Abstract
Homologous recombination (HR) is a major mechanism for eliminating DNA double-strand breaks from chromosomes. In this process, the break termini are resected nucleolytically to form 3′ ssDNA (single-strand DNA) overhangs. A recombinase (i.e., a protein that catalyzes homologous DNA pairing and strand exchange) assembles onto the ssDNA and promotes pairing with a homologous duplex. DNA synthesis then initiates from the 3′ end of the invading strand, and the extended DNA joint is resolved via one of several pathways to restore the integrity of the injured chromosome. It is crucial that HR be carefully orchestrated because spurious events can create cytotoxic intermediates or cause genomic rearrangements and loss of gene heterozygosity, which can lead to cell death or contribute to the development of cancer. In this review, we will discuss how DNA motor proteins regulate HR via a dynamic balance of the recombination-promoting and -attenuating activities that they possess.
DNA motor proteins (e.g., helicases and translocases) regulate homologous recombination via a dynamic balance of recombination-promoting and -attenuating activities.
MECHANISMS OF DNA DOUBLE-STRAND BREAK REPAIR OVERVIEW
DNA double-strand breaks (DSBs) present a major challenge to genome maintenance because, if not handled properly, they can cause gross chromosome rearrangements. Indeed, defects in DSB repair can cause a predisposition to the development of cancer. All kingdoms of life possess two pathways by which DSBs are eliminated from chromosomes—namely, nonhomologous end joining (NHEJ) and homologous recombination (HR). NHEJ involves alignment of DNA ends, minor processing of the ends to rid them of chemical damage, followed by DNA gap filling and end religation (Lieber 2010). NHEJ at “clean” breaks (i.e., those that harbor compatible overhangs and religatable 5′ phosphate and 3′ hydroxyl termini) is usually accurate. Joining of “dirty” breaks with chemically damaged ends that prevent ligation, such as those generated by radiation, requires processing that can result in a loss of genetic information. NHEJ can also lead to chromosome translocations if DNA ends from two different chromosomes are joined. HR, on the other hand, is mostly accurate and mechanistically more complex than NHEJ (see Mehta and Haber 2014). HR involves the engagement of a homologous DNA sequence, usually the sister chromatid but sometimes the homologous chromosome, as a template to guide restorative repair (San Filippo et al. 2008; Mimitou and Symington 2009). The HR reaction is subject to multiple layers of regulatory control that affect the efficiency of the process or the recombinant product types. Here, we will discuss the roles of DNA helicases and translocases in the regulation of the homologous DNA pairing reaction that yields DNA joints between the recombining DNA molecules.
HOMOLOGOUS RECOMBINATION OVERVIEW
After DSB formation, the initial step of HR repair is nucleolytic resection of the 5′ strands from both break ends (Mimitou and Symington 2009; Symington 2014). This serves to generate a pair of 3′ ssDNA tails for the recruitment of the recombinase protein Rad51 and associated ancillary factors. Moreover, the resection reaction helps commit the DNA break to repair by HR (Daley et al. 2005). Based on genetic studies in Saccharomyces cerevisiae, we know that at least three nucleases participate in resection: the endonuclease/exonuclease Mre11, the 5′ to 3′ exonuclease Exo1, and the helicase/endonuclease Dna2 (Gravel et al. 2008; Mimitou and Symington 2008; Zhu et al. 2008). Dna2 functions with the Sgs1 helicase, which is the S. cerevisiae ortholog of human BLM (see below) (Zhu et al. 2008).
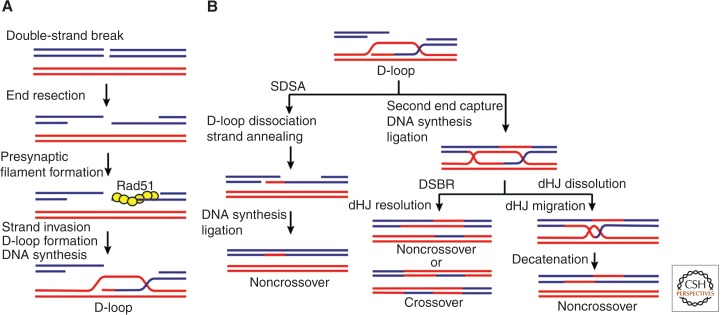
At first, the 3′ tails generated via DNA end resection are bound by the abundant, evolutionarily conserved single-stranded DNA-binding protein RPA (Raderschall et al. 1999). RPA is subsequently exchanged for the Rad51 recombinase, in a process that is facilitated by several recombination mediator proteins, including the Rad52 protein in S. cerevisiae and the tumor suppressor BRCA2 in humans (Sugiyama and Kowalczykowski 2002; Jensen et al. 2010; Liu et al. 2010). The form of Rad51-ssDNA complex that is capable of homologous pairing is a right-handed helical polymer of the recombinase on the ssDNA, with the DNA being held in an extended conformation (Yu et al. 2001). This Rad51-ssDNA complex, commonly referred to as the “presynaptic” filament, samples the incoming duplex DNA for homology (Fig. 1A) (Barzel and Kupiec 2008). On the location of homology in the duplex DNA partner, invasion of the duplex by the 3′-ended tail occurs, yielding a DNA joint known as the displacement loop, or D-loop (San Filippo et al. 2008). Next, the invading strand serves as the primer for DNA synthesis, to result in the extension of the D-loop structure (Fig. 1A).
Figure 1.
DNA double-strand break repair (DSBR) by homologous recombination. (A) Nucleolytic resection of the DNA ends produces 3′ ssDNA overhangs, one of which is engaged by the Rad51 recombinase to form a presynaptic filament capable of locating and invading a homologous sequence. The free end in the resulting D-loop then primes DNA synthesis. (B) The extended D-loop can be resolved by one of several pathways. Synthesis-dependent strand annealing (SDSA), being the most prevalent noncrossover mechanism in mitotic cells, entails D-loop dissociation by a helicase. If the second DNA end is captured, then a double Holliday junction (dHJ) will form. Resolution of the dHJ by a HJ resolvase, as in DSBR, yields either noncrossover or crossover recombinants. Alternatively, the dHJ can be dissolved via convergent migration of the two DNA junctions and DNA decatenation to yield noncrossover products.
As shown in Figure 1B, the extended D-loop can be resolved by one of three mechanistically distinct means that yield different products. In the synthesis-dependent strand annealing (SDSA) pathway, the extended invading strand dissociates from the D-loop and anneals with the complementary 3′ DNA tail of the other break end, followed by gap-filling DNA synthesis and ligation (see Morrical 2014). The SDSA mode of HR produces noncrossover recombinants only. In some instances, the homologous DNA pairing process proceeds to engage the second resected end without the initial invading strand ever dissociating from the D-loop. In this situation, a DNA intermediate that harbors two Holliday junctions is made (Fig. 1B). The double Holliday junction (dHJ) can be resolved by one of several DNA structure–specific endonucleases known as HJ resolvases, with the potential of yielding chromosome arm crossovers (Schwartz and Heyer 2011; Wyatt and West 2014). The resolvase-dependent HR pathway is referred to as the double-strand break repair (DSBR) pathway. Alternatively, the dHJ can be resolved by a process termed dissolution, in which a dedicated helicase-topoisomerase complex disentangles the dHJ (Raynard et al. 2006; Wu et al. 2006; Bizard and Hickson 2014). The dHJ dissolution reaction generates only noncrossover products. It should be noted that crossovers are necessary for tying the homologous chromosome pairs until their segregation in the first meiotic division (see Lam and Keeney 2014; Zickler and Kleckner 2014). However, because of the inherent danger of generating chromosome translocations, the crossover HR pathway is actively suppressed in mitotic DSB repair (see below).
As we will discuss in this review, a number of conserved DNA helicases and translocases have been shown to regulate the stability of the Rad51 presynaptic filament and the DNA strand invasion step of the HR reaction. They also mediate dissociation of the invading strand from the D-loop to avoid crossover formation and to help catalyze dHJ dissolution. We will review what is known about these DNA motor proteins in terms of their genetic characteristics, accessory factors, and mechanisms of action. Table 1 lists the DNA motor proteins addressed in this work. Elsewhere in this collection the readers will find informative articles dedicated to understanding the mechanisms of DNA end resection (Symington 2014), the structure and function of the general recombinases Rad51 and Dmc1 and their orthologs in prokaryotes (Morrical 2014), and the ancillary factors that function with these recombinases (Zelensky et al. 2014).
