ancestral state for sexual reproduction in early land plants was presumably isogamy with oogamous mating evolving in advanced orders (McCourt et al. 2004). The subsequent evolution of aflagellate, desiccation-resistant pollen in advanced land plants freed their male gametes from the requirement for moisture and allowed them to disperse through the air or via animal pollinators.
Macroscopic Complexity in Green Algae Evolved Using Different Strategies
Green algae challenge the notion that equates multicellularity with organismal complexity. Described below are three strategies used in the green lineage to achieve macroscopic complexity, two of which do not involve conventional multicellularity.
Multicellular Algae
Independent transitions to multicellularity or macroscopic forms occurred multiple times in the chlorophyte lineage (Fig. 1). True multicellular forms are characterized by having separate cells, each with its own nucleus, plasma membrane, and cell wall. Streptophytes and many multicellular chlorophyte algae are organized in this manner. It should be noted that connections between neighboring cells (e.g., plasmodesmata) evolved multiple times in the green lineage and provide direct cytoplasmic continuity between cells in many multicellular algae and land plants, thus allowing them some degree of supracellular organization (Raven 1997). Some common multicellular forms are unconnected groups of cells embedded in extracellular matrix, short chains, unbranched and branched filaments with and without differentiated cell types, geometric networks, large leaf-like blades, discoidal colonies, and spheroidal colonies (Fig. 1).
Giant Uninucleate Algae
An alternative form of complex and macroscopic unicellular organization is found in giant unicellular algae that possess a single nucleus. The most well-known species of this type are in the genus Acetabularia (an ulvophyte) that is famous for transplantation experiments demonstrating nuclear control over cell morphology (Mandoli 1998; Mine et al. 2008).
Coenocytic Algae
Coenocytic algae (also referred to as siphonous algae) are large single cells that contain multiple nuclei housed within a common cytoplasm. The sizes of these forms are measured in centimeters and meters making them some of the largest single cells on earth. Despite their lack of multicellular organization, coenocytic algae can form remarkably complex structures with multiple types of specialized subdomains. For example, the multinucleate cells of the marine alga Caulerpa reach meters in size and contain three distinct tissue types that function similarly to roots, stems, and leaves (Jacobs 1970). Moreover, some green algae are both multicellular and coenocytic (termed siphonocladous) because they are composed of multiple cells, each of which contains multiple nuclei (e.g., Cladophora). Cytoplasmic continuity and developmental patterning in coenocytic macroscopic algae are beyond the scope of this review, but are covered in other recent reviews (Baluška et al. 2004; Cocquyt et al. 2010; Niklas et al. 2013).
CHAROPHYTE GREEN ALGAE AND THE ORIGINS OF LAND PLANT MULTICELLULARITY
Overview
Charophyte algae have long been recognized as the closest algal relatives of land plants based on cellular and morphological features (Stewart and Mattox 1975). This view has been corroborated and refined using molecular phylogenetics that have begun to clarify relationships within charophyte algae and with their sister clade, the land plants (embryophytes) (Turmel et al. 2002; Finet et al. 2010; Wodniok et al. 2011; Laurin-Lemay et al. 2012; Timme et al. 2012; Zhong et al. 2013).
A number of land plant multicellular traits originated within the streptophytes (charophyte algae + embryophytes). These include apical meristems, cells that undergo asymmetric division and differentiation, complex branching patterns, matrotrophic support for a multicellular sporophyte, and plasmodesmata that form symplastic connections between cells. Additional cellular and biochemical traits that are streptophyte specific include a hexameric cellulose synthase complex that synthesizes the primary cell wall, the phragmoplast as a mechanism for cytokinesis, synthesis, and transport of phytohormones, and several metabolic specializations (Graham 1996; Graham et al. 2000; Waters 2003; Becker and Marin 2009).
Later changes and innovations that occurred within the embryophytes well after they achieved multicellularity include three-dimensional growth and cell division to produce complex tissues and organs, cuticular waxes, and stomatal pores to control evaporative losses, vascular transport systems, root and shoot apical meristems, transition to a sporophyte dominant life cycle, specialized and complex hormonal signaling, nonflagellated and desiccation-resistant male gametes (i.e., pollen), flowers, and lignified secondary cell walls (Langdale and Harrison 2008). Interestingly, molecular evidence for proteins related to some of these later innovations in embryophytes have been found in charophytes as described below.
It has been argued that the existing charophyte algal orders are actually the remnants of a terrestrial invasion that gave rise to land plants, but whose members were outcompeted by the embryophytes. The remaining extant orders of charophyte algae ended up secondarily aquatic where they were able to compete more effectively than on land (Stebbins and Hill 1980). Although this idea is difficult to test directly, the finding that a nominally aquatic charophyte species, Coleochaete orbicularis, can grow in terrestrial environments and even show adaptive changes in body plan under these conditions supports this idea (Graham et al. 2012). It has also been argued that early streptophytes were preadapted to the colonization of land by being the first algal group to conquer freshwater habitats, whereas chlorophyte algae, which predominate in marine environments, moved into freshwater much later (Becker and Marin 2009).
A closer examination of charophyte evolution may provide an outline for understanding some of the early multicellular innovations in the embryophytes. It is important to note that the extant charophyte lineages are highly diverged from each other and have probably undergone secondary character losses that complicate comparisons and inferences about the progression to multicellularity in the embryophytes (McCourt et al. 2004; Timme et al. 2012; Bowman 2013). Moreover, the fossil record of charophyte algae is incomplete and does not extend earlier than that of land plants, so is not a reliable source of information on timing for emergence of early embryophytes whose earliest fossils appear ∼500 million years ago (Leliaert et al. 2012). As noted above, the charophyte algal order that is most closely related to land plants is unresolved, but it is clear that the most developmentally complex order, Charophyceae (stoneworts), is probably not the closest relative of the embryophyte lineage (Wodniok et al. 2011; Laurin-Lemay et al. 2012; Timme et al. 2012; Zhong et al. 2013). Despite this ambiguity and the likelihood of both character loss and gain in the extant charophyte algal orders, some of the steps leading to multicellularity in the embryophytes can still be inferred.
To date, there are no sequenced charophyte genomes, so molecular evidence regarding genes and pathways that are shared within streptophytes is somewhat piecemeal. Current understanding of molecular evolution of charophytes algae has been guided mostly by expressed sequence tag (EST) sequences and directed investigations of specific gene families (Simon et al. 2006; Timme and Delwiche 2010; Vannerum et al. 2011; Wodniok et al. 2011).
Key Innovations in the Streptophyte Transition to Multicellularity
Given the uncertainties surrounding the phylogeny of charophyte algae and the apparent gain and loss of some traits, the order of topics listed below is meant only to convey a plausible picture of how multicellular innovations were acquired in the embyrophyte lineage as summarized in Figure 2.
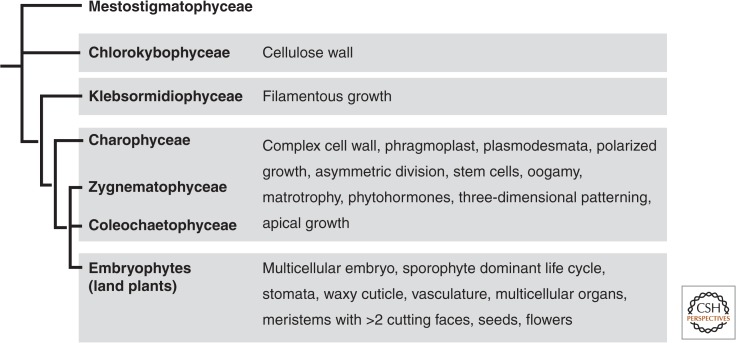
Figure 2.
Multicellular innovations in charophyte algae and embryophytes. Simplified cladogram based on data in Leliaert et al. (2012) showing relationships among major orders of charophyte algae and embryophytes. To the right are shown multicellular innovations associated with different groups. Note that reductions or losses of traits occurred in some orders of advanced charophyte algae (Charophyceae, Zygnematophyceae, Colechaetophyceae), most notably in the Zygnematophyceae (McCourt et al. 2004), so the set of innovations in this group may not apply to all three orders.
The Closest Unicellular Relative of Land Plants
The unicellular ancestor of Streptophytes was a freshwater biflagellate whose most similar extant relative is in the genus Mesostigma (Bhattacharya et al. 1998; Marin and Melkonian 1999; Lemieux et al. 2000, 2007; Rodriguez-Ezpeleta et al. 2006). Mesotigma does not produce cellulose or any type of cell wall, but instead has calcified scales. It divides by furrowing rather than through formation of a phragmoplast (Manton and Ettl 1965). This ancestral form likely had an open mitosis as observed in early diverging charophytes (Stewart and Mattox 1975; Pickett-Heaps 1976). A systematic investigation of its genetic toolkit has not been performed, but several studies have identified streptophyte-specific gene families in Mesostigma (Nedelcu et al. 2006; Petersen et al. 2006; Simon et al. 2006; Grauvogel and Petersen 2007). A genome sequence and development of molecular genetic tools would allow deeper investigation into the cell biology of these unicellular plant relatives and how much of the developmental toolkit for Embyrophytes was present in their most recently diverged unicellular ancestor.
Filamentous Growth and Cytokinesis
The early multicellular forms produced by primitive embyrophytes were probably undifferentiated filaments, a growth habit that is still common in many green algal lineages including extant charophyte algae such as Klebsormidiales, which are among the three most primitive orders of this grade. In all orders except Mesostigmaphyceae, charophyte algae lost motility in the vegetative stage, but with the exception of Zygnematales, retained flagellated zoospores (Stewart and Mattox 1975). The prevalence of filamentous forms in both aquatic and terrestrial environments and diverse algal lineages suggests that filamentation has multiple selective advantages. Filaments can be used to anchor colonies in place efficiently while allowing maximal contact with the surrounding environment along each cell’s surface, a property that is likely to be useful for nutrient exchange in both aquatic and terrestrial habitats (Niklas 2000). Filaments would also serve as protection against grazing predators by being too large to ingest, an idea that has been tested experimentally with filamentous bacteria (Güde 1979; Shikano et al. 1990). A third advantage of filaments is their utility for nutrient foraging because they direct growth along a vector that maximizes exploration of surrounding space. Even without resource sharing between cells (i.e., symplastic connections), the clonal nature of filaments ensures a benefit to growth-directed foraging because all cells in the filament share the same genotype.
Filament formation requires at least one axis of cell polarity to ensure that growth occurs longitudinally and the plane of cytokinesis is perpendicular to the plane of growth. An innovation required for filamentation in charophytes involved the deposition of common cell wall material between daughters. Descriptive and comparative studies have been performed on cytokinesis in primitive filamentous charophytes that occurs after an open mitosis, but does not appear to involve a phragmoplast or phycoplast (Floyd et al. 1972; Pickett-Heaps 1976; Lokhorst and Star 1985; Katsaros et al. 2011). The phragmoplast is still considered an important innovation and originated in the Zygnematales, a transitional group wherein different types of cytokinetic patterns coexist. Some groups in this order divide centripetally (e.g., filamentous desmids) (Hall et al. 2008) whereas others, such as the genus Spirogyra, appear to use a reduced but recognizable phragmoplast (Fowke and Pickett-Heaps 1969; Galway and Hardham 1991), although with some functional distinction from embryophyte phragmoplasts (Sawitzky and Grolig 1995). It is not known whether the shared wall between primitive filamentous charophyte algal cells are specialized in any way, but it is known that their walls lack many of the complex carbohydrate polymers present in advanced charophytes and embryophytes (Sørensen et al. 2010, 2011; Domozych et al. 2012). The more advanced charophyte algae (Charophyceae, Coleochaetophyceae) do have a recognizable phragmoplast that resembles those of embryophytes (Pickett-Heaps 1967; Marchant and Pickett Heaps 1973). Coincident with the evolution of the phragmoplast in advanced charophyte algae is the appearance of what might be a preprophase band of microtubules that marks the future plane of cell division (Galway and Hardham 1991). Detailed molecular information on primitive charophyte cell division is still limited, but these early filamentous divisions in Spirogyra described above may have paved the way for evolution of the phragmoplast and preprophase band as an efficient mechanism for marking division sites and subdividing cells with a rigid wall.
Regulated Division Plane, Cellular Differentiation, and Polarized Growth
Contrasting with simple filamentous forms in which cell growth occurs similarly in all constituent cells, polarized growth involves some form of tissue organization that dictates which cells in a filament or thallus (body) grow, and the directionality of growth. Species from both the Coleochaetophyceae and Charophyceae have spatially regulated division planes. Coleochaetales show diverse vegetative forms that include flat, disk-shaped thalli, branched filaments, and pseudoparenchymatous filaments that are held together within an extracellular matrix (Graham et al. 2009). The discoid species Coleochaete orbicicularis grows outward by a combination of cell expansion and either anti- or periclinal cell divisions along its marginal cells (Cook 2004). Cell division planes are set according to a set of simple rules based on position within the thallus, cell size, and cell shape (Dupuy et al. 2010). Moreover, the division plane orientation behavior of C. orbicularis can be modeled as an energy minimization function that can also be applied to embryophytes where it may be an inherited mechanism that specifies a default pattern of symmetrical cell division (Besson and Dumais 2011).
