Abstract
The links between recombination and replication have been appreciated for decades and it is now generally accepted that these two fundamental aspects of DNA metabolism are inseparable: Homologous recombination is essential for completion of DNA replication and vice versa. This review focuses on the roles that recombination enzymes play in underpinning genome duplication, aiding replication fork movement in the face of the many replisome barriers that challenge genome stability. These links have many conserved features across all domains of life, reflecting the conserved nature of the substrate for these reactions, DNA.
Homologous recombination is essential for completing DNA replication. Recombination enzymes provide a battery of options to aid in replication fork movement and overcome barriers faced by replisomes inside cells.
The interplay between replication and recombination is complex in terms of both mechanism and integration within DNA metabolism. At the heart of this interplay is the requirement for single-stranded DNA (ssDNA), the substrate for DNA-strand-exchange proteins, to initiate recombination (Cox 2007b; San Filippo et al. 2008). Whether, when, and where this ssDNA is generated determines the functional relationship between replication and recombination, a relationship that can operate in both directions. Homologous recombination enzymes are critical for successful completion of genome duplication (Kogoma 1997; Cox et al. 2000) but DNA replication also underpins homologous recombination, as discussed elsewhere in this collection. The links between recombination and replication are therefore intimate and one cannot be considered in isolation from the other. However, involvement of DNA-strand-exchange proteins, regardless of the metabolic context, comes with the unavoidable risk of genome rearrangements. This genome instability can occasionally increase evolutionary fitness but more frequently is deleterious to the viability of the individual.
This review will focus on fundamental aspects of the links between replication and recombination enzymes rather than simply providing a list of known enzymes and reactions. The substrate, DNA, is identical in all of these reactions and this is reflected in the high mechanistic conservation of replication and recombination.
DE NOVO INITIATION OF REPLICATION BY RECOMBINATION
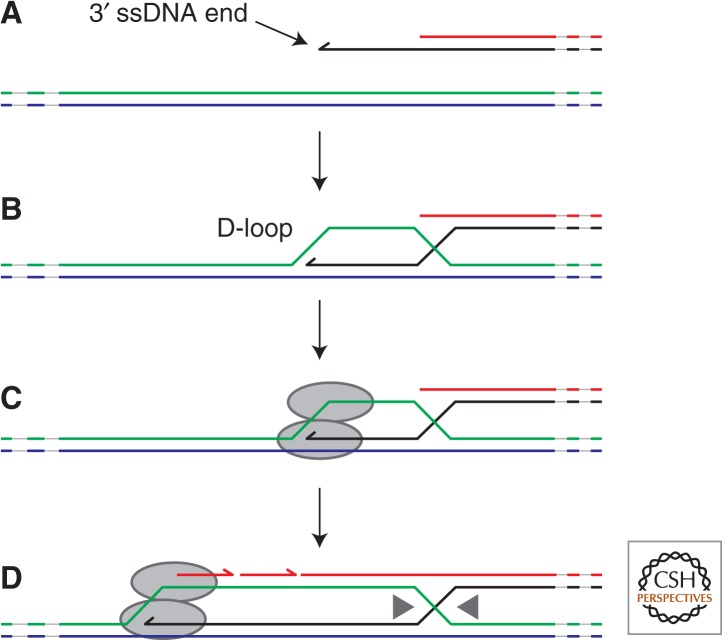
The intimate links between homologous recombination and DNA replication were first noted in the bacteriophage T4 (Luder and Mosig 1982). Early T4 genome duplication occurs at specific origins but once a replisome reaches one of the DNA ends then the inability to complete lagging strand synthesis results in a 3′ ssDNA end (Kreuzer and Brister 2010). Binding of this ssDNA by the bacteriophage DNA-strand-exchange protein, UvsX, results in strand invasion and formation of a D-loop with a homologous region either within the same DNA molecule, directed by terminal redundancy within the T4 genome, or another T4 DNA molecule within the same cell (Liu and Morrical 2010). The D-loop is a central intermediate in the interplay between recombination and replication, providing a specific branched DNA structure that can be recognized by replication initiators and a 3′ DNA end to prime leading strand synthesis (Fig. 1) (McGlynn et al. 1997; Mueser et al. 2000). Loading of the replicative helicase onto the chromosome by these replication initiators acts as a trigger for assembly of other replication enzymes to form the functional replisome. Thus, recombination allows DNA structure-directed replication initiation as opposed to the DNA sequence-directed initiation occurring at specific origins (Heller and Marians 2006a).
Figure 1.
D-loop formation and the initiation of replication. Strand exchange initiated via a 3′ DNA end (A) results in the formation of a D-loop with a homologous duplex (B). This branched DNA structure can act as a target for replication initiation proteins that results in loading of the replisome (C) and consequent leading and lagging strand synthesis. The strand exchange process generates a Holliday junction behind the replication fork that must be removed to allow the sister chromosomes to segregate (D).
Replisome assembly at D-loops appears to be conserved throughout evolution, providing a structural link between recombination and replication initiation (McGlynn and Lloyd 2002; San Filippo et al. 2008). Indeed, recombination-directed formation of D-loops via processing of double-stranded DNA breaks can sustain chromosomal duplication in the absence of normal origin function in bacteria, not just viruses (Kogoma 1997). However, de novo initiation of genome duplication via D-loops demands homologous sequences and so requires a second copy of the genome to be present or terminal redundancy within the genome, as in T4 (Kreuzer and Brister 2010). Recombination-initiated replication thus imposes constraints on ploidy and genome structure that are avoided by the use of specific origins of replication. Specific replication origins also provide greater potential for regulation via the initiator. However, recombination-dependent formation of D- loops, although being supplanted for de novo initiation of genome duplication in nonviral life-forms, still plays key roles in overcoming replicative problems.
THE PROBLEM WITH DNA REPLICATION
DNA lesions, proteins bound to the template, R-loops, DNA secondary structures, and topological strain can all inhibit replisome movement. Replisomes paused at such barriers can continue duplication whether the original block is removed or bypassed (Marians et al. 1998; Guy et al. 2009; Pomerantz and O’Donnell 2010; Yeeles and Marians 2011) but paused replisomes lose activity as a function of time (Marians et al. 1998; McGlynn and Guy 2008; Petermann et al. 2010), implying that pausing increases the probability of loss of replisome function. Oxidative damage to replication enzymes in vivo (Davies 2005) could also result in fork breakdown, a very different form of damage because it is not associated with template blocks and so cannot be bypassed. Such damage might create problems especially when occurring in replication enzymes with low turnover rates in vivo but the extent of this potential problem is unknown.
Determining the frequency with which replisomes pause and subsequently lose activity in vivo is technically difficult. Most pausing events in E. coli do not lead to replisome inactivation but pausing frequency is high, suggesting that even a low probability of paused fork inactivation presents challenges to genome stability (Gupta et al. 2013). Perhaps the most compelling evidence for the challenge posed by loss of replisome activity is the severe loss of viability in the absence of the E. coli replication restart enzyme PriA (Lee and Kornberg 1991; Nurse et al. 1991). PriA binds to DNA structures associated with replisome breakdown such as D-loops and forks (McGlynn et al. 1997), resulting in loading of the replicative helicase and replisome reassembly (Liu et al. 1999; Cadman et al. 2005; Lopper et al. 2007). PriA acts therefore as a DNA structure-specific replication initiator, restarting replication away from oriC in the event of replisome breakdown. The importance of this replisome reassembly is underlined by the inviability of cells lacking PriA and a second structure-specific initiator PriC (Sandler 2000). PriA and PriC have complementary DNA substrate specificities, binding preferentially to forked DNA with and without a leading strand present at the branch point, respectively (Heller and Marians 2005b). The inviability of cells lacking both enzymes provides direct evidence for the low probability of replisomes assembled at oriC being able to complete chromosome duplication.
The multiple origins per chromosome in eukaryotes, including normally dormant but activatable origins, provide a means of completing replication when a replisome breaks down that is not available in bacteria (Blow et al. 2011). However, although multiple origins might reduce the need for replisome reloading, inactivation of paused forks in eukaryotes does present challenges to genome stability (Aguilera and Gomez-Gonzalez 2008; Petermann and Helleday 2010; Petermann et al. 2010; Duch et al. 2012). Forks can lose activity in regions with low origin density, which may reduce the probability of broken fork rescue by adjacent origin firing, a situation in which common fragile sites and associated ultrafine anaphase bridges can occur (Chan et al. 2009; Letessier et al. 2011; Ozeri-Galai et al. 2011; see also Bizard and Hickson 2014). Inactivation of replisomes within unidirectionally replicated sections of the genome (Murray and Carr 2008) will also create a requirement for replisome reassembly. A high density of transcribing RNA polymerases might also present barriers to replisome movement that are so substantial as to inhibit both converging replication forks (Trautinger et al. 2005; Duch et al. 2012; McGlynn et al. 2012). Converging forks are therefore unlikely to provide a safety net for inactivated replisomes that is sufficiently robust to ensure genome stability.
