Significance
Certain oligomeric species generated during the self-assembly of specific proteins into ordered fibrillar aggregates are likely to be key players in the initiation and spreading of neurodegenerative diseases. We have purified stable toxic oligomeric species of α-synuclein and defined and minimized their degree of heterogeneity, which has allowed us to identify distinct subgroups of oligomers and determine their structural properties and three-dimensional molecular architectures. All the oligomeric subgroups possess approximately cylindrical architectures with marked similarities to amyloid fibrils, suggesting that these types of oligomers are kinetically trapped during protein self-assembly. The relative stabilities and inherent pathological roles of different amyloid oligomers are likely to result from the multiplicity of pathways of the misfolding process and the remarkably slow rates of structural conversions.
Keywords: protein misfolding, amyloid aggregation, toxic oligomer, cryoelectron microscopy, neurodegeneration
Abstract
We describe the isolation and detailed structural characterization of stable toxic oligomers of α-synuclein that have accumulated during the process of amyloid formation. Our approach has allowed us to identify distinct subgroups of oligomers and to probe their molecular architectures by using cryo-electron microscopy (cryoEM) image reconstruction techniques. Although the oligomers exist in a range of sizes, with different extents and nature of β-sheet content and exposed hydrophobicity, they all possess a hollow cylindrical architecture with similarities to certain types of amyloid fibril, suggesting that the accumulation of at least some forms of amyloid oligomers is likely to be a consequence of very slow rates of rearrangement of their β-sheet structures. Our findings reveal the inherent multiplicity of the process of protein misfolding and the key role the β-sheet geometry acquired in the early stages of the self-assembly process plays in dictating the kinetic stability and the pathological nature of individual oligomeric species.
Medical disorders associated with amyloid formation, which include Alzheimer’s disease, Parkinson’s disease (PD), and diabetes mellitus type 2, share a common feature, namely the presence of deposits of abnormally aggregated proteins within the body (1, 2). Although the specific protein molecule that is found to be the major component of such deposits varies from one disease to another, the formation of the pathological aggregates appears to occur by a common process of misfolding and self-assembly of a normally soluble polypeptide chain into a series of oligomeric intermediates and, ultimately, into insoluble amyloid fibrils that accumulate within specific organs and tissues.
It has been found that amyloid fibrils can be formed in vitro from a wide range of proteins in addition to those involved in disease and, like fibrils extracted from patients suffering from amyloid-related diseases, they possess a common core architecture, the “cross-β” structure, in which β-strands align perpendicular to the fibril axis and, thus, generate arrays of β-sheets that are oriented parallel to the fibril axis (3). Despite their highly insoluble but noncrystalline nature, which has hampered their detailed structural determination by traditional methods, several structural descriptions of fibrillar assemblies have been reported, some at atomic resolution (4, 5), greatly enhancing our understanding of the different levels of structural complexity inherent in the amyloid cross-β structure. The universality of this architecture has been attributed to the nature of the intramolecular and intermolecular interactions within the β-sheets, which are dominated by hydrogen bonds between the main-chain atoms that are common to all polypeptide chains (6). Information from a variety of techniques suggests that the fibrils typically result from the assembly of a number of protofilaments, each made up of a double layer of β-sheets, which wind around one another to form twisted structures. In most cases, there is evidence for water-filled interfaces between protofilaments (4, 5, 7–10), which have analogies with such interfaces observed in structures of small peptides that assemble into amyloid-like microcrystals (11).
Significant advances have therefore been made toward understanding the structures of fibrillar aggregates, but little detailed structural information (12) is available for the oligomeric species that are frequently observed to accumulate as intermediates in the process of fibril formation, despite increasing evidence that such species can be highly cytotoxic (1, 13, 14) and may be key players not only in the initiation of disease but also in its spreading through cell-to-cell transmission (15). Structural characterization of oligomers is, however, particularly challenging because of their often transient nature and, even more importantly, because of the variability of these species both in terms of size (from dimers to high-order multimers) and structure (in principle, from essentially random coil to a similar degree of β-sheet content to that observed in the fibrillar species). It is therefore of the utmost importance that this intrinsic heterogeneity of oligomeric samples is appropriately controlled and understood if meaningful structural models are to be obtained and related to their toxicity.
Despite these challenges, intense research has elucidated a number of common features between amyloid oligomers formed by different systems (16). In the particular case of α-synuclein (αS), whose deposition is the hallmark of PD, a number of samples enriched in stable oligomeric species has been reported (17–22), which appear to have some common general features, but significant differences in structures, relationships to fibril formation, and proposed mechanisms of toxicity.
We report here the detailed characterization of an ensemble of toxic oligomeric species formed by αS, whose existence and relationship to amyloid fibril formation has been determined by single-molecule studies (23). This characterization has been achieved by the application of a set of complementary biophysical techniques, including a variety of spectroscopic techniques along with analytical ultracentrifugation (AU), atomic force microscopy (AFM), and electron microscopy (EM), to provide a detailed understanding of the structural properties of this type of complex aggregate. The results of this study provide the basis for a more complete understanding of the nature of the self-assembly of polypeptides into β-sheet rich amyloid aggregates, providing a unifying view of the protein misfolding process, and potentially contributing to efforts to identify specific targets for drug discovery.
Results
Preparation and Preliminary Characterization of the Ensemble of αS Oligomers.
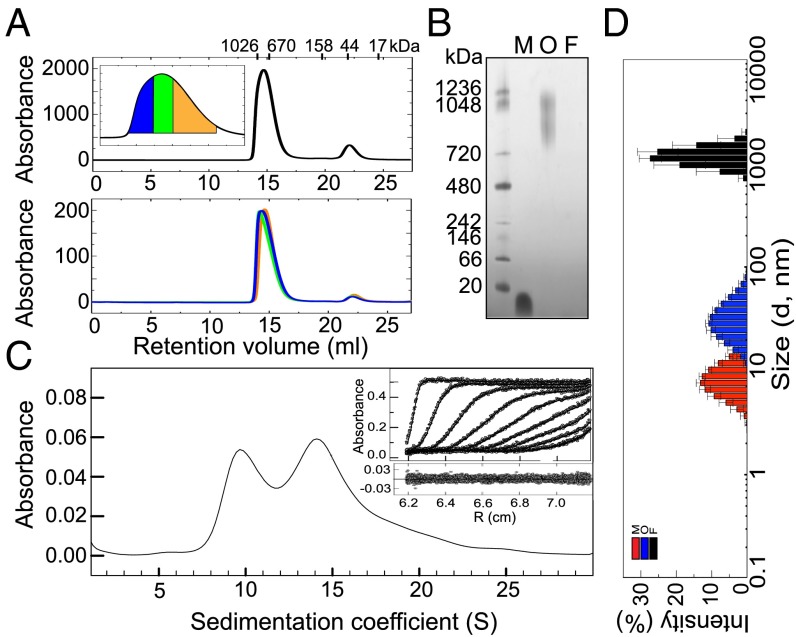
The inherently transient nature of oligomeric intermediates generated during the formation of amyloid fibrils normally results in their presence as a low fraction of all of the protein species in the sample at any time of the reaction. Alternative procedures have, therefore, been used to try to isolate these types of species to characterize them and obtain a better understanding of their structural properties. In the present study, we have made used of lyophilization (an approach widely used previously to generate αS oligomeric samples; refs. 17, 21, 22, and 24–28) to increase significantly the formation of αS oligomeric species, an effect that can be attributed to a combination of factors including a significant increase in the area of the solvent/air interface (29) and a decrease in the intermolecular distances during the vitrification process. The final composition of the purified oligomeric samples used in this study (see SI Appendix, Fig. S1 for the analysis of the kinetic stability of the oligomeric samples) was assessed by HPLC-SEC (size exclusion chromatography) (Fig. 1A) and found to consist of approximately 90% of oligomers with apparent molecular masses of approximately 650–1,100 kDa (similar values, approximately 800–1,200 kDa, were obtained from native-PAGE; Fig. 1B) and 10% of monomers, a feature that is likely to be the result of dissociation of some of the more labile oligomers. A similar level of monomeric protein was also observed in samples subjected to AU and has been accounted for when defining the spectroscopic properties of the oligomeric species. Initial attempts to separate oligomers of different sizes by HPLC-SEC (Fig. 1A) were unsuccessful, suggesting that the oligomeric sample is able to reequilibrate at least to some extent during the time needed for their reanalysis; we were able, however, to tune the size distribution of the oligomers by adding different concentrations of urea to the sample, as discussed below.
Fig. 1.
Determination of the size distribution of the purified αS oligomeric samples. (A) HPLC-SEC analysis of the composition of the oligomeric samples; the oligomeric fraction eluted at 14.5 mL and the monomeric fraction at 22 mL (Top). The elution volumes for each protein standard used for the calibration are shown above (see also SI Appendix, SI Experimental Procedures). The fractions corresponding to the oligomeric peak were collected and separated in three groups: large oligomer HPLC fraction (area depicted in blue in the zoomed-in image of the oligomeric peak), medium-size oligomer HPLC fraction (green area), and small oligomer HPLC fraction (orange area). The three oligomeric solutions were then concentrated and reinjected into the HPLC-SEC column at the same mass concentration for their analysis (Bottom). (B) Native PAGE gel showing the different mobility of fibrillar (F), oligomeric (O), and monomeric (M) αS in comparison with protein markers (first lane). (C) Sedimentation velocity analysis of the αS oligomeric sample. The solid line represents the size distribution of sedimenting species obtained by c(s) analysis (Experimental Procedures). In Inset, the experimental data (symbols), their fit to the model (solid lines), and their residuals (below) are shown. For clarity, only one in every nine scans is represented. (D) Representative DLS-derived size distribution of 45 μM monomeric (red bars), 5 µM oligomeric (blue bars), and 3 µM fibrillar (black bars) αS solutions.