Asymmetric and/or branched histogenetic cell divisions are critical for development in the Coleochatelaes and Charales (Graham 1996; Graham et al. 2000), although little is known how such divisions are controlled. In angiosperms, asymmetric divisions are also crucial for development and regulated by different genetic pathways depending on tissue type (De Smet and Beeckman 2011). Although a homolog of a gene required for asymmetric division in Arabidopsis zygotes, GNOM, has been identified in Coleochaetes, the significance of this finding is unknown (Timme and Delwiche 2010). The relationships between charophyte algal asymmetric cell divisions and those in embryophytes remain to be investigated.
Although more distantly related to land plants than Coleochaetphyceae and Zygnematophyceae, Charaophyceae (e.g., Chara and Nitella) (Fig. 2) show morphologies and growth patterns that more closely resemble those of land plants. These similarities include a long, branched body axis and indeterminate apical meristem cell. Charophyceans do not have multicellular tissues, but their giant multinucleated cells show differentiation into specialized types including long intermodal cells, nodal cells that undergo asymmetric cell divisions to form branches, and rhizoid cells that act as holdfasts and show tip-polarized growth (Cook et al. 1998; Braun and Limbach 2006). Charophyceae also have male (antheridia) and female (oogonia) sexual reproductive structures (see below) that are formed by complex division patterns to generate simple three-dimensional multicellular tissues (Fritsch 1948; Pickett-Heaps 1968; Kwiatkowska 2003; Graham et al. 2009).
Polarized gravitropic growth of the apical meristem and rhizoid cells in Chara and Nitella has been investigated, particularly with respect to actin cytoskeletal dynamics (Braun and Limbach 2006). However, little is known about other developmental patterning cues in charophytes such as light signaling (Rethy 1968; Dring 1988). The discovery of plant hormones in some charophyte algal species (see below) suggests that phytohormones may be an early innovation that enabled the evolution of charophyte multicellularity.
Symplastic Connections and Intercellular Communication
In land plants, plasmodesmata connect adjacent cells allowing metabolites and some macromolecules to pass between them, as well as allow tissue-level interactions among cells (Lucas and Lee 2004). Plasmodesmata are found in two of the advanced orders of charophyte algae, Coleochaetophyceae and Charophyceae, but intercellular connections in algae are widespread and not unique to charophytes (Stewart and Mattox 1975; Raven 1997). In primitive embryophytes, connections between adjacent cells would enable communication, allow metabolite/resource sharing, and promote cell specialization. The multifaceted roles of plasmodesmata in shaping plant development and evolution has been reviewed recently (Lucas and Lee 2004; Hernández-Hernández et al. 2012). Information on how intercellular communication modulates development in charophyte algae is limited, but has been studied in some depth in male reproductive tissue (antheridia) in Chara, in which there is compelling evidence that remodeling of plasmodesmata is important for maintaining and coordinating cell division synchrony and developmental fate (Kwiatkowska 2003). In both embryophytes and charophyte algae, plasmodesmata can be formed during cytokinesis or secondarily in interphase cells by remodeling of walls and fusion of adjacent plasma membranes (Hepler 1982; Franceschi et al. 1994), but the molecular details of plasmodesmata formation in charophyte algae and their degree of similarity to embryophyte plasmodesmata have not been established (Faulkner et al. 2005; Timme and Delwiche 2010).
Hormonal Signaling
Phytohormones are essential regulators of plant development and environmental responses. Besides contributing to developmental plasticity, small diffusible hormones facilitate long-distance communication that may occur on a macroscopic scale to coordinate responses across tissues and cell types in a multicellular organism. The presence of growth-promoting phytohormones in algae has been investigated in diverse taxa (Bradley 1991; Johri 2004; Tarakhovskaya et al. 2007; Stirk et al. 2013a,b), but the molecular bases of phytohormone action in algae is not well understood. Physiological studies on the effects of plant hormones have shown that charophyte algae produce and respond to several phytohormones, indicating an early origin of these chemical signals. Because genome sequences are not available, inferences about the presence of known signaling and biosynthetic machinery for Embyrophyte phytohormones in charophyte algae can only be made from the presence of gene families, but not from their absence.
Indole derivatives (e.g., indole acetic acid) in the auxin family are produced in Chara (Sztein et al. 2000) and are known to affect development by altering shoot elongation, branching, and rhizoid elongation, for example (Imahori and Iwasa 1965; Klämbt et al. 1992; Cooke et al. 2002), and may act through changes in cytoskeletal organization (Jin et al. 2008). Directional transport of auxin in long, giant intermodal cells of Chara has been shown recently, suggesting that this phenomenon may be ancient (Boot et al. 2012; Raven 2013). Some of the auxin biosynthetic enzymes entered the green lineage before the streptophyte–chlorophyte divergence, but the response and transport proteins characteristic of the well-studied auxin-signaling networks in angiosperms first appeared in charophytes, and the full complement of core auxin-signaling proteins became established in early embryophytes (Lau et al. 2009; Prigge et al. 2010; De Smet et al. 2011; Wodniok et al. 2011).
Abscisic acid (ABA) is an isoprenoid-derived stress and drought hormone whose function can be traced back to the earliest embryophyte clade, the bryophytes (Cutler and Rodriguez 2010; Khandelwal et al. 2010; Hauser et al. 2011). Although its function in land plants is associated with the transition to terrestrial life, ABA is also produced by a wide variety of aquatic green algae, although its function in these algae is unknown (Hartung 2010). G protein–coupled receptor G proteins in plants have been linked to ABA signaling in Arabidoposis and have recently been identified in Chara, but not in any chlorophyte algae (Pandey and Assmann 2004; Hackenberg et al. 2013). Studies on spore germination in Chara suggest a possible inhibitory role for ABA in this process that is antagonized by another hormone, gibberrellic acid (GA), which also promotes growth (Imahori and Iwasa 1965; Sabbatini et al. 1987; Sederias and Colman 2007). The apparently antagonistic roles of GA and ABA in Chara are reminiscent of similar roles in Angiosperm seed germination, suggesting an ancient role for this hormonal system in regulating dormancy that may have preceded a role for ABA in drought responses. GA has also been reported to affect growth and development of male reproductive cells (antheridia) in Chara by up-regulating sperm production and endoreplication of support cells (Kwiatkowska et al. 1998). Classical GA-signaling and perception proteins have not been found in charophyte algae (Vandenbussche et al. 2007), although genomic information is unavailable so this negative result awaits confirmation.
Strigolactones (SLs) are cyclic derivatives of carotenoids that have roles in biotic interactions of angiosperms and developmental control of branching architecture (de Saint Germain et al. 2013). In the Bryophyte Physcomitrella, SLs influence branching and sensing of adjacent colonies (Proust et al. 2011). Remarkably, SLs have been identified in the Charophyceae (but not other charophyte algae orders) and assays of SL-treated Chara revealed a developmental response of rhizoid cell elongation (Ruyter-Spira and Bouwmeester 2012).
Much less is known about other land plant hormones in charophyte algae including brassinosteroids, cytokinins, and ethylene. Brassinosteroids and cytokinins have been detected in charophyte algae, and at least one study has reported an effect of cytokinins on growth of Chara (Imahori and Iwasa 1965). Ethylene is reported to bind to Chara (Wang et al. 2006), whereas some ethylene biosynthesis and perception genes were found in charophyte algae ESTs (Timme and Delwiche 2010; Wodniok et al. 2011), but data on the effect of ethylene in charophyte algae growth or physiology is currently unavailable.
Oogamous Sexual Reproduction
In complex multicellular organisms, sexual reproduction is typically oogamous with small motile sperm and large immotile eggs, whereas unicellular or simple multicellular organisms are typically isogamous (having equal-sized gametes). Various explanations for the evolution of anisogamy and oogamy have been put forth (Randerson and Hurst 2001), but one of the earliest by Parker may be particularly relevant to the evolution of multicellularity (Parker et al. 1972). The crux of this argument is the fitness advantage of making a large zygotic cell that is capable of forming a functioning multicellular progeny. Disruptive selection drives one gamete type to be much larger than a single cell so that it can provide resources to the zygote, and the other to be as small and numerous as possible to ensure efficient fertilization.
Sexual reproduction in charophyte algae has been described in the higher orders Charophyceae, Zygnematophyceae, and Coleochaetophyceae, but has not been observed in the lower orders in which it is assumed to be isogamous (McCourt et al. 2004). It is also assumed that charophyte algae have a haploid dominant life cycle similar to most chlorophyte algae and primitive embryophytes, although this assumption has been questioned (Haig 2010). Zygnematales have the best-studied sexual cycles in the charophyte algae, but have secondarily lost flagella and reproduce through isogamous conjugation (Graham et al. 2009; Sekimoto et al. 2012). Both Charophyceae and Coleochaetophyceae have oogamous sexual cycles with specialized egg and sperm cells. Very little is known about the genetics of sex determination in these two orders in which sexual development is usually monoecious (male and female gametes formed in the same organism). As mentioned above, the Charophyceae have particularly striking and elaborate reproductive tissues (male antheridia and female oogonia) that include both gametic cells and support cells (Fritsch 1948; Pickett-Heaps 1968; Sawa and Frame 1974; Beilby and Casanova 2014). Understanding the genetic programs that give rise to reproductive tissues in charophyte algae and how they relate to those of embryophytes may shed light on whether oogamy is a synapomorphic trait within streptophytes.
Matrotrophic Support of the Zygote
A conserved feature in embryophytes is maternal support provided to zygotes and sporophyte embryos after fertilization (Ligrone et al. 1993). This innovation may be related to oogamy because it increases resource allocation to a developing multicellular offspring. The attachment and nourishment of the developing embryo by maternal tissue may have been crucial for establishing a later change in embrophyte evolution, the sporophyte dominant life cycle (Graham and Wilcox 2000). The best-documented occurrence of matrotrophy in charophyte algae comes from studies of Coleochate in which the developing zygote remains associated with the vegetative thallus (Graham 1984). These studies identified regions of the vegetative maternal cells specifically at the zygote interface that appeared specialized for maximal cell–cell contact and nutrient transport (Graham and Wilcox 1983; Delwiche et al. 1989). The molecular details of these junctions and how they form is not known, but their presence in advanced charophyte algae and similarity to embryophyte maternal-zygotic junctions suggests a common origin.
Multicellular Innovations in Land Plants that May Be Rooted in the Green Lineage
The above innovations in the charophyte algae can be directly tied to the transition to multicellularity in the streptophyte lineage. Other important properties of embryophytes that are not directly related to acquisition of multicellularity may also have roots in charophyte algae and the green lineage. Cellulosic walls are found throughout the green algal lineage and in other taxa, but a hexameric cellulose synthase complex evolved uniquely in streptophytes (Tsekos 1999). Other distinctive characteristics of embryophyte cell walls, including complex carbohydrate polymer linkages, first appeared in advanced charophyte green algae (Van Sandt et al. 2007; Popper et al. 2011; Sørensen et al. 2011; Domozych et al. 2012; Proseus and Boyer 2012). The recent discovery of expansin genes in the Zygnematophycean alga Micrasterias suggests that these critical cell wall remodeling proteins from embryophytes originated in charophyte algae (Cosgrove 2000; Vannerum et al. 2011). Lignified secondary walls were a critical innovation in land plants for desiccation tolerance, load-bearing structural integrity, and decay resistance (Boyce et al. 2004). Although charophyte algae do not have secondary walls, lignin or lignin-like molecules have been identified in this group and may contribute to resistance against microbial attack (Gunnison and Alexander 1975; Delwiche et al. 1989; Sørensen et al. 2011). Sporopollenin is a complex molecule found in the walls of angiosperm pollen that protects cells from physical and chemical degradation (Ariizumi and Toriyama 2011; Wallace et al. 2011). However, the origins of sporopollenin-like molecules goes as far back as the charophyte algae in which sporopollenin-like substances were identified in the zygotes of Coleochate (Delwiche et al. 1989). Both lignin- and sporopollenin-like substances are found outside of the streptophyte lineage, so without molecular data about the origins of biosynthetic genes for these compounds in charophyte algae, the possibility of convergence or horizontal transfer into embryophytes cannot be ruled out (Brooks and Shaw 1978; Emiliani et al. 2009; Martone et al. 2009).
Although not directly associated with the transition to multicellularity in land plants, the alteration of generations is a hallmark of the embryophytes in which simple, multicellular sporophytes first appeared (Graham and Wilcox 2000). Interestingly, the genetic program that directs sporophyte development in the bryophyte moss Physcomitrella is directed by a KNOX family homeodomain transcription factor whose homologs in the chlorophyte alga C. reinhardtii direct development of the dormant diploid zygospore formed after fertilization (Lee et al. 2008; Nishimura et al. 2012; Sakakibara et al. 2013). The role of these homeodomain proteins in charophyte algae has not been examined, but they are predicted to participate in zygote development.
How Much of the Embryophyte Molecular Genetic Toolkit Originated in Charophyte Algae?
Although data are incomplete, some progress has been made in identifying charophyte algal homologs of key developmental regulators used by embryophytes (Bowman et al. 2007; Pires and Dolan 2012; Bowman 2013). Without complete genome sequences this list is relatively short and incomplete, but provides a sample of potential discoveries that might be made with increased knowledge of charophyte algal genomes and developmental patterning mechanisms.