Table 1.
Functional attributes of homologous recombination regulatory helicase/translocase proteins
| Protein | Superfamily, helicase/translocase | Biochemical functions | Notable features |
|---|---|---|---|
| Sgs1 (Saccharomyces cerevisiae) | SF2, 3′ to 5′ helicase | 5′ DNA end resection with Dna2 Component of the Sgs1-Top3-Rmi1 (STR) complex dHJ dissolution with Top3-Rmi1 |
Mre11 complex, Top3-Rmi1, and RPA are required for robust 5′ end resection Helicase activity of Sgs1 is required for end resection and dHJ dissolution |
| BLM (Homo sapiens) | SF2, 3′ to 5′ helicase | 5′ DNA end resection with Dna2 Stimulates activity of EXO1 Complex formation with TOPO IIIα-RMI1-RMI2 (BTR complex) dHJ dissolution with TOPO IIIα-RMI1-RMI2 |
MRE11 complex and RPA stimulate end resection by BLM-DNA2 RPA stimulates dHJ dissolution via interaction with RMI1 |
| Srs2 (S. cerevisiae) | SF1, 3′ to 5′ helicase | Antirecombinase function via disassembly of the Rad51-ssDNA filament | Interacts with Rad51, PCNA, and SUMO-PCNA Rad52 and Rad55-Rad57 counteract the antirecombinase activity of Srs2 |
| RECQ5 (H. sapiens) | SF2, 3′ to 5′ helicase | Antirecombinase function via disassembly of the RAD51-ssDNA filament | Interacts with RAD51 and PCNA Complex formation with RNA polymerase II |
| Mph1 (S. cerevisiae) Fml1 (Schizosaccharomyces pombe) FANCM (H. sapiens) |
SF2, 3′ to 5′ helicase (Mph1)/DNA translocase | Mediates D-loop disruption Processes the DNA replication fork and Holliday junction |
FANCM forms a complex with MHF1-MHF2 and FAAP24 |
| RTEL (H. sapiens) | SF2, 5′ to 3′ helicase | Dissociates the D-loop and T-loop | Interacts with RAD51 and PCNA Unable to disassemble the RAD51-ssDNA filament |
| Rad54, Rdh54 (S. cerevisiae) RAD54 (H. sapiens) |
SF2, DNA translocase | Stimulates the Rad51 and Dmc1- mediated D-loop formation Induces topological changes and transient separation of DNA strands in dsDNA Removal of Rad51 from dsDNA Chromatin remodeling |
Members of the Swi2/Snf2 family Interact with Rad51 and Dmc1 |
dHJ, double Holliday junction; RPA, replication protein A; PCNA, proliferating cell nuclear antigen.
REGULATION OF DNA PAIRING OR RECOMBINATION OUTCOME BY DNA HELICASES
The Multifaceted Roles of the Sgs1 and BLM Helicases in HR Mediation and Regulation
Functions in Mitotic HR
The S. cerevisiae Sgs1 protein and its human ortholog BLM belong to the RecQ family of DNA helicases, named after the founding member of this protein family, Escherichia coli RecQ (Ellis et al. 1995; Watt et al. 1996). These proteins share a conserved central region of about 400 residues that contains the helicase motifs, and they all translocate on ssDNA in the 3′ to 5′ direction (Gray et al. 1997; Karow et al. 1997; Shen et al. 1998). Mutations in BLM lead to the cancer-prone disease Bloom syndrome (BS), reflective of the critical role of BLM in the maintenance of genome stability. Whereas Sgs1 is the sole RecQ helicase in S. cerevisiae that has been characterized thus far, human cells possess four other such molecules—namely, RECQ1, WRN, RECQ4, and RECQ5, the last of which also functions in recombination regulation (see sections below). The human RecQ helicases and their orthologs in other organisms all fulfill important roles in DNA replication and repair. Comprehensive recent reviews of the RecQ helicases and their biological functions are available (Chu and Hickson 2009; Daley et al. 2013).
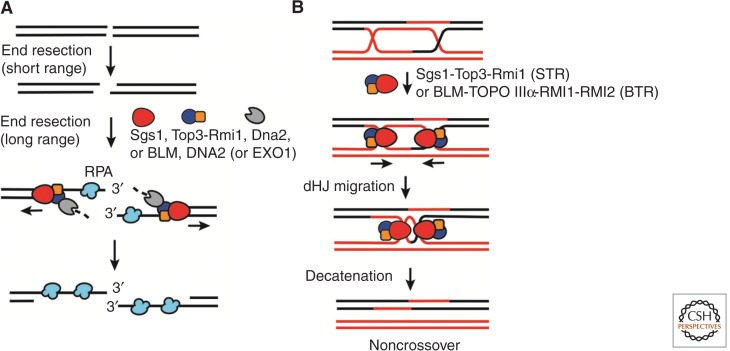
The SGS1 (suppressor of slow growth 1) gene was isolated in a screen for suppressors of the slow growth and genome instability phenotypes of cells lacking TOP3, which codes for a conserved type Ia topoisomerase that physically interacts with Sgs1 protein (Gangloff et al. 1994). The Sgs1-Top3 complex has a partner called Rmi1 (Chang et al. 2005). Likewise, the BLM protein is associated with the orthologs of Top3 and Rmi1—namely, TOPO IIIα and RMI1 (Yin et al. 2005). The BLM complex also harbors a fourth component RMI2 that is absent in the yeast Sgs1 complex (Xu et al. 2008). Extensive genetic analyses have implicated the Sgs1-Top3-Rmi1 (STR) and the BLM-TOPO IIIα-RMI1-RMI2 (BTR) complexes in two distinct steps of the HR reaction. An early role of the Sgs1 protein in the 5′ end resection of DSBs in conjunction with the nuclease Dna2 was revealed in genetic studies performed by two different groups (Mimitou and Symington 2008; Zhu et al. 2008). In biochemical reconstitution experiments, the Sgs1 helicase activity was shown to separate DNA strands, generating 5′ ssDNA tails for incision by Dna2 (Cejka et al. 2010; Niu et al. 2010). Maximal resection activity is dependent on Rmi1-Top3, although, in this regard, the topoisomerase activity of the latter proved to be dispensable both in vitro and in cells (Fig. 2A) (Niu et al. 2010). More limited cytological and genetic analyses have also implicated the BLM helicase in DNA resection in human cells (Gravel et al. 2008). Importantly, biochemical reconstitution has provided direct evidence that BLM functions with DNA2 in a manner analogous to their S. cerevisiae counterparts in DNA end resection (Nimonkar et al. 2011). Interestingly, BLM also enhances the activity of the 5′ to 3′ exonuclease of EXO1, which has been implicated in DNA end resection as well (Fig. 2A) (Nimonkar et al. 2011). This functional relationship is not conserved in S. cerevisiae, in that Sgs1 does not seem to affect the activity of Exo1 (Niu et al. 2010; Cannavo et al. 2013).
Figure 2.
Functions of Sgs1 and BLM in homologous recombination. (A) Sgs1-Top3-Rmi1 (STR), Dna2, and RPA in S. cerevisiae or BLM-DNA2-RPA and BLM-EXO1-RPA in humans perform long-range resection and produce 3′ ssDNA overhangs. Note that the initial, short-range resection is mediated by a protein complex that harbors the conserved Mre11 nuclease (not shown). (B) The STR or BLM-TOPO IIIa-RMI1-RMI2 (BTR) complex dissolves the double Holliday junction (dHJ) to produce noncrossover products.
A hallmark of BLM-deficient cells, such as those derived from BS patients, is a highly elevated level of sister chromatid exchanges, which very likely stems from HR events of the crossover type triggered by replication fork injury or collapse (Bartram et al. 1976). Importantly, genetic ablation of any of the components of the BTR complex yields the same phenotype and in an apparent epistatic fashion to BLM deficiency (Wu and Hickson 2003; Yin et al. 2005). Indeed, S. cerevisiae mutant cells lacking SGS1, TOP3, or RMI1 generate abnormally high levels of crossover recombinants during HR-mediated DSB repair (Ira et al. 2003). Taken together, the aforementioned genetic observations point to a key role of the BTR and STR complexes in regulating HR in favor of the formation of noncrossovers. Such a regulatory role would be consistent with a function of these protein ensembles in the prevention of deleterious chromosome arm translocations associated with crossover HR events.
