Significance
Here, we examine the activity profile of the haloalkanoic acid dehalogenase (HAD) superfamily by screening a customized library against >200 enzymes from a broad sampling of the superfamily. From this dataset, we can infer the function of nearly 35% of the superfamily. Overall, the superfamily was found to show high substrate ambiguity, with 75% of the superfamily utilizing greater than five substrates. In addition, the HAD members with the least amount of structural accessorization of the Rossmann fold were found to be the most specific, suggesting that elaboration of the core domain may have led to increased substrate range of the superfamily.
Keywords: evolution, specificity, phosphatase, substrate screen, promiscuity
Abstract
Large-scale activity profiling of enzyme superfamilies provides information about cellular functions as well as the intrinsic binding capabilities of conserved folds. Herein, the functional space of the ubiquitous haloalkanoate dehalogenase superfamily (HADSF) was revealed by screening a customized substrate library against >200 enzymes from representative prokaryotic species, enabling inferred annotation of ∼35% of the HADSF. An extremely high level of substrate ambiguity was revealed, with the majority of HADSF enzymes using more than five substrates. Substrate profiling allowed assignment of function to previously unannotated enzymes with known structure, uncovered potential new pathways, and identified iso-functional orthologs from evolutionarily distant taxonomic groups. Intriguingly, the HADSF subfamily having the least structural elaboration of the Rossmann fold catalytic domain was the most specific, consistent with the concept that domain insertions drive the evolution of new functions and that the broad specificity observed in HADSF may be a relic of this process.
Since the first genomes were sequenced, there has been an exponential increase in the number of protein sequences deposited into databases worldwide. At the time of this writing the UniProtKB/TrEMBL database contains over 32 million protein sequences. Although this increase in sequence data has dramatically enhanced our understanding of the genomic organization of organisms, as the number of protein sequences grows, the proportion of firm functional assignments diminishes. Traditionally, methods of functional annotation involve comparing sequence identity between experimentally characterized proteins and newly sequenced ones, typically via BLAST (1). In cases where significant sequence similarity cannot be ascertained, proteins are annotated as “hypothetical” or “putative.” Moreover, the decrease in sequence identity leads to an increased uncertainty in functional assignment, especially as the phylogenetic distance between organisms grows, limiting iso-functional ortholog discovery.
As the number of newly sequenced genomes grows larger, more protein sequences are likely to be misannotated, oftentimes resulting in the propagation of incorrect functional annotation across newly identified sequences. To tackle the problem of unannotated or misannotated proteins, newer methods for computational assignment have been created with varying degrees of success (2). Although these methods outperform historical methods, continued improvement is necessary to ensure accurate annotation of function (2). A greater swath of functional space can be covered by screening substrates in a high-throughput manner on multiple enzymes from a family (3, 4). Family-wide substrate profiling offers a data-rich resource. The use of sparse screening of sequence space and a diversified library permits the determination of substrate specificity profiles to provide a family-wide view of the range of substrates and insight into the structure of the prototypical substrate. Where structures are available, correlation between substrate range and structural determinants of specificity can be achieved. In addition, the approach has utility in genomic annotation (inferred function), iso-functional ortholog assignment, and the assignment of in vitro substrate profiles to orphaned PDB entries (enzymes with structure but no function, or SNFs). Here we report the application of in vitro high-throughput functional screening of metabolites and related compounds at the superfamily level. We use as an example prokaryotic members of the haloalkanoic acid dehalogenase superfamily (HADSF), a diverse superfamily of enzymes (5) that catalyze a wide range of reactions involving the formation of a covalent intermediate with an active-site aspartate. Reactions catalyzed by this superfamily include dehalogenation (6) as well as Mg2+-dependent phosphoryltransfer, although the vast majority (∼99%) are phosphotransferases (7). Members of the HADSF share a Rossmannoid fold “core” domain that contains the phosphoryl transfer site (8, 9) and a “cap” domain that provides substrate specificity determinants (10). There are three major types of caps in the HADSF (C0, C1, and C2A/C2B; see SI Appendix, Fig. S1) based on size, position of insert within the Rossmann fold, and overall topology (7). At the time of writing, the HADSF is known to comprise over 120,000 members across the three domains of life with at most 3% associated with an EC identifier (11).
In this study, the functional space of the HADSF was sampled by screening a customized substrate library of 167 compounds against over 200 enzymes from numerous prokaryotic species. The study revealed that a large number of family members show a broad substrate range, with the majority of the HADSF enzymes reacting with five or more substrates. Thus, widespread promiscuity is not incompatible with participation in cell metabolism and may be advantageous to the evolution of new enzyme activities. The activity profiling when applied to putative iso-functional orthologs allowed us to infer annotation for ∼35% of the HADSF. Intriguingly, the HADSF subfamily with the least structural elaboration of the core catalytic Rossmann fold was the most specific with respect to substrate range and number, implying that domain insertions drive the evolution of new functions and that the broad specificity observed in the HADSF may be a relic of this process.
Results
Substrate Library Design and Synthesis.
A structurally diverse library of organophosphate esters and anhydrides was assembled for use in defining the substrate specificity profiles of targeted phosphatases, which together provide comprehensive coverage of the structural and functional diversity of the HADSF. The 167-member library includes 69 compounds derived from commercial sources and 98 synthetic compounds prepared in-house. The majority of the compounds are known metabolites, and most of these are ubiquitous. The universal phosphatase assay reagent para-nitrophenyl phosphate (pNPP) was included in the screen for the purpose of identifying low-level, nonspecific phosphatase activity commonly observed among HADSF phosphomutases, as well as the phosphatases. Other organophosphates that are not presently known to be metabolites yet are structurally related to such were included in the library to further define the elements of substrate specificity while angling for leads for novel metabolites and, hence, novel phosphatase biological function.
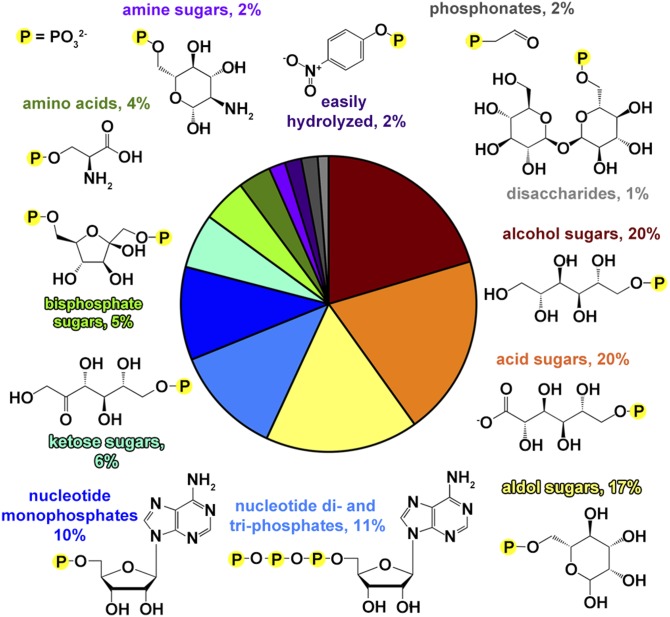
The composition of the library, parsed by designated chemical class, is represented in Fig. 1. The vast majority of the compounds are phosphorylated carbohydrates or carbohydrate derivatives (e.g., nucleotides), thus mirroring the Escherichia coli metabolome, which is heavily weighted toward carbohydrates (12). Comprehensive coverage of three- to seven-carbon phosphorylated d-aldoses and d-ketoses was augmented where possible with the corresponding acid, alcohol, and α-deoxy sugars (2-deoxyaldose and 3-deoxyketose). Secondary metabolites included phosphorylated seven- (d-glycero-d-mannoheptose, “GMH”), eight- (3-deoxy-d-manno-2-octulosonate, “KDO”), and nine-carbon (N-acetylneuraminate, “NeuNAc” and 2-keto-3-deoxy-d-glycero-d-galacto-nonulosonate, “KDN”) sugars. Metabolites trehalose-6-phosphate and sucrose-6-phosphate represent the phosphorylated disaccharides. Naturally occurring pyrimidine, purine, and flavin ribose and/or deoxyribose adducts are well represented. In addition, bis-phosphorylated monosaccharide metabolites were included in the library, as were “unnatural” positional isomers of common phosphorylated monosaccharides (for example, d-glucose-3-phosphate). Phosphorylated amino acids phosphoserine, phosphothreonine, phosphotryosine, and the histidine biosynthetic pathway intermediate histinidol phosphate were included, as were cofactors pyridoxal phosphate, thiamine phosphate, flavin mononucleotide, and CoA. Lipid metabolites phosphoethanolamine, phosphatidylcholine, glycerol-3-phosphate, and prenylphosphate are also represented. Finally, the library phosphonates (exemplified by the HADSF phosphonatase substrate phosphonoacetaldehyde) were used for screening the phosphatases for catalysis of inorganic phosphate formation via hydrolytic P–C bond cleavage.
Fig. 1.
Pie chart showing the chemical composition of the substrate library, separated by designated chemical class of the leaving group. Sections are color-coded with corresponding representative structures and percent occurrence is labeled in a like color.
Despite the biochemical bias of the library toward carbohydrates, at the level of chemical functional groups the library presents sufficient structural diversity to profile the specificity of each phosphatase. In addition to simple visual inspection of the molecular structures, the chemical diversity of the library was analyzed by calculating the Tanimoto coefficient [rapid overlay of chemical structures (ROCS) alignment with electrostatic comparison by EON (www.eyesopen.com)] of the substrate-leaving group (i.e., the PO3 group common to all substrates was removed) (for a recent discussion of the Tanimoto coefficient see ref. 13). The library was found to follow a normal distribution of Tanimoto coefficient (T) values (skewed slightly to higher T values; see SI Appendix, Fig. S2) with an average of T = 1.20 (range 0.23–1.95), showing that the library is diverse, but with an upper quartile of T = 1.42, demonstrating significant similarity of 25% of the substrates, as might be expected from the bias toward sugars.
Validation of the High-Throughput Screen by Determination of Steady-State Kinetic Constants.