Accurate Determination of the Size Distribution of the αS Oligomers.
Although native PAGE and standard SEC techniques were useful to give initial assessments of the sizes and level of heterogeneity of the various protein samples, a more sophisticated method of analysis was needed to establish accurately the distribution of sizes present with such complex mixtures of species. We therefore carried out AU measurements of sedimentation velocities and obtained similar velocity profiles for different batches of freshly prepared purified oligomeric samples (see Figs. 1C and 3A for a comparison), indicating the high reproducibility of our preparation protocol. The data were then analyzed by two complementary approaches (30, 31) (Experimental Procedures), which yielded similar distributions of sedimentation coefficients for the ensemble of particles in the sample.
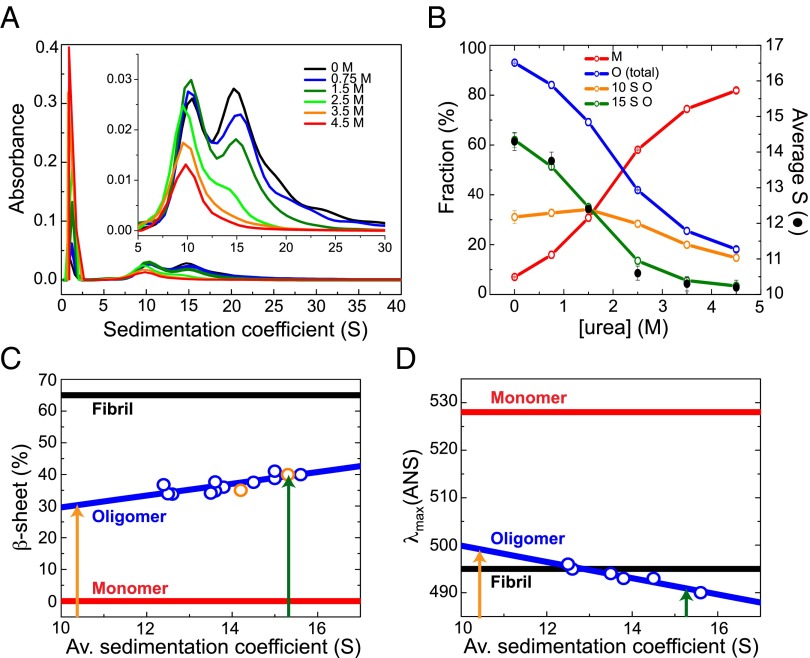
Fig. 3.
Detailed characterization of the individual structural properties of the different sized subgroups of oligomers. (A) AU sedimentation velocity experiments with the oligomeric samples in the presence of different urea concentrations. A zoomed-in view of the size profile of the oligomeric fraction is shown in Inset. (B) Fractions of monomeric (in red) and oligomeric species (in blue) in the oligomeric samples, as a function of urea concentration, estimated by AU. The fraction of the two main oligomeric size subgroups is also represented: the 15S oligomer subgroup in green and the 10S oligomer subgroup in orange. The average sedimentation coefficient of the oligomeric fraction at different urea concentrations is also shown (black circles). The error bars represent experimental errors. Correlation of the secondary structure content (C) and the degree of hydrophobic surface area exposed to the solvent (represented as the wavelength of the maximum fluorescence emission of ANS) (D) with the size of the oligomers (blue symbols represent the experimental data and the line, the correlation function; Sl Appendix, SI Experimental Procedures). Estimates of the β-sheet content by FTIR of two independent oligomeric samples, corresponding to one prepared with freshly purified protein and another prepared with reused flow-through solutions (orange symbols; Sl Appendix, Fig. S4), overlie with the overall trend obtained by far-UV CD analysis (blue symbols). The orange and green arrows indicate the estimated averaged β-sheet content for the 10S and 15S oligomeric subgroup, respectively.
The sedimentation velocity profiles showed a significant polydispersity in size for the oligomeric species, although two broad peaks are clearly distinguishable, with S20,w values of 7.3–13.3 ± 1 S (peak maximum at 10.3 S) and 12.2–18.2 ± 1 S (peak maximum at 15.2 S) and a relative abundance in mass concentration in the sample of 30 ± 3% and 50 ± 3%, respectively (note that the absorbance reflects the relative mass concentration of the particles), and a minor additional group of species (approximately 10% in mass) with larger S20,w values, up to 28 S (Fig. 1C). The S20,w values of these species, together with the corresponding best-fit values of the frictional ratio, f/f0 (a parameter related to the asymmetry of the protein molecules), were then correlated with the values estimated from the cryo-electron microscopy (cryoEM)-derived 3D structures (see sections below) by using HYDROMIC (32). Using this approach, the association state (the average number of protein molecules in an oligomer) for the 10S oligomer subgroup was determined to be 11–25 (an average of 18) (f/f0 of 1.40 ± 0.10) and for the 15S oligomer subgroup to be 19–39 (an average of 29) (f/f0 of 1.32 ± 0.10), corresponding to molecular masses of 160–360 kDa (an average of 260 kDa) and 280–560 kDa (an average of 420 kDa), respectively; the additional larger species present at low levels correspond to particles containing up to 90 protein molecules (molecular masses up to ∼1,300 kDa) and were included in the 15S subgroup for their further structural characterization. The calculated values of the frictional ratios for the two subgroups are consistent with hydrated globular/spherical particles, rather than elongated species.
Definition of the Secondary Structure Content and Hydrophobic Character of the Oligomeric αS Species.
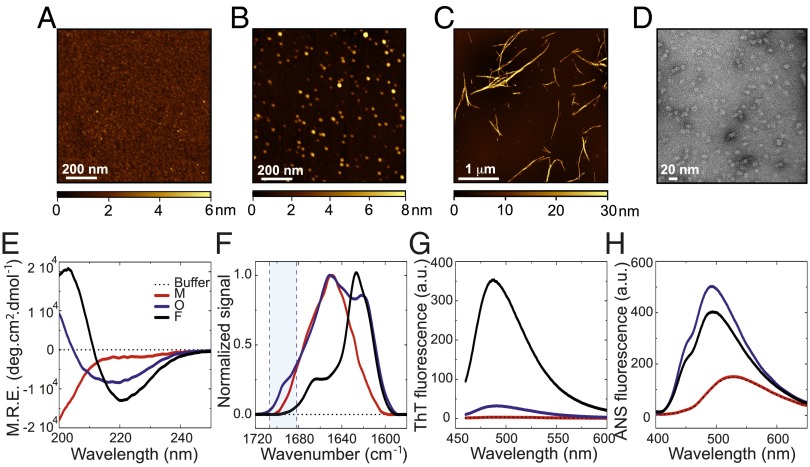
We next set out to characterize the overall morphologies of the oligomeric species in the samples by means of AFM techniques (Fig. 2 A–C). Whereas the fibrillar species appear as uniform thread-like structures of 0.05–3.0 µm in length and 10–35 nm in height (the large variation in heights is attributable to differing degrees of self-association of the fibrils under these conditions), the purified oligomeric species appear to be approximately spherical in the AFM images (in agreement with TEM images; Fig. 2D), with heights ranging between 3 and 16 nm (SI Appendix, Fig. S2), in agreement with a range of previous observations of oligomeric forms of αS (33). The dimensions of the oligomeric species derived from the AFM data are also broadly consistent with the solution-derived size parameters obtained by dynamic light scattering (DLS) (Fig. 1D). Interestingly, all of the oligomeric species, regardless of their size, appear to have similar spherical-like morphologies in the AFM and TEM images.
Fig. 2.
Morphological and structural characterization of the oligomeric αS species. Examples of AFM images of monomeric (A), oligomeric (B), and fibrillar (C) αS species. The color-coding represents the surface topography (height), and the scale bar is shown at the bottom of each image. (D) Representative TEM image of the αS oligomeric samples. Far-UV CD (E), FTIR (F), ThT fluorescence (G), and ANS fluorescence spectra (H) of monomeric (red line), oligomeric (blue line), and fibrillar αS solutions (black line) at the same mass concentration. The spectrum of the buffer is also shown as thin dashed black lines. In F, the position of the aborbance band characteristic of antiparallel β-sheet structure present in the oligomeric species but absent in the fibrillar forms is highlighted in blue.
To gain insight into the structural features of the oligomers, we assessed their secondary structure content and hydrophobicity and compared these properties to those of the monomeric and fibrillar states. Both far-UV circular dichroism (CD) (Fig. 2E) and Fourier transform infrared (FTIR) spectroscopy (Fig. 2F) reveal that the secondary structure content of the oligomers is intermediate between that of the monomeric and the fibrillar species. Deconvolution of the FTIR data in particular (SI Appendix, Fig. S3) indicates that on average, the oligomers contain approximately 35 ± 5% of β-sheet structure, compared with none in the soluble monomers and approximately 65 ± 10% in the fibrils (SI Appendix, Fig. S3); the latter estimate is in good agreement with previous studies of the β-sheet core of the αS fibrils (34). Furthermore, the β-sheet structure of the oligomers appears to be able to interact with thioflavin T (ThT) molecules much less effectively than does that of the fibrils, as the oligomers display approximately 10 times less ThT fluorescence intensity at the maximum wavelength of emission compared with the fibrils at equivalent mass concentrations (Fig. 2G).