What then is the biochemical function of the STR and BTR complexes in the promotion of noncrossover formation? In a seminal study, Wu and Hickson provided compelling biochemical evidence that BLM together with TOPO IIIα untangles the dHJ intermediate that arises during some HR events to form noncrossover recombinants exclusively (Fig. 2B) (Wu and Hickson 2003). This reaction, termed dHJ dissolution, is the subject of Bizard and Hickson (2014).
Role of Sgs1 in Meiotic HR
In meiosis, HR events triggered by programmed DSBs made by the Spo11 protein complex are quite often channeled into the DSBR pathway to yield crossovers (Fig. 1B; Lam and Keeney 2014), which provide a stable linkage between homologous chromosomes to facilitate their disjunction in the first meiotic division. Because the STR complex is expressed in meiotic cells, how is its dHJ dissolution activity constrained to allow sufficient crossovers to occur? Genetic analyses have provided evidence that STR is negatively regulated by a group of meiosis-specific proteins collectively known as ZMM (Borner et al. 2004; Jessop et al. 2006). Interestingly, sgs1 mutants show an improper distribution of crossovers and form complex joint molecules involving multiple chromatids, indicating that Sgs1 plays a crucial role in meiotic HR (Oh et al. 2007). Whether or not this meiotic HR regulatory function of Sgs1 involves dHJ dissolution remains to be determined.
Antirecombinase Srs2 and Its Role in HR Restriction and SDSA Promotion
The S. cerevisiae SRS2 gene encodes a 3′ to 5′ DNA helicase of the SF1 superfamily, and it serves important roles in HR restriction and the promotion of the SDSA pathway of D-loop resolution (Marini and Krejci 2010; Karpenshif and Bernstein 2012). The HR regulatory function of this helicase was first recognized in a screen for mutants that suppress the DNA damage sensitivity phenotype of mutants of RAD6, whose product is an E2 ubiquitin conjugating enzyme needed for postreplicative DNA repair, hence the name SRS2 (suppressor of RAD Six2) (Lawrence and Christensen 1979). Specifically, mutations in SRS2 greatly alleviate the sensitivity of rad6 or rad18 cells (RAD18 codes for an E3 ligase that forms a complex with Rad6) to ultraviolet light and other DNA-damaging agents (Lawrence and Christensen 1979). Further analysis revealed that rad6 or rad18 suppression by srs2 mutations requires a functional HR machinery, and that the genetic ablation of SRS2 in an otherwise wild-type background leads to a hyperrecombination phenotype and an excess of crossover recombinants during DSB repair by HR (Schiestl et al. 1990; Rong et al. 1991; Ira et al. 2003). Altogether, these findings suggested that Srs2 restricts the activity of the HR machinery and promotes the resolution of the D-loop intermediate via SDSA to suppress crossover formation.
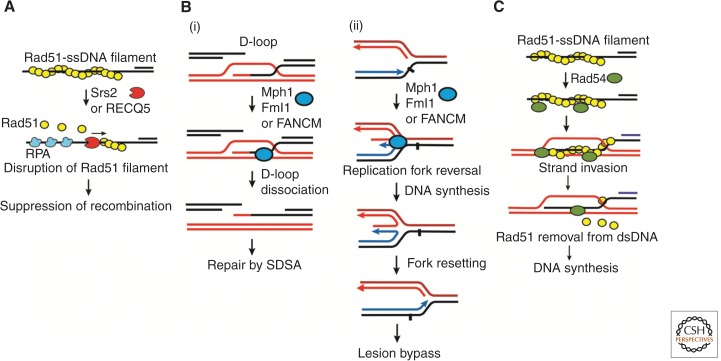
The biochemical function of Srs2 with regard to HR restriction was revealed in studies performed by two groups (Krejci et al. 2003; Veaute et al. 2003). At the expense of ATP hydrolysis, the motor activity of Srs2 displaces Rad51 molecules from ssDNA to disassemble the Rad51 presynaptic filament (Fig. 3A). This function of Srs2 is enhanced by the heterotrimeric ssDNA-binding factor RPA, which prevents the renucleation of Rad51 onto ssDNA. The Srs2-Rad51 interaction is required for this activity, and is thought to trigger ATP hydrolysis by Rad51, which then causes Rad51 to dissociate from the ssDNA (Krejci et al. 2003; Antony et al. 2009). Cytological evidence has been furnished supporting the idea that Srs2 prevents the spurious assembly of Rad51-DNA filaments (Burgess et al. 2009). The suppressive effect of Srs2-RPA on Rad51 filament assembly can be counteracted by Rad52 and the Rad55-Rad57 complex, which are HR mediators that promote presynaptic filament assembly and stability, respectively (Burgess et al. 2009; Liu et al. 2011). In the promotion of SDSA, Srs2 could minimize second DNA end capture and/or limit the size and hence the stability of the D-loop via restriction of the length of the Rad51 presynaptic filament capable of DNA strand invasion. This is consistent with recent genetic evidence that supports a model in which Srs2 promotes noncrossover outcomes by negatively regulating Holliday junction formation (Mitchel et al. 2013).
Figure 3.
Roles of Srs2/RECQ5, Mph1/Fml1/FANCM, and Rad54 in homologous recombination. (A) Srs2 and RECQ5 displace Rad51 from ssDNA, leading to suppression of inappropriate recombination events. (B) Mph1/Fml1/FANCM can (i) release the invading strand from the D-loop to promote SDSA, and (ii) mediate regression of a blocked replication fork to bypass a DNA lesion. (C) Rad54 stimulates Rad51-mediated D-loop formation and facilitates DNA synthesis via the release of Rad51 from the DNA joint.
Interestingly, Srs2 is targeted to DNA replication forks through an interaction with the SUMOylated form of the DNA polymerase processivity clamp PCNA (proliferating cell nuclear antigen) (Armstrong et al. 2012; Kim et al. 2012; Burkovics et al. 2013). It is thought that, within the context of the replication fork, Srs2 helicase prevents ssDNA gaps arising from polymerase blockage from being bound by Rad51, thereby promoting replicative bypass of the lesions.
Although a clear ortholog of Srs2 has not yet been described in higher organisms, there is evidence that its E. coli counterpart UvrD also negatively regulates HR by disrupting presynaptic filaments of RecA (the bacterial ortholog of Rad51) (Morel et al. 1993; Flores et al. 2005; Veaute et al. 2005). As discussed in the next section, the human RECQ5 helicase has functional attributes indicating that it plays a role analogous to Srs2/UvrD in humans.
RECQ5, a Functional Counterpart of Srs2 in Human Cells
To identify a functional ortholog of Srs2 in human cells, an effort was mounted to examine several of the RecQ helicases—namely, RECQ1, RECQ5, and WRN—for their capability to inhibit the RAD51-mediated D-loop reaction and to describe the mechanism by which this occurs (Hu et al. 2007). Importantly, among the three helicases examined, only RECQ5 strongly suppressed D-loop formation in a manner stimulated by RPA. Further analysis revealed that, like Srs2, RECQ5, in conjunction with RPA, efficiently evicts RAD51 molecules from ssDNA to disassemble the presynaptic filament (Fig. 3A). RECQ5 physically interacts with RAD51, and complex formation between the two is important for the efficiency of presynaptic filament disruption (Schwendener et al. 2010; Islam et al. 2012).
Unlike BLM, WRN (defective in Werner syndrome), and RECQ4 (defective in Rothmund-Thomson syndrome), mutations in RECQ5 have not been linked to a human disease. However, RECQ5 knockout mice show increased cancer susceptibility, providing evidence for a tumor suppressor function in RECQ5 (Hu et al. 2007). Cells depleted of RECQ5 show increased RAD51 focus formation, enhanced DSB repair by HR, elevated sister chromatid exchanges (SCEs), and gross chromosomal rearrangements on DNA damage, again indicative of a role in HR regulation. These observations also buttress the validity of the biochemical results revealing an antirecombinase attribute of RECQ5 akin to Srs2. RECQ5 also interacts with PCNA, suggesting that it may be targeted to replication forks to prevent the accumulation of RAD51 on ssDNA gaps arising from fork stalling at a blocking lesion (Kanagaraj et al. 2006).