The substrate library was screened via an end-point inorganic phosphate-release assay using a commercially available malachite-green reagent. Background phosphate was accounted for and a standard curve of phosphate was calculated; all plates were performed in duplicate. A substrate concentration of 1 mM was selected (Km values range from ca. 0.01–1 mM for HADSF phosphatases) to maximize Michealis complex formation while limiting the background reading to <0.2 ± 0.03 OD units (error calculated from the mean variability between duplicates; see Materials and Methods). The library compounds were incubated with 5 μM enzyme in 50 mM Hepes at pH 7.5 and 25 °C for 30 min.
Compounds were considered to have significant substrate activity if the absorbance of the assay solution, corrected for background absorbance, was greater than 0.2 ± 0.03 OD units. The linear response of the assay ceases at net OD readings >0.7. As a consequence, the assay detects enzyme–substrate pairs having kcat values ≥0.004 s−1 but cannot be used to differentiate between enzyme–substrate pairs having kcat values ≥0.014 s−1. Therefore, in practice, the high-throughput screen (HTS) was used to identify active substrates for a given phosphatase, and then for greater precision kcat and kcat/Km values were determined as needed.
To validate the HTS, we tasked it with identifying potential orthologs among previously uncharacterized phosphatases. Because of the high sequence divergence that is characteristic of the HADSF, sequence-based ortholog assignment is limited to matches between phosphatases derived from closely related bacterial taxa. For example, the HTS results obtained for a previously uncharacterized HADSF member (UniProt: Q7M7U5) from Wolinella succinogenes identified l-phosphoserine as a substrate and kinetic characterization showed good catalytic efficiency with kcat = 40 ± 3 s−1, Km = 250 ± 10 μM, and kcat/Km = 1.6 × 105 M−1⋅s−1 (Table 1). This protein is 24% identical (35% similar) to the SerB ortholog from E. coli (14), which is below the level where these proteins would have been confidently assigned as iso-functional orthologs. Similarly, another four possible iso-functional orthologs of phosphoserines phosphatase were identified by their activity against phosphoserine in the screen and they were subsequently characterized. The kinetic constants showed efficient phosphohydrolase activity against phosphoserine with a range of kcat/Km from 2.9 × 104 M−1⋅s−1 to 5.1 × 106 M−1⋅s−1 (Table 1). (Notably, although certain orthologs had no detectable activity with phosphothreonine some had modest activity against this structurally similar amino acid.) Like the enzyme from W. succinogenes, the sequence identity of these enzymes to the characterized E. coli SerB (13–46%) would have precluded their confident assignment without the screen results.
Table 1.
Steady-state kinetic parameters for selected proteins
| UniProt | Protein name | Substrate | kcat, s−1 | Km, μM | kcat/Km, s−1⋅M−1 |
| H0QCK5 | Putative β-PGM | β-Glucose 1,6-BP | 1.3 ± 0.1 | 11 ± 1 | 1.2 × 106 |
| Q7M7U5 | Putative serB | Phosphoserine | 40 ± 3 | 250 ± 30 | 1.6 × 105 |
| R4UY31 | Putative serB | Phosphoserine | 40 ± 1 | 250 ± 9 | 1.6 × 105 |
| Q9K8N3 | BH2972 protein | Phosphoserine | 50 ± 1 | 1,800 ± 80 | 2.8 × 104 |
| Phosphothreonine | 40 ± 3 | 3,500 ± 80 | 1.1 × 104 | ||
| X3MFA4 | Hydrolase | Phosphoserine | 160 ± 10 | 30 ± 1 | 5.1 × 106 |
| Phosphothreonine | 40 ± 5 | 520 ± 10 | 7.3 × 104 | ||
| C3NVP9 | Phosphoserine phosphatase | Phosphoserine | 160 ± 2 | 440 ± 20 | 3.6 × 105 |
| Phosphothreonine | 640 ± 120 | 21,700 ± 6,600 | 3.0 × 104 | ||
| Q81IN0 | Putative phosphoglycolate phosphatase* | Phosphocholine | 3.1 ± 0.3 | 390 ± 100 | 7.9 × 103 |
| Q8A9J5 | Hypothetical protein | O-phosphotyrosine | 74 ± 5 | 230 ± 20 | 3.2 × 105 |
| Q926W0 | Hypothetical protein | l-xylitol 5-P | 0.130 ± 0.004 | 120 ± 20 | 1.1 × 105 |
| d-mannitol 6-P | 4.0 ± 0.5 | 1,500 ± 600 | 2.7 × 103 | ||
| d-mannitol 1-P | 0.56 ± 0.05 | 290 ± 70 | 1.9 × 103 | ||
| d-allitol 6-P | 0.300 ± 0.001 | 170 ± 30 | 1.7 × 103 | ||
| 2′-deoxy 6-phosphoglucitol | 0.9 ± 0.1 | 800 ± 200 | 1.1 × 103 |
This enzyme did not catalyze hydrolysis of phosphoglycolate; data shown are the result of three measurements, error is the SD.
The next protein chosen for validation was annotated as β-phosphoglucomutase (β-PGM, UniProt: H0QCK5) from E. coli K12. This protein is 43% identical (61% similar) to β-PGM from Lactobacillus lactis (UniProt: P71447) studied previously (15), a mutase catalyzing the interconversion of d-glucose 6-phosphate to β-d-glucose 1-phosphate through phosphatase activity on the intermediate of the reaction β-d-glucose 1,6-bisphosphate. High-throughput screening of H0QCK5 revealed the expected phosphatase activity against β-d-glucose 1,6-bisphosphate. Further examination using a mutase assay confirmed H0QCK5 to have kinetic parameters similar to those of the known enzyme from L. lactis (15) (kcat/Km = 7.1 × 106 M−1⋅s−1 and 3.6 × 106 M−1⋅s−1, respectively) (Table 1).
Another approach to validation was to test the identified substrates for selected low- and mid-efficiency enzymes and show that the enzyme–substrate pairs do have the activity indicated by the screen. Q81IN0 was shown by the screen to have a hit against phosphocholine but not phosphoglycolate (although it was annotated as a phosphoglycolate phosphatase); the enzyme was indeed shown to take phosphocholine with low efficiency (kcat/Km = 7.9 × 103 M−1⋅s−1) but not phosphoglycolate (Table 1). Q926W0 is a second example with the various sugar substrates that showed as positive hits in the substrate screen, also showing good to moderate activity as assessed by steady-state kinetic constants with a kcat/Km = 1.1 × 105 M−1⋅s−1 for l-xylitol 5-phosphate and values in the range of ∼103 M−1⋅s−1 for d-mannitol 1-phosphate, d-mannitol 6-phosphate, d-allitol 6-phosphate, and 2′-deoxy 6-phosphoglucitol (Table 1). Overall, the determination of steady-state kinetic constants for individual enzymes supported the validity of the screen in identifying substrates.
Selection and Assay of HADSF Targets.
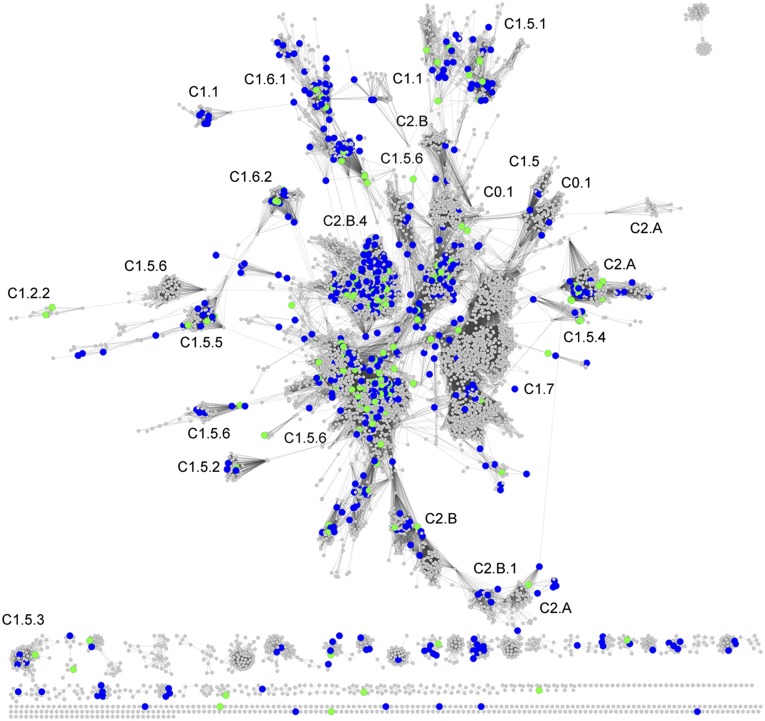
The goal of target selection was to provide a comprehensive sampling of the prokaryotic HAD phosphatase family for function prediction/validation. It is not known how many unique functions are represented by the bacterial/archeal HAD phosphatase sequences. The number of HAD phosphatases varies from one prokaryotic species to another (e.g., Thermatogae species 6–14, Cyanobacteria 11–36, Enterobacteriales 3–35, Actinobacteria 17–45, Methanocaldococcus 4–7, Archaeoglobus 8–12, Thermococcus 15–20, and Pyrobaculum 8–15). Furthermore, even closely related species might share some HAD phosphatases in common while differing in others. E. coli K12 and Salmonella typhimurium LT2 share 25 HAD phosphatases with >70% sequence identity, yet S. typhimurium possesses four HAD phosphatases that are not present in E. coli. Thus, it was critical to select from a broad range of bacteria/archea but also to sample broadly within each organism. The sequence-similarity network (SSN) previously described by our laboratories (16) was used to select targets for functional analysis. The clustering of the network leads to selection of orthologs by sampling within the clusters and selection of diverse functions by sampling across clusters. Briefly, the network comprises 79,778 prokaryotic HADSF sequences from the Structure-Function Linkage Database (SFLD) (17) (Fig. 2) that are organized into representative SSNs (18). In the network, HADSF sequences with greater than 40% identity are organized into metanodes and connected by edges (lines) with BLAST e-value threshold of 10−20 in Cytoscape (Fig. 2).
Fig. 2.
Representative SSN of the HADSF; 79,778 HADSF sequences from the SFLD organized into representative SSNs. HADSF sequences with greater than 40% identity are organized into metanodes and connected by edges with BLAST e-value threshold of 10−20 in Cytoscape. Metanodes with sequences targeted for functional testing are colored blue and those that were successfully screened are colored green. SFLD subgroup clusters are annotated.