More detailed analyis of the FTIR data suggests that the fibrillar conformation of αS is largely composed of parallel β-sheet structure (shown by the presence of a band at 1,620–1,630 cm−1 and the absence of an absorption band at approximately 1,695 cm−1), but that the β-sheet structure in the oligomeric species is predominantly antiparallel (as indicated by a band at approximately 1620–30 cm−1 and a prominent shoulder at approximately 1,695 cm−1, approximately fivefold weaker than the band at 1,620–30 cm−1) (35) (Fig. 2F and SI Appendix, Fig. S3). Interestingly, the detection of a difference in the organization of the β-sheet structure from a dominance of parallel β-sheet structure in the fibrillar form to that of antiparallel β-sheet structure in oligomeric species has been reported previously for αS (17) and for several other amyloidogenic peptides and proteins such as the Aβ-peptide (35), lysozyme (36), a prion-related peptide (37), and β2-microglobulin (38).
Finally, we assessed the extent of hydrophobic surface area exposed to the solvent in each αS species by using the most widely used solvent-sensitive dye, 1-anilinonaphthalene-8-sulfonic acid (ANS). Although the ANS fluorescence spectrum of the monomeric protein shows identical properties to that of the fluorophore in buffer alone (emission maximum at 526.3 ± 0.6 nm in both cases), the increase in its quantum yield (a three- to fourfold enhancement of the fluorescence intensity with respect to the free ANS), with a concomitant blue-shifted emission maximum in the presence of both oligomeric and fibrillar species (emission maximum at 492 ± 1 and 494 ± 2, respectively; Fig. 2H), indicates a greater extent of solvent exposed hydrophobic surface per molecule of αS in the aggregated forms of the protein, being slightly greater in the oligomeric than in the fibrillar state.
Analysis of the Structural Differences Between the Major Subgroups of αS Oligomers.
We have established above that the samples of purified oligomers contain a distribution of particle sizes, but that two distinct major size subgroups of oligomers can be differentiated in the AU analysis: a subgroup of small oligomers, referred as the 10S oligomer subgroup, and a subgroup of larger oligomers, referred as the 15S oligomer subgroup. In addition, further experiments have revealed that the relative proportion of each subgroup of oligomers and, therefore, the overall average sedimentation coefficient of the sample, changes upon addition of chemical denaturants such as urea (Fig. 3A). As the urea concentration was increased from 0 to 3 M, a large shift occurred in the average value of the sedimentation coefficient for the oligomeric species, from approximately 14 S to approximately 10 S; this change can be attributed to the increasing disappearance of the 15S subgroup of oligomers, concomitant with an increase in the fraction of monomeric protein (Fig. 3B). At urea concentrations higher than 3.5 M, the oligomeric distribution is essentially composed of only the 10S subgroup of oligomers. Importantly, we were unable to detect oligomers with S20,w values below approximately 7 S, even with increasing concentrations of urea. This observation indicates that the smaller (10S) oligomers are highly stable and remain within a rather well-defined and specific size range, and suggests that when disaggregation occurs, for example by monomer detachment, oligomers below a specific size limit (corresponding to approximately 200 kDa, i.e., to approximately 14 αS molecules) are no longer stable and rapidly disassemble into monomers.
By combining the analysis of the effects of low concentrations of urea (up to 1.5 M; see SI Appendix, Fig. S5 for the analysis at higher urea concentrations) on oligomeric samples prepared with both freshly purified monomeric protein (Fig. 3 A and B) and the flow-through solutions of previous oligomer purification processes (a procedure that yields a higher fraction of larger oligomeric species in the sample than the oligomeric samples prepared with freshly purified protein in a reproducible manner; SI Appendix, Fig. S4), we were able to perform a detailed analysis of the major overall structural features of the oligomers as a function of their size. We found a strong correlation between the size of the oligomers and both the β-sheet content (Fig. 3C) and the surface hydrophobicity (Fig. 3D): the smaller the oligomers, the lower the β-sheet content and the surface-exposed hydrophobicity. As discussed above, however, there appear to be well-defined limits on the size of stable oligomers and, therefore, on their β-sheet content and extent of hydrophobic surface area exposed to the solvent.
Further analysis of the data for the two types of oligomer subgroups indicates that the 15S oligomers have an average β-sheet content per molecule of approximately 39 ± 3% (ranging from approximately 34–45 according to the AU-derived size distribution: 12.2–18.2 S) and the highest level of exposed hydrophobic surface area of any αS species (including the fibrils), whereas the smaller 10S oligomers have an average β-sheet content of approximately 30 ± 3% (ranging from approximately 25–35 according to the AU-derived size distribution: 7.3–13.3 S) and an exposed hydrophobic surface area that is smaller than both the larger oligomers and the fibrils (Fig. 3 C and D). Using the correlation described above between oligomer size and β-sheet content, we predict the β-sheet content of the largest oligomers detected by AU (28 S) to be approximately 65%. This value is the same as that measured by FTIR for the αS fibrils but, as we discuss in more detail below, the FTIR spectra indicate that the β-sheet geometry is different, the oligomers but not the fibrils having a significant content of antiparallel strands, and the oligomers having, on average, a higher level of exposed hydrophobic surface relative to the fibrils. Indeed, the significant predicted differences in surface hydrophobicity between the 28 S oligomers and that found for the fibrils further indicate that the β-sheet arrangement in these two types of aggregated species is different.
Three-Dimensional Structural Analysis of the Two Major Subgroups of αS Oligomers.
At a fundamental level, the rapid elongation rate of small fibrils in the presence of monomer hampers the study of the intermediate transient oligomeric species generated during the formation of fibrils. The ability to produce and isolate trapped oligomeric forms, such as those described here, opens up the possibility of gaining insights into the nature and structure of these species. To obtain more detailed information concerning the structures of the αS oligomeric species, the samples were analyzed by electron microscopy using both negative stain techniques in TEM (SI Appendix, Fig. S6) and direct visualization of vitrified unstained samples in cryoEM (Fig. 4A). In both cases, the analyses revealed a small number of large aggregates, which were not examined further in this study, and a large number of smaller species that were found in two main orientations (Fig. 4A and SI Appendix, Fig. S6A), one representing a “doughnut” shape, similar to that described in a number of previous reports of TEM images of amyloid oligomers (33, 39), whereas the other orientation has a “cylinder-like” appearance, also described in previous TEM studies (33). Interestingly, both types of images, which appear to correspond to the two main orthogonal orientations of the oligomers, could be observed regardless of the apparent size of the oligomeric species (Fig. 4 B and C and SI Appendix, Fig. S6B).
Fig. 4.
Three-dimensional reconstructions of the two main size subgroups of oligomers of purified oligomeric samples. (A) Example of cryoEM image of an oligomeric sample. Representatives of the two main particle subgroups are highlighted: with arrows, for the 15S oligomeric species; arrowheads, for the 10S oligomers. (B) Typical side view (Top) and end-on view (Bottom) of the small oligomeric subgroup (corresponding to the 10S oligomer subgroup) according to cryoEM. (C) The same views for the large structural group (corresponding to the 15S oligomer subgroup). (D) Two orthogonal views, side (Left) and end-on (Right), of the 3D reconstruction of the average structure for the 10S oligomer subgroup. (E) The same views for the 3D reconstruction of the structure representing the 15S oligomer subgroup.
To investigate these structures further, 7,776 and 17,242 particle images from the unstained (cryoEM) and stained samples, respectively, were selected, processed and classified as described in Experimental Procedures (SI Appendix, Fig. S6 B–D). To understand their significance, we separated the particle images in two size subgroups, which would correspond to the 10S and 15S species previously identified in the AU analysis of the samples. The classification procedure showed clearly the two main types of particle image discussed above (Fig. 4 B and C and SI Appendix, Fig. S6C). We carried out independent 3D reconstructions for each subgroup (Fig. 4 D and E and SI Appendix, Fig. S6E). The results revealed a similar structure for both types of oligomer subgroups, albeit of different size: a cylinder-like structure with a region of low electron density running through the cylinder, giving the appearance of a hollow core. Both structures showed irregularities in the cylindrical shape that are likely to reflect variations in the number and length of the β-strands within the different oligomer particles.
The reconstruction of the 10S oligomer subgroup, which represents about 60% of the particles analyzed, reveals a structure that is approximately 120 Å in length and approximately 90 Å in diameter, with an apparent central cavity of approximately 25 Å in diameter. That of the 15S oligomer subgroup, representing about 40% of the particles analyzed, has a length of approximately 140 Å and a diameter of approximately 100 Å, again with a cavity of approximately 25 Å in diameter. It is interesting that the ratio of the length to the width of both types of oligomer is similar (1.33 and 1.40), as are the dimensions of the central cavity, and that few more elongated structures were observed.
Comparison with αS Oligomers Observed in Single-Molecule Experiments.
We have recently described studies of the aggregation of αS by using single-molecule fluorescence techniques that revealed the presence of two distinct forms of oligomeric species during fibril formation that differed in their fluorescence behavior, kinetics of formation, degree of compactness, and susceptibility to degradation by proteases; we denoted these forms type A and type B, respectively (23). The rate of conversion between the two forms was found to be slow, of the order of tens of hours at 37 °C, and studies using rat primary midbrain neuronal cells showed that the oligomers formed initially during αS aggregation (type A oligomers) were essentially benign, whereas those formed after the conversion process (type B oligomers) were able to induce a significant and aberrant production of reactive oxygen species (ROS).