RECQ5 also associates with RNA polymerase II in a complex distinct from that harboring RAD51 and PCNA (Islam et al. 2010). The RECQ5-RNA Pol II complex has been postulated to prevent transcription-associated genome instability (Islam et al. 2010; Kanagaraj et al. 2010; Li et al. 2011). Although the molecular mechanism by which RECQ5 accomplishes this function is currently unknown, a logical hypothesis would be that RECQ5 remodels DNA structures or nucleoprotein complexes on collision of the transcription and replication machineries.
Roles of Mph1/Fml1/FANCM in Recombination Pathway Choice and DNA Replication Fork Repair
The Hef (helicase-associated endonuclease for fork-structured DNA) family of DNA motor proteins belongs to the SF2 helicase superfamily (Whitby 2010). These proteins translocate on ssDNA with a 3′ to 5′ directionality and are capable of processing DNA structures including D-loops, replication forks, and Holliday junctions (Prakash et al. 2005, 2009). Archaeal Hef is unique within this protein family in that it also possesses a nuclease activity (Komori et al. 2002), whereas the human ortholog FANCM has been at the center of intensive studies owing to its association with the cancer-prone disease Fanconi anemia (Nishino et al. 2003; Meetei et al. 2005).
Genetic analyses in the yeasts S. cerevisiae and S. pombe and also in the plant Arabidopsis thaliana have provided compelling evidence to implicate the Hef helicases in HR regulation and other processes. Cells deficient in the S. cerevisiae MPH1 (mutator phenotype 1) gene that encodes the Hef ortholog are sensitive to DNA-damaging agents and prone to accumulating spontaneous mutations (Scheller et al. 2000; Schurer et al. 2004). Interestingly, deletion of MPH1 leads to an abnormally high level of crossover formation in DSB repair by HR, in a manner that is additive with either the srs2Δ or sgs1Δ mutation, suggesting that Mph1 suppresses crossovers in a mechanism distinct from these helicases (Mitchel et al. 2013). Similarly, S. pombe cells deficient in the Fml1 (FANCM-like 1) protein generate an excess of crossover recombinants during plasmid gap repair (Nandi and Whitby 2012). Additive levels of crossover formation were seen in double mutants of fml1Δ with either srs2Δ or rqh1Δ (equivalent to S. cerevisiae sgs1Δ) mutations (Sun et al. 2008). Importantly, both Mph1 and Fml1 also have a prorecombination function during S phase, specifically in the promotion of replication fork repair via HR (Sun et al. 2008).
Given the importance of the ZMM group of proteins in meiotic crossover formation, ZMM mutants are infertile as they lack a sufficient number of crossovers to maintain proper meiotic chromosome disjunction. A screen for suppressors of the mutation of a ZMM group member, zip4, in A. thaliana led to the identification of FANCM helicase as a major factor that limits meiotic crossover formation (Crismani et al. 2012). The fancm mutant alone has a threefold-increased crossover frequency compared with wild type. These spurious crossovers in the fancm mutant arise not from the pathway that generates most of the crossovers in wild type, but from an alternate pathway. Similarly, in S. pombe, Fml1 helps mediate the formation of noncrossovers in a pathway competitive against a procrossover pathway that is dependent on the nuclease Mus81 (Lorenz et al. 2012).
In vitro studies with purified Mph1, Fml1, and human FANCM have shed light on the mechanisms by which they regulate HR and promote replication fork repair (Prakash et al. 2005). All three proteins have been shown to dissociate D-loops in vitro, and Mph1 is able to do so within the context of the Rad51-catalyzed D-loop reaction (Fig. 3Bi) (Gari et al. 2008; Sun et al. 2008; Prakash et al. 2009; Sebesta et al. 2011). ATP hydrolysis is required for this activity (Gari et al. 2008; Prakash et al. 2009). These data nicely explain how Mph1/Fml1/FANCM suppresses crossover formation, as disassembly of the D-loop forces the cell to use the noncrossover-exclusive SDSA pathway. Thus, these Hef orthologs function in a manner mechanistically distinct from Sgs1 and Srs2 to regulate HR in favor of noncrossover formation via the SDSA pathway. During S phase, it is believed that Mph1, Fml1, and FANCM mediate the regression of injured or stalled replication forks to initiate fork restart (Fig. 3Bii) (Chen et al. 2009; Choi et al. 2010; Chavez et al. 2011).
RTEL, a D-Loop-Disruptive Motor
Besides FANCM, higher organisms have a second DNA motor protein RTEL that disrupts D-loops. This protein was originally identified as a regulator of telomere length, and recent work has shown that it helps to maintain telomere stability by resolving secondary structure in telomeric DNA (Ding et al. 2004; Vannier et al. 2012). In the absence of RTEL, telomeric quadruplex (G4) structures are inappropriately cleaved by the nuclease Slx4, leading to telomere attrition (Vannier et al. 2012). Its ortholog RTEL-1, in the nematode Caenorhabditis elegans, was uncovered in a screen for synthetic lethality with a mutation in the SGS1 ortholog HIM-6 (Barber et al. 2008). Mutant animals lacking RTEL-1 show sensitivity to DNA-damaging agents and increased crossover frequency (Barber et al. 2008; Youds et al. 2010). Loss of RTEL-1 in combination with him-6 leads to accumulation of recombination intermediates and RAD51 foci (Barber et al. 2008). RTEL-1 also functions in meiotic cells, as crossovers in meiosis are up-regulated in rtel-1 mutant worms (Youds et al. 2010). In human cells, RTEL depletion also causes recombination to go up (Barber et al. 2008). RTEL protein has been purified and shown to disrupt RAD51-mediated D-loops in the presence of RPA, but unlike Srs2 or RECQ5, RTEL cannot strip RAD51 from ssDNA (Barber et al. 2008; Youds et al. 2010). Thus, RTEL uses a similar mechanism as the Hef helicases in HR regulation.
PROMOTION OF DNA PAIRING AND REPAIR DNA SYNTHESIS BY RAD54 AND RDH54
As first revealed in genetic studies in S. cerevisiae, the RAD54 gene is crucial for meiotic HR and DSB repair by HR in mitotic cells (Game and Mortimer 1974; Shinohara et al. 1997). The structure and function of the Rad54 protein are conserved among eukaryotes and in some archaeal species (Haseltine and Kowalczykowski 2009; Ceballos and Heyer 2011). Several paralogs of Rad54 protein have been described in various organisms, and one of these, Rdh54 (Rad homolog 54) of S. cerevisiae, will be reviewed here (Mazin et al. 2010; Ceballos and Heyer 2011). Rad54 and Rdh54 proteins belong to the Snf2/Swi2 group of ATP-dependent DNA translocases that are often found in chromatin-remodeling complexes. Rad54 and Rdh54 both have a DNA translocase activity that is fueled by ATP hydrolysis (Petukhova et al. 1998; Nimonkar et al. 2007), and their movement on DNA leads to dynamic topological changes that cause transient separation of the DNA strands (Tan et al. 1999; Petukhova et al. 2000; Van Komen et al. 2000). These proteins are also able to process or dissociate DNA structures, such as the D-loop and branched DNAs including the Holliday junction and to remove Rad51 from dsDNA via their DNA translocase activity (Solinger et al. 2002; Bugreev et al. 2006, 2007). Consistent with their Swi2/Snf2 relatedness, a chromatin-remodeling activity has been found in these proteins (Alexeev et al. 2003; Kwon et al. 2008).