THE STRUCTURE OF BLOCKED REPLICATION FORKS
DNA-strand-exchange proteins, nucleases, and helicases all possess specific DNA structure specificities, and so the structures of blocked forks dictate mechanisms of recombination enzyme-mediated processing.
DNA lesions such as cyclobutane pyrimidine dimers that act as single-strand-specific blocks can inhibit either the leading or the lagging strand polymerase. The discontinuous nature of lagging strand synthesis facilitates repriming of replication downstream from a noncoding lesion, leaving an ssDNA gap that must be repaired subsequently by recombination with the sister duplex or by translesion synthesis (Rupp et al. 1971; Lehmann 1972; Smith 2004; Lopes et al. 2006). Repriming of DNA synthesis can also occur downstream from lesions within the leading strand template, resulting in formation of a ssDNA gap that, as for the lagging strand, must be repaired after passage of the fork (Rupp and Howard-Flanders 1968; Lehmann 1972; Amado and Kuzminov 2006; Elvers et al. 2011; Yeeles and Marians 2011). However, high levels of DNA damage do inhibit replication fork progression (Setlow et al. 1963; Edenberg 1976). Thus, bypass mechanisms might deal efficiently with low, but not high, densities of DNA lesions. However, the relative frequency of lesion bypass versus fork breakdown as a function of lesion density remains unknown. Even when fork breakdown does occur, PriC-directed replisome reassembly might be able to reinitiate replication downstream from leading strand lesions without the need for recombination enzymes in Escherichia coli (Heller and Marians 2005a,b, 2006b). However, it is unknown whether eukaryotes have a PriC-like activity. If both bypass and restart reactions fail, then continued movement of the replicative helicase could allow lagging strand synthesis to continue for some way beyond the terminated leading strand to generate a blocked fork with a gap on the leading strand (Svoboda and Vos 1995; Pagès and Fuchs 2003; Yeeles and Marians 2013).
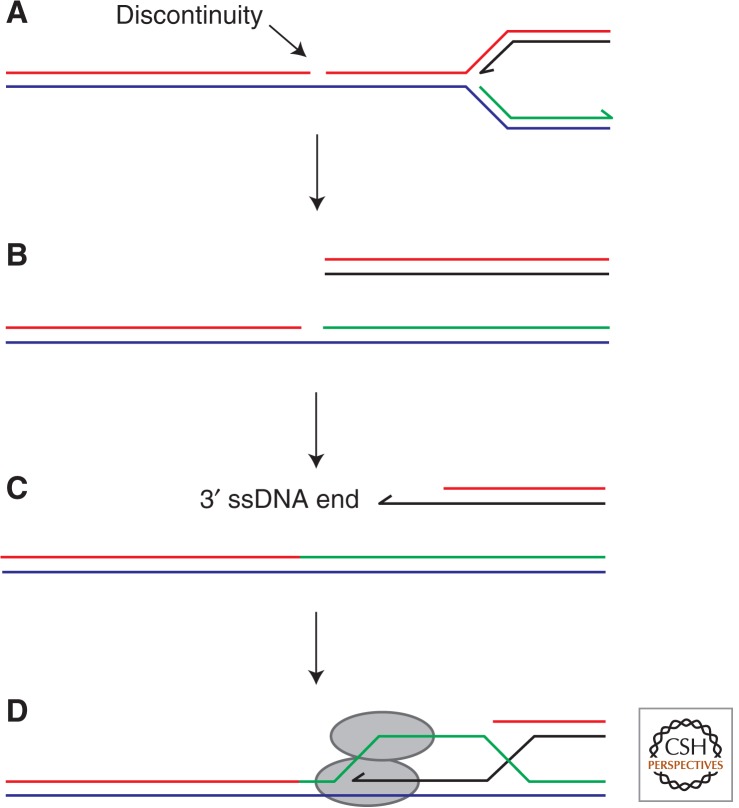
Another type of strand-specific DNA lesion, a discontinuity in the phosphodiester backbone, results in loss of the replication fork structure and generation of a dsDNA end, requiring reintegration of the dsDNA end via recombination and replisome reassembly to reform the fork (Fig. 2) (Hanawalt 1966; Skalka 1974; Kuzminov 2001; Hashimoto et al. 2012). Such breaks in one strand might arise as a result of occasional failure to repair DNA lesions or to ligate adjacent Okazaki fragments (Gottesman et al. 1973; Kouzminova and Kuzminov 2008; Moriel-Carretero and Aguilera 2010). Topoisomerases may also fail to reseal breaks within DNA strands, either when covalent topoisomerase–DNA complexes are stabilized via drugs such as the anticancer agent camptothecin or when topoisomerases act on damaged DNA (Pommier et al. 1998; Pommier 2013). The frequency with which such discontinuities arise is difficult to assess directly. However, the need for efficient double-stranded DNA end processing in E. coli appears to be associated with direct processing of replication forks rather than repair of DNA ends released by collision of forks with strand discontinuities (Capaldo et al. 1974; Seigneur et al. 1998; McGlynn and Lloyd 2000). Single strand breaks might therefore be rare (McGlynn and Lloyd 2000).
Figure 2.
Generation of a dsDNA end by a replisome encountering a discontinuity within a template DNA strand. The discontinuity is depicted within the leading strand template (A) but the same outcome would result from a lagging strand template discontinuity. Processing of the free dsDNA end (B) by exonucleases would result in a 3′ ssDNA substrate (C), which could be used to generate a D-loop via strand exchange (D).
Nucleoprotein complexes, interstrand cross- links, and topological strain require removal of the original block for resumption of replication because such barriers inhibit both polymerases and the helicase, preventing bypass reactions (McGlynn 2013). Saccharomyces cerevisiae forks stalled by the programmed nucleoprotein barrier within the rDNA array possess very little ssDNA on either template strand (Gruber et al. 2000). Similarly, although E. coli forks blocked by the Tus–ter replication barrier in vitro have 50–70 bases of ssDNA on the lagging strand template, there is little on the leading strand template (Hill and Marians 1990).
UNDERPINNING REPLICATION FORK MOVEMENT VIA RECOMBINATION ENZYMES
There are many mechanisms by which recombination enzymes could aid replication fork movement but obtaining good evidence for any of these mechanisms being operative in vivo has proven difficult. Potential mechanisms are often inferred from indirect evidence, raising difficulties in interpretation given the overlap between different pathways of replication repair. It is also important to note that the involvement of recombination enzymes does not necessarily result in exchange of DNA strands between duplexes. Even when strand exchange does occur this may have no genetic consequences if it occurs between identical regions on the sister duplexes generated by the original fork. Conversely, strand exchange between homologous chromosomes can lead to loss of heterozygosity in eukaryotes (St Charles and Petes 2013), whereas nonallelic exchange within the same, or between different, chromosomes can cause gross chromosomal rearrangements (Lambert et al. 2005; Liu et al. 2011; Mizuno et al. 2012).
FORK STABILITY
Recombination enzyme-dependent stabilization of damaged forks implies that DNA at inactivated forks is at risk from nucleolytic degradation. Degradation could be owing to endonucleolytic cleavage of ssDNA or branched structures at the fork, or to exonucleolytic degradation of DNA ends at ssDNA gaps, the fork itself, or via fork regression. Such degradation could facilitate replisome repair (see below) but could also result in loss of genetic information and/or aberrant recombination.
RecA, the bacterial DNA-strand-exchange protein, and RecFOR minimize degradation of DNA in E. coli cells exposed to UV light (Courcelle and Hanawalt 2003). One explanation is that RecFOR promotes RecA binding to SSB-coated ssDNA at the fork, with the resultant nucleoprotein filament inhibiting access of nucleases to the DNA (Courcelle et al. 1997, 2003; Morimatsu and Kowalczykowski 2003). However, RecAFOR-dependent replication restart may prevent degradation of DNA at the fork simply by promoting resumption of replication, possibly via fork regression or the repair of dsDNA ends (see below), rather than directly stabilizing the fork DNA (Rudolph et al. 2007, 2008). RecA-dependent stabilization of blocked forks is therefore still an open question.