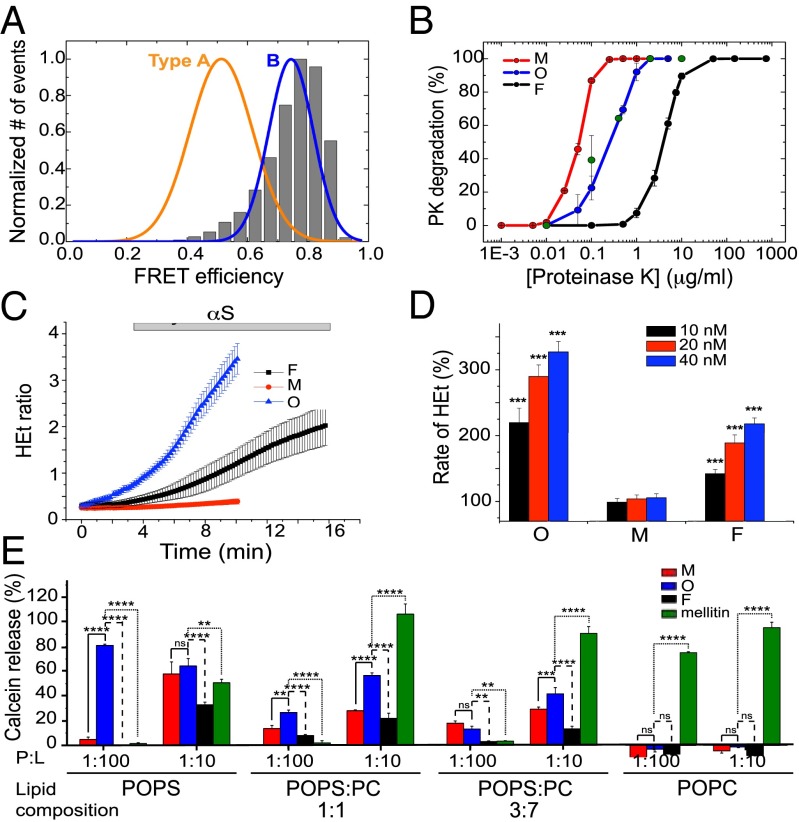
To assess whether the purified oligomers characterized here could be similar to one or the other of these two forms of oligomers previously found to form during αS aggregation (23), we first compared their fluorescent properties, particularly the distribution of fluorescence resonance energy transfer (FRET) efficiencies obtained by single-molecule fluorescence of the purified oligomeric samples described here with the FRET distributions of the type A and type B oligomers (SI Appendix, Experimental Procedures and Fig. S7). The single-molecule FRET distributions of the dual labeled (AlexaFluor-488 as FRET donor and AlexaFluor-647 as FRET acceptor) purified oligomeric samples (and their apparent size distributions; SI Appendix, Fig. S8) were found to overlap completely with the equivalent data obtained in our previous study for the type B oligomers generated during αS fibril formation (Fig. 5A). In addition, both types of oligomers showed identical degrees of resistance against proteinase-K (PK) degradation (Fig. 5B), which were intermediate between that exhibited by the monomeric protein (that is readily degradable because of its intrinsically disordered nature), and the fibrillar form of the protein (that is highly resistant to degradation because of its stable cross–β-sheet structure (34)). Taken together, these findings indicate that the structures of the purified oligomers studied in this work are closely similar to the toxic type B oligomers observed in the single-molecule experiments to be populated during the aggregation reaction. In addition, the similarities between oligomeric forms generated by using the protocol described here and those detected under conditions in which fibrils can form (17, 20–22, 24, 26, 27) indicates that oligomeric structures of similar compactness and overall quaternary structure are formed under a variety of conditions, suggesting a common mechanism for the self-assembly of polypeptide chains into β-sheet–rich aggregates.
Fig. 5.
Comparison of the purified αS oligomers with toxic oligomers previously found during αS fibril formation. (A) The single-molecule FRET efficiency distribution of a purified αS oligomeric sample (shown in gray bars) compared with the FRET distributions of the two main oligomeric species (type A and type B) previously found during αS fibril formation. (B) Proteinase K degradation curves of the different protein species: monomers in red, oligomers in blue, and fibrils in black. The data represented correspond to the average and SE of three different experiments. The degradation profile of type B oligomers is shown by green circles. (C) Time-resolved cytoplasmatic ROS production (HEt: dihydroethidium) in rat midbrain neuronal cultures after exposure to the different αS species: monomers in red, oligomers in blue, and fibrils in black. (D) Dose–response effect of monomers (M), oligomers (O), and fibrils (F) on the rate of cytoplasmatic ROS production. The basal rate of ROS production was taken as 100%. ***P < 0.0001. (E) Calcein release from LUVs of different POPS:POPC ratios after being incubated with monomeric (red bars), oligomeric (blue bars), and fibrillar (black bars) αS, at two different protein:lipid ratios (1:10 and 1:100). The pore-forming peptide, melittin, was used as positive control (green bars). The signal obtained when the detergent Triton X-100 was added to the vesicles was taken as 100%. Data points are averaged triplicates, and the error bars represent the SDs. *P < 0.5; **P < 0.01; ***P < 0.001; ****P < 0.0001; ns, not significant.
Toxicity of the αS Oligomers.
Given the high structural similarities between the purified oligomers characterized here and the type B oligomers generated during the aggregation of αS, we assessed whether they have similar toxic effects on neuronal cells. First, we incubated rat midbrain primary neuronal cultures with 40 nM of purified oligomers and compared the ROS activity, measured by using dihydroethidium (HEt) as a probe to quantify the production of superoxide radicals, with the same mass concentration of the monomeric and fibrillar forms of the protein. We have shown previously that monomeric, oligomeric, and fibrillar αS species are all taken up rapidly by both neurons and astrocytes (within 5–10 min), without significant variations between the different species (23), allowing us to correlate directly the effects of the addition of the same amount of the different protein species to the cells.
The results of these experiments show that the addition of monomeric αS produced a negligible change in ROS production (with maximal values of HEt at 105.1 ± 6.7% of the basal rate; n = 72 cells; Fig. 5C) while αS fibrils were found to elicit a modest increase in ROS response in neuronal cells (218 ± 9%; n = 88, P < 0.001). Addition of solutions of purified oligomers at the same mass concentration (40 nM), however, produced a substantial increase in the rate of cellular ROS formation (327 ± 16%; n = 94 cells, P < 0.0001) (Fig. 5C), significantly higher than that found for the fibrillar sample. To ensure that the effect on neuronal cells upon exposure to the different αS species was specific, we conducted a dose–response assay. Fig. 5D demonstrates that exposing the neuronal cells to increasing concentrations of both oligomeric and fibrillar αS resulted in step-wise increases in cellular ROS production, whereas the monomeric form had no significant effect at any concentration tested. According to our dose–response analysis in the nanomolar protein concentration range, the cells would require four times more protein in the fibrillar state compared with the oligomeric state to be able to produce a similar ROS response. This finding suggests that a transition from αS oligomers to fibrils results in a substantial reduction of ROS-induced cellular damage.
To probe further the toxicity of the oligomers, we used calcein release assays to explore the ability of our oligomeric species to cause membrane disruption in LUVs (large unilamellar vesicles) of lipid compositions similar to those that have been used to examine the effects of αS oligomers (21, 24, 40). It has been shown that monomeric αS interacts with acidic phospholipids, probably through the lysine residues located in the N-terminal region of the protein (41), and a similar initial interaction has been also proposed for some oligomeric conformations (40). For this reason, most of the studies of membrane disruption involving αS oligomeric species have been carried out with a high content of acidic phospholipids, typically 100% phosphatidylglycerol (PG) or phosphatidylserine (PS) (20, 40). It has, however, been found that the effects of αS oligomers on the dynamic properties of synthetic lipidic vesicles depend strongly on the relative proportion of acidic and neutral phospholipids (21, 24). We therefore prepared LUVs containing only either POPS (an acidic phospholipid) or POPC (a neutral phospholipid), and also containing 1:1 and 3:7 POPS-POPC mixtures, with the latter composition being the most physiologically relevant (42). Because of its presence in brain membranes, we chose PS instead of PG to increase the negative charge content of the vesicles.
Because monomeric αS has been found to have its highest helical content and ability to partition into a membrane at protein:lipid (P:L) ratios of approximately 1:100 (43, 44), we compared the extent of calcein release induced by the different αS species at this P:L ratio. In additional experiments, we used a P:L ratio of 1:10 to compare the effect of αS species with the membrane disrupting peptide melittin (the major toxin of honey bee venom), known to induce membrane permeabilization above this threshold ratio (45). A summary of the results is given in Fig. 5E and shows that oligomeric αS causes a higher calcein release from the LUVs at both P:L ratios and at the various lipid compositions tested than either the monomeric or fibrillar forms of the protein. LUVs composed only of POPS showed the highest level of calcein release (more than 60%) in the presence of oligomers at P:L ratios of 1:10 and 1:100. Interestingly, at the low P:L ratio, αS oligomers induce a much higher calcein release in those vesicles than the toxic melittin at any lipid composition tested except for pure POPC LUVs. Because the PS content in vesicles is reduced, less calcein release is observed, with less than 20% leakage at a P:L ratio of 1:100 and less than 50% at a P:L ratio of 1:10 for the more physiologically relevant 3:7 POPS-POPC LUVs. In addition, leakage was not detected with only POPC vesicles. These data are in good agreement with previously reported propensities of various types of αS oligomers to induce disruption of highly negatively charged membranes through formation of defects due to the intrinsic instability of these types of vesicles to protein adsorption (21).