The mechanisms by which Rad54 and Rdh54 help promote HR have been the subject of intensive studies in many laboratories, and a great deal is known in this regard. Both Rad54 and Rdh54 interact physically with Rad51, and protein complex formation serves to target these DNA motor proteins to DSBs (Jiang et al. 1996; Lisby et al. 2004). Functional synergy between Rad54 and Rdh54 with Rad51 has been noted, and this requires complex formation of either motor protein with Rad51. Most notably, the ability of Rad51 to mediate homologous DNA pairing is greatly enhanced by Rad54 (Fig. 3C) or Rdh54, and in turn, the DNA-dependent ATPase and DNA supercoiling activities of these motor proteins are up-regulated by Rad51 (Petukhova et al. 2000; Van Komen et al. 2000). The chromatin-remodeling activity of Rad54 is also similarly stimulated by Rad51 (Alexeev et al. 2003). Importantly, either Rad54 or Rdh54 becomes indispensable when a chromatinized template is used as the information donor in the homologous pairing reaction (Kwon et al. 2008; Sinha and Peterson 2008). As mentioned above, Rad54 and Rdh54 use their translocase activity to remove Rad51 from duplex DNA. This activity likely serves two distinct purposes: namely, (i) to release Rad51 from bulk chromatin to prevent the accumulation of cytotoxic nucleoprotein complexes and to realize intracellular recycling of Rad51 (Shah et al. 2010), and (ii) to free the nascent DNA joint in the D-loop from bound Rad51, so as to permit access of the primer end to a DNA polymerase to initiate repair DNA synthesis (Fig. 3C) (Sugawara et al. 2003; Li and Heyer 2009).
It should be noted that, despite their possession of similar biochemical attributes, the biological functions of Rad54 and Rdh54 are far from redundant. Most notably, Rad54 plays a more prominent role in the repair of certain types of DNA lesions and in the facilitation of repair synthesis, with Rdh54 apparently providing the major function in Rad51 removal from bulk chromatin (Ceballos and Heyer 2011). In both mitotic and meiotic cells, the promotion of HR between homologs is largely dependent on Rdh54, whereas Rad54 is more adept in the mediation of HR events between sister chromatids (Klein 1997; Shinohara et al. 1997; Nimonkar et al. 2012). Interestingly, the activity of the Rad51-Rad54 pair is down-regulated in meiosis via two different means, so as to permit interhomolog HR by the procrossover DSBR pathway. First, the meiosis-specific Mek1 kinase phosphorylates Rad54 at T132 resulting in a decreased affinity for Rad51 (Niu et al. 2009). Second, the meiosis-specific Hed1 protein physically interacts with Rad51 avidly, preventing the latter from complex formation with Rad54 (Busygina et al. 2008).
CONCLUDING REMARKS
As reviewed herein, extensive in vitro and in vivo studies in both yeast and mammalian systems have identified several distinct mechanisms of HR regulation at the DNA pairing stage. In addition to the aforementioned regulators, there is emerging evidence that additional players are involved in HR regulation. Recently, the SWI/SNF-related proteins SMARCAL1 and ZRANB3 have been shown to disrupt RAD51-made D-loops in vitro, suggesting that they may function in a manner analogous to RTEL and Mph1/Fml1/FANCM (Betous et al. 2012; Ciccia et al. 2012). Genetic studies have provided evidence that the SF1 helicase Fbh1, which also possesses ubiquitin E3 ligase activity, may be a negative regulator of HR and act as a functional homolog of Srs2 in humans (Osman et al. 2005; Chiolo et al. 2007; Lorenz et al. 2009). FANCJ, a helicase of the SF2 superfamily associated with Fanconi anemia, has been suggested to affect the stability of the RAD51 presynaptic filament and to interfere with RAD51-mediated D-loop formation in vitro, although genetic evidence thus far is not consistent with the notion that FANCJ opposes recombination in vivo (Litman et al. 2005; Sommers et al. 2009; Xie et al. 2012). On the positive regulatory side, the meiosis-specific Mer3 helicase has been implicated in stabilizing D-loops by promoting branch migration in a direction that extends the nascent DNA joint (Mazina et al. 2004). Indeed, there is much future work to be performed to clarify the roles of these helicases in HR regulation.
ACKNOWLEDGMENTS
The work in our laboratory has been supported by research grants and a program project grant from the National Institutes of Health.
Footnotes
Editors: Stephen Kowalczykowski, Neil Hunter, and Wolf-Dietrich Heyer
Additional Perspectives on DNA Recombination available at www.cshperspectives.org
REFERENCES
*Reference is also in this collection.
- Alexeev A, Mazin A, Kowalczykowski SC 2003. Rad54 protein possesses chromatin-remodeling activity stimulated by the Rad51-ssDNA nucleoprotein filament. Nat Struct Biol 10: 182–186. [DOI] [PubMed] [Google Scholar]
- Antony E, Tomko EJ, Xiao Q, Krejci L, Lohman TM, Ellenberger T 2009. Srs2 disassembles Rad51 filaments by a protein-protein interaction triggering ATP turnover and dissociation of Rad51 from DNA. Mol Cell 35: 105–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armstrong AA, Mohideen F, Lima CD 2012. Recognition of SUMO-modified PCNA requires tandem receptor motifs in Srs2. Nature 483: 59–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barber LJ, Youds JL, Ward JD, McIlwraith MJ, O’Neil NJ, Petalcorin MI, Martin JS, Collis SJ, Cantor SB, Auclair M, et al. 2008. RTEL1 maintains genomic stability by suppressing homologous recombination. Cell 135: 261–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartram CR, Koske-Westphal T, Passarge E 1976. Chromatid exchanges in ataxia telangiectasia, Bloom syndrome, Werner syndrome, and xeroderma pigmentosum. Ann Hum Genet 40: 79–86. [DOI] [PubMed] [Google Scholar]
- Barzel A, Kupiec M 2008. Finding a match: How do homologous sequences get together for recombination? Nat Rev Genet 9: 27–37. [DOI] [PubMed] [Google Scholar]
- Betous R, Mason AC, Rambo RP, Bansbach CE, Badu-Nkansah A, Sirbu BM, Eichman BF, Cortez D 2012. SMARCAL1 catalyzes fork regression and Holliday junction migration to maintain genome stability during DNA replication. Genes Dev 26: 151–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Bizard AH, Hickson ID 2014. The dissolution of double Holliday junctions. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a016477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borner GV, Kleckner N, Hunter N 2004. Crossover/noncrossover differentiation, synaptonemal complex formation, and regulatory surveillance at the leptotene/zygotene transition of meiosis. Cell 117: 29–45. [DOI] [PubMed] [Google Scholar]
- Bugreev DV, Mazina OM, Mazin AV 2006. Rad54 protein promotes branch migration of Holliday junctions. Nature 442: 590–593. [DOI] [PubMed] [Google Scholar]
- Bugreev DV, Hanaoka F, Mazin AV 2007. Rad54 dissociates homologous recombination intermediates by branch migration. Nat Struct Mol Biol 14: 746–753. [DOI] [PubMed] [Google Scholar]