BRCA2-dependent stabilization of binding of Rad51, the eukaryotic DNA-strand-exchange protein, at or near stalled forks, inhibits nucleolytic degradation of nascent DNA (Hashimoto et al. 2010; Schlacher et al. 2011). Fork stabilization could therefore be a conserved feature of DNA-strand-exchange enzymes. There are also multiple mechanisms to stabilize blocked replication forks in eukaryotes that do not require recombination enzymes (Lopes et al. 2001; Tercero and Diffley 2001; Katou et al. 2003; Szyjka et al. 2005; Tittel-Elmer et al. 2009; Vaisica et al. 2011). Perhaps the increased numbers of forks in eukaryotes demands a reduced probability of individual blocked fork instability, requiring multiple stabilization mechanisms. Alternatively, different replicative blocks might require different mechanisms to promote blocked fork stability.
DNA-strand-exchange proteins might also play very different roles in terms of replisome stability as opposed to forked DNA stability. RecA promotes polymerase dissociation from DNA containing an abasic lesion in a reaction that is facilitated by RecFOR when SSB is present, implying that facilitated disassembly may allow subsequent processing and restart of blocked replication forks (McInerney and O’Donnell 2007). Indeed, RecA promotes bypass of damage via activation of translesion DNA polymerases (Schlacher et al. 2006; Indiani et al. 2013), a reaction that might provide a mechanism of last resort if other fork processing pathways fail to restart replication (Courcelle et al. 2006).
Targeting of replication forks by DNA-strand-exchange proteins, regardless of the outcome of this binding, will likely inhibit other types of blocked fork processing and also presents the risk of genome rearrangements occurring. The binding of strand exchange proteins to ssDNA is therefore regulated (Krejci et al. 2012) with turnover of nucleoprotein filaments by specific helicases being a key factor in balancing replication, repair, and recombination (Krejci et al. 2003; Veaute et al. 2003, 2005; Antony et al. 2009; Long et al. 2009). Other, less well-characterized, mechanisms also exist to modulate DNA-strand-exchange protein activity at forks (Moore et al. 2003; Cox 2007a; Bakhlanova et al. 2010). Such control balances the need to repair damaged replication forks with the need to minimize genome instability, raising the possibility that strand exchange protein-mediated remodeling of blocked replication forks might occur only rarely.
BLOCKED FORK PROCESSING WITHOUT FORK REGRESSION
The conceptually easiest link between replication and recombination is the breakdown of replication on encountering a discontinuity in either the leading or the lagging template strand. This would result in release of one of the sister duplexes with a dsDNA end, a substrate for recombination, and would result in D-loop formation with homologous sequences on the sister duplex (Fig. 2) (McGlynn and Lloyd 2002; Aguilera and Gomez-Gonzalez 2008). Assuming that the original discontinuity in the intact sister duplex was repaired, replisome reassembly onto the D-loop would result in restoration of a functional replication fork. PriA in bacteria provides a well-characterized means of such D-loop targeting (Xu and Marians 2003; Heller and Marians 2006a), whereas break-induced replication in eukaryotes (Llorente et al. 2008; Symington and Gautier 2011; see also Mehta and Haber 2014) indicates similar repair of single dsDNA ends is a universal feature of genome duplication.
Other types of block, such as nucleotide damage or protein–DNA complexes, would not lead directly to loss of the branched DNA structure characteristic of replication but may require DNA processing to facilitate repair or bypass of the block and/or replisome reassembly. Potential processing reactions include exonucleolytic degradation of the leading and/or the lagging strands, endonucleolytic cleavage at or behind the fork, and unwinding/rewinding of template and/or nascent DNA strands.
Exonucleolytic processing of blocked forks could be initiated either at the 3′ end of the nascent leading strand or the 5′ end of the lagging strand. Degradation of a nascent strand that has been extended significantly beyond the other strand might promote spontaneous reannealing of the parental DNA although binding of exposed ssDNA by SSBs could inhibit reannealing. Prevention of reannealing of ssDNA might facilitate the loading of strand exchange proteins onto ssDNA at the fork with possible consequences as regards fork remodeling or stability. DNA-strand-exchange proteins bound at the fork may be able to promote regression (Seigneur et al. 2000; Robu et al. 2001). An alternative, but not mutually exclusive, view is that nucleofilament formation results in protection of DNA at the fork from excessive degradation by nucleases, facilitating repair of blocking lesions and resumption of replication (Courcelle et al. 2003, 2006; Bryant et al. 2009).
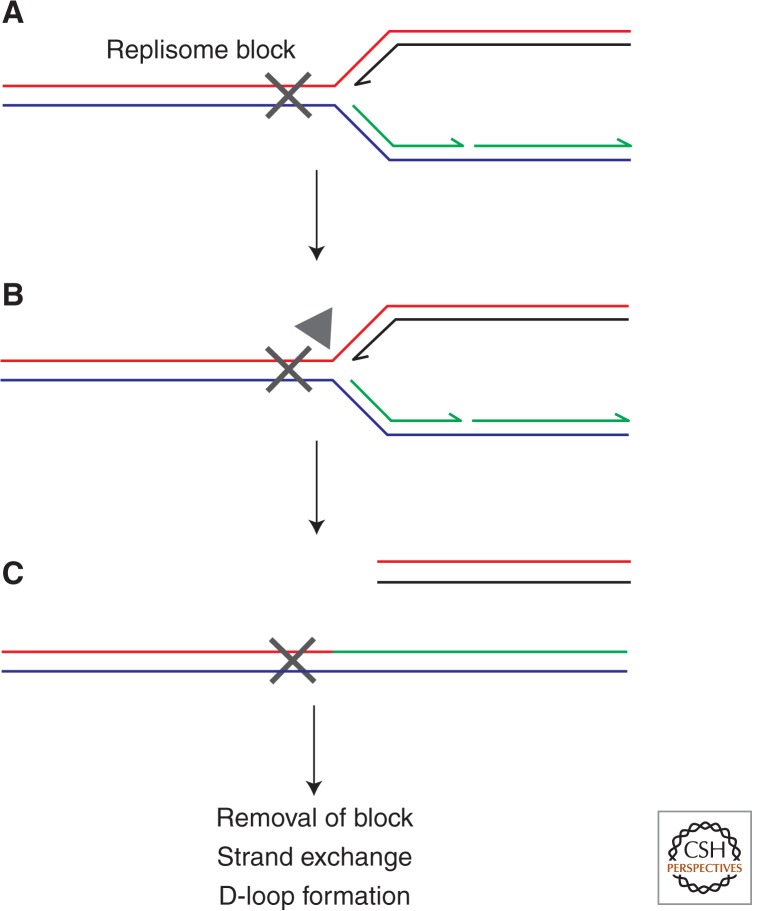
Endonucleolytic processing of blocked replication forks might also occur. Multiple branched DNA-specific endonucleases have been characterized in eukaryotes that cleave DNA flaps, forks, and Holliday junctions (Schwartz and Heyer 2011; Rass 2013). Two of these (Mus81-Mms4 and Slx1-Slx4 in S. cerevisiae) have been implicated in recombination-mediated repair of damaged replication forks (Fricke and Brill 2003; Hanada et al. 2007; Osman and Whitby 2007). Direct cleavage of blocked replication forks by these branched DNA-specific endonucleases has therefore been proposed as one means of promoting replication repair, with cleavage releasing a single dsDNA end that can be recombined back with the sister duplex to provide a D-loop substrate for replisome reloading (Fig. 3) (Ahn et al. 2005; Hanada et al. 2006). However, it is not clear how such direct cleavage followed by recombination would facilitate removal of the original replicative block. Perhaps destruction of the fork facilitates access of excision repair enzymes to the blocking lesion. Recombination-directed restart might also facilitate replication through stochastic blocks such as protein–DNA complexes (Payne et al. 2006). Direct cleavage could also promote recombination in the vicinity of interstrand cross-links, a type of lesion known to require recombination for repair (Hanada et al. 2006; Raschle et al. 2008). Alternatively, mammalian forks stalled for prolonged periods could be rescued by a combination of adjacent origin firing and MUS81-directed cleavage of the original blocked fork (Hanada et al. 2007; Petermann et al. 2010). Homologous recombination from the cleaved fork might then allow completion of duplication of the intervening DNA (Petermann et al. 2010). However, convincing evidence for direct cleavage of replication forks in vivo, as opposed to DNA intermediates formed from the original fork, is lacking. The peak of Mus81 activity also occurs during mitosis rather than S phase, implying that resolution of joint molecules in mitosis rather than cleavage of blocked replication forks during S phase might be the primary function of this endonuclease (Matos et al. 2011). It should also be borne in mind that utilization of these different branched DNA-specific endonucleases varies among different eukaryotes and so it is difficult to generalize concerning the importance of direct cleavage of blocked forks (Schwartz and Heyer 2011).