Sorting by cap type was also performed to ensure structural and functional diversity because the cap domains vary the most in topology among family members and provide the specificity determinants for substrate binding (7, 10). The protein sequences in the network were assigned cap types by comparing sequences with a standard set of HADSF structures using the program CapPredictor (11). In total, 1,319 prokaryotic targets across 401 species were selected for protein production and functional assessment, ensuring that the targets were distributed over cap type, sequence, and structure space using the network (Dataset S1, supplemental spreadsheet 2). The targets were selected in proportion to the naturally occurring distribution of cap type from E. coli, that is, 12% C0, 63% C1, and 25% C2 (4).
Each target that was successfully expressed and purified was assessed for activity against the library. Of the proteins targeted, 217 proteins (17% success rate, a typical success rate for structural genomics consortiums) from 86 species (between 1 and 16 HAD enzymes per organism) were assayed, resulting in a final distribution of cap type of 15% C0, 63% C1, and 21% C2. The results identified enzymes that were specific (or showed limited substrate ambiguity) and that showed either moderate or a high level of ambiguity (Fig. 3). Orthologs of known enzymes were revealed in all these categories. Of the 217 enzymes tested, 13 showed no reactivity against any member of the screening library. Of these, eight localize to a cluster with previously characterized dehalogenases on the SSN (the screen is for phosphatase activity, not dehalogense activity as phosphate release is detected). Of the remaining five, three were from thermophilic organisms and would not be expected to show significant activity at 25 °C. The other inactive proteins may have been unstable in the screening conditions. (The screen for one of the enzymes from a thermophile, UniProt: O29873, was run manually at 75 °C; at this temperature activity was detected for sugar and some nucleotide substrates.) The number of substrates recognized by the remaining 204 enzymes was distributed with a median of 15.5 substrates per enzyme and with a 25th percentile of 5 hits and a 75th percentile of 41 hits. Based on this distribution, we consider an enzyme to be specific if it used 5 substrates or fewer, to show moderate substrate ambiguity if it used between 6 and 40 substrates, and to show high substrate ambiguity if it used greater than 41 substrates. Notably, because the screen uses an end-point assay, it is possible that some enzymes identified as having a broad substrate range actually have much greater catalytic efficiency (as reflected in kcat/Km) for a small number of the identified substrates than for the other identified substrates. Thus, those HAD members identified as having a broad substrate range refers to all possible substrates against which the enzyme bears phosphohydrolase activity. This is because even relatively modest in vivo activity of an enzyme against a substrate could be responsible for physiologically significant metabolism (19). The activities seen in the screening can be attributed solely to the enzyme being screened (rather than to contaminating endogenous E. coli enzymes or to general acid/base catalysis alone). We conclude this because a number of enzymes screened were very specific or showed no activity.
Fig. 3.
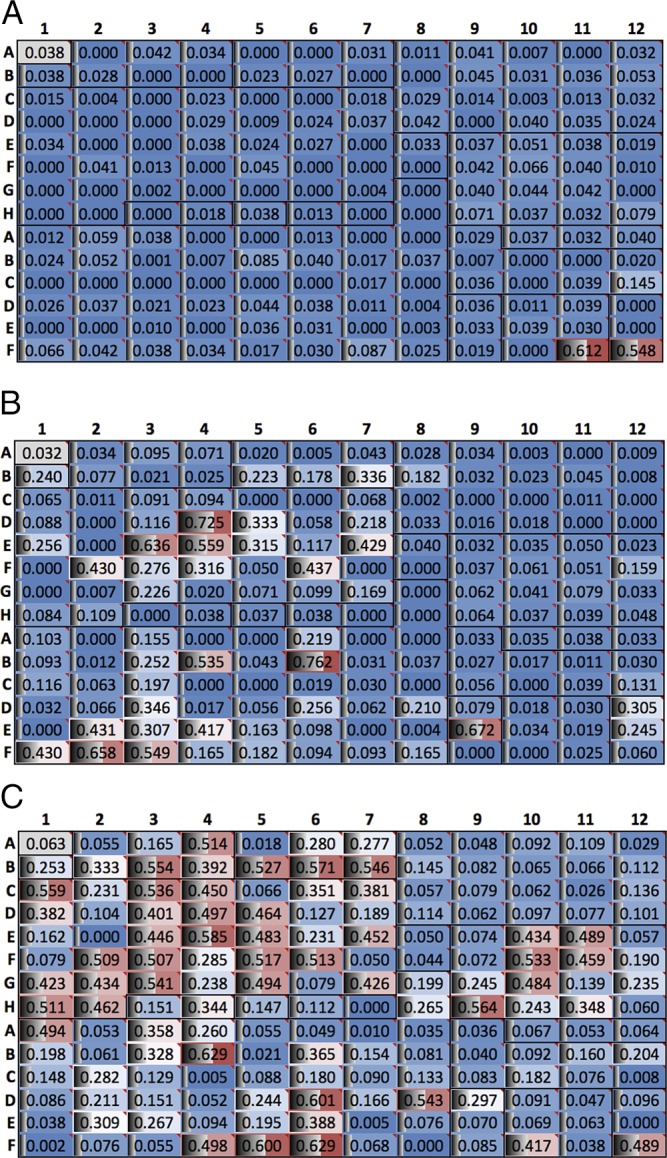
Data from screening of the HADSF. (A) HTS results for specific HAD member (UniProt: Q8A9J5) from B. thetaiotaomicron. Plates show absorbance data, with hits giving higher absorbance readings (red color). Each well is also commented with substrate name (see Dataset S2). (B) HTS results for a moderately ambiguous HAD member (UniProt: X3MXD2) from S. enterica. (C) HTS results for a highly ambiguous HAD member (UniProt: Q88LM3) from P. putida.
Enzymes with Restricted Substrate Range.
Fifty-three enzymes were revealed to be narrow in substrate range, acting on five or fewer substrates. These included orthologs of the previously known enzymes phosphoserine phosphatase, histidinol phosphate phosphatase, β-phosphoglucomutase, d-glycero-d-manno-heptose-1,7(bis)phosphate phosphatase (GmhB), and phosphatases against pyrophosphate, FMN, 2-keto-3-deoxy-9-phosphonononic acid, 2-keto-3-deoxy-8-phospho-octulosonate, phosphoglycolate, and trehalose 6-phosphate. For instance, GmhB has been well characterized in E. coli (20), where the enzyme has high substrate efficiency with the β anomer of d-glycero-d-manno-heptose-1,7-(bis)phosphate (kcat/Km = 7.1 × 106 M−1⋅s−1) (20) and shows discrimination (100-fold) against the α anomer; a range of efficiency ratios between the two anomers is observed (0.17- to 150-fold) depending on the species. Here, we screened GmhB from Pseudomonas putida (UniProt: Q88RS0) and found the enzyme dephosphorylated five substrates including d-glycero-d-manno-α-heptose-1,7(bis)phosphate, fructose 1,6-(bis)phosphate, d-allitol 6-phosphate, 2-deoxy-d-manno-2-octulosonate-8-phosphate, and d-glycero-d-manno-β-heptose-1,7-(bis)phosphate. Anomeric specificity was determined for GmhB orthologs from Vibrio cholerae (UniProt: G7TNF0), Rhodopseudomonas paulstris (UniProt: Q6N2R1), Haemophilus influenza (UniProt: C9MJX1), and Salmonella enterica (UniProt: B5Q1D3). Specific, but previously undescribed, activities were shown against phosphocholine (UniProt: Q81IN0; kcat/Km = 8.0 × 103 M−1⋅s−1), phosphotyrosine (UniProt: Q8A9J5; kcat/Km = 3.2 × 105), bis-phosphorylated compounds (UniProt: Q8DV60), and sugars (UniProt: Q88LY7) (Table 1).
Enzymes with Broad Substrate Range.
A large number of enzymes were found to be moderately ambiguous (101 total), acting on 6–40 substrates. These included enzymes that used five or six carbon sugars only (UniProt: X3NXD2) and 5′-nucleotide monophosphates (UniProt: Q5LGR4), 5′-nucleotide monophosphates (UniProt: Q8P4B4); two to eight carbon sugars, 5′-nucleotide monophosphates, phosphoethanolamine/histidinol-phosphate, and amino acids (UniProt: C7RF86); eight or nine carbon sugars, NADP, FMN, CoA, and 5′-nucleotide monophosphates (UniProt: J7JPC0); and two to six carbon acid sugars (Uniprot: Q049B0).
The remaining 50 enzymes catalyzed the hydrolysis of phosphate from more than 41 substrates (and up to 143 substrates) and were therefore considered to possess a broad substrate range. Notably, none of the proteins screened was predicted to be exported to the periplasm, so in the less specific members periplasmic localization was not the mechanism of substrate selectivity. Enzymes in this class used a broad range of sugars (UniProt: H0QFB3), five or six carbon sugars (UniProt: F0N5C3), five or six carbon sugars and 5′-nucleotide monophosphates (UniProt: Q88LM3), all classes of substrates (UniProt: B5QAY6), and all classes of substrates with the exception of 5′-nucleotide di- and triphosphates (UniProt: M1UCG8). As is true for the specific enzymes, enzymes in this group included iso-functional orthologs of previously characterized enzymes. One such enzyme, NagD, with substrate activity defined by the screen for UMP and GMP, has been previously characterized to be a housekeeping enzyme (showing broad substrate range) involved in ADP ribose catabolism and the recycling of cell-wall metabolites (21).