MADS-box transcription factors are found within and outside the green lineage, but a specific MADS protein subtype, MIKC, defined by the presence of four characteristic domains, is found only in streptophytes and underwent a dramatic expansion in the embryophytes in which members control many aspects of sporophyte developmental patterning (Smaczniak et al. 2012). MIKC-type MADS box genes are also present in advanced charophyte orders in which their expression was analyzed (Tanabe et al. 2005). In Chara, MADS-box gene expression was restricted to reproductive tissues (antheridia and oogonia), whereas expression was detected at multiple stages in a Zygnematophycean, Closterium. In bryophytes, MIKC MADS box genes are incompletely characterized, but appear to have roles in both gametophyte and sporophyte development (Quodt et al. 2007; Singer et al. 2007; Zobell et al. 2010).
Homeodomain leucine zipper (HD-ZIP) transcription factors regulate diverse aspects of angiosperm sporophyte development including shoot apical meristem identity, abaxial-adaxial leaf patterning, and epidermal development (Elhiti and Stasolla 2009). The origin of two major groups of HD-ZIP proteins (class III and class IV) has been traced back to charophyte algae (Floyd et al. 2006; Zalewski et al. 2013). Although HD-ZIP proteins are also found in bryophytes, their function in lower plants is not well characterized (Sakakibara et al. 2001; Prigge and Clark 2006). Interestingly, a microRNA binding site that regulates expression of class III HD-ZIP expression in angiosperms is conserved in lower plants, but not in Chara, suggesting that gene family expansion and regulatory evolution contributed to functional novelty in embryophyte HD-ZIPs (Floyd et al. 2006; Floyd and Bowman 2007). Wuschel-like homeodomain proteins that are important for Angiosperm shoot apical meristem identity are also present in charophyte algae, but their role in this group and bryophytes has not been explored (Timme and Delwiche 2010).
VOLVOCINE ALGAE: A LIVING SNAPSHOT OF MULTICELLUAR EVOLUTION
Overview of Volvocine Algae
Volvocine algae provide an interesting contrast to embryophytes whose body plans are constructed through polarized growth and cell division of indeterminate meristems. Instead, multicellular volvocine algae undergo a single series of embryonic cell divisions and morphogenesis to produce a determinate body plan. How and why did volvocine algae evolve this form of multicellularity? The answer to this question can, potentially, be found by examining the cell biology of unicellular volvocine algae such as C. reinhardtii, which closely approximate the ancestral state of this lineage.
Volvocine algae are particularly well suited for investigating the origins of multicellularity. They are monophyletic and contain genera that are unicellular and multicellular. Two of the key volvocine species, unicellular C. reinhardtii and multicellular Volvox carteri, are model organisms with sequenced genomes and established molecular-genetic toolkits that make them amenable to molecular and genetic experimentation (Merchant et al. 2007; Prochnik et al. 2010; Umen and Olson 2012). V. carteri mutants with developmental phenotypes have been especially illuminating for understanding the molecular genetic origins of multicellularity (Kirk 1998, 2003).
The genus Chlamydomonas has more than 600 species whose properties are typical of unicellular chlorophytes. Although the genus is now recognized as polyphyletic, the model species C. reinhardtii is closely related to multicellular volvocine algae that together form a monophyletic clade with C. reinhardtii as an outgroup (Fig. 3A) (Pröschold et al. 2001, 2005; Herron and Michod 2008; Nakada et al. 2008). The last common ancestor of this clade was estimated to have lived ∼200 million years ago (Herron et al. 2009), but it is worth noting that the single fossil chlorophyte alga used for calibration in the above dating study has produced incongruences in other analyses and may not be part of the taxonomic group it was originally identified with (Berney and Pawlowski 2006). Multicellular or colonial volvocine algal genera can be arranged in a stepwise progression of increasing cell number, colony size, and degree of cell-type specialization (Fig. 3B). This simplistic arrangement that is termed the volvocine lineage hypothesis is probably a good approximation of how complexity evolved within specific sublineages, but does not reflect the fact that some grades of organization such as Volvox arose several times independently (Fig. 3A).
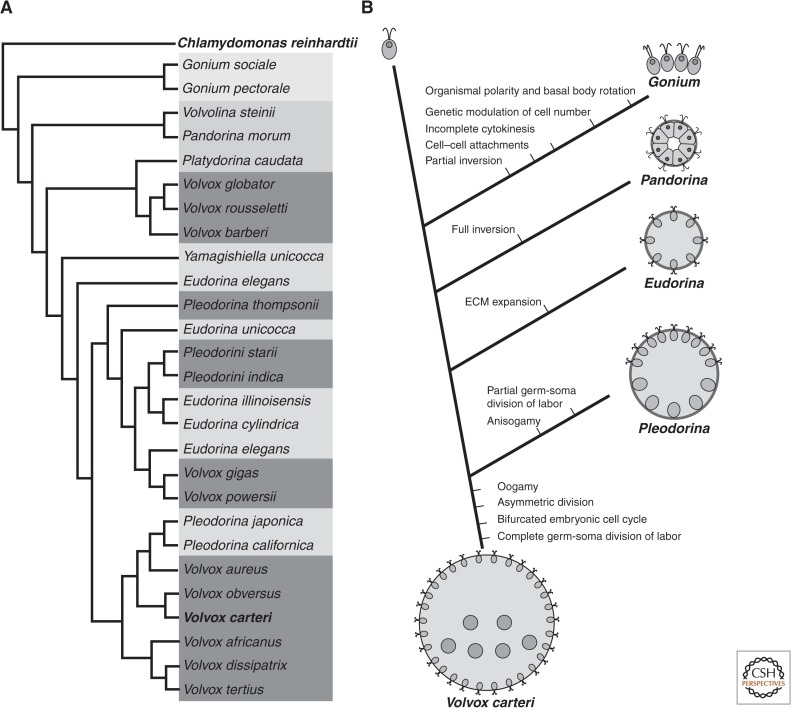
Figure 3.
Multicellular innovations in the volvocine algae. (A) Cladogram based on data in Kirk (2005) and Umen and Olson (2012) showing representative volvocine algal lineages with shading used to highlight different levels of organization in each genus. Note that the genus Volvox is polyphyletic with at least three independently derived clades. (B) Simplified cladogram based on data in Kirk (2005) showing key innovations in the steps leading from C. reinhardtii to V. carteri. Schematic cartoons of each genus are also shown.
The simplest colonial species are in the genus Gonium that form planar colonies of between four and 16 cells that show a slight curvature and have their flagella positioned on the convex side of the colony. The next step in the volvocine lineage was the formation of spheroidal colonies of 16 to 32 cells with a defined interior and exterior as typified in the genus Pandorina. In the genus Eudorina, larger spheroidal colonies of 32 to 64 cells are formed by the addition of an extensive secreted extracellular matrix (ECM) in which cells of the colony are embedded. The addition of ECM in Eudorina allows the colonies to increase in volume without any significant change in cell number or size. Up to the genus Eudorina, all the cells in a colony are reproductive; they grow as flagellated cells to provide motility and then synchronously divide to produce a set of new daughter colonies. Starting with the genus Pleodrina, colonies begin to show germ-soma differentiation. In most species of Pleodorina, there is a clearly visible anteroposterior axis with anterior cells ceasing growth and remaining permanently flagellated to serve a nonreproductive motility function. The posterior cells in a Pleodorina colony show a partial division of labor. They start out as flagellated cells, but continue to grow and, subsequently, retract their flagella and divide into new colonies. The partial germ-soma division of labor shown by Pleodorina becomes a fixed dichotomy in the genus Volvox whose colonies can contain up to 50,000 cells in some species and in which there are dedicated groups of sterile somatic cells and reproductive germ cells. The progression from simpler colonial forms to Volvox was probably stepwise as outlined in the volvocine lineage hypothesis, but variations of this progression led to at least three independent origins of Volvox-like colony organization, each with distinct properties (Fig. 3) (Nozaki et al. 2006b; Herron and Michod 2008; Coleman 2012).
The Cell Biology of Chlamydomonas
The key to understanding the proximal mechanisms underlying multicellularity in Volvocine algae lies in the cell biology of their unicellular ancestor that was similar to C. reinhardtii. C. reinhardtii is a walled biflagellate with a pair of apical flagella and single large cup-shaped chloroplast situated basally that occupies around 2/3 of the cell volume. C. reinhardtii cells have a highly polarized and stereotyped organization centered on their two basal bodies that reside just under the plasma membrane at the cell anterior. The basal bodies nucleate the flagella and organize the internal microtubule cytoskeleton whose principal axes are defined by two- and four-membered microtubule rootlets (Holmes and Dutcher 1989). Basal bodies also coordinate the placement of the cell division plane and the microtubule-based phycoplast used for cytokinesis (Johnson and Porter 1968; Ehler et al. 1995; Ehler and Dutcher 1998). Besides apical-basal polarity, C. reinhardtii cells also have radial polarity that is evident from an eyespot located equatorially and always positioned along the four-membered microtubule rootlet emanating from the younger of the two basal bodies (Holmes and Dutcher 1989). The eyespot allows swimming cells to perceive the direction of illumination and signal to the flagella to enable phototaxis (Hegemann 2008). The basic cellular architecture and polarity of C. reinhardtii cells is conserved in colonial species with some modifications, and is likely to have played a role in establishing organismal-level polarity.
The wall surrounding C. reinhardtii cells is composed mostly of hydroxyproline-rich glycoproteins (HRGPs) that form rod-shaped polyproline-II helices with globular domains at one or both ends (Adair et al. 1987; Lee et al. 2007). As described below, HRGPs are also major components of the extracellular matrix of Volvox and other colonial volvocine algae (Hallmann 2003).
Like many green algae, Chlamydomonas cells and nearly all volvocine algae divide by a mechanism called multiple fission (or palintomy) that is described in the Unique Aspects of Multicellularity in Plants and Green Algae section and whose features are intimately related to multicellularity in volvocine algae. Most important, multiple fission produces clonal clusters of 2n daughter cells within a common mother cell wall at the end of every division cycle. Clonality (versus aggregation) aligns the fitness of each genome in the colony and largely obviates genetic conflicts between cells over reproductive potential (Buss 1987; Queller 2000). Multiple fission also explains why cell number per colony in volvocine algae is usually a power of 2 (i.e., 4,8,16,32,64,128, …).
A second important feature of multiple fission is the spiral-like pattern of successive cleavage divisions caused by rotation of the mitotic spindle axis by 90° in daughters with respect to mothers after each division cycle. As a result, postmitotic daughter cells end up clustered in a configuration with their basal bodies and flagella oriented inward (Johnson and Porter 1968; Kirk et al. 1991). Although this inside-out configuration presents no obstacle for individual C. reinhardtii cells that soon hatch from their mother cell wall and swim their separate ways, it is a topological constraint for colonial species in which cells remain connected through cytoplasmic bridges. The morphogenetic process of inversion that is discussed below is an innovation that overcomes this constraint and is one of the most remarkable aspects of volvocine algal development and evolution.
A third aspect of multiple fission that has impacted the evolution of multicellularity in Volvocine algae is the relationship between cell size and division. In C. reinhardtii, there is inherent flexibility in cell division so that numbers of daughters can range from 2 (1 division) to 16 or even 32 (4 or 5 divisions) depending on mother cell size. A sizer mechanism operates to ensure that the appropriate number of S phase and mitosis cycles is executed to produce daughters of uniform size (Donnan and John 1983; Umen 2005). In multicellular species, cell number per colony is much less variable, so the flexibility seen in C. reinhardtii has been overridden by a more stereotyped relationship between mother cell size and the initiation of cell division to maintain colony uniformity.
As noted above, during division, Chlamydomonas cells resorb their flagella and cannot swim. Although this temporary lack of motility probably does not impact the fitness of Chlamydomonas, it may do so for the larger colonial volvocine genera such as Pleodorina and Volvox that are thought to have evolved germ-soma specialization as a response to their ancestral flagellation constraint (Koufopanou 1994). As colonies grow larger, embryonic cell divisions take longer to complete, meaning that they must endure increasing lengths of time spent immotile and sinking in the water column during mitosis. The evolution of a permanently flagellated caste of somatic cells to provide continuous motility is an innovation that is likely to have enabled the evolution of ever-larger colonies without suffering the cost of lost motility during cell division.
Multicellular Innovations in the Volvocine Lineage Are Modifications of Unicellular Volvocine Algal Cell Biology
In his review of multicellularity and the evolution of Volvox, Kirk (2005) outlines 12 steps that define the minimum changes or innovations required to convert a C. reinhardtii-type cell to a multicellular V. carteri spheroid. These innovations are described below in an updated context that is informed by knowledge of the genomes from these two species and new findings that have been published in the interim. Two additional innovations (anisogamy and oogamy) have also been added (Nishii and Miller 2010).