- Burgess RC, Lisby M, Altmannova V, Krejci L, Sung P, Rothstein R 2009. Localization of recombination proteins and Srs2 reveals anti-recombinase function in vivo. J Cell Biol 185: 969–981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burkovics P, Sebesta M, Sisakova A, Plault N, Szukacsov V, Robert T, Pinter L, Marini V, Kolesar P, Haracska L, et al. 2013. Srs2 mediates PCNA-SUMO-dependent inhibition of DNA repair synthesis. EMBO J 32: 742–755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busygina V, Sehorn MG, Shi IY, Tsubouchi H, Roeder GS, Sung P 2008. Hed1 regulates Rad51-mediated recombination via a novel mechanism. Genes Dev 22: 786–795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannavo E, Cejka P, Kowalczykowski SC 2013. Relationship of DNA degradation by Saccharomyces cerevisiae exonuclease 1 and its stimulation by RPA and Mre11-Rad50-Xrs2 to DNA end resection. Proc Natl Acad Sci 110: E1661–E1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ceballos SJ, Heyer WD 2011. Functions of the Snf2/Swi2 family Rad54 motor protein in homologous recombination. Biochim Biophys Acta 1809: 509–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cejka P, Cannavo E, Polaczek P, Masuda-Sasa T, Pokharel S, Campbell JL, Kowalczykowski SC 2010. DNA end resection by Dna2-Sgs1-RPA and its stimulation by Top3-Rmi1 and Mre11-Rad50-Xrs2. Nature 467: 112–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang M, Bellaoui M, Zhang C, Desai R, Morozov P, Delgado-Cruzata L, Rothstein R, Freyer GA, Boone C, Brown GW 2005. RMI1/NCE4, a suppressor of genome instability, encodes a member of the RecQ helicase/Topo III complex. EMBO J 24: 2024–2033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chavez A, Agrawal V, Johnson FB 2011. Homologous recombination-dependent rescue of deficiency in the structural maintenance of chromosomes (Smc) 5/6 complex. J Biol Chem 286: 5119–5125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen YH, Choi K, Szakal B, Arenz J, Duan X, Ye H, Branzei D, Zhao X 2009. Interplay between the Smc5/6 complex and the Mph1 helicase in recombinational repair. Proc Natl Acad Sci 106: 21252–21257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiolo I, Saponaro M, Baryshnikova A, Kim JH, Seo YS, Liberi G 2007. The human F-Box DNA helicase FBH1 faces Saccharomyces cerevisiae Srs2 and postreplication repair pathway roles. Mol Cell Biol 27: 7439–7450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi K, Szakal B, Chen YH, Branzei D, Zhao X 2010. The Smc5/6 complex and Esc2 influence multiple replication-associated recombination processes in Saccharomyces cerevisiae. Mol Biol Cell 21: 2306–2314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu WK, Hickson ID 2009. RecQ helicases: Multifunctional genome caretakers. Nat Rev Cancer 9: 644–654. [DOI] [PubMed] [Google Scholar]
- Ciccia A, Nimonkar AV, Hu Y, Hajdu I, Achar YJ, Izhar L, Petit SA, Adamson B, Yoon JC, Kowalczykowski SC, et al. 2012. Polyubiquitinated PCNA recruits the ZRANB3 translocase to maintain genomic integrity after replication stress. Mol Cell 47: 396–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crismani W, Girard C, Froger N, Pradillo M, Santos JL, Chelysheva L, Copenhaver GP, Horlow C, Mercier R 2012. FANCM limits meiotic crossovers. Science 336: 1588–1590. [DOI] [PubMed] [Google Scholar]
- Daley JM, Laan RL, Suresh A, Wilson TE 2005. DNA joint dependence of pol X family polymerase action in nonhomologous end joining. J Biol Chem 280: 29030–29037. [DOI] [PubMed] [Google Scholar]
- Daley JM, Niu H, Sung P 2013. Roles of DNA helicases in the mediation and regulation of homologous recombination. Adv Exp Med Biol 767: 185–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding H, Schertzer M, Wu X, Gertsenstein M, Selig S, Kammori M, Pourvali R, Poon S, Vulto I, Chavez E, et al. 2004. Regulation of murine telomere length by Rtel: An essential gene encoding a helicase-like protein. Cell 117: 873–886. [DOI] [PubMed] [Google Scholar]
- Ellis NA, Groden J, Ye TZ, Straughen J, Lennon DJ, Ciocci S, Proytcheva M, German J 1995. The Bloom’s syndrome gene product is homologous to RecQ helicases. Cell 83: 655–666. [DOI] [PubMed] [Google Scholar]
- Flores MJ, Sanchez N, Michel B 2005. A fork-clearing role for UvrD. Mol Microbiol 57: 1664–1675. [DOI] [PubMed] [Google Scholar]
- Game JC, Mortimer RK 1974. A genetic study of x-ray sensitive mutants in yeast. Mutat Res 24: 281–292. [DOI] [PubMed] [Google Scholar]
- Gangloff S, McDonald JP, Bendixen C, Arthur L, Rothstein R 1994. The yeast type I topoisomerase Top3 interacts with Sgs1, a DNA helicase homolog: A potential eukaryotic reverse gyrase. Mol Cell Biol 14: 8391–8398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gari K, Decaillet C, Delannoy M, Wu L, Constantinou A 2008. Remodeling of DNA replication structures by the branch point translocase FANCM. Proc Natl Acad Sci 105: 16107–16112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gravel S, Chapman JR, Magill C, Jackson SP 2008. DNA helicases Sgs1 and BLM promote DNA double-strand break resection. Genes Dev 22: 2767–2772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray MD, Shen JC, Kamath-Loeb AS, Blank A, Sopher BL, Martin GM, Oshima J, Loeb LA 1997. The Werner syndrome protein is a DNA helicase. Nat Genet 17: 100–103. [DOI] [PubMed] [Google Scholar]
- Haseltine CA, Kowalczykowski SC 2009. An archaeal Rad54 protein remodels DNA and stimulates DNA strand exchange by RadA. Nucleic Acids Res 37: 2757–2770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y, Raynard S, Sehorn MG, Lu X, Bussen W, Zheng L, Stark JM, Barnes EL, Chi P, Janscak P, et al. 2007. RECQL5/Recql5 helicase regulates homologous recombination and suppresses tumor formation via disruption of Rad51 presynaptic filaments. Genes Dev 21: 3073–3084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ira G, Malkova A, Liberi G, Foiani M, Haber JE 2003. Srs2 and Sgs1-Top3 suppress crossovers during double-strand break repair in yeast. Cell 115: 401–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Islam MN, Fox D 3rd, Guo R, Enomoto T, Wang W 2010. RecQL5 promotes genome stabilization through two parallel mechanisms—Interacting with RNA polymerase II and acting as a helicase. Mol Cell Biol 30: 2460–2472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Islam MN, Paquet N, Fox D 3rd, Dray E, Zheng XF, Klein H, Sung P, Wang W 2012. A variant of the breast cancer type 2 susceptibility protein (BRC) repeat is essential for the RECQL5 helicase to interact with RAD51 recombinase for genome stabilization. J Biol Chem 287: 23808–23818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen RB, Carreira A, Kowalczykowski SC 2010. Purified human BRCA2 stimulates RAD51-mediated recombination. Nature 467: 678–683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jessop L, Rockmill B, Roeder GS, Lichten M 2006. Meiotic chromosome synapsis-promoting proteins antagonize the anti-crossover activity of sgs1. PLoS Genet 2: e155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang H, Xie Y, Houston P, Stemke-Hale K, Mortensen UH, Rothstein R, Kodadek T 1996. Direct association between the yeast Rad51 and Rad54 recombination proteins. J Biol Chem 271: 33181–33186. [DOI] [PubMed] [Google Scholar]
- Kanagaraj R, Saydam N, Garcia PL, Zheng L, Janscak P 2006. Human RECQ5β helicase promotes strand exchange on synthetic DNA structures resembling a stalled replication fork. Nucleic Acids Res 34: 5217–5231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanagaraj R, Huehn D, MacKellar A, Menigatti M, Zheng L, Urban V, Shevelev I, Greenleaf AL, Janscak P 2010. RECQ5 helicase associates with the C-terminal repeat domain of RNA polymerase II during productive elongation phase of transcription. Nucleic Acids Res 38: 8131–8140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karow JK, Chakraverty RK, Hickson ID 1997. The Bloom’s syndrome gene product is a 3′-5′ DNA helicase. J Biol Chem 272: 30611–30614. [DOI] [PubMed] [Google Scholar]
- Karpenshif Y, Bernstein KA 2012. From yeast to mammals: Recent advances in genetic control of homologous recombination. DNA Repair (Amst) 11: 781–788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SO, Yoon H, Park SO, Lee M, Shin JS, Ryu KS, Lee JO, Seo YS, Jung HS, Choi BS 2012. Srs2 possesses a non-canonical PIP box in front of its SBM for precise recognition of SUMOylated PCNA. J Mol Cell Biol 4: 258–261. [DOI] [PubMed] [Google Scholar]
- Klein HL 1997. RDH54, a RAD54 homologue in Saccharomyces cerevisiae, is required for mitotic diploid-specific recombination and repair and for meiosis. Genetics 147: 1533–1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komori K, Fujikane R, Shinagawa H, Ishino Y 2002. Novel endonuclease in Archaea cleaving DNA with various branched structure. Genes Genet Syst 77: 227–241. [DOI] [PubMed] [Google Scholar]
- Krejci L, Van Komen S, Li Y, Villemain J, Reddy MS, Klein H, Ellenberger T, Sung P 2003. DNA helicase Srs2 disrupts the Rad51 presynaptic filament. Nature 423: 305–309. [DOI] [PubMed] [Google Scholar]