The trend observed for melittin is the opposite of that for αS oligomers, with the greatest perturbation effects with LUVs of low acidic composition, an effect that also reflects the fact that the interaction of the peptide with the bilayer surface highly depends on the surface charge. Although the presence of negatively charged lipids is likely to promote melittin binding, its propensity to induce leakage is more fundamentally governed by its ability to penetrate into the hydrophobic region of the membrane, which becomes more favorable the less electrostatic interactions attract the peptide to the lipidic surface (46). The behavior of melittin with respect to the P:L ratio that we have obtained is also in agreement with the reported threshold P:L ratio value for the effective melittin-induced membrane perturbation (45, 46). Taken together, the calcein release data suggest that the αS oligomers described in the present study are able to disrupt lipid vesicles with a high content of negatively charge headgroups and, consequently, an intrinsic instability of the lipid bilayer. This finding is in agreement with reported effects of αS oligomers on synthetic vesicles with similar lipid composition (21, 24, 40). This effect is greater for the oligomers than for either the monomeric or the fibrillar species, showing the greater ability of the former to affect membrane permeability upon binding. Our data also suggest that the mechanism of disruption is unlikely to be similar to that of the pore-forming melittin.
Discussion
Detailed information on the formation, structure, and mechanisms of toxicity of oligomeric forms of amyloid aggregates is of fundamental importance for developing our understanding of the molecular origins and means of progression of protein misfolding disorders, and as a basis for developing rational means of therapeutic intervention. For this reason, we have generated and purified samples of stable αS oligomers at micromolar concentrations and used a range of biophysical methods to characterize the nature and distributions of their sizes, morphologies, and structural characteristics. All of the information accumulated from these studies indicates that the purified αS oligomers characterized here have a remarkably high degree of similarity, in terms of physico-chemical, structural, and toxic properties, with oligomeric species formed by αS under different conditions and other amyloidogenic peptides and proteins (16, 17, 21, 22, 24–28, 47), and are representative of the most highly compact and stable oligomeric αS species identified to accumulate during fibril formation by means of single-molecule fluorescence techniques (23).
Structural Characteristics of Oligomeric Aggregates.
The oligomers investigated in this work have structural features that are intermediate between the intrinsically disordered monomeric protein and the structurally highly organized mature fibrils, reflected in terms of their size, compactness, β-sheet content, and resistance to proteolytic degradation. The 3D EM reconstructions of the oligomers studied here reveal a broadly cylindrical architecture that appears to be stable over a range of oligomer sizes from approximately 160 to 560 kDa, as estimated by HYDROMIC analysis, with variable β-sheet content. The size range determined for the oligomers present in our experiments agrees well with the sizes of a variety of subgroups of αS oligomers identified in samples generated by using similar approaches to that used in the present study (22, 28), although some species have been reported to differ in their apparent morphologies (22) and stabilities against urea denaturation (26). Two major size subgroups, designated 10S and 15S, with molecular masses of approximately 260 and 420 kDa on average (corresponding to an average of approximately 18 and 29 protein molecules, respectively; note that the overall size distribution of these two subgroups comprises oligomers composed of 10–40 protein molecules), dominate the distribution of oligomers in our sample, which have an average β-sheet content of approximately 30% and 40% respectively, with the rest of the protein being largely disordered, and a significant degree of hydrophobic surface area exposed to the solvent.
In addition, we have found a strong correlation between both the secondary structure content and the degree of exposed hydrophobicity and the size of the oligomers, with the larger oligomers showing a higher level of β-sheet content and a greater area of hydrobobic surface exposed to the solvent. A similar correlation between size and exposed hydrophobicity has been recently observed for a set of oligomeric species of a model amyloidogenic protein generated by mutagenesis (48), suggesting that such a relationship may be a common feature of such species. Interestingly, the surface hydrophobicity of the fibrillar form of αS is estimated to be intermediate between the small and large oligomer subgroups, despite their differences in size, a finding that reflects the differences in the β-sheet geometry between these oligomeric species (predominantly antiparallel β-sheet) and the fibrillar form of the protein (mainly parallel β-sheet) already observed by FTIR spectroscopy.
The cryoEM and TEM image analyses of the αS oligomeric species characterized here reveal essentially two major groups of structural orientations that are independent of the sizes of the oligomers, one with a “doughnut” shape and the other one with a cylindrical appearance, consistent with some previous observations of αS oligomers (33). The high homogeneity of our purified oligomeric sample together with the detailed combined EM-AU analysis have allowed us to carry out 3D reconstructions on the two major size subgroups of oligomers present in our samples. The average 3D reconstructions of the subgroups (10S and 15S) reveal the same type of cylindrical architecture, with dimensions that vary between approximately 120 and 140 Å in length and 90 and 100 Å in diameter in average, but with similar dimensions for the central cavity (approximately 25 Å in diameter). The average thickness of the walls of the cylindrical structures (30 and 40 Å on average for the 10S and the 15S oligomer subgroups, respectively) is too large for a folding core composed of a single β-sheet folded into a barrel structure of the type found for a crystalline hexameric species formed by a short segment of αB crystallin (12). The dimensions of the oligomeric species described here are, instead, much closer to those reported for fibrillar structures of αS from cryo-negative EM studies, which are 80–120 Å in diameter, with a central cavity of approximately 20–30 Å in diameter running through the structure (49) and, indeed, long cylindrical structures with water-filled interfaces have been proposed for a variety of amyloid fibrils (4, 5, 7–10) and similar protein regions as those found in the fibrillar core have been suggested to be involved in intermolecular contacts within αS oligomers prepared by using a similar approach to that we have used here (27).
This comparison suggests that it is possible that the self-assembly of proteins into amyloid aggregates with β-sheet cores generates similar structural architectures at both the fibrillar and oligomeric level, regardless of the β-sheet geometry. Indeed, the 10S oligomeric structures described here are consistent with a folding core composed of a pair of face-to-face β-sheets that are further assembled into its cylindrical structure. Moreover, the large 15S structure appear to be similar but with a higher β-sheet content, probably as a result of the addition of further protein molecules with similar structure (note that the average thickness of the 10S and 15S cylindrical structures of these oligomers is consistent with two and three β-strands per protein molecule, respectively, separated by approximately 10 Å, in agreement with the reported intramolecular β-strand distance common to all amyloid fibrillar structures; ref. 3).
Similar 2D EM images of oligomers as those described here have been reported for a range of amyloid peptides and proteins collectively described as “amyloid pores” (33). Most of these amyloid oligomers have been found to bind the amyloid oligomer-specific A11 antibody (47), and, like them, the αS oligomers described here are also able to bind to A11 (SI Appendix, Fig. S9), suggesting that the underlying architecture we report here can be adopted by different peptides and proteins, regardless of their amino acid composition and sequence, as proposed for the the fibrillar structure (1, 2). The presence of a central cavity in some oligomeric species has led to proposals that this specific “pore-like” structure could be characteristic of some oligomers and a key determinant of their toxicity (47). Based on our findings, we propose that the cavity observed in amyloid oligomers is likely to be an inherent property of the water-mediated face-to-face packing of pairs of β-sheets stabilized by inter-main chain hydrogen bonding networks, as found for the amyloid fibrils, rather than a unique structural feature of the amyloid oligomers.
Relationship Between Oligomeric and Fibrillar Aggregates.
A close similarity between the global architecture of these oligomers and that found to be characteristic of at least some fibrils suggests that similar types of interactions (notably an array of interbackbone hydrogen bonds linking the β-strands as in the fibrillar structures; ref. 6) could stabilize both types of species. Consequently, a high degree of heterogeneity of β-sheet oligomers with the same type of core architecture but different numbers and lengths of β-strands, types of β-sheet arrangements, and permutations of interstrand hydrogen bonding interactions could be expected as has been observed to occur in fibrillar structures, particularly of short peptides (4, 50). Indeed, it is likely that the protein subunits within the same oligomeric species will have different numbers and lengths of β-strands, reflecting imperfections in the packing of the oligomers, as we have already noticed during the EM analysis of the αS oligomeric samples studied here. The accumulation of β-sheet–rich oligomers with a significant degree of heterogeneity in their β-sheet content could well be a consequence of the extremely slow rates of assembly and reorganization of amyloid-like β-sheet structures that we have observed to be orders of magnitude slower than the folding of small proteins into their native functional states (23).
An important difference between the oligomeric forms of αS described here and the fibrils, despite their similarities in overall architecture, is their relative ability to elongate. Whereas the fibrillar structures are readily able to increase in length, the oligomeric species studied here have a much lower tendency to grow by further addition of monomers (at least 3 orders of magnitude slower than fibrils of similar size under the conditions tested; SI Appendix, Fig. S10), a feature that has been reported for samples of αS oligomers prepared in a similar maner (22). This finding can be attributed to the differences in the arrangement of the β-strands in the oligomers and the fibrils. Fibrils containing antiparallel β-sheets have been described, although mainly for relatively short peptides (50, 51), and compared with their parallel counterparts they have been found at least in some cases to be less stable and less efficient in elongating (50). Our findings suggest that a rearrangement of the β-strands from an antiparallel to a parallel configuration would be required for the efficient elongation of these αS oligomers to generate the fibrillar architecture. Such a process is likely to be extremely slow, perhaps involving the partial unfolding and disaggregation of these oligomers by Ostwald ripening as recently observed for the formation of other supramolecular assemblies (52), a fact that could explain the high kinetic stability of these oligomeric species. Moreover, it seems likely that oligomers with a parallel β-sheet architecture, which our results suggest posseses lower degree of surface-exposed hydrophobicity and, therefore, a lower level of intrinsic toxicity, are also formed in the early stages of αS aggregation but are able to elongate rapidly and generate fibrils. This conclusion highlights the role β-sheet geometry plays in the process of misfolding and self-association of amyloid proteins, and the importance of the rates (and energy barriers) of the structural conversions between different β-sheet geometries for dictating the kinetic stability of the different aggregated forms.