Six of the 12 innovations outlined by Kirk (2005) occur in the initial multicellular transition from Chlamydomonas to Gonium. The fact that half of the innovations in the lineage occurred at this step suggests that forming small integrated functional colonies might be the most difficult aspect of the transition to multicellularity. The idea of a bottleneck at this step is consistent with the finding that the simplest colonial forms are all basal to the more complex volvocine genera. This basal positioning can be interpreted to mean that Gonium-like colony organization evolved successfully just once, and the more complex volvocine genera arose just once from among the Goniaceae (Herron and Michod 2008; Hayama et al. 2010). The apparent difficulty of the transition between unicellular Chlamydomonas and colonial Gonium appears to contradict findings of experimental evolution in which unicellular yeasts and Chlamydomonas could be converted to facultative colonial forms in a short number of generations by artificial selection (Ratcliff et al. 2012, 2013). However, this discrepancy is not so paradoxical considering the degree of evolved functionality that arose in Gonium that distinguishes it from the disorganized cell aggregates produced during experimental evolution. It may be easy to form such aggregates, but they may not truly recapitulate the first steps that led to the evolution of Gonium-like colonies.
Some of the innovations below are grouped together in cases in which they are related, so the numbering does not correspond to that described by Kirk (2005), but the progression is the same.
-
1.
Genetic modulation of cell number. As described above, Chlamydomonas cells show inherent flexibility in mother cell size and number of mitotic cell divisions, producing two daughters under poor growth conditions and up to 16 daughters under favorable growth conditions (Donnan and John 1983; John 1987). Colonial species, on the other hand, have more stereotyped numbers of divisions to produce colonies that typically vary in cell number by a factor of 2 (Coleman 2012). Moreover, unlike Chlamydomonas cells that are competent to divide as soon as they double in size, cells in colonial species must grow large enough between reproductive cycles so that new colonies can be formed with the appropriate number of cells. In Chlamydomonas, the retinoblastoma tumor suppressor pathway controls cell size and division number (Umen and Goodenough 2001; Umen 2005; Fang et al. 2006), and may have been modified in colonial species to increase the minimum size threshold at which division can occur.
-
2.
Incomplete cytokinesis. Incomplete cytokinesis is observed in all colonial volvocine species and can result in either temporary or perduring cytoplasmic connections or bridges between postmitotic cells. Ultrastructural studies show that the bridges are spatially organized within and between cells, and contain specialized substructures that may represent mechanical reinforcements and/or facilitate intercellular communication much like plasmodesmata in plants. Bridges can also be removed or resolved as occurs postembryonically in some species (Green et al. 1981; Coleman 2012).
-
3 and 4.
Organismal polarity and basal body rotation. 16-cell Gonium colonies that have a planar 4 × 4 arrangement of cells show polarity that is evident by comparing the arrangements of basal bodies and flagella of interior versus peripheral cells. The four cells on the interior of the colony have their basal bodies oriented with 180° rotational symmetry and beat in opposite directions in a breast stroke motion just as they do in Chlamydomonas. The 12 cells on the edge, however, rotate their basal bodies and flagella to a parallel orientation that cause the colony to rotate in a pinwheel-like fashion as it swims forward (Greuel and Floyd 1985). The basal body reorientation observed in Gonium peripheral cells occurs in all of the motile cells of spheroidal volvocine genera and is crucial for coordinated swimming (Hoops 1984; Ueki et al. 2010). Visible signs of organismal polarity are present in other aspects of colony development in larger volvocine genera that appear during inversion, germ-soma differentiation, and asymmetric cell division as described below.
-
5 and 6.
Partial and complete inversion. As a result of the spiral division pattern of volvocine algal cells described above, postmitotic embryos have their apical ends and basal bodies clustered on the interior of the embryo (or the concave surface for Gonium). This configuration of interior-facing basal bodies and flagella must be reversed by inversion for the embryo to swim. Inversion involves a coordinated series of cell-shape changes resulting in a complete reversal of embryonic curvature. Although inversion has been best studied in Volvox, it has also been observed in all other colonial volvocine genera (Hallmann 2006a). The partial inversion process that causes a reversal of curvature in Gonium embryos is presumably related to the process of full inversion in spheroidal volvocine embryos in which a hollow ball of cells turns itself completely inside out.
Full inversion of spheroidal colonies has been examined carefully in V. carteri and V. globator (Viamontes and Kirk 1977; Nishii and Ogihara 1999; Nishii 2003; Ueki and Nishii 2009; Höhn and Hallmann 2011), which use different but related mechanisms to invert. In both species, inversion is driven by coordinated cell-shape changes that move through the spheroid along the anteroposterior axis and drive cell sheet bending and involution. Inversion in many ways parallels gastrulation in animals in which similar cell-shape changes are used to remodel the hollow, ball-like blastula and generate primordial embryonic germ layers of the gastrula (Solnica-Krezel and Sepich 2012). The sequence of events and types of cell-shape changes that drive inversion in V. carteri (termed type A inversion) and V. globator (termed type B inversion) differ substantially (Höhn and Hallmann 2011) and underscore the fact that these two species belong to independent clades of Volvox (Fig. 3A). In both types of inversion, cytoplasmic bridges are crucial for maintaining mechanical integrity of the cell sheets and fixed relationships between neighboring cells.
V. carteri inversionless mutants have provided insights into the processes governing type A inversion. One such mutant, invA, affects a volvocine algal kinesin that localizes to the cytoplasmic bridge region of cells (Nishii 2003). The function of InvA protein is postulated to involve moving the cytoplasmic bridges relative to the rest of the cell along longitudinal microtubules to effect cell-shape changes and tissue bending. It is noteworthy that the highly similar C. reinhardtii ortholog of invA, IAR1, can functionally substitute for invA in Volvox despite C. reinhardtii lacking any known process analogous to inversion (Nishii and Miller 2010). Additional inv loci define a second extraembryonic function required for inversion associated with enlargement of the glycoprotein vesicle that encapsulates each embryo. When this vesicle fails to enlarge as it does in invB and invC mutants, the inverting embryo is physically constrained and becomes stuck midway through inversion (Ueki and Nishii 2008, 2009). invB and invC encode enzymes involved in sugar transport or metabolism and both have C. reinhardtii orthologs. It is likely that the C. reinhardtii orthologs of invB and invC proteins are involved in cell wall biosynthesis, but this idea remains to be tested.
-
7 and 8.
Transformation of the cell wall into ECM and ECM expansion. Although individual Gonium cell walls have a tripartite structure similar to that of Chlamydomonas, they also have an added outer layer that maintains colony cohesion and contains specialized regions of electron-dense material at cell–cell attachment points (Nozaki 1990).
Further modifications of the ECM are found in the more complex colonial genera. These include formation of a colony boundary layer in spheroidal genera (Pandorina, Eudorina, Pleodorina, and Volvox) that resembles the tripartite structure of the cell wall surrounding individual Chlamydomonas and Gonium cells. In Eudorina, Pleodorina, and Volvox, the ECM is expanded to fill large spaces between cells and within the colony interior (Coleman 2012). Although Volvox colonies have more than one thousand cells, ECM expansion accounts for >99% of their volume and makes them six orders of magnitude larger than an individual C. reinhardtii cell. ECM expansion in volvocine algae has some parallels with vacuolar expansion in land plants that also allows massive organismal growth without the addition of new cells or significant protein biomass (Robaglia et al. 2004). Although the ECM in volvocine algal colonies appears transparent to the eye, it has an elaborate internal structure that has been investigated in detail for V. carteri in which each cell has its own ECM domain and the colony as a whole has distinguishable layers of ECM (Kirk et al. 1986; Hallmann 2003). Paralleling this ECM expansion has been a notable increase in the number of ECM genes from two families in V. carteri relative to counterparts in C. reinhardtii (Prochnik et al. 2010). The expanded families are pherophorins that have structural and signaling roles (Hallmann 2006b) and the VMPs (Volvox metalloproteinases) that function in ECM remodeling (Heitzer and Hallmann 2002). There are dozens of other uncharacterized volvocine algal protein families that are differentially expanded in V. carteri that may also play a role in multicellularity (Prochnik et al. 2010).
-
9 and 10.
Partial and complete germ-soma differentiation. Perhaps the most well-studied multicellular innovation in volvocine algae is the differentiation of germ and somatic cell types that is observed in the genera Pleodorina and Volvox.
The genetic basis for germ-soma differentiation has been investigated in V. carteri in which developmental mutants exist that alter cell fate. In regA (somatic regenerator) mutants, somatic cells form normally after embryogenesis, but then redifferentiate into gonidia; they withdraw their flagella, grow, and subsequently divide into new embryos (Starr 1970; Huskey and Griffin 1979). regA is a nuclear protein that is related to transcriptional repressors and expressed in somatic cells and required to maintain their fate (Kirk et al. 1999; Stark et al. 2001). regA belongs to a family of VARL (Volvocine algae regA-like) proteins with 14 members in Volvox and 12 in Chlamydomonas (Duncan et al. 2006, 2007). Phylogenies of VARL proteins indicate complex patterns of gene duplication and loss in both algal lineages, with no ortholog of regA present in C. reinhardtii. It has been hypothesized that VARL proteins evolved in unicellular and simple colonial species to coordinate temporal cell differentiation with environmental cues (Nedelcu and Michod 2006).
A second class of mutants in V. carteri is at lag loci (late gonidia), whose gene products have not been identified. In lag mutants, cells that are normally destined to form gonidia instead adopt a somatic or quasi-somatic cell fate and become flagellated and stop growing. Eventually, however, lag mutant gonidia withdraw their flagella and redifferentiate as reproductive cells (Kirk 1988).
The phenotypes of regA and lag mutants suggest that establishment and maintenance of cell fate in V. carteri are separable functions controlled by multiple loci (Kirk 1988). Continued work on these mutants and isolation of new regulatory loci promises to further clarify the origins of cell-type specification in this lineage.
-
11 and 12.
Asymmetric cell division and a bifurcated cell division program. As described above, the genus Volvox is polyphyletic with germ-soma differentiation established by different mechanisms in separate groups (Desnitski 1995; Coleman 2012). In V. carteri and its close relatives, the vegetative germ line is established early in embryonic development by means of asymmetric cell divisions. At embryonic division 6 (32 → 64 cell stage), a subset of 16 anterior cells in the embryo divides asymmetrically to produce a large and small daughter. The large daughters divide asymmetrically one or two more times and then cease dividing, whereas the smaller daughters continue dividing five or six more times along with the remaining embryonic cells that divide symmetrically. Asymmetric cell divisions combined with early exit from the cell cycle of the large daughters results in an embryo with approximately 2000 small cells and 16 large cells. The 16 large cells will mature and differentiate as gonidia, whereas the small cells become somatic (Kirk et al. 1991).
Mutants that disrupt asymmetric cell division have been identified and at least one locus, glsA, has been cloned (Miller and Kirk 1999). glsA is in a conserved family of eukaryotic proteins that contain a dnaJ-like chaperone domain that mediates an essential interaction with HSP70-type heat shock proteins (Cheng et al. 2005), as well as SANT domains that mediate chromatin association in V. carteri (Pappas and Miller 2009). Although glsA is distributed uniformly through the embryo, its HSP70A partner is distributed in a decreasing anterior-to-posterior concentration gradient at the time of asymmetric division and may provide positional cues for localizing asymmetric cell divisions to the anterior of the embryo (Cheng et al. 2005). During mitosis, glsA localizes to the spindle suggesting that it might be directly involved in spindle positioning, but this localization was insufficient to mediate its function that may also involve transcriptional or translational regulation (Cheng et al. 2005; Pappas and Miller 2009). Although it is not known to divide asymmetrically, C. reinhardtii has a glsA ortholog, GAR1, that can complement a glsA mutant (Cheng et al. 2003). The function of GAR1 in C. reinhardtii is not known, but the above finding suggests that at least some of the machinery for asymmetric cell division existed in the unicellular ancestor of colonial volvocine algae.
Elegant experiments involving manipulation of embryonic division and daughter cell size showed that cell size (rather than position within the embryo) is the primary determinant of cell fate in V. carteri (Kirk et al. 1993). The nature of this cell-size signal and how it controls subsequent differentiation is unknown, but it may be coupled to the retinoblastoma pathway that functions in Chlamydomonas to sense and control cell size (Fang et al. 2006). Interestingly, comparative genomic studies of cell cycle genes revealed an expansion of the D-type cyclins in V. carteri compared with C. reinhardtii (Prochnik et al. 2010). D-type cyclins in plants and animals are activating subunits for cyclin-dependent kinases that regulate retinoblastoma-related proteins through phosphorylation (Gutzat et al. 2012). Whether the expanded D-cyclin repertoire in V. carteri enabled modifications of the embryonic cell division program or contributes to germ-soma differentiation remains to be determined.
-
13 and 14.
Anisogamy and oogamy. Volvocine algae recapitulate the transition from isogamy to anisogamy to oogamy and are a model system in which to investigate the evolution of these traits (Nozaki et al. 2000; Umen 2011). C. reinhardtii is isogamous with two mating types, plus and minus, that are determined by a complex, multigenic mating locus (MT), with MT+ and MT− haplotypes (Umen 2011; De Hoff et al. 2013). Gonium and Pandorina are also isogamous, whereas Eudorina and related genera show a mixture with some isogamous species and anisogamous species (Coleman 2012). The larger and more complex spheroids in the genus Pleodorina are anisogamous, whereas all Volvox species are oogamous (Nozaki 1996). Other than C. reinhardtii MT, V. carteri is the only other volvocine algal mating locus characterized to date. V. carteri MT is syntenic with Chlamydomonas MT, but its male and female haplotypes are genetically more complex and share several properties with sex chromosomes (Ferris et al. 2010). The developmental and evolutionary bases for oogamy and anisogamy that are controlled by MT remain to be determined, but may involve the transcription factor MID (minus dominance) that specifies sexual differentiation in Chlamydomonas and is found in either the minus mating type or males of all volvocine algae examined to date (Ferris and Goodenough 1997; Nozaki et al. 2006a; Hamaji et al. 2008, 2013).