Figure 3.
Direct cleavage of blocked replication forks. Cleavage (gray triangle) of template DNA at blocked replication forks (A,B) can release a free dsDNA end (C). Strand exchange-mediated D-loop formation could then restore a functional replisome allowing completion of genome duplication assuming that the original block was removed.
BLOCKED FORK PROCESSING WITH FORK REGRESSION
Reannealing of the parental DNA strands at a fork, together with annealing of the nascent strands, would result in formation of a four-stranded structure that resembles a classical Holliday junction (Figs. 4A,B and 5A,B) (Hotchkiss 1974). Genetic and physical evidence that fork regression can occur has been reviewed elsewhere in detail (Atkinson and McGlynn 2009). The frequency of regression appears low as judged by electrophoretic and electron microscopic techniques (Sogo et al. 2002; Pohlhaus and Kreuzer 2006) indicating that fork regression might be a pathological event. However, many genetic observations can be explained by invoking fork regression with such models being supported by the identification of enzymes able to catalyze regression in vitro (Aguilera and Gomez-Gonzalez 2008; Atkinson and McGlynn 2009). Blocked fork regression would not have to be a frequent event for it to be an important method of fork processing with respect to viability and genetic assays might provide sensitive readouts of such events. It is therefore difficult to unequivocally state the importance of fork regression based on one set of observations alone.
Figure 4.
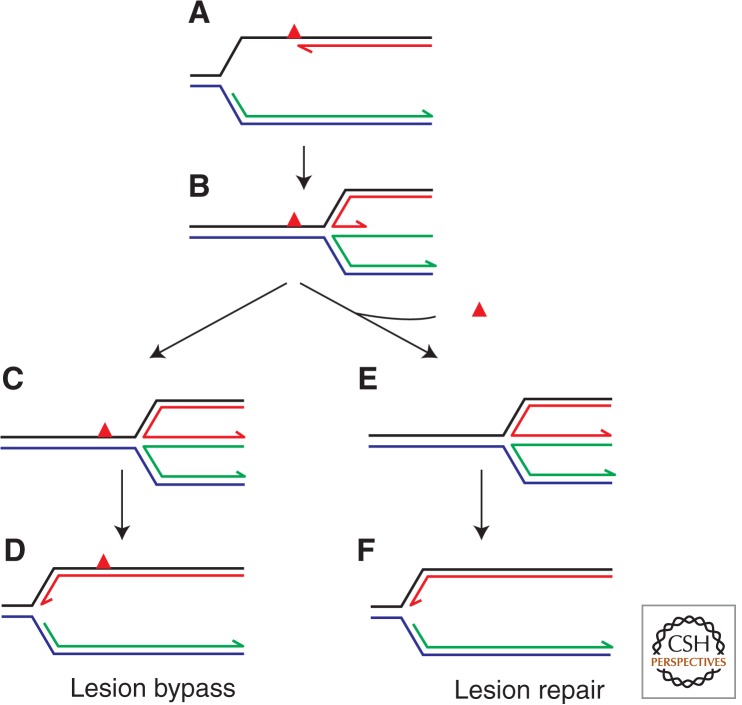
Template switching may provide the means to bypass or repair strand-specific DNA lesions. Fork regression (A,B) could allow the lagging strand to provide a template for extension of the blocked leading strand (B,C). Subsequent reversal of the original regression would result in bypass of the lesion and reinitiation of replication at the reconstituted fork structure (D). Note that such fork structures resemble the branched DNA found within D-loops and, at least in bacteria, replication reinitiation proteins can reload the replication apparatus back onto these forks. Alternatively, fork regression could reposition the lesion opposite an intact lagging strand template, promoting excision repair (B,E). Reversal of fork regression would again reform the fork to allow replication reinitiation (F).
Figure 5.
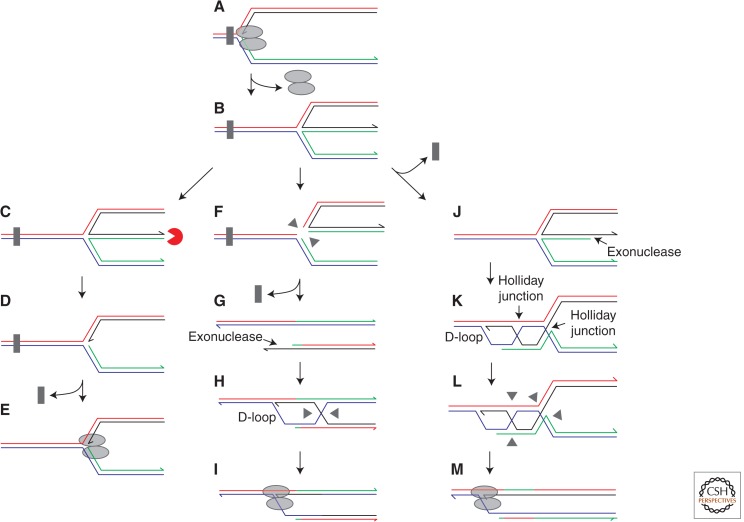
Fork regression coupled with exo- or endonuclease-mediated processing could provide multiple pathways for replication fork restart. Fork regression after blockage of the replisome (A,B) could generate a dsDNA end that could be fully degraded by exonucleases (C,D) to reconstitute a fork structure onto which the replisome can be reloaded (E). Alternatively, Holliday junction endonucleases might cleave the regressed fork (F) to generate a dsDNA end that can catalyze D-loop formation and replisome reloading (G–I). Endonucleolytic cleavage could also occur after strand exchange between the DNA end formed by fork regression and the homologous sequence within the parental duplex, providing an intramolecular recombination reaction that might limit the potential for aberrant recombination (J–M).
What might drive fork regression? Positive torsional strain ahead of the fork can promote rewinding of the parental DNA strands and hence regression (McGlynn et al. 2001; Postow et al. 2001). Several helicases/translocases have also been shown to catalyze fork regression in vitro (McGlynn and Lloyd 2001; Machwe et al. 2006; Ralf et al. 2006; Webb et al. 2007; Betous et al. 2012). Regression also appears to be a universal catalytic property of DNA-strand-exchange proteins in vitro, a property that can explain the in vivo functions of RecA and Rad51 in the face of certain replicative blocks (Seigneur et al. 2000; Robu et al. 2001; Courcelle et al. 2003; Kadyrov and Drake 2004; Yoon et al. 2004). However, the initial binding of strand exchange proteins requires ssDNA, implying that access of these proteins to blocked forks will depend on the nature of the replicative block and processing of the blocked fork by exonucleases (see above).
How might fork regression facilitate genome duplication? DNA lesions within the leading strand template, if not bypassed by repriming, may be refractory to excision repair owing to the absence of a complementary DNA strand and so continue to block leading strand synthesis. Fork regression might allow extension of the blocked leading strand using the nascent lagging strand as a template (Fig. 4B) (Fujiwara and Tatsumi 1976; Higgins et al. 1976). This template switching reaction might then allow bypass of the original lesion on reversal of fork regression and reloading of the replication apparatus at the fork (Fig. 4C,D). Postreplicative bypass mechanisms are emerging as important in eukaryotes (Blastyak et al. 2007; Hishida et al. 2009) but perhaps the best evidence to date for template switching is the reconstitution of this reaction in vitro using bacteriophage T4 enzymes (Manosas et al. 2012).
Regression followed by direct reversal might also promote repair, as opposed to bypass, of DNA lesions. Regression of forks halted by strand-specific lesions would reposition the lesion opposite a complementary DNA strand, facilitating excision repair followed by replisome reassembly on reversal of regression (Fig. 4B,E,F) (Courcelle et al. 2003, 2005). Degradation of the extruded duplex arm, as opposed to reversal of fork regression, might provide an alternative means of replication restart after fork regression (Fig. 5C) (Seigneur et al. 1998; Michel et al. 2004). Regression followed by degradation of the dsDNA end and restart could also promote replication through stochastic blocks such as protein–DNA complexes.
Regressed forks might also be cleaved by endonucleases rather than be removed by direct reversal or degradation of the extruded dsDNA arm. Cleavage of the four-stranded structure by a Holliday junction endonuclease would generate one intact sister duplex, after ligation of the phosphodiester backbone, and a second duplex with a dsDNA end (Fig. 5F,G) (Seigneur et al. 1998). This cleavage would destroy, rather than remodel, the original fork structure, necessitating strand exchange between the dsDNA end and the intact sister duplex to reform the fork via D-loop formation (Fig. 5G–I). Like regression followed by degradation of the extruded dsDNA arm, recreating an active replication fork upstream of the original block could promote access of repair enzymes to the block or provide a second opportunity for the replisome to translocate through stochastic blocks. However, the need for recombination subsequent to cleavage of the regressed fork would create a risk of inaccurate recombination, raising questions as to the frequency of such processing relative to degradation of the extruded arm under normal circumstances (Seigneur et al. 1998; Flores et al. 2001; Michel et al. 2004).