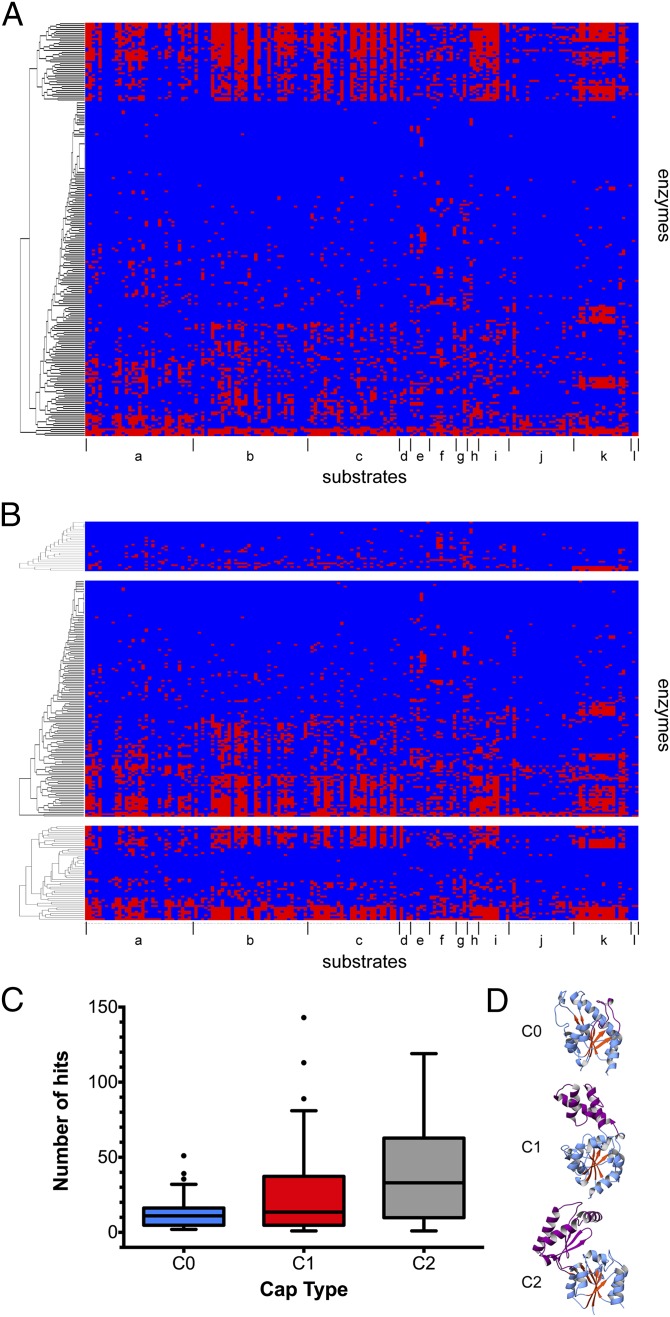
To examine the activity across the targets, the functional profiles were visualized using heat maps. The results were clustered by functional profile (Fig. 4) as well as by cap type. As described, the HADSF can be subcategorized into C0, C1, and C2A/C2B classes by the domain inserted in the Rossmann fold (see SI Appendix, Fig. S1). Bioinformatics analyses suggest the last universal common ancestor contained the progenitors for all cap types and diversification and expansion of substrate specificities probably evolved later (7, 11). To assess any correlation between substrate specificity and cap type, functional profiles were clustered based on their corresponding cap type (11) (Fig. 4B). As discussed, 75% of the enzymes tested were found to be ambiguous in their substrate preference. Intriguingly, this substrate ambiguity was dominated by HADSF members with C1 or C2 cap types (Fig. 4C). C0 members were more specific for their corresponding substrates with an average substrate count of 13.3 vs. 24.9 and 39.1 (see SI Appendix, Table S2) for C1 and C2 members, respectively. Under the null hypothesis that the median number of substrate “hits” of all groups are equal, we performed the Kruskal–Wallis test to determine the statistical significance of these differences. Our data result in χ2 = 11.6621 with a P value of 0.002935, establishing statistical significance and suggesting the three groups have markedly different means.
Fig. 4.
Heat maps showing activity profiles for screened enzymes. Each row represents a screened enzyme and each column a specific substrate. (A) Enzymes hierarchically clustered by substrate profile and (B) clustered by cap type C0, C1, and C2 (top to bottom). Substrates are clustered together and marked (a, acid sugars; b, alcohol sugars; c, aldolase sugars; d, amine sugars; e, amino acids; f, bisphosphate sugars; g, disaccharides; h, easily hydrolyzed; i, ketose sugars; j, nucleotide di- and triphosphates; k, nucleotide monophosphates; l, phosphonates). (C) Boxplot of number of hits per enzyme arranged by cap type shows C1 and C2 members act upon more substrates than C0 members. (D) Inset depicting C0, C1, and C2 members (PDB ID codes: 3L8E, 1ZOL, and 2FUC).
The question might be raised as to whether the C0 members are simply less efficient enzymes and therefore the lower limit set for observed activity against a substrate should be different for this class. However, a sampling of kcat/Km values from this analysis (Table 1; kcat/Km = 1.8 × 106 M−1⋅s−1 for Q88RS0 d-glycero-d-manno-heptose 1β,7-bisphosphate against d-glycero-β-d-manno 1,7-bisphosphate) and from the literature shows that the C0 members are often moderately to highly efficient with kcat/Km ∼104–107 M−1⋅s−1 (20, 22–24), similar to those of C1 and C2 enzymes (21, 22, 25–27). Overall, the finding that C0 members, which have the most minimal cap inserts, are equally efficient and more specific than the other subfamilies suggests that incorporation of additional domains leads to the expansion of the substrate repertoire in the HADSF.
To analyze the activity profiles of enzymes with a narrow versus a moderate or a very broad substrate range, the distribution of chemical similarity using the Tanimoto coefficient (T) for all substrate hits for each group of enzymes was compared against all hits and was compared against the distribution of T values for the entire library (see SI Appendix, Fig. S2). Overall, no significant difference was observed between the substrate sets of the narrow range versus the broad range enzymes based on chemical diversity in terms of shape and electrostatics. Given the range of substrate diversity present in our library (and reported by T) ambiguous and specific enzymes could not be distinguished by the structure and electrostatic profiles of the substrates alone. It can be inferred that the structural basis of substrate specificity versus substrate ambiguity requires information about the interaction of enzyme and substrate and cannot be based solely on substrate structure.
Functional Discovery.
An important consequence of screening large numbers of enzymes is the identification of new functions not observed previously in the HADSF. Screening of an enzyme previously annotated as a hypothetical protein from Listeria innocua (UniProt: Q926W0) revealed specificity for five or six carbon alcohol sugars, with the highest activities for d-mannitol 1-phosphate, d-mannitol 6-phosphate, and l-xylitol 5-phosphate (kcat/Km = 1.9 ×103, 2.7 ×103, and 1.1 ×105 M−1⋅s−1, respectively, Table 1). Examination of the genome neighborhood revealed the presence of several conserved proteins involved in carbohydrate metabolic processes (mannitol-specific enzyme IIA, mannitol-specific enzyme IIBC, ROK transcription factor, and an MviM sugar dehydrogenase). These proteins are part of a versatile phophoenolpyruvate:carbohydrate phosphotransferases system (PTS) (28) that has evolved to allow microorganisms to use diverse substrates available in ever-changing environments. These PTS systems transfer a phosphate from phosphoenolpyruvate to a carbohydrate during import (28). PTS systems have been shown to be important in regulating carbohydrate flux in bacterial cells (29) including enteroinvasive E. coli and S. enterica serovar Typhimurium. In this particular system, the HADSF member likely dephosphorylates the imported xylitol, mannitol, or similar phosphosugar for metabolic utilization, although high activity with other alcohol sugars may point to substrate variability in this PTS system. Further work is necessary to fully elucidate the role of this hydrolase in the PTS system.
Included among the targets selected for screening were enzymes with SNFs. Currently, there are ∼150 nonredundant protein structures from the HADSF in the PDB; roughly half of these are SNFs. One such enzyme, BT0820 from Bacteroides thetaiotaomicron (UniProt: Q8A9J5), was screened and found to have activity against phosphotyrosine. The structure (PDB ID code: 2OBB) revealed a short C0 cap insert and crystal packing and standardized size-exclusion chromatography performed herein was consistent with a monomeric protein. HADSF enzymes with C0 cap types have been associated with histidine biosynthesis (30), inner core lipopolysaccharide biosynthesis (20), glycolipid and glycoprotein decoration for cell surface display (31, 32), tRNA repair (33), and phosphoprotein phosphatase activity (23, 34). As is the case with Q8A9J5 the HADSF polynucleotide phosphatases and phosphoprotein phosphatases are monomeric [in published cases Fcp1, Scp1, dullard, MDP-1, and T4 polynucleotide phosphatase/kinase (23, 33–36)], apparently allowing recognition of large polymeric (i.e., protein and nucleic acid) substrates. Steady-state kinetic assay of Q8A9J5 revealed specificity for phosphotyrosine with kcat/Km = 3.2 × 105 M−1⋅s−1. The shorter C0 cap, monomeric nature, and phosphotyrosine phosphatase activity point to a role as a putative phosphoprotein phosphatase, although further work is necessary to identify the protein/macromolecular substrate.
Inferred Functional Annotation.
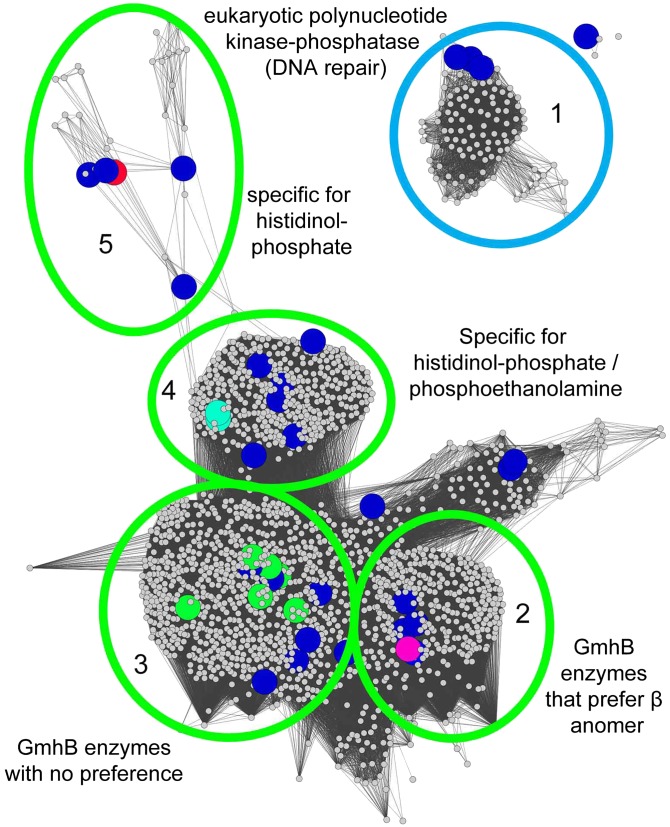
The data were manually assessed to ascribe in vitro function by identifying the substrate “hits” for each enzyme (e.g., specific for phosphoserine, ambiguous for five or six carbon sugars, broadly ambiguous, etc.). The resulting annotations were mapped onto the full SSNs for each of the subfamilies listed in the SFLD (C0.1, C1.1, etc.) with the exception of the ATPases (C1.7) (Dataset S1, supplemental spreadsheet 3, Fig. 5, and SI Appendix, Figs. S3–S21). Separated clusters are expected to be isofunctional and annotations are made by propagation of the assignments from screened enzymes to the subcluster in which they reside, as has been described for the proline racemase superfamily (37). For example, the C1.5.5 subfamily consisting of 1,994 members was previously broadly annotated as a subfamily of heptose bisphosphate phosphate-like phosphatases. To examine this subfamily in greater detail, a full SSN was constructed with a BLAST e-value threshold of 10−25 (Fig. 5). Mapping of the HTS data to the subfamily revealed a range of substrate utilization across the subfamily including d-glycero-d-manno-α-heptose 1,7-(bis)phosphate, d-glycero-d-manno-β-heptose 1,7-(bis)phosphate, phosphoethanolamine/histidinol-phosphate, and histidinol-phosphate. A cluster was also present that contained a previously characterized polynucleotide kinase/phosphatase involved in DNA repair (Fig. 5). In this fashion putative substrates could be extrapolated to most clusters in the HADSF. Whereas 4,884 enzymes could be annotated by our manual propagation of assignments from structure–function studies reported in the literature to their corresponding cluster in the expanded SSN (Fig. 4 and SI Appendix, Figs. S3–S21), the function of a total of 27,392 enzymes are now inferred based on propagation of our HTS results together with those from the literature (Dataset S1, supplemental spreadsheet 3).