Future Perspectives for Volvocine Algae and Multicellularity
Volvocine algae allow detailed inferences to be made regarding the cell biological and developmental events required for the transition to multicellularity, but have only just begun to be exploited to gain mechanistic insights into how underlying processes evolved. The finding that developmental mutants in Volvox can be complemented with orthologous genes from Chlamydomonas should spur further research into how these genes are used in Chlamydomonas and what has allowed their function to be altered for development in Volvox. The genomes of other volvocine algae will also soon be sequenced and help clarify the origins of gene families that contribute to developmental innovations in the lineage. The identification and characterization of new developmental mutants and investigation of existing mutants in both V. carteri and C. reinhardtii promises to further deepen our understanding of how multicellularity and evolutionary innovation occur at an unprecedented level of detail.
CONCLUDING REMARKS
The specific mechanisms used to achieve multicellular innovation differ between lineages, but some of the emerging themes and principles discovered in charophyte and volvocine algae may be more universal. First, it is clear that understanding the transition to multicellularity requires a detailed understanding of the life cycle, cell biology, and ecology of simpler ancestor(s) that gave rise to multicellular descendant lineages. Indeed, research into the origins of animal multicellularity have been transformed by investigating their closest unicellular ancestors including choanoflagellates and others (King et al. 2008; Fairclough et al. 2013; Sebe-Pedros et al. 2013; Alegado and King 2014). Gene family expansion and novel protein domain organization are observed in metazoans and embryophytes and associated with some key innovations (Floyd and Bowman 2007; Degnan et al. 2009; Srivastava et al. 2010). However, wholesale changes in numbers or types of protein families and domain structures need not underlie the innovations required to achieve multicellularity as evidenced by the similarity between the C. reinhardtii and V. carteri genomes (Prochnik et al. 2010), and shared origins with C. reinhardtii of key developmental regulators in V. carteri. Similar ideas have emerged from investigating the origins of developmental pathways from land plants that arose in charophyte algae or the earliest embryophyte lineages (Preston et al. 2011; Pires and Dolan 2012; Bowman 2013). Third, lineage-specific protein families such as the Chlamydomonas cell wall/Volvox ECM proteins may provide a particularly rich source of raw material for evolutionary innovation. Although very little is known about such proteins in the volvocine lineage, there are many examples of proteins whose origins are unique to the animal/choanoflagellate lineage that play key roles in metazoan development (King et al. 2008; Fairclough et al. 2013). It can be anticipated that full genome sequences of charophyte algae will reveal additional examples of such proteins that contribute to the evolution of land plants.
ACKNOWLEDGMENTS
I thank Gavriel Matt and Patrick Keeling for comments and editorial suggestions on this manuscript, and Joe Heitman for helping to involve me in this collection. Research on volvocine algal cell biology and evolution in my laboratory is supported by National Institutes of Health grants R01GM092744 and R01GM078376.
Footnotes
Editors: Patrick J. Keeling and Eugene V. Koonin
Additional Perspectives on The Origin and Evolution of Eukaryotes available at www.cshperspectives.org
REFERENCES
*Reference is also in this collection.
- Adair WS, Steinmetz SA, Mattson DM, Goodenough UW, Heuser JE 1987. Nucleated assembly of Chlamydomonas and Volvox cell walls. J Cell Biol 105: 2373–2382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adl SM, Simpson AGB, Lane CE, Lukeš J, Bass D, Bowser SS, Brown MW, Burki F, Dunthorn M, Hampl V, et al. 2012. The revised classification of Eukaryotes. J Eukaryot Microbiol 59: 429–514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Alegado RA, King N 2014. Bacterial influences on animal origins. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a016162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ariizumi T, Toriyama K 2011. Genetic regulation of sporopollenin synthesis and pollen exine development. Annu Rev Plant Biol 62: 437–460. [DOI] [PubMed] [Google Scholar]
- Baluška F, Volkmann D, Barlow PW 2004. Eukaryotic cells and their cell bodies: Cell theory revised. Ann Bot 94: 9–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beardall J, Allen D, Bragg J, Finkel Z, Flynn K, Quigg A, Rees T, Richardson A, Raven J 2009. Allometry and stoichiometry of unicellular, colonial and multicellular phytoplankton. New Phytologist 181: 295–309. [DOI] [PubMed] [Google Scholar]
- Becker B, Marin B 2009. Streptophyte algae and the origin of embryophytes. Ann Bot 103: 999–1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beilby MJ, Casanova MT 2014. The physiology of Characean cells. Springer, Berlin. [Google Scholar]
- Bell G 1985. The origin and early evolution of germ cells as illustrated by the Volvocales. In The origin and evolution of sex (ed. Halvorson HO, Monroy A), pp. 221–256 Alan R. Liss, New York. [Google Scholar]
- Berney C, Pawlowski J 2006. A molecular time-scale for eukaryote evolution recalibrated with the continuous microfossil record. Proc Biol Sci 273: 1867–1872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besson S, Dumais J 2011. Universal rule for the symmetric division of plant cells. Proc Natl Acad Sci 108: 6294–6299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharya D, Weber K, An SS, Berning-Koch W 1998. Actin phylogeny identifies Mesostigma viride as a flagellate ancestor of the land plants. J Mol Evol 47: 544–550. [DOI] [PubMed] [Google Scholar]
- Boot KJM, Libbenga KR, Hille SC, Offringa R, van Duijn B 2012. Polar auxin transport: An early invention. J Exp Bot 63: 4213–4218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boraas ME, Seale DB, Boxhorn JE 1998. Phagotrophy by a flagellate selects for colonial prey: A possible origin of multicellularity. Evol Ecol 12: 153–164. [Google Scholar]
- Bowman JL 2013. Walkabout on the long branches of plant evolution. Curr Opin Plant Biol 16: 70–77. [DOI] [PubMed] [Google Scholar]
- Bowman JL, Floyd SK, Sakakibara K 2007. Green genes-comparative genomics of the green branch of life. Cell 129: 229–234. [DOI] [PubMed] [Google Scholar]
- Boyce CK, Zwieniecki MA, Cody GD, Jacobsen C, Wirick S, Knoll AH, Holbrook NM 2004. Evolution of xylem lignification and hydrogel transport regulation. Proc Natl Acad Sci 101: 17555–17558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradley PM 1991. Plant hormones do have a role in controlling growth and development of algae. J Phycol 27: 317–321. [Google Scholar]
- Braun M, Limbach C 2006. Rhizoids and protonemata of characean algae: Model cells for research on polarized growth and plant gravity sensing. Protoplasma 229: 133–142. [DOI] [PubMed] [Google Scholar]
- Brooks J, Shaw G 1978. Sporopollenin: A review of its chemistry, palaeochemistry and geochemistry. Grana 17: 91–97. [Google Scholar]
- Buss LW 1987. The evolution of individuality. Princeton University Press, Princeton. [Google Scholar]
- Cheng Q, Fowler R, Tam L-W, Edwards L, Miller SM 2003. The role of GlsA in the evolution of asymmetric cell division in the green alga Volvox carteri. Dev Genes Evol 213: 328–335. [DOI] [PubMed] [Google Scholar]
- Cheng Q, Pappas V, Hallmann A, Miller SM 2005. Hsp70A and GlsA interact as partner chaperones to regulate asymmetric division in Volvox. Dev Biol 286: 537–548. [DOI] [PubMed] [Google Scholar]
- Cocquyt E, Verbruggen H, Leliaert F, De Clerck O 2010. Evolution and cytological diversification of the green seaweeds (Ulvophyceae). Mol Biol Evol 27: 2052–2061. [DOI] [PubMed] [Google Scholar]
- Coleman AW 2012. A comparative analysis of the Volvocaceae (Chlorophyta). J Phycol 48: 491–513. [DOI] [PubMed] [Google Scholar]
- Cook ME 2004. Cytokinesis in Coleochaete orbicularis (Charophyceae): An ancestral mechanism inherited by plants. Am J Bot 91: 313–320. [DOI] [PubMed] [Google Scholar]
- Cook ME, Graham LE, Lavin CA 1998. Cytokinesis and nodal anatomy in the charophycean green alga Chara zeylanica. Protoplasma 203: 65–74. [Google Scholar]
- Cooke TJ, Poli D, Sztein AE, Cohen JD 2002. Evolutionary patterns in auxin action. Plant Mol Biol 49: 319–338. [PubMed] [Google Scholar]
- Cosgrove DJ 2000. Loosening of plant cell walls by expansins. Nature 407: 321–326. [DOI] [PubMed] [Google Scholar]
- Cutler SR, Rodriguez PL 2010. Abscisic acid: Emergence of a core signaling network. Annu Rev Plant Biol 61: 651–679. [DOI] [PubMed] [Google Scholar]
- Dayel MJ, Alegado RA, Fairclough SR, Levin TC, Nichols SA, McDonald K, King N 2011. Cell differentiation and morphogenesis in the colony-forming choanoflagellate Salpingoeca rosetta. Dev Biol 357: 73–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Clerck O, Bogaert KA, Leliaert F 2012. Diversity and evolution of algae. In Advances in botanical research, Vol. 64, pp. 55–86 Elsevier, Amsterdam. [Google Scholar]
- Degnan BM, Vervoort M, Larroux C, Richards GS 2009. Early evolution of metazoan transcription factors. Curr Opin Genet Dev 19: 591–599. [DOI] [PubMed] [Google Scholar]
- De Hoff PL, Ferris P, Olson BJSC, Miyagi A, Geng S, Umen JG 2013. Species and population level molecular profiling reveals cryptic recombination and emergent asymmetry in the dimorphic mating locus of C. reinhardtii. PLoS Genet 9: e1003724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delwiche CF, Graham LE, Thomson N 1989. Lignin-like compounds and sporopollenin coleochaete, an algal model for land plant ancestry. Science 245: 399–401. [DOI] [PubMed] [Google Scholar]
- Derelle E, Ferraz C, Rombauts S, Rouzé P, Worden AZ, Robbens S, Partensky F, Degroeve S, Echeynié S, Cooke R, et al. 2006. Genome analysis of the smallest free-living eukaryote Ostreococcus tauri unveils many unique features. Proc Natl Acad Sci 103: 11647–11652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Saint Germain A, Bonhomme S, Boyer F-D, Rameau C 2013. Novel insights into strigolactone distribution and signalling. Curr Opin Plant Biol 16: 583–589. [DOI] [PubMed] [Google Scholar]
- De Smet I, Beeckman T 2011. Asymmetric cell division in land plants and algae: The driving force for differentiation. Nat Rev Mol Cell Biol 12: 177–188. [DOI] [PubMed] [Google Scholar]
- De Smet I, Voss U, Lau S, Wilson M, Shao N, Timme RE, Swarup R, Kerr I, Hodgman C, Bock R, et al. 2011. Unraveling the evolution of auxin signaling. Plant Physiol 155: 209–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desnitski A 1995. A review on the evolution of development in Volvox-morphological and physiological aspects. Eur J Protist 31: 241–247. [Google Scholar]
- Domozych DS, Ciancia M, Fangel JU, Mikkelsen MD, Ulvskov P, Willats WG 2012. The cell walls of green algae: A journey through evolution and diversity. Front Plant Sci 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donnan L, John PC 1983. Cell cycle control by timer and sizer in Chlamydomonas. Nature 304: 630–633. [DOI] [PubMed] [Google Scholar]
- Dring MJ 1988. Photocontrol of development in algae. Annu Rev Plant Physiol Plant Mol Biol 39: 157–174. [Google Scholar]
- Duncan L, Nishii I, Howard A, Kirk D, Miller SM 2006. Orthologs and paralogs of regA, a master cell-type regulatory gene in Volvox carteri. Curr Genet 50: 61–72. [DOI] [PubMed] [Google Scholar]
- Duncan L, Nishii I, Harryman A, Buckley S, Howard A, Friedman NR, Miller SM 2007. The VARL gene family and the evolutionary origins of the master cell-type regulatory gene, regA, in Volvox carteri. J Mol Evol 65: 1–11. [DOI] [PubMed] [Google Scholar]
- Dupuy L, Mackenzie J, Haseloff J 2010. Coordination of plant cell division and expansion in a simple morphogenetic system. Proc Natl Acad Sci 107: 2711–2716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehler LL, Dutcher SK 1998. Pharmacological and genetic evidence for a role of rootlet and phycoplast microtubules in the positioning and assembly of cleavage furrows in Chlamydomonas reinhardtii. Cell Motil Cytoskeleton 40: 193–207. [DOI] [PubMed] [Google Scholar]