The Toxicity of the Oligomeric Species.
One of the most commonly reported measurements of the toxicity of amyloid oligomers is the extent to which they disrupt lipid membranes; the ability to generate such disruption appears to be a general feature of all amyloid oligomers (16). In agreement with previously reported data on the ability of specific αS oligomers to disrupt synthetic lipid vesicles (21, 24, 40), we have observed that the oligomers characterized in the present study are much more efficient in permeabilizing lipid vesicles than are monomeric or fibrillar forms of αS at the same mass concentration. Indeed, the oligomers are more efficient than melittin, a peptide toxin that acts through pore formation (45), when the vesicles are primarily composed of negatively charged phopholipids; when the content of acidic phospholipids in the vesicles is reduced, we observed a gradual decrease of this effect. Our data are fully consistent with the results of a previous systematic analysis of the influence of the stability of synthetic lipid vesicles on the degree to which they are perturbed by αS oligomers (21), and suggest that the accessibility of the hydrophobic core of the bilayer depends on intrinsic defects in its structure, such as those caused by a high content of negatively charged phospholipids, and that such defects seem to be crucial for the ability of αS oligomers to disrupt the lipid bilayer.
We also report here that the purified αS oligomers have the ability to induce an aberrant production of ROS in primary neuronal cells even at protein concentrations in the nanomolar range. Excessive generation of free radicals has itself been reported to trigger pathological production of misfolded proteins, abnormal mitochondrial function, and the stimulation of apoptotic pathways in neuronal cells (53). We find that αS fibrils would require much higher protein concentrations to produce similar levels of ROS than the oligomeric species studied here, whereas monomeric protein molecules do not appear to stimulate detectable levels of ROS production in neuronal cells. The differences in ROS production between the oligomers and fibrils, normalized for the number of protein molecules in each species, are likely to be a result of the greater surface-to-volume ratio in the former species or of differences in subcellular localization.
The fact that certain oligomeric species formed during amyloid aggregation can induce cellular ROS production has been reported for several systems (23, 54, 55), and is particularly relevant in the context of PD, because in this disorder the link between oxidative stress and the development of disease is well established (56). Furthermore, in the context of αS, it has been reported that overexpression of αS increases the vulnerability of neurons to dopamine-induced cell death through excess intracellular ROS generation (57). Interestingly, increased ROS and raised oxidative stress levels have been reported to cause damage to neuronal membranes (58) and, indeed, to promote further aggregation of αS (59), reflecting the fact that positive feedback can occur between the different types of pathological processes in PD.
Multiplicity of Misfolding Pathways and Its Significance for Disease.
The high kinetic stability of the oligomers studied here can be attributed to the antiparallel nature of at least some of the β-sheets within their core structures; because the fibrillar form of aggregated αS is characterized by a parallel arrangement of β-strands within the core structure, the rate of rearrangement of this type of oligomer into species capable of efficient elongation is likely to be extremely slow. By contrast, those oligomers within the heterogeneous mixture of species formed early in the self-assembly process that have assembled into structures containing parallel β-sheets, probably through reorganization of initially more amorphous aggregates (23), are likely to be able to elongate efficiently to form fibrils without the need for a major structural reorganization and, hence, to be transient in nature. An interesting feature of this mechanism of multiple misfolding pathways is that it is directly analogous to the multiplicity of parallel pathways observed in the productive folding of a range of proteins and shown to result from an initial collapse to disordered structures followed by subsequent reorganizational events (60, 61). As proposed here for misfolding and aggregation processes, in such cases, some pathways lead to rapid acquisition of stable structure and others to the accumulation of metastable intermediates before the slower accumulation of the more stable state.
The concept of a multiplicity of assembly steps resulting in an ensemble of oligomers with differing β-sheet arrangements, rates of elongation, and inherent toxicities leads to the interesting possibility that protein misfolding and aggregation process in the cell can generate species with different pathological roles; the elongation prone, fibril-like oligomers with parallel β-sheet arrangement could act as key pathogenic species for the spreading and transmission of the disease, whereas oligomers with an antiparallel β-sheet arrangement, such as that described here, could accumulate within cells and, being highly hydrophobic and slow to degrade because of their inherent resistance to proteolysis, act as potent toxins.
Experimental Procedures
Preparation of Purified αS Oligomeric Samples.
αS oligomeric samples were prepared on the basis of previous protocols (17, 21, 22, 24, 25, 27, 40, 62, 63). Briefly, 6 mg of lyophilized protein was resuspended in PBS buffer, pH 7.4, to give a final concentration of approximately 800 μM (12 mg/mL) and passed through a 0.22-μm cutoff filter immediately before incubation at 37 °C for 20–24 h without agitation or the application of any other process that could induce shear and, hence, accelerate the conversion of monomers and oligomers into fibrils (23, 29). During this time, a small number of fibrillar species were observed to form and removed by ultracentrifugation for 1 h at 90,000 rpm (using a TLA-120.2 Beckman rotor; 288,000 × g). The excess of monomeric protein, and the low levels of small oligomers, were removed by means of multiple filtration steps (using 100-kDa cutoff membranes) to enrich the sample in pure oligomeric species of αS (see SI Appendix, SI Experimental Procedures for a full description of the protocol). The oligomeric samples were found to remain stable for days (SI Appendix, Fig. S1) and were used within the first two days after their production. The concentrations of the final solutions of oligomers were estimated from the absorbance at 275 nm by using a molar extinction coefficient of 5,600 M−1⋅cm−1 (no significant changes in the molar extinction coefficient value were found for the oligomeric species relative to the monomeric protein). The concentration values given in this work represent the total mass concentration of protein, i.e., the total concentration in monomer equivalents.
We noted that there was a substantial enrichment in stable αS oligomers when lyophilized protein stock solutions were used compared with nonlyophilized protein samples, and these oligomeric species have been shown to have a high degree of similarity to oligomeric species formed when freshly prepared protein is incubated under standard conditions that lead to the formation of amyloid fibrils (see refs. 17, 21, 22, 24, 27, 40, and 63 and Results). These oligomeric species are thermodynamically and kinetically stable (Fig. 3 and SI Appendix, Fig. S1), a property attributable to their amyloid-like structural architecture and their antiparallel β-sheet arrangement, and can be isolated and remain stable for days after they are produced, even in the presence of monomeric protein (SI Appendix, Fig. S1).
Sedimentation Velocity Experiments by Analytical Ultracentrifugation.
Sedimentation velocity measurements were performed at 20 °C, 38,000–43,000 rpm (106,750–136,680 × g) by using a Beckman-Coulter Optima XL-I analytical ultracentrifuge equipped with UV-visible absorbance optics and an An50Ti rotor. All protein samples (40–80 μM) were incubated in their respective buffers for at least 6 h before the start of the sedimentation velocity experiments. The sedimentation coefficient distributions, corrected to standard conditions by using the SEDNTERP program (64), were calculated via least-squares boundary modeling of sedimentation velocity data using the c(s) and ls-g*(s) methods, as implemented in the SEDFIT program (www.analyticalultracentrifugation.com/default.htm).
Cryoelectron Microscopy and Image Processing.
Protein aliquots were applied to glow-discharged, holey carbon grids (carbon-coated Quantifoil R 1.2/ R1.3 300 mesh grids) containing an additional continuous thin layer of carbon and plunged into liquid ethane. Images were acquired under minimal dose conditions with a Tecnai F20 transmission electron microscope at 200 kV. The images were taken at a magnification of ∼50,000× by using a 16 megapixel (Mpx) FEI Eagle CCD camera with a step size of 15 μm; thus, the original pixel size of the acquired images was 2.74 Å. Individual particles were selected manually and extracted by using XMIPP software (65). Particle classification was carried out by using maximum-likelihood multireference refinement approaches, and the resulting class averages and their corresponding assigned particles were visually separated into two main size groups (as suggested by the AU analysis of the samples) on the basis of their overall dimensions, and then subjected to an iterative procedure consisting of several rounds of 2D classification to enable a clearer separation of the two size populations (SI Appendix, Fig. S6). For the 3D reconstruction of the two populations that were resolved, several starting reference models and initial 3D reconstruction steps based on iterative angular refinement were performed by using the EMAN software package (66) (see SI Appendix for more details). The different strategies converged to similar solutions, and one of the models for each population was selected to complete the refinement. The resolution of the 3D reconstructions determined by the FSC 0.5 criterion was 18 Å and 19 Å for the small and large oligomer population, that would correspond to the10S and 15S oligomer subgroups identified by AU, repectively. A detailed description of the EM methodology and the rest of the experimental procedures can be found in SI Appendix, Experimental Procedures.