- Kwon Y, Seong C, Chi P, Greene EC, Klein H, Sung P 2008. ATP-dependent chromatin remodeling by the Saccharomyces cerevisiae homologous recombination factor Rdh54. J Biol Chem 283: 10445–10452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Lam I, Keeney S 2014. Mechanism and regulation of meiotic recombination initiation. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a016634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence CW, Christensen RB 1979. Metabolic suppressors of trimethoprim and ultraviolet light sensitivities of Saccharomyces cerevisiae rad6 mutants. J Bacteriol 139: 866–876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Heyer WD 2009. RAD54 controls access to the invading 3′-OH end after RAD51-mediated DNA strand invasion in homologous recombination in Saccharomyces cerevisiae. Nucleic Acids Res 37: 638–646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Xu X, Liu Y 2011. The SET2-RPB1 interaction domain of human RECQ5 is important for transcription-associated genome stability. Mol Cell Biol 31: 2090–2099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieber MR 2010. The mechanism of double-strand DNA break repair by the nonhomologous DNA end-joining pathway. Annu Rev Biochem 79: 181–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisby M, Barlow JH, Burgess RC, Rothstein R 2004. Choreography of the DNA damage response: Spatiotemporal relationships among checkpoint and repair proteins. Cell 118: 699–713. [DOI] [PubMed] [Google Scholar]
- Litman R, Peng M, Jin Z, Zhang F, Zhang J, Powell S, Andreassen PR, Cantor SB 2005. BACH1 is critical for homologous recombination and appears to be the Fanconi anemia gene product FANCJ. Cancer Cell 8: 255–265. [DOI] [PubMed] [Google Scholar]
- Liu J, Doty T, Gibson B, Heyer WD 2010. Human BRCA2 protein promotes RAD51 filament formation on RPA-covered single-stranded DNA. Nat Struct Mol Biol 17: 1260–1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Renault L, Veaute X, Fabre F, Stahlberg H, Heyer WD 2011. Rad51 paralogues Rad55-Rad57 balance the antirecombinase Srs2 in Rad51 filament formation. Nature 479: 245–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenz A, Osman F, Folkyte V, Sofueva S, Whitby MC 2009. Fbh1 limits Rad51-dependent recombination at blocked replication forks. Mol Cell Biol 29: 4742–4756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenz A, Osman F, Sun W, Nandi S, Steinacher R, Whitby MC 2012. The fission yeast FANCM ortholog directs non-crossover recombination during meiosis. Science 336: 1585–1588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marini V, Krejci L 2010. Srs2: The “Odd-Job Man” in DNA repair. DNA Repair (Amst) 9: 268–275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazin AV, Mazina OM, Bugreev DV, Rossi MJ 2010. Rad54, the motor of homologous recombination. DNA Repair (Amst) 9: 286–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazina OM, Mazin AV, Nakagawa T, Kolodner RD, Kowalczykowski SC 2004. Saccharomyces cerevisiae Mer3 helicase stimulates 3′-5′ heteroduplex extension by Rad51; implications for crossover control in meiotic recombination. Cell 117: 47–56. [DOI] [PubMed] [Google Scholar]
- Meetei AR, Medhurst AL, Ling C, Xue Y, Singh TR, Bier P, Steltenpool J, Stone S, Dokal I, Mathew CG, et al. 2005. A human ortholog of archaeal DNA repair protein Hef is defective in Fanconi anemia complementation group M. Nat Genet 37: 958–963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Mehta A, Haber JE 2014. Sources of DNA double-strand breaks and models for recombinational DNA repair. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a016428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mimitou EP, Symington LS 2008. Sae2, Exo1 and Sgs1 collaborate in DNA double-strand break processing. Nature 455: 770–774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mimitou EP, Symington LS 2009. Nucleases and helicases take center stage in homologous recombination. Trends Biochem Sci 34: 264–272. [DOI] [PubMed] [Google Scholar]
- Mitchel K, Lehner K, Jinks-Robertson S 2013. Heteroduplex DNA position defines the roles of the Sgs1, Srs2, and Mph1 helicases in promoting distinct recombination outcomes. PLoS Genet 9: e1003340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morel P, Hejna JA, Ehrlich SD, Cassuto E 1993. Antipairing and strand transferase activities of E. coli helicase II (UvrD). Nucleic Acids Res 21: 3205–3209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Morrical SW 2014. DNA pairing and annealing processes in homologous recombination and homology-directed repair. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a016444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nandi S, Whitby MC 2012. The ATPase activity of Fml1 is essential for its roles in homologous recombination and DNA repair. Nucleic Acids Res 40: 9584–9595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nimonkar AV, Amitani I, Baskin RJ, Kowalczykowski SC 2007. Single molecule imaging of Tid1/Rdh54, a Rad54 homolog that translocates on duplex DNA and can disrupt joint molecules. J Biol Chem 282: 30776–30784. [DOI] [PubMed] [Google Scholar]
- Nimonkar AV, Genschel J, Kinoshita E, Polaczek P, Campbell JL, Wyman C, Modrich P, Kowalczykowski SC 2011. BLM-DNA2-RPA-MRN and EXO1-BLM-RPA-MRN constitute two DNA end resection machineries for human DNA break repair. Genes Dev 25: 350–362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nimonkar AV, Dombrowski CC, Siino JS, Stasiak AZ, Stasiak A, Kowalczykowski SC 2012. Saccharomyces cerevisiae Dmc1 and Rad51 proteins preferentially function with Tid1 and Rad54 proteins, respectively, to promote DNA strand invasion during genetic recombination. J Biol Chem 287: 28727–28737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishino T, Komori K, Ishino Y, Morikawa K 2003. X-ray and biochemical anatomy of an archaeal XPF/Rad1/Mus81 family nuclease: Similarity between its endonuclease domain and restriction enzymes. Structure 11: 445–457. [DOI] [PubMed] [Google Scholar]
- Niu H, Wan L, Busygina V, Kwon Y, Allen JA, Li X, Kunz RC, Kubota K, Wang B, Sung P, et al. 2009. Regulation of meiotic recombination via Mek1-mediated Rad54 phosphorylation. Mol Cell 36: 393–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu H, Chung WH, Zhu Z, Kwon Y, Zhao W, Chi P, Prakash R, Seong C, Liu D, Lu L, et al. 2010. Mechanism of the ATP-dependent DNA end-resection machinery from Saccharomyces cerevisiae. Nature 467: 108–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh SD, Lao JP, Hwang PY, Taylor AF, Smith GR, Hunter N 2007. BLM ortholog, Sgs1, prevents aberrant crossing-over by suppressing formation of multichromatid joint molecules. Cell 130: 259–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osman F, Dixon J, Barr AR, Whitby MC 2005. The F-Box DNA helicase Fbh1 prevents Rhp51-dependent recombination without mediator proteins. Mol Cell Biol 25: 8084–8096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petukhova G, Stratton S, Sung P 1998. Catalysis of homologous DNA pairing by yeast Rad51 and Rad54 proteins. Nature 393: 91–94. [DOI] [PubMed] [Google Scholar]
- Petukhova G, Sung P, Klein H 2000. Promotion of Rad51-dependent D-loop formation by yeast recombination factor Rdh54/Tid1. Genes Dev 14: 2206–2215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prakash R, Krejci L, Van Komen S, Anke Schurer K, Kramer W, Sung P 2005. Saccharomyces cerevisiae MPH1 gene, required for homologous recombination-mediated mutation avoidance, encodes a 3′ to 5′ DNA helicase. J Biol Chem 280: 7854–7860. [DOI] [PubMed] [Google Scholar]
- Prakash R, Satory D, Dray E, Papusha A, Scheller J, Kramer W, Krejci L, Klein H, Haber JE, Sung P, et al. 2009. Yeast Mph1 helicase dissociates Rad51-made D-loops: Implications for crossover control in mitotic recombination. Genes Dev 23: 67–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raderschall E, Golub EI, Haaf T 1999. Nuclear foci of mammalian recombination proteins are located at single-stranded DNA regions formed after DNA damage. Proc Natl Acad Sci 96: 1921–1926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raynard S, Bussen W, Sung P 2006. A double Holliday junction dissolvasome comprising BLM, topoisomerase IIIα, and BLAP75. J Biol Chem 281: 13861–13864. [DOI] [PubMed] [Google Scholar]