Fig. 5.
Functional Annotation of the HADSF by SSN expansion. Individual SFLD subgroups were expanded to annotate function based on either high-throughput screening or information from previously characterized enzymes. Clusters colored blue were EFI targets that were not screened. The remaining colors represent the annotations below. Five clusters of subgroup 1.5.5 were annotated as follows: (1) eukaryotic polynucleotide kinase/phosphatases involved in DNA repair, (2) GmhB enzymes that prefer the alpha anomer of d-d-heptose-1,7-bisphosphate, (3) GmhB enzymes that take either the alpha or beta anomer, (4) bifunctional HisB enzymes that show specificity for histidinol-phosphate or phosphoethanolamine, and (5) enzymes that are highly specific for histidinol-phosphate. The annotation list is available in Dataset S2.
Substrate Utilization in the HADSF.
To examine the propensity of HADSF members for hydrolyzing the substrates in the library, the frequency of each substrate being considered a “hit” was computed. Markedly, pNPP was among the most common substrates (ranked eighth) and was used by 30.4% of the enzymes tested. This is instructive, given the fact that pNPP is commonly used to test for generic phosphatase activity. The four most commonly used substrates in the screen were 2′-deoxy 6-phosphoglucitol, 2′-deoxyribitol 5-phosphate, 2′-deoxyribose 5-phosphate, and ribitol 5-phosphate (used by 33.6, 32.3, 31.8, and 31.3% of all enzymes tested, respectively, Table 2). If the classes are ranked, the most commonly dephosphorylated substrate classes were two to six carbon sugars and 5′-nucleotide monophosphates. Notably the most common substrates were neither the most chemically reactive in the library, nor did they possess structural features that make the phosphoryl groups more accessible to the phosphatases.
Table 2.
Substrate utilization in the HADSF
| Most commonly used substrates | Least commonly used substrates | ||
| Substrate | % using | Substrate | % using |
| 2-deoxy 6-phosphoglucitol | 33.6 | CTP | 3.7 |
| d-2-deoxyribitol 5-P | 32.3 | CoA | 3.7 |
| 2-deoxyribose 5-P | 31.8 | d-mannitol 5-P | 3.2 |
| d-ribitol 5-P | 31.3 | d-xylonate 3-P | 3.2 |
| Acetyl phosphate | 30.9 | d-gluconate 3-P | 3.2 |
| d-3-deoxygluconate 6-P | 30.4 | d-mannose 2-P | 3.2 |
| Glycerol phosphate | 30.4 | Inosine 5′-diphosphate | 3.2 |
| Paranitrophenyl phosphate | 30.4 | Guanosine 5′-triphosphate | 3.2 |
| l-xylitol 5-P | 30.0 | l-lyxose 5-P | 2.8 |
| d-sorbitol 1-P | 30.0 | d-glucuronate 5-P | 2.8 |
| Imidodiphosphate | 29.5 | Pyrophosphate | 2.8 |
| d-3-deoxyglucose 6-P | 29.0 | Thymidine 5′-triphosphate | 2.8 |
| l-arabitol 1-P | 28.6 | l-xylitol 3-P | 2.3 |
| d-lyxose 5-P | 28.6 | l-xylonate 3-P | 2.3 |
| Riboflavin 5-P (FMN) | 28.1 | l-glucitol 3-P | 2.3 |
| l-xylonate-5-P | 27.6 | d-glucose 3-P | 2.3 |
| Arabinose 5-P | 27.2 | d-mannitol 2-P | 2.3 |
| d-3-deoxysorbitol 6-P | 27.2 | l-glucose 3-P | 2.3 |
| Meso-erythritol 4-P | 27.2 | d-glucitol 3-P | 1.8 |
| 2-deoxy-d-glucose 6-P | 26.7 | d-galactonate 6-P | 1.8 |
| Pyridoxal 5-P | 26.7 | d-arabitol 1-P | 1.8 |
| d-mannitol 6-P | 25.8 | dl-2-amino-3-phosphonopropionic acid | 1.8 |
| d-allitol 6-P | 25.8 | Phosphonoformic acid | 1.8 |
| d-2-deoxyribonate 5-P | 25.3 | d-allonate 3-P | 1.4 |
| d-ribonate 5-P | 25.3 | l-gluconate 3-P | 0.9 |
Discussion
Systematic study of function across enzyme families has only recently been attempted and has not previously been applied to a representative sampling of an entire superfamily on this scale. A small, uncharacterized enzyme family, DUF849, was studied by Bastard et al. (3) using an integrated strategy for evaluating functional diversity. Based on screening of a set of representative enzymes (124 out of 900) against 17 putative substrates this study found that the family generally catalyzes the condensation of a β-keto acid with acetyl-CoA producing acetoacetate and CoA ester. Widespread catalytic promiscuity was described in the metallo-β-lactamase superfamily through the analyses of 24 enzymes against 10 catalytically distinct hydrolytic reactions where enzymes catalyzed on average 1.5 reactions in addition to their native activity (38). In an orthogonal study, screening of all 23 soluble HADSF members within a single species (E. coli) was performed with a set of 80 commercially available phosphorylated substrates (4). The results annotated the E. coli enzymes and demonstrated that HADSF members possess the capacity to hydrolyze a wide range of phosphorylated compounds, with 19 of the 29 enzymes tested showing a broad substrate range.
Our findings demonstrate, through a representative sampling of prokaryotic organisms in the HADSF, that the degree of substrate ambiguity is great and ubiquitous. This leads to the question, How might the presence of enzymes with broad substrate range contribute to the fitness of individual organisms and to complex microbial communities? Accumulation of metabolites may disrupt metabolic flux or metabolites themselves may prove toxic to organisms (39). Broad substrate overlap between HADSF members may provide a safeguard against such deleterious metabolic effects. Overlap in function may serve to provide sufficient turnover (without optimizing efficiency of any one enzyme) to metabolize large pools of similar substrates. Substrate overlap may also aid in the evolution of new metabolic function in response to environmental challenges. Promiscuity increases flexibility during evolution in response to changes in the substrate pool. During evolution, if there is a variation in the substrate pool, it is more facile for mutations to change the substrate specificity spectrum of broad, overlapping enzymes—that is, divergence before gene duplication (40, 41).
The finding that HADSF members with a minimal or no cap insertion (type C0) are more specific than those with domain insertions (types C1, C2A, or C2B, Fig. 4 B and C) is surprising because the cap domain provides the specificity determinants in the HADSF (10). The expectation might be that in C0 members there are few enzyme residues that interact with the substrate-leaving group and thus the leaving group could vary in size, shape, and electrostatic surface, producing an enzyme with broad specificity. However, an alternative model explains the observation made here. The substrate-leaving group structure may be limited by the necessity of acting as a barrier between bulk solvent and the active-site Mg2+ cofactor, nucleophilic Asp, and Asp general acid/base, replacing the cap domain in this role. Indeed, structures of the C0 HADSF members Scp-1 (35) and GmhB (42) show that the fit between enzyme and substrate does not allow a probe the size of water to enter the active site (see SI Appendix, Fig. S22). Conversely, in enzymes with cap domains there is an enclosed active site and a number of enzyme residues from the cap specificity loop(s) are available to make interactions with different substrates, allowing evolution of activity against multiple substrates. Additionally, the mobility of the cap domain with respect to the core domain may allow access to different substrates of varying size.
The concept that domain insertions provide the raw material for the evolution of specificity determinants (including the residues that allow substrate ambiguity) is supported by recent findings. The directed evolution and bioinformatics analysis of a lactamase supports the model that enzymes are more capable of catalyzing multiple reactions when the active site is composed of loops juxtaposed to, but separate from, the core scaffold (43). Domain insertion can be considered an extreme example of this observation. The facilitation of substrate ambiguity via domain insertion can be a widespread phenomenon in protein evolution, because it has been estimated that ∼10% of insertion events are domain insertions (44). Overall, our findings are consistent with the concept that domain insertions act to drive the evolution of new functions. The widespread substrate ambiguity in the C1- and C2-type HADSF members observed herein may be a vestige of the evolutionary process. Previous work has shown that in the process of in vitro evolution selection pressure toward increasing one activity results in “generalists” that show multiple activities that were not selected for (45).
Materials and Methods
Cloning.
Genes of interest were amplified from genomic DNA and PCR performed using KOD Hot Start DNA Polymerase (Novagen). The conditions were 2 min at 95 °C, followed by 40 cycles of 30 s at 95 °C, 30 s at 66 °C, and 30 s at 72 °C. The amplified fragments were cloned into either a pET-28–based N-terminal TEV cleavable 6x-His-tag–containing vector, pNIC28-Bsa4, or a pET-30–based C-terminal TEV-cleavable StrepII-6xHis-tag vector, CHS30, by ligation-independent cloning (46).