Supplementary Material
Acknowledgments
We thank Dr. Katherine Stott, from the Biophysics Facility, Department of Biochemistry, University of Cambridge, for her assistance in using these facilities. This work was supported by the Agency for Science, Technology and Research, Singapore (S.W.C.), the “La Caixa” foundation (S.D.), Wellcome/MRC (Medical Research Council) Parkinson’s Disease Consortium Grant WT089698 (to E.D. and N.W.W.), National Institute for Health Research Biomedical Research Centres funding at University College London (to N.W.W.), the BBSRC through Grants BB/H003843/1 (to M.O.) and BB/E019927/1 (to C.M.D.), the Spanish Ministry of Economy and Competitiveness through Grants SAF 2012-39720 (to C.R.), BFU2013-44202 (to J.M.V.), and BIO2011-28941-C03-03 (to C.A. and G.R.), the Spanish Ministry of Health with cofunding by The European Regional Development Fund through Grant CP10/00527 (to C.R.), the Madrid Regional Government through Grant S2013/MIT-2807 (to J.M.V.), Parkinson’s UK through Grant H-0903 (to T.G.), the Wellcome Trust, the Leverhulme Trust, the European Commission through project LSHM-CT-2006-037525 (to C.M.D.), the Medical Research Council through Grant MRC G1002272 (to E.J.D.-G. and C.M.D.), and the Engineering and Physical Sciences Research Council (C.M.D.). A.Y.A. was a Parkinson’s UK Senior Research Fellow. N.C. is a Royal Society Research Fellow and also acknowledges financial support by the Human Frontier Science Program from Long-Term Fellowship LT000795/2009.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1421204112/-/DCSupplemental.
References
- 1.Chiti F, Dobson CM. Protein misfolding, functional amyloid, and human disease. Annu Rev Biochem. 2006;75:333–366. doi: 10.1146/annurev.biochem.75.101304.123901. [DOI] [PubMed] [Google Scholar]
- 2.Eisenberg D, Jucker M. The amyloid state of proteins in human diseases. Cell. 2012;148(6):1188–1203. doi: 10.1016/j.cell.2012.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sunde M, et al. Common core structure of amyloid fibrils by synchrotron X-ray diffraction. J Mol Biol. 1997;273(3):729–739. doi: 10.1006/jmbi.1997.1348. [DOI] [PubMed] [Google Scholar]
- 4.Fitzpatrick AW, et al. Atomic structure and hierarchical assembly of a cross-β amyloid fibril. Proc Natl Acad Sci USA. 2013;110(14):5468–5473. doi: 10.1073/pnas.1219476110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang R, et al. Interprotofilament interactions between Alzheimer’s Abeta1-42 peptides in amyloid fibrils revealed by cryoEM. Proc Natl Acad Sci USA. 2009;106(12):4653–4658. doi: 10.1073/pnas.0901085106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Knowles TP, Vendruscolo M, Dobson CM. The amyloid state and its association with protein misfolding diseases. Nat Rev Mol Cell Biol. 2014;15(6):384–396. doi: 10.1038/nrm3810. [DOI] [PubMed] [Google Scholar]
- 7.Jiménez JL, et al. Cryo-electron microscopy structure of an SH3 amyloid fibril and model of the molecular packing. EMBO J. 1999;18(4):815–821. doi: 10.1093/emboj/18.4.815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Serpell LC, et al. Examination of the structure of the transthyretin amyloid fibril by image reconstruction from electron micrographs. J Mol Biol. 1995;254(2):113–118. doi: 10.1006/jmbi.1995.0604. [DOI] [PubMed] [Google Scholar]
- 9.Serpell LC, Smith JM. Direct visualisation of the beta-sheet structure of synthetic Alzheimer’s amyloid. J Mol Biol. 2000;299(1):225–231. doi: 10.1006/jmbi.2000.3650. [DOI] [PubMed] [Google Scholar]
- 10.Serpell LC, et al. The protofilament substructure of amyloid fibrils. J Mol Biol. 2000;300(5):1033–1039. doi: 10.1006/jmbi.2000.3908. [DOI] [PubMed] [Google Scholar]
- 11.Sawaya MR, et al. Atomic structures of amyloid cross-beta spines reveal varied steric zippers. Nature. 2007;447(7143):453–457. doi: 10.1038/nature05695. [DOI] [PubMed] [Google Scholar]
- 12.Laganowsky A, et al. Atomic view of a toxic amyloid small oligomer. Science. 2012;335(6073):1228–1231. doi: 10.1126/science.1213151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bucciantini M, et al. Inherent toxicity of aggregates implies a common mechanism for protein misfolding diseases. Nature. 2002;416(6880):507–511. doi: 10.1038/416507a. [DOI] [PubMed] [Google Scholar]
- 14.Stefani M, Dobson CM. Protein aggregation and aggregate toxicity: New insights into protein folding, misfolding diseases and biological evolution. J Mol Med (Berl) 2003;81(11):678–699. doi: 10.1007/s00109-003-0464-5. [DOI] [PubMed] [Google Scholar]
- 15.Nath S, et al. Spreading of neurodegenerative pathology via neuron-to-neuron transmission of β-amyloid. J Neurosci. 2012;32(26):8767–8777. doi: 10.1523/JNEUROSCI.0615-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bemporad F, Chiti F. Protein misfolded oligomers: Experimental approaches, mechanism of formation, and structure-toxicity relationships. Chem Biol. 2012;19(3):315–327. doi: 10.1016/j.chembiol.2012.02.003. [DOI] [PubMed] [Google Scholar]
- 17.Celej MS, et al. Toxic prefibrillar α-synuclein amyloid oligomers adopt a distinctive antiparallel β-sheet structure. Biochem J. 2012;443(3):719–726. doi: 10.1042/BJ20111924. [DOI] [PubMed] [Google Scholar]
- 18.Paslawski W, Mysling S, Thomsen K, Jørgensen TJ, Otzen DE. Co-existence of two different α-synuclein oligomers with different core structures determined by hydrogen/deuterium exchange mass spectrometry. Angew Chem Int Ed Engl. 2014;53(29):7560–7563. doi: 10.1002/anie.201400491. [DOI] [PubMed] [Google Scholar]
- 19.Hong DP, Fink AL, Uversky VN. Structural characteristics of alpha-synuclein oligomers stabilized by the flavonoid baicalein. J Mol Biol. 2008;383(1):214–223. doi: 10.1016/j.jmb.2008.08.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim HY, et al. Structural properties of pore-forming oligomers of alpha-synuclein. J Am Chem Soc. 2009;131(47):17482–17489. doi: 10.1021/ja9077599. [DOI] [PubMed] [Google Scholar]
- 21.van Rooijen BD, Claessens MM, Subramaniam V. Lipid bilayer disruption by oligomeric alpha-synuclein depends on bilayer charge and accessibility of the hydrophobic core. Biochim Biophys Acta. 2009;1788(6):1271–1278. doi: 10.1016/j.bbamem.2009.03.010. [DOI] [PubMed] [Google Scholar]
- 22.Lorenzen N, et al. The role of stable α-synuclein oligomers in the molecular events underlying amyloid formation. J Am Chem Soc. 2014;136(10):3859–3868. doi: 10.1021/ja411577t. [DOI] [PubMed] [Google Scholar]
- 23.Cremades N, et al. Direct observation of the interconversion of normal and toxic forms of α-synuclein. Cell. 2012;149(5):1048–1059. doi: 10.1016/j.cell.2012.03.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Volles MJ, et al. Vesicle permeabilization by protofibrillar alpha-synuclein: Implications for the pathogenesis and treatment of Parkinson’s disease. Biochemistry. 2001;40(26):7812–7819. doi: 10.1021/bi0102398. [DOI] [PubMed] [Google Scholar]
- 25.Danzer KM, et al. Different species of alpha-synuclein oligomers induce calcium influx and seeding. J Neurosci. 2007;27(34):9220–9232. doi: 10.1523/JNEUROSCI.2617-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Paslawski W, et al. High stability and cooperative unfolding of α-synuclein oligomers. Biochemistry. 2014;53(39):6252–6263. doi: 10.1021/bi5007833. [DOI] [PubMed] [Google Scholar]
- 27.Gallea JI, Celej MS. Structural insights into amyloid oligomers of the Parkinson disease-related protein α-synuclein. J Biol Chem. 2014;289(39):26733–26742. doi: 10.1074/jbc.M114.566695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zijlstra N, Blum C, Segers-Nolten IM, Claessens MM, Subramaniam V. Molecular composition of sub-stoichiometrically labeled α-synuclein oligomers determined by single-molecule photobleaching. Angew Chem Int Ed Engl. 2012;51(35):8821–8824. doi: 10.1002/anie.201200813. [DOI] [PubMed] [Google Scholar]
- 29.Campioni S, et al. The presence of an air-water interface affects formation and elongation of α-Synuclein fibrils. J Am Chem Soc. 2014;136(7):2866–2875. doi: 10.1021/ja412105t. [DOI] [PubMed] [Google Scholar]