It is also possible that the recombination event could occur before any cleavage (Fig. 5J–M). The reversal followed by degradation model assumes that exonucleases would degrade the extruded duplex arm completely (Fig. 5C,D). However, this exonucleolytic degradation could also generate ssDNA that could act as a substrate for recombination (Louarn et al. 1991), with strand exchange occurring potentially with the homologous sequence within the reformed parental duplex (Fig. 5K) (Seigneur et al. 1998; Ahn et al. 2005). Reassembly of the replisome at the D-loop would generate a replication fork with two upstream Holliday junctions, both of which would require resolution by endonucleases or helicases/topoisomerases (Fig. 5L). Although such a reaction would again result in the risk of inaccurate homologous recombination, this risk might be reduced by physical association of the dsDNA end and the target donor sequence. However, although each enzymatic step of this potential pathway has been characterized within other contexts, the only direct evidence for such a mechanism comes from the early stages of bacteriophage T4 infection (Long and Kreuzer 2008).
WHEN REPLICATION FORKS COLLIDE—A BLOCK IN ALL BUT NAME
Work on replicative problems has focused on forks running into barriers to progression. Most eukaryotic and bacterial forks must also collide with a converging fork during every cell cycle. What happens when these collisions occur is poorly understood. Hyperrecombination is a feature of the E. coli replication termination zone (Bierne et al. 1991; Louarn et al. 1991) and depends on dsDNA end processing (Horiuchi et al. 1994). Colliding E. coli replisomes might also be prone to overreplication (Rudolph et al. 2009a,b, 2010, 2013), possibly owing to the replicative helicase continuing to unwind the 3′ end of the opposing nascent leading strand (Hiasa and Marians 1994). These putative 3′ ssDNA flaps could result in binding of RecA, stimulating pathological recombination events if not degraded rapidly by exonucleases (Rudolph et al. 2010, 2013). Although even less is known about replication termination in eukaryotes, the broad zones in which replisomes collide in S. cerevisiae act as hot spots of mitotic recombination (St Charles and Petes 2013). X-shaped intermediates, chromosome breaks, and enhanced genome rearrangements also become detectable within termination zones when replisome fusion is perturbed (Fachinetti et al. 2010). Given the number of replication termination events in eukaryotes, recombination might therefore be an undesirable, unavoidable side reaction during fork fusion.
CONCLUDING REMARKS
The wide variety of potential barriers encountered by replication forks result in the generation of many different types of blocked fork structures. Blockage does not necessarily lead to replisome inactivation but there are many potential circumstances when clearance/bypass of the block by the original replisome cannot occur. Under such circumstances remodeling of the DNA may be needed together with reassembly of the replication apparatus. Recombination enzymes, either with or without DNA-strand exchange occurring, provide a battery of options to accommodate replication breakdown. We are currently at the point of understanding many of the potential reactions that can occur with the complement of replication and recombination enzymes present in vivo. The challenge now lies in determining which of these pathways operate and how they interface under normal circumstances.
ACKNOWLEDGMENTS
Work in the authors' laboratory on the links between recombination and replication is funded by BBSRC Grant BB/J014826/1.
Footnotes
Editors: Stephen Kowalczykowski, Neil Hunter, and Wolf-Dietrich Heyer
Additional Perspectives on DNA Recombination available at www.cshperspectives.org
REFERENCES
*Reference is also in this collection.
- Aguilera A, Gomez-Gonzalez B 2008. Genome instability: A mechanistic view of its causes and consequences. Nat Rev Genet 9: 204–217. [DOI] [PubMed] [Google Scholar]
- Ahn JS, Osman F, Whitby MC 2005. Replication fork blockage by RTS1 at an ectopic site promotes recombination in fission yeast. EMBO J 24: 2011–2023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amado L, Kuzminov A 2006. The replication intermediates in Escherichia coli are not the product of DNA processing or uracil excision. J Biol Chem 281: 22635–22646. [DOI] [PubMed] [Google Scholar]
- Antony E, Tomko EJ, Xiao Q, Krejci L, Lohman TM, Ellenberger T 2009. Srs2 disassembles Rad51 filaments by a protein-protein interaction triggering ATP turnover and dissociation of Rad51 from DNA. Mol Cell 35: 105–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atkinson J, McGlynn P 2009. Replication fork reversal and the maintenance of genome stability. Nucleic Acids Res 37: 3475–3492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakhlanova IV, Dudkina AV, Baitin DM, Knight KL, Cox MM, Lanzov VA 2010. Modulating cellular recombination potential through alterations in RecA structure and regulation. Mol Microbiol 78: 1523–1538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betous R, Mason AC, Rambo RP, Bansbach CE, Badu-Nkansah A, Sirbu BM, Eichman BF, Cortez D 2012. SMARCAL1 catalyzes fork regression and Holliday junction migration to maintain genome stability during DNA replication. Genes Dev 26: 151–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bierne H, Ehrlich SD, Michel B 1991. The replication termination signal terB of the Escherichia coli chromosome is a deletion hot spot. EMBO J 10: 2699–2705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Bizard AH, Hickson ID 2014. The dissolution of double Holliday junctions. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a016477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blastyak A, Pinter L, Unk I, Prakash L, Prakash S, Haracska L 2007. Yeast Rad5 protein required for postreplication repair has a DNA helicase activity specific for replication fork regression. Mol Cell 28: 167–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blow JJ, Ge XQ, Jackson DA 2011. How dormant origins promote complete genome replication. Trends Biochem Sci 36: 405–414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryant HE, Petermann E, Schultz N, Jemth AS, Loseva O, Issaeva N, Johansson F, Fernandez S, McGlynn P, Helleday T 2009. PARP is activated at stalled forks to mediate Mre11-dependent replication restart and recombination. EMBO J 28: 2601–2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cadman CJ, Lopper M, Moon PB, Keck JL, McGlynn P 2005. PriB stimulates PriA helicase via an interaction with single-stranded DNA. J Biol Chem 280: 39693–39700. [DOI] [PubMed] [Google Scholar]
- Capaldo FN, Ramsey G, Barbour SD 1974. Analysis of the growth of recombination-deficient strains of Escherichia coli K-12. J Bacteriol 118: 242–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan KL, Palmai-Pallag T, Ying S, Hickson ID 2009. Replication stress induces sister-chromatid bridging at fragile site loci in mitosis. Nat Cell Biol 11: 753–760. [DOI] [PubMed] [Google Scholar]
- Courcelle J, Hanawalt PC 2003. RecA-dependent recovery of arrested DNA replication forks. Annu Rev Genet 37: 611–646. [DOI] [PubMed] [Google Scholar]
- Courcelle J, Carswell-Crumpton C, Hanawalt PC 1997. recF and recR are required for the resumption of replication at DNA replication forks in Escherichia coli. Proc Natl Acad Sci 94: 3714–3719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courcelle J, Donaldson JR, Chow KH, Courcelle CT 2003. DNA damage-induced replication fork regression and processing in Escherichia coli. Science 299: 1064–1067. [DOI] [PubMed] [Google Scholar]
- Courcelle CT, Belle JJ, Courcelle J 2005. Nucleotide excision repair or polymerase V-mediated lesion bypass can act to restore UV-arrested replication forks in Escherichia coli. J Bacteriol 187: 6953–6961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courcelle CT, Chow KH, Casey A, Courcelle J 2006. Nascent DNA processing by RecJ favors lesion repair over translesion synthesis at arrested replication forks in Escherichia coli. Proc Natl Acad Sci 103: 9154–9159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox MM 2007a. Regulation of bacterial RecA protein function. Crit Rev Biochem Mol Biol 42: 41–63. [DOI] [PubMed] [Google Scholar]