- Rong L, Palladino F, Aguilera A, Klein HL 1991. The hyper-gene conversion hpr5-1 mutation of Saccharomyces cerevisiae is an allele of the SRS2/RADH gene. Genetics 127: 75–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- San Filippo J, Sung P, Klein H 2008. Mechanism of eukaryotic homologous recombination. Annu Rev Biochem 77: 229–257. [DOI] [PubMed] [Google Scholar]
- Scheller J, Schurer A, Rudolph C, Hettwer S, Kramer W 2000. MPH1, a yeast gene encoding a DEAH protein, plays a role in protection of the genome from spontaneous and chemically induced damage. Genetics 155: 1069–1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiestl RH, Prakash S, Prakash L 1990. The SRS2 suppressor of rad6 mutations of Saccharomyces cerevisiae acts by channeling DNA lesions into the RAD52 DNA repair pathway. Genetics 124: 817–831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schurer KA, Rudolph C, Ulrich HD, Kramer W 2004. Yeast MPH1 gene functions in an error-free DNA damage bypass pathway that requires genes from Homologous recombination, but not from postreplicative repair. Genetics 166: 1673–1686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz EK, Heyer WD 2011. Processing of joint molecule intermediates by structure-selective endonucleases during homologous recombination in eukaryotes. Chromosoma 120: 109–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwendener S, Raynard S, Paliwal S, Cheng A, Kanagaraj R, Shevelev I, Stark JM, Sung P, Janscak P 2010. Physical interaction of RECQ5 helicase with RAD51 facilitates its anti-recombinase activity. J Biol Chem 285: 15739–15745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sebesta M, Burkovics P, Haracska L, Krejci L 2011. Reconstitution of DNA repair synthesis in vitro and the role of polymerase and helicase activities. DNA Repair (Amst) 10: 567–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah PP, Zheng X, Epshtein A, Carey JN, Bishop DK, Klein HL 2010. Swi2/Snf2-related translocases prevent accumulation of toxic Rad51 complexes during mitotic growth. Mol Cell 39: 862–872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen JC, Gray MD, Oshima J, Loeb LA 1998. Characterization of Werner syndrome protein DNA helicase activity: Directionality, substrate dependence and stimulation by replication protein A. Nucleic Acids Res 26: 2879–2885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinohara M, Shita-Yamaguchi E, Buerstedde JM, Shinagawa H, Ogawa H, Shinohara A 1997. Characterization of the roles of the Saccharomyces cerevisiae RAD54 gene and a homologue of RAD54, RDH54/TID1, in mitosis and meiosis. Genetics 147: 1545–1556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinha M, Peterson CL 2008. A Rad51 presynaptic filament is sufficient to capture nucleosomal homology during recombinational repair of a DNA double-strand break. Mol Cell 30: 803–810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solinger JA, Kiianitsa K, Heyer WD 2002. Rad54, a Swi2/Snf2-like recombinational repair protein, disassembles Rad51:dsDNA filaments. Mol Cell 10: 1175–1188. [DOI] [PubMed] [Google Scholar]
- Sommers JA, Rawtani N, Gupta R, Bugreev DV, Mazin AV, Cantor SB, Brosh RM Jr 2009. FANCJ uses its motor ATPase to destabilize protein-DNA complexes, unwind triplexes, and inhibit RAD51 strand exchange. J Biol Chem 284: 7505–7517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugawara N, Wang X, Haber JE 2003. In vivo roles of Rad52, Rad54, and Rad55 proteins in Rad51-mediated recombination. Mol Cell 12: 209–219. [DOI] [PubMed] [Google Scholar]
- Sugiyama T, Kowalczykowski SC 2002. Rad52 protein associates with replication protein A (RPA)-single-stranded DNA to accelerate Rad51-mediated displacement of RPA and presynaptic complex formation. J Biol Chem 277: 31663–31672. [DOI] [PubMed] [Google Scholar]
- Sun W, Nandi S, Osman F, Ahn JS, Jakovleska J, Lorenz A, Whitby MC 2008. The FANCM ortholog Fml1 promotes recombination at stalled replication forks and limits crossing over during DNA double-strand break repair. Mol Cell 32: 118–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Symington LS 2014. Processing of DNA breaks: Mechanism and regulation. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a016436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan TL, Essers J, Citterio E, Swagemakers SM, de Wit J, Benson FE, Hoeijmakers JH, Kanaar R 1999. Mouse Rad54 affects DNA conformation and DNA-damage-induced Rad51 foci formation. Curr Biol 9: 325–328. [DOI] [PubMed] [Google Scholar]
- Van Komen S, Petukhova G, Sigurdsson S, Stratton S, Sung P 2000. Superhelicity-driven homologous DNA pairing by yeast recombination factors Rad51 and Rad54. Mol Cell 6: 563–572. [DOI] [PubMed] [Google Scholar]
- Vannier JB, Pavicic-Kaltenbrunner V, Petalcorin MI, Ding H, Boulton SJ 2012. RTEL1 dismantles T loops and counteracts telomeric G4-DNA to maintain telomere integrity. Cell 149: 795–806. [DOI] [PubMed] [Google Scholar]
- Veaute X, Jeusset J, Soustelle C, Kowalczykowski SC, Le Cam E, Fabre F 2003. The Srs2 helicase prevents recombination by disrupting Rad51 nucleoprotein filaments. Nature 423: 309–312. [DOI] [PubMed] [Google Scholar]
- Veaute X, Delmas S, Selva M, Jeusset J, Le Cam E, Matic I, Fabre F, Petit MA 2005. UvrD helicase, unlike Rep helicase, dismantles RecA nucleoprotein filaments in Escherichia coli. EMBO J 24: 180–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watt PM, Hickson ID, Borts RH, Louis EJ 1996. SGS1, a homologue of the Bloom’s and Werner’s syndrome genes, is required for maintenance of genome stability in Saccharomyces cerevisiae. Genetics 144: 935–945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitby MC 2010. The FANCM family of DNA helicases/translocases. DNA Repair (Amst) 9: 224–236. [DOI] [PubMed] [Google Scholar]
- Wu L, Hickson ID 2003. The Bloom’s syndrome helicase suppresses crossing over during homologous recombination. Nature 426: 870–874. [DOI] [PubMed] [Google Scholar]
- Wu L, Bachrati CZ, Ou J, Xu C, Yin J, Chang M, Wang W, Li L, Brown GW, Hickson ID 2006. BLAP75/RMI1 promotes the BLM-dependent dissolution of homologous recombination intermediates. Proc Natl Acad Sci 103: 4068–4073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Wyatt HDM, West SC 2014. Holliday junction resolvases. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a016485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie J, Peng M, Guillemette S, Quan S, Maniatis S, Wu Y, Venkatesh A, Shaffer SA, Brosh RM Jr, Cantor SB 2012. FANCJ/BACH1 acetylation at lysine 1249 regulates the DNA damage response. PLoS Genet 8: e1002786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu D, Guo R, Sobeck A, Bachrati CZ, Yang J, Enomoto T, Brown GW, Hoatlin ME, Hickson ID, Wang W 2008. RMI, a new OB-fold complex essential for Bloom syndrome protein to maintain genome stability. Genes Dev 22: 2843–2855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin J, Sobeck A, Xu C, Meetei AR, Hoatlin M, Li L, Wang W 2005. BLAP75, an essential component of Bloom’s syndrome protein complexes that maintain genome integrity. EMBO J 24: 1465–1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youds JL, Mets DG, McIlwraith MJ, Martin JS, Ward JD, NJ ON, Rose AM, West SC, Meyer BJ, Boulton SJ 2010. RTEL-1 enforces meiotic crossover interference and homeostasis. Science 327: 1254–1258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X, Jacobs SA, West SC, Ogawa T, Egelman EH 2001. Domain structure and dynamics in the helical filaments formed by RecA and Rad51 on DNA. Proc Natl Acad Sci 98: 8419–8424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Zelensky A, Kanaar R, Wyman C 2014. Mediators of homologous DNA pairing. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a016451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Z, Chung WH, Shim EY, Lee SE, Ira G 2008. Sgs1 helicase and two nucleases Dna2 and Exo1 resect DNA double-strand break ends. Cell 134: 981–994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Zickler D, Kleckner N 2014. Recombination, pairing, and synapsis of homologs during meiosis. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a016626. [DOI] [PMC free article] [PubMed] [Google Scholar]