- 30.Schuck P. Size-distribution analysis of macromolecules by sedimentation velocity ultracentrifugation and lamm equation modeling. Biophys J. 2000;78(3):1606–1619. doi: 10.1016/S0006-3495(00)76713-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schuck P, Rossmanith P. Determination of the sedimentation coefficient distribution by least-squares boundary modeling. Biopolymers. 2000;54(5):328–341. doi: 10.1002/1097-0282(20001015)54:5<328::AID-BIP40>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- 32.García de la Torre J, Llorca O, Carrascosa JL, Valpuesta JM. HYDROMIC: Prediction of hydrodynamic properties of rigid macromolecular structures obtained from electron microscopy images. Eur Biophys J. 2001;30(6):457–462. doi: 10.1007/s002490100176. [DOI] [PubMed] [Google Scholar]
- 33.Lashuel HA, et al. Alpha-synuclein, especially the Parkinson’s disease-associated mutants, forms pore-like annular and tubular protofibrils. J Mol Biol. 2002;322(5):1089–1102. doi: 10.1016/s0022-2836(02)00735-0. [DOI] [PubMed] [Google Scholar]
- 34.Miake H, Mizusawa H, Iwatsubo T, Hasegawa M. Biochemical characterization of the core structure of alpha-synuclein filaments. J Biol Chem. 2002;277(21):19213–19219. doi: 10.1074/jbc.M110551200. [DOI] [PubMed] [Google Scholar]
- 35.Cerf E, et al. Antiparallel beta-sheet: A signature structure of the oligomeric amyloid beta-peptide. Biochem J. 2009;421(3):415–423. doi: 10.1042/BJ20090379. [DOI] [PubMed] [Google Scholar]
- 36.Zou Y, Li Y, Hao W, Hu X, Ma G. Parallel β-sheet fibril and antiparallel β-sheet oligomer: New insights into amyloid formation of hen egg white lysozyme under heat and acidic condition from FTIR spectroscopy. J Phys Chem B. 2013;117(15):4003–4013. doi: 10.1021/jp4003559. [DOI] [PubMed] [Google Scholar]
- 37.Natalello A, et al. Conformational plasticity of the Gerstmann-Sträussler-Scheinker disease peptide as indicated by its multiple aggregation pathways. J Mol Biol. 2008;381(5):1349–1361. doi: 10.1016/j.jmb.2008.06.063. [DOI] [PubMed] [Google Scholar]
- 38.Fabian H, et al. Early stages of misfolding and association of beta2-microglobulin: Insights from infrared spectroscopy and dynamic light scattering. Biochemistry. 2008;47(26):6895–6906. doi: 10.1021/bi800279y. [DOI] [PubMed] [Google Scholar]
- 39.Quist A, et al. Amyloid ion channels: A common structural link for protein-misfolding disease. Proc Natl Acad Sci USA. 2005;102(30):10427–10432. doi: 10.1073/pnas.0502066102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lorenzen N, Lemminger L, Pedersen JN, Nielsen SB, Otzen DE. The N-terminus of α-synuclein is essential for both monomeric and oligomeric interactions with membranes. FEBS Lett. 2014;588(3):497–502. doi: 10.1016/j.febslet.2013.12.015. [DOI] [PubMed] [Google Scholar]
- 41.Davidson WS, Jonas A, Clayton DF, George JM. Stabilization of alpha-synuclein secondary structure upon binding to synthetic membranes. J Biol Chem. 1998;273(16):9443–9449. doi: 10.1074/jbc.273.16.9443. [DOI] [PubMed] [Google Scholar]
- 42.Sastry PS. Lipids of nervous tissue: Composition and metabolism. Prog Lipid Res. 1985;24(2):69–176. doi: 10.1016/0163-7827(85)90011-6. [DOI] [PubMed] [Google Scholar]
- 43.Shvadchak VV, Yushchenko DA, Pievo R, Jovin TM. The mode of α-synuclein binding to membranes depends on lipid composition and lipid to protein ratio. FEBS Lett. 2011;585(22):3513–3519. doi: 10.1016/j.febslet.2011.10.006. [DOI] [PubMed] [Google Scholar]
- 44.Ouberai MM, et al. α-Synuclein senses lipid packing defects and induces lateral expansion of lipids leading to membrane remodeling. J Biol Chem. 2013;288(29):20883–20895. doi: 10.1074/jbc.M113.478297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Huang HW. Molecular mechanism of antimicrobial peptides: The origin of cooperativity. Biochim Biophys Acta. 2006;1758(9):1292–1302. doi: 10.1016/j.bbamem.2006.02.001. [DOI] [PubMed] [Google Scholar]
- 46.Benachir T, Lafleur M. Study of vesicle leakage induced by melittin. Biochim Biophys Acta. 1995;1235(2):452–460. doi: 10.1016/0005-2736(95)80035-e. [DOI] [PubMed] [Google Scholar]
- 47.Kayed R, et al. Common structure of soluble amyloid oligomers implies common mechanism of pathogenesis. Science. 2003;300(5618):486–489. doi: 10.1126/science.1079469. [DOI] [PubMed] [Google Scholar]
- 48.Mannini B, et al. Toxicity of protein oligomers is rationalized by a function combining size and surface hydrophobicity. ACS Chem Biol. 2014;9(10):2309–2317. doi: 10.1021/cb500505m. [DOI] [PubMed] [Google Scholar]
- 49.Vilar M, et al. The fold of alpha-synuclein fibrils. Proc Natl Acad Sci USA. 2008;105(25):8637–8642. doi: 10.1073/pnas.0712179105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Qiang W, Yau WM, Luo Y, Mattson MP, Tycko R. Antiparallel β-sheet architecture in Iowa-mutant β-amyloid fibrils. Proc Natl Acad Sci USA. 2012;109(12):4443–4448. doi: 10.1073/pnas.1111305109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Petkova AT, et al. Solid state NMR reveals a pH-dependent antiparallel beta-sheet registry in fibrils formed by a beta-amyloid peptide. J Mol Biol. 2004;335(1):247–260. doi: 10.1016/j.jmb.2003.10.044. [DOI] [PubMed] [Google Scholar]
- 52.Levin A, et al. Ostwald’s rule of stages governs structural transitions and morphology of dipeptide supramolecular polymers. Nat Commun. 2014;5:5219. doi: 10.1038/ncomms6219. [DOI] [PubMed] [Google Scholar]
- 53.Nakamura T, Lipton SA. Molecular mechanisms of nitrosative stress-mediated protein misfolding in neurodegenerative diseases. Cell Mol Life Sci. 2007;64(13):1609–1620. doi: 10.1007/s00018-007-6525-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Canevari L, Abramov AY, Duchen MR. Toxicity of amyloid beta peptide: Tales of calcium, mitochondria, and oxidative stress. Neurochem Res. 2004;29(3):637–650. doi: 10.1023/b:nere.0000014834.06405.af. [DOI] [PubMed] [Google Scholar]
- 55.Cecchi C, et al. Differing molecular mechanisms appear to underlie early toxicity of prefibrillar HypF-N aggregates to different cell types. FEBS J. 2006;273(10):2206–2222. doi: 10.1111/j.1742-4658.2006.05234.x. [DOI] [PubMed] [Google Scholar]
- 56.Gandhi S, Abramov AY. Mechanism of oxidative stress in neurodegeneration. Oxid Med Cell Longev. 2012;2012:428010. doi: 10.1155/2012/428010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Junn E, Mouradian MM. Human alpha-synuclein over-expression increases intracellular reactive oxygen species levels and susceptibility to dopamine. Neurosci Lett. 2002;320(3):146–150. doi: 10.1016/s0304-3940(02)00016-2. [DOI] [PubMed] [Google Scholar]
- 58.Valko M, Morris H, Cronin MT. Metals, toxicity and oxidative stress. Curr Med Chem. 2005;12(10):1161–1208. doi: 10.2174/0929867053764635. [DOI] [PubMed] [Google Scholar]
- 59.Goodwin J, Nath S, Engelborghs Y, Pountney DL. Raised calcium and oxidative stress cooperatively promote alpha-synuclein aggregate formation. Neurochem Int. 2013;62(5):703–711. doi: 10.1016/j.neuint.2012.11.004. [DOI] [PubMed] [Google Scholar]
- 60.Dinner AR, Sali A, Smith LJ, Dobson CM, Karplus M. Understanding protein folding via free-energy surfaces from theory and experiment. Trends Biochem Sci. 2000;25(7):331–339. doi: 10.1016/s0968-0004(00)01610-8. [DOI] [PubMed] [Google Scholar]
- 61.Dobson CM. Protein folding and misfolding. Nature. 2003;426(6968):884–890. doi: 10.1038/nature02261. [DOI] [PubMed] [Google Scholar]
- 62.Roodveldt C, et al. A rationally designed six-residue swap generates comparability in the aggregation behavior of α-synuclein and β-synuclein. Biochemistry. 2012;51(44):8771–8778. doi: 10.1021/bi300558q. [DOI] [PubMed] [Google Scholar]
- 63.Zhang H, Griggs A, Rochet JC, Stanciu LA. In vitro study of α-synuclein protofibrils by cryo-EM suggests a Cu(2+)-dependent aggregation pathway. Biophys J. 2013;104(12):2706–2713. doi: 10.1016/j.bpj.2013.04.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Laue TM, Shah BD, Ridgeway TM, Pelletier SL. Computer-aided interpretation of analytical sedimentation data for proteins. In: Harding SE, Rowe AJ, Horton JC, editors. Analytical Ultracentrifugation in Biochemistry and Polymer Science. Roy Soc Chem; Cambridge, UK: 1992. pp. 90–125. [Google Scholar]
- 65.Marabini R, et al. San Martin MC de la Fraga LG Xmipp: An image processing package for electron microscopy. J Struct Biol. 1996;116(1):237–240. doi: 10.1006/jsbi.1996.0036. [DOI] [PubMed] [Google Scholar]
- 66.Ludtke SJ, Baldwin PR, Chiu W. EMAN: Semiautomated software for high-resolution single-particle reconstructions. J Struct Biol. 1999;128(1):82–97. doi: 10.1006/jsbi.1999.4174. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.