- Cox MM 2007b. Motoring along with the bacterial RecA protein. Nat Rev Mol Cell Biol 8: 127–138. [DOI] [PubMed] [Google Scholar]
- Cox MM, Goodman MF, Kreuzer KN, Sherratt DJ, Sandler SJ, Marians KJ 2000. The importance of repairing stalled replication forks. Nature 404: 37–41. [DOI] [PubMed] [Google Scholar]
- Davies MJ 2005. The oxidative environment and protein damage. Biochim Biophys Acta 1703: 93–109. [DOI] [PubMed] [Google Scholar]
- Duch A, Felipe-Abrio I, Barroso S, Yaakov G, Garcia-Rubio M, Aguilera A, de Nadal E, Posas F 2012. Coordinated control of replication and transcription by a SAPK protects genomic integrity. Nature 493: 116–119. [DOI] [PubMed] [Google Scholar]
- Edenberg HJ 1976. Inhibition of DNA replication by ultraviolet light. Biophys J 16: 849–860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elvers I, Johansson F, Groth P, Erixon K, Helleday T 2011. UV stalled replication forks restart by re-priming in human fibroblasts. Nucleic Acids Res 39: 7049–7057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fachinetti D, Bermejo R, Cocito A, Minardi S, Katou Y, Kanoh Y, Shirahige K, Azvolinsky A, Zakian VA, Foiani M 2010. Replication termination at eukaryotic chromosomes is mediated by Top2 and occurs at genomic loci containing pausing elements. Mol Cell 39: 595–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flores MJ, Bierne H, Ehrlich SD, Michel B 2001. Impairment of lagging strand synthesis triggers the formation of a RuvABC substrate at replication forks. EMBO J 20: 619–629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fricke WM, Brill SJ 2003. Slx1-Slx4 is a second structure-specific endonuclease functionally redundant with Sgs1-Top3. Genes Dev 17: 1768–1778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiwara Y, Tatsumi M 1976. Replicative bypass repair of ultraviolet damage to DNA of mammalian cells: Caffeine sensitive and caffeine resistant mechanisms. Mutat Res 37: 91–110. [DOI] [PubMed] [Google Scholar]
- Gottesman MM, Hicks ML, Gellert M 1973. Genetics and function of DNA ligase in Escherichia coli. J Mol Biol 77: 531–547. [DOI] [PubMed] [Google Scholar]
- Gruber M, Wellinger RE, Sogo JM 2000. Architecture of the replication fork stalled at the 3′ end of yeast ribosomal genes. Mol Cell Biol 20: 5777–5787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta MK, Guy CP, Yeeles JT, Atkinson J, Bell H, Lloyd RG, Marians KJ, McGlynn P 2013. Protein–DNA complexes are the primary sources of replication fork pausing in Escherichia coli. Proc Natl Acad Sci 110: 7252–7257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guy CP, Atkinson J, Gupta MK, Mahdi AA, Gwynn EJ, Rudolph CJ, Moon PB, van Knippenberg IC, Cadman CJ, Dillingham MS, et al. 2009. Rep provides a second motor at the replisome to promote duplication of protein-bound DNA. Mol Cell 36: 654–666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanada K, Budzowska M, Modesti M, Maas A, Wyman C, Essers J, Kanaar R 2006. The structure-specific endonuclease Mus81-Eme1 promotes conversion of interstrand DNA crosslinks into double-strands breaks. EMBO J 25: 4921–4932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanada K, Budzowska M, Davies SL, van Drunen E, Onizawa H, Beverloo HB, Maas A, Essers J, Hickson ID, Kanaar R 2007. The structure-specific endonuclease Mus81 contributes to replication restart by generating double-strand DNA breaks. Nat Struct Mol Biol 14: 1096–1104. [DOI] [PubMed] [Google Scholar]
- Hanawalt PC 1966. The U.V. sensitivity of bacteria: Its relation to the DNA replication cycle. Photochem Photobiol 5: 1–12. [PubMed] [Google Scholar]
- Hashimoto Y, Ray Chaudhuri A, Lopes M, Costanzo V 2010. Rad51 protects nascent DNA from Mre11-dependent degradation and promotes continuous DNA synthesis. Nat Struct Mol Biol 17: 1305–1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto Y, Puddu F, Costanzo V 2012. RAD51- and MRE11-dependent reassembly of uncoupled CMG helicase complex at collapsed replication forks. Nat Struct Mol Biol 19: 17–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heller RC, Marians KJ 2005a. Unwinding of the nascent lagging strand by Rep and PriA enables the direct restart of stalled replication forks. J Biol Chem 280: 34143–34151. [DOI] [PubMed] [Google Scholar]
- Heller RC, Marians KJ 2005b. The disposition of nascent strands at stalled replication forks dictates the pathway of replisome loading during restart. Mol Cell 17: 733–743. [DOI] [PubMed] [Google Scholar]
- Heller RC, Marians KJ 2006a. Replisome assembly and the direct restart of stalled replication forks. Nat Rev Mol Cell Biol 7: 932–943. [DOI] [PubMed] [Google Scholar]
- Heller RC, Marians KJ 2006b. Replication fork reactivation downstream of a blocked nascent leading strand. Nature 439: 557–562. [DOI] [PubMed] [Google Scholar]
- Hiasa H, Marians KJ 1994. Tus prevents overreplication of oriC plasmid DNA. J Biol Chem 269: 26959–26968. [PubMed] [Google Scholar]
- Higgins NP, Kato K, Strauss B 1976. A model for replication repair in mammalian cells. J Mol Biol 101: 417–425. [DOI] [PubMed] [Google Scholar]
- Hill TM, Marians KJ 1990. Escherichia coli Tus protein acts to arrest the progression of DNA replication forks in vitro. Proc Natl Acad Sci 87: 2481–2485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hishida T, Kubota Y, Carr AM, Iwasaki H 2009. RAD6-RAD18-RAD5-pathway-dependent tolerance to chronic low-dose ultraviolet light. Nature 457: 612–615. [DOI] [PubMed] [Google Scholar]
- Horiuchi T, Fujimura Y, Nishitani H, Kobayashi T, Hidaka M 1994. The DNA replication fork blocked at the Ter site may be an entrance for the RecBCD enzyme into duplex DNA. J Bacteriol 176: 4656–4663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hotchkiss RD 1974. Models of genetic recombination. Annu Rev Microbiol 28: 445–468. [DOI] [PubMed] [Google Scholar]
- Indiani C, Patel M, Goodman MF, O’Donnell ME 2013. RecA acts as a switch to regulate polymerase occupancy in a moving replication fork. Proc Natl Acad Sci 110: 5410–5415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadyrov FA, Drake JW 2004. UvsX recombinase and Dda helicase rescue stalled bacteriophage T4 DNA replication forks in vitro. J Biol Chem 279: 35735–35740. [DOI] [PubMed] [Google Scholar]
- Katou Y, Kanoh Y, Bando M, Noguchi H, Tanaka H, Ashikari T, Sugimoto K, Shirahige K 2003. S-phase checkpoint proteins Tof1 and Mrc1 form a stable replication-pausing complex. Nature 424: 1078–1083. [DOI] [PubMed] [Google Scholar]
- Kogoma T 1997. Stable DNA replication: Interplay between DNA replication, homologous recombination, and transcription. Microbiol Mol Biol Rev 61: 212–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouzminova EA, Kuzminov A 2008. Patterns of chromosomal fragmentation due to uracil-DNA incorporation reveal a novel mechanism of replication-dependent double-stranded breaks. Mol Microbiol 68: 202–215. [DOI] [PubMed] [Google Scholar]
- Krejci L, Van Komen S, Li Y, Villemain J, Reddy MS, Klein H, Ellenberger T, Sung P 2003. DNA helicase Srs2 disrupts the Rad51 presynaptic filament. Nature 423: 305–309. [DOI] [PubMed] [Google Scholar]
- Krejci L, Altmannova V, Spirek M, Zhao X 2012. Homologous recombination and its regulation. Nucleic Acids Res 40: 5795–5818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreuzer KN, Brister JR 2010. Initiation of bacteriophage T4 DNA replication and replication fork dynamics: A review in the Virology Journal series on bacteriophage T4 and its relatives. Virol J 7: 358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuzminov A 2001. Single-strand interruptions in replicating chromosomes cause double-strand breaks. Proc Natl Acad Sci 98: 8241–8246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambert S, Watson A, Sheedy DM, Martin B, Carr AM 2005. Gross chromosomal rearrangements and elevated recombination at an inducible site-specific replication fork barrier. Cell 121: 689–702. [DOI] [PubMed] [Google Scholar]
- Lee EH, Kornberg A 1991. Replication deficiencies in priA mutants of Escherichia coli lacking the primosomal replication n′ protein. Proc Natl Acad Sci 88: 3029–3032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehmann AR 1972. Postreplication repair of DNA in ultraviolet-irradiated mammalian cells. J Mol Biol 66: 319–337. [DOI] [PubMed] [Google Scholar]