- Ehler LL, Holmes JA, Dutcher SK 1995. Loss of spatial control of the mitotic spindle apparatus in a Chlamydomonas reinhardtii mutant strain lacking basal bodies. Genetics 141: 945–960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elhiti M, Stasolla C 2009. Structure and function of homodomain-leucine zipper (HD-Zip) proteins. Plant Signal Behav 4: 86–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emiliani G, Fondi M, Fani R, Gribaldo S 2009. A horizontal gene transfer at the origin of phenylpropanoid metabolism: A key adaptation of plants to land. Biol Direct 4: 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fairclough SR, Chen Z, Kramer E, Zeng Q, Young S, Robertson HM, Begovic E, Richter DJ, Russ C, Westbrook MJ, et al. 2013. Premetazoan genome evolution and the regulation of cell differentiation in the choanoflagellate Salpingoeca rosetta. Genome Biol 14: R15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang S-C, de los Reyes C, Umen JG 2006. Cell size checkpoint control by the retinoblastoma tumor suppressor pathway. PLoS Genet 2: e167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faulkner CR, Blackman LM, Cordwell SJ, Overall RL 2005. Proteomic identification of putative plasmodesmatal proteins from Chara corallina. Proteomics 5: 2866–2875. [DOI] [PubMed] [Google Scholar]
- Ferris PJ, Goodenough UW 1997. Mating type in Chlamydomonas is specified by mid, the minus-dominance gene. Genetics 146: 859–869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferris P, Olson BJSC, De Hoff PL, Douglass S, Casero D, Prochnik S, Geng S, Rai R, Grimwood J, Schmutz J, et al. 2010. Evolution of an expanded sex-determining locus in Volvox. Science 328: 351–354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finet C, Timme RE, Delwiche CF, Marlétaz F 2010. Multigene phylogeny of the green lineage reveals the origin and diversification of land plants. Curr Biol 20: 2217–2222. [DOI] [PubMed] [Google Scholar]
- Floyd SK, Bowman JL 2007. The ancestral developmental tool kit of land plants. Int J Plant Sci 168: 1–35. [Google Scholar]
- Floyd GL, Stewart KD, Mattox KR 1972. Cellular organization, mitosis, and cytokinesis in the Ulotrichalean Alga, Klebsormidium. J Phycol 8: 176–184. [Google Scholar]
- Floyd SK, Zalewski CS, Bowman JL 2006. Evolution of class III homeodomain-leucine zipper genes in streptophytes. Genetics 173: 373–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fowke LC, Pickett-Heaps JD 1969. Cell division in Spirogyra: II. Cytokinesis. J Phycol 5: 273–281. [DOI] [PubMed] [Google Scholar]
- Franceschi VR, Ding B, Lucas WJ 1994. Mechanism of plasmodesmata formation in characean algae in relation to evolution of intercellular communication in higher plants. Planta 192: 347–358. [Google Scholar]
- Fritsch FE 1948. The structure and reproduction of the algae. Cambridge University Press, Cambridge. [Google Scholar]
- Galway ME, Hardham AR 1991. Immunofluorescent localization of microtubules throughout the cell cycle in the green alga Mougeotia (Zygnemataceae). Am J Bot 78: 451–461. [Google Scholar]
- Graham LE 1984. Coleochaete and the origin of land plants. Am J Bot 71: 603–608. [Google Scholar]
- Graham LE 1996. Green algae to land plants: An evolutionary transition. J Plant Res 109: 241–251. [Google Scholar]
- Graham LE, Wilcox LW 1983. The occurrence and phylogenetic significance of putative placental transfer cells in the green alga Coleochaete. Am J Bot 70: 113–120. [DOI] [PubMed] [Google Scholar]
- Graham LK, Wilcox LW 2000. The origin of alternation of generations in land plants: A focus on matrotrophy and hexose transport. Philos Trans R Soc Lond B Biol Sci 355: 757–766; discussion 766–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham LE, Cook ME, Busse JS 2000. The origin of plants: Body plan changes contributing to a major evolutionary radiation. Proc Natl Acad Sci 97: 4535–4540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham LE, Graham JM, Wilcox LW 2009. Algae. Benjamin-Cummings, San Francisco. [Google Scholar]
- Graham LE, Arancibia-Avila P, Taylor WA, Strother PK, Cook ME 2012. Aeroterrestrial Coleochaete (Streptophyta, Coleochaetales) models early plant adaptation to land. Am J Bot 99: 130–144. [DOI] [PubMed] [Google Scholar]
- Grauvogel C, Petersen J 2007. Isoprenoid biosynthesis authenticates the classification of the green alga Mesostigma viride as an ancient streptophyte. Gene 396: 125–133. [DOI] [PubMed] [Google Scholar]
- Green KJ, Viamontes GI, Kirk DL 1981. Mechanism of formation, ultrastructure, and function of the cytoplasmic bridge system during morphogenesis in Volvox. J Cell Biol 91: 756–769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greuel BT, Floyd GL 1985. Development of the flagellar apparatus and flagellar orientation in the colonial green alga Gonium pectorale (Volvocales). J Phycol 21: 358–371. [Google Scholar]
- Grosberg R, Strathmann R 2007. The evolution of multicellularity: A minor major transition? Annu Rev Ecol Evol Syst 38: 621–654. [Google Scholar]
- Güde H 1979. Grazing by protozoa as selection factor for activated sludge bacteria. Microb Ecol 5: 225–237. [DOI] [PubMed] [Google Scholar]
- Gunnison D, Alexander M 1975. Basis for the resistance of several algae to microbial decomposition. Appl Microbiol 29: 729–738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutzat R, Borghi L, Gruissem W 2012. Emerging roles of RETINOBLASTOMA-RELATED proteins in evolution and plant development. Trends Plant Sci 17: 139–148. [DOI] [PubMed] [Google Scholar]
- Hackenberg D, Sakayama H, Nishiyama T, Pandey S 2013. Characterization of the heterotrimeric G-protein complex and its regulator from the green alga Chara braunii expands the evolutionary breadth of plant G-protein signaling. Plant Physiol 163: 1510–1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haig D 2010. What do we know about Charophyte (Streptophyta) life cycles? J Phycol 46: 860–867. [Google Scholar]
- Hall JD, McCourt RM, Delwiche CF 2008. Patterns of cell division in the filamentous Desmidiaceae, close green algal relatives of land plants. Am J Bot 95: 643–654. [DOI] [PubMed] [Google Scholar]
- Hallmann A 2003. Extracellular matrix and sex-inducing pheromone in Volvox. Int Rev Cytol 227: 131–182. [DOI] [PubMed] [Google Scholar]
- Hallmann A 2006a. Morphogenesis in the family Volvocaceae: Different tactics for turning an embryo right-side out. Protist 157: 445–461. [DOI] [PubMed] [Google Scholar]
- Hallmann A 2006b. The pherophorins: Common, versatile building blocks in the evolution of extracellular matrix architecture in Volvocales. Plant J 45: 292–307. [DOI] [PubMed] [Google Scholar]
- Hamaji T, Ferris PJ, Coleman AW, Waffenschmidt S, Takahashi F, Nishii I, Nozaki H 2008. Identification of the minus-dominance gene ortholog in the mating-type locus of Gonium pectorale. Genetics 178: 283–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamaji T, Ferris PJ, Nishii I, Nishimura Y, Nozaki H 2013. Distribution of the sex-determining gene MID and molecular correspondence of mating types within the isogamous genus Gonium (Volvocales, Chlorophyta). PLoS ONE 8: e64385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartung W 2010. The evolution of abscisic acid (ABA) and ABA function in lower plants, fungi and lichen. Funct Plant Biol 37: 806. [Google Scholar]
- Hauser F, Waadt R, Schroeder JI 2011. Evolution of abscisic acid synthesis review and signaling mechanisms. Curr Biol 21: R346–R355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayama M, Nakada T, Hamaji T, Nozaki H 2010. Morphology, molecular phylogeny and taxonomy of Gonium maiaprilis sp. nov. (Goniaceae, Chlorophyta) from Japan. Phycologia 49: 221–234. [Google Scholar]
- Hegemann P 2008. Algal sensory photoreceptors—Annual review of plant biology. Annu Rev Plant Biol 59: 167. [DOI] [PubMed] [Google Scholar]
- Heitzer M, Hallmann A 2002. An extracellular matrix-localized metalloproteinase with an exceptional QEXXH metal binding site prefers copper for catalytic activity. J Biol Chem 277: 28280–28286. [DOI] [PubMed] [Google Scholar]
- Hepler PK 1982. Endoplasmic reticulum in the formation of the cell plate and plasmodesmata. Protoplasma 111: 121–133. [Google Scholar]
- Hernández-Hernández V, Niklas KJ, Newman SA, Benítez M 2012. Dynamical patterning modules in plant development and evolution. Int J Dev Biol 56: 661–674. [DOI] [PubMed] [Google Scholar]
- Herron MD, Michod RE 2008. Evolution of complexity in the volvocine algae: Transitions in individuality through Darwin’s eye. Evolution 62: 436–451. [DOI] [PubMed] [Google Scholar]
- Herron MD, Hackett JD, Aylward FO, Michod RE 2009. Triassic origin and early radiation of multicellular volvocine algae. Proc Natl Acad Sci 106: 3254–3258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Höhn S, Hallmann A 2011. There is more than one way to turn a spherical cellular monolayer inside out: Type B embryo inversion in Volvox globator. BMC Biol 9: 89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes JA, Dutcher SK 1989. Cellular asymmetry in Chlamydomonas reinhardtii. J Cell Sci 94: 273–285. [DOI] [PubMed] [Google Scholar]
- Hoops HJ 1984. Somatic cell flagellar apparatuses in two species of Volvox (Chlorophyceae). J Phycol 20: 20–27. [Google Scholar]
- Huskey RJ, Griffin BE 1979. Genetic control of somatic cell differentiation in Volvox analysis of somatic regenerator mutants. Dev Biol 72: 226–235. [DOI] [PubMed] [Google Scholar]
- Imahori K, Iwasa K 1965. Pure culture and chemical regulation of the growth of charophytes. Phycologia 4: 127–134. [Google Scholar]
- Ispolatov I, Ackermann M, Doebeli M 2012. Division of labour and the evolution of multicellularity. Proc Biol Sci 279: 1768–1776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs WP 1970. Development and regeneration of the algal giant coenocyte Caulerpa. Ann NY Acad Sci 175: 732–748. [Google Scholar]
- Jin Q, Scherp P, Heimann K, Hasenstein KH 2008. Auxin and cytoskeletal organization in algae. Cell Biol Int 32: 542–545. [DOI] [PubMed] [Google Scholar]
- John P 1987. Control points in the Chlamydomonas cell cycle. In Algal development (ed. Wiessner W, Robinson DG, Starr RC), Springer, Berlin. [Google Scholar]
- Johnson UG, Porter KR 1968. Fine structure of cell division in Chlamydomonas reinhardi. Basal bodies and microtubules. J Cell Biol 38: 403–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johri MM 2004. Possible origin of hormonal regulation in green plants. Proc Natl Acad Sci India B 70: 335–365. [Google Scholar]
- Katsaros CI, Varvarigos V, Gachon CMM, Brand J, Motomura T, Nagasato C, Küpper FC 2011. Comparative immunofluorescence and ultrastructural analysis of microtubule organization in Uronema sp., Klebsormidium flaccidum, K. subtilissimum, Stichococcus bacillaris and S. chloranthus (Chlorophyta). Protist 162: 315–331. [DOI] [PubMed] [Google Scholar]
- Khandelwal A, Cho SH, Marella H, Sakata Y, Perroud P-F, Pan A, Quatrano RS 2010. Role of ABA and ABI3 in desiccation tolerance. Science 327: 546. [DOI] [PubMed] [Google Scholar]
- King N, Westbrook MJ, Young SL, Kuo A, Abedin M, Chapman J, Fairclough S, Hellsten U, Isogai Y, Letunic I, et al. 2008. The genome of the choanoflagellate Monosiga brevicollis and the origin of metazoans. Nature 451: 783–788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirk DL 1988. The ontogeny and phylogeny of cellular differentiation in Volvox. Trends Genet 4: 32–36. [DOI] [PubMed] [Google Scholar]
- Kirk DL 1998. Volvox. Cambridge University Press, Cambridge. [Google Scholar]
- Kirk DL 2003. Seeking the ultimate and proximate causes of Volvox multicellularity and cellular differentiation. Integr Comp Biol 43: 247–253. [DOI] [PubMed] [Google Scholar]
- Kirk DL 2005. A twelve-step program for evolving multicellularity and a division of labor. BioEssays 27: 299–310. [DOI] [PubMed] [Google Scholar]
- Kirk DL, Birchem R, King N 1986. The extracellular matrix of Volvox: A comparative study and proposed system of nomenclature. J Cell Sci 80: 207–231. [DOI] [PubMed] [Google Scholar]