Expression and Purification of Targets in pNIC-Bsa4.
pNIC28-Bsa4 vector containing the insert of interest was transformed into BL21(DE3) E. coli containing the pRIL plasmid (Stratagene) and used to inoculate an overnight culture containing 25 μg/mL kanamycin and 34 μg/mL chloramphenicol. The culture was allowed to grow overnight at 37 °C in a shaking incubator. The overnight culture (1 mL) was used to inoculate 1 L of PASM-5052 auto-induction media (47) containing 100 μg/mL kanamycin and 34 μg/mL chloramphenicol. The culture was placed in a LEX48 airlift fermenter and incubated at 37 °C for 4 h and then at 22 °C overnight. The culture was harvested and pelleted by centrifugation and stored at −80 °C until time of use.
Cells were suspended in lysis buffer [20 mM Hepes (pH 7.5), 500 mM NaCl, 20 mM imidazole, and 10% (vol/vol) glycerol] and lysed by sonication. The lysate was clarified by centrifugation at 35,000 × g for 30 min. Clarified lysate was loaded onto an ÄKTAxpress FPLC (GE Healthcare). Lysate was loaded onto a 1 mL HisTrap FF column (GE Healthcare), washed with 10 column volumes of lysis buffer, and eluted in buffer containing 20 mM Hepes (pH 7.5), 500 mM NaCl, 500 mM imidazole, and 10% glycerol. The purified sample was loaded onto a HiLoad S200 16/60 PR gel filtration column that was equilibrated with SECB buffer [20 mM Hepes (pH 7.5), 150 mM NaCl, 10% (vol/vol) glycerol, and 5 mM DTT]. Peak fractions were collected and protein was analyzed by SDS/PAGE, snap-frozen in liquid nitrogen, and stored at −80 °C (48, 49).
Expression and Purification of Targets in CHS30.
CHS30 vector containing the insert of interest was transformed into BL21(DE3) E. coli containing the pRIL plasmid (Stratagene) and used to inoculate an overnight culture containing 25 μg/mL kanamycin and 34 μg/mL chloramphenicol. The culture was allowed to grow overnight at 37 °C in a shaking incubator. The overnight culture (1 mL) was used to inoculate 1 L of PASM-5052 auto-induction media (47) containing 100 μg/mL kanamycin and 34 μg/mL chloramphenicol. The culture was placed in a LEX48 airlift fermenter and incubated at 37 °C for 4 h and then at 22 °C overnight. The culture was harvested and pelleted by centrifugation and stored at −80 °C until time of use.
Cells were suspended in lysis buffer [20 mM Hepes (pH 7.5), 500 mM NaCl, 20 mM imidazole, and 10% (vol/vol) glycerol] and lysed by sonication. The lysate was clarified by centrifugation at 35,000 × g for 30 min. Clarified lysate was loaded onto an ÄKTAxpress FPLC (GE Healthcare). Lysate was loaded onto a 5-mL Strep-Tactin column (IBA), washed with five column volumes of lysis buffer, and eluted in StrepB buffer [20 mM Hepes (pH 7.5), 500 mM NaCl, 20 mM imidazole, 10% (vol/vol) glycerol, and 2.5 mM desthiobiotin]. The eluent was loaded onto a 1-mL HisTrap FF column (GE Healthcare), washed with 10 column volumes of lysis buffer, and eluted in buffer containing 20 mM Hepes (pH 7.5), 500 mM NaCl, 500 mM imidazole, and 10% (vol/vol) glycerol. The purified sample was loaded onto a HiLoad S200 16/60 PR gel filtration column that was equilibrated with SECB buffer [20 mM Hepes (pH 7.5), 150 mM NaCl, 10% (vol/vol) glycerol, and 5 mM DTT]. Peak fractions were collected and protein was analyzed by SDS/PAGE, snap-frozen in liquid nitrogen, and stored at −80 °C (48, 49).
Library Synthesis.
The structures of the sugar phosphate substrates subjected to screening are given in SI Appendix, Tables S2–S6 and the syntheses of these substances are briefly described below and in detail in SI Appendix and SI Appendix, Schemes S1–S25. The spectroscopic properties of previously prepared sugar phosphates (references given below) match those of the substances synthesized in this effort. All sugar phosphates that have not been prepared previously were found to have spectroscopic properties that matched those expected for their structures.
Synthesis using kinase-catalyzed reactions and ATP regeneration (sugar phosphates 1–7, SI Appendix, Scheme S1).
Many chemical methods for the preparation of phosphate esters of sugars have been described (50–55). However, most rely on the use of sophisticated protecting group protocols that enable selective phosphorylation of a specific hydroxyl group and, as a result, they lack generality. In contrast, enzyme catalyzed reactions, especially those promoted by kinases, can be used to generate sugar phosphates in a direct manner and without the need for protective group manipulations. Nevertheless, two main limitations of kinase catalyzed phosphorylation reactions do exist when the goal is to produce targets in large quantities. One is associated with the narrow substrate scope of kinases and the other is that high concentrations of substrates, products, and cofactors are not readily tolerated in these enzymatic processes. However, kinase-promoted reactions can be adapted to large-scale synthesis when cofactors (e.g., ATP) are regenerated in the reaction mixtures (56–59). We have developed a general method for conducting kinase-catalyzed reactions that directly transform sugars to their monophosphate analogs. The procedure takes advantage of ATP regeneration through reaction of the product ADP with phosphoenol pyruvate catalyzed by pyruvate kinase. Processes based on this strategy, outlined in SI Appendix, Scheme S1, were used to prepare sugar phosphates 1–7.
Synthesis by combined use of kinase-catalyzed and other enzyme-promoted reactions (sugar phosphates 8–20, SI Appendix, Schemes S2–S4).
Under many circumstances, the starting sugar scaffolds themselves are difficult to prepare. However, other relevant enzymatic reactions can be coupled with kinase-promoted phosphorylation to produce target sugar phosphates. Processes based on this strategy, outlined in SI Appendix, Scheme S2, were used to prepare sugar phosphates 8–12. Several procedures have been described for the synthesis of KDPG (12) using the aldolase-promoted reaction of pyruvate and d-glyceraldehyde-3-phosphatehosphate (60–62). However, the 6-phosphogluconate dehydratase catalyzed reaction of d-gluconate-6-phosphate (SI Appendix, Scheme S3) serves as a more efficient (80%) method to prepare this sugar phosphate (63). In addition, sugar phosphates 13–15 were synthesized by the processes displayed in SI Appendix, Scheme S3. Finally, sequences were developed (SI Appendix, Scheme S4) to prepare the sugar phosphates 16–20.
Chemical synthesis of rare five-carbon sugar phosphates (sugar phosphates 21–27, SI Appendix, Schemes S5–S11).
Not all sugar phosphates can be prepared using enzymatic phosphoryl-transfer reactions. To remedy this problem, synthetic approaches relying purely on organic reactions were used to prepare a series of rare, five-carbon sugar phosphates. By using these approaches, outlined in SI Appendix, Schemes S5–S11, we prepared sugar phosphates 21–27.
Utilization of organic chemical methods (sugar phosphates 28–34, SI Appendix, Schemes S12–S18).
Chemical approaches (SI Appendix, Schemes S12–S18) were used to prepare sugar phosphates 28–34.
Simple chemical transformations for synthesis (sugar phosphates 35–65, SI Appendix, Schemes S19–S21).
The library was expanded to include alditol and aldonic acid analogs 35–61 that were prepared by performing bromine oxidation (64) and sodium borohydride reduction (65) reactions of aldehyde moieties of sugar phosphates (SI Appendix, Schemes S19 and S20). In addition, aldonic acid phosphate 45, 47, and 62–65 were prepared by performing oxidation reactions on basic solutions of 2-keto-sugar phosphates under an oxygen atmosphere (SI Appendix, Scheme S21) (66).
Synthesis using a variety of methods (sugar phosphates 66–97, SI Appendix, Schemes S22–S25).
Sugar phosphates 66–97 were prepared using a variety of the methods described and outlined in SI Appendix, Schemes S22–S24.
Target Screening.
A library was prepared consisting of 167 commercially sourced or synthesized phosphorylated compounds encompassing 16 substrate classes including sugars (mono- and bis-phosphates), nucleotides (mono, di-, and triphosphates), disaccharides, phosphonates, amino acids, and amino sugars (Dataset S1, supplementary spreadsheet 1) were collected and organized. The compounds were prepared by dilution in double-distilled H2O to a concentration of 20 mM and stored in small aliquots in PCR plates at −80 °C. Before screening, the substrates were diluted 10-fold in an assay buffer consisting of 25 mM Hepes (pH 7.5), 50 mM NaCl, 5 mM MgCl2, and 2 mM DTT. Target proteins were removed from storage, thawed on ice, and diluted to 10 μM concentration in assay buffer. Protein and substrate (each 2.5 μL) were mixed robotically (Biomek FX; Beckman Coulter) in duplicate to 384-well plates (3540; Corning), resulting in final enzyme or substrate concentrations of 5 μM or 1 mM, respectively. After a 30-min incubation at room temperature, 13.5 μL of BioMol Green (Enzo Life Sciences) was robotically (Evolution P3 liquid handling system) added to each well to detect free phosphate. After a 30-min incubation, the absorbance of each well was measured at λ = 650 nM using a microplate reader (EnVision Multilabel Reader). Background phosphate (or phosphate present in the compounds) was corrected for by incubating substrate and buffer (no enzyme present) for 30 min followed by the addition of BioMol Green. This background absorbance was subtracted from the experimental assays. Substrate stability was assessed by following background phosphate after freeze–thaw and over time. No appreciable increase in background phosphate occurred during the study (approximately a 3-y duration).
Data Analysis.
Plate absorbance data for each enzyme was loaded into separate tabs in Microsoft Excel and corrected for background. Conditional formatting was used to color cells based on absorbance using a gradient (Fig. 3). Cells with the highest absorbance were colored red and lower-valued cells were colored blue. A bar was also added to each cell to compare relative activity across all plates (0.0–1.0 for the highest absorbance identified in the screen). In addition, cells are “commented” with the substrate name. The combination of conditional formatting and commenting allows for rapid, visual identification of substrate hits for each enzyme, while also indicating the relative activity of the enzyme toward the substrates. A lookup table was used to label each plate with EFI ID, GI number, hypothetical protein name, species, predicted cap type (11) (see below), and closest PDB structure. Each enzyme was manually annotated to describe the substrates that were used. To account for variability between duplicates, the difference between each was calculated. From this, the mean and SEM were determined for the entire screen (0.0307 ± 0.0003). This signifies that the screen is highly reproducible.