- Letessier A, Millot GA, Koundrioukoff S, Lachages AM, Vogt N, Hansen RS, Malfoy B, Brison O, Debatisse M 2011. Cell-type-specific replication initiation programs set fragility of the FRA3B fragile site. Nature 470: 120–123. [DOI] [PubMed] [Google Scholar]
- Liu J, Morrical SW 2010. Assembly and dynamics of the bacteriophage T4 homologous recombination machinery. Virol J 7: 357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Xu L, Sandler SJ, Marians KJ 1999. Replication fork assembly at recombination intermediates is required for bacterial growth. Proc Natl Acad Sci 96: 3552–3555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu P, Erez A, Nagamani SC, Dhar SU, Kolodziejska KE, Dharmadhikari AV, Cooper ML, Wiszniewska J, Zhang F, Withers MA, et al. 2011. Chromosome catastrophes involve replication mechanisms generating complex genomic rearrangements. Cell 146: 889–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llorente B, Smith CE, Symington LS 2008. Break-induced replication: What is it and what is it for? Cell Cycle 7: 859–864. [DOI] [PubMed] [Google Scholar]
- Long DT, Kreuzer KN 2008. Regression supports two mechanisms of fork processing in phage T4. Proc Natl Acad Sci 105: 6852–6857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long JE, Renzette N, Sandler SJ 2009. Suppression of constitutive SOS expression by recA4162 (I298 V) and recA4164 (L126 V) requires UvrD and RecX in Escherichia coli K-12. Mol Microbiol 73: 226–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopes M, Cotta-Ramusino C, Pellicioli A, Liberi G, Plevani P, Muzi-Falconi M, Newlon CS, Foiani M 2001. The DNA replication checkpoint response stabilizes stalled replication forks. Nature 412: 557–561. [DOI] [PubMed] [Google Scholar]
- Lopes M, Foiani M, Sogo JM 2006. Multiple mechanisms control chromosome integrity after replication fork uncoupling and restart at irreparable UV lesions. Mol Cell 21: 15–27. [DOI] [PubMed] [Google Scholar]
- Lopper M, Boonsombat R, Sandler SJ, Keck JL 2007. A hand-off mechanism for primosome assembly in replication restart. Mol Cell 26: 781–793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louarn JM, Louarn J, Francois V, Patte J 1991. Analysis and possible role of hyperrecombination in the termination region of the Escherichia coli chromosome. J Bacteriol 173: 5097–5104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luder A, Mosig G 1982. Two alternative mechanisms for initiation of DNA replication forks in bacteriophage T4: Priming by RNA polymerase and by recombination. Proc Natl Acad Sci 79: 1101–1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machwe A, Xiao L, Groden J, Orren DK 2006. The Werner and Bloom syndrome proteins catalyze regression of a model replication fork. Biochemistry 45: 13939–13946. [DOI] [PubMed] [Google Scholar]
- Manosas M, Perumal SK, Croquette V, Benkovic SJ 2012. Direct observation of stalled fork restart via fork regression in the T4 replication system. Science 338: 1217–1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marians KJ, Hiasa H, Kim DR, McHenry CS 1998. Role of the core DNA polymerase III subunits at the replication fork: α is the only subunit required for processive replication. J Biol Chem 273: 2452–2457. [DOI] [PubMed] [Google Scholar]
- Matos J, Blanco MG, Maslen S, Skehel JM, West SC 2011. Regulatory control of the resolution of DNA recombination intermediates during meiosis and mitosis. Cell 147: 158–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGlynn P 2013. Helicases at the replication fork. Adv Exp Med Biol 767: 97–121. [DOI] [PubMed] [Google Scholar]
- McGlynn P, Guy CP 2008. Replication forks blocked by protein-DNA complexes have limited stability in vitro. J Mol Biol 381: 249–255. [DOI] [PubMed] [Google Scholar]
- McGlynn P, Lloyd RG 2000. Modulation of RNA polymerase by (p)ppGpp reveals a RecG-dependent mechanism for replication fork progression. Cell 101: 35–45. [DOI] [PubMed] [Google Scholar]
- McGlynn P, Lloyd RG 2001. Action of RuvAB at replication fork structures. J Biol Chem 276: 41938–41944. [DOI] [PubMed] [Google Scholar]
- McGlynn P, Lloyd RG 2002. Recombinational repair and restart of damaged replication forks. Nat Rev Mol Cell Biol 3: 859–870. [DOI] [PubMed] [Google Scholar]
- McGlynn P, Al-Deib AA, Liu J, Marians KJ, Lloyd RG 1997. The DNA replication protein PriA and the recombination protein RecG bind D-loops. J Mol Biol 270: 212–221. [DOI] [PubMed] [Google Scholar]
- McGlynn P, Lloyd RG, Marians KJ 2001. Formation of Holliday junctions by regression of nascent DNA in intermediates containing stalled replication forks: RecG stimulates regression even when the DNA is negatively supercoiled. Proc Natl Acad Sci 98: 8235–8240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGlynn P, Savery NJ, Dillingham MS 2012. The conflict between DNA replication and transcription. Mol Microbiol 85: 12–20. [DOI] [PubMed] [Google Scholar]
- McInerney P, O’Donnell M 2007. Replisome fate upon encountering a leading strand block and clearance from DNA by recombination proteins. J Biol Chem 282: 25903–25916. [DOI] [PubMed] [Google Scholar]
- *.Mehta A, Haber JE 2014. Sources of DNA double-strand breaks and models for recombinational DNA repair. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a016428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michel B, Grompone G, Flores MJ, Bidnenko V 2004. Multiple pathways process stalled replication forks. Proc Natl Acad Sci 101: 12783–12788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuno K, Miyabe I, Schalbetter SA, Carr AM, Murray JM 2012. Recombination-restarted replication makes inverted chromosome fusions at inverted repeats. Nature 493: 246–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore T, McGlynn P, Ngo HP, Sharples GJ, Lloyd RG 2003. The RdgC protein of Escherichia coli binds DNA and counters a toxic effect of RecFOR in strains lacking the replication restart protein PriA. EMBO J 22: 735–745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moriel-Carretero M, Aguilera A 2010. A postincision-deficient TFIIH causes replication fork breakage and uncovers alternative Rad51- or Pol32-mediated restart mechanisms. Mol Cell 37: 690–701. [DOI] [PubMed] [Google Scholar]
- Morimatsu K, Kowalczykowski SC 2003. RecFOR proteins load RecA protein onto gapped DNA to accelerate DNA strand exchange: A universal step of recombinational repair. Mol Cell 11: 1337–1347. [DOI] [PubMed] [Google Scholar]
- Mueser TC, Jones CE, Nossal NG, Hyde CC 2000. Bacteriophage T4 gene 59 helicase assembly protein binds replication fork DNA. The 1.45 Å resolution crystal structure reveals a novel α-helical two-domain fold. J Mol Biol 296: 597–612. [DOI] [PubMed] [Google Scholar]
- Murray JM, Carr AM 2008. Smc5/6: A link between DNA repair and unidirectional replication? Nat Rev Mol Cell Biol 9: 177–182. [DOI] [PubMed] [Google Scholar]
- Nurse P, Zavitz KH, Marians KJ 1991. Inactivation of the Escherichia coli priA DNA replication protein induces the SOS response. J Bacteriol 173: 6686–6693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osman F, Whitby MC 2007. Exploring the roles of Mus81-Eme1/Mms4 at perturbed replication forks. DNA Repair (Amst) 6: 1004–1017. [DOI] [PubMed] [Google Scholar]
- Ozeri-Galai E, Lebofsky R, Rahat A, Bester AC, Bensimon A, Kerem B 2011. Failure of origin activation in response to fork stalling leads to chromosomal instability at fragile sites. Mol Cell 43: 122–131. [DOI] [PubMed] [Google Scholar]
- Pagès V, Fuchs RP 2003. Uncoupling of leading- and lagging-strand DNA replication during lesion bypass in vivo. Science 300: 1300–1303. [DOI] [PubMed] [Google Scholar]
- Payne BT, van Knippenberg IC, Bell H, Filipe SR, Sherratt DJ, McGlynn P 2006. Replication fork blockage by transcription factor-DNA complexes in Escherichia coli. Nucleic Acids Res 34: 5194–5202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petermann E, Helleday T 2010. Pathways of mammalian replication fork restart. Nat Rev Mol Cell Biol 11: 683–687. [DOI] [PubMed] [Google Scholar]
- Petermann E, Orta ML, Issaeva N, Schultz N, Helleday T 2010. Hydroxyurea-stalled replication forks become progressively inactivated and require two different RAD51-mediated pathways for restart and repair. Mol Cell 37: 492–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pohlhaus JR, Kreuzer KN 2006. Formation and processing of stalled replication forks—Utility of two-dimensional agarose gels. Methods Enzymol 409: 477–493. [DOI] [PubMed] [Google Scholar]