- Kirk DL, Kaufman MR, Keeling RM, Stamer KA 1991. Genetic and cytological control of the asymmetric divisions that pattern the Volvox embryo. Dev Suppl 1: 67–82. [PubMed] [Google Scholar]
- Kirk MM, Ransick A, McRae SE, Kirk DL 1993. The relationship between cell size and cell fate in Volvox carteri. J Cell Biol 123: 191–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirk MM, Stark K, Miller SM, Müller W, Taillon BE, Gruber H, Schmitt R, Kirk DL 1999. regA, a Volvox gene that plays a central role in germ-soma differentiation, encodes a novel regulatory protein. Development 126: 639–647. [DOI] [PubMed] [Google Scholar]
- Klämbt D, Knauth B, Dittmann I 1992. Auxin dependent growth of rhizoids of Chara globularis. Physiol Plant 85: 537–540. [Google Scholar]
- Knoll AH 2011. The multiple origins of complex multicellularity. Annu Rev Earth Planet Sci 39: 217–239. [Google Scholar]
- Koufopanou V 1994. The evolution of soma in the Volvocales. Am Nat 143: 907–931. [Google Scholar]
- Kwiatkowska M 2003. Plasmodesmal changes are related to different developmental stages of antheridia of Chara species. Protoplasma 222: 1–11. [DOI] [PubMed] [Google Scholar]
- Kwiatkowska M, Wojtczak A, Poptoriska K 1998. Effect of GA3 treatment on the number of spermatozoids and endopolyploidy levels of non-generative cells in Antheridia of Chara vulgaris L. Plant Cell Physiol 39: 1388–1390. [Google Scholar]
- Langdale JA, Harrison CJ 2008. Developmental transitions during the evolution of plant form. In Evolving pathways: Key themes in evolutionary developmental biology (ed. Minelli A), pp. 299–316 Cambridge University Press, Cambridge. [Google Scholar]
- Lau S, Shao N, Bock R, Jurgens G, De Smet I 2009. Auxin signaling in algal lineages: Fact or myth? Trends Plant Sci 14: 182–188. [DOI] [PubMed] [Google Scholar]
- Laurin-Lemay S, Brinkmann H, Philippe H 2012. Origin of land plants revisited in the light of sequence contamination and missing data. Curr Biol 22: R593–R594. [DOI] [PubMed] [Google Scholar]
- Lee J-H, Lin H, Joo S, Goodenough U 2008. Early sexual origins of homeoprotein heterodimerization and evolution of the plant KNOX/BELL family. Cell 133: 829–840. [DOI] [PubMed] [Google Scholar]
- Lee J-H, Waffenschmidt S, Small L, Goodenough U 2007. Between-species analysis of short-repeat modules in cell wall and sex-related hydroxyproline-rich glycoproteins of Chlamydomonas. Plant Physiol 144: 1813–1826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leliaert F, Smith DR, Moreau H, Herron MD, Verbruggen H, Delwiche CF, De Clerck O 2012. Phylogeny and molecular evolution of the green algae. Crit Rev Plant Sci 31: 1–46. [Google Scholar]
- Lemieux C, Otis C, Turmel M 2000. Ancestral chloroplast genome in Mesostigma viride reveals an early branch of green plant evolution. Nature 403: 649–652. [DOI] [PubMed] [Google Scholar]
- Lemieux C, Otis C, Turmel M 2007. A clade uniting the green algae Mesostigma viride and Chlorokybus atmophyticus represents the deepest branch of the Streptophyta in chloroplast genome-based phylogenies. BMC Biol 5: 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis LA, McCourt RM 2004. Green algae and the origin of land plants. Am J Bot 91: 1535–1556. [DOI] [PubMed] [Google Scholar]
- Ligrone R, Duckett JG, Renzaglia KS 1993. The gametophyte-sporophyte junction in land plants. In Advances in Botanical Research, Vol. 19, pp. 231–318 Elsevier, Amsterdam. [Google Scholar]
- Lokhorst GM, Star W 1985. Ultrastructure of mitosis and cytokinesis in Klebsormidium Mucosum nov. comb., formerly Ulothrix Verrucosa (Chlorophyta). J Phycol 21: 466–476. [Google Scholar]
- Lucas WJ, Lee J-Y 2004. Plasmodesmata as a supracellular control network in plants. Nat Rev Mol Cell Biol 5: 712–726. [DOI] [PubMed] [Google Scholar]
- Mandoli DF 1998. Elaboration of body plan and phase change during development of Acetabularia: How is the complex architecture of a giant unicell built? Annu Rev Plant Physiol Plant Mol Biol 49: 173–198. [DOI] [PubMed] [Google Scholar]
- Manton I, Ettl H 1965. Observations on the fine structure of Mesostigma viride lauterborn. Bot J Linn Soc 59: 175–184. [Google Scholar]
- Marchant HJ, Pickett Heaps JD 1973. Mitosis and cytokinesis in Coleochaete scutata. J Phycol 9: 461–471. [Google Scholar]
- Marin B, Melkonian M 1999. Mesostigmatophyceae, a new class of streptophyte green algae revealed by SSU rRNA sequence comparisons. Protist 150: 399–417. [DOI] [PubMed] [Google Scholar]
- Martone PT, Estevez JM, Lu F, Ruel K, Denny MW, Somerville C, Ralph J 2009. Discovery of lignin in seaweed reveals convergent evolution of cell-wall architecture. Curr Biol 19: 169–175. [DOI] [PubMed] [Google Scholar]
- McCourt RM, Delwiche CF, Karol KG 2004. Charophyte algae and land plant origins. Trends Ecol Evol (Amst) 19: 661–666. [DOI] [PubMed] [Google Scholar]
- Merchant SS, Prochnik SE, Vallon O, Harris EH, Karpowicz SJ, Witman GB, Terry A, Salamov A, Fritz-Laylin LK, Maréchal-Drouard L, et al. 2007. The Chlamydomonas genome reveals the evolution of key animal and plant functions. Science 318: 245–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michod RE, Roze D 2001. Cooperation and conflict in the evolution of multicellularity. Heredity 86: 1–7. [DOI] [PubMed] [Google Scholar]
- Miller SM, Kirk DL 1999. glsA, a Volvox gene required for asymmetric division and germ cell specification, encodes a chaperone-like protein. Development 126: 649–658. [DOI] [PubMed] [Google Scholar]
- Mine I, Menzel D, Okuda K 2008. Morphogenesis in giant-celled algae. Int Rev Cell Mol Biol 266: 37–83. [DOI] [PubMed] [Google Scholar]
- Nakada T, Misawa K, Nozaki H 2008. Molecular systematics of Volvocales (Chlorophyceae, Chlorophyta) based on exhaustive 18S rRNA phylogenetic analyses. Mol Phylogenet Evol 48: 281–291. [DOI] [PubMed] [Google Scholar]
- Nedelcu AM, Michod RE 2006. The evolutionary origin of an altruistic gene. Mol Biol Evol 23: 1460–1464. [DOI] [PubMed] [Google Scholar]
- Nedelcu AM, Borza T, Lee RW 2006. A land plant-specific multigene family in the unicellular Mesostigma argues for its close relationship to Streptophyta. Mol Biol Evol 23: 1011–1015. [DOI] [PubMed] [Google Scholar]
- Niklas KJ 2000. The evolution of plant body plans—A biomechanical perspective. Ann Bot 85: 411–438. [Google Scholar]
- Niklas KJ, Kutschera U 2009. The evolution of the land plant life cycle. New Phytologist 185: 27–41. [DOI] [PubMed] [Google Scholar]
- Niklas KJ, Newman SA 2013. The origins of multicellular organisms. Evol Dev 15: 41–52. [DOI] [PubMed] [Google Scholar]
- Niklas KJ, Cobb ED, Crawford DR 2013. The evo-devo of multinucleate cells, tissues, and organisms, and an alternative route to multicellularity. Evol Dev 15: 466–474. [DOI] [PubMed] [Google Scholar]
- Nishii I 2003. A kinesin, InvA, plays an essential role in Volvox morphogenesis. Cell 113: 743–753. [DOI] [PubMed] [Google Scholar]
- Nishii I, Miller SM 2010. Volvox: Simple steps to developmental complexity? Curr Opin Plant Biol 13: 646–653. [DOI] [PubMed] [Google Scholar]
- Nishii I, Ogihara S 1999. Actomyosin contraction of the posterior hemisphere is required for inversion of the Volvox embryo. Development 126: 2117–2127. [DOI] [PubMed] [Google Scholar]
- Nishimura Y, Shikanai T, Nakamura S, Kawai-Yamada M, Uchimiya H 2012. Gsp1 triggers the sexual developmental program including inheritance of chloroplast DNA and mitochondrial DNA in Chlamydomonas reinhardtii. Plant Cell 24: 2401–2414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nozaki H 1990. Ultrastructure of the extracellular matrix of Gonium (Volvocales, Chlorophyta)*. Phycologia 29: 1–8. [Google Scholar]
- Nozaki H 1996. Morphology and evolution of sexual reproduction in the Volvocaceae (Chlorophyta). J Plant Res 109: 353–361. [Google Scholar]
- Nozaki H, Misawa K, Kajita T, Kato M, Nohara S, Watanabe MM 2000. Origin and evolution of the colonial volvocales (Chlorophyceae) as inferred from multiple, chloroplast gene sequences. Mol Phylogenet Evol 17: 256–268. [DOI] [PubMed] [Google Scholar]
- Nozaki H, Mori T, Misumi O, Matsunaga S, Kuroiwa T 2006a. Males evolved from the dominant isogametic mating type. Curr Biol 16: R1018–R1020. [DOI] [PubMed] [Google Scholar]
- Nozaki H, Ott FD, Coleman AW 2006b. Morphology, molecular phylogeny and taxonomy of two new species of Pleodorina (Volvoceae, Chlorophyceae). J Phycol 42: 1072–1080. [Google Scholar]
- Pandey S, Assmann SM 2004. The Arabidopsis putative G protein-coupled receptor GCR1 interacts with the G protein αsubunit GPA1 and regulates abscisic acid signaling. Plant Cell 16: 1616–1632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pappas V, Miller SM 2009. Functional analysis of the Volvox carteri asymmetric division protein GlsA. Mech Dev 126: 842–851. [DOI] [PubMed] [Google Scholar]
- Parker GA, Baker RR, Smith VG 1972. The origin and evolution of gamete dimorphism and the male-female phenomenon. J Theor Biol 36: 529–553. [DOI] [PubMed] [Google Scholar]
- Petersen J, Teich R, Becker B, Cerff R, Brinkmann H 2006. The GapA/B gene duplication marks the origin of Streptophyta (charophytes and land plants). Mol Biol Evol 23: 1109–1118. [DOI] [PubMed] [Google Scholar]
- Pickett-Heaps JD 1967. Ultrastructure and differentiation in Chara sp: II. Mitosis. Aust J Biol Sci 20: 883–894. [Google Scholar]
- Pickett-Heaps JD 1968. Ultrastructure and differentiation in Chara sp: III. Formation of the antheridium. Aust J Biol Sci 21: 255–274. [Google Scholar]
- Pickett-Heaps JD 1975. Green alage, structure, reproduction and evolution in selected genera. Sinauer, Sunderland, MA. [Google Scholar]
- Pickett-Heaps J 1976. Cell division in eucaryotic algae. BioScience 26: 445–450. [Google Scholar]
- Pires ND, Dolan L 2012. Morphological evolution in land plants: New designs with old genes. Philos Trans R Soc Lond B Biol Sci 367: 508–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popper ZA, Michel G, Hervé C, Domozych DS, Willats WGT, Tuohy MG, Kloareg B, Stengel DB 2011. Evolution and diversity of plant cell walls: From algae to flowering plants. Annu Rev Plant Biol 62: 567–590. [DOI] [PubMed] [Google Scholar]
- Preston JC, Hileman LC, Cubas P 2011. Reduce, reuse, and recycle: Developmental evolution of trait diversification. Am J Bot 98: 397–403. [DOI] [PubMed] [Google Scholar]
- Prigge MJ, Clark SE 2006. Evolution of the class III HD-Zip gene family in land plants. Evol Dev 8: 350–361. [DOI] [PubMed] [Google Scholar]
- Prigge MJ, Lavy M, Ashton NW, Estelle M 2010. Physcomitrella patens auxin-resistant mutants affect conserved elements of an auxin-signaling pathway. Curr Biol 20: 1907–1912. [DOI] [PubMed] [Google Scholar]
- Prochnik SE, Umen J, Nedelcu AM, Hallmann A, Miller SM, Nishii I, Ferris P, Kuo A, Mitros T, Fritz-Laylin LK, et al. 2010. Genomic analysis of organismal complexity in the multicellular green alga Volvox carteri. Science 329: 223–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pröschold T, Marin B, Schlösser UG, Melkonian M 2001. Molecular phylogeny and taxonomic revision of Chlamydomonas (Chlorophyta): I. Emendation of Chlamydomonas Ehrenberg and Chloromonas Gobi, and description of Oogamochlamys gen. nov. and Lobochlamys gen. nov. Protist 152: 265–300. [DOI] [PubMed] [Google Scholar]
- Pröschold T, Harris EH, Coleman AW 2005. Portrait of a species: Chlamydomonas reinhardtii. Genetics 170: 1601–1610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proseus TE, Boyer JS 2012. Pectate chemistry links cell expansion to wall deposition in Chara corallina. Plant Signal Behav 7: 1490–1492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proust H, Hoffmann B, Xie X, Yoneyama K, Schaefer DG, Yoneyama K, Nogue F, Rameau C 2011. Strigolactones regulate protonema branching and act as a quorum sensing-like signal in the moss Physcomitrella patens. Development 138: 1531–1539. [DOI] [PubMed] [Google Scholar]
- Queller DC 2000. Relatedness and the fraternal major transitions. Philos Trans R Soc Lond B Biol Sci 355: 1647–1655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quodt V, Faigl W, Saedler H, Münster T 2007. The MADS-domain protein PPM2 preferentially occurs in gametangia and sporophytes of the moss Physcomitrella patens. Gene 400: 25–34. [DOI] [PubMed] [Google Scholar]