Custom scripts were used to extract data from the spreadsheets such as substrate hits, manual annotations, and absorbance data. As discussed in Results, an absorbance value of greater than 0.2 ± 0.03 for a substrate was considered a hit. Custom python scripts were used to compute (i) the number of hits for each enzyme and (ii) the number of times a substrate was hydrolyzed by any enzyme. The absorbance data were visualized as a heat map using the gplots package in statistical programming software R. Each protein in the analysis was annotated with a cap type class (C0, C1, or C2) using CapPredictor (11). Briefly, CapPredictor uses a gold-standard, structure-based sequence profile to identify large domain insertions.
Similarity analysis for the library was performed using RDKit (www.rdkit.org/) to generate and optimize (with the Universal Force Field) the 3D model for each substrate using distance geometry. OpenEye ROCS (www.eyesopen.com) was implemented to align the pairs of substrate conformations based on shape and optimize the alignment to maximize the overlap volume. OpenEye EON was used on the alignments from ROCS to compare the electrostatic potential maps and report T scores that represent both shape and electrostatic similarity.
Steady-State Kinetics.
For steady-state kinetic characterization, purified enzyme was diluted to an appropriate concentration in a buffer consisting of 25 mM Hepes (pH 7.5), 50 mM NaCl, and 5 mM MgCl2. The steady-state kinetic parameters (Km and kcat) for substrates of interest were determined from initial reaction velocities measured at varying substrate concentrations (0.5–5 × Km) using the EnzCheck Phosphate Assay Kit (Invitrogen). Absorbance measurements were performed with a Beckman DU800 spectrophotometer using quartz cuvettes (Starna Cells, 18B-Q-10) and 250-μL volumes in triplicate. Data were fit to the following using SigmaPlot Enzyme Kinetics Module:
Here, vo is the initial velocity, Vmax the maximum velocity, [S] the substrate concentration, and Km the Michaelis–Menten constant calculated for substrates of interest. The value for kcat was calculated from the following:
where [E] is the enzyme concentration in the assay. The steady-state kinetic constants for all enzymes tested are reported in Table 1.
For the mutase reaction, the assay was performed using β d-glucose 1-phosphate as substrate and β d-glucose 1,6-bisphosphate as cofactor as previously reported (15).
Functional Annotation.
Manual annotations from screening were appended to both superfamily-wide representative SSNs and subgroup expanded sequence similarity networks in Cytoscape. The resulting annotations within the subgroups were examined to ascertain substrate similarity among proteins that clustered together. Clustered proteins with similar functions were labeled (Fig. 5 and SI Appendix, Figs. S3–S21) and annotations were applied to all proteins within the cluster. If no screening data were available for a particular cluster, the clusters were examined for previously characterized proteins. If a previously characterized protein was present, the annotation was applied to all proteins in the same cluster. If no proteins were examined by either screening or contained previously characterized data, the clusters remained unannotated.
Supplementary Material
Acknowledgments
We thank Yu (Brandon) Xia for useful discussions on protein evolution and Adrian Whitty for helpful discussions and help in reviewing the manuscript. We acknowledge funding from National Institutes of Health to the Enzyme Function Initiative (Grant U54-GM093342 to K.N.A., D.D.-M., and S.C.A.) and the New York Structural Genomics Research Consortium (Grant U54-GM094662 to S.C.A.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1423570112/-/DCSupplemental.
References
- 1.Altschul SF, et al. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997;25(17):3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Radivojac P, et al. A large-scale evaluation of computational protein function prediction. Nat Methods. 2013;10(3):221–227. doi: 10.1038/nmeth.2340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bastard K, et al. Revealing the hidden functional diversity of an enzyme family. Nat Chem Biol. 2014;10(1):42–49. doi: 10.1038/nchembio.1387. [DOI] [PubMed] [Google Scholar]
- 4.Kuznetsova E, et al. Genome-wide analysis of substrate specificities of the Escherichia coli haloacid dehalogenase-like phosphatase family. J Biol Chem. 2006;281(47):36149–36161. doi: 10.1074/jbc.M605449200. [DOI] [PubMed] [Google Scholar]
- 5.Koonin EV, Tatusov RL. Computer analysis of bacterial haloacid dehalogenases defines a large superfamily of hydrolases with diverse specificity. Application of an iterative approach to database search. J Mol Biol. 1994;244(1):125–132. doi: 10.1006/jmbi.1994.1711. [DOI] [PubMed] [Google Scholar]
- 6.Motosugi K, Esaki N, Soda K. Purification and properties of a new enzyme, DL-2-Haloacid dehalogenase, from Pseudomonas sp. J Bacteriol. 1982;150(2):522–527. doi: 10.1128/jb.150.2.522-527.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Burroughs AM, Allen KN, Dunaway-Mariano D, Aravind L. Evolutionary genomics of the HAD superfamily: Understanding the structural adaptations and catalytic diversity in a superfamily of phosphoesterases and allied enzymes. J Mol Biol. 2006;361(5):1003–34. doi: 10.1016/j.jmb.2006.06.049. [DOI] [PubMed] [Google Scholar]
- 8.Lu Z, Dunaway-Mariano D, Allen KN. The catalytic scaffold of the haloalkanoic acid dehalogenase enzyme superfamily acts as a mold for the trigonal bipyramidal transition state. Proc Natl Acad Sci USA. 2008;105(15):5687–5692. doi: 10.1073/pnas.0710800105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cho H, et al. BeF(3)(-) acts as a phosphate analog in proteins phosphorylated on aspartate: Structure of a BeF(3)(-) complex with phosphoserine phosphatase. Proc Natl Acad Sci USA. 2001;98(15):8525–8530. doi: 10.1073/pnas.131213698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lahiri SD, Zhang G, Dai J, Dunaway-Mariano D, Allen KN. Analysis of the substrate specificity loop of the HAD superfamily cap domain. Biochemistry. 2004;43(10):2812–2820. doi: 10.1021/bi0356810. [DOI] [PubMed] [Google Scholar]
- 11.Pandya C, Dunaway-Mariano D, Xia Y, Allen KN. Structure-guided approach for detecting large domain inserts in protein sequences as illustrated using the haloacid dehalogenase superfamily. Proteins. 2014;82(9):1896–1906. doi: 10.1002/prot.24543. [DOI] [PubMed] [Google Scholar]
- 12.Nobeli I, Ponstingl H, Krissinel EB, Thornton JM. A structure-based anatomy of the E.coli metabolome. J Mol Biol. 2003;334(4):697–719. doi: 10.1016/j.jmb.2003.10.008. [DOI] [PubMed] [Google Scholar]
- 13.Maggiora G, Vogt M, Stumpfe D, Bajorath J. Molecular similarity in medicinal chemistry. J Med Chem. 2014;57(8):3186–3204. doi: 10.1021/jm401411z. [DOI] [PubMed] [Google Scholar]
- 14.Neuwald AF, Stauffer GV. DNA sequence and characterization of the Escherichia coli serB gene. Nucleic Acids Res. 1985;13(19):7025–7039. doi: 10.1093/nar/13.19.7025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dai J, Wang L, Allen KN, Radstrom P, Dunaway-mariano D. Conformational cycling in beta-phosphoglucomutase catalysis: Reorientation of the beta-D-glucose 1,6-(bis)phosphate intermediate. Biochemistry. 2006;45(25):7818–7824. doi: 10.1021/bi060136v. [DOI] [PubMed] [Google Scholar]
- 16.Pandya C, et al. Consequences of domain insertion on sequence-structure divergence in a superfold. Proc Natl Acad Sci USA. 2013;110(36):E3381–E3387. doi: 10.1073/pnas.1305519110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Akiva E, et al. The Structure-Function Linkage Database. Nucleic Acids Res. 2014;42(Database issue):D521–D530. doi: 10.1093/nar/gkt1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Atkinson HJ, Morris JH, Ferrin TE, Babbitt PC. Using sequence similarity networks for visualization of relationships across diverse protein superfamilies. PLoS ONE. 2009;4(2):e4345. doi: 10.1371/journal.pone.0004345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Copley SD. Evolution of a metabolic pathway for degradation of a toxic xenobiotic: The patchwork approach. Trends Biochem Sci. 2000;25(6):261–265. doi: 10.1016/s0968-0004(00)01562-0. [DOI] [PubMed] [Google Scholar]
- 20.Wang L, et al. Divergence of biochemical function in the HAD superfamily: D-glycero-D-manno-heptose-1,7-bisphosphate phosphatase (GmhB) Biochemistry. 2010;49(6):1072–1081. doi: 10.1021/bi902018y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tremblay LW, Dunaway-Mariano D, Allen KN. Structure and activity analyses of Escherichia coli K-12 NagD provide insight into the evolution of biochemical function in the haloalkanoic acid dehalogenase superfamily. Biochemistry. 2006;45(4):1183–1193. doi: 10.1021/bi051842j. [DOI] [PubMed] [Google Scholar]