- Pomerantz RT, O’Donnell M 2010. Direct restart of a replication fork stalled by a head-on RNA polymerase. Science 327: 590–592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pommier Y 2013. Drugging topoisomerases: Lessons and challenges. ACS Chem Biol 8: 82–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pommier Y, Pourquier P, Fan Y, Strumberg D 1998. Mechanism of action of eukaryotic DNA topoisomerase I and drugs targeted to the enzyme. Biochim Biophys Acta 1400: 83–105. [DOI] [PubMed] [Google Scholar]
- Postow L, Ullsperger C, Keller RW, Bustamante C, Vologodskii AV, Cozzarelli NR 2001. Positive torsional strain causes the formation of a four-way junction at replication forks. J Biol Chem 276: 2790–2796. [DOI] [PubMed] [Google Scholar]
- Ralf C, Hickson ID, Wu L 2006. The Bloom’s syndrome helicase can promote the regression of a model replication fork. J Biol Chem 281: 22839–22846. [DOI] [PubMed] [Google Scholar]
- Raschle M, Knipsheer P, Enoiu M, Angelov T, Sun J, Griffith JD, Ellenberger TE, Scharer OD, Walter JC 2008. Mechanism of replication-coupled DNA interstrand crosslink repair. Cell 134: 969–980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rass U 2013. Resolving branched DNA intermediates with structure-specific nucleases during replication in eukaryotes. Chromosoma 122: 499–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robu ME, Inman RB, Cox MM 2001. RecA protein promotes the regression of stalled replication forks in vitro. Proc Natl Acad Sci 98: 8211–8218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudolph CJ, Upton AL, Lloyd RG 2007. Replication fork stalling and cell cycle arrest in UV-irradiated Escherichia coli. Genes Dev 21: 668–681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudolph CJ, Upton AL, Lloyd RG 2008. Maintaining replication fork integrity in UV-irradiated Escherichia coli cells. DNA Repair (Amst) 7: 1589–1602. [DOI] [PubMed] [Google Scholar]
- Rudolph CJ, Upton AL, Harris L, Lloyd RG 2009a. Pathological replication in cells lacking RecG DNA translocase. Mol Microbiol 73: 352–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudolph CJ, Upton AL, Lloyd RG 2009b. Replication fork collisions cause pathological chromosomal amplification in cells lacking RecG DNA translocase. Mol Microbiol 74: 940–955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudolph CJ, Mahdi AA, Upton AL, Lloyd RG 2010. RecG protein and single-strand DNA exonucleases avoid cell lethality associated with PriA helicase activity in Escherichia coli. Genetics 186: 473–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudolph CJ, Upton AL, Stockum A, Nieduszynski CA, Lloyd RG 2013. Avoiding chromosome pathology when replication forks collide. Nature 500: 608–611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rupp WD, Howard-Flanders P 1968. Discontinuities in the DNA synthesized in an excision-defective strain of Escherichia coli following ultraviolet irradiation. J Mol Biol 31: 291–304. [DOI] [PubMed] [Google Scholar]
- Rupp WD, Wilde CE III, Reno DL, Howard-Flanders P 1971. Exchanges between DNA strands in ultraviolet-irradiated Escherichia coli. J Mol Biol 61: 25–44. [DOI] [PubMed] [Google Scholar]
- Sandler SJ 2000. Multiple genetic pathways for restarting DNA replication forks in Escherichia coli K-12. Genetics 155: 487–497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- San Filippo J, Sung P, Klein H 2008. Mechanism of eukaryotic homologous recombination. Annu Rev Biochem 77: 229–257. [DOI] [PubMed] [Google Scholar]
- Schlacher K, Cox MM, Woodgate R, Goodman MF 2006. RecA acts in trans to allow replication of damaged DNA by DNA polymerase V. Nature 442: 883–887. [DOI] [PubMed] [Google Scholar]
- Schlacher K, Christ N, Siaud N, Egashira A, Wu H, Jasin M 2011. Double-strand break repair-independent role for BRCA2 in blocking stalled replication fork degradation by MRE11. Cell 145: 529–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz EK, Heyer WD 2011. Processing of joint molecule intermediates by structure-selective endonucleases during homologous recombination in eukaryotes. Chromosoma 120: 109–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seigneur M, Bidnenko V, Ehrlich SD, Michel B 1998. RuvAB acts at arrested replication forks. Cell 95: 419–430. [DOI] [PubMed] [Google Scholar]
- Seigneur M, Ehrlich SD, Michel B 2000. RuvABC-dependent double-strand breaks in dnaBts mutants require recA. Mol Microbiol 38: 565–574. [DOI] [PubMed] [Google Scholar]
- Setlow RB, Swenson PA, Carrier WL 1963. Thymine dimers and inhibition of DNA synthesis by ultraviolet irradiation of cells. Science 142: 1464–1466. [DOI] [PubMed] [Google Scholar]
- Skalka A 1974. A replicator's view of recombination (and repair). In Mechanisms in recombination (ed. Grell RF), pp. 421–432 Plenum, New York. [Google Scholar]
- Smith KC 2004. Recombinational DNA repair: The ignored repair systems. Bioessays 26: 1322–1326. [DOI] [PubMed] [Google Scholar]
- Sogo JM, Lopes M, Foiani M 2002. Fork reversal and ssDNA accumulation at stalled replication forks owing to checkpoint defects. Science 297: 599–602. [DOI] [PubMed] [Google Scholar]
- St Charles J, Petes TD 2013. High-resolution mapping of spontaneous mitotic recombination hotspots on the 1.1 Mb arm of yeast chromosome IV. PLoS Genet 9: e1003434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svoboda DL, Vos JM 1995. Differential replication of a single, UV-induced lesion in the leading or lagging strand by a human cell extract: Fork uncoupling or gap formation. Proc Natl Acad Sci 92: 11975–11979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Symington LS, Gautier J 2011. Double-strand break end resection and repair pathway choice. Annu Rev Genet 45: 247–271. [DOI] [PubMed] [Google Scholar]
- Szyjka SJ, Viggiani CJ, Aparicio OM 2005. Mrc1 is required for normal progression of replication forks throughout chromatin in S. cerevisiae. Mol Cell 19: 691–697. [DOI] [PubMed] [Google Scholar]
- Tercero JA, Diffley JF 2001. Regulation of DNA replication fork progression through damaged DNA by the Mec1/Rad53 checkpoint. Nature 412: 553–557. [DOI] [PubMed] [Google Scholar]
- Tittel-Elmer M, Alabert C, Pasero P, Cobb JA 2009. The MRX complex stabilizes the replisome independently of the S phase checkpoint during replication stress. EMBO J 28: 1142–1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trautinger BW, Jaktaji RP, Rusakova E, Lloyd RG 2005. RNA polymerase modulators and DNA repair activities resolve conflicts between DNA replication and transcription. Mol Cell 19: 247–258. [DOI] [PubMed] [Google Scholar]
- Vaisica JA, Baryshnikova A, Costanzo M, Boone C, Brown GW 2011. Mms1 and Mms22 stabilize the replisome during replication stress. Mol Biol Cell 22: 2396–2408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veaute X, Jeusset J, Soustelle C, Kowalczykowski SC, Le Cam E, Fabre F 2003. The Srs2 helicase prevents recombination by disrupting Rad51 nucleoprotein filaments. Nature 423: 309–312. [DOI] [PubMed] [Google Scholar]
- Veaute X, Delmas S, Selva M, Jeusset J, Le Cam E, Matic I, Fabre F, Petit MA 2005. UvrD helicase, unlike Rep helicase, dismantles RecA nucleoprotein filaments in Escherichia coli. EMBO J 24: 180–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webb MR, Plank JL, Long DT, Hsieh TS, Kreuzer KN 2007. The phage T4 protein UvsW drives Holliday junction branch migration. J Biol Chem 282: 34401–34411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L, Marians KJ 2003. PriA mediates DNA replication pathway choice at recombination intermediates. Mol Cell 11: 817–826. [DOI] [PubMed] [Google Scholar]
- Yeeles JT, Marians KJ 2011. The Escherichia coli replisome is inherently DNA damage tolerant. Science 334: 235–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeeles JT, Marians KJ 2013. Dynamics of leading-strand lesion skipping by the replisome. Mol Cell 52: 855–865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon D, Wang Y, Stapleford K, Wiesmuller L, Chen J 2004. p53 inhibits strand exchange and replication fork regression promoted by human Rad51. J Mol Biol 336: 639–654. [DOI] [PubMed] [Google Scholar]