- Randerson JP, Hurst LD 2001. The uncertain evolution of the sexes. Trends Ecol Evol 16: 571–579. [Google Scholar]
- Ratcliff WC, Denison RF, Borrello M, Travisano M 2012. Experimental evolution of multicellularity. Proc Natl Acad Sci 109: 1595–1600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratcliff WC, Herron MD, Howell K, Pentz JT, Rosenzweig F, Travisano M 2013. Experimental evolution of an alternating uni- and multicellular life cycle in Chlamydomonas reinhardtii. Nat Commun 4: 2742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raven JA 1997. Miniview: Multiple origins of plasmodesmata. Eur J Phycol 32: 95–101. [Google Scholar]
- Raven JA 2013. Polar auxin transport in relation to long-distance transport of nutrients in the Charales. J Exp Bot 64: 1–9. [DOI] [PubMed] [Google Scholar]
- Rethy R 1968. Red (R), far-red (FR) photoreversible effects on the growth of Chara sporelings. Z Pflanzenphysiol 59: 100–102. [Google Scholar]
- Robaglia C, Menand B, Lei Y, Sormani R, Nicolaï M, Gery C, Teoulé E, Deprost D, Meyer C 2004. Plant growth: The translational connection. Biochem Soc Trans 32: 581–584. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Ezpeleta N, Philippe H, Brinkmann H, Becker B, Melkonian M 2006. Phylogenetic analyses of nuclear, mitochondrial, and plastid multigene data sets support the placement of Mesostigma in the Streptophyta. Mol Biol Evol 24: 723–731. [DOI] [PubMed] [Google Scholar]
- Rokas A 2008. The origins of multicellularity and the early history of the genetic toolkit for animal development. Annu Rev Genet 42: 235–251. [DOI] [PubMed] [Google Scholar]
- Ruyter-Spira C, Bouwmeester H 2012. Strigolactones affect development in primitive plants. The missing link between plants and arbuscular mycorrhizal fungi? New Phytologist 195: 730–733. [DOI] [PubMed] [Google Scholar]
- Sabbatini MR, Argüello JA, Fernández OA, Bottini RA 1987. Dormancy and growth-inhibitor levels in oospores of Chara contraria A. Braun ex Kütz. (Charophyta). Aquat Bot 28: 189–194. [Google Scholar]
- Sakakibara K, Nishiyama T, Kato M, Hasebe M 2001. Isolation of homeodomain-leucine zipper genes from the moss Physcomitrella patens and the evolution of homeodomain-leucine zipper genes in land plants. Mol Biol Evol 18: 491–502. [DOI] [PubMed] [Google Scholar]
- Sakakibara K, Ando S, Yip HK, Tamada Y, Hiwatashi Y, Murata T, Deguchi H, Hasebe M, Bowman JL 2013. KNOX2 genes regulate the haploid-to-diploid morphological transition in land plants. Science 339: 1067–1070. [DOI] [PubMed] [Google Scholar]
- Sawa T, Frame PW 1974. Comparative anatomy of Charophyta: I. Oogonia and oospores of Tolypella—With special reference to the sterile oogonial cell. Bull Torrey Bot Club 136–144. [Google Scholar]
- Sawitzky H, Grolig F 1995. Phragmoplast of the green alga Spirogyra is functionally distinct from the higher plant phragmoplast. J Cell Biol 130: 1359–1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sebe-Pedros A, Irimia M, del Campo J, Parra-Acero H, Russ C, Nusbaum C, Blencowe BJ, Ruiz-Trillo I 2013. Regulated aggregative multicellularity in a close unicellular relative of metazoa. Elife 2: e01287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sederias J, Colman B 2007. The interaction of light and low temperature on breaking the dormancy of Chara vulgaris oospores. Aquat Bot 87: 229–234. [Google Scholar]
- Sekimoto H, Abe J, Tsuchikane Y 2012. New insights into the regulation of sexual reproduction in Closterium. Int Rev Cell Mol Biol 297: 309–338. [DOI] [PubMed] [Google Scholar]
- Šetlík I, Zachleder V 1984. The multiple fission cell reproductive patterns in algae. In The microbial cell cycle (ed. Nurse P, Streiblová E), pp. 253–279 CRC, Boca Raton, FL. [Google Scholar]
- Shikano S, Luckinbill LS, Kurihara Y 1990. Changes of traits in a bacterial population associated with protozoal predation. Microb Ecol 20: 75–84. [DOI] [PubMed] [Google Scholar]
- Simon A, Glöckner G, Felder M, Melkonian M, Becker B 2006. EST analysis of the scaly green flagellate Mesostigma viride (Streptophyta): Implications for the evolution of green plants (Viridiplantae). BMC Plant Biol 6: 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer SD, Krogan NT, Ashton NW 2007. Clues about the ancestral roles of plant MADS-box genes from a functional analysis of moss homologues. Plant Cell Rep 26: 1155–1169. [DOI] [PubMed] [Google Scholar]
- Smaczniak C, Immink RGH, Angenent GC, Kaufmann K 2012. Developmental and evolutionary diversity of plant MADS-domain factors: Insights from recent studies. Development 139: 3081–3098. [DOI] [PubMed] [Google Scholar]
- Smolarkiewicz M, Dhonukshe P 2013. Formative cell divisions: Principal determinants of plant morphogenesis. Plant Cell Physiol 54: 333–342. [DOI] [PubMed] [Google Scholar]
- Solnica-Krezel L, Sepich DS 2012. Gastrulation: Making and shaping germ layers. Annu Rev Cell Dev Biol 28: 687–717. [DOI] [PubMed] [Google Scholar]
- Sørensen I, Domozych D, Willats WGT 2010. How have plant cell walls evolved? Plant Physiol 153: 366–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sørensen I, Pettolino FA, Bacic A, Ralph J, Lu F, O’Neill MA, Fei Z, Rose JKC, Domozych DS, Willats WGT 2011. The charophycean green algae provide insights into the early origins of plant cell walls. Plant J 68: 201–211. [DOI] [PubMed] [Google Scholar]
- Srivastava M, Simakov O, Chapman J, Fahey B, Gauthier MEA, Mitros T, Richards GS, Conaco C, Dacre M, Hellsten U, et al. 2010. The Amphimedon queenslandica genome and the evolution of animal complexity. Nature 466: 720–726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanley SM 1973. An ecological theory for the sudden origin of multicellular life in the late precambrian. Proc Natl Acad Sci 70: 1486–1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark K, Kirk DL, Schmitt R 2001. Two enhancers and one silencer located in the introns of regA control somatic cell differentiation in Volvox carteri. Genes Dev 15: 1449–1460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starr RC 1970. Control of differentiation in Volvox. Symp Soc Dev Biol 29: 59–100. [DOI] [PubMed] [Google Scholar]
- Stebbins GL, Hill G 1980. Did multicellular plants invade the land? Am Nat 115: 342–353. [Google Scholar]
- Stewart KD, Mattox KR 1975. Comparative cytology, evolution and classification of the green algae with some consideration of the origin of other organisms with chlorophylls a and b. Botan Rev 41: 104–135. [Google Scholar]
- Stirk WA, Bálint P, Tarkowská D, Novák O, Strnad M, Ördög V, van Staden J 2013a. Hormone profiles in microalgae: Gibberellins and brassinosteroids. Plant Physiol Biochem 70: 348–353. [DOI] [PubMed] [Google Scholar]
- Stirk WA, Ördög V, Novák O, Rolčík J, Strnad M, Bálint P, van Staden J 2013b. Auxin and cytokinin relationships in 24 microalgal strains 1. J Phycol 49: 459–467. [DOI] [PubMed] [Google Scholar]
- Szathmáry E, Smith JM 1995. The major evolutionary transitions. Nature 374: 227–232. [DOI] [PubMed] [Google Scholar]
- Sztein AE, Cohen JD, Cooke TJ 2000. Evolutionary patterns in the auxin metabolism of green plants. Int J Plant Sci 161: 849–859. [Google Scholar]
- Tanabe Y, Hasebe M, Sekimoto H, Nishiyama T, Kitani M, Henschel K, Münster T, Theissen G, Nozaki H, Ito M 2005. Characterization of MADS-box genes in charophycean green algae and its implication for the evolution of MADS-box genes. Proc Natl Acad Sci 102: 2436–2441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarakhovskaya ER, Maslov YI, Shishova MF 2007. Phytohormones in algae. Russ J Plant Physiol 54: 163–170. [Google Scholar]
- Timme RE, Bachvaroff TR, Delwiche CF 2012. Broad phylogenomic sampling and the sister lineage of land plants. PLoS ONE 7: e29696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timme RE, Delwiche CF 2010. Uncovering the evolutionary origin of plant molecular processes: Comparison of Coleochaete (Coleochaetales) and Spirogyra (Zygnematales) transcriptomes. BMC Plant Biol 10: 96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsekos I 1999. The sites of cellulose synthesis in algae: Diversity and evolution of cellulose-synthesizing enzyme complexes. J Phycol 35: 635–655. [Google Scholar]
- Turmel M, Ehara M, Otis C, Lemieux C 2002. Phylogenetic relationships among streptophytes as inferred from chloroplast small and large subunit rRNA gene sequences. J Phycol 38: 364–375. [Google Scholar]
- Ueki N, Nishii I 2008. Idaten is a new cold-inducible transposon of Volvox carteri that can be used for tagging developmentally important genes. Genetics 180: 1343–1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueki N, Nishii I 2009. Controlled enlargement of the glycoprotein vesicle surrounding a Volvox embryo requires the InvB nucleotide-sugar transporter and is required for normal morphogenesis. Plant Cell 21: 1166–1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueki N, Matsunaga S, Inouye I, Hallmann A 2010. How 5000 independent rowers coordinate their strokes in order to row into the sunlight: Phototaxis in the multicellular green alga Volvox. BMC Biol 8: 1–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umen JG 2005. The elusive sizer. Curr Opin Cell Biol 17: 435–441. [DOI] [PubMed] [Google Scholar]
- Umen JG 2011. Evolution of sex and mating loci: An expanded view from Volvocine algae. Curr Opin Microbiol 14: 634–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umen JG, Goodenough UW 2001. Control of cell division by a retinoblastoma protein homolog in Chlamydomonas. Genes Dev 15: 1652–1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umen JG, Olson BJSC 2012. Genomics of volvocine algae. In Genomic insights into the biology of algae (ed. Piganeau G), Vol. 64, pp. 185–243 Elsevier, Amsterdam. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandenbussche F, Fierro AC, Wiedemann G, Reski R, Van Der Straeten D 2007. Evolutionary conservation of plant gibberellin signalling pathway components. BMC Plant Biol 7: 65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vannerum K, Huysman MJJ, De Rycke R, Vuylsteke M, Leliaert F, Pollier J, Lütz-Meindl U, Gillard J, De Veylder L, Goossens A, et al. 2011. Transcriptional analysis of cell growth and morphogenesis in the unicellular green alga Micrasterias (Streptophyta), with emphasis on the role of expansin. BMC Plant Biol 11: 128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Sandt VST, Stieperaere H, Guisez Y, Verbelen J-P, Vissenberg K 2007. XET activity is found near sites of growth and cell elongation in bryophytes and some green algae: New insights into the evolution of primary cell wall elongation. Ann Bot 99: 39–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viamontes G, Kirk D 1977. Cell shape changes and the mechanism of inversion in Volvox. J Cell Biol 75: 719–730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace S, Fleming A, Wellman CH, Beerling DJ 2011. Evolutionary development of the plant spore and pollen wall. AoB Plants 2011: plr027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Esch JJ, Shiu S-H, Agula H, Binder BM, Chang C, Patterson SE, Bleecker AB 2006. Identification of important regions for ethylene binding and signaling in the transmembrane domain of the ETR1 ethylene receptor of Arabidopsis. Plant Cell 18: 3429–3442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters ER 2003. Molecular adaptation and the origin of land plants. Mol Phylogenet Evol 29: 456–463. [DOI] [PubMed] [Google Scholar]
- Wodniok S, Brinkmann H, Glöckner G, Heidel AJ, Philippe H, Melkonian M, Becker B 2011. Origin of land plants: do conjugating green algae hold the key? BMC Evol Biol 11: 104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zalewski CS, Floyd SK, Furumizu C, Sakakibara K, Stevenson DW, Bowman JL 2013. Evolution of the class IV HD-Zip gene family in streptophytes. Mol Biol Evol 30: 2347–2365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong B, Liu L, Yan Z, Penny D 2013. Origin of land plants using the multispecies coalescent model. Trends Plant Sci 18: 492–495. [DOI] [PubMed] [Google Scholar]
- Zobell O, Faigl W, Saedler H, Munster T 2010. MIKC* MADS-Box proteins: Conserved regulators of the gametophytic generation of land plants. Mol Biol Evol 27: 1201–1211. [DOI] [PubMed] [Google Scholar]