- 22.Dai J, et al. Analysis of the structural determinants underlying discrimination between substrate and solvent in beta-phosphoglucomutase catalysis. Biochemistry. 2009;48(9):1984–1995. doi: 10.1021/bi801653r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wu R, Garland M, Dunaway-Mariano D, Allen KN. Homo sapiens dullard protein phosphatase shows a preference for the insulin-dependent phosphorylation site of lipin1. Biochemistry. 2011;50(15):3045–3047. doi: 10.1021/bi200336b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Eaton JM, et al. Lipin 2 binds phosphatidic acid by the electrostatic hydrogen bond switch mechanism independent of phosphorylation. J Biol Chem. 2014;289(26):18055–18066. doi: 10.1074/jbc.M114.547604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Farelli JD, et al. Structure of the trehalose-6-phosphate phosphatase from Brugia malayi reveals key design principles for anthelmintic drugs. PLoS Pathog. 2014;10(7):e1004245. doi: 10.1371/journal.ppat.1004245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huang H, et al. Divergence of structure and function in the haloacid dehalogenase enzyme superfamily: Bacteroides thetaiotaomicron BT2127 is an inorganic pyrophosphatase. Biochemistry. 2011;50(41):8937–8949. doi: 10.1021/bi201181q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Silvaggi NR, et al. The X-ray crystal structures of human alpha-phosphomannomutase 1 reveal the structural basis of congenital disorder of glycosylation type 1a. J Biol Chem. 2006;281(21):14918–14926. doi: 10.1074/jbc.M601505200. [DOI] [PubMed] [Google Scholar]
- 28.Postma PW, Lengeler JW, Jacobson GR. Phosphoenolpyruvate:carbohydrate phosphotransferase systems of bacteria. Microbiol Rev. 1993;57(3):543–594. doi: 10.1128/mr.57.3.543-594.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gabor E, et al. The phosphoenolpyruvate-dependent glucose-phosphotransferase system from Escherichia coli K-12 as the center of a network regulating carbohydrate flux in the cell. Eur J Cell Biol. 2011;90(9):711–720. doi: 10.1016/j.ejcb.2011.04.002. [DOI] [PubMed] [Google Scholar]
- 30.Rangarajan ES, et al. Structural snapshots of Escherichia coli histidinol phosphate phosphatase along the reaction pathway. J Biol Chem. 2006;281(49):37930–37941. doi: 10.1074/jbc.M604916200. [DOI] [PubMed] [Google Scholar]
- 31.Lu Z, Wang L, Dunaway-Mariano D, Allen KN. Structure-function analysis of 2-keto-3-deoxy-D-glycero-D-galactonononate-9-phosphate phosphatase defines specificity elements in type C0 haloalkanoate dehalogenase family members. J Biol Chem. 2009;284(2):1224–1233. doi: 10.1074/jbc.M807056200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Daughtry KD, et al. Structural basis for the divergence of substrate specificity and biological function within HAD phosphatases in lipopolysaccharide and sialic acid biosynthesis. Biochemistry. 2013;52(32):5372–5386. doi: 10.1021/bi400659k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Galburt EA, Pelletier J, Wilson G, Stoddard BL. Structure of a tRNA repair enzyme and molecular biology workhorse: T4 polynucleotide kinase. Structure. 2002;10(9):1249–1260. doi: 10.1016/s0969-2126(02)00835-3. [DOI] [PubMed] [Google Scholar]
- 34.Peisach E, Selengut JD, Dunaway-Mariano D, Allen KN. X-ray crystal structure of the hypothetical phosphotyrosine phosphatase MDP-1 of the haloacid dehalogenase superfamily. Biochemistry. 2004;43(40):12770–12779. doi: 10.1021/bi0490688. [DOI] [PubMed] [Google Scholar]
- 35.Zhang Y, et al. Determinants for dephosphorylation of the RNA polymerase II C-terminal domain by Scp1. Mol Cell. 2006;24(5):759–770. doi: 10.1016/j.molcel.2006.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang M, Cho EJ, Burstein G, Siegel D, Zhang Y. Selective inactivation of a human neuronal silencing phosphatase by a small molecule inhibitor. ACS Chem Biol. 2011;6(5):511–519. doi: 10.1021/cb100357t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhao S, et al. Prediction and characterization of enzymatic activities guided by sequence similarity and genome neighborhood networks. eLife. 2014;3:1–32. doi: 10.7554/eLife.03275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Baier F, Tokuriki N. Connectivity between catalytic landscapes of the metallo-β-lactamase superfamily. J Mol Biol. 2014;426(13):2442–2456. doi: 10.1016/j.jmb.2014.04.013. [DOI] [PubMed] [Google Scholar]
- 39.Kormish JD, McGhee JD. The C. elegans lethal gut-obstructed gob-1 gene is trehalose-6-phosphate phosphatase. Dev Biol. 2005;287(1):35–47. doi: 10.1016/j.ydbio.2005.08.027. [DOI] [PubMed] [Google Scholar]
- 40.Conant GC, Wolfe KH. Turning a hobby into a job: How duplicated genes find new functions. Nat Rev Genet. 2008;9(12):938–950. doi: 10.1038/nrg2482. [DOI] [PubMed] [Google Scholar]
- 41.Soskine M, Tawfik DS. Mutational effects and the evolution of new protein functions. Nat Rev Genet. 2010;11(8):572–582. doi: 10.1038/nrg2808. [DOI] [PubMed] [Google Scholar]
- 42.Nguyen HH, et al. Structural determinants of substrate recognition in the HAD superfamily member D-glycero-D-manno-heptose-1,7-bisphosphate phosphatase (GmhB) Biochemistry. 2010;49(6):1082–1092. doi: 10.1021/bi902019q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dellus-Gur E, Toth-Petroczy A, Elias M, Tawfik DS. What makes a protein fold amenable to functional innovation? Fold polarity and stability trade-offs. J Mol Biol. 2013;425(14):2609–2621. doi: 10.1016/j.jmb.2013.03.033. [DOI] [PubMed] [Google Scholar]
- 44.Aroul-Selvam R, Hubbard T, Sasidharan R. Domain insertions in protein structures. J Mol Biol. 2004;338(4):633–641. doi: 10.1016/j.jmb.2004.03.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Matsumura I, Ellington AD. In vitro evolution of beta-glucuronidase into a beta-galactosidase proceeds through non-specific intermediates. J Mol Biol. 2001;305(2):331–339. doi: 10.1006/jmbi.2000.4259. [DOI] [PubMed] [Google Scholar]
- 46.Aslanidis C, de Jong PJ. Ligation-independent cloning of PCR products (LIC-PCR) Nucleic Acids Res. 1990;18(20):6069–6074. doi: 10.1093/nar/18.20.6069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Studier FW. Protein production by auto-induction in high density shaking cultures. Protein Expr Purif. 2005;41(1):207–234. doi: 10.1016/j.pep.2005.01.016. [DOI] [PubMed] [Google Scholar]
- 48.Savitsky P, et al. High-throughput production of human proteins for crystallization: The SGC experience. J Struct Biol. 2010;172(1):3–13. doi: 10.1016/j.jsb.2010.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sauder MJ, et al. High throughput protein production and crystallization at NYSGXRC. Methods Mol Biol. 2008;426:561–575. doi: 10.1007/978-1-60327-058-8_37. [DOI] [PubMed] [Google Scholar]
- 50.Mann KM, Lardy HA. Phosphoric esters of biological importance. V. The synthesis of L-sorbose-1-phosphate and L-sorbose-6-phosphate. J Biol Chem. 1950;187(1):339–348. [PubMed] [Google Scholar]
- 51.Anderson BRL, Wengeg WC. D-galactose 6-phosphate and D-tagatose 6-phosphate. Methods Enzymol. 1982;89:93–98. [Google Scholar]
- 52.Ballou CE. Preparation and properties of D-erythrose-4-phosphate. Methods Enzymol. 1963;6:479–484. [Google Scholar]
- 53.MacDonald DL, Wong RY. A chemical synthesis of Trehalose-6-phosphate. Biochim Biophys Acta. 1964;86:390–392. doi: 10.1016/0304-4165(64)90066-2. [DOI] [PubMed] [Google Scholar]
- 54.Cox RJ, de Andrés-Gómez A, Godfrey CRA. Rapid and flexible synthesis of 1-deoxy-D-xylulose-5-phosphate, the substrate for 1-deoxy-D-xylulose-5-phosphate reductoisomerase. Org Biomol Chem. 2003;1(18):3173–3177. doi: 10.1039/b307314a. [DOI] [PubMed] [Google Scholar]
- 55.Meyer O, Hoeffler J-F, Grosdemange-Billiard C, Rohmer M. Practical synthesis of 1-deoxy-d-xylulose and 1-deoxy-d-xylulose 5-phosphate allowing deuterium labelling. Tetrahedron. 2004;60:12153–12162. [Google Scholar]
- 56.Avigad G, Englard S. 5-keto-D-fructose and its phosphate esters-1. Methods Enzymol. 1974;41:84–90. doi: 10.1016/s0076-6879(75)41020-5. [DOI] [PubMed] [Google Scholar]
- 57.Pollak A, Baughn RL, Adalsteinsson O, Whitesides GM. Large-scale enzyme-catalyzed synthesis of ATP from adenosine and acetyl phosphate. Regeneration of ATP from AMP. J Am Chem Soc. 1978;100:304–306. [Google Scholar]
- 58.Kameda A, et al. A novel ATP regeneration system using polyphosphate-AMP phosphotransferase and polyphosphate kinase. J Biosci Bioeng. 2001;91(6):557–563. doi: 10.1263/jbb.91.557. [DOI] [PubMed] [Google Scholar]
- 59.Zhao H, van der Donk WA. Regeneration of cofactors for use in biocatalysis. Curr Opin Biotechnol. 2003;14(6):583–589. doi: 10.1016/j.copbio.2003.09.007. [DOI] [PubMed] [Google Scholar]
- 60.Bolt A, Berry A, Nelson A. Directed evolution of aldolases for exploitation in synthetic organic chemistry. Arch Biochem Biophys. 2008;474(2):318–330. doi: 10.1016/j.abb.2008.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Shelton MC, et al. 2-Keto-3-deoxy-6-phosphogluconate aldolases as catalysts for stereocontrolled carbon-carbon bond formation. J Am Chem Soc. 1996;118:2117–2125. [Google Scholar]
- 62.Meloche HP, Wood WA. 2-keto-3-deoxy-6-phosphogluconate. Methods Enzymol. 1966;9:51–53. [Google Scholar]
- 63.O’Connell EL, Meloche HP. Enzymatic synthesis of 2-keto-3-deoxygluconate 6-phosphate using 6-phosphogluconate dehydratase. Methods Enzymol. 1982;89:98–101. [Google Scholar]
- 64.Isbell HS. Oxidation of the alpha and beta forms of the sugars. J Am Chem Soc. 1932;54:1692–1693. [Google Scholar]
- 65.Wolfrom ML, Anno K. Sodium borohydride as a reducing agent in the sugar series. J Am Chem Soc. 1983;74(22):5583–5584. [Google Scholar]
- 66.Chirgwin JM, Noltmann EA. The enediolate analogue 5-phosphoarabinonate as a mechanistic probe for phosphoglucose isomerase. J Biol Chem. 1975;250(18):7272–7276. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.